Structured Abstract
Objectives
Osterix (Osx)-expressing mesenchymal cells are progenitors for tooth root forming cells. The aim of this study was to reveal the fates of Osx-expressing cells during and after root formation using a lineage tracing experiment.
Material and Methods
To reveal the fates of Osx-expressing dental mesenchymal progenitors, we took advantage of tamoxifen-inducible Cre reporter system. Osx-creER; R26R-tdTomato mice received tamoxifen (0.1 mg/body) at postnatal day 3 (P3). In this system, Osx-expressing at P3 (Osx-P3) cells undergo recombination, and they and their descendants continue to express Tomato red fluorescence protein permanently. Mandibles were dissected at serial time points ranging from P4 to P116 to investigate how Osx-P3 cells participated in root formation. Tomato+ cells on frozen sections were imaged under fluorescence microscopy.
Results
Osx-P3 cells and their descendants differentiated into all kinds of cells that contributed to the root and periodontal tissues, such as odontoblasts, cementoblasts, alveolar bone osteoblasts and periodontal ligament (PDL) cells during root formation. Even after root formation was completed, they persisted in dental pulp and PDL to provide progenitor cells for odontoblasts and cementoblasts.
Conclusion
Osx-expressing cells play important roles in the entire processes of tooth root formation; their progeny continue to contribute to maintenance of tooth root even after root formation is complete.
Keywords: cementum, dental root formation, osteoblast, Osterix, periodontal ligament
1 | INTRODUCTION
Dental morphogenesis is composed of two distinct stages of crown formation and root formation. Epithelial-mesenchymal interactions are necessary in both stages and are achieved by well-orchestrated functions of multiple growth and transcription factors.1,2 After the completion of crown formation, the inner and outer enamel epithelium proliferate apically and then form a bilayered epithelial structure called Hertwig’s epithelial root sheath (HERS).3 Meanwhile, dental mesenchymes—dental papilla and dental follicle—are derived from cranial neural crest cell (CNC).4 During root formation, dental papilla cells differentiate into dental pulp cells and odontoblasts; dental follicle cells differentiate into cementoblasts, PDL cells and alveolar bone osteoblasts in cooperation with HERS.5,6
Runx2 and Osterix (Osx) were originally identified as essential regulators of osteoblast differentiation during bone formation.7,8 Both osteoblasts and odontoblasts are originated from mesenchymal cells and share several common characteristics. Dental mesenchymes express Runx2 and Osx during cap and bell stages. Afterward, Runx2 expression is downregulated in dental papilla cells and odontoblasts. Contrastingly, Osx expression in these cells remains higher during odontoblast differentiation.9 Although dentin matrix is similar to bone matrix, it is still unclear how Osx-expressing cells contribute to odontoblast differentiation and dentin formation in vivo. Several studies reported the role of Osx in root formation. Targeted ablation of Osx in odontoblast using Col1a1-Cre, Osxfl/fl mice and Osteocalcin-Cre, Osxfl/fl mice leads to short molar root and thin interradicular dentin.10 Overexpression of Osx using a Col1a1 promoter accelerates the formation of cellular cementum.11 We also reported that the deletion of parathyroid hormone-related peptide (PTHrP) receptor in Osx-expressing cells leads to failure of tooth eruption and significantly truncated root lacking periodontal ligaments.12 However, the fates of these Osx-expressing cells during and after completion of root formation have not been completely studied.
In this study, we administered tamoxifen at postnatal day 3 and chased them for a long term to investigate how cells initially expressing Osx participate in root formation and what fate they follow after completion of root formation. Our findings will reveal what roles Osx-expressing mesenchymal cells play in root formation and maintenance of periodontal tissues.
2 | MATERIAL AND METHODS
2.1 | Mice
The mice used in this study have been described in the following references: Osx-creER.13 Rosa26-loxP-stop-loxP-tdTomato (R26R-tdTomato, JAX007914) mice were acquired from the Jackson laboratory. All procedures were conducted in compliance with the Guidelines for the Care and Use of Laboratory Animals approved by Institutional Animal Care and Use of Laboratory Animals of the University of Michigan. Mice were used for analysis regardless of the sex and sacrificed by over-dosage of carbon dioxide or decapitation under inhalation anaesthesia in a drop jar. For lineage-tracing experiments, 3-day- old mice received 0.1 mg of tamoxifen intraperitoneally. Tamoxifen (Sigma T5648, St. Louis, MO, USA) was dissolved first in 100% ethanol and then in sunflower seed oil (Sigma S5007, St. Louis, MO, USA) overnight at 60°C. Mice were genotyped by PCR and fluorescence visualization using a BLS miner’s lamp.
2.2 | Histology
Mandibles were dissected and fixed in 4% paraformaldehyde/PBS (Affymetrix/USB), overnight at 4°C and then decalcified in 15% EDTA for 1–14 days. Decalcified samples were cryoprotected in 30% sucrose/PBS followed by 30% sucrose/PBS:optimal cutting temperature (OCT) compound (Tissue-Tek, Sakura Finetek USA, CA, USA; 1:1) solution, each overnight at 4°C. Samples were embedded in OCT compound under a dissecting microscope (Nikon SMZ-800N, Nikon Instruments, NY, USA) to ensure the parallelism of sectional planes, and cryosectioned at 12 mm thickness (Leica CM1850, Leica Biosystems, IL, USA). Images were captured with a fluorescence microscope (Zeiss Axio Observer, Carl Zeiss Microscopy, NY, USA) with prefigured triple-band filter settings for DAPI/FITC/TRITC and merged with Zen Blue Software. Corresponding band-pass filters for DAPI (Ex.405 nm, BP435-485), GFP (Ex.488 nm, BP500-550), tdTomato (Ex.543 nm, BP570-640) were utilized.
3 | RESULTS
3.1 | Root-forming mesenchymal progenitors express Osterix
To understand how Osterix-expressing cells contribute to dental root formation, we used a tamoxifen-inducible genetic lineage tracing system. In this system Osterix-CreER, R26R-tdTomato mice received tamoxifen at P3 when dental root formation begins. Only actively Osx-expressing cells at P3 undergo recombination and Cre recombinase removes stop codons flanked by loxP sequences in the Rosa26 locus. After recombination, cells expressing Osx and their descendant continue to express tdTomato red fluorescence protein permanently, described previously.12
Twenty-four hours after a single tamoxifen injection, odontoblasts immediately adjacent to the enamel (Figure 1A, yellow arrowheads), some mesenchymal cells in dental papilla (Figure 1B, white arrowheads) and dental follicle (Figure 1B, asterisk) began to express tdTomato protein, indicating that Osx marks dental mesenchymal cells both in the dental pulp and in the dental follicle. We subsequently chased the fates of Osx-expressing cells (Osx-P3 cells).
FIGURE 1.
Root-forming mesenchymal progenitors express Osterix. (A) P4 mandibular first molar (M1) of Osx-creER; R26RtdTomato/+ is shown. Tomato+ odontoblasts were observed beneath enamel (yellow arrowheads). (B) High magnification of rectangle area. DP, dental papilla; DF, dental follicle. P11 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+ (C) and high magnification of rectangle area (D, E). P18 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+ (F) and high magnification of rectangle area (G, H). Scale bars represent 200 µm (upper panels), 50 µm (lower panels) and 20 µm ((E) panel)
After 8 days of chase at P11, when dental root is actively developing, the number of Osx-P3 cells increased. We detected many tdTomato+ cells in the dental papilla (Figure 1C, asterisks). In higher magnifications, Osx-P3 cells differentiated into PDL cells (Figure 1D, white arrowheads), cementoblasts (Figure 1D, asterisks) and odontoblasts (Figure 1D, yellow arrowheads) at the middle of root, indicating that Osx-P3 cells are progenitors of these cells. At the apical area, tdTomato+ red cells in dental follicle could be found immediately outside of Hertwig’s epithelial root sheath (HERS), whereas red cells in dental papilla were also observed inside of HERS, indicating that Osx-P3 cells and their descendants actively engaged in root formation (Figure 1E).
After 15 days of chase at P18, when greater than half of the dental root was formed, Osx-P3 cells became odontoblasts and dental pulp cells (Figure 1F, asterisks: odontoblasts, sharps: dental pulp cells). Additionally, Osx-P3 cells differentiated into periodontal ligament cells attached to the surface of cementum at the middle of root and apical area (Figure 1G,H, white arrowheads). At P18, weaning stage yet and still experienced a non-functional occlusion, the tdTomato+ PDL cells were still not developed and merely limited to the surface of cementum (Figure 1G,H, white arrowheads).
3.2 | Osx-P3 cells persist in dental pulp and on root surface cells even after completion of root formation
After 22 days of chase at P25, when the M1 molars had erupted into oral cavity promoting functional occlusion, odontoblasts and dental pulp cells continued to express tdTomato proteins. (Figure 2A, odontoblasts: asterisk, dental pulp cells: sharp) Moreover, tdTomato+ PDL cells were visible surrounding almost the entire root (Figure 2A). The length and density of diagonal fibres markedly increased (Figure 2B, sharp), indicating that PDL supported the occlusal force by attaching to both bone and cementum on the root surface (Figure 2B, yellow asterisks).14 We also found that the cementum on root surface became thicker than before functioning; tdTomato+ Osx-P3 descendants differentiated into cementoblasts secreting cementum matrix onto the root surface (Figure 2C, asterisks).
FIGURE 2.
Osx-P3 cells exist in dental pulp as odontoblasts and on root surface as PDL cells even after completion of root formation. (A) P25 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+. Tomato+ PDL cells were surrounding the almost entire root. High magnification of rectangle area (B, C) (D: dentin, C: cementum). (D) P35 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+. (E) Odontoblast in dental pulp continued to express tdTomato proteins (asterisk). (F) Tomato+ PDL cells differentiated into cementoblast, and some embedded cementocytes were found at apical area (yellow arrowheads). (G) P46 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+ and high magnification of rectangle area (H, I). Scale bars represent 200 µm (upper panels) and 50 µm (lower panels)
After 32 days of chase at P35, although the number of tdTomato+ PDL cells became smaller than those in P25, they still persisted on the root surface. Odontoblasts in dental pulp continued to express tdTomato proteins (Figure 2D,E, asterisks). At the apical area, Osx-P3 cells actively differentiated into cementoblasts secreting matrix onto the root surface. Interestingly, some embedded tdTomato+ cementocytes could be found in the newly formed cementum (Figure 2F yellow arrow heads), suggesting they were derived from Osx-P3 cells.
After 43 days of chase at P46, dental pulp cavity became narrower, suggesting that tdTomato+ odontoblasts in the dental pulp slowly supplied the secondary dentin on top of the primary dentin (Figure 2G asterisk). On the other hand, the density of tdTomato+ PDL cells seemingly became even smaller (Figure 2H,I, yellow arrowheads).
3.3. | Osx-P3 cells and their progeny provide long-term odontoblasts and cementoblasts after root formation completed
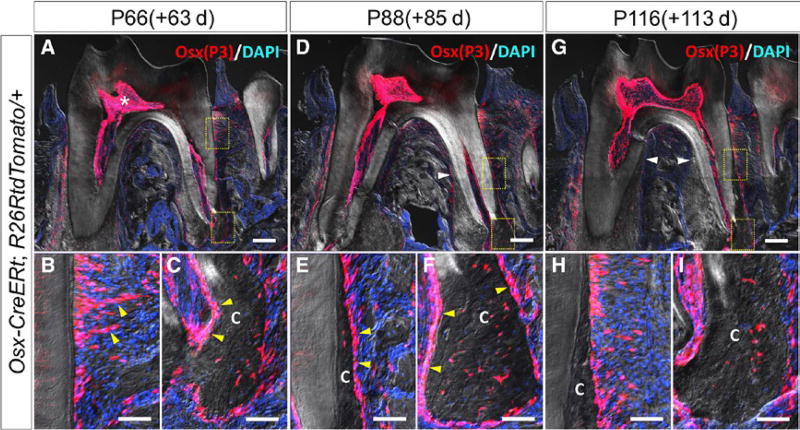
After 63 days of chase at P66, the thickness of cementum in the apical area grew and dental pulp cavity became even narrower (Figure 3A, asterisk). Tomato+ PDL cells also existed in the middle of root (Figure 3B, yellow arrowheads). At the apical area, as the cellular cementum became much thicker, more tdTomato+ cementocytes were observed in the cementum, suggesting that cementoblasts derived from Osx-P3 cells added cellular cementum on the surface of cementum (Figure 3C, yellow arrowheads).
FIGURE 3.
Osx-P3 cells existing in dental pulp and PDL provide progenitors for odontoblasts and cementoblasts after root formation. P66 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+ (A) and high magnification of rectangle area (B, C) (C: cementum). P88 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+ (D) and high magnification of rectangle area (E, F). P116 mandibular first molar (M1) section of Osx-creER; R26RtdTomato/+ (G) and high magnification of rectangle area (H, I). Scale bars represent 200 µm (upper panels) and 50 µm (lower panels)
After 85 days of chase at P88, the increase in cementum thickness expanded to the middle of root (Figure 3D, white arrowhead). We found many tdTomato+ cementocytes embedded in the newly added cementum both in the middle and apical portion of the root. Also, the newly formed cementum was surrounded by tdTomato+ PDL cells and cementoblasts (Figure 3E,F, yellow arrowheads).
After 113 days of chase at P116, the trend observed at P88 continued. Thickened cellular cementum invaded coronally. More than two-third of the root length was covered by the cellular cementum (Figure 3G, white arrowheads). Taken together, Osx-P3 cells and their descendants continued to provide odontoblasts and cementoblasts for a long term during the maintenance phase of the dental root.
4 | DISCUSSION
Despite previous studies, the characteristics and fates of mesenchymal progenitors for dental root formation have not been fully understood. Using a lineage-tracing strategy, we followed the fate of cells initially expressing Osx and their descendants during and after root formation. In terms of root formation, Osx-P3 cells and their descendants thoroughly participated in root morphogenesis by providing all types of root forming cells including odontoblasts, osteoblasts, dental pulp cells, cementoblasts and PDL cells. Even after completion of the root formation, they continue to exist in the dental pulp and on the root surface. Descendants of Osx-P3 cells that remained on the root surface are likely to contain progenitor cells for PDL cells and cementoblasts. On the other hand, Osx-P3 cells in the dental pulp are likely to contain progenitor cells for odontoblasts. Previous studies reported the role of Osx in the tooth root formation. Osx is not required for crown dentin formation, but required for dentin formation during root development.15 The number of Osx-expressing cells in the PDL increased in 4-to 6-week- old mice, but few Osx-expressing cells were observed in the PDL of 6-month- old mice.11 These findings are indicating that Osx expression is upregulated in stage-specific manner for root formation. They also reported that overexpression of Osx accelerates the cementum formation.11 Osx inhibits proliferation of dental papilla cells, whereas it promotes odontoblastic differentiation and mineralization, and ALP activity of dental papilla cells.16
Several recent studies showed the important roles of various signalling pathways for root formation. Knockout mice of Smad4 in odontoblasts using Osteocalcin-Cre showed disruption of root development.17 NFIC has been shown to be the key regulator of postnatal root formation and its deletion leads to short and abnormal root due to suppression of odontoblast proliferation and differentiation, as well as inducing subsequent apoptosis of aberrant odontoblasts.18–20 A recent study reported that Smad4-Shh- Nfic signalling cascade is important for normal root formation.21 In addition to the importance of these signalling pathways, we previously reported the crucial roles of parathyroid/parathyroid-related protein (PTH/PTHrP) and its receptor (PPR) signalling in tooth root formation and eruption.12 In our model, deletion of PPR signalling in Osx-expressing progenitors suppresses their proliferation and differentiation, results in truncated roots and irregular cementum formation. Dental root formation is a complicated process requiring epithelial and mesenchymal interactions. To elucidate the mechanism for dental root formation, further studies are needed.
From this study, we demonstrated that Osx-expressing cells at the early postnatal stage include dental mesenchymal progenitors and their descendants differentiate into cells of the dental root and the supporting periodontal tissue. Even after root formation is complete, they continue to exist in the dental pulp and the PDL to supply progenitor cells for odontoblasts and cementoblasts for a long term.
5 | CONCLUSIONS
Osterix-expressing mesenchymal cells play an important role in tooth root formation by differentiating into all kinds of cells capable of forming the root and periodontal tissues and continue to provide progeny to support the dental root for a long term.
Acknowledgments
This study was supported by American Association of Orthodontists Foundation Grant to W.O., National Institution of Health Grant DE022564 to N.O., and University of Michigan MCubed 2.0 Grant to N.O. and W.O.
Funding information
National Institute of Dental and Craniofacial Research, Grant/Award Number: DE022564; American Association of Orthodontists Foundation; National Institution of Health; University of Michigan
Footnotes
CONFLICT OF INTERESTS
The authors have no conflict of interests to report.
References
- 1.Jussila M, Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb Perspect Biol. 2012;4:a008425. doi: 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003;116:1647–1648. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Bringas P, Jr, Slavkin HC, Chai Y. Fate of HERS during tooth root development. Dev Biol. 2009;334:22–30. doi: 10.1016/j.ydbio.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 5.Morsczeck C, Gotz W, Schierholz J, et al. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24:155–165. doi: 10.1016/j.matbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Yao S, Pan F, Prpic V, Wise GE. Differentiation of stem cells in the dental follicle. J Dent Res. 2008;87:767–771. doi: 10.1177/154405910808700801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 8.Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Gluhak-Heinrich J, Wang YH, et al. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TH, Bae CH, Lee JC, et al. Osterix regulates tooth root formation in a site-specific manner. J Dent Res. 2015;94:430–438. doi: 10.1177/0022034514565647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Z, Zhang H, Zhou X, et al. Genetic evidence for the vital function of Osterix in cementogenesis. J Bone Miner Res. 2012;27:1080–1092. doi: 10.1002/jbmr.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono W, Sakagami N, Nishimori S, Ono N, Kronenberg HM. Parathyroid hormone receptor signalling in osterix-expressing mesenchymal progenitors is essential for tooth root formation. Nat Commun. 2016;7:11277. doi: 10.1038/ncomms11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maes C, Kobayashi T, Selig MK, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rios HF, Ma D, Xie Y, et al. Periostin is essential for the integrity and function of the periodontal ligament during occlusal loading in mice. J Periodontol. 2008;79:1480–1490. doi: 10.1902/jop.2008.070624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Jiang Y, Qin C, Liu Y, Ho SP, Feng JQ. Essential role of osterix for tooth root but not crown dentin formation. J Bone Miner Res. 2015;30:742–746. doi: 10.1002/jbmr.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang G, Li X, Yuan G, Liu P, Fan M. The effects of osterix on the proliferation and odontoblastic differentiation of human dental papilla cells. J Endod. 2014;40:1771–1777. doi: 10.1016/j.joen.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, Yang G, Weng T, et al. Disruption of Smad4 in odontoblasts causes multiple keratocystic odontogenic tumors and tooth malformation in mice. Mol Cell Biol. 2009;29:5941–5951. doi: 10.1128/MCB.00706-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee DS, Park JT, Kim HM, et al. Nuclear factor I-C is essential for odontogenic cell proliferation and odontoblast differentiation during tooth root development. J Biol Chem. 2009;284:17293–17303. doi: 10.1074/jbc.M109.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TY, Lee DS, Kim HM, et al. Disruption of Nfic causes dissociation of odontoblasts by interfering with the formation of intercellular junctions and aberrant odontoblast differentiation. J Histochem Cytochem. 2009;57:469–476. doi: 10.1369/jhc.2009.952622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JC, Herr Y, Kim HJ, Gronostajski RM, Cho MI. Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J Periodontol. 2007;78:1795–1802. doi: 10.1902/jop.2007.060363. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Xu X, Bringas P, Jr, Hung YP, Chai Y. Smad4-Shh-Nfic signalling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 2010;25:1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]