Abstract
During gastrulation, cell types from all three germ layers are specified and the basic body plan is established1. However, molecular analysis of this key developmental stage has been hampered by limited cell numbers and a paucity of markers. Single cell RNA sequencing circumvents these problems, but has so far been limited to specific organ systems2. Here we report single-cell transcriptomic characterisation of over 20000 cells immediately following gastrulation at E8.25 of mouse development. We identify 20 major cell types, which frequently contain sub-structure, including three distinct signatures in early foregut cells. Pseudospace ordering of somitic progenitor cells identifies dynamic waves of transcription and candidate regulators, which are validated by molecular characterisation of spatially resolved regions of the embryo. Within the endothelial population, cells that transition from haemogenic endothelial to erythro-myeloid progenitors specifically express Alox5 and its co-factor Alox5ap, which control leukotriene production. Functional assays using mouse embryonic stem cells demonstrate that leukotrienes promote haematopoietic progenitor cell generation. This comprehensive single cell map therefore can be exploited to reveal previously unrecognised pathways contributing to tissue development.
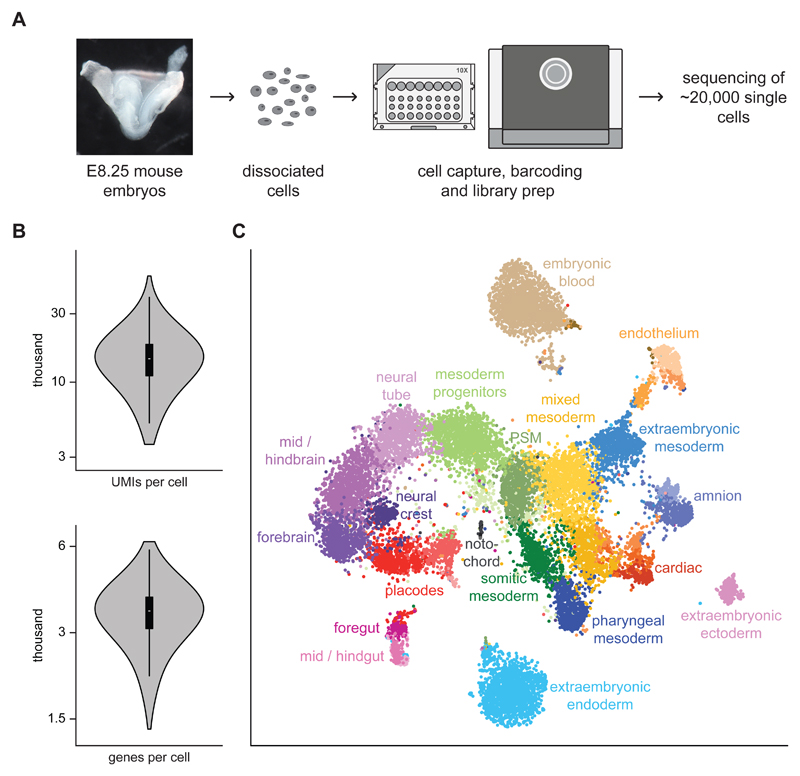
During mouse gastrulation, epiblast cells differentiate into the three germ layers endoderm, mesoderm and ectoderm. This process is followed by rapid differentiation into organ-specific cell types so that, by embryonic day E8.25, precursor cells of major organs have been formed1. To characterise the full complement of cell types present at this stage, we collected C57BL/6 mouse embryos at E8.25, including their extraembryonic tissues. Following dissociation, embryos were pooled and processed on a 10X microfluidic chip, and the resulting libraries were sequenced on an Illumina HiSeq 2500 (Fig. 1A). Following filtering of low quality samples (Methods), 19,396 cells were retained for downstream analyses. On average, 15,073 unique transcripts were captured and around 3,518 genes were detected in a typical cell (Fig. 1B).
Fig. 1. Single-cell RNA-seq of whole mouse E8.25 embryos identifies 20 major cell types.
A) E8.25 whole mouse embryos were dissociated and processed with the 10X genomics platform to capture single cells and produce libraries for RNA sequencing. A representative image of the sequenced embryos is shown. B) Violin plots indicating the number of UMIs and genes obtained per cell. A boxplot is shown on the inside (center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; n = 19,396 cells). C) t-SNE plot of all the cells that passed quality control (19,396) computed from highly variable genes; the first two dimensions are shown. Cells with similar transcriptional profiles were clustered into 33 different groups, as indicated by the different colours. Each cluster was annotated based on the expression of marker genes into 20 major different cell types. Several cell types are composed of two or more clusters. PSM = presomitic mesoderm.
Following identification of genes with highly variable expression across the dataset, we assigned cells into 33 different groups (Methods). We then used the expression of previously annotated marker genes to infer each clusters’ identity. We annotated 20 major cell populations, several of which comprised two or more clusters (Fig. 1C). Cluster identification was consistent between cell populations from one sample captured on different 10X chip channels, and between two independent samples (Supplementary Fig. 1A-B). The proportions of cells from each sample were generally consistent with the expected proportions based on the overall dataset (Supplementary Table 1), suggesting that the capture rate is unbiased across experiments.
Next, to assess the stability of our classification, we repeated the experiment using embryos from an F2 cross of mixed genetic background (C57BL/6 and CBA). In this case we sequenced ~7,000 cells from three individual embryos. Remarkably, the results from the clustering analysis were almost identical and all major cell types were identified in both datasets, except for the extraembryonic ectoderm, which was removed when dissecting the F2 embryos (Supplementary Fig. 1C-D). Thus, we conclude that we have captured the heterogeneity in cell populations present in mouse embryos of different genetic backgrounds at this stage of development.
We observed cell types from all three germ layers (Fig. 1C), characterised by the expression of 869, 240 and 159 genes preferentially upregulated in endodermal, mesodermal or ectodermal cells respectively (false discovery rate < 5%, fold-change > 2; Supplementary Fig. 1E; Supplementary Table 2). This included well-established markers, such as Sox17, Epcam and Foxa1/2 for endoderm3, Pdgfra, Tbx6 and Brachyury (T) for mesoderm4 and Sox1, Pax6 and Pou3f1 for ectoderm5. We also identified germ layer specific genes that have not been described in the context of embryo development including Gm2694 and Mir124-2hg, which show specific expression in ectoderm (Supplementary Fig. 1F). Furthermore, many other genes showed restricted expression to one or a few of our defined cell types (Supplementary Fig. 1E), providing valuable candidate markers for defining and potentially programming populations of cells toward specific lineages (for visualisation see http://marionilab.cruk.cam.ac.uk/organogenesis/).
Closer inspection of specific clusters revealed that most exhibited additional, subtle sub-structure. We hypothesised that such sub-structure could shed light on early regulatory processes that drive fine-grained specification of cell fate. For example, between E8.0 and E9.0 the endoderm undergoes a series of morphogenetic changes that turn it from a flat sheet into a tube where the domains of major organs like the liver and lung arise6. While ventral folding and formation of the foregut pouch is already induced at E8.256, the earliest stages of foregut endoderm diversification remain ill-defined at the molecular level.
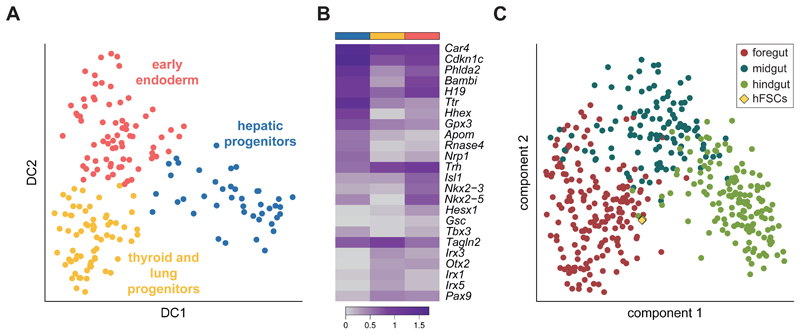
To explore this further, we considered cells in the foregut cluster (Fig. 1C) and used a diffusion map approach7 to visualise three sub-clusters (Fig. 2A and Supplementary Fig. 2A). We then identified differentially expressed genes (Fig. 2B; Supplementary Table 3) and contrasted these with in situ images from the literature to assign cluster identities. The red cluster expressed markers of early endodermal cells including Gsc, Trh and Otx28, 9. In contrast, the blue cluster expressed Ttr, Hhex and Tbx310, all markers of hepatic progenitors, while the yellow cluster was characterised by Irx1/3/5 and Pax911, 12, typical of the thyroid anlage and lung specification.
Fig. 2. Sub-structure within the E8.25 mouse foregut.
A) Diffusion map of the foregut endoderm cells (Fig. 1C; n = 185); the first two diffusion components (DC) are shown. The different colours correspond to three sub-clusters detected by the k-branch algorithm. Based on their expression pattern (see panel B), likely identities of early endoderm cells (red), hepatic progenitors (blue) and thyroid and lung progenitors (yellow) were assigned. B) Heatmap showing the average expression of the top 5 most differentially expressed genes in each of the three sub-clusters (indicated by the coloured bars on top) along with well-characterised marker genes. The colour gradient is log10(normalised counts + 1). C) Principal Component Analysis of the foregut, midgut and hindgut cells from the mouse (circles; n = 437) and human pluripotent stem cell derived foregut progenitor cells (diamonds; n = 3); the first two components are shown. The human samples are closest to the mouse foregut cells.
Lineage tracing studies have followed the movement of endodermal cells in embryos from the one to ten somite stages and revealed that cells from different regions of the gut populate different organs later in development13. Our findings suggest that regionalization is also evident at the molecular level as early as the 4-somite stage. Importantly, this included potential markers of early foregut lineage specification. For example, Hesx1 is a homeodomain transcription factor involved in the development of the forebrain and the pituitary gland14; in our data, it is restricted to the early endoderm cluster suggesting a possible role in regulating foregut development. Overall, our analysis illustrates how domain specific knowledge can be used to allocate biological identity in the context of sparse scRNA-seq data.
The molecular processes driving differentiation cannot be readily studied in human embryos. This poses difficulties for the validation of protocols that aim to produce authentic cell types from human induced pluripotent stem cells. We compared the transcriptome of human foregut progenitor cells – induced from human pluripotent stem cells (Methods) – to our mouse data. We used the pairs classifier, a classification algorithm that is robust to confounding effects due to differences in experimental protocols and normalisation15, to map the human foregut-like cell samples onto our single cell endoderm atlas. All replicates were assigned a foregut identity when compared with the mouse data for fore-, mid- and hindgut (Fig. 2C and Supplementary Fig. 2B). Thus, our single cell mouse embryo dataset provides a valuable in vivo reference that can be used to assess the identity of in vitro derived cell populations.
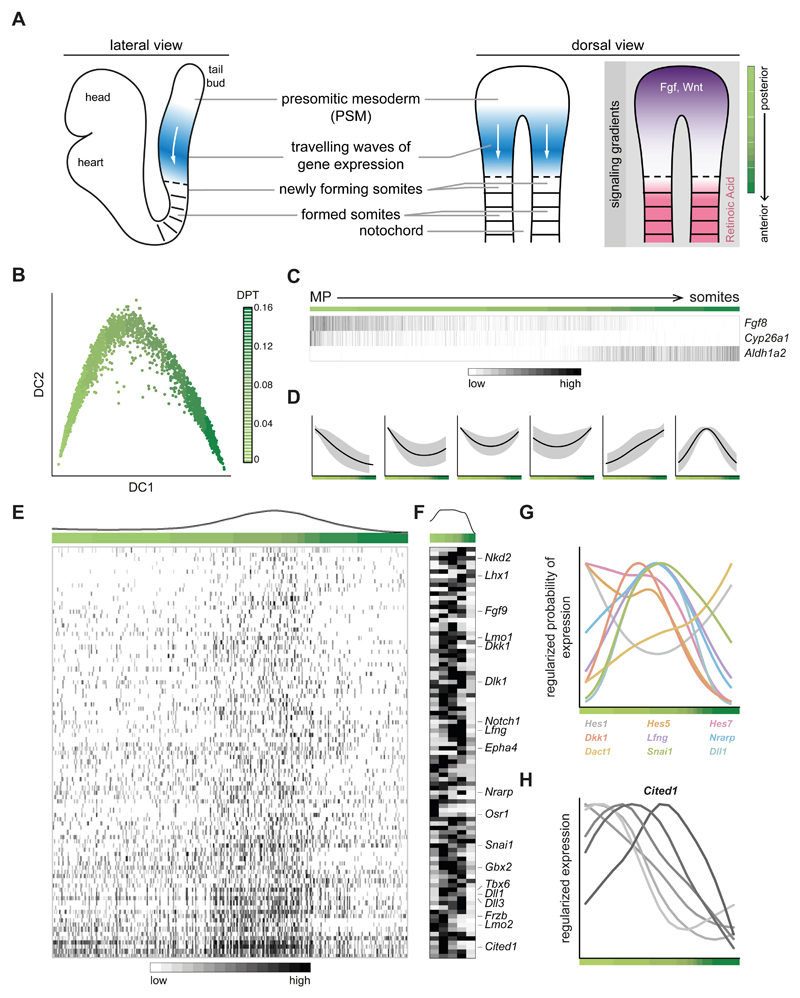
As a snapshot measure, scRNA-seq data seems ill-suited to recover dynamic information on cell fate specification. However, when entry into a defined differentiation program is desynchronised across a cell sub-population, dynamic information can be recovered through the “chromatographic” segregation of the molecular profile. Motivated by this, we focused on the process of somitogenesis, which involves the segmentation of the developing embryonic body axis into somites and is guided by oscillating genes, which create waves of expression that travel across the presomitic mesoderm (PSM) from posterior to anterior16 (Fig. 3A). Upon arrival of a wave at the PSM's anterior end, a new somite is formed. The posterior end of the PSM is marked by high levels of Wnt and FGF signalling while somites show high levels of retinoic acid (RA)16 (Fig. 3A).
Fig. 3. Oscillating patterns of gene expression during somitogenesis can be inferred from scRNA-seq data.
A) Schematic of mouse somitogenesis, which proceeds along the anteroposterior (AP) axis. From the tail-bud (posterior) extends the presomitic mesoderm (PSM) which gives rise to somites (anterior). On the right, travelling waves of gene expression of oscillatory genes are shown along with signalling gradients on the AP axis; FGF and Wnt are posterior-high while retinoic acid (RA) has the opposite pattern. B) Diffusion map of the cells from the mesoderm progenitors (MP), presomitic and somitic mesoderm clusters (n = 2999), ordered based on the expression of genes correlated with Fgf8 expression; the first two diffusion components (DC) are shown. The colour gradient indicates the trajectory from MP to somites as a pseudo-space measurement. C) Heatmap of the genes involved in establishing signalling gradients. Aldh1a2 is the enzyme that synthesises RA while Cyp26a1 degrades RA. Cells have been ordered in pseudo-space on the x-axis. Each gene is regularised so that expression values are within [0,1]. D) Expression changes along the pseudo-space trajectory can be clustered into six groups, one of which (last) shows a wave-like pattern consistent with oscillatory expression. E) Heatmap of the expression of all genes in the last cluster from D. Cells have been ordered in pseudo-space on the x-axis. Each gene is regularised so that expression values are within [0,1]. F) Representative heatmap of the same genes on the dissected PSM of an embryo that was split into five segments from posterior to anterior, as schematised at the far right in A. Six biological replicates were analysed, all with similar results; the other five replicates are presented in Supplementary Fig. 3B. G) Regularised logistic fit of the expression across the pseudo-space for genes with well-characterised oscillatory expression16. Most show a wave-like pattern. H) Expression pattern of Cited1 in dissected segments of PSM from most posterior to most anterior, for six different biological replicates. The gene shows a wave-like pattern, and different embryos peak at different regions of the PSM.
To explore whether coherent patterns of gene expression could be resolved from our snapshot data, we analysed the cells from the mesoderm progenitors, presomitic and somitic mesoderm clusters (Fig. 1C). We first ordered cells along a putative anteroposterior (AP) axis by using genes highly correlated with Fgf8, which serves as a positional landmark16 (Fig. 3B). The inferred pseudo-space axis recapitulated the expected signalling gradients, from the highest expression of Fgf8 to the highest expression of Aldh1a2, the synthesizing enzyme of RA (Fig. 3C). Next, we modelled gene expression along this pseudo-space axis to identify genes characterised by a localised wave-like peak within the PSM.
The expression profiles of a thousand genes were inconsistent with constant expression across pseudo-space (Fig. 3D; Methods); 93 of these showed wave-like expression that peaked along the pseudo-space trajectory (Fig. 3E), and included several well-known regulators of somitogenesis such as Hes5, Lfng and Dll116. Indeed, when examining the expression across pseudo-space of experimentally characterised oscillating genes, most showed wave-like expression (Fig. 3G). Moreover, we identified several genes where oscillatory activity has not been reported (Fig. 3E), but that behave similarly to classic oscillating genes. One of these, Cited1 (Fig. 3E), has been identified as being expressed within the PSM17 and is known to block epithelial differentiation in the kidney18. We thus hypothesise a possible role during somitogenesis, where the interior of the somite remains mesenchymal whereas the somite boundary undergoes a mesenchymal-to-epithelial transition19.
To validate these findings, we dissected the PSM of four different mouse embryos – keeping the left and right sides separate – and divided each into five segments from posterior to anterior (Supplementary Fig. 3A and Supplementary Video 1). We then performed RNA sequencing on each segment, for six biological replicates. The expression dynamics across the AP axis of the 93 genes we defined as oscillatory (Fig. 3E) were well correlated to the single-cell data (median Pearson’s correlation, interquartile range for all genes = 0.51-0.78; Fig. 3F and Supplementary Fig. 3B), and so were the profiles of well-characterised oscillatory genes (Supplementary Fig. 3C). Furthermore, the expression profile of Cited1 showed a wave-like pattern in five out of the six replicates, peaking at distinct locations along the AP axis, consistent with embryo-specific wave progression (Fig. 3H). Together, these findings show that static snapshots of single cell molecular profiles provide a promising strategy to identify candidate genes that contribute to developmental processes driven by oscillatory gene expression.
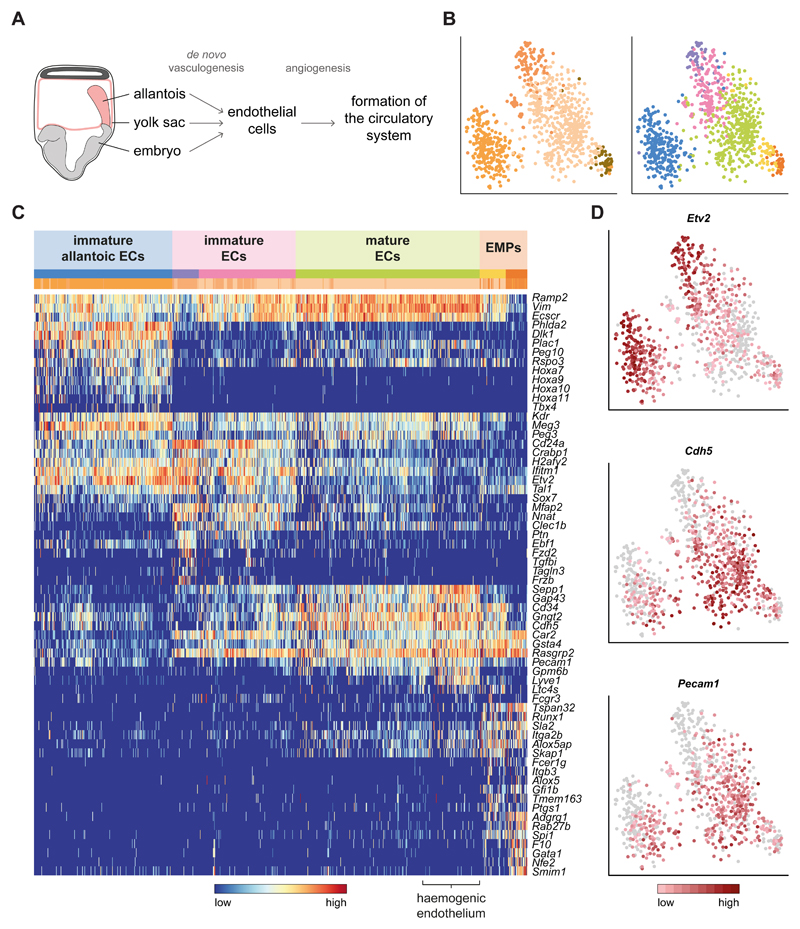
While many of the cells captured in this study are primarily found in a specific organ within the adult, endothelial cells will be distributed across the whole body. Endothelial cells (ECs) originate by de novo vasculogenesis from at least three sites within the embryo during E7.0-E8.0: the yolk sac, the allantois, and intra-embryonically in the aortic primordia20 (Fig. 4A). All subsequently proliferate by angiogenesis and converge at the base of the allantois, giving rise to the circulatory system at around E8.520 (Fig. 4A). Unsupervised clustering of the four populations annotated as ECs (Fig. 1C) revealed substantial substructure, identifying six distinct sub-clusters (Fig. 4B). Interestingly, some ECs had an underlying allantoic signature (Fig. 4C, blue cluster) characterised by expression of Tbx4, Hoxa10 and Hoxa1121.
Fig. 4. The endothelium can be subdivided based on maturity and location of origin.
A) Schematic diagram of how endothelial cells (ECs) and the circulatory system are formed in the embryo. B) t-SNE plot of the cells in the four endothelial clusters (n = 871). Left: original clusters coloured as in Fig. 1C. Right: colours correspond to the redefined subclusters. The first two dimensions are shown. C) Heatmap of the top 5 differentially expressed genes across subclusters, along with well-characterised genes for the endothelium. Coloured bars indicate the new cluster (top) and original cluster (bottom) they belong to. Each gene is regularised so that expression values are within [0,1]. D) Expression patterns of the endothelial markers Etv2, Cdh5 and Pecam1 on the t-SNE from B. The colour gradient is log10(normalised counts + 1). ECs: endothelial cells; EMPs: erythroid-myeloid progenitors.
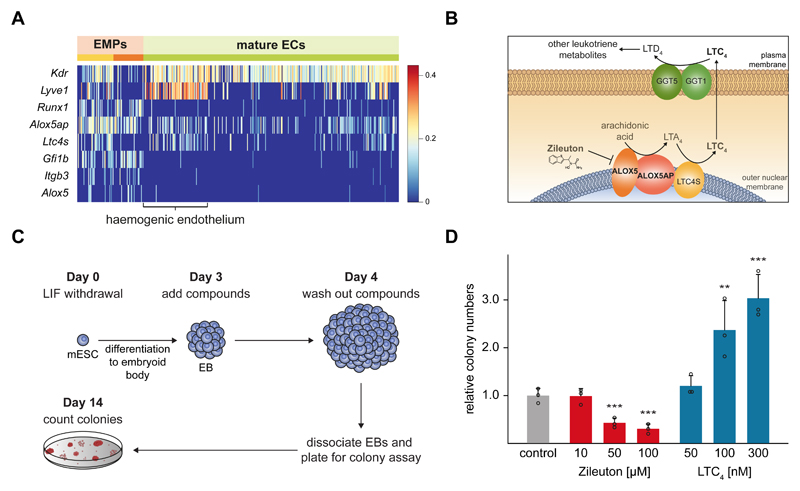
Within the non-allantoic clusters, cells could be clearly distinguished by their level of maturity. Elevated levels of Etv2 pointed towards more immature cells20 in the purple and pink clusters, while the mature EC markers Cdh5 and Pecam122 showed increased expression in the green subgroup (Fig.4D). Due to the developmental stage analysed, we consider that many of the non-allantoic mature ECs may be of yolk sac (YS) origin22. Accordingly, we noted a subset of cells with high levels of Lyve1, which has recently been reported as a marker for yolk sac haemogenic endothelium (HE; Fig.4C)23. Furthermore, adjacent to the HE, two other clusters - yellow and orange - expressed the haematopoietic progenitor markers Runx1, Spi1 (PU.1) and Gfi1b. This transcriptional profile corresponds to the second wave of haematopoiesis, where definitive erythroid-myeloid progenitors (EMPs) emerge in the YS by endothelial to haematopoietic transition (EHT)24. Although these haemogenic cells still expressed an endothelial signature (Cdh5 and Pecam1), the orange cluster displayed lower levels of these markers, indicating their more mature blood phenotype. This latter group also expressed erythroid (Gata1, Nfe2) and megakaryocytic (F10) markers, supporting this notion (Fig.4C).
Next we analysed in more detail the transcriptomes of the HE and EMP cells. Interestingly, we found that Alox5 and Alox5ap were upregulated in these cells, compared to the rest of the ECs (Fig. 5A), a finding also recapitulated in single endothelial cells sorted based on Flk1 expression25 (Supplementary Fig. 4A). The Alox5 enzyme and its cofactor Alox5ap occupy a central position in the production of leukotrienes from arachidonic acid (Fig. 5B). Thus, we hypothesised that this pathway might be important in early blood development. To further investigate this, we used mouse embryonic stem cell (ESC) differentiation assays that recapitulate the formation of HE and EMP cells in vitro.
Fig. 5. The leukotriene biosynthesis pathway drives blood formation.
A) Heatmap showing the characteristic genes of erythro-myeloid progenitors (EMPs) and haemogenic endothelium within the non-allantoic mature endothelial cell (EC) cluster (Fig. 4C). The colour gradient is log10(normalised counts + 1). See also Supplementary Fig. 4A. B) Schematic diagram of the leukotriene biosynthesis pathway, highlighting the functions of ALOX5, ALOX5AP and the position of the leukotriene C4 (LTC4). C) Experimental setup for embryonic stem cell (ESC) differentiation to embryoid bodies (EBs) and haematopoietic colony formation assays. D) Bar plot showing the fold change in number of colonies relative to carrier control when EBs were treated with the indicated concentrations of Zileuton or LTC4 for 24 hours. Bars represent the mean plus standard deviation of n=3 biological replicates. The individual data points are shown as open circles. Statistically significant changes compared to controls were tested with a one-tail Student’s t test (p-value = 0.004 for Zileuton-50µM; 0.002 for Zileuton-100µM; 0.027 for LTC4-100µM; 0.007 for LTC4-300µM;).
Mouse ESCs were differentiated into embryoid bodies (EBs) and exposed to the Alox5 inhibitor Zileuton or to leukotriene C4 (LTC4), between days three to four of differentiation. EBs were then dissociated, the compounds washed out, and the number of haematopoietic progenitor cells assessed using colony forming assays (Fig. 5C). While addition of Zileuton caused a dose-dependent reduction in colony numbers, LTC4 resulted in a reciprocal increase of up to 3-fold (Fig. 5D). This demonstrates that the leukotriene pathway plays a previously unrecognised role in modulating the formation of early blood progenitor cells.
Recent advances in single cell expression profiling technology are having a major impact across almost all areas of biomedical research. In contrast to previous studies, which have been restricted to small and well-defined populations of cells, we performed an unbiased sampling of cells from the entire embryo and thus generated a rich resource for the developmental biology community across all major mammalian organ systems.
The identification of subtle sub-structure within the endothelial and endodermal cell populations suggests that other clusters also contain cryptic and small subgroups of cells. One key challenge moving forward will be to identify and characterise these populations in an automated way. This will be particularly critical for small and rare sub-populations, where discriminating between genuine biological signal and technical noise will be challenging. Primordial germ cells (PGCs) represent a well-characterised yet exceedingly rare cell population in the developing embryo26. Our processing pipeline did not identify a separate cell cluster for PGCs; however, targeted interrogation of the dataset revealed 25 cells that expressed high levels of the very specific PGC marker gene Stella (Dppa3) along with several other genes expressed in PGCs26 (Supplementary Fig. 5A-B and Supplementary Table 4).
Additional challenges come from the somewhat philosophical question of how to define a cell type: here the boundaries can quickly get blurred, especially in dynamically developing systems where the concept of a continuum of cellular states may be more appropriate than rigid cell type categorizations. This concept is well illustrated in the context of somitogenesis, where our data shows a smooth continuum along the differentiation path from mesodermal progenitors to somitic cells. By ordering cells in a trajectory, we identified and validated spatially-restricted patterns of wave-like expression, including additional candidate regulators such as Cited1.
The endothelial cells from our dataset could be divided both by maturity and by their location of origin within the embryo. Macrophages are also found across the entire adult organism, and are thought to acquire tissue-specific molecular signatures following migration, presumably driven by distinct microenvironmental signals. Interestingly, we observed that endothelial cells could be partitioned into two major groups based on a gene expression signature specific to allantoic mesoderm. In contrast to macrophages, endothelium may therefore have a tissue-of-origin signature from very early in development. It will be fascinating to explore how this initial patterning may influence the diverse range of endothelial functions.
Blood cells develop in close association with other mesodermal tissues, in particular the endothelium, where flat endothelial cells undergo a profound change in cell shape to give rise to round blood progenitor cells, through an endothelial to haematopoietic transition (EHT)24. Although EHT has been recognised as a key step that will require optimization to achieve robust in vitro production of blood cells from pluripotent stem cells27, much remains to be learned about the underlying molecular processes. We identified the haemogenic endothelial cells as well as the blood progenitors they give rise to; access to the full transcriptomes of these key developmental populations in vivo allowed the subsequent identification of the leukotriene biosynthesis pathway as a regulator of early blood development.
Unlike the previously identified transcriptional regulators of EHT such as Runx1 or Gfi1/Gfi1b28, the leukotriene pathway will be easier to exploit in a translational setting because of the ready availability of small molecule agonists and antagonists. Leukotrienes are produced in a multi-step process from arachidonic acid, which can be metabolised into a number of distinct functionally active molecules, all with their own receptors and spectrum of biological activities, including the fine-tuning of haematopoietic stem cell activity at the time of their first emergence in the aorta-gonad-mesonephros region at E11.529. Of note, the arachidonic acid derivatives prostaglandin and epoxyeicosatrienoic acid have been identified in small molecule screens for compounds that can amplify blood stem and progenitor cells30, 31, and have already entered clinical trials to enhance blood stem cell transplantation32. However, there is as yet no evidence to suggest that prostaglandin or epoxyeicosatrienoic acid function endogenously during early blood progenitor development. It will be intriguing to decipher how the leukotriene pathway promotes the formation of blood progenitor cells, and to incorporate its manipulation into current protocols for in vitro production of blood cells for regenerative medicine and drug development applications.
In summary, our analyses have characterised all major cell populations (both embryonic and extra-embryonic) present in a post-gastrulation mammalian embryo. Our results provide a rich resource for the scientific community that can be used for different purposes. For instance, by combining our reference atlas with data from in vitro differentiation protocols it is possible to rigorously assess the ability to efficiently generate a particular lineage. Additionally, our dataset facilitates both hypotheses generation and the identification of marker genes to isolate specific populations for further study. To this end, we have created a tool to browse the expression of any gene, including those we have identified as potential markers for specific lineages (http://marionilab.cruk.cam.ac.uk/organogenesis/).
Online Methods
Embryo collection and single-cell RNA sequencing
All mice were bred and maintained at the University of Cambridge, in microisolator cages with sterile bedding; sterile food and water were provided ad libitum. All animals were kept in specified pathogen-free conditions. All procedures were performed in strict accordance to the United Kingdom Home Office regulations for animal research under project number PPL70/8406. This work complies with all relevant ethical regulations pertaining to animal experiments. Timed matings were set up between C57BL/6 mice. Upon dissection, only embryos staged as 4-somite pair embryos (Theiler Stage 12) according to the morphologic criteria of Downs and Davies were kept. Suspensions of cells were prepared by incubating the embryos with TrypLE Express dissociation reagent (Life Technologies) at 37°C for 10 minutes and quenching with heat inactivated serum.
For the first sample, 16 embryos were pooled together whereas a second sample consisted of 7 independent embryos. The first sample was run in two independent channels of the Chromium 10X Genomics to generate single-cell libraries for high throughput sequencing; the second sample was processed in a single channel, at a later date. All samples were multiplexed together and sequenced across two flow cells of an Illumina HiSeq 2500, to generate paired-end 100bp data.
For the replication experiment, timed matings were set up between C57BL/6 x CBA F1 mice. Embryos were processed in the same way as above, except in this case single embryos were used and the extraembryonic ectoderm was removed upon dissection. Each sample was run in two independent channels of the Chromium 10X Genomics. All samples were multiplexed together and sequenced across six lanes of an Illumina Hi-Seq 2500.
Data processing with the Cell Ranger package and quality control
Sequencing data was processed with the Cell Ranger 1.1.0 software to align, filter and count UMIs per sample. Data was mapped to the mouse reference genome GRCm38.p4 and the transcriptome annotation from the Ensembl database, version 84 (http://mar2016.archive.ensembl.org/index.html). The resulting data comprised 20,819 cells from all three samples. Data from all samples were consolidated into a single dataset using the cellranger aggr program, which downsamples the depth of different samples to make it equivalent across the whole dataset. We removed all cells that expressed less than a thousand genes or that had more than 3% of their transcripts mapped to mitochondrial genes. We further removed any cells that expressed both Xist and any of Kdm5d, Eif2s3y, Gm29650, Uty or Ddx3y (genes in the Y chromosome) as these are likely to be doublets. We identified 400 cells that could be affected by index swapping (since they share the same cell barcode with another cell), even though the rates of this phenomenon are very low for the HiSeq 2500. However, these were scattered across the whole tSNE and there was no difference in their library size or number of genes expressed. Therefore, these cells were not removed.
Data normalisation
The data were normalised for cell-specific biases using the method proposed in Lun et al. (2016)33 and implemented in the Bioconductor package scran34. To calculate size factors, genes with mean expression lower than 0.1 were filtered out; the quickCluster function was used to obtain the initial clustering of the cells (method igraph). The estimated size factors were used to normalise all genes expressed in at least one cell. Normalised counts are provided with the ArrayExpress submission.
Identification of highly variable genes and dimensionality reduction
For downstream analyses we filtered out all genes with mean expression lower than 0.01. To identify highly variable genes, we implemented the distance-to-median (DM) method proposed in Kolodziejczyk et al. (2015)35, and called as highly variable those with the 20% highest DM values. We discarded all genes from the Y chromosome, Xist, haemoglobins and ribosomal protein genes. Spearman’s correlation coefficient was computed from this set of genes and then used to build a distance matrix defined as √((1-ρ)/2). A t-SNE plot was constructed from the distance matrix, using the Rtsne package36.
Clustering of cells into distinct populations
To classify cells into different clusters we used hierarchical clustering on the distance matrix (see above; hclust function in R, with average method) followed by the dynamic hybrid cut algorithm (dynamicTreeCut package37) to define clusters (cutreeDynamic function in R with the hybrid method and a minimum cluster size of 60 cells). Cells that were outliers and could not be assigned to any cluster by the algorithm were removed. This resulted in the definition of 20 clusters.
We further searched for substructure in each of these clusters. For each cluster, we defined the set of highly variable genes and computed the distance matrix as detailed above. We then used hierarchical clustering and the dynamic hybrid cut algorithm (minimum cluster size of 40 cells) to define clusters. In cases where more than one cluster were identified, we performed a stability analysis by subsampling the number of cells and genes to 2/3 of the total and identifying clusters with the same procedure; we then used the Jaccard coefficient to assess the similarity of the obtained clusters with the full and subsampled data. This procedure was repeated a hundred times and clusters with a median Jaccard index of at least 0.5 were split. This resulted in 33 clusters that could not be stably subdivided further.
To annotate each cluster we examined the expression of well-characterised marker genes. Several groups of clusters that were adjacent in the t-SNE plot were all annotated as the same cell type; whereas they differ in the expression of subsets of genes, they share the core of gene markers that characterise them as a single population. We annotated 20 distinct cell types. We tested whether the proportions of cells from each sample were different for each of these 20 subpopulations with a Pearson’s chi-squared test (p-values corrected for multiple testing using the Benjamini & Hochberg method; Supplementary Table 1). Only five were significantly different, three of which were the extraembryonic subpopulations; this is consistent with extraembryonic tissues being more susceptible to biased recovery upon dissection of the embryos.
Identification of germ layer marker genes
To identify genes that had specific expression in particular populations of cells, we used edgeR38 to perform differential expression analysis. For this, we used scran’s function convertTo to create a DGElist object with the data and the appropriate size factors for normalisation. We then defined the groups to test by classifying each cluster from Fig. 1C into endoderm, mesoderm or ectoderm (as indicated in Supplementary Fig. 1E). Finally, we used generalised linear models to test each pairwise comparison (through glmFit and glmLRT) and corrected the returned p-values for multiple testing using the Benjamini & Hochberg method.
To identify genes that are preferentially expressed in a given germ layer, we first computed the third quartile for each gene across the 20 cell populations (Fig. 1C). We excluded all genes with a value greater than zero in more than 10 populations; this ensures that the genes to be analysed are not ubiquitously expressed. For each germ layer, we required significant adjusted p-values (FDR < 5%) in their comparisons against the other two germ layers, and a positive log-fold-change, to retain the genes significantly upregulated. The resulting gene lists can be found in Supplementary Table 2.
Characterisation of early specification of foregut cells
To characterise the substructure within the foregut cells, we recomputed the set of highly variable genes as described above, and selected those that were highly correlated among them. We then constructed a diffusion map on the log-transformed matrix of expression of these genes in the foregut cells (DiffusionMap function with default options, destiny R package39). To find sub-clusters, we used the k-branches algorithm40 on the first two diffusion components (kbranches.global function in kbranches R package; the parameter fixed_centre was set to the averages of DC1 and DC2). The gap statistics (performed with clusGap function in cluster package41) suggested the existence of three sub-clusters (Supplementary Fig. 2A). We identified differentially expressed genes between these three sub-clusters in an analogous way as described above for the germ layers. The resulting gene lists can be found in Supplementary Table 3.
Induction of human pluripotent stem cells into foregut progenitors
Human embryonic stem cells were differentiated towards foregut using chemically defined media as described in42, and harvested at day 7 of differentiation. Three biological replicate samples were analysed by bulk RNA-seq using standard Illumina protocols. Reads were mapped to Ensembl GRCh38, release 77 (http://oct2014.archive.ensembl.org/index.html), of the human genome using TopHat 2.0.1043. We supplied TopHat with the gene model annotations and known transcripts using the option ’-GTF’; all other parameters were left with their default values. Only read alignments with mapping quality score MAPQ>10 were kept for further processing. Finally, we used featureCounts44 from the Subread package to count the number of reads mapping uniquely to exons.
Comparison of induced human foregut progenitors to the mouse cell atlas
First, we recomputed the highly variable genes for the foregut and mid-hindgut subpopulations and computed the distance matrix as described earlier. We found three clusters using a dynamic tree cut algorithm (minimum cluster size of 30); based on the expression of marker genes we annotated these as foregut, midgut and hindgut.
We then ran the “pairs” classifier15 implemented in the scran34 R package to compare the human foregut stem cell samples to the mouse endodermal cells from the fore-, mid- and hindgut clusters (Supplementary Fig. 2B). The classifier was trained on the mouse data with the sandbag function, by considering only genes with a 1:1 human ortholog (as annotated in the Ensembl database) that were differentially expressed between the three clusters of mouse gut cells.
For the Principal Component Analysis shown in Fig. 2C we used the top 200 genes that were differentially expressed between the three clusters of mouse gut cells, further restricted to 1:1 orthologs in human. In order to reduce confounding effects due to technical reasons, quantile normalisation was performed jointly on the mouse and human data.
Pseudo-space ordering of presomitic and somitic cells
The mesoderm progenitors, presomitic and somitic mesoderm cells are split into four clusters. We noted that the smallest cluster of presomitic mesoderm (light green in Fig. 1C) is scattered across the tSNE and, also, that these cells have a significantly higher number of genes expressed compared to the rest of the dataset; this might indicate the presence of doublets. Thus we excluded this cluster from downstream analyses. To order the remaining cells along the anteroposterior (AP) embryo axis, we reasoned we could use the information provided by the Fgf8 signalling gradient, which decreases as cells become more anterior. When visualised in a tSNE plot, the three remaining clusters showed a trajectory correlated to Fgf8 expression levels. However, there was a group of cells negative for Fgf8 at the start of the trajectory that instead expressed markers of the adjacent neural tube cluster. We thus identified the substructure in the mesoderm progenitors cluster and removed the subpopulation of cells that did not express Fgf8.
To order cells along the Fgf8 gradient, we first identified the top 300 genes significantly correlated (both positively and negatively) with Fgf8 using the correlatePairs function from scran34. We visually inspected this set of genes and removed any that did not increase or decrease monotonically, retaining 260 genes. We then used the expression data of these genes to construct a diffusion map (DiffusionMap function in the destiny package39). Finally, we calculated the diffusion pseudotime with the DPT function to order the cells along the inferred trajectory. We refer to this quantity as pseudo-space, since the cells were ordered along the embryo’s AP axis.
Identifying genes that have dynamic expression along pseudo-space
To identify genes that change their expression levels along the pseudo-space trajectory we regressed the binarised expression levels (1=expressed; 0=not expressed) along the pseudo-space of all genes with mean expression of at least 0.1. For this we fitted a constant or a degree 2 model using local logistic regression (locfit function, with nn set to 1 and binomial as the family) and calculated Akaike’s information criterion (AIC) for each. We selected genes that were better fitted by the degree 2 model by computing the difference (ΔAIC) of the AIC of the degree 2 model minus the AIC of the null model. We retained all genes with a ΔAIC < -25.
To cluster the genes into different patterns of expression we predicted, for each gene, the values of the degree 2 model fit along the pseudo-space axis, and standardised each to be contained within [0,1]. We then computed Spearman’s correlation matrix and transformed it into a dissimilarity matrix by using the transformation √((1-ρ)/2). Finally, we used hierarchical clustering (method average) on the distance of the dissimilarity matrix, followed by the dynamic hybrid cut algorithm37 (minimum cluster size of 80) to define groups.
Validation of dynamic expression along the presomitic mesoderm
To confirm that the genes we identified as possible oscillating genes in the PSM were indeed cycling, we collected additional C57BL/6 embryos to isolate the PSM. Upon dissection, only pre-turned embryos were kept. Embryos were dissected in M2 media. The mesoderm was separated from the other germ layers after treatment with pancreatin for one minute at 37°C. The left and right sides of the PSM were finely dissected using tungsten needles, and each was cut into five segments along the anteroposterior axis. Each segment was collected in 15µl of lysis buffer (0.2% Triton X100 plus 1:20 RNase inhibitor (Clonetech)) that had been prepared fresh at the start of the dissections. Samples were vortexed, centrifuged and frozen on dry ice.
To prepare libraries for RNA-seq, samples were first processed with the Smart-seq2 protocol as described previously45; libraries were prepared using the Illumina Nextera XT DNA preparation kit. All libraries were pooled and sequenced on the Illumina HiSeq 4000 platform.
Data were aligned to the same mouse genome and annotation as used for the single cell data, with STAR 2.5.2a46. The numbers of fragments mapped to each gene were counted with the program featureCounts44 from the Subread package. Samples with fewer than three million reads were discarded. The remaining data was normalised for differences in depth of sequencing by using the method implemented in DESeq247. To model the expression pattern across the AP axis (segments 1 -> 5), we fitted a degree 2 model using local linear regression (locfit function, with nn set to 1); then we used this model to predict the expression levels across 17 regularly spaced intervals from most posterior to most anterior, to generate smoother profiles (Fig. 3H and Supplementary Fig. 3C).
Characterisation of molecular signatures within endothelial cells
For the endothelium study, we selected all cells in the four clusters annotated as endothelium (Fig. 1C). First, we re-calculated the highly variable genes and computed the distance matrix and tSNE as described above. We used hierarchical clustering (method average) followed by the dynamic hybrid cut algorithm37 (minimum cluster size of 20) to define groups. To characterise each subcluster, we used the findMarkers function from scran34 to identify genes that were preferentially expressed in a given group of cells; we removed genes with a median expression above zero in all subclusters. For the heatmap in Fig. 4C, we selected the top 5 differentially expressed genes for each cluster plus other informative markers based on the literature. We used these to annotate each cluster’s identity.
Assessing the role of the leukotriene pathway on blood production
HM-1 murine embryonic stem (ES) cells (kindly provided by David Melton) were grown in Knock Out DMEM (Gibco) supplemented with 15% serum batch tested for maintenance of pluripotency (Hyclone), 1000 U/ml leukaemia inhibitory factor (LIF) (Millipore), 2 mM L glutamine/100 U/ml penicillin/100 μg/ml streptomycin (Gibco), 0.1 mM β-mercaptoethanol (Gibco) at 37°C, 5% CO2, on gelatinised plates (Falcon, Corning) at a plating density of ~2×104 cells/cm2. Pluripotency was validated by their ability to differentiate into derivatives of the three germ layers. Cells were split every 2-3 days as necessary. ES cells were validated by their ability to differentiate into derivatives of the three germ layers and tested negative for mycoplasma contamination.
ES cells were harvested and plated on gelatinised dishes at a density of 4×104 cells/cm2 in standard ES growth medium (described above). 24 hours later the cells were dissociated and plated on gelatinised dishes at a density of 4×104 cells/cm2. 24 hours later the cells were dissociated again and washed once with PBS to remove all remaining ES medium and LIF. The cells were resuspended in IMDM based in vitro differentiation (IVD) medium containing 15% serum batch tested for EB differentiation (Gibco), 10% protein free hybridoma medium II (Gibco), 2 mM L-glutamine/100 U/ml penicillin/100 μg/ml streptomycin, 0.15 mM MTG, 180 μg/ml human transferrin (Roche) and 50 μg/ml L-ascorbic acid (Sigma) at a density of 104 cells/ml. The cells were plated in Costar low adherence 6-well plates (Corning) and incubated for 4 days at 37°C / 5% CO2 to form EBs. Zileuton (Sigma), LTC4 (abcam) or carrier were added on day 3 at the indicated concentrations (Fig. 5D). The EB suspension was harvested on day 4, transferred to appropriate tubes and the EBs were left to settle by gravity for 10 minutes. The medium was discarded, the EBs were washed with PBS and left to settle again by gravity. PBS was removed and the EBs were completely dissociated by addition of 1 ml TryplE and gentle pipetting. TryplE was inactivated by adding 10 ml IMDM containing 20% EB serum. The cells were counted, centrifuged at 300×g for 5 minutes at room temperature and resuspended in IVD medium. 4×104 cells were transferred in 4 ml of Methocult GF M3434 (Stem cell technologies) supplemented with 100 U/ml penicillin / 100 μg/ml streptomycin (Gibco). 1 ml aliquots were plated in triplicate in 35 mm low adherence dishes (Corning). Colonies were counted on day 14 and differences in colony numbers were tested with a two-tailed Student’s t-test.
To ensure that treatment with Zileuton or LTC4 does not affect the proliferation of the mESCs, 106 cells were harvested by centrifugation after dissociation of EBs on day 4 and washed in PBS. The cell pellet was resuspended in residual volume and fixed by dropwise addition of ice cold 70% methanol. The cells were incubated at 4°C for 1 hour and then washed twice with PBS. The cells were resuspended in 300μl of propidium iodide (PI) staining buffer (200μg/ml RNaseA, 20μg/ml propidium iodide, 0.1% Triton X100 in PBS) and stained at room temperature for 1 hour. The cells were analysed on a BD Fortessa. Post-acquisition analysis was performed with the FlowLogic suite (Supplementary Fig. 4B-C).
To ensure that treatment with Zileuton or LTC4 does not affect the viability of the mESCs, 106 cells were harvested by centrifugation after dissociation of EBs on day 4 and washed in PBS. The cells were resuspended in 100μl Annexin binding buffer (10 mM HEPES, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2) containing 5μl Annexin V APC (BD Biosciences; Cat. no. 550474; Lot. 16808) and 1 μg/ml DAPI. The cells were diluted up to 400 μl with Annexin binding buffer and analysed on a BD Fortessa cytometer. Post-acquisition analysis was performed with the FlowLogic suite (Supplementary Fig. 4D).
Statistics and reproducibility
Statistical test were performed in R and the details of the tests and p-values are stated in the text and figure legends. We have performed two independent single-cell sequencing experiments with animals from different genetic backgrounds, and reproduced all results in both. The first experiment used C57BL/6 embryos and consisted of two biological replicates; for one of these we performed two technical replicates. The second experiment used F2 embryos with a mixed genetic background from C57BL/6 and CBA, and consisted of three biological replicates, with two technical replicates each. For the validation experiments for the presomitic mesoderm section we performed the experiment in four different embryos; we kept separate the left and right portions of the PSM and each represents a biological replicate. We sequenced six biological replicates with similar results in all.
Supplementary Material
Acknowledgments
The authors would like to thank Aaron Lun for help with the cell ranger tool and the CRUK Cambridge Institute Genomics and Bioinformatics Cores for supporting the DNA sequencing and demultiplexing of the data. Research in the authors' laboratories is supported by the MRC, CRUK, Bloodwise, the Leukemia and Lymphoma Society, NIH-NIDDK, the Sanger-EBI Single Cell Centre and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust - MRC Cambridge Stem Cell Institute and by core funding from Cancer Research UK and the European Molecular Biology Laboratory. WJ is a Wellcome Trust Clinical Research Fellow. AS is supported by the Sanger-EBI Single Cell Centre. This work was funded as part of the Wellcome Trust Strategic Award 105031/D/14/Z "Tracing early mammalian lineage decisions by single cell genomics" awarded to Drs Wolf Reik, Sarah Teichmann, Jenny Nichols, Ben Simons, Thierry Voet, Shankar Srinivas, Ludovic Vallier, Bertie Göttgens and John Marioni.
Footnotes
Author contributions
W.J., B.P.-S., V.L., R.T., F.J.C.-N, C.M., J.N. and S.S. performed experiments. X.I.-S., B.P.-S. and A.S. analysed data. W.J., D.J.J., L.V. and B.D.S. provided expertise. X.I.-S., W.J., B.P.-S., A.S., D.J.J., L.V., B.G. and J.C.M. interpreted results. B.G. and J.C.M. conceived the project. X.I.-S., B.P.-S., A.S., B.G. and J.C.M. wrote the manuscript. All authors read and approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
Code availability
The R code used is available on request.
Data availability
The processed data reported in this paper have been tabulated and are available together with the raw data in ArrayExpress under accession numbers E-MTAB-6153 for the C57BL/6 dataset; E-MTAB-5728 for the C56BL/6 x CBA dataset; and E-MTAB-6155 for the validation of oscillating genes in the presomitic mesoderm dataset. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Kaufman M, Bard J. The Anatomical Basis of Mouse Development. Academic Press; San Diego, CA: 1999. [Google Scholar]
- 2.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Molecular cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grapin-Botton A. Endoderm specification. In: The Stem Cell Research Community, editor. StemBook. StemBook; 2008. [Google Scholar]
- 4.Inoue-Yokoo T, Tani K, Sugiyama D. Mesodermal and hematopoietic differentiation from ES and iPS cells. Stem cell reviews. 2013;9:422–434. doi: 10.1007/s12015-012-9388-1. [DOI] [PubMed] [Google Scholar]
- 5.Tang K, Peng G, Qiao Y, Song L, Jing N. Intrinsic regulations in neural fate commitment. Development, growth & differentiation. 2015;57:109–120. doi: 10.1111/dgd.12204. [DOI] [PubMed] [Google Scholar]
- 6.Spence JR, Lauf R, Shroyer NF. Vertebrate intestinal endoderm development. Developmental dynamics. 2011;240:501–520. doi: 10.1002/dvdy.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nature methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 8.Yap C, Goh HN, Familari M, Rathjen PD, Rathjen J. The formation of proximal and distal definitive endoderm populations in culture requires p38 MAPK activity. Journal of cell science. 2014;127:2204–2216. doi: 10.1242/jcs.134502. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, et al. A systematic screen for genes expressed in definitive endoderm by Serial Analysis of Gene Expression (SAGE) BMC developmental biology. 2007;7:92. doi: 10.1186/1471-213X-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Developmental cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Becker MB, Zulch A, Bosse A, Gruss P. Irx1 and Irx2 expression in early lung development. Mechanisms of development. 2001;106:155–158. doi: 10.1016/s0925-4773(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 12.Mou H, et al. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell stem cell. 2012;10:385–397. doi: 10.1016/j.stem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin V, et al. Regionalisation of the endoderm progenitors and morphogenesis of the gut portals of the mouse embryo. Mechanisms of development. 2008;125:587–600. doi: 10.1016/j.mod.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Andoniadou CL, et al. Lack of the murine homeobox gene Hesx1 leads to a posterior transformation of the anterior forebrain. Development. 2007;134:1499–1508. doi: 10.1242/dev.02829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Oates AC, Morelli LG, Ares S. Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development. 2012;139:625–639. doi: 10.1242/dev.063735. [DOI] [PubMed] [Google Scholar]
- 17.Dunwoodie SL, Rodriguez TA, Beddington RS. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mechanisms of development. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 18.Plisov S, et al. Cited1 is a bifunctional transcriptional cofactor that regulates early nephronic patterning. Journal of the American Society of Nephrology. 2005;16:1632–1644. doi: 10.1681/ASN.2004060476. [DOI] [PubMed] [Google Scholar]
- 19.Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nature reviews. Genetics. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- 20.De Val S, Black BL. Transcriptional control of endothelial cell development. Developmental cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scotti M, Kmita M. Recruitment of 5' Hoxa genes in the allantois is essential for proper extra-embryonic function in placental mammals. Development. 2012;139:731–739. doi: 10.1242/dev.075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- 23.Lee LK, et al. LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage. Cell reports. 2016;17:2286–2298. doi: 10.1016/j.celrep.2016.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGrath KE, et al. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell reports. 2015;11:1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scialdone A, et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535:289–293. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- 27.Slukvin I. Generating human hematopoietic stem cells in vitro -exploring endothelial to hematopoietic transition as a portal for stemness acquisition. FEBS letters. 2016;590:4126–4143. doi: 10.1002/1873-3468.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thambyrajah R, et al. New insights into the regulation by RUNX1 and GFI1(s) proteins of the endothelial to hematopoietic transition generating primordial hematopoietic cells. Cell cycle. 2016;15:2108–2114. doi: 10.1080/15384101.2016.1203491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, et al. Let-7 microRNA-dependent control of leukotriene signaling regulates the transition of hematopoietic niche in mice. Nature communications. 2017;8:128. doi: 10.1038/s41467-017-00137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, et al. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature. 2015;523:468–471. doi: 10.1038/nature14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cutler C, et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood. 2013;122:3074–3081. doi: 10.1182/blood-2013-05-503177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome biology. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolodziejczyk AA, et al. Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell stem cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krijthe J. Rtsne: T-Distributed Stochastic Neighbor Embedding using Barnes-Hut Implementation. R package version 0.11. 2015 [Google Scholar]
- 37.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 40.Chlis NK, Alexander Wolf F, Theis FJ. Model-based branching point detection in single-cell data by K-Branches clustering. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. cluster: Cluster Analysis Basics and Extensions. R package version 2.0.5. 2016 [Google Scholar]
- 42.Hannan NR, Segeritz CP, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nature protocols. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 45.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 46.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.