Abstract
Rotavirus, a leading cause of severe gastroenteritis and diarrhoea in young children, accounts for around 215,000 deaths annually worldwide1. Rotavirus specifically infects the intestinal epithelial cells in the host small intestine and has evolved strategies to antagonize interferon and NF-κB signalling2–5, raising the question as to whether other host factors participate in antiviral responses in intestinal mucosa. The mechanism by which enteric viruses are sensed and restricted in vivo, especially by NOD-like receptor (NLR) inflammasomes, is largely unknown. Here we uncover and mechanistically characterize the NLR Nlrp9b that is specifically expressed in intestinal epithelial cells and restricts rotavirus infection. Our data show that, via RNA helicase Dhx9, Nlrp9b recognizes short double-stranded RNA stretches and forms inflammasome complexes with the adaptor proteins Asc and caspase-1 to promote the maturation of interleukin (Il)-18 and gasdermin D (Gsdmd)-induced pyroptosis. Conditional depletion of Nlrp9b or other inflammasome components in the intestine in vivo resulted in enhanced susceptibility of mice to rotavirus replication. Our study highlights an important innate immune signalling pathway that functions in intestinal epithelial cells and may present useful targets in the modulation of host defences against viral pathogens.
Previous studies suggested that anti-rotavirus antibody isotype switching depends on Il-1β and Il-18 (ref.6), and that recombinant Il-18 prevents and resolves rotavirus infection7. However, the precise role of inflammasome signalling that mediates the maturation of these cytokines in the context of enteric virus infections is largely unknown. To directly interrogate whether inflammasomes participate in the restriction of enteric viruses, we orally inoculated suckling pups with mouse rotavirus. Notably, rotavirus infection potently induced caspase-1 (Casp1) p10 cleavage, indicative of inflammasome activation (Fig. 1a). In addition, mice deficient in Asc or Casp1, two universal inflammasome components, exhibited higher viral loads in the small intestine, increased fecal shedding of viral antigens, and more frequent incidences of diarrhoea compared to their wild-type littermates (Fig. 1b–d, Extended Data Fig. 1a), suggesting that inflammasome signalling protects against rotavirus infection.
Figure 1. Inflammasome signalling protects against rotavirus infection.
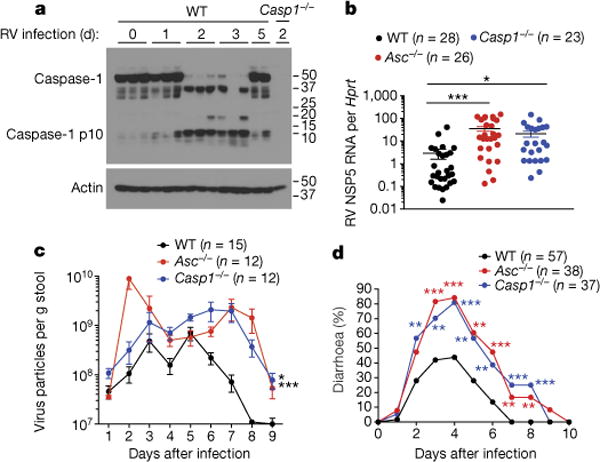
a, Western blots of Casp1 p10 cleavage. Each lane is representative of ileal tissues from an individual mouse with or without rotavirus (RV) infection. b–d, Rotavirus viral loads in ileum (mean ± s.e.m., Student’s t-test, **P < 0.01, ***P < 0.001) (b) or rotavirus fecal antigen shedding (mean ± s.e.m., two-way ANOVA, **P < 0.01) (c) or diarrhoea duration and percentage of mice with diarrhoea (score ≥ 2) (χ2 test, **P < 0.01, ***P < 0.001) (d), from rotavirus-infected wild-type (WT), Asc−/− and Casp1−/− mice. Representative of three (a) or combined from three, two, and four independent experiments, respectively (b, c, d).
The heightened susceptibility of Asc−/− (also known as Pycard−/−) mice to rotavirus infection implies a role of certain NLR(s) in this antiviral response. Therefore, we examined a panel of knockout mice deficient in Nlrp3, Nlrp6, Nlrc4 and Aim2, all of which are known to activate inflammasomes in the intestine8–11. With the exception of Nlrp6, none of these strains had increased susceptibility to rotavirus infection comparable to Asc−/− or Casp1−/− mice (Extended Data Fig. 1b, c). Given that mouse rotavirus has a specific tropism for the small intestine12, we systemically examined the transcription levels of all NLRs by RNA sequencing of ileum intestinal epithelial cells (IECs). Interestingly, as well as Nlrp6, Nlrc4 and Naips9,13–15, we discovered a novel NLR termed Nlrp9b that has not yet been characterized (Fig. 2a). Nlrp9b, but not the closely related Nlrp9a and Nlrp9c, was detected in different sections of the small and large intestines, highlighting its unique expression pattern16.
Figure 2. Nlrp9b deficiency results in increased susceptibility to rotavirus infection.
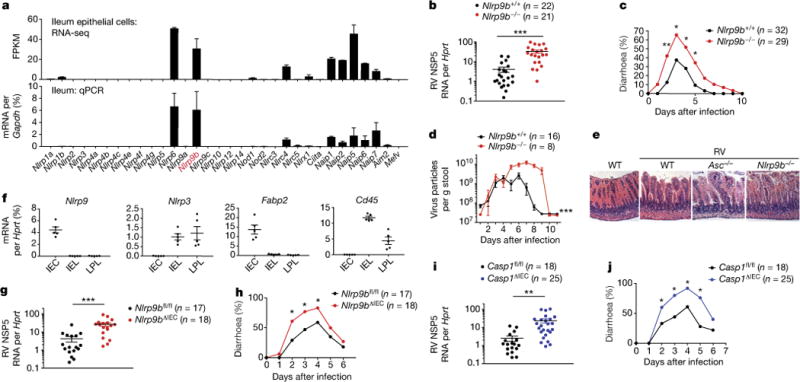
a, RNA sequencing and qPCR analysis of indicated NLR expression. b, c, Viral loads (mean ± s.e.m., Student’s t-test, ***P < 0.001) in ileum (b) and diarrhoea development (score ≥ 2) (χ2 test, *P < 0.05, **P < 0.01) (c) or rotavirus fecal antigen shedding (mean ± s.e.m., two-way ANOVA, ***P < 0.001) (d) from wild-type (Nlrp9b+/+) and Nlrp9b-knockout (Nlrp9b−/−) mice. e, Haematoxylin and eosin staining of rotavirus-infected ileal sections. f, qPCR analysis of gene expression in indicated cell populations. Fabp2, IEC marker; Cd45 (also known as Ptprc), haematopoietic marker. g–j, Viral loads in ileum (mean ± s.e.m., Student’s t-test, **P < 0.01, ***P < 0.001) (g, i) and diarrhoea development (score ≥ 2) (χ2 test, *P < 0.05) (h, j) in Nlrp9bfl/fl (WT), Nlrp9bΔIEC (Nlrp9bfl/fl Villin-Cre) (g, h), or Casp1fl/fl (WT), Casp1ΔIEC (Casp1fl/fl Villin-Cre) mice (i, j). a, f, qPCR experiment representative of three independent experiments; b, c, combined from three independent experiments; d, g–j, combined from two independent experiments; e repeated twice.
To functionally examine the potential anti-rotavirus role of Nlrp9b, we generated Nlrp9b−/− mice (Extended data Fig. 2c, d). Nlrp9b−/− mice resembled Asc−/− and Caspi−/− mice and also exhibited elevated viral load in IECs, fecal antigen shedding, diarrhoea occurrence and more severe pathology compared to wild-type pups (Fig. 2b–e). Notably, loss of Nlrp9b did not affect normal gut homeostasis (Extended data Fig. 2e, f). 16S ribosomal DNA sequencing further indicated that the microbiota composition in Nlrp9b−/− mice was not different from that in wild-type mice (Extended data Fig. 2f). Taken together, these findings strongly suggest that Nlrp9b, along with other inflammasome components, is involved in the host innate immune defence against rotavirus infection, independent of microbiota effects.
We next isolated both IECs and intraepithelial lymphocytes (IELs) from lamina propria and found Nlrp9b expression only in the EDTA-dissociated fraction, consisting of 80–90% IECs and <20% IELs (Extended Data Fig. 3a). A further separation using Villin-Cre-tomato+ reporter mice (Extended Data Fig. 3b) revealed that Nlrp9b and Il-18 were exclusively expressed in Villin-Cre-tomato+ IECs (Fig. 2f, Extended Data Fig. 3c). To definitively confirm an IEC-specific role of Nlrp9b, we generated conditional knockout mice with specific depletion of Nlrp9b in IECs (Nlrp9bΔIEC) (Extended Data Fig. 3d, e). Paralleling whole-body Nlrp9b−/− mice, Nlrp9bΔIEC mice were more susceptible to rotavirus infection compared to control (Nlrp9bfl/fl) mice (Fig. 2g, h). Likewise, Casp1ΔIEC mice with Casp1 deletion in IECs also showed enhanced susceptibility to rotavirus infection compared to control (Casp1fl/fl) mice (Fig. 2i, j). Taken together, these data strongly suggest that the Nlrp9b inflammasome functions only in IECs and restricts rotavirus infection.
To further dissect the molecular mechanisms underlying Nlrp9b protection against rotavirus infection, we examined the two downstream signals of inflammasome activation, the induction of pyroptosis and the secretion of Il-1β and Il-18. Consistent with previous publications17,18, we found disruption of the IEC monolayer and induction of cell death (Extended Data Fig. 4). Notably, rotavirus-induced IEC death at an early stage was largely dependent on Nlrp9b and Casp1 (Fig. 3a), whereas Aim2 was critical for radiation-induced death11. Using a mouse intestinal organoid model, we also found that Il-18 secretion from rotavirus infection was noticeably reduced in the absence of Nlrp9b, Casp1/11 or Asc (Fig. 3b, Extended Data Fig. 5), suggesting that Nlrp9b accounts for both downstream events. By contrast, interferons (IFNs) and other pro-inflammatory cytokines were comparably expressed in wild-type and Nlrp9b−/− mice after infection (Extended Data Fig. 5b, c).
Figure 3. Gsdmd-induced pyroptosis mediates the anti-rotavirus function of Nlrp9b inflammasome.
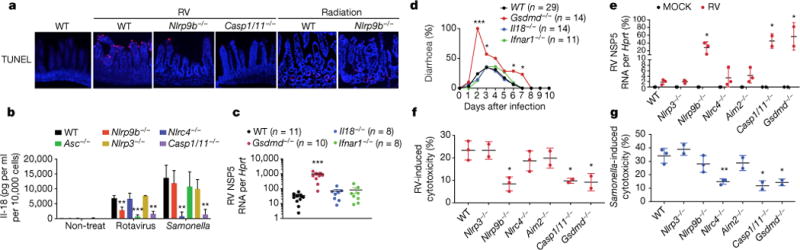
a, TUNEL staining of rotavirus-infected wild-type, Nlrp9b−/− and Casp1−/− ileal sections. b, Il-18 secretion from rotavirus- or Salmonella-infected intestinal organoids (mean ± s.e.m., Student’s t-test, **P < 0.01, ***P < 0.001). c, d, Viral loads (mean ± s.e.m., Student’s t-test, ***P < 0.001) (c) and diarrhoea development (score ≥ 2) (χ2 test, *P < 0.05, ***P < 0.001) (d) in Gsdmd−/−, Il18−/− and Ifnar1−/− mice. e, Viral loads of rotavirus-infected organoids (mean ± s.e.m., Student’s t-test, *P < 0.05). f, g, Cytotoxicity of rotavirus- or Salmonella-infected organoids (mean ± s.e.m., Student’s t-test, *P < 0.05, **P < 0.01). a, e–g, Representative of three experiments; b, c, combined from two independent experiments; d, combined from three independent experiments.
To assess the relative contribution of pyroptosis versus Il-18, we generated mice lacking gasdermin D (Gsdmd) (Extended Data Fig. 6a–d), a central mediator of pyroptosis19,20, and highly expressed in the small intestine (Extended Data Fig. 6e, f). Notably, mice deficient in Gsdmd but not Il-18 displayed marked susceptibility to rotavirus infection compared to wild-type and Ifnar1−/− mice (Fig. 3c, d), previously shown to minimally restrict homologous rotavirus infection21. Moreover, we observed more robust rotavirus replication and reduced IEC death from Nlrp9b-, Casp1/11- and Gsdmd-deficient intestinal organoids (Fig. 3e, f). In contrast, Nlrp9b deficiency had no effect on Salmonella infection (Fig. 3g), highlighting its specific role in RNA virus infections. Collectively, our results support that Nlrp9b partners with adaptors Asc and Casp1 to assemble the inflammasome and mediate IEC pyroptosis to confine rotavirus replication.
We next performed in vitro experiments to examine how rotavirus infection triggers the Nlrp9b inflammasome. Using HEK293T cells reconstituted with Flag-tagged human NLRP9 and HA-tagged ASC, we observed an interaction between the two only during rotavirus infection (Fig. 4a). Such binding was not detected for mouse Nlrp9b, probably owing to low expression. We hypothesized that rotavirus segmented double-stranded RNA (dsRNA) genome or other RNA byproducts during replication may activate cell-intrinsic innate immunity22. Indeed, despite the fact that the majority of rotavirus RNA is localized at the viroplasm23, we observed weak yet detectable levels of viral RNA in the cytoplasm during infection (Extended Data Fig. 7a). This is consistent with previous observations24 and suggests that rotavirus dsRNA might be recognized by and activate NLRP9. To this end, we performed assays to determine the ability of different sensors to bind to rotavirus RNA and found that NLRP9 co-precipitated with viral RNA more strongly than the controls NLRP6 and RIG-I (Fig. 4b). Interestingly, as opposed to NLRP6, which preferably bound to high molecular weight poly(I:C), a long dsRNA mimetic25, and RIG-I, with an affinity for 5′-triphosphate RNA26, NLRP9 preferentially bound to short dsRNA stretches, represented by low molecular weight poly(I:C) and weakly to 5′-triphosphate RNA (Fig. 4c). Intestinal organoid and in vivo studies corroborated our conclusion that Nlrp9b is not involved in the control of encephalomyocarditis virus (EMCV) (Extended Data Fig. 7b), which predominantly produces long dsRNA during infection27.
Figure 4. NLRP9 inflammasome recognizes rotavirus RNA pathogen-associated molecular pattern through DHX9.
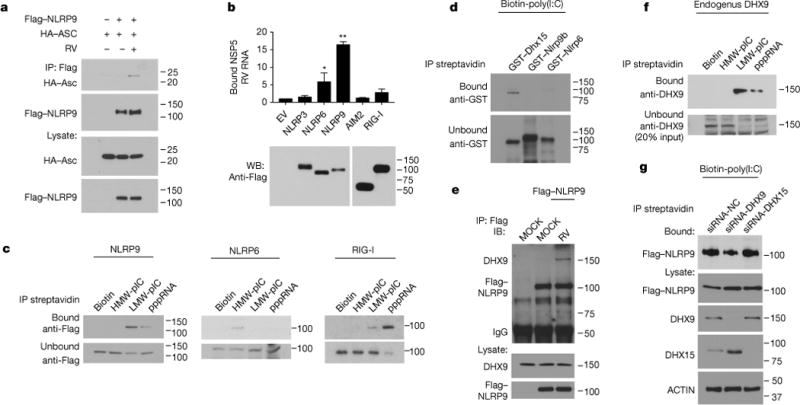
a, Rotavirus-infected HEK293T cells were co-transfected with Flag-NLRP9 and HA-ASC, and analysed by immunoprecipitation using the indicated antibodies. b, Rotavirus-infected HEK293T cells were transfected with indicated plasmids, immunoprecipitated and analysed by qPCR to detect bound rotavirus RNA (mean ± s.e.m., Student’s t-test, *P < 0.05, **P < 0.01). c, HEK293T cells transfected with indicated plasmids were immunoprecipitated with biotin-labelled RNA analogues. d, GST-tagged proteins were immunoprecipitated with biotin-labelled poly(I:C) (HMW+LMW). e, Rotavirus-infected HEK293T cells were transfected with Flag–NLRP9 and analysed by immunoprecipitation using indicated antibodies. f, Immunoprecipitation of DHX9 with biotin-labelled RNA analogs in HEK293T cells. g, HEK293T cells were transfected with Flag–NLRP9 and indicated siRNA and immunoprecipitated with biotin-labelled poly(I:C). a–c, Representative of three experiments; d–g, repeated twice.
We further tested whether Nlrp9b directly interacts with viral RNA. Although GST-tagged recombinant Dhx15 co-precipitated with biotinylated poly(I:C) as previously reported25, we did not observe any interaction for Nlrp9b (Fig. 4d), suggesting the potential involvement of other RNA binding proteins. To identify such factors in an unbiased manner, we performed immunoprecipitation–mass spectrometry for Flag-tagged NLRP9 and biotin-labelled viral RNA, respectively, and identified RNA helicase Dhx9 as a common hit in both data sets. Importantly, NLRP9 co-precipitated with endogenous DHX9 upon rotavirus infection (Fig. 4e) and the RNA-binding pattern of DHX9 also revealed a unique affinity for short dsRNA, similar to that of NLRP9 (Fig. 4c). Functionally, small interfering RNA (siRNA) knockdown of DHX9 but not DHX15 or RIG-I impaired the RNA-binding ability of NLRP9 (Fig. 4g, Extended Data Fig. 8a), and rotavirus-induced inflammasome formation (Extended Data Fig. 8b), in support of the hypothesis that the host RNA helicase DHX9 serves as a direct RNA-binding protein that mediates NLRP9 recognition of viral RNA. Consistently, rotavirus-infected Dhx9-deficient intestinal organoids were defective in both pyroptosis and Il-18 production (Extended Data Fig. 8c–f).
We next generated Flag-tag knock-in mice to study Nlrp9b regulation better (Extended Data Fig. 9a). Whereas Nlrp9b protein levels increased at 12 h.p.i (Extended Data Fig. 9b), Nlrp9b mRNA and protein levels decreased at 36 h.p.i. (Extended Data Fig. 9b, c), probably owing to negative feedback loop. We also measured Nlrp9b expression in ileum and colon at different days after birth. Intriguingly, we observed an age-dependent increase in Nlrp9b and Asc expression, but not Nlrp3, Casp1 and Rig-I (Extended Data Fig. 9d). Such a pattern was reported for Tlr3 (ref.28) (Extended Data Fig. 9d) and is particularly interesting as the upregulation of Nlrp9b and Asc levels coincides with the decreased susceptibility to rotavirus infection with age29,30.
Finally, we sought to examine a physiological relevance of NLRP9 in the human intestine. Compared to peripheral blood mononuclear cells and lamina propria lymphocytes, NLRP9 was only detected in the IEC fraction, whereas ASC and CASP1 were expressed in both populations (Extended Data Fig. 10a–c). We further validated that both NLRP6 and NLRP9 are highly expressed exclusively in primary human IEC organoids but not transformed intestinal epithelial cell lines (Extended Data Fig. 10d).
Using rotavirus infection as a model, our study comprehensively characterizes and functionally dissects the role of a novel NLR, Nlrp9b, with expression that is highly specific to IECs, in the host defence against enteric virus infection. Upon Dhx9 recognition of short dsRNA stretches, the Nlrp9b inflammasome is activated and promotes Il-18 release and Gsdmd-mediated pyroptosis. The latter is especially important for the host to restrict rotavirus replication by inducing premature death of infected IECs and maintaining intestinal homeostasis. Future studies on the Nlrp9b inflammasome regulation may enable the rational design of new therapeutics for infectious diseases.
METHODS
Mice
To generate Nlrp9b−/− mice, exon 2 and part of exon 3 of the Nlrp9b gene were targeted by two sgRNAs using CRISPR-Cas9 technology (Extended Data Fig. 2c). The deleted region contains the common translation-initiation codon ATG of two different isoforms. Genotyping of Nlrp9b−/− mice was performed using primers: 5′-TGGCTGACCTGGAACTCTCT-3′ and 5′-GCGTTACCAGGCTTTCTTTG-3′ for the wild-type allele; primers: 5′-GATCTCTGGCTGCCTCTGAC-3′ and 5′-CTTCCTCCAGTCATTGCACA-3′ for the targeted allele.
To generate Gsdmd−/− mice, exon 2 and part of exon 8 of the Gsdmd gene were targeted by two sgRNAs using CRISPR-Cas9 technology (Extended Data Fig. 6a). Genotyping of Gsdmd−/− mice was performed using primers: 5′-TCCTGAGTGCTGGAGGAACT-3′ and 5′-TGAATAACGGGGTTTCCAGA-3′ for the wild-type allele; primers: 5′-TCCTGAGTGCTGGAGGAACT-3′ and 5′-CCTCTAAGGCTTCCATGCTG-3′ for the targeted allele; and primers: 5′-TCCTGAGTGCTGGAGGAACT-3′ and 5′-TGAATAACGGGGTTTCCAGA-3′ for mutations generated by 5′-sgRNA.
These two knockout mice were both generated on C57BL/6N background, originally obtained from NCI.
To generate Nlrp9b-floxed mice, Nlrp9b-tm1a (KOMP) Wtsi ES cells (JM8A agouti) were purchased from and analysed by KOMP. Then ES cells were microinjected into C57BL/6 blastocysts. Chimaeric offsprings were backcrossed to C57BL/6 mice, and germline transmission was confirmed by PCR of tail genomic DNA. Founder mice were then crossed to Flpo mice to deplete the FRT-Neo-FRT cassette to generate Nlrp9b-floxed pups (Extended Data Fig. 3d). Genotyping of Nlrp9b-floxed mice was performed using universal primers 5′-ACAACCAACTGAGGGTCTGG-3′ and 5′-CACCTGATGCAGGTTTTCCT-3′, with amplicons of a 615-bp product from the wild-type allele, and a 770-bp product from the targeted allele.
Asc−/− (ref.31), Casp1−/− (refs11,32), Nlrp6−/− (refs9,33), Nlrp3−/− (ref.31) and Nlrc4−/− (ref.34) mice were previously generated and characterized in our laboratory. The Aim2−/− (ref.35) strain was provided by K. Fitzgerald (University of Massachusetts). Casp1/11−/− mice were purchased from the Jackson Laboratory.
Age and sex-matched C57BL/6 (CD45.2) mice from the National Cancer Institute were used as wild-type controls where indicated. Mice used in this study were on a C57BL/6 background. Mice were specific pathogen-free, maintained under a strict 12 h light cycle (lights on at 07:00 and off at 19:00), and given a regular chow diet (Harlan, diet 2018) ad libitum. All protocols used in this study were approved by the Yale Institutional Animal Care and Use Committee.
One set of experiments using wild-type C57BL/6, Asc−/− and Casp1−/− mice were conducted at Veterinary Medical Unit of Palo Alto VA Health Care System (PAVAHCS). The Institutional Animal Care Committee at the VAPAHCS approved these studies.
Human subjects
Informed consent was obtained from all human subjects enrolled in the study for biopsy sampling per protocol approved by the institutional review board at Yale University. Biopsies were obtained from the descending colon of four individuals undergoing screening colonoscopy. The experiments in this study involving human intestinal organoids (from two individuals) does not require submission to the Institutional Review Board at Stanford University because it does not contain individually identifiable private information.
Reagents
ELISA kits were purchased from eBioscience (mouse IL-18, mouse IL-1β, and mouse IFN-λ), and PBL (mouse IFN-β). Antibody pairs for ELISA detection of cytokines were purchased from BD Pharmingen (IFN-γ, IL-6) or from eBioscience (TNF).
Plasmids: human NLRP9 was cloned from colon biopsy cDNA, mouse AIM2, and ASC were cloned from intestine tissue cDNA, tagged with Flag or HA, and then inserted into a pcDNA3.1 vector. Flag-tagged NLRP3, NLRP6 and RIG-I were described previously25.
Tunnel kit for the in situ staining of intestinal epithelial cell death was purchased from Roche. Annexin V/7-AAD staining kit was purchased from eBioscience.
Cell culture: HEK293T (ATCC CRL-3216), HEK293 (ATCC CRL-1573) and Caco-2 (ATCC HTB-37) cells were cultured in DMEM with 10% FBS; HT-29 (ATCC HTB-38) cells were cultured in DMEM supplemented with F12 nutrients with 10% FBS. THP-1 (ATCC TIB-202) cells were cultured in RPMI medium with 10% FBS. These cell lines are not listed in the database of commonly misidentified cell lines maintained by ICLAC, and not authenticated or tested for mycoplasma contamination.
Virus infections
Rotavirus EW is a non-cell culture-adapted wild-type murine rotavirus strain. EW stock virus was prepared by infecting 5-day-old B6 mice, and harvesting crude centrifugation-clarified intestinal homogenate as previously described36. The virus titration of EW was expressed as 50% diarrhoea dose (DD50) defined as the highest dilution that causes diarrhoea in 50% of suckling B6 mice37.
For all rotavirus infection except indicated elsewhere, 8-day-old wild-type mice, or genetically deficient mice were orally inoculated by gavage with 1 DD50 EW virus in 50 μl PBS. The appearance of diarrhoea was monitored over time by changes in colour and consistency of feces. Stool samples were collected at different time points post inoculation. Fecal rotavirus antigen shedding was measured by ELISA assay as previously described38. At 5 days after infection, mice were killed and small intestine collected. EW in intestinal tissues were detected by RT–qPCR using a TaqMan assay based on rotavirus gene 11 (NSP5) sequences as previously described39. To monitor pyroptosis induction and Nlrp9b regulation during rotavirus infection, 8-day-old wild-type or Nlrp9b Flag-tagged mice were orally inoculated by gavage with 100 DD50 EW virus in 50 μl PBS, and intestinal tissues were harvested for TUNEL staining at 1.5–2 days after infection or western blot.
For all rotavirus infection experiments, diarrhoea was documented, and fecal and tissue samples were collected and measured in a double-blinded manner. The percentage and severity of diarrhoea among the littermates during the course of infection was recorded as previously described40. In brief, diarrhoea was scored on the basis of colour, consistency and amount, and numbered as follows: 0 = normal; 1 = pasty; 2 = semi-liquid; 3 = liquid, and consider score ≥ 2 as diarrhoea. Fecal specimens were collected into pre-weighed eppendorf tubes and stored at −80 °C.
Transfected HEK293T cells were infected with human rotavirus WA strain (MOI = 2) for 6 h (Fig. 4a) or 16 h (Fig. 4b) before collection for immunoprecipitation and western blot analysis.
EMCV (VR-129B) was purchased from American Type Culture Collection (ATCC) (Manassas, VA20110 USA). This method was previously described in ref.25.
Isolation of epithelial cells and lymphocytes in the intestine
Small intestines were excised and flushed thoroughly three times with PBS. They were opened longitudinally, transferred into HBSS with 2 mM EDTA, and shaken for 20 min at 37 °C. Supernatants were collected and the intestine washed 3 times with PBS. Supernatants were collected through a 100-mm cell strainer to get single-cell suspensions. Cells were collected as the IEC fraction. When Villin-Cre-tomato reporter mice were used, tomato-positive cells (IECs) and CD45.2-positive cells (IELs) were sorted. The lamina propia was then digested for 45 min at 37 °C with digestion solution (DMEM containing 2% FCS, 1 mg ml−1 collagenase, and 0.5 mg ml−1 DNaseI). Single-cell suspensions were obtained by grinding through a 100 mm cell strainer and then subjected to percoll gradient isolation. The interface cells were aspirated and spun down as LPL cells. The same approach was used with human colon biopsies to separate IEC fraction and lamina propria fraction.
Mouse intestinal epithelial cell (IEC) organoid culture
IntestiCult Organoid Growth Medium (Mouse) (STEMCELL, cat. 06005) was use for the establishment and maintenance of mouse intestinal organoids. Mice were killed according to approved institutional guidelines and approximately 15 cm small intestine was harvested from each mouse. The intestine was flushed gently with cold PBS and cut into 2 mm segments. A 10 ml serological pipette was used to wash the intestinal pieces by pipetting up and down 3 times. Then intestinal pieces settled by gravity and supernatant was removed. This wash procedure was repeated until supernatant was clear. Next, the supernatant was removed and intestinal pieces were suspended in 20 ml Gentle Cell Dissociation Reagent (STEMCELL, cat. 07174) in a shaker at 20 r.p.m. for 15 min at room temperature. Then supernatant was removed and the intestinal pieces were suspended in 10 ml cold PBS + 0.1% bovine serum albumin (BSA). The supernatant was removed and passed through a 70-μm filter into a 50 ml conical tube. This step was repeated for additional three times to generate four fractions. Then the specific fraction that was enriched for intestinal crypts was centrifuged at 290g for 5 min at 4 °C. The supernatants were discarded and each pellet was suspended in 10 ml of cold PBS + 0.1% BSA. The suspension was transferred to a 15 ml conical tube and centrifuged at 200g for 3 min at 4 °C. Then the supernatants were discarded and each crypt pellet was resuspended in 10 ml cold DMEM/F12. The quality of the suspensions was assessed by using an inverted microscope. The crypts were aliquoted into 15 ml conical tubes in volumes containing 20–50 crypts for each dome and centrifuged at 200g for 5 min at 4 °C. Then the supernatant was discarded and 150 μl complete IntestiCult Organoid Growth Medium at room temperature was added to each pellet. Next, 150 μl undiluted Matrigel Matrix was added to each tube and pipetted up and down to suspend the pellet. Then 50 μl suspension for each dome was pipetted into 24-well plate. These samples formed domes in the middle of each well. Next, the plate was incubated at 37 °C for 5–10 min until the Matrigel was solidified. Finally, 750 μl of complete IntestiCult Organoid Growth Medium at room temperature was added to each well and incubated at 37 °C and 5% CO2. The culture medium was exchanged every 3 days.
Mouse IEC organoid infection by rotavirus and Salmonella
Organoids were treated with LPS (1 μg μl−1) for 3 h and were dissociated by 1 ml warm TrypLE Express (Gibco, 12604013) for 5 min. After pipetting up and down 20 times, the suspension was centrifuged at 290g for 5 min. Then the supernatant was discarded and the pellets was resuspended in 1 ml DMEM/F-12 containing rotavirus (5 × 105 DD50, an estimated MOI of 5) or Salmonella (an estimated MOI of 10) and incubate at 37 °C for 1 h. After a spin down at 290g for 5 min, the supernatant was discarded and the pellets were resuspended with 200 μl complete IntestiCult Organoid Growth Medium. After an incubation of 3 h, the supernatant and cells were subjected to analysis of ELISA, qPCR and LDH release assay.
Human IEC organoid culture
Primary human intestinal organoids were provided by C. J. Kuo (Stanford University). Culturing methods were similar to a previous publication41. In brief, 3D culture of IEC organoids in Matrigel (Trevigen) was maintained in advanced DMEM/F12 media supplemented with epidermal growth factor, noggin, R-spondin, Wnt3A, nicotinamide, gastrin I, SB202190, B27, N2 and acetylcysteine. For measurement of NLRP9 and other gene expression, IEC organoids were harvested for RNA extraction followed by RT–qPCR analysis.
Relative gene expression analysis
Tissues were preserved in RNAlater solution (Life Technologies) and subsequently homogenized in Trizol reagent (Life Technologies). Cells subjected to FACS were resuspended in Trizol reagent. RNA was purified according to the manufacturer’s instructions. One microgram of total RNA was used to generate cDNA with Superscript III (Life Technologies). RT-PCR was performed using gene-specific primer/probe sets (Applied Biosystems) and Kapa Probe Fast qPCR kit (Kapa Biosystems) on a 7500 Fast Real Time PCR instrument (Applied Biosystems). Data were analysed using the Sequence Detection Software according the ΔCt method. All results were normalized to Hprt or Gapdh quantified in parallel amplification reactions.
Immunoprecipitation and western blot analysis
HEK293T cells transfected with different combinations of plasmids were harvested and resuspended in 1 ml ice-cold Pierce IP buffer (87787) containing protease inhibitors (Complete Mini EDTA-free, Roche and PMSF) and phosphatase inhibitor (NaF and NaVO3). Cells were lysed for 30 min at 4 °C and lysates were spun for 30 min at maximum speed at 4 °C. 20–40 μl anti-Flag M2 beads (Sigma, M8823) were added to the cell lysate and incubated overnight. Beads were washed five times with immunoprecipitation buffer and, finally, bound proteins eluted by boiling in loading buffer. Samples were separated on 4–12% Tris-Bis gels (Life Technology) and transferred onto PVDF membranes (Whatman). Western blot analysis was performed using antibodies against Flag (Sigma, F1804 or F7425), HA (Sigma, H3663 or Abcam, ab9110), Casp1 p10 (Santa Cruz, sc-514), DHX9 (Abcam, ab26271), DHX15 (Abcam, ab70454 or ab13311), Casp3 p17/p19 (Cell Signaling, 9661), LC3 (Cell Signaling, 2775), or β-actin (Sigma, A1978).
Statistical analysis
The sample size chosen for our animal experiments in this study was estimated based on our prior experience of performing similar sets of experiments. All animal results were included and no method of randomization was applied. For all the bar graphs, data were expressed as mean ± s.e.m. Statistical analyses were performed using GraphPad Prism 6. Survival curves were analysed using a log-rank (Mantel–Cox) test. Diarrhoea incidence was calculated using chi-square test. Fecal antigen shedding was analysed using a two-way ANOVA test. For other data, a standard two-tailed unpaired Student’s t-test or paired Student’s t-test was performed using GraphPad Prism 6. To compare two non-parametric data sets, a Mann-Whitney U-test was used. P values ≤ 0.05 were considered significant. The sample sizes (biological replicates), specific statistical tests used, and the main effects of our statistical analyses for each experiment were detailed in each figure legend.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq data sets have been deposited in GEO under the accession number GSE98388.
Extended Data
Extended Data Figure 1. Increase of rotavirus infection in Casp1-deficient but not NLR-deficient mice.
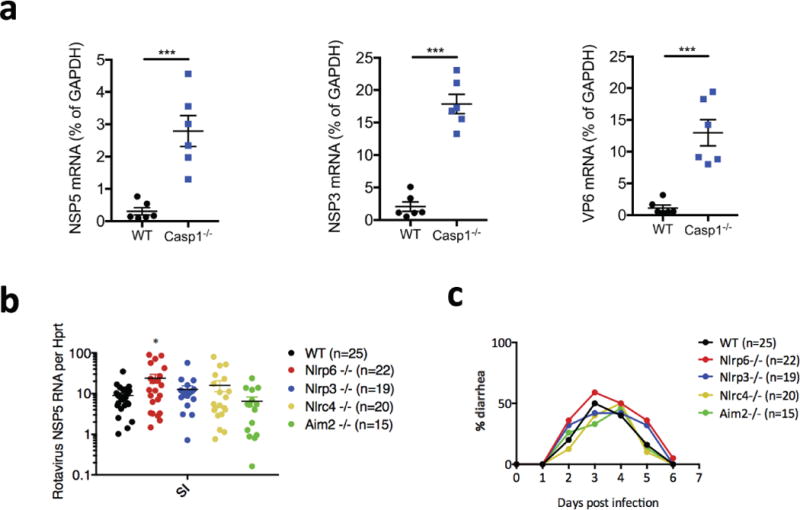
a, qPCR detection of viral loads in ileum by measuring different virus transcripts, non-structural protein 5 (NSP5), viral protein 6 (VP6) or non-structural protein 3 (NSP3) (mean ± s.e.m., Student’s t-test, ***P < 0.001) from wild-type and Casp1−/− suckling mice which were orally inoculated with rotavirus EW strain with diarrhoea dose 1 (DD)50. b, c, Viral loads in ileum (mean ± s.e.m., Student’s t-test, *P < 0.05) (b) or diarrhoea duration and percentage of diarrhoea (c) from wild-type and different inflammasome-forming NLRs or other receptor-knockout mice, which were orally inoculated with rotavirus EW strain with DD50. b, c, Representative of two independent experiments.
Extended Data Figure 2. Tissue-specific Nlrp9b expression and characterization of Nlrp9b-deficient mice.
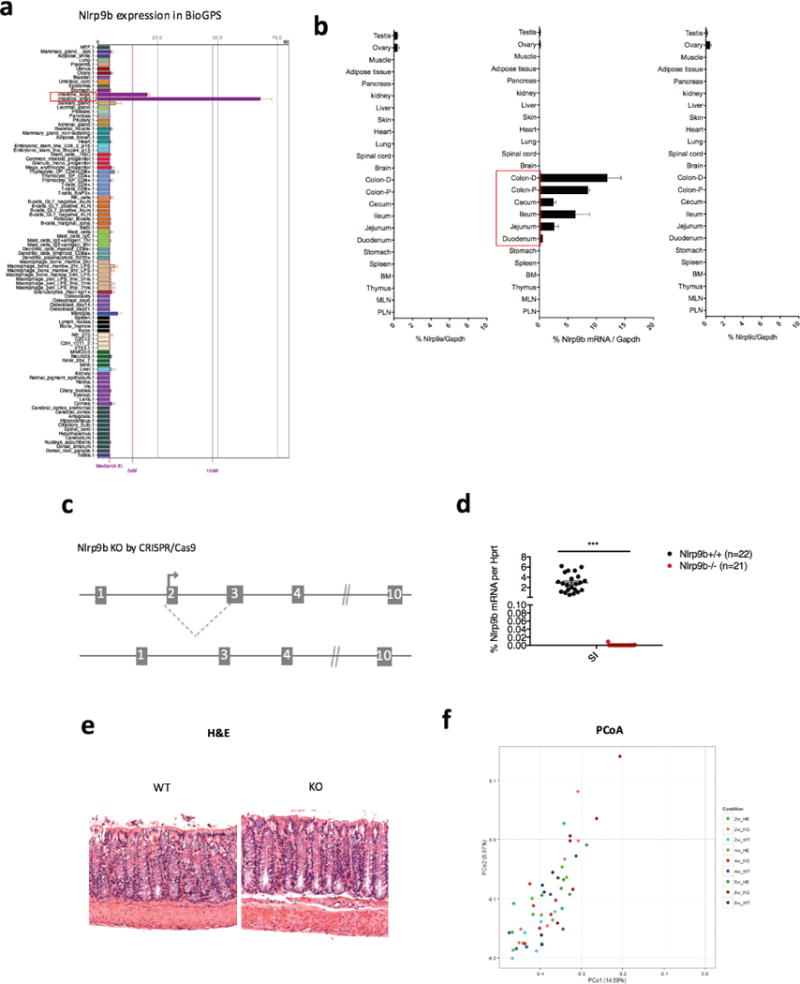
a, Nlrp9b expression in different mouse tissues as per BioGPS. b, qPCR verification of Nlrp9. a–c, Expression in different mouse tissues. c, Illustration of the CRISPR–Cas9 strategy to generate Nlrp9b-deficient mice. d, qPCR validation of Nlrp9b expression in the intestine tissues from littermate wild-type and Nlrp9b−/− mice (mean ± s.e.m., Student’s t-test, ***P < 0.001). e, Haematoxylin-and-eosin staining of ileal sections from wild-type and Nlrp9b-knockout mice in steady state. f, Unweighted UniFrac PCoA analysis followed by bacterial 16S rRNA sequencing of fecal microbiota harvested from littermate wild-type mice and Nlrp9b-knockout mice separated by indicated time points. b, Representative of three independent experiments.
Extended Data Figure 3. Specific Nlrp9b expression in IECs and generation of Nlrp9b IEC conditional knockout mice.
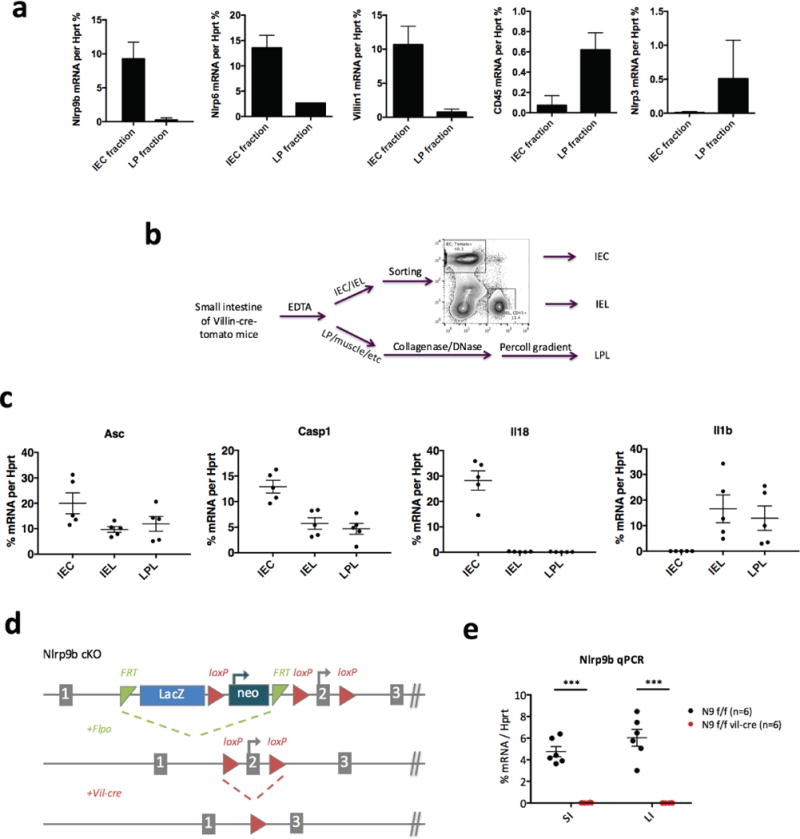
a, qPCR analysis of NLR inflammasome components in IEC fraction and IEL fraction isolated by EDTA. Vil1 is an IEC marker and Cd45 is a haematopoietic marker. b, Method to isolate IEC, IEL and LPL from intestine of Villin-Cre-tomato mice for experiment in Fig. 2f. c, qPCR analysis of NLR inflammasome components in intestinal epithelial cells (IECs), intraepithelial lymphocytes (IELs), lamina propria lymphocytes (LPLs), isolated according to the method showed in b. d, Workflow to generate Nlrp9b IEC conditional knockout mice. e, qPCR validation of Nlrp9b expression in IECs from Nlrp9b IEC conditional knockout and the littermate control (mean ± s.e.m., Student’s t-test, ***P < 0.001). SI, small intestine; LI, large intestine. a, c, Representative of three independent experiments.
Extended Data Figure 4. Rotavirus induction of cell death in the small intestine.
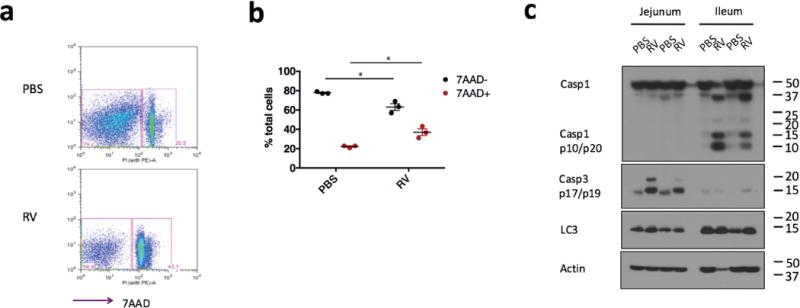
a, Representative cell death staining in IECs from wild-type mice orally inoculated with PBS or rotavirus. b, Summary of cell death and survival from the experiment in a (mean ± s.e.m., Student’s t-test, *P < 0.05). c, Western blots of different cell death indicators, Casp1 p10 (pyroptosis), Casp3 p17/p19 (apoptosis), and Lc3 I/II (autophagy), in the IECs from rotavirus infected ileum. a–c, Representative of two independent experiments.
Extended Data Figure 5. Establishment of intestinal organoid culture and the role of Nlrp9b in cytokine secretion.
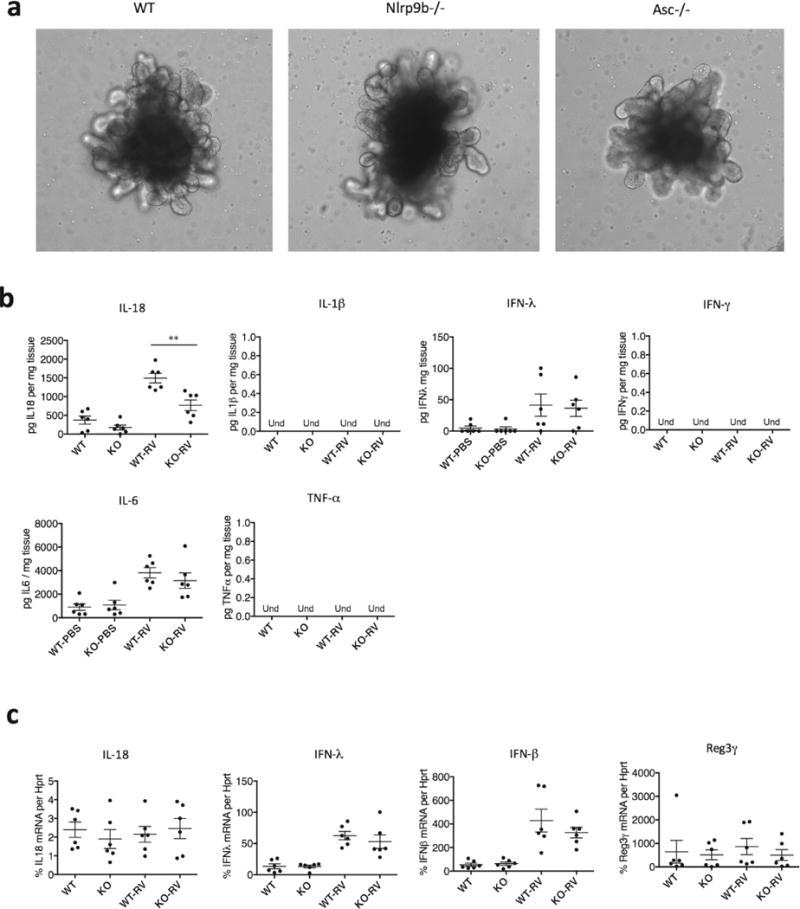
a, Single organoid (minigut) culture. Crypts are isolated from the small intestine of indicated mouse strains and cultured in Matrigel with IntestiCult growth medium from Stem Cell Technology for 3 weeks. b, ELISA measurement of cytokines and interferons (IFNs) production or c. QPCR analysis of cytokine, IFNs and anti-microbial peptides in the explants of wild-type and Nlrp9b−/− mice treated with rotavirus. (mean ± s.e.m., Student’s t-test, **P < 0.01). b, c, Representative of three independent experiments.
Extended Data Figure 6. Gsdmd expression in different tissues and generation of Gsdmd deficient mice.
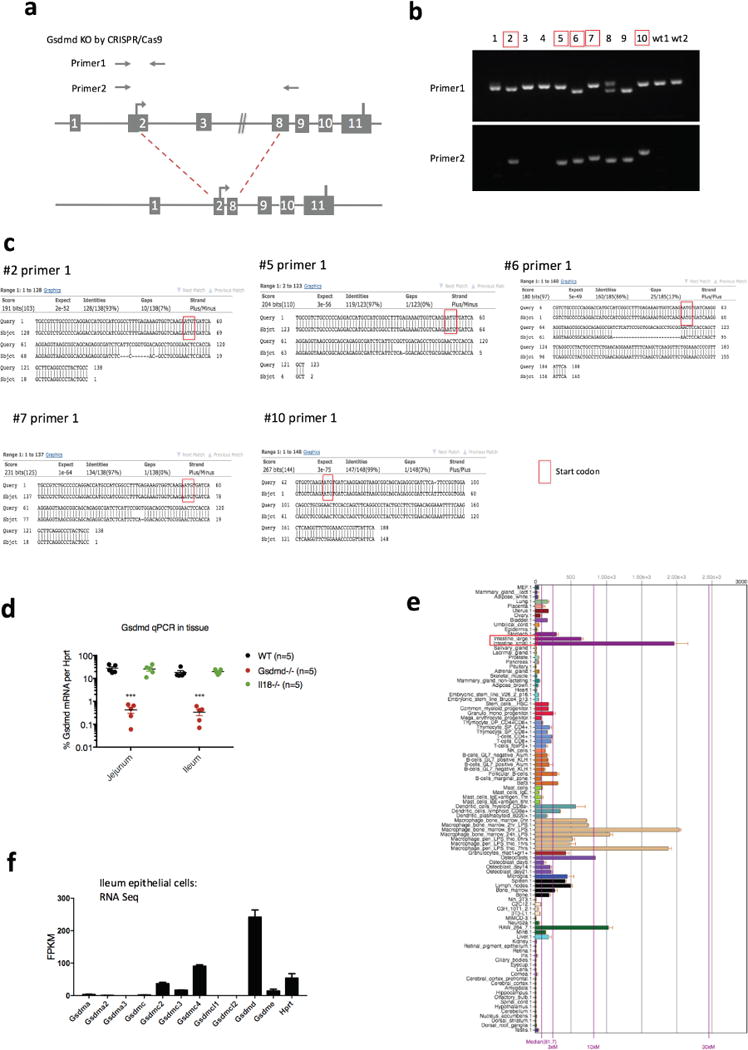
a, Illustration of the CRISPR–Cas9 strategy to generate Nlrp9b-deficient mice and primer design used in b. b, Genotyping of the Gsdmd mutant pups. The mice chosen for experiments shown in Fig. 3c, d are shown in red, with a deletion in one allele sequenced by primer 2, as well as mutations that result in frame shift in another allele sequenced by primer 1. c, Sequencing of redlabelled mice with primer 1. d, qPCR validation of Gsdmd expression in the intestine tissues from these five red-labelled Gsdmd mutant mice, comparing with wild-type mice and Il18−/− mice (mean ± s.e.m., Student’s t-test, ***P < 0.001). e, Gsdmd expression in different mouse tissues as per BioGPS. f, RNA sequencing data reveals the expression of gasdermin family members and housekeeping gene Hprt in the ileal epithelial cells of wild-type mice. b–d, Representative of two experiments and the data was combined in Fig. 3c and d.
Extended Data Figure 7. Rotavirus RNA distribution in the cytoplasm, and role of Nlrp9b in anti-EMCV response in the intestine.
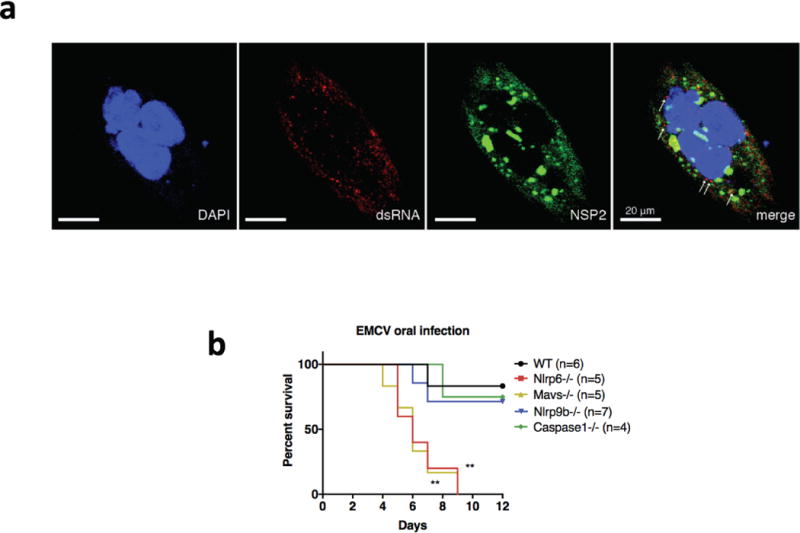
a, Human intestinal epithelial cell line HT-29 cells were seeded into chamber slides and infected with RRV at MOI = 1 for 24 h. Infected HT-29 cells were stained sequentially with anti-dsRNA (J2 mouse monoclonal Ab, 1:60), anti-mouse Alexa-594 secondary antibody (1:200) and Alexa-488-conjugated anti-NSP2 (clone 191, mouse monoclonal Ab, 1:250). Cells were mounted with anti-fade DAPI and processed for confocal microscopy (LSM710). b, Survival curve (Log-rank (Mantel-Cox) test, **P < 0.01) of different mouse strains challenged by EMCV (105 pfu) orally. b, Representative of two independent experiments.
Extended Data Figure 8. Role of Dhx9 in Nlrp9b-mediated antirotavirus response.
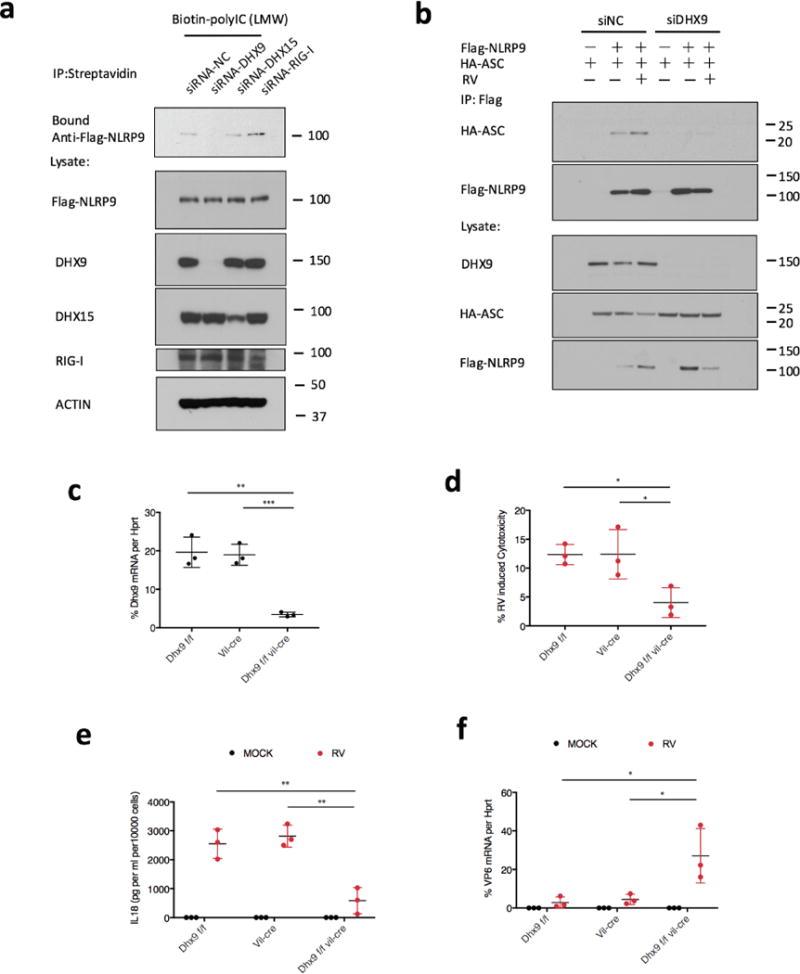
a, Biotin-labelled low molecular weight poly(I:C) were incubated with whole-cell lysates from HEK293T cells transfected with Flag-NLRP9 and indicated siRNA. b, HEK293T cells were transfected with DHX9-targeting siRNA or control siRNA for 24 h, co-transfected with Flag-NLRP9 and HA-ASC for another 24h, and infected with rotavirus human rotavirus WA strain (estimated MOI = 2) for 6 h. Cell lysates were co-immunoprecipitated with anti-Flag antibody and analysed by western blot with the indicated antibodies. c, Validation of Dhx9 conditional knockout mice. Dhx9 mRNA expression in intestinal epithelial cells from indicated mouse strains (mean ± s.e.m., Student’s t-test, **P < 0.01, ***P < 0.001). d, Cytotoxicity measured by lactate dehydrogenase (LDH) release in organoid cells from the indicated mouse strains infected with rotavirus EW strain (5 × 105 DD50, estimated MOI = 5) for 3 h (mean ± s.e.m., Student’s t-test, *P < 0.05). e, ELISA measurement of Il-18 secretion upon treating intestinal organoid cells from the indicated mouse strains with rotavirus for 3 h (mean ± s.e.m., Student’s t-test, **P < 0.01). f, qPCR detection of viral loads by measuring virus transcript VP6 in organoid cells from the indicated mouse strains infected with rotavirus for 6 h (mean ± s.e.m., Student’s t-test, *P < 0.05).
Extended Data Figure 9. Nlrp9b regulation during rotavirus infection and murine intestine.
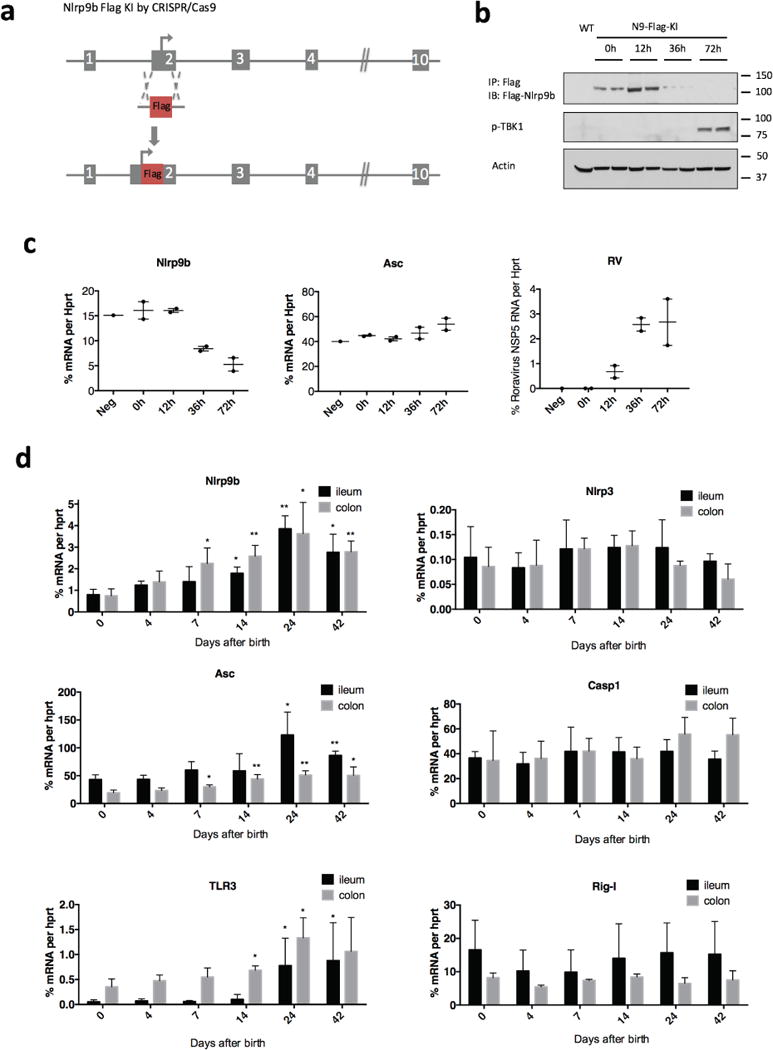
a, Workflow to generate Flag–Nlrp9 -knock-in mice. b, Immunoprecipitation and western blots detection of Flag-Nlrp9b protein, c, qPCR analysis of Nlrp9b, Asc and rotavirus transcripts, in IECs samples from Flag-Nlrp9b-knock-in mice that were infected with 100 DD50 of rotavirus by the indicated time points. d, Expression of select innate immune receptors or adaptors were measured by qPCR in different intestine sections isolated from mice with different ages as indicated in the figure. (mean ± s.e.m., Student’s t-test compare to expression levels in day 0, *P < 0.05, **P < 0.01.) Experiments in b and c were replicated twice.
Extended Data Figure 10. Human NLRP9 expression in intestinal epithelial cells.
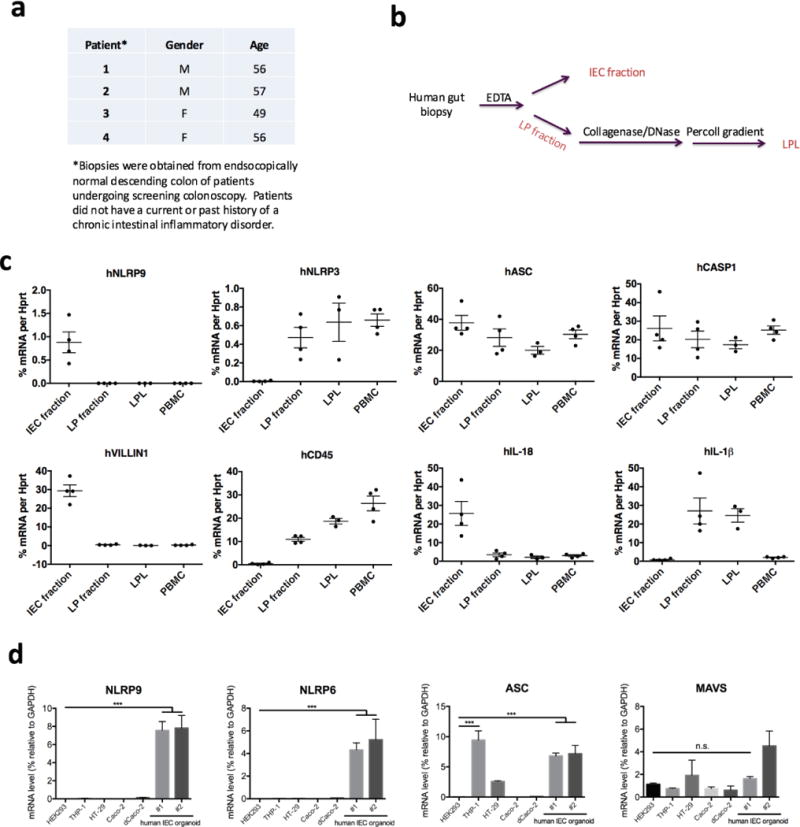
a, Clinical information of patients from whom the gut biopsies were collected. b, Method to isolate IEC fraction, lamina propria fraction and LPL from human gut biopsies. c, qPCR analysis of human NLRP9 inflammasome components in IEC fraction, lamina propria fraction, LPL and PBMC, which are isolated according to b. VILLIN1 is an IEC marker, and CD45 is a haematopoietic marker. d, Expression of indicated genes were measured by qPCR in human IEC organoids and select cell lines (mean ± s.e.m., Student’s t-test, ***P < 0.001).
Supplementary Material
Acknowledgments
We would like to thank C. Jin, B. Hu, A. Solis, P. Bielecki, R. Nowarski, R. Jackson, W. Bailis, A. Rongvaux, J. Henao-Mejia, A. Williams, E. Elinav, T Strowig, J. Alderman, C. Lieber, J. Stein, C. Hughes, L. Evangelisti, L. Borelli, P Ranney and other members of Flavell laboratory for technical help and helpful discussion. S.Z. is supported by a fellowship from Helen Hay Whitney Foundation-Howard Hughes Medical Institute. S.D. is supported by the Walter V and Idun Berry Postdoctoral Fellowship and an Early Career Award from the Thrasher Research Fund. P.W. is supported by R21AI103807. C.A. is supported by R01DK099097, R01DK106593 and R01AI120369. H.B.G. is supported by VA Merit Review grant I01BX000158, and NIH grants R01AI021362 and U19AI116484. R.A.F. and E.F. are investigators of the Howard Hughes Medical Institute, and are supported by U19AI089992.
Footnotes
Online Content: Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions: S.Z. wrote the manuscript, designed, performed and interpreted experiments with technical assistance from S.D. and P.H.W; S.D. and H.B.G. provided rotavirus related reagents, S.D. performed the rotavirus fecal shedding assay, qPCR and immunostaining experiments; P.H.W, W.P and Y.Z performed RNA binding experiments; Z.W. and H.C. performed infections in intestinal organoids; Y.Y. and H.Y performed TUNEL staining; H.-B.L. and K.A.J. helped with DHX9 functional studies; G.W., X.L. and Y.Z. assisted in the characterization of Nlrp9b mice; N.P., M.Z., and J.Z. performed 16s sequencing and PcoA analysis; N.F. performed the human IEC organoid experiment; C.A. provided human gut biopsies; S.D., P.H.W., N.W.P., W.P., L.S., Y.K., J.L., C.A., E.F., H.B.G. and R.A.F. provided critical comments and suggestions; R.A.F. supervised the project.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer Information: Nature thanks I. Goodfellow, C. Reis e Sousa and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Tate JE, et al. Effectiveness of pentavalent rotavirus vaccine under conditions of routine use in Rwanda. Clin Infect Dis. 2016;62(Suppl 2):S208–S212. doi: 10.1093/cid/civ1016. [DOI] [PubMed] [Google Scholar]
- 2.Sen A, Rott L, Phan N, Mukherjee G, Greenberg HB. Rotavirus NSP1 protein inhibits interferon-mediated STAT1 activation. J Virol. 2014;88:41–53. doi: 10.1128/JVI.01501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding S, et al. Comparative proteomics reveals strain-specific β-TrCP degradation via rotavirus NSP1 hijacking a host cullin-3–Rbx1 complex. PLoS Pathog. 2016;12:e1005929. doi: 10.1371/journal.ppat.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graff JW, Ettayebi K, Hardy ME. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 2009;5:e1000280. doi: 10.1371/journal.ppat.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barro M, Patton JT. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci USA. 2005;102:4114–4119. doi: 10.1073/pnas.0408376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. MyD88-mediated TLR signaling protects against acute rotavirus infection while inflammasome cytokines direct Ab response. Innate Immun. 2014;21:416–428. doi: 10.1177/1753425914547435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, et al. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346:861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirota SA, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordlander S, Pott J, Maloy KJ. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunol. 2014;7:775–785. doi: 10.1038/mi.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu B, et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354:765–768. doi: 10.1126/science.aaf7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng N, Yasukawa LL, Sen A, Greenberg HB. Permissive replication of homologous murine rotavirus in the mouse intestine is primarily regulated by VP4 and NSP1. J Virol. 2013;87:8307–8316. doi: 10.1128/JVI.00619-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy M, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, Macleod I, Su AI. BioGPS and MyGene. info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halasz P, Holloway G, Coulson BS. Death mechanisms in epithelial cells following rotavirus infection, exposure to inactivated rotavirus or genome transfection. J Gen Virol. 2010;91:2007–2018. doi: 10.1099/vir.0.018275-0. [DOI] [PubMed] [Google Scholar]
- 18.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 20.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 21.Lin JD, et al. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 2016;12:e1005600. doi: 10.1371/journal.ppat.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deal EM, Jaimes MC, Crawford SE, Estes MK, Greenberg HB. Rotavirus structural proteins and dsRNA are required for the human primary plasmacytoid dendritic cell IFNα response. PLoS Pathog. 2010;6:e1000931. doi: 10.1371/journal.ppat.1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvestri LS, Taraporewala ZF, Patton J. T Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol. 2004;78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rojas M, Arias CF, López S. Protein kinase R is responsible for the phosphorylation of eIF2α in rotavirus infection. J Virol. 2010;84:10457–10466. doi: 10.1128/JVI.00625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 27.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pott J, et al. Age-dependent TLR3 expression of the intestinal epithelium contributes to rotavirus susceptibility. PLoS Pathog. 2012;8:e1002670. doi: 10.1371/journal.ppat.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eydelloth RS, Vonderfecht SL, Sheridan JF, Enders LD, Yolken RH. Kinetics of viral replication and local and systemic immune responses in experimental rotavirus infection. J Virol. 1984;50:947–950. doi: 10.1128/jvi.50.3.947-950.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheridan JF, Eydelloth RS, Vonderfecht SL, Aurelian L. Virus-specific immunity in neonatal and adult mouse rotavirus infection. Infect Immun. 1983;39:917–927. doi: 10.1128/iai.39.2.917-927.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Case CL, et al. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc Natl Acad Sci USA. 2013;110:1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng N, et al. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J Virol. 2008;82:7578–7590. doi: 10.1128/JVI.00391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns JW, et al. Analyses of homologous rotavirus infection in the mouse model. Virology. 1995;207:143–153. doi: 10.1006/viro.1995.1060. [DOI] [PubMed] [Google Scholar]
- 38.Feng N, Burns JW, Bracy L, Greenberg HB. Comparison of mucosal and systemic humoral immune responses and subsequent protection in mice orally inoculated with a homologous or a heterologous rotavirus. J Virol. 1994;68:7766–7773. doi: 10.1128/jvi.68.12.7766-7773.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen A, Pruijssers AJ, Dermody TS, García-Sastre A, Greenberg HB. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J Virol. 2011;85:3717–3732. doi: 10.1128/JVI.02634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ball JM, Tian P, Zeng CQ, Morris AP, Estes M. K Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 41.Saxena K, et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol. 2015;90:43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq data sets have been deposited in GEO under the accession number GSE98388.


