Abstract
KT/HAK/KUP (KUP) family is responsible for potassium ion (K+) transport, which plays a vital role in the response of plants to abiotic stress by maintaining osmotic balance. However, our understanding of the functions of the KUP family in the drought-resistant crop cassava (Manihot esculenta Crantz) is limited. In the present study, 21 cassava KUP genes (MeKUPs) were identified and classified into four clusters based on phylogenetic relationships, conserved motifs, and gene structure analyses. Transcriptome analysis revealed the expression diversity of cassava KUPs in various tissues of three genotypes. Comparative transcriptome analysis showed that the activation of MeKUP genes by drought was more in roots than that in leaves of Arg7 and W14 genotypes, whereas less in roots than that in leaves of SC124 variety. These findings indicate that different cassava genotypes utilize various drought resistance mechanism mediated by KUP genes. Specific KUP genes showed broad upregulation after exposure to salt, osmotic, cold, H2O2, and abscisic acid (ABA) treatments. Taken together, this study provides insights into the KUP-mediated drought response of cassava at transcription levels and identifies candidate genes that may be utilized in improving crop tolerance to abiotic stress.
Keywords: cassava, drought stress, gene expression, identification, KUP family
Introduction
Potassium ion (K+) is an essential nutrient for various plant physiological functions, such as maintaining intracellular osmolality, cell turgor, and pH homeostasis. Although K+ is abundant on earth, the concentrations of K+ on the root surface is often lower than that in soil solution. Thus, plants depend on various K+ transport systems that mediate K+ uptake and transport to different plant tissues (Gierth et al., 2005; Ashley et al., 2006; Amrutha et al., 2007; Very et al., 2014). K+ transporters in plants can be classified into four major families: Trk/HKT, KT/HAK/KUP (KUP), CHX (cation/hydrogen exchanger), and KEA (K+ efflux antiporter) (Gupta et al., 2008). The KT/HAK/KUP family is the largest K+ transporter family and is responsible for K+ transport across membranes in bacteria, fungi, and plants (Li et al., 2017). The plant KT/KUP/HAK transporters were first isolated from barley (HAK1) and Arabidopsis (KUP1/KT1 and KUP2/KT2) according to their homologs to fungal HAK and bacterial KUP. Thus, the composite name of KT/HAK/KUP is used to widely represent the entire family in plants (Very et al., 2014). Mutation analysis has revealed that the 8th transmembrane domain and the C-terminus of KT/HAK/KUP play crucial roles in determining K+ transport capacity (Mangano et al., 2008). Moreover, plant KT/HAK/KUPs have different K+ affinity and are involved in cation influx and efflux (Nieves-Cordones et al., 2016). These KT/HAK/KUPs contain 10–15 transmembrane domains with both N- and C-termini at the intracellular side of the membrane, the latter being much longer (Gierth and Maser, 2007; Nieves-Cordones et al., 2016). Molecular evolution analysis has indicated that segmental duplications occurred 35.89–62.77 million years ago, resulting in its expansion in tomato (Hyun et al., 2014). Phylogenetic analysis has grouped plant KT/HAK/KUPs into four clusters (Rubio et al., 2000; Banuelos et al., 2002). Most of the KUPs in cluster I function in high-affinity K+ uptake, whereas cluster II are involved in low-affinity K+ transport (Gupta et al., 2008). To date, 13 and 27 KT/HAK/KUP gene family members have been identified from Arabidopsis and rice, respectively (Rubio et al., 2000; Maser et al., 2001; Banuelos et al., 2002; Gupta et al., 2008). Expression analysis suggests that most members of the Arabidopsis KT/HAK/KUP family expressed in the roots, siliques, leaves, and flowers. AtHAK5 expression is induced under conditions of K+ deprivation. Ten AtKT/KUPs expressed in root hairs and five of them expressed in root tip cells, thereby implying their role in K+ uptake (Ahn et al., 2004). In rice, transcripts of 26 OsHAK genes were detected in at least 1 of the 27 tested tissues, and five genes were observed to be expressed in all tissues in all three genotypes (Gupta et al., 2008). Moreover, the expression of KT/HAK/KUP genes in other plant species also supported their possible role in K+-mediated multiple biological processes, such as tissue development and abiotic stress responses (Su et al., 2002; Grabov, 2007; Song et al., 2015).
Some KT/HAK/KUP genes are essential for plant growth and development (Ahn et al., 2004). Knocking out AtKT3/KUP4 results in tiny root hairs, suggesting its function in cell expansion (Rigas et al., 2001). A mutation in AtKT/KUP2 (shy3-1) induces a dwarf phenotype, which results from a reduction in cell size (Elumalai et al., 2002). ARF2 directly bind to the promoter of HAK5, regulating root hair elongation (Zhao et al., 2016). Rice phloem has relatively high OsHAK5 transcript levels that regulates K+/Na+ ratio during shoot growth (Yang et al., 2014). OsHAK1 transcript abundance is elevated in the roots of K+-starved rice and OsHAK1 mutants exhibit a reduction in root and shoot growth (Chen et al., 2015). The GhKT1 was found to be associated with the expansion of cotton fibers in turgor-dependent growth (Ruan et al., 2001). VvKUP1 and VvKUP2 play a role in K+-mediated cell expansion in grape (Davies et al., 2006). These findings highlight the importance of KT/HAK/KUP transporters in plant development and K+ uptake.
Members of KT/HAK/KUP family also participate in stress-related responses. OsHAK1 expression is induced by K+ deficiency or salt stress and it confers salt tolerance by regulating K+ uptake and K+/Na+ ratio (Chen et al., 2015). Knocking out OsHAK21 decreases the K+/Na+ ratio and salt tolerance (Shen et al., 2015). Constitutive overexpression of OsHAK5 in tobacco improves K+ accumulation during salt stress and confers increased salt resistance (Horie et al., 2011; Yang et al., 2014). KUP2/6/8 plays a positive role in drought stress response by regulating osmotic homeostasis and the abscisic acid (ABA) response in Arabidopsis (Osakabe et al., 2013). Recently, INTEGRIN-LINKED KINASE1 (ILK1) was found to interact with and promote HAK5 accumulation and positively regulate osmotic stress tolerance in Arabidopsis (Brauer et al., 2016). Together, these studies reveal the crucial role of KT/HAK/KUPs in K+-mediated abiotic stress response.
Cassava (Manihot esculenta Crantz) is considered as a food crop and potential biofuel crop because of its high starch production (Zidenga et al., 2012). Cassava is highly resistant to abiotic stresses, such as drought and low nitrogen (Xu et al., 2013). Abiotic stress resistance in other crops may be improved using gene resources from cassava. However, the mechanism underlying cassava resistance to abiotic stress remains less known. Advancements in sequencing technologies have facilitated gene identification and expression analysis. We previously sequenced the genomes of different cassava subspecies (including wild ancestor species and modern cultivated species; Wang et al., 2014), which allows subsequent analysis of whole gene families in cassava.
To date, members of the KT/HAK/KUP family have been well-characterized in Arabidopsis, rice, peach, tomato, Physcomitrella patens, Selaginella moellendorffii and poplar by genome-wide analyses (Rubio et al., 2000; Maser et al., 2001; Banuelos et al., 2002; Gupta et al., 2008; Gomez-Porras et al., 2012; He et al., 2012; Song et al., 2015; Nieves-Cordones et al., 2016). In the present study, we identified 21 KUP genes (MeKUPs) from the cassava genome and analyzed their phylogenetic relationship, gene structure, conserved domain, and expression profiles in response to drought, salt, osmotic, cold, H2O2, and ABA treatments. Our analyses reveal the transcriptional control of MeKUP genes in different genotypes and candidate KUP genes that may be potentially utilized in improving crop resistance to abiotic stress.
Materials and methods
Plant materials and treatments
W14 (M. esculenta ssp. flabellifolia), a wild subspecies, is the nearest ancestor of cultivated cassava. It shows low photosynthesis rate, tuberous root yield, and starch content in tuberous root, but robust resistance to drought stress (Wang et al., 2014). KU50 is a representative cultivar of the cultivated cassava because of its high root yield and high starch content in tuberous root and extensively used in commercial plantations in East Asia (Utsumi et al., 2012; Wang et al., 2014). Arg7 is a variety containing elite agronomic traits, including a certain level of growth under moderate drought stress (Zhao et al., 2015). SC124, a widely planted cassava cultivar in China, can survive in prolonged severe drought stress (Zhao et al., 2015). Arg7, KU50, and W14 were used to study the expression profiles of KUP genes in different organs to get some clues on cassava organ development. W14 was confirmed to show stronger drought resistance than Arg7 and SC124 in our previous study (Hu et al., 2016). These three genotypes were selected to investigate the expression patterns of KUP genes in response to drought stress. Segments of cassava stems from mother plants were cultured in pots filled with soil and vermiculite (1:1) in growth room with a 16 h/35°C day and 8 h/20°C night regime, and a relative humidity of 70%. Thereafter, 90-day-old stems, 90-day-old leaves, 90-day-old tuberous roots (early), 150-day-old tuberous roots (middle), and 270-day-old tuberous roots (late) were acquired from KU50, Arg7, and W14 under normal conditions to study the expression levels of MeKUPs in distinct organs. To detect the transcriptional changes of MeKUPs in response to drought, leaves, and roots were collected from Arg7, SC124, and W14, respectively, under drought conditions for 12 d. For osmotic, salt, cold, ABA, and H2O2 treatments, 2-month-old seedlings of Arg7 were challenged with 200 mM mannitol for 2 h, 6 h, 3 d, 14 d, 18 d, and 24 d, 300 mM NaCl for 2 h, 6 h, 3 d, 14 d, 18 d, and 24 d, low temperature (4°C) for 2, 5, 15 h, 48 h following 7 and 14 d recovery, 100 μM abscisic acid (ABA) for 2, 6, 10, 24, 48, and 72 h, 10% H2O2 for 2, 6, 10, 24, 48, and 72 h, respectively.
Identification and evolutionary analysis
The protein sequences of AtKUPs in Arabidopsis and OsHAKs in rice were downloaded from UniPort and RGAP, respectively (Kawahara et al., 2013; The UniProt Consortium, 2015). The whole protein and nucleotide sequences of cassava were downloaded from the cassava genome database (Prochnik et al., 2012). The known KUP protein sequences were used to build HMM profiles that were employed to query the cassava dataset using HMMER software (Eddy, 2011; Finn et al., 2011). The identified KUPs from cassava were also validated by BLAST with KUPs from rice and Arabidopsis as queries. With the PFAM and CDD databases, the identified cassava KUPs were subjected to conserved domains validation (Marchler-Bauer et al., 2015; Finn et al., 2016). The evolutionary trees were constructed with the KUPs proteins from Arabidopsis, rice, and cassava using MEGA 5.0 and Clustal X2.0 softwares (bootstrap values for 1,000 replicates) (Larkin et al., 2007; Tamura et al., 2011).
Sequence analysis
ExPASy proteomics server was used to predict the molecular weight (MW) and isoelectric points (pI) of cassava KUP family proteins (Gasteiger et al., 2003). MEME program was employed to identify the conserved protein motifs of MeKUPs, which were further annotated with InterProScan (Mulder and Apweiler, 2007; Brown et al., 2013). The gene structures were assessed with the GSDS software (Hu B. et al., 2015; Hu W. et al., 2015). The cis-elements in the 1500 bp sequences upstream of the coding sequences were analyzed by PlantCARE databases. Those elements (ABRE, DRE, LTRE, ERE, MBS, and GARE) related to abiotic stress response were subjected to further analysis (Shinwari et al., 1998; Narusaka et al., 2003; Gou et al., 2010; Yun et al., 2010).
Transcriptome analyses
Total RNA of each sample was extracted with plant RNA extraction kit (TIANGEN, China) and used for cDNA library construction. The sequencing was performed with an Illumina GAII following manufacturer's instructions. Adapter sequences were removed with FASTX-toolkit. Clean reads were generated by removing low quality sequences using FastQC. Tophat v.2.0.10 was used to map the clean reads to the cassava genome. Using cufflinks, the transcriptome data was assembled (Trapnell et al., 2012). Reads Per Kilobase of exon model per Million mapped reads (FPKM) was employed to calculate gene expression levels. The transcriptiomic data was submitted to NCBI and the accession number was listed in Table S1.
Quantitative real-time PCR (qRT-PCR) analysis
qRT-PCR analysis was run on StratageneMx3000P (Stratagene, CA, USA) instrument using SYBR®Premix Ex Taq™ (TaKaRa). The relative expression of the tested MeKUP genes under different treatments was measured according to 2−ΔΔCt method (Livak and Schmittgen, 2001). The primer pairs were examined by melting curve, agarose gel electrophoresis, and sequencing PCR products (Table S2). The amplification efficiency was in the range of 0.92–1.04. The relative expression of MeKUP genes in each time point was calculated according to the control and treated samples that consist of three independent experiments.
Results
Identification of the KUP gene family in Manihot esculenta
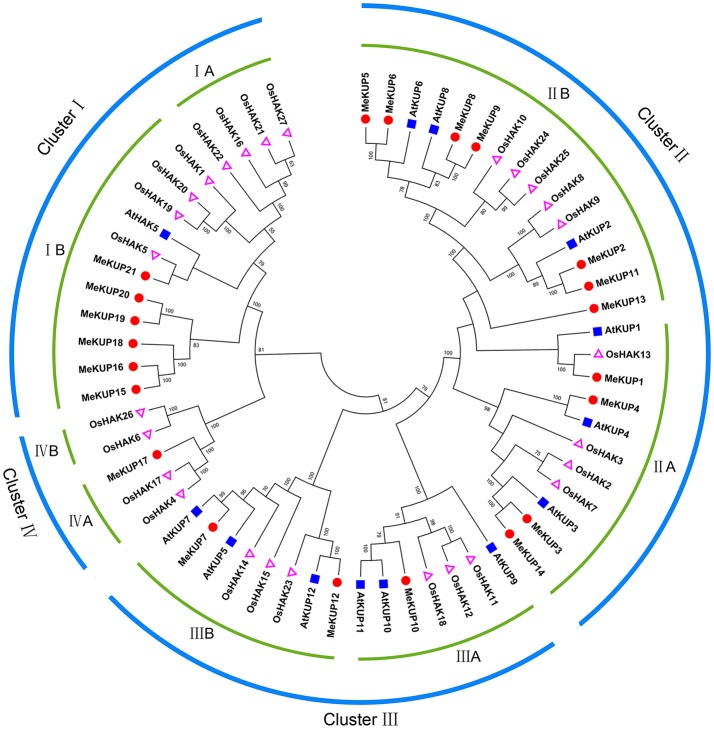
BLAST and Hidden Markov Model searches were conducted to extensively identify cassava KUPs using Arabidopsis and rice KUP protein sequences as queries. Twenty-one predicted full-length MeKUPs were identified in the M. esculenta genome, which were designated as MeKUP1-MeKUP21 based on their phylogenetic relationship with Arabidopsis. Conserved domain analysis further confirmed that all KUPs contain one K+ potassium transporter domain, which is hallmark of the KUP family (Table S3). The number of amino acid residues of the predicted MeKUPs ranged from 572 to 840, and their relative molecular mass varied from 63.97 to 87.93 kDa (Table S4). Phylogenetic analysis of the 21 MeKUPs together with 13 AtKUPs and 27 OsHAKs showed that the KUP family could be classified into four clusters (from I to IV). Cluster I included MeKUP15,-16,-18,-19,-20, and -21; Cluster II consisted of MeKUP1,-2,-3,-4,-5,-6,-8,-9,-11,-13, and -14; Cluster III comprised MeKUP7,-10, and -12; and Cluster IV included MeKUP17 (Figure 1; Table S5). The constructed dendrogram showed that MeKUPs were generally most closely related to the KUPs of Arabidopsis than those of rice, which coincides with current established plant evolutionary relationships.
Figure 1.
Phylogenetic analysis of KUPs from cassava, rice, and Arabidopsis using the complete protein sequences. The Neighbor-joining (NJ) tree was reconstructed using Clustal X 2.0 and MEGA 5.0 softwares with the pair-wise deletion option. One thousand bootstrap replicates were used to assess tree reliability.
Conserved motif analysis
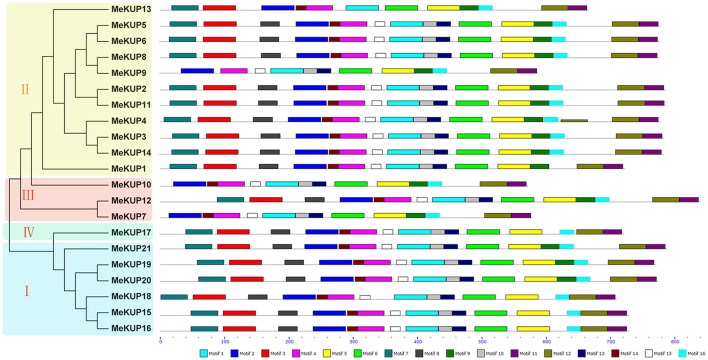
To study the structural features of the MeKUPs, conserved motifs were identified based on their evolutionary relationships. MEME database search identified 16 conserved motifs (Figure 2). After InterProScan search, motifs 1, 2, 3, 4, 6, 7, and 8 were annotated as K+ potassium transporter motif (Table S6). As shown in Figure 2, all the identified MeKUPs contained motifs 1, 2, 4, 5, 6, 11, and 12, suggesting that at least four K+ potassium transporter motifs existed in all 21 MeKUPs. The KUPs in Cluster I and IV harbored motifs 1–8 and 10–16. Cluster II KUPs showed motifs 1–16, except for MeKUP1, MeKUP9, and MeKUP13. Cluster III KUPs featured motifs 1–2, 4–6, and 9–16. Although some homologous KUPs had distinct motifs structures, such as MeKUP8/9 and MeKUP7/12, most of the homologous KUPs showed the same motif structure, including MeKUP5/6, MeKUP2/11, MeKUP3/14, MeKUP15/16, and MeKUP19/20. Together, these results indicate that all the identified MeKUPs have typical motifs of K+ potassium transporter, and each subgroup shares similar motif features, further supporting the phylogenetic classification of KUP family.
Figure 2.
The conserved motifs of cassava KUPs according to phylogenetic relationship. All motifs were identified by MEME database with the complete amino acid sequences of cassava KUPs.
Gene structure and promoter analysis
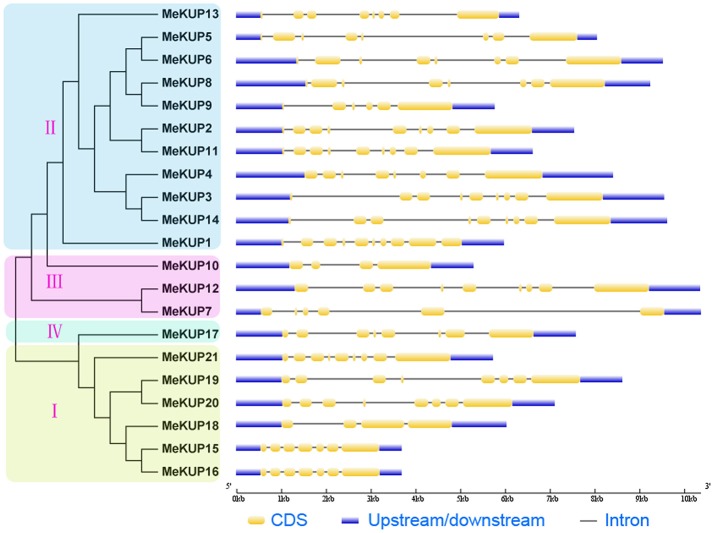
As shown in Figure 3, although two MeKUP genes (MeKUP10 and MeKUP18) had only four exons, the majority of the MeKUP genes harbored 6 to 10 exons. Additionally, some MeKUP genes in the same cluster had the same amount of exons such as MeKUP19,-20, and -21 in Cluster I, and MeKUP4,-5,-6,-8, and -13 in Cluster II. Furthermore, 1,500 bp upstream sequences of coding sequence from MeKUPs were identified, and the stress responsive cis-elements, including ABA-responsive element (ABRE), dehydration-responsive element (DRE), low temperature-responsive element (LTRE), ethylene-responsive element (ERE), MYB-binding site (MBS), and gibberellin-responsive element (GARE) in the MeKUP gene promoters were analyzed. The results revealed that 38.1% of MeKUPs contained ABRE, 23.8% contained LTRE, 19.0% contained ERE, 71.4% contained MBS, 28.6% contained GARE, and DRE was not found in all MeKUPs. From the above results, 85% of the MeKUPs (except for MeKUP7, MeKUP18, and MeKUP20) contained at least one of the tested elements in their promoter regions, suggesting the possible involvement of these genes in responses to different abiotic stressors.
Figure 3.
Gene structure analyses of cassava KUPs according to phylogenetic relationship. Exon-intron structure analyses were performed by GSDS database. The blue boxes, yellow boxes, and the black lines indicate upstream/downstream, exons, and introns, respectively.
Expression profiles of MeKUP genes in different tissues
To study the expression profiles of MeKUP genes in different tissues, transcriptome analyses of the leaves, stems, and storage roots in a wild subspecies (W14) and two cultivated varieties (Arg7 and KU50) were performed (Figure 4; Table S7). Fifteen MeKUP genes expressed in the tested tissues of the three genotypes. Moreover, MeKUP3,-4,-8,-12, and -14 showed high expression levels (Log2 based value >2) in all organs of the three cassava varieties. However, MeKUP1,-19, and -20 were highly expressed only in the leaves of W14. In addition, MeKUP5 had broadly high expression in W14, and only highly expressed in the stems of Arg7 and the leaves of KU50. These results implied the differential roles of these genes in tissue development in different genotypes.
Figure 4.
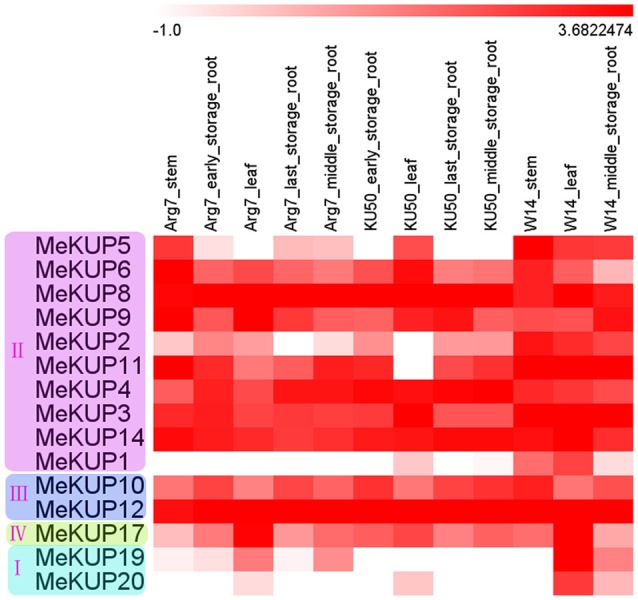
Expression profiles of cassava KUPs in different tissues of Arg7, KU50, and W14. Log2 based fold change was used to create the heat map. Fold changes in gene expression are shown in color as the scale.
Expression profiles of MeKUP genes in response to drought stress
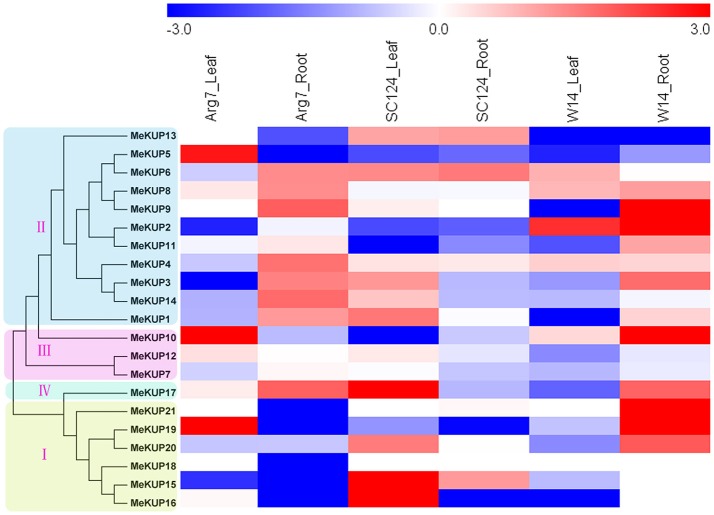
Because previous reports have revealed that the member of KT/HAK/KUP family participate in osmotic adjustment and drought stress response, the expression profiles of MeKUP genes in response to drought stress were further detected in three cassava genotypes by transcriptome analysis (Figure 5; Table S8). In the Arg7 variety, 3 and 8 of the 21 MeKUP genes were induced (Log2 based fold change >1) in leaves and roots after drought treatment. In the SC124 variety, 8 and 3 of the 21 MeKUP genes were upregulated (Log2 based fold change >1) in leaves and roots after drought treatment. In the W14 accession, 1 and 10 of the 21 MeKUP genes showed induction (Log2 based fold change >1) in leaves and roots after drought treatment. These results indicated that a higher number of MeKUP genes were upregulated in the roots in response to drought than that in leaves of Arg7 and W14, whereas fewer genes in roots than that in leaves of SC124 were induced after drought exposure. Generally, MeKUP genes show similar expression profiles in Arg7 and W14, which differs from that in SC124 after drought treatment. MeKUP3 showed repression (Log2 based fold change < 1) in the leaves of Arg7 and W14, whereas induction (Log2 based fold change >1) in the leaves of SC124. MeKUP-3,-8,-9, and -17 were upregulated (Log2 based fold change >1) in the roots of W14 and Arg7, whereas downregulated or no response in the roots of SC124.
Figure 5.
Expression profiles of cassava KUPs in response to drought stress in leaves and roots of Arg7, SC124, and W14. Log2 based fold change was used to create the heat map. Fold changes in gene expression are shown in color as the scale.
Notably, 3 (MeKUP-5,-10, and -19), 3 (MeKUP-15,-16, and -17), and 1 (MeKUP2) KUP genes showed strong induction (Log2 based fold change >2) after drought stress in the leaves of Arg7, SC124, and W14 respectively. In contrast, 5 KUP genes (MeKUP-2,-9,-10,-19, and -21) were strongly upregulated (Log2 based fold change >2) in the roots of W14 after drought treatment, whereas no KUP genes were strongly induced after drought treatment in the roots of Arg7 and SC124. Thus, the total number of strongly upregulated MeKUP genes was higher in W14 than in Arg7 and SC124.
Differential expression of MeKUP genes under abiotic stress and signal molecule treatments
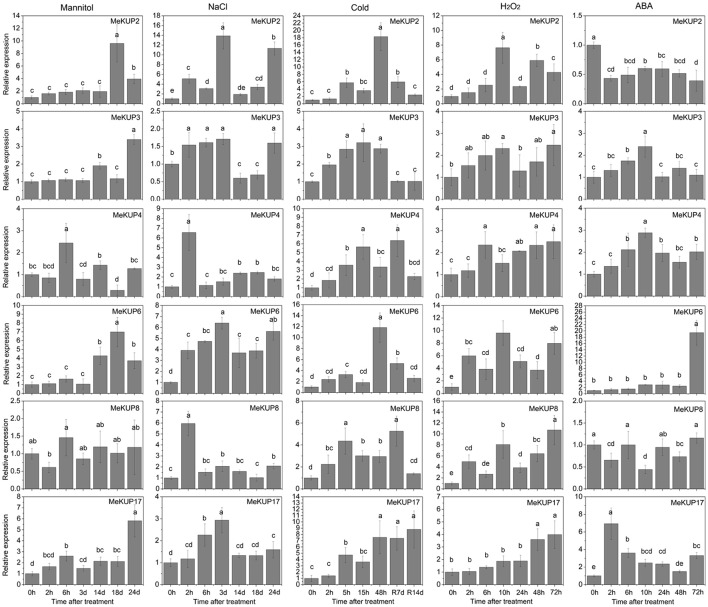
Based on the RNA-seq data, some MeKUP genes were upregulated in different cassava genotypes such as MeKUP2 in the leaves and roots of W14, MeKUP3 and MeKUP17 in the roots of Arg7 and W14 and the leaves of SC124, MeKUP4 in the roots of Arg7 and W14, MeKUP6 in the roots of Arg7 and SC124 and leaves of SC124, and MeKUP8 in the roots of Arg7 and W14. To investigate the response of MeKUP genes to abiotic stresses and related signaling at the transcriptional level, these six genes (MeKUP-2,-3,-4,-6,-8, and -17) were selected for further expression profiling after salt, osmotic, cold, H2O2, and ABA treatments (Figure 6). MeKUP2 was upregulated after 18–24 d of exposure to osmotic stress, 2–24 d of exposure to salt stress, 5–48 h of cold treatment followed by 7 d of recovery, and 10–72 h of H2O2 treatment, whereas it was downregulated by ABA treatment. MeKUP3 was upregulated after 2–3 d and 24 d of salt treatment, 14 and 24 d of osmotic treatment, 2–48 h of cold treatment, 2–10 h and 48–72 h of H2O2 treatment, and 2–10 h of ABA treatment. MeKUP4 was induced after 6 h and 14 d of osmotic treatment, and upregulated by salt, cold, H2O2, and ABA treatments at most of the tested time points. MeKUP8 was upregulated after $2 h-14 d and 24 d of salt treatment, 2–48 h of cold treatment and after recovery, and 2–72 h of H2O2 treatment, whereas downregulated after 2, 10, and 48 h of ABA treatment. In addition, MeKUP6 and MeKUP17 showed induction after all the treatments at several time points.
Figure 6.
Expression patterns of MeKUPs after osmotic, NaCl, cold, H2O2, and ABA treatments treatment in cassava. The mean fold changes of each gene between treated and control samples at each time point were used to calculate its relative expression levels. NTC indicates no treatment controls (mean value = 1). Data are means ± SD of n = 3 biological replicates. Means denoted by the same letter do not significantly differ at P < 0.05 as determined by Duncan's multiple range test.
Discussion
KUP genes play important roles in plant growth, development, and response to abiotic stresses (Osakabe et al., 2013; Zhao et al., 2016). Currently, our understanding of the role of KUP family in the drought-resistant crop cassava is limited. Here, we identified 21 KUPs from the cassava genome, which was classified into four clusters based on their evolutionary relationships (Figure 1). This is consistent with the previous classification of KUPs in Arabidopsis and rice (Rubio et al., 2000; Maser et al., 2001; Banuelos et al., 2002; Gupta et al., 2008). Moreover, the classification of cassava KUPs was further supported by conserved motif and gene structure analyses with each subgroup sharing similar motifs and exon-intron structures (Figures 2, 3). Conserved motif analysis suggested that all the identified KUPs had at least four typical motifs of K+ potassium transporters (Figure 2). In rice, all the 27 KUP members, except for OsHAK22, showed three conserved motifs (Gupta et al., 2008). Besides, some KUP proteins harbored a truncated K+ potassium transporter motif (He et al., 2012). This evidence indicates protein structure divergence among KUP family members over the course of evolution. Gene structure analysis showed that most of the MeKUP genes contained 6-10 exons, and the last exon exhibited the maximum length, which coincides with the exon-intron structure of KUP genes from Arabidopsis and poplar (Ahn et al., 2004; He et al., 2012). This indicates the conservation of the gene structure of the KUP family.
Organ expression profile analyses revealed 15 differentially expressed MeKUP genes in the stems, leaves, and storage roots of three cassava genotypes, including five (MeKUP3,-4,-8,-12, and -14) that showed high expression levels in all organs of the three cassava genotypes (Figure 4). In Arabidopsis, differentially expressed KUP genes were detected in the roots, older leaves, younger leaves, developing siliques, and flowers (Ahn et al., 2004). In rice, five KUP genes (OsHAK2, OsHAK10, OsHAK15, OsHAK23, and OsHAK25) expressed in all tissues of three genotypes (Gupta et al., 2008). In peach, KT/HAK/KUP genes expressed in nine tested tissues (Song et al., 2015). Additionally, three MeKUP genes (MeKUP1,-19, and -20) showed high expression only in the leaves of W14 (Figure 4). This phenomenon has also been observed in Arabidopsis and rice, with OsHAK5, OsHAK16, OsHAK25, AtKUP6, AKUP8, and AtKUP9 showing high expression in the leaves. Besides, the fruits and leaves of peach showed the highest number of expressed KT/HAK/KUP genes (Song et al., 2015). K+ is an essential nutrient for various physiological processes, particularly plant growth, development, and responses to abiotic stress (Ruan et al., 2001; Ahn et al., 2004; Osakabe et al., 2013). The high expression of KUP genes in leaves suggests its involvement in K+ transport during leaf development or environment adaptation.
Previous studies have revealed the fundamental role of osmotic adjustment in plant response to drought stress. Cellular osmotic balance is affected by various substances, including amino acids, sugars, and K+ (Osakabe et al., 2013). K+ uptake and efflux involve various types of channels and transporters that regulate water potential and turgor during osmotic adjustment (Very and Sentenac, 2003). Biochemical and genetic studies further support that the KUP/HAK/KT family transporters, including AtKUP2/6/8 and OsHAK1/5/21, positively regulate drought and osmotic resistances by influencing the K+-mediated ABA response, stomatal behavior, and osmotic homeostasis (Gierth and Maser, 2007; Grabov, 2007; Horie et al., 2011; Osakabe et al., 2013; Yang et al., 2014; Chen et al., 2015; Shen et al., 2015; Brauer et al., 2016). In the present study, we observed that drought stress induced the upregulation of several cassava KUP genes in the roots and leaves of different genotypes, suggesting their possible roles drought stress response. Generally, MeKUP genes showed similar expression profiles in Arg7 and W14, which differed from that in SC124 after drought treatment. The number of MeKUP genes upregulated by drought was significantly higher in the roots than that in the leaves of Arg7 and W14, whereas fewer genes were upregulated in the roots than in the leaves of SC124 (Figure 5). Previous studies have demonstrated that KUP genes positively regulate ABA response during lateral root formation and K+ efflux-mediated stomatal closure in leaves, thereby increasing plant resistance to drought and osmotic stresses (Osakabe et al., 2013). Based on this evidence, the expression diversity of KUP genes under drought stress implies differences in its roles in drought response in various cassava genotypes. For the Arg7 and W14 genotypes, a higher number of MeKUP genes are involved in drought-induced ABA responses in roots, whereas in SC124, more MeKUP genes participate in K+ efflux-mediated stomatal closure in the leaves under drought stress. Both functions contribute to cassava resistance to drought stress.
Additionally, the total number of strongly upregulated MeKUP genes (Log2 based fold change >2) was higher in W14 than in Arg7 and SC124 (Figure 5). W14 has greater drought resistance than Arg7 and SC124 (Hu et al., 2016). Previous studies have demonstrated that KUP genes play a positive role in drought or osmotic stress response by affecting ABA response, stomatal behavior, and osmotic homeostasis (Gierth and Maser, 2007; Grabov, 2007; Osakabe et al., 2013). These drought or osmotic responses involve KUP-mediated K+ uptake from the roots, transport from vascular tissues, and efflux from the leaves (Li et al., 2017). Thus, a higher number of strongly upregulated MeKUP genes in W14 may contribute to its drought resistance.
Previous studies have demonstrated the positive role of KUP genes in osmotic, drought, salt, or ABA responses, such as, AtKUP2, AtKUP6, and AtKUP8 in Arabidopsis, and OsHAK1, OsHAK5, and OsHAK21 in rice (Horie et al., 2011; Osakabe et al., 2013; Chen et al., 2015). Transcriptome analysis has identified several MeKUP genes that were responsive to drought stress (Figure 5). Thus, there is a need to investigate the expression patterns of MeKUP genes under various abiotic stress and stress-related signal molecule treatments. In the present study, we observed that all the tested genes (MeKUP-2,-3,-4,-6,-8, and -17) showed induction after salt and osmotic treatments at several time points, which coincides with the expression patterns of AtKUP2/6/8 in Arabidopsis (Osakabe et al., 2013). Notably, MeKUP2/6/8 is the homologs of AtKUP2/6/8 according to the evolutionary analysis, thereby suggesting that it may also be involved in abiotic stress response. All tested cassava genes were induced after cold and H2O2 treatments, which supplies a clue for further investigation of KUP-mediated cold and H2O2 responses. MeKUP6 and MeKUP17 were upregulated after all the treatments, indicating that these genes may play a role in multiple stress signaling pathways (Figure 6). Based on the importance of K+-mediated osmotic adjustment in plant response to abiotic stress, the observed response of MeKUP genes under abiotic stress, and the nature of cassava resistance to drought, it may be important to functionally characterize these genes.
In conclusion, this study identified 21 KUPs in cassava and investigated their classification, protein motifs, and gene structure. Transcriptome analysis revealed the potential role of MeKUP genes against drought stress in different cassava genotypes. Additionally, we identified several MeKUP genes that may be utilized as candidates for improving crop resistances to multiple stresses. Further studies are required to reveal the molecular mechanisms of MeKUPs in response to abiotic stress at translational and post-translational levels by biochemical and genetic approaches.
Author contributions
SZ, KL, and WH: conceived the study; WO, XM, CH, WT, YY, ZD, CW, ZX, and WW: performed the experiments and carried out the analysis; WH, WO, XM, and CH: designed the experiments and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31771859), the Natural Science Foundation of Hainan Province (317255), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052016005, 1630052016006, 1630052017021), the Central Public-interest Scientific Institution Basal Research Fund for Innovative Research Team Program of CATAS (17CXTD-28), the earmarked fund for Modern Agro-industry Technology Research System (CARS-11), and the Foundation of Hubei Provincial Department of Education (Q20143008).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00017/full#supplementary-material
References
- Ahn S. J., Shin R., Schachtman D. P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134, 1135–1145. 10.1104/pp.103.034660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrutha R. N., Sekhar P. N., Varshney R. K., Kishor P. B. K. (2007). Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 172, 708–721. 10.1016/j.plantsci.2006.11.019 [DOI] [Google Scholar]
- Ashley M. K., Grant M., Grabov A. (2006). Plant responses to potassium deficiencies: a role for potassium transport proteins. J. Exp. Bot. 57, 425–436. 10.1093/jxb/erj034 [DOI] [PubMed] [Google Scholar]
- Banuelos M. A., Garciadeblas B., Cubero B., Rodriguez-Navarro A. (2002). Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130, 784–795. 10.1104/pp.007781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer E. K., Ahsan N., Dale R., Kato N., Coluccio A. E., Pi-eros M. A., et al. (2016). The RAF-like kinase ILK1 and the high affinity K+ transporter HAK5 are required for innate immunity and abiotic stress response. Plant Physiol. 171, 1470–1484. 10.1104/pp.16.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P., Baxter L., Hickman R., Beynon J., Moore J. D., Ott S., et al. (2013). MEME-LaB: motif analysis in clusters. Bioinformatics 29, 1696–1697. 10.1093/bioinformatics/btt248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Hu Q., Luo L., Yang T., Zhang S., Hu Y., et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38, 2747–2765. 10.1111/pce.12585 [DOI] [PubMed] [Google Scholar]
- Davies C., Shin R., Liu W., Thomas M. R., Schachtman D. P. (2006). Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J. Exp. Bot. 57, 3209–3216. 10.1093/jxb/erl091 [DOI] [PubMed] [Google Scholar]
- Eddy S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elumalai R. P., Nagpal P., Reed J. W. (2002). A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14, 119–131. 10.1105/tpc.010322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A., et al. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M., Maser P. (2007). Potassium transporters in plants–involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 581, 2348–2356. 10.1016/j.febslet.2007.03.035 [DOI] [PubMed] [Google Scholar]
- Gierth M., Maser P., Schroeder J. I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ up take and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137, 1105–1114. 10.1104/pp.104.057216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Porras J. L., Ria-o-Pachón D. M., Benito B., Haro R., Sklodowski K., Rodríguez-Navarro A., et al. (2012). Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 3:167. 10.3389/fpls.2012.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J., Strauss S. H., Tsai C. J., Fang K., Chen Y., Jiang X., et al. (2010). Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22, 623–639. 10.1105/tpc.109.073239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A. (2007). Plant KT/KUP/HAK potassium transporters: single family - multiple functions. Ann. Bot. 99, 1035–1041. 10.1093/aob/mcm066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Qiu X., Wang L., Xie W., Zhang C., Xiong L., et al. (2008). KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genomics 280, 437–452. 10.1007/s00438-008-0377-7 [DOI] [PubMed] [Google Scholar]
- He C., Cui K., Duan A., Zeng Y., Zhang J. (2012). Genome-wide and molecular evolution analysis of the Poplar KT/HAK/KUP potassium transporter gene family. Ecol. Evol. 2, 1996–2004. 10.1002/ece3.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Sugawara M., Okada T., Taira K., Kaothien-Nakayama P., Katsuhara M., et al. (2011). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 111, 346–356. 10.1016/j.jbiosc.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G., et al. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Hou X., Xia Z., Yan Y., Wei Y., Wang L., et al. (2016). Genome-wide survey and expression analysis of the calcium-dependent protein kinase gene family in cassava. Mol. Genet. Genom. 291, 241–253. 10.1007/s00438-015-1103-x [DOI] [PubMed] [Google Scholar]
- Hu W., Xia Z., Yan Y., Ding Z., Tie W., Wang L., et al. (2015). Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front. Plant Sci. 6:914. 10.3389/fpls.2015.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun T. K., Rim Y., Kim E., Kim J. S. (2014). Genome-wide and molecular evolution analyses of the KT/HAK/KUP family in tomato (Solanum lycopersicum L.). Genes Genomics 36, 365–374. 10.1007/s13258-014-0174-0 [DOI] [Google Scholar]
- Kawahara Y., de la Bastide M., Hamilton J. P., Kanamori H., McCombie W. R., Ouyang S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Li W., Xu G., Alli A., Yu L. (2017). Plant HAK/KUP/KT K+ transporters: function and regulation. Semin. Cell Dev. Biol. [Epub ahead of print]. 10.1016/j.semcdb.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mangano S., Silberstein S., Santamaría G. E. (2008). Point mutations in the barley hvhak1 potassium transporter lead to improved K+-nutrition and enhanced resistance to salt stress. FEBS Lett. 582, 3922–3928. 10.1016/j.febslet.2008.10.036 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P., Thomine S., Schroeder J. I., Ward J. M., Hirschi K., Sze H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. 10.1104/pp.126.4.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N., Apweiler R. (2007). InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. 396, 59–70. 10.1007/978-1-59745-515-2_5 [DOI] [PubMed] [Google Scholar]
- Narusaka Y., Nakashima K., Shinwari Z. K., Sakuma Y., Furihata T., Abe H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. 10.1046/j.1365-313X.2003.01708.x [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M., Ródenas R., Chavanieu A., Rivero R. M., Martinez V., Gaillard I., et al. (2016). Uneven HAK/KUP/KT protein diversity among angiosperms: species distribution and perspectives. Front. Plant Sci. 7:127. 10.3389/fpls.2016.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Arinaga N., Umezawa T., Katsura S., Nagamachi K., Tanaka H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. 10.1105/tpc.112.105700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik S., Marri P. R., Desany B., Rabinowicz P. D., Kodira C., Mohiuddin M., et al. (2012). The cassava genome: current progress, future directions. Trop. Plant Biol. 5, 88–94. 10.1007/s12042-011-9088-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S., Debrosses G., Haralampidis K., Vicente-Agullo F., Feldmann K. A., Grabov A., et al. (2001). TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13, 139–151. 10.1105/tpc.13.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.-L., Llewellyn D. J., Furbank R. T. (2001). The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13, 47–60. 10.1105/tpc.13.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F., Santa-María G. E., Rodríguez-Navarro A. (2000). Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 109, 34–43. 10.1034/j.1399-3054.2000.100106.x [DOI] [Google Scholar]
- Shen Y., Shen L., Shen Z., Jing W., Ge H., Zhao J., et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 38, 2766–2779. 10.1111/pce.12586 [DOI] [PubMed] [Google Scholar]
- Shinwari Z. K., Nakashima K., Miura S., Kasuga M., Seki M., Yamaguchi-Shinozaki K., et al. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250, 161–170. 10.1006/bbrc.1998.9267 [DOI] [PubMed] [Google Scholar]
- Song Z. Z., Ma R. J., Yu M. L. (2015). Genome-wide analysis and identification of KT/HAK/KUP potassium transporter gene family in peach (Prunus persica). Genet. Mol. Res. 14, 774–787. 10.4238/2015.January.30.21 [DOI] [PubMed] [Google Scholar]
- Su H., Golldack D., Zhao C., Bohnert H. J. (2002). The expression of hak-type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 129, 1482–1493. 10.1104/pp.001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S., et al. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi Y., Tanaka M., Morosawa T., Kurotani A., Yoshida T., Mochida K., et al. (2012). Transcriptome analysis using a high-density oligomicroarray under drought stress in various genotypes of cassava: an important tropical crop. DNA Res. 19, 335–345. 10.1093/dnares/dss016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Very A. A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171, 748–769. 10.1016/j.jplph.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Very A. A., Sentenac H. (2003). Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 54, 575–603. 10.1146/annurev.arplant.54.031902.134831 [DOI] [PubMed] [Google Scholar]
- Wang W., Feng B., Xiao J., Xia Z., Zhou X., Li P., et al. (2014). Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 5:5110. 10.1038/ncomms6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Duan X., Yang J., Beeching J. R., Zhang P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161, 1517–1528. 10.1104/pp.112.212803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Zhang S., Hu Y., Wu F., Hu Q., Chen G., et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166, 945–959. 10.1104/pp.114.246520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K. Y., Park M. R., Mohanty B., Herath V., Xu F., Mauleon R., et al. (2010). Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 10:16. 10.1186/1471-2229-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Liu P., Shao J., Li C., Wang B., Guo X., et al. (2015). Analysis of different strategies adapted by two cassava cultivars in response to drought stress: ensuring survival or continuing growth. J. Exp. Bot. 66, 1477–1488. 10.1093/jxb/eru507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Zhang M. L., Ma T. L., Wang Y. (2016). Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28, 3005–3019. 10.1105/tpc.16.00684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidenga T., Leyva-Guerrero E., Moon H., Siritunga D., Sayre R. (2012). Extending cassava root shelf life via reduction of reactive oxygen species production. Plant Physiol. 159, 1396–1407. 10.1104/pp.112.200345 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.