Abstract
Wheat bran fibers are considered beneficial to human health through their impact on gut microbiota composition and activity. Here, we assessed the prebiotic potential of selected bran fractions by performing a series of fecal slurry anaerobic fermentation experiments using aleurone as well as total, ultrafine, and soluble wheat bran (swb) as carbon sources. By combining amplicon-based community profiling with a fluorescent in situ hybridization (FISH) approach, we found that incubation conditions favor the growth of Proteobacteria such as Escherichia and Bilophila. These effects were countered in all but one [total wheat bran (twb)] fermentation experiments. Growth of Bifidobacterium species was stimulated after fermentation using ultrafine, soluble, and twb, in the latter two as part of a general increase in bacterial load. Both ultrafine and swb fermentation resulted in a trade-off between Bifidobacterium and Bilophila, as previously observed in human dietary supplementation studies looking at the effect of inulin-type fructans on the human gut microbiota. Aleurone selectively stimulated growth of Dorea and butyrate-producing Roseburia. All fermentation experiments induced enhanced gas production; increased butyrate concentrations were only observed following soluble bran incubation. Our results open perspectives for the development of aleurone as a complementary prebiotic selectively targeting colon butyrate producers.
Keywords: wheat bran, aleurone, prebiotic, in vitro, fermentation, microbiome
Introduction
Although amplicon sequencing approaches are nowadays applied routinely to study the bacterial composition of the gut microbiota (Valles-Colomer et al., 2016), their application in prebiotic food ingredient research has been lagging behind (Hutkins et al., 2016). Only recently, 16S rRNA gene sequencing was applied to map the compositional changes induced by a dietary intervention using inulin-type fructans (Vandeputte et al., 2017). A community-wide analysis estimated the effect size of this prebiotic intervention in relative microbiome composition to 1.2% (Vandeputte et al., 2017) – modest, but comparable to the impact of top covariates of microbiome variation such as whole bread consumption (Falony et al., 2016). Unexpectedly, the study confirmed selective stimulation of a limited number of bacterial taxa upon inulin consumption, in line with the original definition of a prebiotic substrate (Gibson and Roberfroid, 1995). This observation has challenged the emerging scientific consensus regarding a community-level response of the gut microbiota to prebiotic interventions (Claus, 2017). It will most likely revive efforts to identify complementary food ingredients, selectively stimulating growth or activity of distinct sets of intestinal bacteria with potential beneficial properties.
A key aspect in maintaining gastrointestinal health lies in the consumption of dietary fiber. Fiber does not only accelerate intestinal transit (Burkitt et al., 1972), it also provides a variety of fermentable substrates to the intestinal microbiota (Sonnenburg and Sonnenburg, 2014). Fermentation of fiber polysaccharides increases microbiota production of short chain fatty acids (De Filippis et al., 2016), including butyrate. The latter not only represents the main energy source for colon epithelial cells (Roediger, 1982), it also affects cellular differentiation processes and has been shown to exert anti-inflammatory effects (Hamer et al., 2008; Louis et al., 2014). Moreover, the availability of readily fermentable, fiber-derived polysaccharides reduces potentially deleterious proteolytic fermentation processes (De Preter et al., 2010) and restrains microbial erosion of the mucus layer (Desai et al., 2016). By promoting growth and activity of commensal micro-organisms, dietary fiber consumption also reduces the risk of pathogenic invasion, both by enhancing colonization resistance and decreasing luminal pH (Lawley and Walker, 2013).
Wheat bran represents one of the main contributors to daily fiber intake in Western diets (Stevenson et al., 2012). Specific components of bran fiber such as arabinoxylans (Broekaert et al., 2011) have been shown to induce a prebiotic effect in the gut microbial ecosystem. Here, we contribute to the pursuit of novel potential prebiotics by assessing compositional and metabolic changes in fecal slurries upon fermentation of selected wheat bran fractions.
Results
Fecal Fermentations Impact Microbiota Composition
To assess the prebiotic potential of different wheat bran fractions, a series of 18 fecal fermentations experiments was set up using fecal material donated by six healthy volunteers (Supplementary Table S1). Selected bran fractions comprised total wheat bran (twb), ultrafine wheat bran (uwb), soluble wheat bran (swb), and aleurone (alr) (Table 1). Fermentations were carried out in triplicate; each replicate was inoculated with fecal material from a different donor. In parallel, cellulose incubations were set up with fecal material from each volunteer. Given its limited fermentability by colon bacteria (Mudgil and Barak, 2013), in these control cellulose fermentations, the effect of experimental conditions was expected to dominate over the impact of substrate fermentation. Hence, they allowed assessing the impact of the experimental set-up on the fecal microbiota composition. Overall, 24 h cellulose fermentation resulted in a decrease of genus richness when compared to donor material [paired t-test, effect size (ES) = -0.66, p-value = 8.1 × 10-3; Supplementary Figure S1]. Changes in taxa abundances included the increased relative abundances of Escherichia (paired t-test, ES = 0.97, FDR = 7.0 × 10-5), Bilophila (ES = 0.92, FDR = 3.3 × 10-4), and Sutterella (ES = 0.62, FDR = 2.8 × 10-2; Supplementary Table S2). In contrast, proportions of Roseburia (paired t-test, ES = -0.95, FDR = 1.6 × 10-4), Bacteroides (ES = -0.61, FDR = 2.8 × 10-2), Faecalibacterium (ES = -0.54, FDR = 4.9 × 10-2), and Blautia (ES = -0.47, FDR = 6.4 × 10-2) were reduced (Supplementary Table S2). Hence, in summary, experimental conditions were shown to result in a decrease of bacterial richness, mostly due to proportional blooming of Proteobacteria.
Table 1.
Chemical composition of total wheat bran, ultrafine wheat bran, wheat aleurone, and soluble wheat bran (in %; mean values, rsd < 5%).
| Total wheat bran | Ultrafine wheat bran | Aleurone | Soluble wheat bran | |
|---|---|---|---|---|
| Moisture (%) | 11 | 7.7 | 6.6 | 2.1 |
| Total protein (%) | 17 | 18 | 17 | 0.5 |
| Total fat (%) | / | / | 5.5 | / |
| Starch (%) | 26 | 27 | 6.3 | / |
| Ash (%) | 5.2 | 5.1 | 7.2 | 0.4 |
| Fiber | 38 | 37 | 56 | 79 |
| Insoluble fiber | 34 | 33 | 48 | / |
Donor and Substrate Diversify the Outcomes of Fecal Slurry Fermentations
Next, we assessed donor- and substrate-specific variation through the analysis of sample microbiome dissimilarity after 24 h of fermentation. A principal component analysis (PCoA) was used to visualize the between-samples dissimilarity in terms of microbiota composition (Bray–Curtis dissimilarity), revealing separate clusters of cellulose incubations and fermentations inoculated with donor 3 (D3) fecal material (Figure 1). These observations were confirmed by a hierarchical clustering approach, with the cellulose and D3 subgroups branching out at higher levels. D3 sample grouping clearly originated from Acidaminococcus blooming. Even though only representing a minor fraction of the donor microbiota, D3 fecal material was characterized by elevated Acidaminococcus relative abundances [unpaired t-test, (ES) = 0.99, p-value = 5.7 × 10-9; overall donor variation in core taxa (present in 80% of samples, abundance > 5% in at least one sample) is summarized in Supplementary Table S3 and Figure S2]. Clustering of cellulose incubations did suggest substrate-driven microbiome differentiation of bran fraction fermentations. Overall, substrate and donor explained, respectively, between 25–48 and 6–40% of microbiome variation during incubation experiments (Supplementary Table S4).
FIGURE 1.
Genus-level microbiome community variation of fecal slurry incubation experiments after 24 h, represented by principal coordinates analysis (Bray–Curtis dissimilarity PCoA). Samples (n = 18) were colored and shaped by donor and substrate, respectively. Genera that displayed significant variation among samples (db-RDA, FDR < 0.10) were scaled according to contribution and plotted on the ordination. The percentage of variance explained by the two first PCoA dimensions are reported on the axes.
Bran Fraction Fermentations Do Not Differentially Alter Community Richness
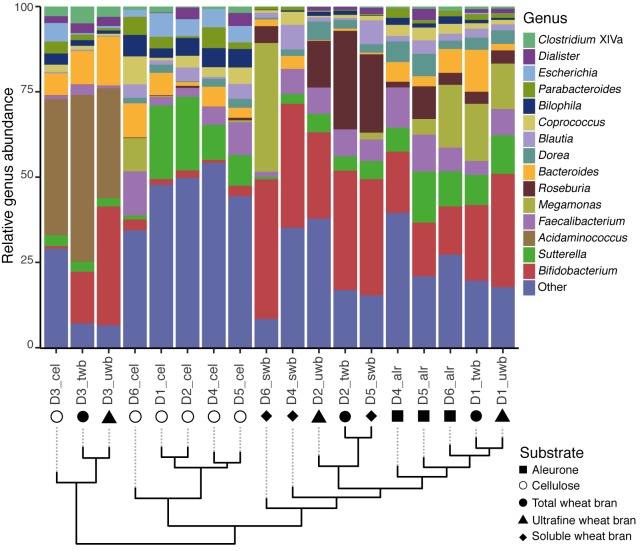
Fecal microbiome richness has been put forward as a read-out of colonic microbiota stability or resilience (Vieira-Silva et al., 2016), with reduced estimates thought to be indicative for ecosystem dysbiosis (Qin et al., 2012; Le Chatelier et al., 2013). However, some recent findings have associated high community richness indices to hard stools (Vandeputte et al., 2016), prolonged transit times (Roager et al., 2016), and enhanced proteolytic fermentation (Macfarlane et al., 1989; Roager et al., 2016), suggesting a less straightforward association between richness and host health than generally assumed. Given the interest in prebiotic modulation of microbiota richness (Druart et al., 2014; Bindels et al., 2015; Vandeputte et al., 2017), we assessed the impact of substrate variation on the number of observed genera following 24 h fecal slurry incubations (Figure 2). We did not observe any shift in richness associated with incubation of cellulose or any of the selected wheat bran fractions (Kruskal–Wallis test, p-value = 0.21; Supplementary Figure S3). In contrast, donor material did moderately affect the outcome of fermentation experiments (Kruskal–Wallis, p-value = 6.3 × 10-2; Supplementary Figure S4) – which could, however, be attributed to reduced richness in D3 incubations (Dunn’s test; Supplementary Table S5).
FIGURE 2.
Genus level relative microbiome composition of fecal slurry incubation experiments. Samples are clustered based on Bray–Curtis dissimilarity. The top 15 classified genera are depicted, with all others pooled into ‘Other.’ Clustering analysis was performed by hierarchical clustering with mean linkage.
Bran Fraction Fermentations Suggest Specific Prebiotic Effects on Gut Microbiota
Using an in vitro incubation approach, the prebiotic potential of a substrate can only be deduced from its ability to stimulate beneficial bacteria upon fermentation. To identify substrate-specific responsive genera, we compared taxa relative abundances after 24 h of bran fermentations with the outcome of cellulose incubations in matching donor fecal slurries (Supplementary Table S6). On genus level, wheat bran fractions were found to induce specific changes in slurry microbiota compositions. While all bran fraction fermentations resulted in increased Bifidobacterium relative abundances (paired t-test, twb, ES = 0.99, FDR = 2.6 × 10-2; uwb, ES = 0.91, FDR = 9.8 × 10-2; swb, ES = 0.90, FDR = 5.4 × 10-2; alr, ES = 0.77, FDR = 9.5 × 10-2), alr microbiome profiles were additionally characterized by higher proportions of both Roseburia (ES = 0.92, FDR = 5.9 × 10-2) and Dorea (ES = 0.80, FDR = 9.5 × 10-2). In contrast, the genera Escherichia (swb, ES = -0.98, FDR = 1.9 × 10-2; alr, ES = -1.0, FDR = 7.9 × 10-4), Parabacteroides (swb, ES = -0.99, FDR = 9.7 × 10-3; alr, ES = -0.88, FDR = 6.6 × 10-2), and Bilophila (swb, ES = -0.94, FDR = 3.9 × 10-2; alr, ES = -0.91, FDR = 5.9 × 10-2) were underrepresented following swb and alr fermentation. While a similar decrease in Parabacteroides relative abundances could be observed in twb fermentation (ES = -0.97, FDR = 3.3 × 10-2), uwb fermentation outcomes only mirrored reduced Bilophila populations (ES = -0.98, FDR = 3.8 × 10-2). In general, the addition of wheat bran fractions to fecal slurries resulted in increased Bifidobacterium relative abundances – mostly at the expense of the Proteobacteria, shown to be favored by incubation conditions. Of all substrates under investigation, alr displayed the broadest impact range on microbiota composition. Remarkably, the Bifidobacterium/Bilophila trade-off observed in uwb, swb, and alr incubations matched particularly well microbiota fluctuations following inulin consumptions observed in vivo (Vandeputte et al., 2017).
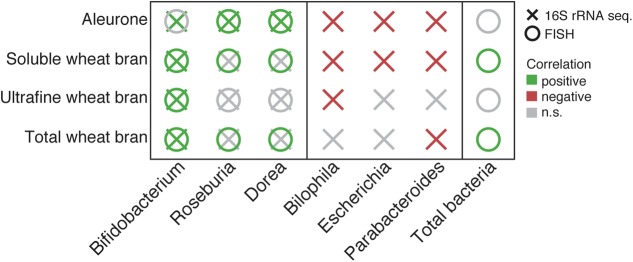
FISH Taxon Enumeration Confirms Bifidogenic Effect of Bran Fractions
Given the compositional nature of microbiome data, comparative analyses such as described above cannot reveal absolute directionality of observed abundance fluctuation. To bypass this problem, we performed a validation experiment using a FISH approach (Figure 3 and Supplementary Table S7). Given pretreatment and nature of the substrates studied, overall increases in total bacterial abundances were mostly limited (Supplementary Table S1). FISH enumeration data for bifidobacteria were found to correlate surprisingly well with relative abundances obtained through amplicon sequencing (Pearson, r = 0.83, p-value = 2.4 × 10-5). Except for alr, absolute quantification allowed us to confirm the bifidogenic effect of twb (paired t-test, ES = 0.90, p-value = 1.9 × 10-2), uwb (ES = 0.97, p-value = 4.8 × 10-3), and swb (ES = 0.81, p-value = 4.0 × 10-2; Supplementary Table S7). The association observed between alr and Roseburia/Dorea relative abundances was also validated (ES = 0.87, p-value = 2.5 × 10-2) based on the abundances detected using the FISH Eubacterium rectale/Clostridium coccoides spp (EREC) probe (Duncan et al., 2007). Of note, total and soluble bran fermentations also resulted in stimulated growth of EREC taxa (twb, ES = 0.75, p-value = 5.9 × 10-2; swb, ES = 0.94, p-value = 1.1 × 10-2), which remained unnoticed using a compositional sequencing approach – most probably due to the overall increase of bacterial numbers following both twb (ES = 0.65, p-value = 9.8 × 10-2) and swb (swb, ES = 0.81, p-value = 4.0 × 10-2) incubation.
FIGURE 3.
Summary of significant changes in core taxa relative abundances following bran fraction incubation experiments compared to cellulose fermentations as detected through 16S rRNA gene amplicon sequencing. Quantitative validation of results was performed using a FISH approach (18 samples; paired t-test, statistics are included in Supplementary Tables S6, S7).
Incubation of Soluble Wheat Bran Stimulates Butyrate Production
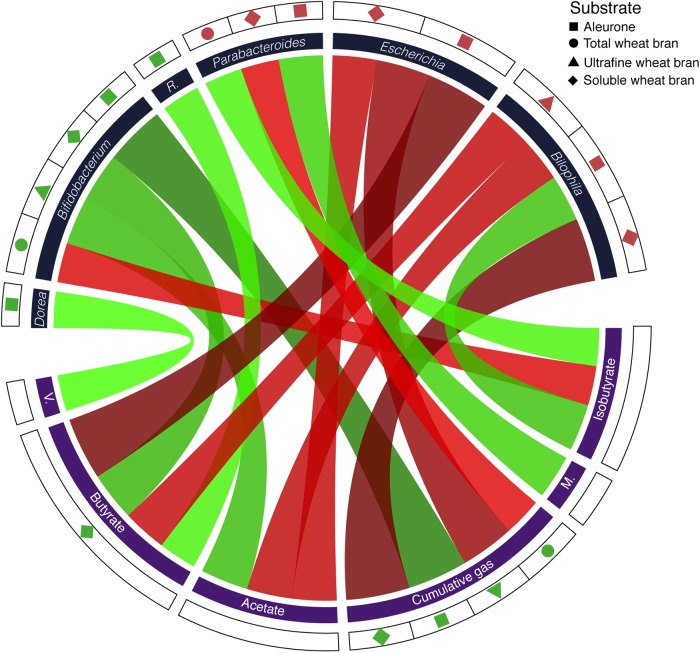
In all fermentation experiments, we quantified acetate, propionate, butyrate, lactate, valerate, isobutyrate, and methylbutyrate concentrations as well as total gas production after 24 h of substrate fermentation. Based on the metabolite profiles, dissimilarity between wheat bran and cellulose fermentations was assessed (Euclidean distance). Metabolite dissimilarity was found strongly correlated to sample microbiome differentiation (Bray–Curtis) (Mantel test, r = 0.24, p-value = 1.6 × 10-2). Hence, changes in taxa relative abundances were reflected in the metabolic output of fecal slurry fermentation processes (Supplementary Figure S5). Compared to cellulose, fermentations of selected wheat bran fractions gave rise to increased gas production (twb, paired t-test, ES = 0.99, p-value = 2.2 × 10-3; uwb, ES = 0.98, p-value = 3.8 × 10-3; swb, ES = 0.98, p-value = 4.1 × 10-3; alr, ES = 0.83, p-value = 3.7 × 10-2; Supplementary Table S8). Moreover, swb fermentations displayed elevated butyrate concentrations (ES = 0.88, p-value = 2.3 × 10-2), matching FISH findings regarding EREC cluster absolute counts (Hold et al., 2003). Interestingly, independent of donor or substrate variation, relative abundances of Bifidobacterium, Bilophila, Escherichia, and Parabacteroides could be correlated with several metabolites, including acetate, butyrate, isobutyrate, and cumulative gasses (Supplementary Table S9). In contrast, Roseburia could only be associated to butyrate, in line with the genus’ metabolic profile (Duncan et al., 2002). Dorea, on the other hand, was exclusively linked with valerate concentrations – although the taxon is known as a major gas producer (Rajilić-Stojanović and de Vos, 2014). An overview of substrate, taxon, metabolite associations observed in bran fraction incubation experiments is presented in Figure 4.
FIGURE 4.
Circos plot depicting correlations between substrates, metabolites, and relative genus abundances [FDR < 10% for core taxon (n = 14) – substrate (n = 4) associations; FDR < 10% for core taxon (n = 4; only correlations of taxa with increased relative abundances following bran fermentations were included) – metabolite (n = 8) correlations; p-value < 5% for metabolite (n = 8) – substrate (n = 4) associations]. Green and red ribbons represent positive and negative relationships, respectively. Ribbons are sized and colored according to the strength of the association (effect size). R, Roseburia; M, methylbutyrate; V, valerate.
Discussion
Our in vitro incubation analyses largely confirmed the bifidogenic effect of total, ultrafine, and swb fractions (Cloetens et al., 2010; Neyrinck et al., 2011; François et al., 2012; Maki et al., 2012). The strategies and mechanisms applied by Bifidobacterium spp. to degrade non-digestible carbohydrates have previously been described in detail (Rivière et al., 2014, 2016). Moreover, wheat fiber is constituted primarily of arabinoxylans (Grootaert et al., 2007; Hughes et al., 2007; Vardakou et al., 2008) with known dose-dependent bifidogenic properties (Cloetens et al., 2010; Neyrinck et al., 2011; François et al., 2012; Maki et al., 2012). Although end-products of bifidobacterial carbohydrate metabolism are mainly limited to lactate, acetate, formate, and ethanol (Rivière et al., 2016), the genus has been shown to sustain a broad range of gut microbial commensals through cross-feeding interactions (Duncan et al., 2004; Belenguer et al., 2006; Falony et al., 2006; Moens et al., 2016, 2017). The latter would expand the metabolic impact of stimulated Bifidobacterium growth to include the observed increase in butyrate and gas production (De Vuyst and Leroy, 2011).
Given its putative health-promoting properties (Scheppach and Weiler, 2004), the enhancement of colonic butyrate production has been a long-standing target of prebiotic research. Here, we noted increased abundance of Roseburia spp. following fermentation of both swb and aleurone. Butyrate-producing Roseburia spp. have been shown to be able to grown on inulin (Scott et al., 2014), xylans (Duncan et al., 2002; Chassard et al., 2007), and arabinoxylans (Sheridan et al., 2016), as well as on intermediates of primary polysaccharide degradation (Belenguer et al., 2006; Falony et al., 2006). While increased abundance of Roseburia following soluble fiber incubation was part of a more generalized stimulation of total bacterial growth, the effect induced by aleurone appeared more taxon-specific. Aleurone has previously been attributed bifidogenic properties (Brouns et al., 2012). Here, using a community-wide analytical approach, we demonstrate that it could potentially be applied for targeted stimulation of Clostridium cluster IVa bacteria, encompassing several colon butyrate producers.
In all but one (twb) incubation experiments, fermentation of bran fractions resulted in decreased Proteobacteria taxa when compared to cellulose. Besides the fact that these genera are known to thrive under in vitro conditions, their proteolytic or aminoacidolytic nature provides them with a selective advantage when incubated in the absence of readily fermentable carbohydrates. While their decrease in relative abundances could partially result from compositionality effects, it also demonstrates the potential of refined bran fractions to extend saccharolytic fermentation to more distal gut regions. Given the production of potentially deleterious components resulting from proteolytic fermentation (Hamer et al., 2011), the latter is considered a desirable property of functional food ingredients targeting the microbial gut ecosystem (Macfarlane et al., 2006). The effects of ultrafine or swb incubations on Bilophila relative abundances matched remarkably well previous in vivo findings regarding the prebiotic properties of inulin (Vandeputte et al., 2017). The most prominent intestinal Bilophila isolate is Bilophila wadsworthia, an asaccharolytic sulfate reducer that has been characterized as an opportunistic pathogen (Baron et al., 1989; Baron, 1997; da Silva et al., 2008). Our in vitro findings suggest that the trade-off between the Bifidobacterium and Bilophila taxa is a consequence of direct bacterial interactions (e.g., competition or metabolite production) rather than a host-mediated response to a dietary intervention or intervention-induced primary shifts in microbiota composition as suggested previously (Vandeputte et al., 2017). Overall, the results presented indicate that similar bacterial interactions make up the core of the microbiota’s colonization resistance against non-commensal intruders – a phenomenon that appears to be strengthened by the availability of fermentable substrates, as suggested by the restrained blooming of Proteobacteria in bran incubations. The fermentable fibers present in the wheat bran fractions studied provide the commensal microbiota with a competitive advantage, hampering settlement of opportunistic colonizers. In this pH-controlled setting, we observed colonization resistance to be independent of total bacterial abundance. The role of bacterial metabolites produced upon fiber fermentations in this process remains to be elucidated.
Finally, bran supplementation also altered the outcome of niche competition between saccharolytic taxa. Parabacteroides spp., part of a normal large-intestinal microbiota but often associated with opportunistic infections (Nakano et al., 2011), have been described to bloom on resistant starches (Martínez et al., 2010) rather than on complex non-starch polysaccharides (Sakamoto and Benno, 2006). Except for uwb, fecal fermentations of bran fractions provided a growth advantage to saccharolytic competitors such as bifidobacteria, allowing them to dominate over the Parabacteroides fraction.
Materials and Methods
Donor Fecal Material
Fecal material was collected from six healthy, male subjects aged between 30 and 47, not receiving antibiotic treatment for at least 3 months, not consuming pre- or probiotic containing supplements prior to experimentation, and without history of intestinal disorders. Fecal slurry was prepared under anaerobic conditions by homogenizing fresh human fecal material in ten times the volume of pre-reduced phosphate buffered saline (PBS; 8 g/L NaCl, 0.2 g/L KCl, 1.15 g/L Na2PO4, and 0.2 g/L KH2HPO4).
Substrates
Total wheat bran (twb), uwb, swb, and aleurone (alr) were used. Uwb was obtained through mechanical milling of twb, resulting in particles with a size < 100 μm. Swb was obtained through enzymatic treatment of wheat bran, reducing arabinoxylan chain length into shorter oligosaccharides (arabinoxylan-oligosaccharide content = 79%). Aleurone is a single cell layer of wheat located between the starchy endosperm and the outer bran layers. The aleurone cells were separated from the pericarp layer and isolated to obtain a standard aleurone fraction. Cellulose (methyl-cellulose, Sigma–Aldrich) was used as a negative control substrate. All substrates were added to the fermentation medium at a concentration of 1% (wt/vol).
Fecal Fermentation Experiments
Anaerobic (N2-sparged) batch fermentations were performed in triplicate using 10% fecal slurry (1% fecal inoculum) under controlled conditions [water-jacket vessels (Soham Scientific, Soham, United Kingdom), pH 6.8, temperature 37°C]. Basal medium contained per liter: 2 g peptone (Oxoid, Basingstoke, United Kingdom), 2 g yeast extract (Oxoid), 0.1 g NaCl (Fisher Scientific, Fair Lawn, NJ, United States), 0.04 g K2HPO4 (BDH, Toronto, ON, Canada), 0.04 g KH2PO4 (BDH), 0.01 g MgSO47H2O (BDH), 0.01 g CaCl26H2O (Honeywell, Morris Plains, NY, United States), 2 g NaHCO3 (Oxoid), 2 mL Tween 80 (Sigma–Aldrich, Oakville, ON, Canada), 0.05 g Hemin (Sigma–Aldrich) dissolved in 1 mL of 4 M NaOH (Fisher Scientific), 10 μL Vitamin K (Sigma–Aldrich), 0.5 g l-Cysteine HCL (Sigma–Aldrich), 0.5 g Bile Salts (Oxoid), and 4 mL of Resazurin (Sigma–Aldrich) (0.025 g/100 mL). Vessels were dosed with the substrates (1% wt/vol) after simulated in vitro upper gastrointestinal digestion and dialysis (Mandalari et al., 2008) and inoculated with 10% fecal slurry. The final volume of each culture was 200 mL. Samples were harvested at time points 0 (immediately after incubation) and 24 h.
Fecal Microbiota Phylogenetic Profiling
Samples taken at T0 and originating from a same donor were pooled for further analysis. Briefly, DNA was extracted from 1 mL aliquots of fermentation effluent using the Fast DNA spin kit for feces (MP Biomedicals, Santa Ana, CA, United States). Fecal microbiota profiling was performed as described previously (Falony et al., 2016). The V4 region of the 16S rRNA gene was amplified with primer pair 515F/ 806R (GTGYCAGCMGCCGCGGTAA/GGACTACNVGGGTWTCTAAT, respectively) modified to contain a barcode sequence between each primer and the Illumina adaptor sequences to produce dual-barcoded libraries (Tito et al., 2017). Sequencing was performed on the Illumina MiSeq platform (MiSeq Reagent Kit v2, 500- cycles, 20% PhiX; Illumina, San Diego, CA, United States) according to the manufacturer’s specifications to generate paired-end reads of 250 bases in length in each direction. After de-multiplexing, fastq sequences were merged using FLASH (Magoč and Salzberg, 2011) software with default parameters, except for –min-overlap and –max-overlap which were set to 140 and 230, respectively. Successfully combined reads were filtered based on quality using seqtk trimfq with default parameters1. Chimeras were removed with the uchime2_ref algorithm of USEARCH (version 9.2.64) (Edgar et al., 2011). The taxonomy of reads was assigned using RDP classifier 2.12 (Wang et al., 2007) to generate phylum to genus level composition matrices. Bootstrap values from the RDP classifier were used to identify sequences with high-confidence genus assignments (bootstrap value > 0.8), while sequences classified with lower confidence were binned to the family assignment (labeled unclassified_family). To compare the different samples, sample counts were rarefied to 20,000 reads by random selection of reads and trimmed for the consequently absent OTUs with the phyloseq package (McMurdie and Holmes, 2013) in R version 3.3.0. In total, 18 samples, retrieved after 24 h of incubation, were analyzed covering 161 genera with an average of 65 genera per sample.
Fluorescence in Situ Hybridization (FISH)
Genus-specific 16S rRNA targeted oligonucleotide probes labeled with the fluorescent dye Cy3 were used for enumerating bacteria. Fecal batch culture samples (375 μL) were fixed using cold 4% paraformaldehyde (Sigma–Aldrich) (pH 7.2) at a ratio of 1:3 (vol/vol) in a 1.5 mL Eppendorf tube and stored at 4°C between 4 and 16 h. Samples were centrifuged at 13,000 g for 5 min and washed twice (resuspending the pellet in 1 mL filtered PBS and subsequent centrifuging). The washed pellet was resuspended in a filtered-sterilized PBS/ethanol mix (1:1 vol/vol) and stored at -20°C for up to 3 months. The enumeration of microbial populations was carried out as described previously (Daims et al., 2005) using FISH-technique. Oligonucleotide probes used were Bif164 [specific for the Bifidobacterium genus (Langendijk et al., 1995)]; Bac303 [Bacteroides and Prevotella (Manz et al., 1996)], Chis150 [Clostridium histolyticum subgroup (Franks et al., 1998)], Erec482 [Ruminococcus–Eubacterium–Clostridium (EREC) cluster (Franks et al., 1998)], and Fpra655 [Faecalibacterium (Hold et al., 2003)]. Oligonucleotide EUB388 mix (Amann et al., 1990) was used for total bacteria enumeration, using 4′-6-diamidine-2-phenylindole (DAPI) staining as a control. Slides were enumerated using an Olympus microscope (Olympus, Shinjuku-ku, Tokyo, Japan) fitted with an EPI-fluorescence attachment and 15 randomized views (0,025 mm2, 100×) were counted for each sample.
Gas Chromatography
Short-chain fatty acids (acetate, propionate, and butyrate) and branched short-chain fatty acids (isobutyrate, valerate, and methylbutyrate) were analyzed as described by Fava et al. (2012) with slight modifications to the method. Samples were acidified to pH 2–3 with 6 M HCl, centrifuged at 13,000 × g for 5 min, and filtered through a 0.2 mm polycarbonate syringe filter. Standard solutions containing 20, 10, 5, 1, and 0.5 mM external standards and 2 mM of internal standard (2-ethylbutyric acid) were used. Fatty acids were determined by gas chromatography on a Hewlett Packard (Agilent) 5890 Series II GC system (HP, Crawley, United Kingdom), fitted with a FFAP column (30 m × 0.53 mm, diameter 0.50 mm, J&W Scientific, Agilent Technologies, Ltd., South Queensferry, United Kingdom), and a flame-ionization detector. Glass wool was inserted in the injection port. The injected sample volume was 1 mL. Helium was used as carrier gas. The head pressure was set at 10 psi and the split ratio was 10:1. The flow rate of total gas was 140 mL/min. Injector and detector temperature were set at 280 and 300°C, respectively. The initial oven temperature was 100°C, maintained for 0.5 min, raised to 150°C at 81°C per min, then increased to 250°C at 50°C per min, and finally held at 250°C for 2 min. Fatty acid concentrations were calculated by peak integration using Atlas Lab managing software (Thermo Lab Systems, Mainz, Germany) and expressed as mM.
Lactate Analyses Using Enzymatic Assays
Lactate was measured using the Lactate Assay Kit (Sigma–Aldrich) according to manufacturer’s instructions. Briefly, samples were centrifuged at 13,000 × g for 10 min and the supernatants were stored at -80°C, thawed on ice and filtered through 10 kDa MW cut-off columns (Millipore Amicon Ultra, Merck, Darmstadt, Germany). Two standard curves were used for each sample reading. Results are reported in mM.
Gas Measurements
Gas production was measured through five replicate measures in separately conducted batch cultures (utilizing fecal samples from the same donors) and under anaerobic conditions using airtight serum bottles. Growth medium (pH controlled) [according to Rycroft et al. (2001)] was inoculated with freshly (anaerobic) prepared feces (1% wt/vol) and incubated anaerobically at 37°C for 24 h. Gas volume and pressure were measured at 3, 6, 9, 12, and 24 h using a transducer (Gems Sensors, Basingstoke, United Kingdom) according to the manufacturer’s instructions (Sarbini et al., 2011).
Microbiome Analysis
Statistical analyses were performed in R version 3.3.0 (R Core Team, 2013). Observed richness was calculated with the R package phyloseq (McMurdie and Holmes, 2013). Microbiome variation between samples was determined by principal coordinates analysis (PCoA) using Bray–Curtis dissimilarity on the genus-level relative abundance matrix with the R package vegan (Oksanen et al., 2015) and visualized using the R package ggplot2 (Wickham, 2009). Clustering analysis was performed based on hierarchical clustering with mean linkage using the R package stats (R Core Team, 2013). To assess microbiome variation between bran fraction and cellulose incubations, and between donors, (un)paired t-tests with Welch’s correction for unequal variances were carried out on log(1+×) transformed relative genus abundance data. Analyses were performed on core taxa, identified as annotated genera, present in at least 80% of samples, with abundance > 5% in at least one sample. The corresponding correlation effect sizes were calculated in R using the lsr (Navarro, 2015) and compute.es (Del Re, 2013) packages. Correction for multiple testing [Benjamini–Hochberg method (Benjamini and Hochberg, 1995), FDR] was performed. For the microbiome variation between bran fraction and cellulose incubations, FDR was applied for each substrate separately. Differences in observed richness between the different substrate incubation regimes (five-level categorical data) were assessed using non-parametric ANOVA (Kruskal–Wallis test using R package stats) and post hoc Dunn’s test (using R package FSA) for all pairs of comparisons between groups, with Benjamini–Hochberg adjustment for multiple testing (FDR).
Donor/Substrate Effect Sizes in Microbiome Variation
Variation partitioning by stepwise distance-based redundancy analysis (dbRDA) was performed to determine how much of the microbial community profiles variation (Bray–Curtis dissimilarity) could be explained by the cumulative and individual contributions of substrate and donor, with significance calculated with a permutation test.
Analysis of Metabolite Production
A Mantel test, based on Spearman’s rank correlation rho, was performed to test whether microbiome Bray–Curtis and metabolome Euclidean distance-based between-sample dissimilarities were correlated (1,000,000 permutations, R packages vegan and ecodist). Metadata was fitted on the PCA ordination (Supplementary Figure S5) using the prcomp function in the R package stats.
Normality of quantitative FISH and metabolite concentrations was assessed using the car package in R. As these metadata were normally distributed, no data transformation was applied. Differences between bran fraction and cellulose incubations were calculated using paired t-tests with Welch’s correction for FISH data and metabolite concentrations as described before. Correlations between relative genus abundances and metabolite data were analyzed using non-parametric Spearman tests for which correction for multiple testing [Benjamini–Hochberg method (Benjamini and Hochberg, 1995), FDR] was applied. The circos plot was constructed using the R package circlize.
Conclusion
Here, we assessed the prebiotic potential of selected wheat bran fractions by performing a series of fecal slurry fermentations. We confirmed the bifidogenic effect of wheat bran fractions. The increase in Bifidobacterium spp. following ultrafine and soluble bran incubations was paired with a decrease in Bilophila relative abundances, matching in vivo observations on the prebiotic effect of inulin. In contrast with the more generalized effects observed upon pericarp bran fraction supplementation, aleurone fermentations selectively stimulated growth of butyrate-producing Roseburia, opening perspectives for its future development as a complementary prebiotic.
Authors Note
The 16S rRNA gene amplicon sequencing results are available upon reasonable request.
Author Contributions
LC, FF, GF, JV, and KT performed the conception and design of the study. LC and FF performed the data collection and experimental work. KD and SV-S performed the data preparation. KD, SV-S, and GF performed the statistical analysis, data analysis, and interpretation. KD, SV-S, GF, and JR drafted the manuscript. All authors performed the critical revision of the article and approved the final version for publication.
Conflict of Interest Statement
Study products were provided by Cargill and JV is a Cargill employee. Cargill funded fermentation experiments and metabolite/microbiota analyses at FEM and sequencing/statistical analyses at VIB.
Acknowledgments
The authors would like to thank Vicenta Garcia Campayo (Cargill R&D Minneapolis) for her contribution to the interpretation of project results, Leen Rymenans and Chloë Verspecht for library preparation, and all members of the Raes Lab for participating in scientific discussions regarding the manuscript.
Funding. KD and SV-S are funded by a (post-) doctoral fellowship from Research Foundation Flanders (FWO-Vlaanderen) (11T1616N and 12K5116N).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00031/full#supplementary-material
References
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56 1919–1925. 10.1111/j.1469-8137.2004.01066.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron E., Summanen P., Downes J., Roberts M. C., Wexler H., Finegold S. M. (1989). Bilophila wadsworthia, gen. nov. and sp. nov., a unique gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J. Gen. Microbiol. 135 3405–3411. 10.1099/00221287-135-12-3405 [DOI] [PubMed] [Google Scholar]
- Baron E. J. (1997). Bilophila wadsworthia: a unique Gram-negative anaerobic rod. Anaerobe 3 83–86. 10.1006/anae.1997.0075 [DOI] [PubMed] [Google Scholar]
- Belenguer A., Duncan S. H., Calder A. G., Holtrop G., Louis P., Lobley G. E., et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72 3593–3599. 10.1128/AEM.72.5.3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Bindels L. B., Delzenne N. M., Cani P. D., Walter J. (2015). Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 12 303–310. 10.1038/nrgastro.2015.47 [DOI] [PubMed] [Google Scholar]
- Broekaert W. F., Courtin C. M., Verbeke K., Van de Wiele T., Verstraete W., Delcour J. A. (2011). Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit. Rev. Food Sci. Nutr. 51 178–194. 10.1080/10408390903044768 [DOI] [PubMed] [Google Scholar]
- Brouns F., Hemery Y., Price R., Anson N. M. (2012). Wheat aleurone: separation, composition, health aspects, and potential food use. Crit. Rev. Food Sci. Nutr. 52 553–568. 10.1080/10408398.2011.589540 [DOI] [PubMed] [Google Scholar]
- Burkitt D. P., Walker A. R., Painter N. S. (1972). Effect of dietary fibre on stools and the transit-times, and its role in the causation of disease. Lancet 2 1408–1412. 10.1016/S0140-6736(72)92974-1 [DOI] [PubMed] [Google Scholar]
- Chassard C., Goumy V., Leclerc M., Del’homme C., Bernalier-Donadille A. (2007). Characterization of the xylan-degrading microbial community from human faeces. FEMS Microbiol. Ecol. 61 121–131. 10.1111/j.1574-6941.2007.00314.x [DOI] [PubMed] [Google Scholar]
- Claus S. P. (2017). Inulin prebiotic: Is it all about bifidobacteria? Gut 66 1883–1884. 10.1136/gutjnl-2017-313800 [DOI] [PubMed] [Google Scholar]
- Cloetens L., Broekaert W. F., Delaedt Y., Ollevier F., Courtin C. M., Delcour J. A., et al. (2010). Tolerance of arabinoxylan-oligosaccharides and their prebiotic activity in healthy subjects: a randomised, placebo-controlled cross-over study. Br. J. Nutr. 103 703–713. 10.1017/S0007114509992248 [DOI] [PubMed] [Google Scholar]
- da Silva S. M., Venceslau S. S., Fernandes C. L. V., Valente F. M. A., Pereira I. A. C. (2008). Hydrogen as an energy source for the human pathogen Bilophila wadsworthia. Antonie Van Leeuwenhoek 93 381–390. 10.1007/s10482-007-9215-x [DOI] [PubMed] [Google Scholar]
- Daims H., Stoecker K., Wagner M. (2005). “Fluorescence in situ hybridization for the detection of prokaryotes,” in Advanced Methods in Molecular Microbial Ecology eds Osborn A., Smith C. (Abingdon: Taylor & Francis group; ) 213–239. [Google Scholar]
- De Filippis F., Pellegrini N., Vannini L., Jeffery I. B., La Storia A., Laghi L., et al. (2016). High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65 1812–1821. 10.1136/gutjnl-2015-309957 [DOI] [PubMed] [Google Scholar]
- De Preter V., Falony G., Windey K., Hamer H. M., De Vuyst L., Verbeke K. (2010). The prebiotic, oligofructose-enriched inulin modulates the faecal metabolite profile: an in vitro analysis. Mol. Nutr. Food Res. 54 1791–1801. 10.1002/mnfr.201000136 [DOI] [PubMed] [Google Scholar]
- De Vuyst L., Leroy F. (2011). Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int. J. Food Microbiol. 149 73–80. 10.1016/j.ijfoodmicro.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Del Re A. C. (2013). compute.es: Compute Effect Sizes. R Package Version 0.2-2. [Google Scholar]
- Desai M. S., Seekatz A. M., Koropatkin N. M., Kamada N., Hickey C. A., Wolter M., et al. (2016). A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167 1339.e21–1353.e21. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart C., Alligier M., Salazar N., Neyrinck A. M., Delzenne N. M. (2014). Modulation of the gut microbiota by nutrients. Adv. Nutr. 5 624–633. 10.3945/an.114.005835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. H., Belenguer A., Holtrop G., Johnstone A. M., Flint H. J., Lobley G. E. (2007). Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73 1073–1078. 10.1128/AEM.02340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. H., Hold G. L., Barcenilla A., Stewart C. S., Flint H. J. (2002). Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52 1615–1620. [DOI] [PubMed] [Google Scholar]
- Duncan S. H., Louis P., Flint H. J. (2004). Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70 5810–5817. 10.1128/AEM.70.10.5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., et al. (2016). Population-level analysis of gut microbiome variation. Science 352 560–564. 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- Falony G., Vlachou A., Verbrugghe K., De Vuyst L. (2006). Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 72 7835–7841. 10.1128/AEM.01296-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava F., Gitau R., Griffin B. A., Tuohy K. M., Gibson G., Lovegrove J. (2012). The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short-chain fatty acid excretion in a metabolic syndrome ‘at-risk’ population syndrome. Int. J. Obes. 37 216–223. 10.1038/ijo.2012.33 [DOI] [PubMed] [Google Scholar]
- François I. E. J. A., Lescroart O., Veraverbeke W. S., Marzorati M., Possemiers S., Evenepoel P., et al. (2012). Effects of a wheat bran extract containing arabinoxylan oligosaccharides on gastrointestinal health parameters in healthy adult human volunteers: a double-blind, randomised, placebo-controlled, cross-over trial. Br. J. Nutr. 108 2229–2242. 10.1017/S0007114512000372 [DOI] [PubMed] [Google Scholar]
- Franks A. H., Harmsen H. J. M., Raangs G. C., Jansen G. J., Schut F., Welling G. W. (1998). Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64 3336–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. R., Roberfroid M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125 1401–1412. [DOI] [PubMed] [Google Scholar]
- Grootaert C., Verstraete W., Van de Wiele T. (2007). Microbial metabolism and prebiotic potency of arabinoxylan oligosaccharides in the human intestine. Trends Food Sci. Technol. 18 64–71. 10.1016/j.tifs.2006.08.004 19508502 [DOI] [Google Scholar]
- Hamer H. M., De Preter V., Windey K., Verbeke K. (2011). Functional analysis of colonic bacterial metabolism: relevant to health? Am. J. Physiol. Gastrointest. Liver Physiol. 302 G1–G9. 10.1152/ajpgi.00048.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer H. M., Jonkers D., Venema K., Vanhoutvin S., Troost F. J., Brummer R.-J. (2008). Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27 104–119. 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- Hold G. L., Schwiertz A., Aminov R. I., Blaut M., Flint H. J. (2003). Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69 4320–4324. 10.1128/AEM.69.7.4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. A., Shewry P. R., Li L., Gibson G. R., Sanz M. L., Rastall R. A. (2007). In vitro fermentation by human fecal microflora of wheat arabinoxylans. J. Agric. Food Chem. 55 4589–4595. 10.1021/jf070293g [DOI] [PubMed] [Google Scholar]
- Hutkins R. W., Krumbeck J. A., Bindels L. B., Cani P. D., Fahey G., Goh Y. J., et al. (2016). Prebiotics: Why definitions matter. Curr. Opin. Biotechnol. 37 1–7. 10.1016/j.copbio.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langendijk P. S., Schut F., Jansen G. J., Raangs G. C., Kamphuis G. R., Wilkinson M. H. F., et al. (1995). Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61 3069–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Walker A. W. (2013). Intestinal colonization resistance. Immunology 138 1–11. 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500 541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G. L., Flint H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12 661–672. 10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- Macfarlane G. T., Cummings J. H., Macfarlane S., Gibson G. R. (1989). Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous culture system. J. Appl. Bacteriol. 67 521–527. 10.1111/j.1365-2672.1989.tb02524.x [DOI] [PubMed] [Google Scholar]
- Macfarlane S., Macfarlane G. T., Cummings J. H. (2006). Review article: prebiotics in the gastrointestinal tract. Aliment. Pharmacol. Ther. 24 701–714. 10.1111/j.1365-2036.2006.03042.x [DOI] [PubMed] [Google Scholar]
- Magoč T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki K. C., Gibson G. R., Dickmann R. S., Kendall C. W. C., Chen C. Y. O., Costabile A., et al. (2012). Digestive and physiologic effects of a wheat bran extract, arabino-xylan-oligosaccharide, in breakfast cereal. Nutrition 28 1115–1121. 10.1016/j.nut.2012.02.010 [DOI] [PubMed] [Google Scholar]
- Mandalari G., Nueno-Palop C., Bisignano G., Wickham M. S. J., Narbad A. (2008). Potential prebiotic properties of almond (Amygdalus communis L.) seeds. Appl. Environ. Microbiol. 74 4264–4270. 10.1128/AEM.00739-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W., Amann R., Ludwig W., Vancanneyt M., Schleifer K. H. (1996). Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142 1097–1106. 10.1099/13500872-142-5-1097 [DOI] [PubMed] [Google Scholar]
- Martínez I., Kim J., Duffy P. R., Schlegel V. L., Walter J. (2010). Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLOS ONE 5:e15046. 10.1371/journal.pone.0015046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens F., Verce M., De Vuyst L. (2017). Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int. J. Food Microbiol. 241 225–236. 10.1016/j.ijfoodmicro.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Moens F., Weckx S., De Vuyst L. (2016). Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 231 76–85. 10.1016/j.ijfoodmicro.2016.05.015 [DOI] [PubMed] [Google Scholar]
- Mudgil D., Barak S. (2013). Composition, properties and health benefits of indigestible carbohydrate polymers as dietary fiber: a review. Int. J. Biol. Macromol. 61 1–6. 10.1016/j.ijbiomac.2013.06.044 [DOI] [PubMed] [Google Scholar]
- Nakano V., Nascimento e Silva A., Merino V. R. C., Wexler H. M., Avila-Campos M. J. (2011). Antimicrobial resistance and prevalence of resistance genes in intestinal Bacteroidales strains. Clinics 66 543–547. 10.1590/S1807-59322011000400004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro D. J. (2015). Learning Statistics with R: A Tutorial for Psychology Students and Other Beginners. (Version 0.5). Adelaide, SA: University of Adelaide. [Google Scholar]
- Neyrinck A. M., Possemiers S., Druart C., van de Wiele T., de Backer F., Cani P. D., et al. (2011). Prebiotic effects of wheat Arabinoxylan related to the increase in bifidobacteria, roseburia and bacteroides/prevotella in diet-induced obese mice. PLOS ONE 6:e20944. 10.1371/journal.pone.0020944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., et al. (2015). vegan: Community Ecology Package. R Package Version 2.2-1. [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490 55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rajilić-Stojanović M., de Vos W. M. (2014). The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 38 996–1047. 10.1111/1574-6976.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière A., Moens F., Selak M., Maes D., Weckx S., De Vuyst L. (2014). The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl. Environ. Microbiol. 80 204–217. 10.1128/AEM.02853-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière A., Selak M., Lantin D., Leroy F., De Vuyst L. (2016). Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roager H. M., Hansen L. B. S., Bahl M. I., Frandsen H. L., Carvalho V., Gøbel R. J., et al. (2016). Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 1:16093. 10.1038/nmicrobiol.2016.93 [DOI] [PubMed] [Google Scholar]
- Roediger W. E. (1982). Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 83 424–429. [PubMed] [Google Scholar]
- Rycroft C. E., Jones M. R., Gibson G. R., Rastall R. A. (2001). A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91 878–887. 10.1046/j.1365-2672.2001.01446.x [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Benno Y. (2006). Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 56 1599–1605. 10.1099/ijs.0.64192-0 [DOI] [PubMed] [Google Scholar]
- Sarbini S. R., Kolida S., Naeye T., Einerhand A., Brison Y., Remaud-Simeon M., et al. (2011). In vitro fermentation of linear and alpha-12-branched dextrans by the human fecal microbiota. Appl. Environ. Microbiol. 77 5307–5315. 10.1128/AEM.02568-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheppach W., Weiler F. (2004). The butyrate story: old wine in new bottles? Curr. Opin. Clin. Nutr. Metab. Care 7 563–567. 10.1097/00075197-200409000-00009 [DOI] [PubMed] [Google Scholar]
- Scott K. P., Martin J. C., Duncan S. H., Flint H. J. (2014). Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 87 30–40. 10.1111/1574-6941.12186 [DOI] [PubMed] [Google Scholar]
- Sheridan P. O., Martin J. C., Lawley T. D., Browne H. P., Harris H. M., Bernalier-Donadille A., et al. (2016). Polysaccharide utilization loci and nutritional specialisation in a dominant group of butyrate-producing human colonic Firmicutes. Microb. Genomics 2:e000043. 10.1099/mgen.0.000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg E. D., Sonnenburg J. L. (2014). Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 20 779–786. 10.1016/j.cmet.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson L., Phillips F., O’Sullivan K., Walton J. (2012). Wheat bran: its composition and benefits to health, a European perspective. Int. J. Food Sci. Nutr. 63 1001–1013. 10.3109/09637486.2012.687366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito R. Y., Cypers H., Joossens M., Varkas G., Van Praet L., Glorieus E., et al. (2017). Brief Report: Dialister as a microbial marker of disease activity in spondyloarthritis. Arthritis Rheumatol. 69 114–121. 10.1002/art.39802 [DOI] [PubMed] [Google Scholar]
- Valles-Colomer M., Darzi Y., Vieira-Silva S., Falony G., Raes J., Joossens M. (2016). Meta-omics in inflammatory bowel disease research: applications, challenges, and guidelines. J. Crohns Colitis 10 735–746. 10.1093/ecco-jcc/jjw024 [DOI] [PubMed] [Google Scholar]
- Vandeputte D., Falony G., Vieira-Silva S., Tito R. Y., Joossens M., Raes J. (2016). Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65 57–62. 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte D., Falony G., Vieira-Silva S., Wang J., Sailer M., Theis S., et al. (2017). Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66 1968–1974. 10.1136/gutjnl-2016-313271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardakou M., Palop C. N., Christakopoulos P., Faulds C. B., Gasson M. A., Narbad A. (2008). Evaluation of the prebiotic properties of wheat arabinoxylan fractions and induction of hydrolase activity in gut microflora. Int. J. Food Microbiol. 123 166–170. 10.1016/j.ijfoodmicro.2007.11.007 [DOI] [PubMed] [Google Scholar]
- Vieira-Silva S., Falony G., Darzi Y., Lima-Mendez G., Garcia Yunta R., Okuda S., et al. (2016). Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1:16088. 10.1038/nmicrobiol.2016.88 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag; 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.