Abstract
The β-barrel assembly machinery (BAM) is a conserved multicomponent protein complex responsible for the biogenesis of β-barrel outer membrane proteins (OMPs) in Gram-negative bacteria. Given its role in the production of OMPs for survival and pathogenesis, BAM represents an attractive target for the development of therapeutic interventions, including drugs and vaccines against multidrug-resistant bacteria such as Neisseria gonorrhoeae. The first structure of BamA, the central component of BAM, was from N. gonorrhoeae, the etiological agent of the sexually transmitted disease gonorrhea. To aid in pharmaceutical targeting of BAM, we expanded our studies to BamD and BamE within BAM of this clinically relevant human pathogen. We found that the presence of BamD, but not BamE, is essential for gonococcal viability. However, BamE, but not BamD, was cell-surface–displayed under native conditions; however, in the absence of BamE, BamD indeed becomes surface-exposed. Loss of BamE altered cell envelope composition, leading to slower growth and an increase in both antibiotic susceptibility and formation of membrane vesicles containing greater amounts of vaccine antigens. Both BamD and BamE are expressed in diverse gonococcal isolates, under host-relevant conditions, and throughout different phases of growth. The solved structures of Neisseria BamD and BamE share overall folds with Escherichia coli proteins but contain differences that may be important for function. Together, these studies highlight that, although BAM is conserved across Gram-negative bacteria, structural and functional differences do exist across species, which may be leveraged in the development of species-specific therapeutics in the effort to combat multidrug resistance.
Keywords: BAM complex, BamD, BamE, cell envelope, Neisseria gonorrhoeae
Introduction
Gram-negative bacteria, mitochondria, and plastids, such as chloroplasts, contain integral β-barrel outer membrane proteins (OMPs)4 that play a myriad of pivotal physiological and structural functions, including nutrient acquisition, secretion, signal transduction, outer membrane biogenesis, and motility (1, 2). In pathogenic bacteria, OMPs additionally assist in virulence by facilitating host colonization and exploiting immune responses as well as drug extrusion (3–5). Therefore, understanding the mechanisms that direct targeting and folding of OMPs is critical for development of pharmaceutical interventions to combat clinically important pathogens, including the recent emergence of multidrug-resistant Neisseria gonorrhoeae, the etiologic agent of gonorrhea. This sexually transmitted infection remains a major public health problem globally, and 78 million cases were estimated in 2012 (6). Recent quantitative proteomic investigations of OMPs in N. gonorrhoeae yielded new insights into cell envelope composition and identified new vaccine/drug protein targets that include LptD, TamA, TamB, and the BAM complex as well as a plethora of uncharacterized lipoproteins (7, 8).
OMPs are first synthesized in the cytoplasm with an N-terminal leader sequence that routes them across the inner membrane into the periplasm by the Sec system (4, 9, 10). Periplasmic chaperones, such as SurA, FkpA, and/or Skp, then bind the nascent OMPs and escort them to the outer membrane where the β-barrel assembly machinery (BAM) then folds and/or inserts them into the outer membrane (3–5, 9, 10). BAM has been primarily investigated in Escherichia coli and Neisseria meningitidis (3, 4, 11, 12). In E. coli, BAM is a five-protein complex consisting of BamA, an OMP itself, and four accessory lipoproteins, BamB, BamC, BamD, and BamE (3, 5, 11). In contrast, no BamB ortholog is present in Neisseria genomes, whereas RmpM has been identified as an additional accessory protein (11). BamA is the central component of BAM, and its removal results in loss of viability. Similarly, a bamD deletion causes lethality in E. coli and N. meningitidis, whereas lack of other accessory Bam lipoproteins results in various degrees of growth phenotypes and more subtle effects on cell envelope composition and integrity (3, 11, 13–16). In vitro, all lipoproteins are required for fully efficient folding of OmpT, a model OMP substrate of BAM (17, 18). BamB–E are found within the periplasm anchored to the periplasmic leaflet of the outer membrane via a lipid moiety at their N terminus. Albeit still controversial, particularly with the recently reported structures of BAM (19–22), the helix-grip domains of BamC have also been shown to be surface-exposed (23).
Over the past decade, the individual structures of all the Bam proteins have been reported from E. coli, and more recently, several groups reported the structure of fully assembled E. coli BAM (20–22, 24–39). These structures have provided molecular details about how the individual Bam proteins interact with one another and revealed that the barrel domain of BamA undergoes a large conformational change within the membrane not previously observed in OMPs (19–22, 30, 38–40). BamB and BamD were found to interact directly with BamA with BamD also interacting with the barrel domain. BamE was found to not only interact with BamD as shown previously (14) but also with BamA, bridging an additional interaction of BamA with BamD. These structures have contributed significantly toward our understanding of the architecture and dynamics of BAM; however, exactly how BAM functions in E. coli remains unknown. In Neisseria, only the structure of full-length BamA (N. gonorrhoeae) has been reported for BAM (30) along with the structure of RmpM (N. meningitidis) (41).
Growing lines of evidence build an appreciation for the existence of significant differences in homologous protein function, structure, and localization, often despite a close relatedness of the organisms. For instance, factor H–binding protein, which is incorporated into the BEXSERO meningococcal B vaccine, is a surface-localized protein in N. meningitidis but not in N. gonorrhoeae (42). Furthermore, the protein responsible for transporting lipopolysaccharide in E. coli and lipooligosaccharide in Neisseria to the cell surface, LptD, is non-essential in N. meningitidis but essential in E. coli and N. gonorrhoeae (7, 43, 44).
To gain additional insight into the role of BamD and BamE within BAM in Neisseria and to aid in future therapeutic development, here we have performed mutagenesis and knockout studies to assay the effects on Neisseria growth and OMP assembly. Our work shows that BamD, but not BamE, is essential for viability and that BamE, but not BamD, is surface-exposed, similar to what has been observed for BamC in E. coli. However, in the absence of BamE, BamD did become surface-exposed with a concomitant increase in antibiotic susceptibility and production of membrane vesicles with altered OMP composition, providing further evidence that BamE may be a new vaccine target against N. gonorrhoeae. Furthermore, to better guide future investigations on Neisseria BAM and to assist in structure-based therapeutic methods, we have determined the X-ray crystal structures of both BamD and BamE from N. gonorrhoeae. These studies show that Neisseria BamD and BamE share overall folds with their E. coli orthologs, but there are differences that may be functionally important.
Results
The sequences of bamE and bamD loci in N. gonorrhoeae
BLAST searches for the gonococcal homolog of BamE protein, BamEGC, using the genome sequence of N. gonorrhoeae strain FA1090 and sequence comparisons with Clustal Omega (1.2.; http://www.clustal.org/omega/5; Ref. 76) resulted in identification of locus NGO1780 with 28.7 and 92.7% sequence identity to the E. coli and N. meningitidis MC58 counterparts, respectively (Fig. S1). The homolog of BamD, NGO0277 (hereafter BamDGC), was previously identified in N. gonorrhoeae and named ComL (45). The deduced amino acid sequence of the N. gonorrhoeae FA1090 BamDGC shares 33.7% sequence identity with its E. coli counterpart and 96.3% sequence identity with the meningococcal homolog (Fig. S1). Conservation of BamE (locus NEIP0196) and BamD (locus NEIP0653) was analyzed by comparing DNA sequences between 42,412 Neisseria isolates deposited to the PubMLST database (https://pubmlst.org/neisseria/5; Ref. 77) as of April 18, 2017 and demonstrated the presence of 179 alleles with 89 single nucleotide polymorphisms (SNPs) for BamE and 269 alleles with 186 SNPs for BamD (Fig. S2).
BamDGC is an essential BAM component in N. gonorrhoeae
In E. coli and N. meningitidis, both BamA and BamD are essential proteins, and their depletion results in OMP folding, stability, and assembly defects (11, 46–48). Recently, we demonstrated the essential nature of BamAGC in N. gonorrhoeae (8), but bamDGC was designated dispensable due to the successful generation of a transposon mutant (45). This mutant, however, showed pleiotropic phenotypes, including smaller cell size, cratered and crinkled colony morphology, and reduced transformation competence.
Based on studies in E. coli and N. meningitidis (11, 46), we reasoned that BamDGC is also essential for N. gonorrhoeae viability. To test this hypothesis, we first constructed a strain in the N. gonorrhoeae FA1090 background that carried an additional copy of bamDGC placed under an isopropyl β-d-1-thiogalactopyranoside (IPTG)–inducible promoter in a different site on the chromosome. We next insertionally inactivated the bamDGC gene in its chromosomal locus using the kanamycin resistance cassette. The resulting strain, ΔbamDGC/Plac::bamDGC, formed robust colonies in the presence of the inducer but failed to grow when subcultured on solid medium lacking IPTG (Fig. 1A). To deplete BamD during growth in liquid medium, we applied an experimental strategy utilized previously to diminish levels of the N. gonorrhoeae essential proteins BamA, Obg, and GmhA (8, 49, 50). After harvesting from solid medium supplemented with IPTG, bamD::kan/Plac::bamD was transferred to broth with or without IPTG. Following 3-h incubation, both cultures were adjusted to the same optical density (shown as time 0 h on the graph in Fig. 1B) and cultured under permissive and non-permissive conditions for another 6 h. Omitting IPTG in the liquid medium prevented bacterial growth (Fig. 1B), which was concomitant with BamDGC depletion as shown by immunoblotting with anti-BamDGC antiserum (Fig. 1C). Together, these studies confirmed the essential nature of BamD in N. gonorrhoeae.
Figure 1.
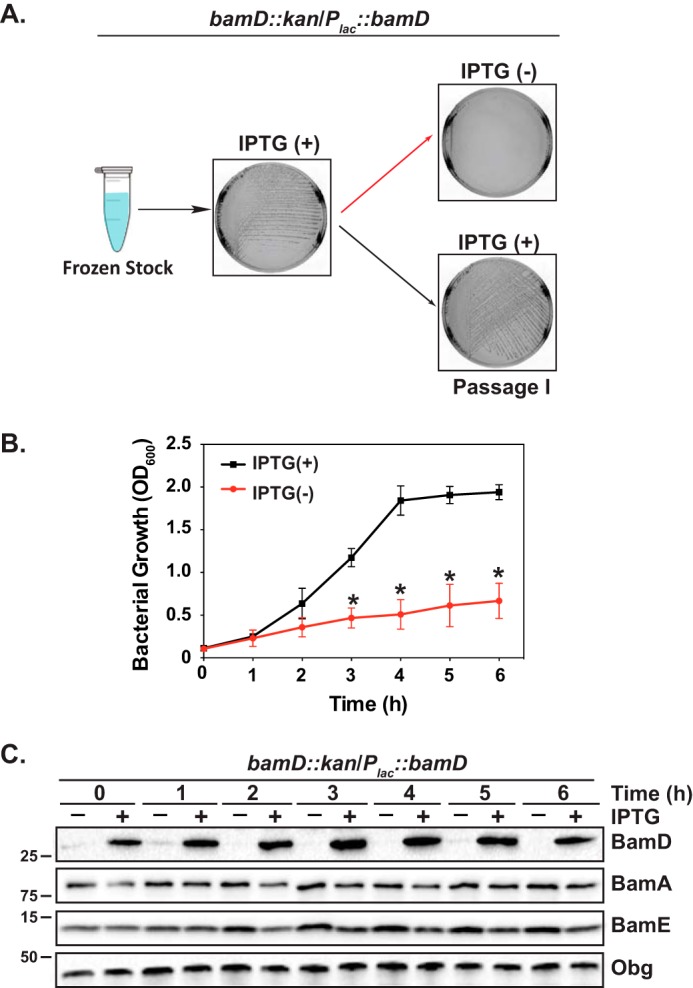
Depletion of BamDGC is lethal for N. gonorrhoeae. A, the N. gonorrhoeae FA1090 bamDGC::kan/Plac::bamDGC was streaked from frozen stock on solid medium supplemented with kanamycin and IPTG. Following incubation at standard conditions, bacteria were passaged onto solid medium with (+) or without (−) IPTG, and the colonies were examined after 22 h. Representative photographs are shown. B and C, non-piliated colonies of N. gonorrhoeae bamDGC::kan/Plac::bamDGC obtained after permissive growth in the presence of IPTG were suspended in liquid medium to an A600 of 0.1, washed, divided, and cultured in the presence or absence of IPTG. After 3 h (shown as time 0 h on the graph), cultures were back-diluted in fresh medium to the same turbidity (A600 of 0.1) and cultured under permissive (black squares) and non-permissive (red circles) conditions for an additional 6 h. At every hour, bacterial growth was examined by measuring A600, and samples were withdrawn for immunoblotting analysis (C) with anti-BamDGC, anti-BamAGC, anti-BamEGC, and anti-ObgGC antisera. Migration of a molecular mass marker (kDa) is indicated on the left. Error bars in B represent S.D.
N. gonorrhoeae lacking BamEGC displays a slower growth rate in liquid medium under standard conditions
To characterize the role of BamEGC, we cloned and purified a construct lacking the predicted lipoprotein signal peptide and tagged with a C-terminal His6 tag and used the purified protein to raise polyclonal rabbit antiserum. The purified protein migrated in SDS-PAGE according to a predicted molecular mass of ∼12 kDa, corresponding to the recombinant protein. Subsequently, we created a null bamEGC mutant, ΔbamEGC, and a complemented strain ΔbamEGC/Plac::bamEGC, in N. gonorrhoeae FA1090. The anti-BamEGC antiserum cross-reacted with wildtype whole-cell lysates. As expected, no signal was detected in the bamEGC knockout strain, whereas the complemented strain expressed BamEGC at levels proportional to the concentrations of IPTG added (Fig. 2A). The use of 20 μm IPTG resulted in amounts of BamEGC closely resembling the native protein pool in the wildtype strain and consequently was chosen in further complementation experiments.
Figure 2.
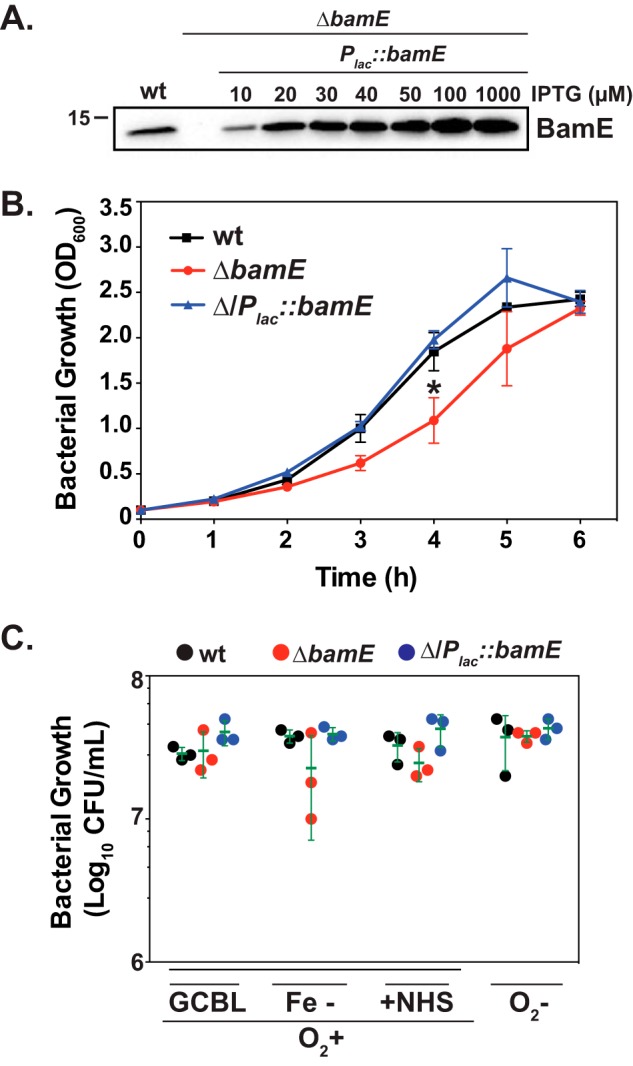
Fitness assessment of the N. gonorrhoeae null bamEGC mutant. A, cells of N. gonorrhoeae FA1090 wildtype, isogenic ΔbamEGC, and ΔbamEGC/Plac::bamEGC were harvested from solid medium supplemented with IPTG (as indicated) and subjected to SDS-PAGE followed by immunoblotting with anti-BamEGC antiserum. Migration of a molecular mass marker (kDa) is indicated on the left. B and C, non-piliated colonies of FA1090 wildtype, isogenic ΔbamEGC, and ΔbamEGC/Plac::bamEGC were suspended in liquid medium with IPTG to an A600 of 0.1 and incubated under standard aerobic conditions for 3 h. Then, cultures were either back-diluted in fresh medium and cultured for an additional 6 h, and growth was monitored at the indicated time points by measuring A600 (B), or bacteria were serially diluted and plated for fitness assessments under standard growth conditions on solid medium (GCB), during iron limitation (Fe−), in the presence of 7.5% normal human serum (+NHS), and under anaerobic conditions (O2−) (C). Experiments were performed in three biological replicates, and means with corresponding S.D. (error bars) are presented. cfu values were enumerated after 22 h of aerobic and 48 h of anaerobic growth.
The colony size of ΔbamEGC was similar to that in a parental strain, but a slower proliferation rate was observed during midlogarithmic growth in liquid medium. At the end of the experiment, however, the mutant culture reached the same density as the wildtype, and the apparent lag in growth was fully rescued in ΔbamEGC/Plac::bamEGC (51) (Fig. 2B). Culture supernatants derived from ΔbamEGC contained increased amounts of cytoplasmic protein markers such as Zwf, Obg, and GmhA, suggesting increased cell lysis (51). Interestingly, culturing the mutant in chemically defined Graver-Wade medium significantly reduced this phenotype. Slower growth in liquid medium was also observed in the Caulobacter crescentus ΔbamE but not in E. coli, N. meningitidis, Pseudomonas aeruginosa, or Salmonella enterica serovar Typhimurium (14, 52–54). In contrast, no significant fitness differences, as measured by counts of colony-forming units (cfu), were noted when the wildtype, ΔbamEGC, and ΔbamEGC/Plac::bamEGC were maintained on solid medium under standard aerobic conditions as well as conditions mimicking different microecological niches in the human host, including iron deprivation, presence of normal human serum, and anaerobiosis (Fig. 2C).
Expression of BamEGC and BamDGC
Limited information is available regarding expression of BAM components as most research efforts have focused on understanding the architecture and protein interactions of this protein complex. Therefore, to further characterize the accessory lipoproteins BamDGC and BamEGC, we examined their expression patterns in the wildtype FA1090 throughout growth in liquid medium, during exposure to environmental stimuli relevant to different infections sites in the human host, and in a panel of 36 different gonococcal isolates.
Both BamDGC and BamEGC were continuously expressed throughout all stages of N. gonorrhoeae growth (Figs. 1A and 3A). A similar expression pattern was reported for BamAGC (8). Moreover, although levels of BamDGC remained unchanged upon exposure of N. gonorrhoeae to aerobic and anaerobic conditions, upon iron limitation and in the presence of normal human serum, expression of BamEGC was noticeably induced during anaerobiosis (Fig. 3B). This could suggest an additional requirement for BamEGC during anoxia, but this potential role seems not to be sufficiently significant as fitness of N. gonorrhoeae lacking bamEGC was not affected under these conditions (Fig. 2C).
Figure 3.
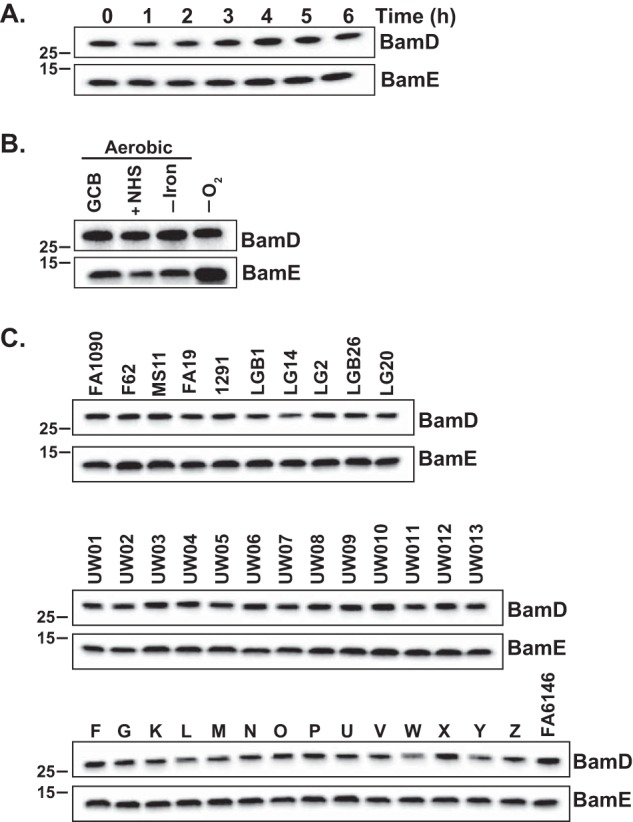
Expression studies of the accessory lipoproteins BamDGC and BamEGC in N. gonorrhoeae. A, wildtype N. gonorrhoeae FA1090 was cultured in liquid medium, and at the indicated time points, samples were withdrawn and processed for SDS-PAGE and immunoblotting analysis. B, quantities of BamDGC and BamEGC in wildtype FA1090 during in vitro conditions relevant to different infection sites (standard growth under aerobic conditions on solid medium (GCB), in the presence of normal human serum (+NHS), during iron deprivation (−Iron), and under anaerobiosis (−O2)) were assessed by probing the whole-cell lysates with respective antibodies. C, 37 strains of N. gonorrhoeae, as indicated above the immunoblots, were grown concurrently on solid medium for 20 h in 5% CO2 at 37 °C, and bacteria were collected, lysed, and processed for immunoblotting. In all experiments, samples containing the whole-cell lysates were matched by equivalent A600 units, resolved in a 4–20% Tris-glycine gel, and transferred onto nitrocellulose. Immunoblot analysis was performed using polyclonal rabbit antisera against BamDGC and BamEGC. Migration of a molecular mass marker (kDa) is indicated on the left.
Analysis of SNPs showed high conservation of both BamEGC and BamDGC among different N. gonorrhoeae isolates (Fig. S2). Corroborating this observation, the anti-BamEGC and anti-BamDGC antiserum cross-reacted with whole-cell lysates derived from common laboratory strains (FA1090, F62, MS11, FA19, and 1291) and temporally and geographically diversified clinical isolates, including the 2016 World Health Organization (WHO) reference strains (Fig. 3C). In addition, these experiments demonstrated that, similarly to BamAGC (8), the accessory lipoproteins BamEGC and BamDGC are ubiquitously expressed in a highly diverse pool of gonococcal isolates, further underscoring the potential of BAM as a target for new therapeutic interventions.
Subcellular localization studies of BamDGC and BamEGC
Using a quantitative proteomic approach, we have previously identified BamAGC, BamDGC, and BamEGC in both cell envelopes and naturally released membrane vesicles derived from four different N. gonorrhoeae strains (7, 55). Intriguingly, differences in homologous proteins' subcellular localization between even closely related organisms such as N. meningitidis and N. gonorrhoeae have been reported (42). Furthermore, recent findings showed that E. coli BamC is exposed on the surface and accessible to antibodies and proteases, which challenges the dogma for the architecture of BAM (3, 23). Therefore, we first confirmed the outer membrane localization and surface exposure of BamAGC (8). Subsequently, we sought to examine the subcellular location of BamDGC and BamEGC. Subproteome fractions isolated from wildtype N. gonorrhoeae FA1090 were separated by SDS-PAGE and probed with antisera against BAM proteins as well as a cytoplasmic protein marker, ObgGC. As expected, BamAGC and both lipoproteins, BamEGC and BamDGC, were detected in the cell envelope and membrane vesicle fractions but not in the cytoplasm or culture supernatants, whereas ObgGC was primarily localized to the cytoplasmic compartment (Fig. 4A).
Figure 4.
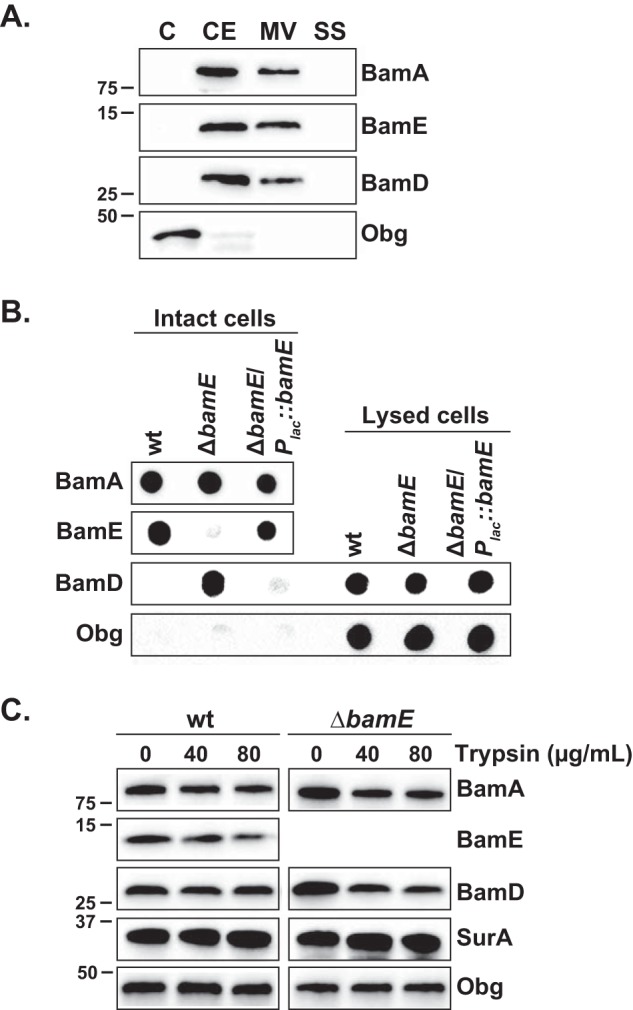
Assessment of subcellular localization of BamDGC and BamEGC. A, wildtype N. gonorrhoeae FA1090 harvested during the midlogarithmic phase was subjected to proteome extraction to separate cytoplasmic proteins (C), cell envelopes (CE), naturally released membrane vesicles (MV), and soluble proteins in culture supernatants (SS). Subproteome fractions, normalized based on the total amount of protein, were resolved by SDS-PAGE and probed with polyclonal antisera against the indicated proteins. B, N. gonorrhoeae strains, as shown above the graphs, were cultured in liquid medium, harvested, and suspended to the same A600 of 2.0. Intact as well as lysed cells were spotted onto nitrocellulose membranes and probed with polyclonal antisera against BamEGC, BamAGC, BamDGC, and ObgGC. C, N. gonorrhoeae FA1090 wildtype and isogenic ΔbamEGC cultures at an A600 of ∼1.0 were harvested, suspended in sterile PBS, and incubated at 37 °C for 1 h without or with increasing concentrations of trypsin as indicated above the immunoblot. The reaction was stopped with the addition of PMSF, cells were washed, and individual protein profiles were analyzed by immunoblotting with specific antisera against BamEGC, BamAGC, BamDGC, SurAGC, and ObgGC. Migration of a molecular mass marker (kDa) is indicated on the left.
Furthermore, to assess the surface exposure of BamEGC and BamDGC, dot blotting and protease treatment experiments using whole cells were performed according to optimized protocols for N. gonorrhoeae that ensure intactness of the cells (8). Anti-BamEGC antiserum cross-reacted with intact wildtype and ΔbamEGC/Plac::bamEGC cells but not with cells of the ΔbamEGC mutant. In contrast, BamDGC was not recognized on wildtype cells by anti-BamDGC antiserum unless the cells were lysed (Fig. 4B). Corroborating these findings, exposing intact gonococci to increasing concentrations of trypsin resulted in detection of decreased amounts of BamEGC, similar to the surface-exposed BamAGC, whereas levels of BamDGC; a periplasmic marker, SurA; and ObgGC remained unchanged (Fig. 4C). These studies suggested that at least part of the cellular pool of BamEGC is localized on the outside of the cell, whereas BamDGC faces the periplasmic side of the outer membrane. In addition, in the absence of BamEGC, BamDGC became accessible to antibodies and susceptible to protease treatment. The levels of SurA remained unchanged in ΔbamEGC in comparison with wildtype cells, excluding the possibility that surface localization of BamDGC in the ΔbamEGC mutant is solely attributable to altered outer membrane integrity (Fig. 4C). It has been suggested that both BamC and BamE stabilize the BamA-BamD interaction. In E. coli, upon BamE depletion, BamA becomes dramatically susceptible to exogenously added protease, whereas periplasmic proteins, including SurA and BamD, are completely unaffected (56). In contrast, our studies suggest that loss of BamEGC weakens the BamAGC-BamDGC interaction, causing a surface exposure of BamDGC with no significant increase in the protease sensitivity of BamAGC. Furthermore, although the absence of BamEGC had no significant impact on the steady-state-levels of any other BAM components in the cell envelopes of E. coli (56) and N. gonorrhoeae (Fig. 5C), depletion of BamDGC resulted in an increase in the cellular pool of both BamAGC and BamEGC (Fig. 5C).
Figure 5.
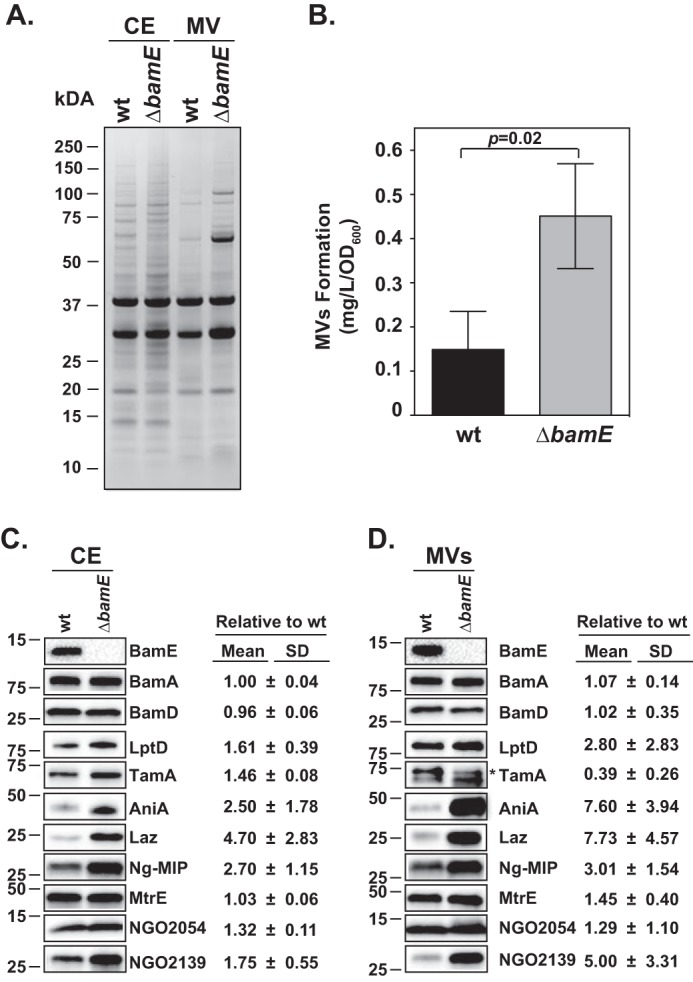
Loss of BamEGC has significant effect on the N. gonorrhoeae cell envelope. Wildtype N. gonorrhoeae FA1090 (wt) and ΔbamE strains were harvested during midlogarithmic phase and subjected to proteome extraction to separate cell envelopes (CE) and naturally released membrane vesicles (MV). A, C, and D, samples containing purified cell envelopes and membrane vesicles were normalized by total protein concentration, separated by SDS-PAGE and either visualized by Coomassie Brilliant Blue G-250 staining (A) or transferred onto nitrocellulose membrane and probed with the indicated antibodies, and the tested protein's abundance was quantified by densitometry (C and D). B, quantification of membrane vesicles (MVs) was achieved by calculating the protein content in the membrane vesicles to 1.0 liter original culture volume/OD unit (mg liter−1 OD unit−1). Experiments were performed in three biological replicates. Mean values and corresponding S.D. (error bars) are presented. Migration of a molecular mass marker (kDa) is indicated on the left. * denotes TamA (8).
Lack of BamEGC affects the proteome of cell envelopes and membrane vesicles
In E. coli, loss of BamB, but not BamC or BamE, results in significant alterations in the barrier function of the outer membrane, manifested by elevated sensitivity to several antibiotics (56). In contrast, BamE-depleted cells of N. meningitidis, C. crescentus, and S. enterica are deficient in both outer membrane assembly and integrity (11, 52, 53). To examine the impact of BamEGC on membrane permeability, we used several different methods and growth media, including disc diffusion, E-tests, and phenotypic microarrays (51) (Tables 1 and 2). In disc diffusion assays, 14 different conditions were tested with a total of 10 compounds, including detergents and antibiotics (vancomycin, carbenicillin, and polymyxin B), significantly impacting the ΔbamEGC strain in comparison with the wildtype (Table 1). Furthermore, E-tests demonstrated significantly lowered minimal inhibitory concentrations (MICs) for cefuroxime, azithromycin, ciprofloxacin, and polymyxin B (Table 2). Previously applied phenotypic microarrays with 1,056 conditions and performed in defined liquid medium showed three and six conditions uniquely beneficial and detrimental, respectively, to the ΔbamEGC mutant in comparison with six null mutants in novel gonorrhea vaccine candidates (51). The three beneficial compounds were osmolytes, whereas sodium benzoate and chromium chloride attenuated the growth of ΔbamEGC. The mutant was also negatively affected by nalidixic acid, rifampicin, doxycycline, and cefsulodin, which all exert different mechanisms to kill bacteria. Similarly, loss of BamE caused increased sensitivity to nalidixic acid, rifampicin, and carbenicillin in C. crescentus (52).
Table 1.
Disc diffusion assays were performed using different chemical probes and N. gonorrhoeae FA1090 isogenic wildtype, ΔbamEGC, and ΔbamEGC/Plac::bamEGC
| Compound | Wildtypea |
ΔbamEGC |
ΔbamEGC/Plac::bamEGC |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | n | Mean | S.D. | n | Mean | S.D. | n | |
| Tween (10%) | 10.31 | 1.1 | 8 | 12.94* | 0.98 | 8 | 10.94 | 1.74 | 8 |
| Triton X-100 (10%) | 10.00 | 1.51 | 8 | 14.75* | 3.32 | 8 | 13.50 | 3.62 | 8 |
| SDS (5%) | 27.50 | 1.91 | 4 | 28.50 | 1.00 | 4 | 27.50 | 1.91 | 4 |
| Bile salts (10%) | 11.50 | 1.29 | 4 | 11.75 | 0.95 | 4 | 11.00 | 1.41 | 4 |
| EDTA (0.5 m) | 16.67 | 0.57 | 3 | 17.33 | 0.57 | 3 | 15.67 | 1.52 | 3 |
| Oxytetracycline (1 mg/ml) | 30.75 | 1.70 | 4 | 33.25 | 1.25 | 4 | 29.25 | 1.70 | 4 |
| Oxytetracycline (2 mg/ml) | 33.80 | 2.58 | 5 | 35.60 | 1.94 | 5 | 33.60 | 3.84 | 5 |
| Chloramphenicol (0.5 mg/ml) | 31.00 | 1.68 | 4 | 32.75 | 0.48 | 4 | 26.25 | 1.03 | 4 |
| Chloramphenicol (1 mg/ml) | 35.33 | 2.88 | 3 | 37.33 | 1.15 | 3 | 30.00 | 3.46 | 3 |
| Polymyxin B (100,000 units) | 10.17 | 1.83 | 6 | 13.17* | 2.22 | 6 | 10.67 | 2.22 | 6 |
| Vancomycin (30 mg/ml) | 23.50 | 1.73 | 4 | 27.40* | 2.80 | 5 | 25.40 | 1.67 | 5 |
| Carbenicillin (1 mg/ml) | 31.00 | 2.64 | 3 | 34.00 | 2.00 | 3 | 30.00 | 1.00 | 3 |
| Carbenicillin (2.5 mg/ml) | 33.00 | 2.64 | 3 | 35.67 | 2.08 | 3 | 34.00 | 0.00 | 3 |
| Carbenicillin (5 mg/ml) | 36.29 | 1.89 | 7 | 38.86* | 0.94 | 7 | 34.86 | 1.12 | 7 |
a N. gonorrhoeae FA1090 wildtype, isogenic ΔbamE, and complemented strain ΔbamE/Plac::bamE were collected from solid medium and suspended in GCBL to an A600 of 0.2, and 100 μl of cell suspensions was plated on GCB supplemented with IPTG. Subsequently, sterile paper discs immersed in 10 μl of various chemicals, as indicated, were placed on the surface of the agar. Inhibition zone was recorded (mm) after 22 h of incubation. The data are presented as mean values with corresponding S.D. values and the number of biological replicates (n).
* indicates statistically significant differences with p < 0.05.
Table 2.
Minimal inhibitory concentration in the absence of BamEGC measured using E-test
| Antibiotic | Wildtypea | ΔbamEGC |
|---|---|---|
| Cefuroxime | 0.016 | <0.016 |
| Cefotaxime | 0.004 | 0.004 |
| Azithromycin | 0.064 | 0.032 |
| Tetracycline | 0.125 | 0.125 |
| Ciprofloxacin | 0.004 | 0.002 |
| Polymyxin B | 128 | 64 |
| Ampicillin | 0.125 | 0.125 |
| Benzylpenicillin | 0.125 | 0.125 |
a The N. gonorrhoeae wildtype and ΔbamEGC cells were suspended to McFarland standard of 0.5, spread on solid medium, and allowed to dry. E-test strips with different antibiotics were placed on the top of the agar, and bacteria were incubated for 22 h. The following day, the MICs of tested antibiotics were determined based on the zone of bacterial growth inhibition.
The sensitivity phenotype of ΔbamEGC suggested defects in membrane permeability, but examination of the general cell envelope protein profile did not show apparent alterations (Fig. 5A). We therefore analyzed naturally released membrane vesicles from wildtype and ΔbamEGC. There was an increase in the abundance of several protein species as revealed by SDS-PAGE and Coomassie staining (Fig. 5A) and a 3-fold higher shedding of membrane vesicles from ΔbamEGC in comparison with the wildtype (Fig. 5B). Outer membrane vesiculation is a well-recognized indicator of cell envelope stress (57). To gain further insights into the scale of OMP defects associated with loss of BamEGC, immunoblotting with antisera specific to 10 OMPs was performed on cell envelopes and membrane vesicles derived from the wildtype and ΔbamEGC. These studies demonstrated elevated levels of seven OMPs within the cell envelope fraction with the most significant alterations observed for AniA, Laz, and Ng-MIP (Fig. 5C). This effect was exacerbated in the membrane vesicles where the amounts of AniA and Laz increased over 7-fold, NGO2139 (MetQ) increased 5-fold, and LptD increased almost 3-fold (Fig. 5D). In contrast, the other Bam components and MtrE remained unaltered, whereas TamA was about 2-fold depleted. These results suggest a specific contribution of BamEGC to cell envelope biogenesis.
The structures of N. gonorrhoeae BamD and BamE
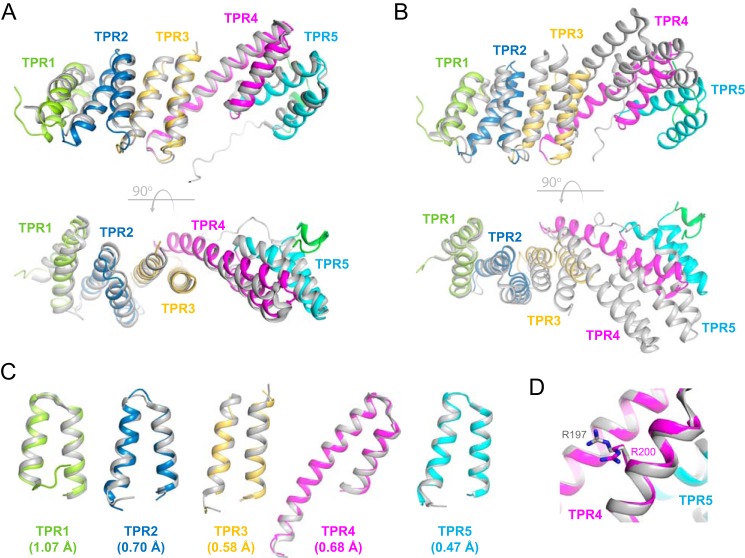
To gain insights into the structure and function of BamDGC and BamEGC and to facilitate the future targeting of BAM with small molecule inhibitors, we obtained recombinant BamDGC and BamEGC for structural studies as described under “Experimental procedures.” The structure of BamDGC was solved by molecular replacement to 2.5-Å resolution with final R/Rfree values of 0.24/0.29 and contained one molecule per asymmetric unit. The BamDGC structure closely resembles that from E. coli, consisting of five tetratricopeptide repeat (TPR) domains and having an overall root mean square deviation (r.m.s.d.) of 2.05 Å (residues 30–257) (35, 36, 58) (Fig. 6A). The shape of BamDGC is slightly more bent than the E. coli ortholog, which is best observed when aligning both structures along TPR1 only (Fig. 6B). Upon closer inspection, the individual TPR domains are more conserved than the overall r.m.s.d. suggests with r.m.s.d. values for TPR1 alone of BamDGC calculated to be 1.07 Å (residues 30–66), for TPR2 0.704 Å (residues 67–104), for TPR3 0.583 Å (residues 105–159), for TPR4 0.675 Å (residues 160–210), and for TPR5 0.471 Å (residues 211–245) (Fig. 6C). Additionally, the conserved arginine residue of BamDGC (Arg-200), important for forming a salt bridge interaction with a conserved glutamate residue of BamA (Glu-373) (19, 21, 22, 31, 59), was also found to be well-conserved and perfectly positioned to serve the same role in Neisseria (Fig. 6D).
Figure 6.
The crystal structure of BamD from N. gonorrhoeae. A, the structure of Neisseria BamD (color) superimposed with the E. coli ortholog (Protein Data Bank code 2YHC) (gray), having an overall r.m.s.d. of 2.05 Å. The bottom panel is an orthogonal view with respect to the top. B, a similar alignment as shown in A except the structures have been superimposed along only TPR1 to illustrate the overall compression of the TPR domains. C, superposition of the individual TPR domains shows a higher degree of structural conservation when compared with the overall r.m.s.d.. Individual r.m.s.d. values are shown for each TPR domain in parentheses. D, the conserved arginine in BamD (Arg-200) was found to align well with that observed in E. coli (Arg-197), indicating it would serve the same role in Neisseria BAM by mediating a salt bridge with Glu-373 of BamA.
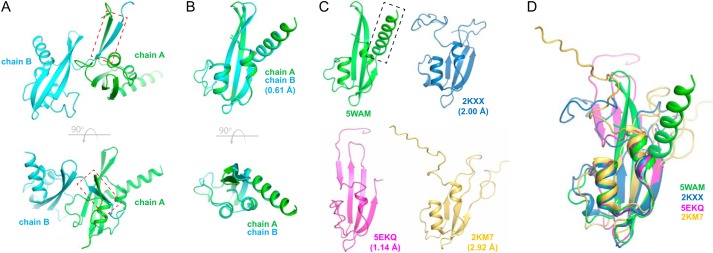
The BamEGC structure was solved by selenium single-wavelength anomalous diffraction to 2.45-Å resolution with final R/Rfree values of 0.20/0.24 and contained two molecules per asymmetric unit with each monomer interacting with the other through a β-β interaction along residues 30–36 (Fig. 7A). Each monomer contained the core ααβββ fold found in other reported BamE structures with a calculated r.m.s.d. between chain A and chain B of 0.61 Å (Fig. 7B). Our structure of Neisseria BamEGC reported here aligned reasonably well with the core (residues 39–106) of the other reported BamE structures, particularly with BamE found in the E. coli BAM crystal structure (Protein Data Bank code 5EKQ), having an r.m.s.d. of 1.14 Å. r.m.s.d. values were lower for the E. coli structures solved by NMR with calculated values of 2.00 (Protein Data Bank code 2KXX) and 2.92 Å (Protein Data Bank code 2KM7) (Fig. 7, C and D). Interestingly, BamEGC contains an additional C-terminal helix not observed in the other reported BamE structures.
Figure 7.
The crystal structure of BamE from N. gonorrhoeae. A, the asymmetric unit of the BamEGC crystal structure shows the two chains interacting through a β-β interaction along residues 30–36 (red dashed box). The bottom panel is an orthogonal view. B, alignment of chain A and chain B of BamEGC showing minor variations between the two chains, having an r.m.s.d. of 0.61 Å along the core of the structure. The bottom panel is an orthogonal view. C, side-by-side comparison of BamEGC (Protein Data Bank code 5WAM) with the orthologous structures from E. coli. Protein Data Bank code 5EKQ is from the crystal structure of E. coli BAM, whereas Protein Data Bank codes 2KXX and 2KM7 are single states from the reported NMR structures. The additional helix observed in BamEGC is indicated by the black dashed box. r.m.s.d. values are shown in parentheses compared with BamEGC. D, a structural alignment of BamEGC (Protein Data Bank code 5WAM) with the reported structures of E. coli orthologs (Protein Data Bank codes 2KXX, 5EKQ chain E, and 2KM7).
Discussion
BAM is an essential multicomponent complex that resides in the outer membranes of Gram-negative bacteria, making it an attractive target for engineering a novel class of antibiotics that do not have to cross cellular membranes. Recent studies show that the composition of BAM varies by bacterial species with E. coli having at least five components within the core complex, N. meningitidis and N. gonorrhoeae having four BAM proteins (lacking a BamB ortholog) and RmpM, and C. crescentus having BamF instead of BamC (11, 41, 60).
The structures of all the components of BAM in E. coli have been solved, including the full complex (19–22). We have previously reported the structure of the central and essential component of BAM, BamA, from N. gonorrhoeae, and here we report the structures of BamDGC and BamEGC (Figs. 6 and 7). The structure of BamDGC most closely matches that of the E. coli ortholog; however, there are small structural differences. The structure of Neisseria BamEGC, however, deviates more significantly compared with the E. coli ortholog with an additional C-terminal helix not previously observed in BamE. The possible role of this helix will be explored in future studies to determine whether it is important for BAM function.
In addition to the structures, we studied the roles of BamDGC and BamEGC within BAM in N. gonorrhoeae. We found that BamDGC is essential for viability, whereas BamEGC was dispensable, aligning well with what has been observed previously for N. meningitidis (11) and E. coli (14, 46, 47). Analogous to the presence of BamC on the surface of E. coli (23), we found that BamEGC, but not BamD, was surface-displayed in our surface labeling and protease shaving experiments (Figs. 4, B and C, and 8A). Intriguingly, we also observed BamDGC on the surface of gonococci but only in BamEGC-depleted cells (Figs. 4, B and C, and 8B). This suggests that the absence of BamEGC leads to destabilization of BAM, possibly weakening the interaction between BamAGC and BamDGC and resulting in surface exposure of BamDGC. There was, however, no significant increase in the protease sensitivity of BamAGC. It is possible that BamCGC and (an)other yet unrecognized protein(s) docked into the BAM complex influence BamAGC conformation. In E. coli, both BamE and BamC are important for stabilizing the BamA-BamD interaction, and BamE may also modulate the conformational state of BamA (56).
Figure 8.
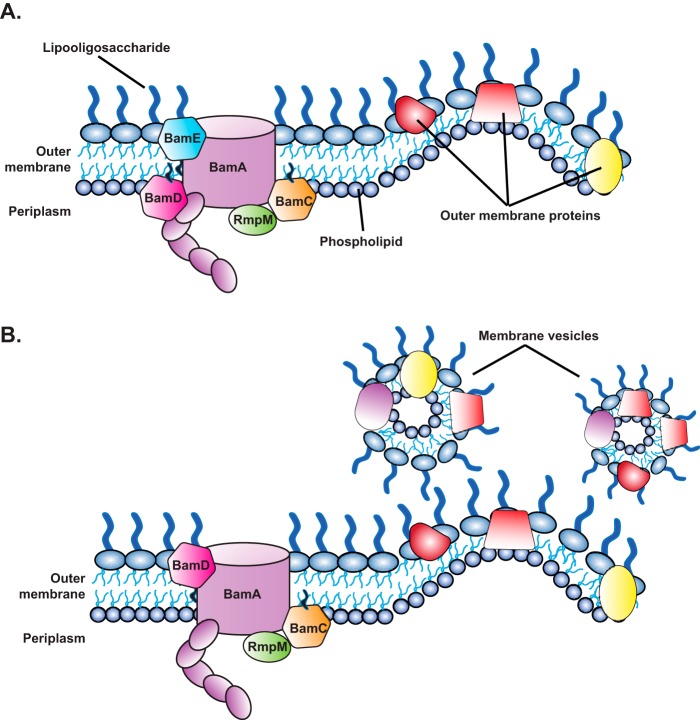
Model of the N. gonorrhoeae BAM in the presence and absence of BamEGC. A, under wildtype BAM conditions, BamEGC, but not BamDGC, was found localized at the cell surface of gonococci. B, in the absence of BamEGC, topological changes in BAM occur, including BamDGC becoming surface-exposed, which is also accompanied by an elevation in the release of membrane vesicles with altered OMP composition.
The lack of BamEGC was also accompanied by an increase in antibiotic susceptibility (Tables 1 and 2 and Ref. 51) and a significantly greater release of membrane vesicles containing altered levels of new vaccine antigens (Fig. 8B). Cumulatively, our experiments confirmed that BamDGC and BamEGC localized to the cell envelope and membrane vesicles; showed the ubiquitous expression of BamAGC (8), BamDGC, and BamEGC in a diverse pool of gonococcal isolates, further underscoring the potential of BAM as a target for novel antibiotics and vaccines against Neisseria; and, importantly, revealed additional interspecies differences existing within BAM, illuminating the need for parallel studies in different organisms to enhance our understanding of cell envelope biogenesis. Together, these studies indicate that, although BAM is conserved across all Gram-negative bacteria, structural and functional differences do exist across bacterial species and may be utilized in the development of species-specific antibiotics and vaccines in the effort to combat multidrug resistance.
Experimental procedures
Bacterial strains and growth conditions
N. gonorrhoeae utilized in this study include strain FA1090 (61) and 35 temporally and geographically diverse clinical isolates (8, 62). E. coli strains MC1061 (63), BL21(DE3) (New England Biolabs), and B834(DE3) (Millipore Sigma) were used for molecular cloning and production of recombinant proteins, respectively. N. gonorrhoeae strains were maintained on either gonococcal base solid medium (GCB; Difco), GCB without Kellogg's Supplement II and with deferoxamine mesylate salt (Desferal, Sigma) at 5 μm final concentration (iron-limited conditions), GCB with the addition of 7.5% normal human serum (NHS), or GCB with 1.2 μm nitrite as a terminal electron acceptor (anaerobic conditions) (8) or were cultured under standard growth conditions in gonococcal base liquid medium (GCBL) supplemented with sodium bicarbonate at a final concentration of 0.042% and Kellogg's Supplements I (1:100) and II (1:1000) (64). Gonococci were streaked from glycerol stocks maintained at −80 °C onto GCB and incubated for 22 h at 37 °C with 5% atmospheric CO2. Piliated or non-piliated colonies were passaged onto fresh GCB for an additional 22-h incubation. Piliated colonies were used for transformation, whereas non-piliated variants were used in all other experiments. E. coli strains were grown either on Luria-Bertani agar (LBA; Difco) or cultured in Luria-Bertani broth (LB; Difco) at 37 °C. Antibiotics were used in the following concentrations: for N. gonorrhoeae, kanamycin, 40 μg/ml; and erythromycin, 0.5 μg/ml; for E. coli, kanamycin, 50 μg/ml; erythromycin, 250 μg/ml; and carbenicillin, 50 or 100 μg/ml as specified in the text.
Genetic manipulations
Oligonucleotides used in this study (Table S1) were designed using SnapGene software version 2.8 (GSL Biotech LLC) based on the genomic sequence of N. gonorrhoeae FA1090 (NC_002946). Primers were synthesized by Integrated DNA Technologies. Genomic DNA of N. gonorrhoeae FA1090 was purified with the Wizard Genomic DNA Purification kit (Promega) or purchased directly from ATCC and used as template in PCRs with applicable oligonucleotides and Q5® high-fidelity DNA polymerase (New England Biolabs). PCR products and plasmid DNA were purified using a QIAprep Spin Miniprep kit (Qiagen). Obtained genetic constructs were verified by Sanger sequencing at the Center for Genomic Research and Biocomputing at Oregon State University and USA Macrogen. Transformation of N. gonorrhoeae was performed as described previously (55).
The N. gonorrhoeae FA1090 conditional bamD knockout, bamD::kan/Plac::bamD, was constructed according to the following steps. First, an additional copy of the bamD gene (NGO0277) under lac regulatory sequences, Plac::bamD (8), was placed at an unlinked chromosomal locus between the lctP and aspC genes using the Neisseria Insertional Complementation System, pGCC4 (65), to yield the FA1090 Plac::bamD strain. Subsequently, a 536-bp DNA fragment containing the N-terminal part of the bamD gene and upstream DNA region was amplified with primers BamD-Up-F and BamD-Up-R, digested with EcoRI/KpnI, and introduced into similarly treated pUC18K (66), yielding pUC18K-BamD-Up. Next, the downstream DNA fragment for allelic replacement (562 bp) was amplified with primers BamD-Down-F/BamD-Down-R, digested with BamHI/HindIII, and ligated into BamHI/HindIII–cleaved pUC18K-BamD-Up. The pUC18K-ΔbamD::kan was linearized with HindIII and introduced into FA1090 Plac::bamD. Transformants were selected on GCB supplemented with kanamycin and 0.05 mm IPTG and verified for disruption of the bamD gene with the kanamycin resistance cassette by PCR with primers BamD-Ver-F/BamD-Ver-R and immunoblotting analysis using BamD antiserum (8).
To generate a clean deletion of bamE in N. gonorrhoeae FA1090, the upstream region of NGO1780 was amplified with primers BamE-Up-F/BamE-Up-R. The 757-bp product was cleaved with SacI/KpnI and cloned into similarly treated pUC18K, yielding pUC18K-BamE-Up. Subsequently, the downstream region from the gene encoding BamE was amplified with primers BamE-Down-F/BamE-Down-R, and the obtained 724-bp product was cloned into BamHI/HindIII–treated pUC18K-BamE-Up. The final product, pUC18K-ΔbamE, was used for an allelic exchange of bamE with the kanamycin resistance cassette as described above. Deletion of bamE was confirmed by PCR with primers BamE-Ver-F/BamE-Ver-R using chromosomal DNA isolated from wildtype FA1090 as controls and by probing the whole-cell lysates of wildtype and ΔbamE with antiserum against BamE.
To complement the N. gonorrhoeae FA1090 ΔbamE mutant, first the Plac::bamE construct was generated by amplification of bamE with native ribosome-binding site using primers cBamE-F/cBamE-R. The 41-bp PCR product was digested with FseI and inserted into ScaI/FseI–cleaved pGCC4, yielding pGCC4-BamE. Next, pGCC4-BamE was introduced into the ΔbamE mutant by transformation, and clones were selected on GCB containing erythromycin and validated by PCR with primers pGCC4-Ver-F/pGCC4-Rev-R as well as immunoblotting with anti-BamE antiserum.
To obtain pET28-rBamE used for production of recombinant BamE (rBamE), which lacked the native signal peptide and contained a C-terminal His6 tag, the ngo1780 gene was amplified with primers rBamE-F/rBamE-R and cloned into NcoI/HindIII–digested pET28a.
The gene encoding SurA (ngo1714) lacking the DNA encoding signal peptide was amplified using primers SurA-F/SurA-R. The subsequent PCR product was digested with NcoI/HindIII and ligated into similarly cut pRSF-NT to create a TEV protease–cleavable C-terminal His6-tagged fusion. For structural studies, the BamD and BamE coding regions, starting after the N-terminal cysteine, were amplified from N. gonorrhoeae strain FA1090 genomic DNA (ATCC) and subcloned into the pHIS-parallel2 vector using NcoI and XhoI restriction sites. All sequences were verified by sequencing analysis (primers available upon request).
Growth assays
Depletion of BamD was achieved by applying an experimental strategy utilized previously to diminish levels of N. gonorrhoeae BamA, Obg, and GmhA (8, 49, 50). N. gonorrhoeae FA1090 bamD::kan/Plac::bamD was harvested from GCB with 0.05 mm IPTG (permissive conditions), and the A600 was adjusted to 0.1. Bacteria were washed twice and cultured in GCBL with or without IPTG at a final concentration of 0.05 mm. After 3 h, both cultures were diluted to an A600 of 0.1 and cultured in fresh GCBL with or without IPTG for another 6 h. At every hour, A600 measurements were taken, and samples were withdrawn for immunoblotting analysis. Three biological replicates of the experiment were performed. Mean values and corresponding S.D. are reported.
The growth kinetics of FA1090 wildtype, ΔbamE, and ΔbamE/Plac::bamE were performed in GCBL under standard growth conditions. Bacteria were collected from GCB and suspended in GCBL to an A600 of 0.1. Media for growing ΔbamE/Plac::bamE were supplemented with 0.02 mm IPTG. Following 3 h of incubation at 37 °C with aeration (220 rpm), bacterial cultures were back-diluted to an A600 of 0.1 in fresh GCBL and cultured for an additional 6 h. Samples were withdrawn for A600 measurements every hour (n = 3; mean ± S.D.).
To assess the viability of N. gonorrhoeae lacking bamE during host-relevant in vitro growth conditions, colonies of FA1090 wildtype, ΔbamE, and ΔbamE/Plac::bamE were collected from GCB, suspended in GCBL to an A600 of 0.1, and cultured for 3 h at 37 °C with aeration. Subsequently, the cultures were normalized to an A600 of 0.2, serially diluted, and plated on solid medium for standard growth conditions (GCB), iron-limiting conditions, NHS, and anaerobic conditions as described above. All media were additionally supplemented with 0.02 mm IPTG. The cfu values were scored after 22 and 48 h for aerobic and anaerobic conditions, respectively. Experiments were performed on three separate occasions, and mean cfu values with corresponding S.D. are reported.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was tested using a slightly modified Kirby-Bauer (disk diffusion) method (67) and E-test. In disc diffusion experiments, FA1090 wildtype, isogenic ΔbamE, and ΔbamE/Plac::bamE strains were collected from GCB, and the suspensions were adjusted in GCBL to match an A600 of 0.2. Cell suspensions (100 μl) were immediately plated on GCB, and 6-mm filter paper disks (Whatman) impregnated with 10 μl of tested compounds, as indicated below, were placed on the surface of the agar. The zones of inhibition in mm were measured after 22 h. Experiments were performed in biological triplicates, and mean values with S.D. are presented.
MICs for cefuroxime, cefotaxime, azithromycin, tetracycline, ciprofloxacin, polymyxin B, ampicillin, and benzylpenicillin were determined using an E-test (Biomerieux) according to the manufacturer's recommendations. Each determination was performed on three separate occasions using fresh bacterial cultures, and the consensus MIC obtained in at least two trials was reported.
Protein localization assays
Subcellular fractionations, immunodotting, and trypsin accessibility studies were performed following procedures described previously (8). Briefly, N. gonorrhoeae FA1090 wildtype and ΔbamE at the midlogarithmic phase of growth were used to extract proteins from the cytosolic, cell envelope, membrane vesicle, and soluble supernatant fractions. Cell envelopes were separated from cytoplasmic proteins by a sodium carbonate extraction method and differential centrifugation, whereas culture supernatants were subjected to filtration and ultracentrifugation to separate naturally released membrane vesicles from soluble proteins. Quantification of membrane vesicles was achieved by calculating the protein content in the membrane vesicles to 1.0 liter of original culture volume/OD unit (mg liter−1 OD unit−1) as described (68).
In immunodotting and protease susceptibility studies, intact bacterial cells were used (8). For immunodotting, bacteria were suspended in GCBL to an A600 of 0.1, cultured with aeration for 3 h, harvested, and spotted as 5-μl suspensions onto a nitrocellulose membrane after adjusting the A600 to 2.0. The samples were dried at room temperature for 15 min and subjected to immunoblotting.
In trypsin shaving assays, gonococci were subcultured in GCBL for 3 h after collecting from solid medium, diluted to an A600 of 0.1, and cultured until an A600 of ∼1.0 was reached. Bacteria were gently harvested and suspended in sterile PBS, pH 8.0, to an A600 of 2.5, and 500-μl suspensions were incubated for 1 h at 37 °C with trypsin at final concentrations of 0, 40, or 80 μg/ml. To deactivate trypsin, 10 μl of 50 mm phenylmethylsulfonyl fluoride (PMSF) was added, bacteria were washed with GCBL and subjected to SDS-PAGE, and trypsin accessibility of selected proteins was detected by immunoblotting with polyclonal antiserum.
Purification of rBamE and rSurA and preparation of polyclonal antisera
An overnight culture of E. coli BL21(DE3) carrying either pET28a-rBamE or pET28a-rSurA was back-diluted into 3.0 or 1.0 liter of LB broth, respectively, supplemented with kanamycin and incubated with aeration at 37 °C. The production of rBamE and rSurA was induced with 0.1 and 1 mm IPTG, respectively, during midlogarithmic growth. Bacterial cells were collected by centrifugation 3 h after induction, suspended in lysis buffer (500 mm NaCl, 10 mm imidazole, 20 mm Tris-HCl, pH 8.0, and Complete EDTA-free protease inhibitor tablet (Roche Applied Science)), and lysed by passaging through a French pressure cell at 12,000 p.s.i. Unbroken cells and cell debris were removed by centrifugation at 16,000 × g for 30 min at 4 °C. The cell-free lysate was passed through a 0.22-μm filter unit (VWR International) and applied onto Bio-Scale Mini Profinity immobilized metal affinity chromatography cartridges (Bio-Rad). Loosely bound proteins were removed with 10 column volumes of wash buffer (20 mm Tris, pH 8.0, 500 mm NaCl, 40 mm imidazole), and proteins were eluted with a 40–250 mm imidazole gradient using an NGC Purification System (Bio-Rad). Fractions containing rBamE were pooled and dialyzed against 20 mm Tris, pH 8.0, 10% glycerol, whereas fractions containing eluted rSurA were incubated overnight at 4 °C with TEV protease in a 1:20 ratio to remove the His6 tag. After concentrating the sample to 5 ml using Vivaspin 20 centrifuge concentrators (GE Healthcare), proteins were subjected to size exclusion chromatography using a HiLoad 16/600 Superdex 75 prep grade column (GE HealthCare) with phosphate-buffered saline (PBS) as running buffer. Finally, fractions containing rSurA were concentrated using a Vivaspin 20 centrifuge concentrator.
Polyclonal antisera against purified rSurA and rBamE were prepared by Pacific Immunology Corp. using a 13-week antibody production protocol and two New Zealand White rabbits under Animal Protocol 1 approved by the Institutional Animal Care and Use Committee and the National Institutes of Health Animal Welfare Assurance Program (A4182-01) in a certified animal facility (United States Department of Agriculture 93-R-283). The rabbit polyclonal anti-BamD antiserum were obtained and evaluated previously (8).
Expression and purification of recombinant N. gonorrhoeae BamD and BamE
For expression of BamDGC and BamEGC for structural studies, each construct was introduced into E. coli BL21(DE3) chemically competent cells, then plated onto LBA supplemented with carbenicillin (100 μg/ml), and incubated overnight at 37 °C. For native expression, a single colony was used to inoculate a 5-ml LB with carbenicillin (50 μg/ml) starter culture, which was cultured to an A600 of ∼1.0. The cells were then washed with fresh LB and inoculated into 2 liters of 2× YT (16 g/liter Tryptone, 10 g/liter yeast extract, 5.0 g/liter NaCl) medium supplemented with carbenicillin (50 μg/ml). Bacteria were cultured at 37 °C until an A600 of ∼0.8 was reached, expression was induced with 0.2 mm IPTG, and bacteria were grown an additional 8 h at 25 °C before harvesting.
For selenomethionine-substituted BamEGC, E. coli B834(DE3) cells were transformed and plated onto LBA overnight. A single colony was used to inoculate a 25-ml LB with carbenicillin (50 μg/ml) culture and allowed to grow at 37 °C to an A600 of 0.8–1.0. The cells were then centrifuged, washed three times with minimal medium lacking methionine, and resuspended in 6 ml of wash medium, and then 1 ml was added to six flasks containing 1 liter each of minimal medium supplemented with selenomethionine (40 mg/liter) and carbenicillin (50 μg/ml). These cultures were grown at 37 °C until the A600 was between 0.6 and 0.8, then induced with 0.5 mm IPTG, and allowed to grow an additional 24 h before harvesting.
For protein purification, cells were resuspended in PBS (12 mm phosphate buffer, pH 7.4, 137 mm NaCl, 2.7 mm KCl) at 5 ml/g of cell paste and supplemented with DNase I (10 μg/ml) and PMSF (0.2 mm). The cells were then lysed by two passages through an Avestin C3 Emulsiflex, and the lysates were centrifuged at 39,000 × g for 45 min. The supernatants were then applied to a pre-equilibrated 5-ml nickel-nitrilotriacetic acid resin column. The column was washed with at least 5 column volumes of PBS, a pre-elution was performed using PBS with 25 mm imidazole, and a final elution was done with PBS containing 250 mm imidazole. To remove the N-terminal His tag, TEV protease was added to the final protein sample along with 1 mm DTT and 0.5 mm EDTA and dialyzed overnight at 4 °C in PBS. The dialyzed samples were then again applied to a 2-ml pre-equilibrated nickel-nitrilotriacetic acid resin column. The filtrate was collected, concentrated, and then applied to a HiPrep Sephacryl S-100 HR gel filtration column (GE Healthcare) in PBS as a final purification step. Peak fractions were confirmed by SDS-PAGE analysis, pooled, and concentrated to 10 mg/ml.
Crystallization and structure determination of BamD and BamE from N. gonorrhoeae
For crystallization, the samples were screened using commercial sparse-matrix crystallization screens on a TTP LabTech Mosquito Crystal crystallization robot using the hanging-drop vapor-diffusion method with a drop ratio of 1:1 (protein:well solution), and lead conditions were further optimized. BamDGC was crystallized in final conditions of 1.0 m LiCl, 100 mm HEPES, pH 7.0, 10% PEG 6000. Native and selenomethionine-substituted BamEGC crystals were grown in final conditions of 10 mm zinc sulfate heptahydrate, 100 mm MES monohydrate, pH 6.5, 25% PEG monomethyl ether 550. Crystals were harvested directly from their drops, cryoprotected in mother liquor solution containing 20% glycerol, and flash frozen in liquid nitrogen until data collection.
Data sets for native BamDGC and BamEGC were collected at the GM/CA-CAT beamline 23ID at the Advanced Photon Source, Argonne National Laboratory. Data sets for selenomethionine-substituted BamEGC were collected at beamline X25 at the National Synchrotron Light Source, Brookhaven National Laboratory. All data were processed using HKL2000 (69) or xia2 (70). The BamDGC structure was solved by molecular replacement with PHASER within PHENIX (71) in space group P43212 to 2.5-Å resolution using the E. coli BamD structure (Protein Data Bank code 2YHC) as a search model. The BamEGC structure was solved by selenium single-wavelength anomalous diffraction in space group P21212 to 2.45-Å resolution with AutoSol within PHENIX (71). A single selenium site per monomer was used to calculate initial phases followed by AutoBuild (71). All manual model building was performed with Coot (72), and refinement was performed with phenix.refine within PHENIX (71). Structure validation was performed using MolProbity within PHENIX (71) using the online server (73). r.m.s.d. analysis and rigid body fitting were performed within PyMOL (Schrödinger). Data collection and refinement statistics are summarized in Table 3. Minimization of the partial BAM model was performed using Chiron (74). All figures were made using PyMOL (Schrödinger) and annotated and finalized in Adobe Photoshop/Illustrator.
Table 3.
Data collection and refinement statistics
r.m.s., root mean square.
| BamDGC | BamEGC | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.0332 | 0.9794 |
| Space group | P43212 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 64.42, 64.42, 166.39 | 62.80, 86.08, 49.92 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 45.55–2.50 (2.59–2.50)a | 43.18–2.45 (2.54–2.45) |
| Rsym | 0.104 (2.30) | 0.160 (0.795) |
| Rpim | 0.040 (0.813) | 0.088 (0.439) |
| CC½b | (0.500) | (0.491) |
| I/σI | 17.5 (1.12) | 7.2 (1.48) |
| Completeness (%) | 99.9 (100) | 98.1 (99.5) |
| Multiplicity | 8.1 (8.8) | 4.2 (4.3) |
| Refinement | ||
| Resolution (Å) | 43.93–2.50 | 43.04–2.45 |
| No. reflections (total/free) | 12,772/1,277 | 10,276/521 |
| Rwork/Rfree | 0.236/0.285 | 0.198/0.240 |
| Number of atoms | ||
| Protein | 1,733 | 1,589 |
| Ligand/ion | 1 | |
| Water | 13 | 89 |
| B-factors | ||
| Protein | 91.6 | 41.8 |
| Ligand/ion | 32.0 | |
| Water | 69.3 | 37.8 |
| All atoms | 91.5 | 41.6 |
| Wilson B | 74.7 | 31.9 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.002 |
| Bond angles (°) | 0.39 | 0.45 |
| Ramachandran distributionc (%) | ||
| Favored | 96.6 | 97.5 |
| Allowed | 3.4 | 2.5 |
| Outliers | 0 | 0 |
| Protein Data Bank code | 5WAQ | 5WAM |
a Values in parentheses are for the highest-resolution shell.
b CC½: Pearson's correlation coefficient (CC) between intensity estimates from half data sets. Primary indicator for use for selecting high resolution cutoff for data processing.
c Calculated using MolProbity.
SDS-PAGE and immunoblotting
Samples of whole-cell lysates, protein fractions, intact cells, or purified proteins were normalized based on either A600 or protein concentration as specified. Protein concentration was measured using the DC Protein Assay (Bio-Rad). Proteins were visualized by staining with Coomassie Brilliant Blue G-250 or transferred onto nitrocellulose membrane using a Trans-blot Turbo (Bio-Rad) and detected by immunoblotting as described previously (8) using the following dilutions of primary polyclonal rabbit antisera: anti-BamEGC, 1:20,000; anti-BamAGC, 1:20,000 (8); anti-BamDGC, 1:10,000 (8); anti-ObgGC, 1:10,000 (49); anti-SurAGC, 1:10,000; anti-LptD, 1:5,000; anti-TamA, 1:5,000; anti-AniA, 1:10,000; anti-Laz, 1:20,000; anti-Ng-MIP, 1:10,000; anti-MtrE, 1:10,000; anti-NGO2054, 1:10,000; and anti-NGO2139, 1: 20,000 (8).
Densitometry analysis
Protein abundance was quantified by densitometry using the Image Lab 5.0 software (Bio-Rad) volume tool (rectangle), local background subtraction, and linear regression method as described previously (49, 75). Experiments were performed in biological triplicates and are shown in Fig. S3. The relative protein levels are presented as mean values and S.D.
Statistical analysis
GraphPad Prism's built-in t test was utilized to determine statistically significant differences between experimental results. A confidence level of 95% was used for all analyses.
Author contributions
A. E. S., I. H. W., S. K. B., and N. N. conceptualization; A. E. S., and N. N. resources; A. E. S., I. H. W., R. A. Z., R. F. R., K. V. K., S. K. B., and N. N. data curation; A. E. S., I. H. W., R. A. Z., R. F. R., K. V. K., and N. N. formal analysis; A. E. S., S. K. B., and N. N. supervision; A. E. S., S. K. B., and N. N. funding acquisition; A. E. S. and N. N. validation; A. E. S., I. H. W., R. A. Z., R. F. R., K. V. K., S. K. B., and N. N. investigation; A. E. S. and N. N. visualization; A. E. S., I. H. W., R. A. Z., R. F. R., K. V. K., S. K. B., and N. N. methodology; A. E. S., I. H. W., R. A. Z., R. F. R., K. V. K., and N. N. writing-original draft; A. E. S. and N. N. project administration; A. E. S., I. H. W., R. A. Z., R. F. R., K. V. K., S. K. B., and N. N. writing-review and editing.
Supplementary Material
Acknowledgments
We thank the respective staffs at the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility (GM/CA) beamline at the Advanced Photon Source (APS), Argonne National Laboratory, and at the X25 beamline at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, for assistance. Use of the APS was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38, and use of the NSLS was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 5WAQ and 5WAM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- OMP
- outer membrane protein
- BAM
- β-barrel assembly machinery
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MIC
- minimal inhibitory concentration
- TPR
- tetratricopeptide repeat
- r.m.s.d.
- root mean square deviation
- GCB
- gonococcal base solid medium
- NHS
- normal human serum
- GCBL
- gonococcal base liquid medium
- LBA
- Luria-Bertani agar
- rBamE
- recombinant BamE
- TEV
- tobacco etch virus
- rSurA
- recombinant SurA
- Ng-MIP
- Neisseria gonorrhoeae Macrophage infectivity potentiator.
References
- 1. Noinaj N., Easley N. C., Oke M., Mizuno N., Gumbart J., Boura E., Steere A. N., Zak O., Aisen P., Tajkhorshid E., Evans R. W., Gorringe A. R., Mason A. B., Steven A. C., and Buchanan S. K. (2012) Structural basis for iron piracy by pathogenic Neisseria. Nature 483, 53–58 10.1038/nature10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fairman J. W., Noinaj N., and Buchanan S. K. (2011) The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr. Opin. Struct. Biol. 21, 523–531 10.1016/j.sbi.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noinaj N., Gumbart J. C., and Buchanan S. K. (2017) The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204 10.1038/nrmicro.2016.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hagan C. L., Silhavy T. J., and Kahne D. (2011) β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 10.1146/annurev-biochem-061408-144611 [DOI] [PubMed] [Google Scholar]
- 5. Rollauer S. E., Sooreshjani M. A., Noinaj N., and Buchanan S. K. (2015) Outer membrane protein biogenesis in Gram-negative bacteria. Philos. Trans. R Soc. Lond. B Biol. Sci. 370, 20150023 10.1098/rstb.2015.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Newman L., Rowley J., Vander Hoorn S., Wijesooriya N. S., Unemo M., Low N., Stevens G., Gottlieb S., Kiarie J., and Temmerman M. (2015) Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10, e0143304 10.1371/journal.pone.0143304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zielke R. A., Wierzbicki I. H., Weber J. V., Gafken P. R., and Sikora A. E. (2014) Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol. Cell. Proteomics 13, 1299–1317 10.1074/mcp.M113.029538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zielke R. A., Wierzbicki I. H., Baarda B. I., Gafken P. R., Soge O. O., Holmes K. K., Jerse A. E., Unemo M., and Sikora A. E. (2016) Proteomics-driven antigen discovery for development of vaccines against gonorrhea. Mol. Cell. Proteomics 15, 2338–2355 10.1074/mcp.M116.058800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ricci D. P., and Silhavy T. J. (2012) The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084 10.1016/j.bbamem.2011.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knowles T. J., Scott-Tucker A., Overduin M., and Henderson I. R. (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 10.1038/nrmicro2069 [DOI] [PubMed] [Google Scholar]
- 11. Volokhina E. B., Beckers F., Tommassen J., and Bos M. P. (2009) The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191, 7074–7085 10.1128/JB.00737-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gentle I. E., Burri L., and Lithgow T. (2005) Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58, 1216–1225 10.1111/j.1365-2958.2005.04906.x [DOI] [PubMed] [Google Scholar]
- 13. Voulhoux R., and Tommassen J. (2004) Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155, 129–135 10.1016/j.resmic.2003.11.007 [DOI] [PubMed] [Google Scholar]
- 14. Sklar J. G., Wu T., Gronenberg L. S., Malinverni J. C., Kahne D., and Silhavy T. J. (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 6400–6405 10.1073/pnas.0701579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruiz N., Falcone B., Kahne D., and Silhavy T. J. (2005) Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317 10.1016/j.cell.2005.02.014 [DOI] [PubMed] [Google Scholar]
- 16. Wimley W. C. (2003) The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13, 404–411 10.1016/S0959-440X(03)00099-X [DOI] [PubMed] [Google Scholar]
- 17. Hagan C. L., Kim S., and Kahne D. (2010) Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 10.1126/science.1188919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roman-Hernandez G., Peterson J. H., and Bernstein H. D. (2014) Reconstitution of bacterial autotransporter assembly using purified components. Elife 3, e04234 10.7554/eLife.04234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bakelar J., Buchanan S. K., and Noinaj N. (2016) The structure of the β-barrel assembly machinery complex. Science 351, 180–186 10.1126/science.aad3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iadanza M. G., Higgins A. J., Schiffrin B., Calabrese A. N., Brockwell D. J., Ashcroft A. E., Radford S. E., and Ranson N. A. (2016) Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat. Commun. 7, 12865 10.1038/ncomms12865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gu Y., Li H., Dong H., Zeng Y., Zhang Z., Paterson N. G., Stansfeld P. J., Wang Z., Zhang Y., Wang W., and Dong C. (2016) Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 10.1038/nature17199 [DOI] [PubMed] [Google Scholar]
- 22. Han L., Zheng J., Wang Y., Yang X., Liu Y., Sun C., Cao B., Zhou H., Ni D., Lou J., Zhao Y., and Huang Y. (2016) Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 192–196 10.1038/nsmb.3181 [DOI] [PubMed] [Google Scholar]
- 23. Webb C. T., Selkrig J., Perry A. J., Noinaj N., Buchanan S. K., and Lithgow T. (2012) Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J. Mol. Biol. 422, 545–555 10.1016/j.jmb.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gatzeva-Topalova P. Z., Walton T. A., and Sousa M. C. (2008) Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16, 1873–1881 10.1016/j.str.2008.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., and Kahne D. (2007) Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 10.1126/science.1143993 [DOI] [PubMed] [Google Scholar]
- 26. Kim K. H., and Paetzel M. (2011) Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J. Mol. Biol. 406, 667–678 10.1016/j.jmb.2010.12.020 [DOI] [PubMed] [Google Scholar]
- 27. Kim K. H., Aulakh S., and Paetzel M. (2011) Crystal structure of β-barrel assembly machinery BamCD protein complex. J. Biol. Chem. 286, 39116–39121 10.1074/jbc.M111.298166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim K. H., Kang H. S., Okon M., Escobar-Cabrera E., McIntosh L. P., and Paetzel M. (2011) Structural characterization of Escherichia coli BamE, a lipoprotein component of the β-barrel assembly machinery complex. Biochemistry 50, 1081–1090 10.1021/bi101659u [DOI] [PubMed] [Google Scholar]
- 29. Noinaj N., Fairman J. W., and Buchanan S. K. (2011) The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J. Mol. Biol. 407, 248–260 10.1016/j.jmb.2011.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Noinaj N., Kuszak A. J., Gumbart J. C., Lukacik P., Chang H., Easley N. C., Lithgow T., and Buchanan S. K. (2013) Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 10.1038/nature12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Neil P. K., Rollauer S. E., Noinaj N., and Buchanan S. K. (2015) Fitting the pieces of the β-barrel assembly machinery complex. Biochemistry 54, 6303–6311 10.1021/acs.biochem.5b00852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heuck A., Schleiffer A., and Clausen T. (2011) Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J. Mol. Biol. 406, 659–666 10.1016/j.jmb.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 33. Jansen K. B., Baker S. L., and Sousa M. C. (2015) Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine. J. Biol. Chem. 290, 2126–2136 10.1074/jbc.M114.584524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bergal H. T., Hopkins A. H., Metzner S. I., and Sousa M. C. (2016) The structure of a BamA-BamD fusion illuminates the architecture of the β-barrel assembly machine core. Structure 24, 243–251 10.1016/j.str.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Albrecht R., and Zeth K. (2011) Structural basis of outer membrane protein biogenesis in bacteria. J. Biol. Chem. 286, 27792–27803 10.1074/jbc.M111.238931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sandoval C. M., Baker S. L., Jansen K., Metzner S. I., and Sousa M. C. (2011) Crystal structure of BamD: an essential component of the β-barrel assembly machinery of Gram-negative bacteria. J. Mol. Biol. 409, 348–357 10.1016/j.jmb.2011.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knowles T. J., Browning D. F., Jeeves M., Maderbocus R., Rajesh S., Sridhar P., Manoli E., Emery D., Sommer U., Spencer A., Leyton D. L., Squire D., Chaudhuri R. R., Viant M. R., Cunningham A. F., et al. (2011) Structure and function of BamE within the outer membrane and the β-barrel assembly machine. EMBO Rep. 12, 123–128 10.1038/embor.2010.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Albrecht R., Schütz M., Oberhettinger P., Faulstich M., Bermejo I., Rudel T., Diederichs K., and Zeth K. (2014) Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr. D Biol. Crystallogr. 70, 1779–1789 10.1107/S1399004714007482 [DOI] [PubMed] [Google Scholar]
- 39. Ni D., Wang Y., Yang X., Zhou H., Hou X., Cao B., Lu Z., Zhao X., Yang K., and Huang Y. (2014) Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J. 28, 2677–2685 10.1096/fj.13-248450 [DOI] [PubMed] [Google Scholar]
- 40. Noinaj N., Kuszak A. J., Balusek C., Gumbart J. C., and Buchanan S. K. (2014) Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062 10.1016/j.str.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grizot S., and Buchanan S. K. (2004) Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 51, 1027–1037 10.1111/j.1365-2958.2003.03903.x [DOI] [PubMed] [Google Scholar]
- 42. Jongerius I., Lavender H., Tan L., Ruivo N., Exley R. M., Caesar J. J., Lea S. M., Johnson S., and Tang C. M. (2013) Distinct binding and immunogenic properties of the gonococcal homologue of meningococcal factor h binding protein. PLoS Pathog. 9, e1003528 10.1371/journal.ppat.1003528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Braun M., and Silhavy T. J. (2002) Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302 10.1046/j.1365-2958.2002.03091.x [DOI] [PubMed] [Google Scholar]
- 44. Bos M. P., Tefsen B., Geurtsen J., and Tommassen J. (2004) Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 10.1073/pnas.0402340101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fussenegger M., Facius D., Meier J., and Meyer T. F. (1996) A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol. Microbiol. 19, 1095–1105 10.1046/j.1365-2958.1996.457984.x [DOI] [PubMed] [Google Scholar]
- 46. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., and Kahne D. (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 10.1016/j.cell.2005.02.015 [DOI] [PubMed] [Google Scholar]
- 47. Malinverni J. C., Werner J., Kim S., Sklar J. G., Kahne D., Misra R., and Silhavy T. J. (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 10.1111/j.1365-2958.2006.05211.x [DOI] [PubMed] [Google Scholar]
- 48. Onufryk C., Crouch M.-L., Fang F. C., and Gross C. A. (2005) Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 187, 4552–4561 10.1128/JB.187.13.4552-4561.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zielke R. A., Wierzbicki I. H., Baarda B. I., and Sikora A. E. (2015) The Neisseria gonorrhoeae Obg protein is an essential ribosome-associated GTPase and a potential drug target. BMC Microbiol. 15, 129 10.1186/s12866-015-0453-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wierzbicki I. H., Zielke R. A., Korotkov K. V., and Sikora A. E. (2017) Functional and structural studies on the Neisseria gonorrhoeae GmhA, the first enzyme in the glycero-manno-heptose biosynthesis pathways, demonstrate a critical role in lipooligosaccharide synthesis and gonococcal viability. Microbiologyopen 6, e00432 10.1002/mbo3.432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baarda B. I., Emerson S., Proteau P. J., and Sikora A. E. (2017) Deciphering function of new gonococcal vaccine antigens using phenotypic microarrays. J. Bacteriol. 199, e00037–17 10.1128/JB.00037-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ryan K. R., Taylor J. A., and Bowers L. M. (2010) The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane β-barrel proteins in Caulobacter crescentus. Microbiology 156, 742–756 10.1099/mic.0.035055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lewis C., Skovierova H., Rowley G., Rezuchova B., Homerova D., Stevenson A., Sherry A., Kormanec J., and Roberts M. (2008) Small outer-membrane lipoprotein, SmpA, is regulated by σE and has a role in cell envelope integrity and virulence of Salmonella enterica serovar Typhimurium. Microbiology 154, 979–988 10.1099/mic.0.2007/011999-0 [DOI] [PubMed] [Google Scholar]
- 54. Ochsner U. A., Vasil A. I., Johnson Z., and Vasil M. L. (1999) Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 181, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zielke R. A., and Sikora A. E. (2014) Isolation of cell envelopes and naturally released membrane vesicles of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. 34, 4A.3.1–17 10.1002/9780471729259.mc04a03s34 [DOI] [PubMed] [Google Scholar]
- 56. Rigel N. W., Schwalm J., Ricci D. P., and Silhavy T. J. (2012) BamE modulates the Escherichia coli β-barrel assembly machine component BamA. J. Bacteriol. 194, 1002–1008 10.1128/JB.06426-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kulp A., and Kuehn M. J. (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dong C., Hou H. F., Yang X., Shen Y. Q., and Dong Y. H. (2012) Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr. D Biol. Crystallogr. 68, 95–101 10.1107/S0907444911051031 [DOI] [PubMed] [Google Scholar]
- 59. Ricci D. P., Hagan C. L., Kahne D., and Silhavy T. J. (2012) Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. U.S.A. 109, 3487–3491 10.1073/pnas.1201362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anwari K., Poggio S., Perry A., Gatsos X., Ramarathinam S. H., Williamson N. A., Noinaj N., Buchanan S., Gabriel K., Purcell A. W., Jacobs-Wagner C., and Lithgow T. (2010) A modular BAM complex in the outer membrane of the α-proteobacterium Caulobacter crescentus. PLoS One 5, e8619 10.1371/journal.pone.0008619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Connell T. D., Black W. J., Kawula T. H., Barritt D. S., Dempsey J. A., Kverneland K. Jr, Stephenson A., Schepart B. S., Murphy G. L., and Cannon J. G. (1988) Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol. Microbiol. 2, 227–236 10.1111/j.1365-2958.1988.tb00024.x [DOI] [PubMed] [Google Scholar]
- 62. Unemo M. G., D; Grad Y., Limnios A; Wi T., Lahra M., and Harris S. (2015) Phenotypic, genetic and genomic characterisation of the WHO Neisseria gonorrhoeae reference strains for quality assurance of laboratory investigations globally. Sex. Transm. Infect. 91, Suppl. 2, A111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Casadaban M. J., and Cohen S. N. (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138, 179–207 10.1016/0022-2836(80)90283-1 [DOI] [PubMed] [Google Scholar]
- 64. Spence J. M., Wright L., and Clark V. L. (2008) Laboratory maintenance of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. Chapter 4, Unit 4A.1 10.1002/9780471729259.mc04a01s8 [DOI] [PubMed] [Google Scholar]
- 65. Mehr I. J., Long C. D., Serkin C. D., and Seifert H. S. (2000) A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ménard R., Sansonetti P. J., and Parsot C. (1993) Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175, 5899–5906 10.1128/jb.175.18.5899-5906.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bauer A. W., Kirby W. M., Sherris J. C., and Turck M. (1966) Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496 [PubMed] [Google Scholar]
- 68. Roier S., Zingl F. G., Cakar F., and Schild S. (2016) Bacterial outer membrane vesicle biogenesis: a new mechanism and its implications. Microb. Cell 3, 257–259 10.15698/mic2016.06.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 70. Winter G. (2010) xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 10.1107/S0021889809045701 [DOI] [Google Scholar]
- 71. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ramachandran S., Kota P., Ding F., and Dokholyan N. V. (2011) Automated minimization of steric clashes in protein structures. Proteins 79, 261–270 10.1002/prot.22879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Park B. R., Zielke R. A., Wierzbicki I. H., Mitchell K. C., Withey J. H., and Sikora A. E. (2015) A metalloprotease secreted by the type II secretion system links Vibrio cholerae with collagen. J. Bacteriol. 197, 1051–1064 10.1128/JB.02329-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jolley K. A., and Maiden M. C. (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.