Abstract
Triple-negative breast cancer (TNBC) comprises ∼20% of all breast cancers and is the most aggressive mammary cancer subtype. Devoid of the estrogen and progesterone receptors, along with the receptor tyrosine kinase ERB2 (HER2), that define most mammary cancers, there are no targeted therapies for patients with TNBC. This, combined with a high metastatic rate and a lower 5-year survival rate than for other breast cancer phenotypes, means there is significant unmet need for new therapeutic strategies. Herein, the anti-neoplastic effects of the electrophilic fatty acid nitroalkene derivative, 10-nitro-octadec-9-enoic acid (nitro-oleic acid, NO2-OA), were investigated in multiple preclinical models of TNBC. NO2-OA reduced TNBC cell growth and viability in vitro, attenuated TNFα-induced TNBC cell migration and invasion, and inhibited the tumor growth of MDA-MB-231 TNBC cell xenografts in the mammary fat pads of female nude mice. The up-regulation of these aggressive tumor cell growth, migration, and invasion phenotypes is mediated in part by the constitutive activation of pro-inflammatory nuclear factor κB (NF-κB) signaling in TNBC. NO2-OA inhibited TNFα-induced NF-κB transcriptional activity in human TNBC cells and suppressed downstream NF-κB target gene expression, including the metastasis-related proteins intercellular adhesion molecule-1 and urokinase-type plasminogen activator. The mechanisms accounting for NF-κB signaling inhibition by NO2-OA in TNBC cells were multifaceted, as NO2-OA (a) inhibited the inhibitor of NF-κB subunit kinase β phosphorylation and downstream inhibitor of NF-κB degradation, (b) alkylated the NF-κB RelA protein to prevent DNA binding, and (c) promoted RelA polyubiquitination and proteasomal degradation. Comparisons with non-tumorigenic human breast epithelial MCF-10A and MCF7 cells revealed that NO2-OA more selectively inhibited TNBC function. This was attributed to more facile mechanisms for maintaining redox homeostasis in normal breast epithelium, including a more favorable thiol/disulfide balance, greater extents of multidrug resistance protein-1 (MRP1) expression, and greater MRP1-mediated efflux of NO2-OA–glutathione conjugates. These observations reveal that electrophilic fatty acid nitroalkenes react with more alkylation-sensitive targets in TNBC cells to inhibit growth and viability.
Keywords: breast cancer, cancer chemoprevention, drug action, NF-kappaB, proliferation, reactive nitrogen species (RNS), reactive oxygen species (ROS), signaling, tumor immunology, tumor necrosis factor (TNF)
Introduction
Triple-negative breast cancer (TNBC)4 is characterized by an absence of estrogen receptor (ER), progesterone receptor, and human epidermal growth factor receptor-2 expression (2, 3). TNBC accounts for up to 20% of breast cancer incidence and is the subtype with the worst prognosis (4). The majority of TNBC tumors are “basal-like”, with 5-year survival rates lower than for all other breast cancer phenotypes (∼77% versus ∼93%, respectively) (3). TNBC patients are also at greater risk for relapse during the first 5 years post-chemotherapy. The recurrent tumors are more aggressive and invasive (5, 6), resulting in a life expectancy of 3–22 months after reappearance (7, 8). Consequently, there is an urgent unmet need for new therapeutic strategies for TNBC, beyond the limited options of standard chemotherapy, ionizing radiation, and surgery.
Activation of nuclear factor-κB (NF-κB) is strongly linked with TNBC development and progression (9–11), with NF-κB signaling constitutively activated in ER-negative breast cancer cell lines and primary tumors (10–13). The inhibition of NF-κB activation, induced by overexpression of the non-degradable inhibitor of NF-κB (IκBα) superrepressor (Ser-32/36 mutations of IκBα), significantly inhibits the growth of several TNBC cell lines (13). The pro-inflammatory cytokine TNFα also contributes significantly to this complex inflammatory microenvironment that promotes tumor progression. TNFα activates tumor metastasis and invasion through NF-κB–mediated up-regulation of extracellular matrix degradation enzymes and adhesion molecule expression (14). Notably, a meta-analysis revealed that TNBC patients with elevated TNFα expression have an increased risk of tumor metastasis to distant organs (15). Thus, NF-κB activation and the downstream signaling actions of its pro-inflammatory mediators play a critical role in TNBC malignancy. This motivates the development of novel NF-κB inhibition strategies as a chemotherapeutic approach for countering metastatic TNBC.
Electrophilic fatty acid nitroalkene derivatives (NO2-FA) are endogenously formed by the acidic conditions of digestion and the complex redox milieu that is up-regulated during inflammation. These environments facilitate the reaction of the nitric oxide (•NO) and nitrite (NO2−)-derived nitrating species nitrogen dioxide (•NO2) (16) with biological targets, such as unsaturated fatty acids. Basal plasma and urinary NO2-FA concentrations in healthy humans range from 2 to 20 nm, with additional pools of NO2-FA present as (a) Michael addition products with the abundant biological nucleophiles present in tissues and fluids and (b) esterified species in complex neutral and polar lipids (17, 18). Tissue NO2-FA levels are affected by both dietary lipid and nitrogen oxide concentrations and during metabolic stress can rise to concentrations as high as 1 μm (19, 20).
The unique electrophilic character of fatty acid nitroalkene substituents promotes kinetically rapid and reversible Michael addition with nucleophilic Cys and, to a lesser extent, His residues of proteins (21, 22). This reversible protein adduction by fatty acid nitroalkenes decreases the potential for toxicity stemming from the accumulation of Schiff's base and Michael addition products characteristic of other lipid electrophiles, such as α,β-unsaturated oxo (or keto) and cyclopentanone derivatives (21, 23, 24). Through transient post-translational modification (PTM) reactions with hyperreactive protein thiols, NO2-FA modulate signaling pathways involved in cell proliferation and inflammatory responses. This occurs as a result of the alkylation of functionally significant Cys residues in transcriptional regulatory proteins, including the Kelch-like ECH-associated protein-1 (Keap1) regulator of nuclear factor (erythroid-derived-2)-like 2 (Nrf2) signaling, the nuclear lipid receptor peroxisome proliferator–activated receptor γ (PPARγ), and NF-κB (25–27). Of relevance to the present study, NO2-FA inhibit NF-κB–mediated signaling in diverse cell and murine models of metabolic and inflammatory stress to cardiovascular, pulmonary, and renal systems (27–29).
NO2-FA specifically alkylate Cys-38 of the RelA subunit of NF-κB, a functionally significant, lipid electrophile-reactive thiol located in the DNA-binding domain of RelA. Redox-dependent PTMs of RelA Cys-38 inhibit DNA binding and downstream pro-inflammatory mediator gene expression (27). Current data indicate that other electrophilic species, such as the isothiocyanate derivative sulforaphane, mediate therapeutic actions in preclinical models of breast cancer (30, 31), thus motivating the present studies. Moreover, the pleiotropic signaling actions of NO2-FA include the activation of angiogenesis via up-regulation of HIF-1α signaling during hypoxia (32). Because these effects may potentially modulate cancer cell and tumor properties, it was important to test the impact of an electrophilic NO2-FA in both in vitro and in vivo models of an aggressive cancer phenotype, TNBC.
This study reports the inhibition of TNBC (MDA-MB-231 and MDA-MB468) cell proliferation, invasion, and metastasis by a synthetic homolog of an endogenous electrophilic NO2-FA found in species ranging from plants to humans (10-nitro-octadec-9-enoic acid, termed nitro-oleic acid and NO2-OA). NO2-OA displayed lower cytotoxic and anti-proliferative effects on non-tumorigenic breast ductal epithelium (MCF-10A and MCF7) versus triple-negative human breast ductal epithelial cells, due to the more stable mechanisms for maintaining redox homeostasis in MCF-10A and MCF7 cells. NO2-OA also attenuated TNFα-induced TNBC cell migration and invasion via inhibition of NF-κB signaling. Two newly discovered mechanisms also accounted for NO2-OA inhibition of TNBC NF-κB transcriptional activity. First, NO2-OA alkylated the inhibitor of NF-κB subunit kinase β (IKKβ), leading to inhibition of its kinase activity and downstream IκBα phosphorylation. Second, NO2-OA alkylated NF-κB RelA protein, a reaction that not only inhibited DNA binding, but also promoted proteasomal RelA degradation. As a consequence, NO2-OA inhibited the expression of two NF-κB–regulated, TNFα-induced genes that are central to tumor metastasis, intercellular adhesion molecule-1 (ICAM-1) and urokinase-type plasminogen activator (uPA). Finally, in a nude mouse xenograft model, NO2-OA reduced the growth of established MDA-MB-231 tumors. In aggregate, these findings reveal that electrophilic NO2-FA can mediate chemotherapeutic actions in treating TNBC and possibly other inflammation-related cancers.
Results
NO2-OA inhibits TNBC cell growth and viability
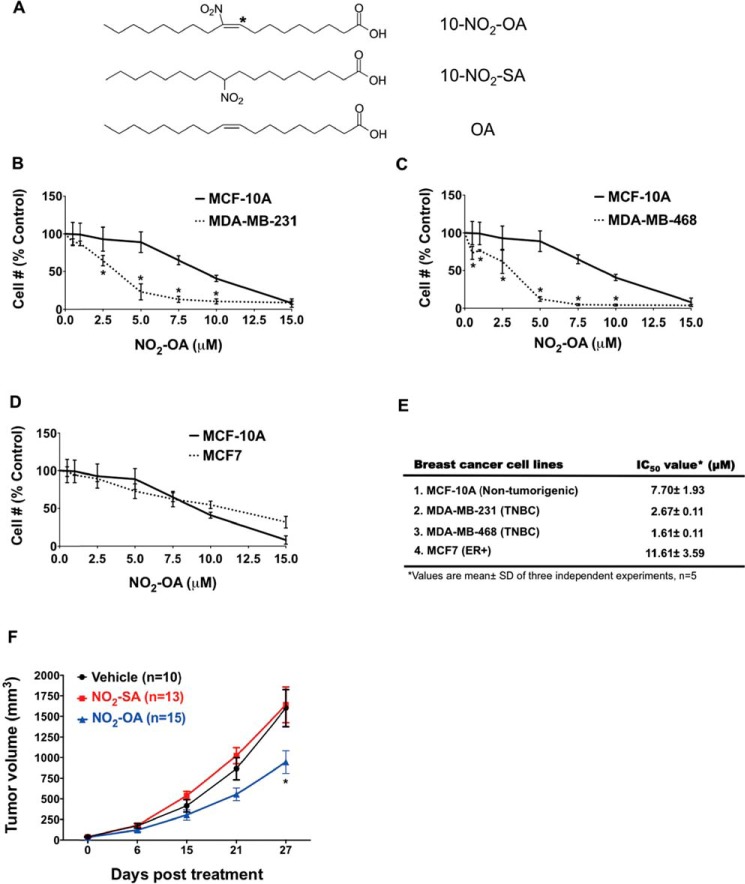
The endogenously occurring lipid electrophile NO2-OA and its non-electrophilic control fatty acids (NO2-SA and OA) were evaluated for their impact on normal and cancerous breast ductal epithelial cell growth and signaling responses (Fig. 1A). To examine whether NO2-OA preferentially inhibited TNBC cell growth, Hoechst 33258 was used for counting non-tumorigenic breast epithelial cells (MCF-10A), an ER+ breast cancer cell line (MCF7), and two TNBC cell lines (MDA-MB-231 and MDA-MB-468). Each cell line was treated with a range of NO2-OA concentrations (0–15 μm) for 48 h. NO2-OA significantly inhibited the growth of both TNBC cell lines but not ER+ or MCF-10A cells (Fig. 1, B, C, and D). The IC50 for NO2-OA was significantly greater for non-cancerous MCF-10A cells (7.7 ± 1.93 μm) and MCF7 (11.61 ± 3.59 μm), as opposed to TNBC MDA-MB-231 (2.7 ± 0.11 μm) and MDA-MB-468 (1.6 ± 0.11 μm) cells (Fig. 1E). In addition to preferential TNBC cell growth inhibition, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) detection of intact cell electron transfer mechanisms revealed that NO2-OA also significantly reduced the viability of both MDA-MB-231 and MDA-MB-468 cells, but not MCF7 or MCF-10A cells (Fig. S1, A–C). No cytotoxicity was detectable in any cell line for up to 24 h at the 5 μm NO2-OA concentrations typically used for subsequent cell signaling and functional studies that had durations ranging from 1 to 8 h. Non-electrophilic NO2-SA, structurally related to NO2-OA (Fig. 1A), did not affect TNBC cell growth (Fig. S2), affirming that NO2-OA–mediated TNBC cell growth inhibition is attributable to the electrophilic nitroalkene moiety.
Figure 1.
NO2-OA inhibits TNBC cell growth in vitro and in vivo. A, chemical structures of NO2-OA and the non-electrophilic NO2-SA and OA. *, electrophilic carbon (35). The effect of NO2-OA on the growth of MDA-MB-231 (B), MDA-MB-468 (C), and MCF7 (D) was compared with the effect on MCF-10A cells. Data are shown as a percentage of untreated control cells (mean ± S.D. (error bars)). *, p < 0.05 indicates significant difference between two cell types within each treatment. Three independent experiments were performed (n = 5 each). E, IC50 values of NO2-OA in each breast cancer cell line. F, effect of NO2-OA (7.5 mg/kg daily) on MDA-MB-231 xenograft tumor growth (mean ± S.E. (error bars)). *, p < 0.05 versus vehicle group within treatment time. Significance was determined by two-way analysis of variance followed by Tukey's post hoc test.
NO2-OA reduces MDA-MB-231 xenograft tumor growth
Given that TNBC cell growth and viability are inhibited by NO2-OA, the efficacy of NO2-OA on tumor growth was examined in a murine xenograft model of TNBC. MDA-MB-231 cells were injected into the fourth inguinal mammary fat pad of 6-week-old female athymic nude mice. Oral gavage with NO2-OA (7.5 mg/kg/day), NO2-SA (7.5 mg/kg/day), or sesame oil (vehicle control) was initiated and continued for 4 weeks after the average tumor sizes reached between 50 and 100 mm3. There was significantly reduced tumor growth in the mice treated with NO2-OA versus vehicle controls and NO2-SA–treated mice at 27 days post-treatment (Fig. 1F). During the course of treatment, there was no weight loss in NO2-OA-treated or control mice (Fig. S3). These results indicate that NO2-OA mediates in vivo growth suppression of MDA-MB-231 cells with no overt toxic effects.
NO2-OA induces cell cycle arrest and apoptotic cell death in TNBC cells
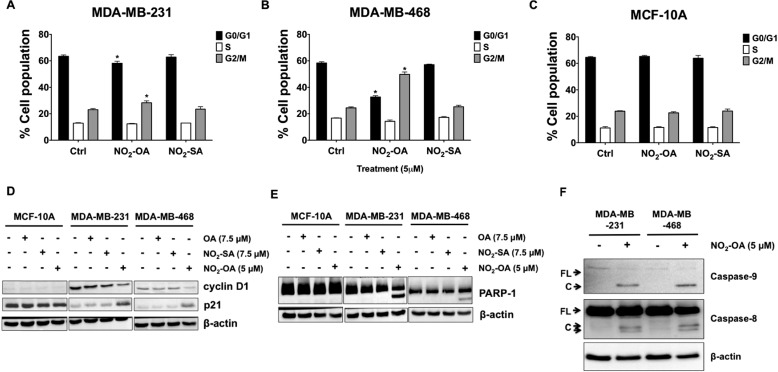
To determine whether the decreased cell numbers were due to NO2-OA–induced cell cycle alterations, FACS analysis was performed. NO2-OA significantly increased the percentage of cells at G2/M phase and decreased the percentage of cells in G0/G1 upon 24-h treatment in MDA-MB-231 and MDA-MB-468 cells (Fig. 2, A and B). Notably, all cell cycle phase populations (G0/G1, S, and G2/M) of MCF-10A cells were not affected by NO2-OA (Fig. 2C). The cell cycle inhibition by NO2-OA was accompanied by an increase in p21 and a decrease in cyclin D1 protein expression in both MDA-MB-231 and MDA-MB-468 cells, but not MCF-10A cells (Fig. 2D). Consistent with the lack of an effect on cell growth and viability (Fig. S2), NO2-SA did not affect cell cycle populations or the expression of cell cycle–regulatory proteins in MCF-10A, MDA-MB-231, and MDA-MB-468 cells (Fig. 2D). The gene expression of cyclin D1 and p21 was also determined by quantitative RT-PCR. NO2-OA down-regulated cyclin D1 and up-regulated p21 gene expression after 24 h treatment of MDA-MB-231 and MDA-MB-468 cells, but not MCF-10A cells (Fig. S4). These results indicate that NO2-OA selectively induced cell cycle arrest in TNBC cells.
Figure 2.
NO2-OA promotes cell cycle arrest and apoptosis in TNBC cells. Percentages of the cell population in each phase of the cell cycle (G0/G1, S, and G2/M) are shown for MDA-MB-231 (A), MDA-MB-468 (B), and MCF-10A cells (C) treated with NO2-OA (5 μm) for 24 h. Cells were harvested and analyzed by fluorescence-activated cell sorting. Significance was determined by one-way analysis of variance followed by Tukey post hoc test. Data are mean ± S.D. (error bars) (n = 3). *, p < 0.05 versus control. D, immunoblot analysis of cyclin D1 and p21 in MCF-10A, MDA-MB-231, and MDA-MB-468 cells that were treated with OA (7.5 μm), NO2-SA (7.5 μm), or NO2-OA (5 μm) for 24 h. E, immunoblot analysis of PARP-1 cleavage in MCF-10A, MDA-MB-231, and MDA-MB-468 cells treated with OA (7.5 μm), NO2-SA (7.5 μm), or NO2-OA (5 μm) for 24 h. F, immunoblot analysis of caspase-8 and caspase-9 cleavage in MDA-MB-231 and MDA-MB-468 cells treated with or without NO2-OA (5 μm) for 24 h. β-actin was used as loading control. Data in D–F are representative of three independent experiments.
Increased sub-G1 cell populations were apparent in both MDA-MB-231 and MDA-MB-468 cells 24 h after NO2-OA treatment (Fig. S5). To determine whether the effect of NO2-OA on sub-G1 cells in TNBC cells was apoptosis-mediated, cleavage of poly(ADP-ribose) polymerase-1 (PARP-1) was examined by Western blotting. Treatment with NO2-OA for 24 h promoted caspase-3–mediated cleavage of PARP-1 (Fig. 2E) in MDA-MB-231 and MDA-MB-468 cells, but not in MCF-10A cells, indicating that NO2-OA preferentially induced TNBC apoptosis through caspase-3 activation. Also, it is possible that the increase in p21 blocks cell cycle entry into the S phase, resulting in the increase in sub-G1 cells. To further investigate apoptotic signaling responses to NO2-OA in TNBC cells, the activation of initiator caspases (caspase-8 for the extrinsic pathway and caspase-9 for the intrinsic pathway) was analyzed using antibodies that detect both the pro-caspase and activated (cleaved) forms of these initiator caspases. NO2-OA treatment increased cleavage of caspase-8 and caspase-9 in both MDA-MB-231 and MDA-MB-468 cells, suggesting that NO2-OA induced apoptosis through both intrinsic (mitochondria-dependent) and extrinsic (death receptor-dependent) apoptotic signaling mechanisms in TNBC cells (Fig. 2F). In aggregate, these results confirm that NO2-OA selectively modulates cell cycle arrest and apoptosis in TNBC cells versus MCF-10A cells.
Extracellular NO2-OA–glutathione adduct efflux is linked with multidrug resistance protein-1 (MRP1) expression
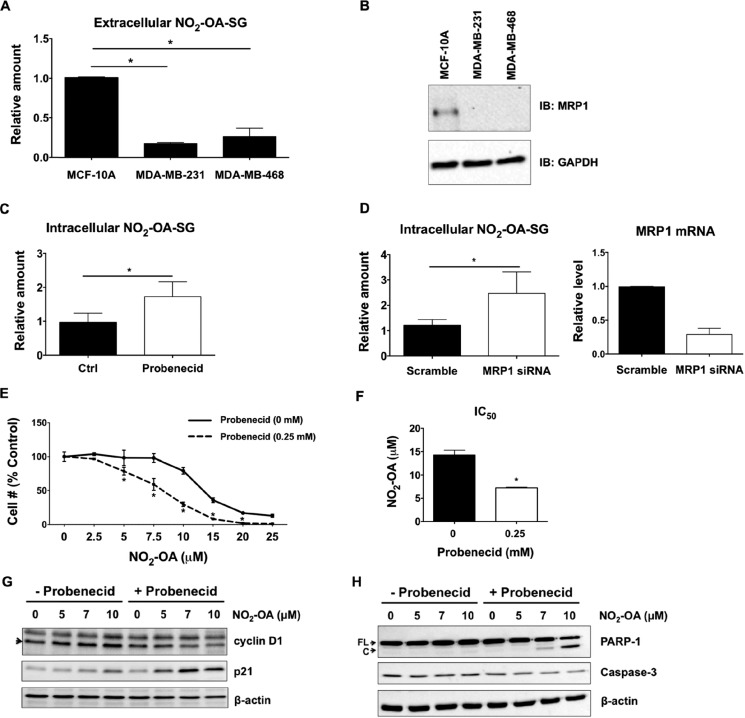
In the intracellular compartment, GSH and its reactive Cys moiety is more abundant than protein thiols; thus, GSH and other low-molecular weight thiols are the primary targets for oxidation and alkylation by free radicals, oxidants, and electrophiles (33). In the case of NO2-OA, which readily diffuses and gains access to the intracellular compartment and subcellular organelle protein targets (26, 34), GSH conjugates (NO2-OA-SG) are formed that can be actively transported from cells by the GSH-conjugate efflux pump MRP1 (1). This phenomenon was further investigated by measuring concentrations of extracellular NO2-OA-SG in the media of MCF-10A, MDA-MB-231, and MDA-MB-468 cells after a 1-h treatment with 5 μm NO2-OA. There were significantly lower levels of NO2-OA-SG being exported into the media of both MDA-MB-231 and MDA-MB-468 cells, as opposed to that released by MCF-10A cells (Fig. 3A). This 4–5-fold difference in extracellular NO2-OA-SG levels produced by MCF-10A and TNBC cells prompted comparison of the relative extents of expression of MRP1 protein and the GSH and GSSG content of TNBC and non-cancerous cell lines. Western blot analysis detected MRP1 protein expression in MCF-10A cells, but MRP1 was undetectable in both TNBC cell lines (Fig. 3B). MRP4 mRNA was detected at low levels in all three cell types, but protein expression was not evident by Western blotting (not shown).
Figure 3.
MRP1 influences NO2-OA trafficking and signaling in TNBC cells. A, the export of NO2-OA-SG by MCF-10A, MDA-MB-231, and MDA-MB-468 cells was measured by LC-MS/MS analysis. The relative extent of NO2-OA-SG export is reported as a ratio of NO2-OA-SG to an externally added 15NO2-d4-OA-SG standard. *, p < 0.05 versus MCF-10A, n = 4 (Mann–Whitney U test). B, representative immunoblot of endogenous MRP1 protein expression in MCF-10A, MDA-MB-231, and MDA-MB-468 cells. Suppression of MRP1 activity (C) and MRP1 expression (D) increased intracellular NO2-OA-SG adduct concentrations in MCF-10A cells. The relative amount represents the relative abundance of NO2-OA-SG to 15NO2-d4-OA-SG standard, normalized to protein concentrations from each NO2-OA–treated sample divided by the abundance of control (Ctrl) or scrambled sample. *, p < 0.05 versus control (n = 6) or scrambled (n = 9) was determined by Mann–Whitney U test. The siRNA knockdown efficiency of MRP1 was evaluated by real-time qPCR (n = 4). E, the effect of probenecid on NO2-OA growth inhibition of MCF-10A cells. Cells were pretreated with or without probenecid (0.25 mm) for 1 h and then combined with 0–25 μm NO2-OA for 48 h. A FluoReporter dsDNA stain assay was performed to measure cell numbers. Data are shown as percentage of untreated control cells (n = 3); *, p < 0.05 (0 mm versus 0.25 mm probenecid between treatments, two-way analysis of variance followed by Tukey post test). F, the average IC50 values of NO2-OA in MCF-10A cells treated with or without probenecid. *, p < 0.05, n = 3 (unpaired Student's t test). G, immunoblot analysis of cyclin D1 and p21 in MCF-10A cells treated with NO2-OA (5 μm) in the presence or absence of probenecid (1 mm used for this 24-h incubation). H, immunoblot analysis of caspase-3 and PARP-1 cleavage in MCF-10A cells treated with NO2-OA (5 μm) in the presence or absence of probenecid (1 mm) for 24 h. The full-length (FL) and cleaved (C) forms of PARP-1 and pro-caspase-3 protein level are shown. All data are mean ± S.D. (error bars). All immunoblots are representative of three independent experiments.
MRP1 influences NO2-OA bioactivity in MCF-10A cells
Two strategies, use of the organic anion transport inhibitor probenecid, often used as an MRP inhibitor, and siRNA knockdown of MRP1, facilitated investigation of the role of MRP1 in cellular responses to NO2-OA. Both probenecid and MRP1 siRNA knockdown (about 70% knockdown efficiency) enhanced intracellular levels of NO2-OA-SG adducts in MCF-10A cells (Fig. 3, C and D). Notably, probenecid also significantly enhanced MCF-10A cell growth inhibition by NO2-OA (Fig. 3E). The IC50 of NO2-OA (7.23 ± 0.15 μm) was decreased 2-fold in MCF-10A cells pretreated with probenecid, compared with only NO2-OA treatment (14.23 ± 1.05 μm; Fig. 3F). Moreover, probenecid increased the extent of NO2-OA–induced cell cycle arrest of MCF-10A cells, as reflected by increased p21 levels and a concomitant decrease in cyclin D1 expression (Fig. 3G). Probenecid also enhanced NO2-OA–induced apoptosis in MCF-10A cells in the context of increased caspase-3 activation and PARP-1 cleavage (Fig. 3H). These observations are consistent with both the intracellular concentrations and the cell growth/cell survival signaling actions of NO2-OA being influenced by the extents of NO2-OA reaction with GSH and subsequent MRP1 export of NO2-OA-SG.
GSH and GSSG responses to NO2-OA in MCF-10A cells versus TNBC cells
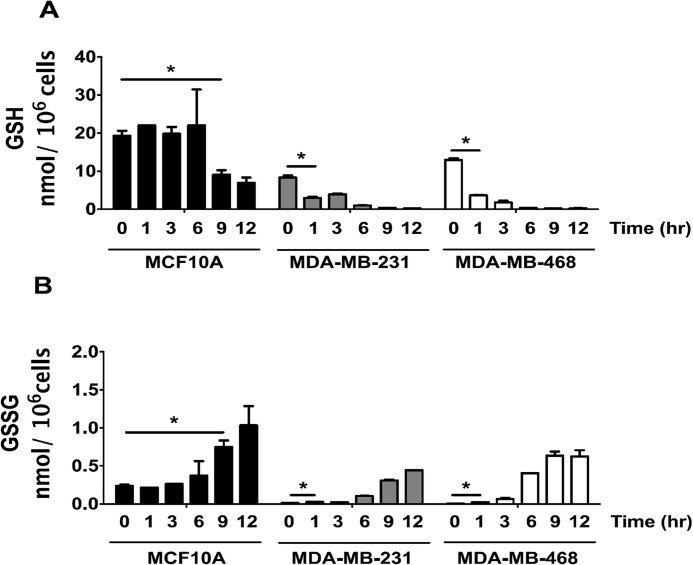
LC-MS quantitation of GSH and GSSG from 0 to 12 h after treatment with 5 μm NO2-OA revealed that basal GSH levels in MCF-10A cells (19.3 ± 1.9 nmol/106 cells) were >2-fold that of MDA-MB-231 (8.3 ± 0.8 nmol/106 cells) and ∼1.5-fold greater than MDA-MB-468 cells (12.9 ± 0.5 nmol/106 cells) (Fig. 4A). GSSG levels (Fig. 4B) at time 0 were greater in MCF-10A cells, resulting in an initial GSH/GSSG ratio of 82 ± 16 compared with 653 ± 68 for MDA-MB-231 cells and 2003 ± 163 in MDA-MB-468 cells. MCF-10A cells maintained the GSH/GSSG ratio over the first 6 h after NO2-OA treatment, whereas the GSH/GSSG ratio rapidly decreased in MDA-MB-231 and MDA-MB-468 cells due to decreased GSH concentrations. In aggregate, the data in Figs. 3 and 4 indicate that there will be a more extensive reaction expected between NO2-OA and cellular protein targets in TNBC cells because of the more favorable pharmacokinetics (greater intracellular concentration and longer t0.5) lent by the lower GSH concentrations and the suppression of NO2-OA-SG export by the MRP1-deficient TNBC cell phenotype. In MCF-10A cells, NO2-OA will be more readily glutathionylated and exported, thus limiting reactions with signaling pathway proteins.
Figure 4.
NO2-OA depletes GSH levels and enhances GSSG formation in TNBC cells. The response of cellular GSH (A) and GSSG (B) to NO2-OA in MCF-10A (black bars), MDA-MB-231 (gray bars), and MDA-MB-468 (white bars) cells is shown. Cells were treated with NO2-OA (5 μm) for the indicated times (h). GSH and GSSG were extracted from cells (3 × 106 cells/ml) and quantitated by LC-MS/MS. *, p < 0.05 versus 0 h via unpaired two-tailed Student's t test. Data are presented as mean ± S.D. (error bars) (n = 5).
NO2-OA inhibits TNFα-induced TNBC cell migration and invasion
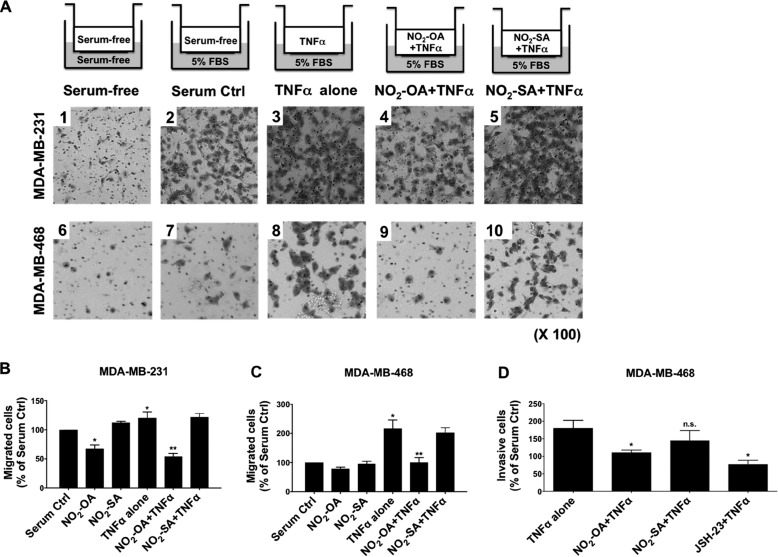
Inflammatory stimuli, such as TNFα, induce responses in the tumor microenvironment that promote TNBC tumor metastasis and invasion (14). Because electrophilic NO2-FAs mediate anti-inflammatory signaling actions (27, 28), the impact of NO2-OA on TNFα-induced TNBC cell migration was evaluated. Boyden chamber migration analyses indicated that TNFα augmented migration of both MDA-MB-231 and MDA-MB-468 cells (Fig. 5A, images 3 and 8), compared with basal conditions (Fig. 5A, images 2 and 7). NO2-OA significantly inhibited both MDA-MB-231 and MDA-MB-468 cell migration induced by TNFα (Fig. 5, A (images 4 and 9), B, and C). NO2-OA modestly inhibited the basal, non-stimulated migration of MDA-MB-231 and MDA-MB-468 cells (Fig. 5, B and C). Next, cells were placed in Transwell permeable supports coated with Matrigel for invasion assays to assess the potential effect of NO2-OA on the invasive phenotype of TNBC cells. TNFα-induced invasion was significantly inhibited by NO2-OA treatment of MDA-MB-468 cells, whereas the non-electrophilic control fatty acid (NO2-SA) displayed no significant effect on tumor cell invasion (Fig. 5D). The inhibitory actions of NO2-OA on MDA-MB-468 invasion were compared with cell responses to the NF-κB inhibitor JSH-23, which inhibits nuclear translocation of the RelA subunit (36). Similar to JSH-23, NO2-OA inhibited TNFα-induced invasion in MDA-MB-468 cells (Fig. 5D).
Figure 5.
NO2-OA inhibits TNFα-induced TNBC cell migration and invasion. A, experimental schemes and representative images of crystal violet-stained migrating MDA-MB-231 or MDA-MB-468 cells. Cells (1 × 105) were placed in the upper chamber with serum-free medium under the indicated treatment conditions. Migrating cells were photographed using a light microscope at ×100. B and C, quantitation of migrated cells from Fig. 4A was performed by solubilization of crystal violet and spectrophotometric analysis at A573 nm. The percentage of migrating cells in each treatment group was compared with numbers of migrating cells in the absence of TNFα stimulation (Serum Ctrl). *, p < 0.05 versus in the absence of TNFα stimulation; **, p < 0.05 versus TNFα alone. D, to test the impact of NO2-OA on TNBC cell invasion, MDA-MB-468 cells were incubated in serum-free medium containing 20 ng/ml TNFα combined with NO2-OA (5 μm), NO2-SA (5 μm), or JSH-23 (10 μm), and then invasion was determined by the extents of cell migration through the Matrigel matrix toward a 5% FBS chemoattractant for 24 h. The percentage of invading cells in each treatment was relative to the number of migrating cells in the absence of TNFα stimulation. *, p < 0.05 versus TNFα alone n.s., not significant. Significance was determined by one-way analysis of variance followed by Tukey post hoc test. All data are mean ± S.D. (error bars).
NO2-OA inhibits TNFα-induced NF-κB transcriptional activity in TNBC cells
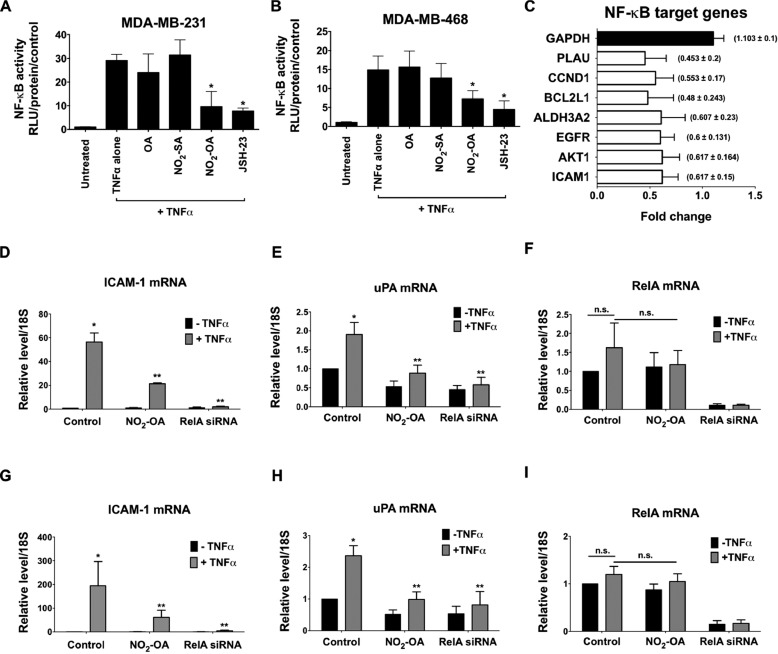
The inhibition of MDA-MB-468 cell invasion by JSH-23 (Fig. 5D) suggests that NO2-OA may also inhibit TNFα-induced breast cancer cell mobility due to a capacity to inhibit NF-κB signaling. To test this concept, the effect of NO2-OA on TNFα-activated NF-κB transcriptional activity in TNBC cells was examined. MDA-MB-231 and MDA-MB-468 cells were transiently transfected with an NF-κB luciferase reporter plasmid and treated with 5 μm NO2-OA for 2 h, followed by activation with 20 ng/ml TNFα for 4 h. In addition to NO2-OA, the non-electrophilic lipid controls NO2-SA (5 μm) and OA (5 μm) were also examined. NO2-OA significantly inhibited NF-κB–dependent transcription of luciferase in both TNBC cell lines, compared with TNFα alone, whereas NO2-SA and OA had no effect. Moreover, the extent of inhibition of NF-κB–dependent luciferase expression by NO2-OA was similar to that induced by the NF-κB inhibitor JSH-23 (20 μm; Fig. 6, A and B). These data indicate that the electrophilic reactivity of NO2-OA accounts for the inhibition of TNFα-induced NF-κB transcriptional activity in TNBC cells.
Figure 6.
NO2-OA inhibits TNFα-induced NF-κB transcriptional activity in TNBC cells. The effect of NO2-OA on TNFα-induced activation of NF-κB–dependent reporter gene transcription was measured in NF-κB-luciferase reporter–transfected MDA-MB-231 (A) or MDA-MB-468 (B) cells. *, p < 0.05 versus TNFα alone (n = 3). Significance was determined by Kruskal–Wallis test followed by Dunn's post test with Bonferroni corrections for multiple comparisons. C, determination of NF-κB target genes down-regulated by NO2-OA in MDA-MB-468 cells using a human NF-κB target PCR array. Histograms represent the fraction of mRNA expression in NO2-OA–treated versus untreated cells. GAPDH was used as an internal control (black bar). Shown is the effect of NO2-OA on expression of ICAM-1 (D), uPA (E), or RelA (F) genes in TNFα-induced MDA-MB-231 cells. Similarly, the effect of NO2-OA on expression of ICAM-1 (G), uPA (H), or RelA (I) genes in TNFα-induced MDA-MB-468 cells is shown. The -fold increase relative to untreated controls is presented. *, p < 0.05 versus untreated control; **, p < 0.05 versus TNFα alone. n.s., not significant. Significance was determined by one-way analysis of variance followed by Tukey post test. All data are presented as mean ± S.D. (error bars) (n = 5).
NO2-OA inhibits NF-κB–regulated gene expression linked with TNBC tumor metastasis
Inhibition of NF-κB transcriptional activity by NO2-OA suggested that the expression of metastasis-related downstream target genes may be decreased. To investigate this, key NF-κB target genes were evaluated via RT2 profiler PCR array analysis of MDA-MB-468 cells treated with NO2-OA (5 μm) for 24 h. The expression levels of NF-κB target genes that were regulated by NO2-OA were compared with untreated MDA-MB-468 cells as a control. Data revealed that treatment with NO2-OA decreased the mRNA expression of multiple NF-κB target genes, including ICAM-1 and uPA, two critical mediators of tumor progression and metastasis (Fig. 6C). TNFα induces the expression of both ICAM-1 and uPA in MDA-MB-231 cells (37, 38). To more directly examine whether NO2-OA suppressed TNFα-induced expression of ICAM-1 and uPA in TNBC cells, MDA-MD-231 or MDA-MD-468 cells were treated with 5 μm NO2-OA and 20 ng/ml TNFα. Simultaneous treatment with either NO2-OA or RelA siRNA led to suppression of TNFα-induced expression of ICAM-1 and uPA genes in TNBC cells (Fig. 6, D, E, G, and H). The impact of NO2-OA and RelA siRNA on RelA-dependent target gene expression was further evaluated by real-time qPCR (Fig. 6, F and I). RelA mRNA levels were suppressed by RelA siRNA treatment, but not NO2-OA. Both NO2-OA and RelA siRNA inhibited gene expression of TNFα-induced ICAM-1 and uPA gene expression via NF-κB–dependent mechanisms. To determine whether NO2-OA suppressed TNFα-induced pro-metastatic ICAM-1 and uPA gene expression during cell migration, transcript levels of ICAM-1 and uPA genes were evaluated in MDA-MB-468 cells being studied in Boyden chamber migration assays (Fig. 5C). Under these conditions, NO2-OA significantly inhibited TNFα-induced expression of ICAM-1 and uPA in migrating tumor cells (Fig. S6, A and B), again indicating that NO2-OA inhibited expression of NF-κB–regulated genes involved in metastasis.
NO2-OA suppresses TNFα-induced IKKβ/IκBα signaling in TNBC
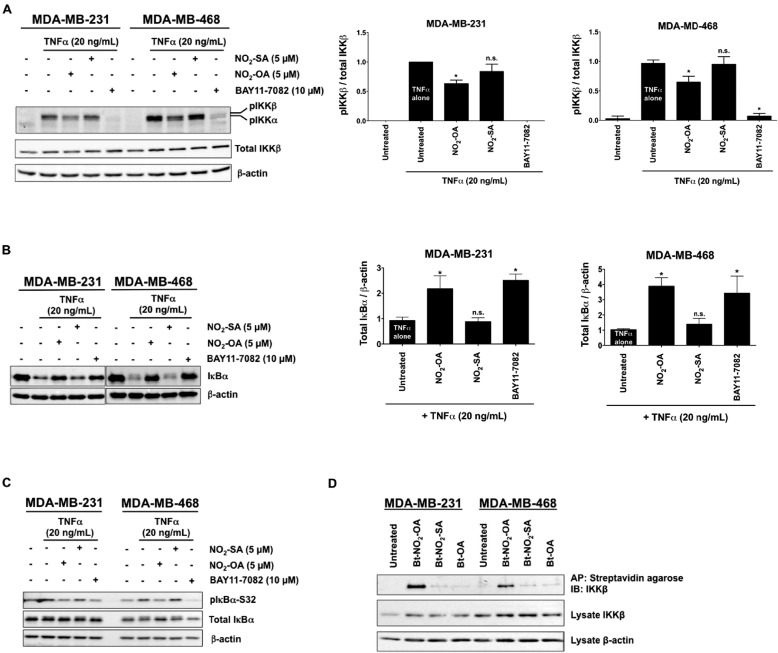
To better define mechanisms accounting for NO2-OA inhibition of TNFα-activated NF-κB signaling, MDA-MB-231 or MDA-MB-468 cells were pretreated with NO2-OA (5 μm) or the IKK inhibitor BAY11-7082 (10 μm) for 2 h before TNFα stimulation (20 ng/ml, 5 min). TNFα-induced IKKβ phosphorylation was diminished by both NO2-OA and BAY11-7082 (Fig. 7A). Both NO2-OA and BAY11-7082 also inhibited the degradation of IκBα following TNFα stimulation (20 ng/ml, 10 min; Fig. 7B). Moreover, decreased IκBα phosphorylation occurred in cells pretreated with NO2-OA or BAY11-7082 and the proteasome inhibitor MG-132 (10 μm; Fig. 7C). This indicates that NO2-OA suppresses TNFα-induced IKKβ phosphorylation and IκBα degradation, with these actions in turn inhibiting downstream NF-κB signaling in TNBC cells.
Figure 7.
NO2-OA inhibits TNFα-induced IKKβ phosphorylation and IκBα degradation and covalently adducts IKKβ. MDA-MB-231 and MDA-MB-468 cells were used in all studies. A, representative immunoblot of IKKβ (Ser-180) phosphorylation, total IKKβ levels, and relative phosphorylated IKKβ levels. Then all phosphorylated IKKβ levels normalized to total IKKβ were quantified. B, representative immunoblot of IκBα protein levels is shown, and the relative total IκBα levels (normalized to total β-actin) are quantified in response to NO2-SA, NO2-OA, and the NF-κB inhibitor BAY11-7082. C, representative immunoblots of IκBα (Ser-32) phosphorylation and total IκBα are shown in response to NO2-SA, NO2-OA, and the NF-κB inhibitor BAY11-7082. D, NO2-OA alkylates TNBC IKKβ protein. Biotinylated NO2-OA, NO2-SA, and OA and adducted proteins were affinity-purified by streptavidin-agarose beads from cell lysates. Pulled-down IKKβ protein was then detected by immunoblotting. IKKβ and control β-actin immunoblots from the same input lysates used for affinity purification are shown below the panel. *, p < 0.05 versus TNFα alone. n.s., not significant. Significance was determined by one-way analysis of variance followed by Tukey post test.
NO2-OA alkylates IKKβ and RelA proteins
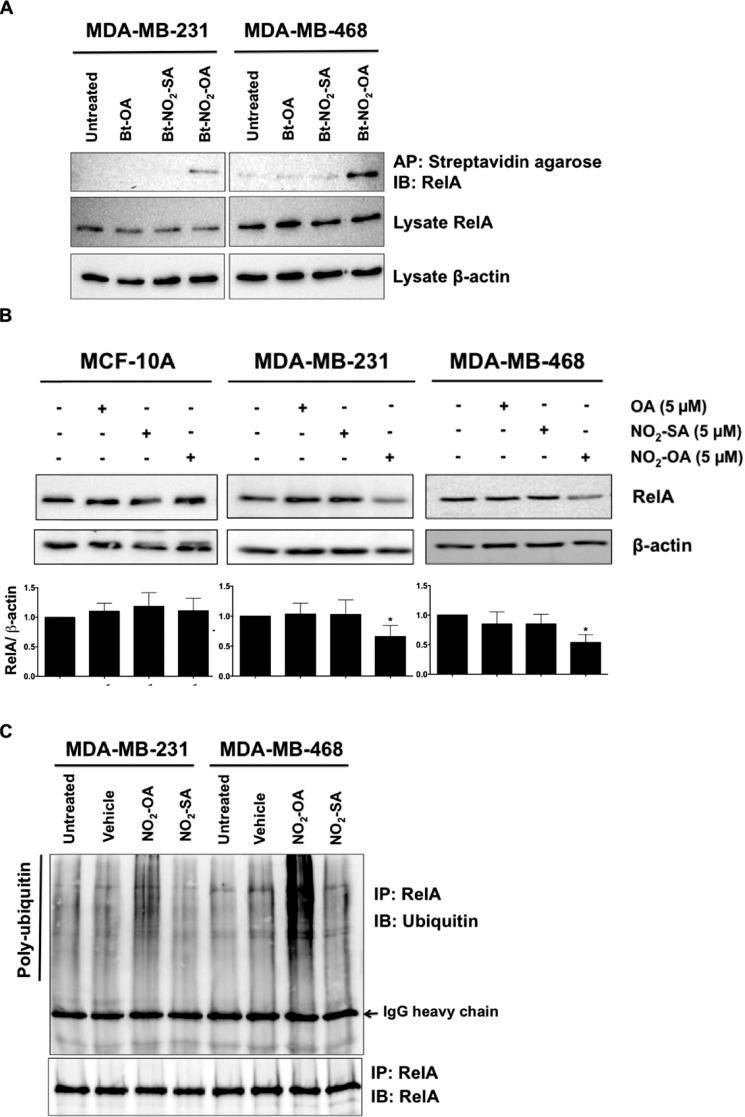
Cys-179, located in the activation loop of IKKβ, is a target for oxidation and electrophile alkylation reactions (39, 40). Because NO2-OA suppresses TNFα-induced phosphorylation of IKKβ and IκBα in TNBC cells (Fig. 7, A and C), the potential for NO2-OA to alkylate IKKβ was investigated. Biotinylated lipids (Bt-NO2-OA, Bt-NO2-SA, and Bt-OA) were synthesized (Fig. S7) to facilitate affinity capture–mediated measurement of NO2-OA and control fatty acid adduction of IKKβ. MDA-MB-231 or MDA-MB-468 cells were treated with 5 μm Bt-NO2-OA, Bt-NO2-SA, or Bt-OA for 2 h, and then all alkylated proteins were pulled down from whole-cell lysates using streptavidin-conjugated beads. Western blotting revealed that IKKβ was pulled down by Bt-NO2-OA, but not by non-electrophilic control fatty acids (Fig. 7D). Similarly, Bt-NO2-OA (but not control fatty acids) promoted the pull-down NF-κB RelA (Fig. 8A).
Figure 8.
NO2-OA alkylates and destabilizes NF-κB RelA protein in TNBC cells. A, MDA-MB-231 or MDA-MB-468 cells were treated with 5 μm Bt-NO2-OA, Bt-NO2-SA, or Bt-OA for 2 h. After cell lysis, biotinylated NO2-FAs with adducts were affinity-purified (AP) using streptavidin-agarose beads. Pulled-down RelA protein was then detected by immunoblotting (IB). RelA and control β-actin immunoblots from the same input lysates used for affinity purification are shown below the panel. B, endogenous RelA protein levels were detected by immunoblotting probed with anti-RelA antibody using β-actin as a loading control. The relative total RelA levels (normalized by total β-actin) compared with untreated controls were quantified. *, p < 0.05 versus untreated control. Significance was determined by one-way analysis of variance followed by Tukey post test. C, MDA-MB-231 or MDA-MB-468 cells were treated with vehicle (methanol), NO2-OA (5 μm), or NO2-SA (5 μm) for 6 h, and then cell lysates were harvested and immunoprecipitated (IP) by anti-RelA antibody followed by immunoblotting. Pulldown level of immunoprecipitated RelA proteins is shown below the panel.
NO2-OA inhibits LPS-induced NF-κB transcriptional activity, in part a consequence of the alkylation of RelA Cys-38 and inhibition of RelA DNA binding (27). LC-MS/MS proteomic analysis showed that RelA Cys-105 was also alkylated by NO2-OA (Fig. S8), with the functional significance of the NO2-OA alkylation of RelA Cys-105 undefined. In aggregate, Bt-NO2-OA promotes the pulldown of IKKβ and RelA, and direct proteomic analysis revealed the NO2-OA alkylation of RelA. These observations underscore that NO2-OA mediates PTMs that inhibit multiple facets of pro-inflammatory NF-κB signaling.
NO2-OA stimulates RelA protein proteasomal degradation
Proteolytic degradation of NF-κB contributes to the termination of its signaling. Thiol-alkylating and nitrosating agents induce the degradation of NF-κB subunit p50 via the PTM of Cys-62 in both HT29 and HCT116 tumor cell lines (41). Because NO2-OA covalently adducts RelA in both MDA-MB-231 and MDA-MB-468 cells (Fig. 8A), the impact of NO2-OA PTMs on RelA protein stability was investigated. To validate this putative mechanism, we first examined whether endogenous RelA protein expression responded to NO2-OA. MDA-MB-231, MDA-MB-468, and MCF-10A cells were treated with 5 μm NO2-OA or control lipids (NO2-SA and OA) for 24 h. NO2-OA decreased the abundance of RelA in TNBC cells, whereas NO2-SA and OA had no effect (Fig. 8B). In contrast, RelA protein levels in MCF-10A cells were not altered by NO2-OA (Fig. 8B). In all three cell lines, RelA mRNA levels were not altered by NO2-OA (Fig. S9). These data indicate that NO2-OA impacts RelA protein stability via alkylation of RelA in TNBC cells. RelA is regulated by ubiquitin- and proteasome-dependent degradation signals that govern NF-κB activation (42–44). To determine whether RelA modification by NO2-OA induced ubiquitination of endogenous RelA in TNBC cells, MDA-MB-231 or MDA-MB-468 cells were treated with 5 μm NO2-OA or NO2-SA for 5 h. RelA protein was immunoprecipitated, and its polyubiquitination was detected by anti-ubiquitin. NO2-OA, but not NO2-SA, promoted polyubiquitination of RelA in both TNBC cell lines (Fig. 8C). This indicates that NO2-OA interacts with RelA and destabilizes RelA protein by promoting ubiquitination and proteasomal degradation in TNBC cells.
Discussion
Compared with other breast cancer phenotypes, TNBC is an aggressive subtype with a poor prognosis (3). Patients are 4 times more likely to show visceral metastases to the lung, liver, and brain within 5 years after diagnosis (45). Because TNBC does not respond to endocrine therapy or other more targeted chemotherapeutic agents, DNA damage–inducing strategies, such as ionizing radiation, cisplatin, and doxorubicin, remain mainstay treatments. Adverse systemic responses to DNA-directed chemotherapeutic agents, including cardiac and renal toxicity, limit chemotherapy options because of cytotoxic effects on non-cancerous cells (46–48). Herein, NO2-OA inhibited cultured TNBC cell viability, motility, and tumor cell proliferation–related signaling reactions to an extent where in vivo tumor growth in MDA-MB-231 xenografted mice was attenuated by oral administration of NO2-OA. This initial observation motivates more detailed dose-timing, dose-response, and structure-function studies of nitroalkene-based drug candidates, with respect to effects on tumor growth and metastasis of multiple breast cancer phenotypes, both in vitro and in preclinical animal models.
At lower concentrations, there was selective cytotoxicity of NO2-OA toward TNBC cells, compared with non-tumorigenic MCF-10A breast ductal epithelial cells. One significant explanation for this selectivity of action stemmed from the analysis of both basal GSH levels and the formation and fate of NO2-OA-SG adducts in control and TNBC cells. Because of the abundance and reactivity of the GSH thiolate, GSH is a primary intracellular reaction target of endogenously generated and exogenously administered oxidants and electrophilic species (49). The rate of MRP1-mediated efflux of GSH-adducted electrophiles from cells is important, as it contributes to defining the net intracellular concentration, half-life, alternative reactions with target proteins, and thus the net cellular and tissue responses to lipid electrophiles (1, 50, 51). MRP1 was highly expressed in MCF-10A cells compared with TNBC cells, motivating the LC-MS/MS determination of extracellular NO2-OA-SG levels in the media of NO2-OA–treated MCF-10A versus MDA-MB-231 and MDA-MB-468 cells. Consistent with the relative extents of MRP1 expression, MCF-10A cells formed and exported 4–5-fold greater amounts of NO2-OA-SG adducts into the extracellular compartment compared with TNBC cells (Fig. 3A). This more extensive export of NO2-OA-SG by MCF-10A cells, relative to MDA-MB-231 and MDA-MB-468 cells, was also notable because basal GSH concentrations and the GSH/GSSG ratio of MCF-10A cells were more stable after treatment with NO2-OA. In contrast, the GSH concentrations and GSH/GSSG ratio in MDA-MB-231 and MDA-MB-468 cells quickly decreased after treatment with NO2-OA (Fig. 4). These results indicate that MRP1 export of NO2-OA-SG and the more sufficient antioxidant capacity of the MCF10A cell line, as opposed to TNBC cells (52), plays a role in defining the vulnerability of TNBC cells to NO2-OA signaling actions. Another electrophile, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), displays antitumor activity by inducing apoptosis in a variety of cancers. CDDO rapidly decreases mitochondrial GSH and induces increased generation of reactive species in pancreatic cancer cells (53, 54). In contrast, NO2-OA did not significantly impact cellular rates of H2O2 production after both short-term and extended (6-h) treatment of TNBC cells, indicating that NO2-OA inhibition of TNBC cell growth and viability are not due to induction of oxidative stress (Fig. S10).
When the MRP1 transport activity of MCF-10A cells was inhibited by the organic acid probenecid (55), a more TNBC-like phenotype was conferred in the context of sensitivity to NO2-OA. For example, the impact of NO2-OA on cell growth arrest and killing (Fig. 3, C and D), cell cycle arrest (cyclin D1, p21), and apoptosis-regulating mediators (PARP-1, caspase-3) all supported the concept that NO2-OA signaling actions are enhanced in MRP1-depleted cells because of more favorable pharmacokinetics in the intracellular compartment. This affirms that the cellular concentrations of GSH, the reaction of GSH with NO2-OA, and the subsequent MRP1 export of NO2-OA-SG all influence downstream responses to NO2-OA. It is possible that other mechanisms, yet to be described, are also responsible for this differentiation of breast epithelial cell responses.
Anti-proliferative actions of NO2-OA on macrophages, vascular smooth muscle cells, and fibroblasts are observed in models of chronic vascular and pulmonary disease (56–61), but the impact of fatty acid nitroalkenes on cancer cell proliferation had not been considered. This motivated experimental consideration, because there are a limited number of reports suggesting that the up-regulation of Nrf2 signaling may result in intrinsic or acquired chemoresistance (62). In contrast, we observed the in vitro and in vivo inhibition of TNBC growth by NO2-OA (Fig. 1, B–E). This growth inhibition of TNBC cells was the result of alterations in signaling responses that were specific to TNBC cells and not non-transformed MCF-10A cells. Increased p21 and decreased cyclin D1 expression (Fig. 2D) were observed, along with an increase in the sub-G1 population of TNBC cells (Fig. 2, A–C). Two distinct pathways of apoptotic signaling were engaged by NO2-OA in TNBC cells, initiated by both mitochondria-regulated (caspase-9 activation) and death receptor–regulated (caspase-8 activation; Fig. 2F) mechanisms. In aggregate, these data reveal that NO2-OA displays pleiotropic anti-cancer properties via the inhibition of cell proliferation and induction of apoptosis in TNBC. At this point, more detailed mechanisms of NO2-OA–induced apoptotic cell death remain to be defined; however, the electrophilic thiocyanate sulforaphane also decreases Bcl-2 expression, activates cytochrome c release from the mitochondria, and increases FasL expression in TNBC cells (30). These actions imply that electrophilic fatty acid nitroalkene derivatives might mediate similar actions in the regulation of apoptosis.
The inhibition of NF-κB signaling by NO2-OA also limits TNBC cell migration and invasion. Pro-inflammatory cytokines, such as TNFα, enhance the metastatic potential of TNBC, with the up-regulation of TNFα expression and activity in TNBC patients strongly linked with tumor metastasis phenotype (63). TNFα stimulates the expression of the epithelial–mesenchymal transition and chemokine genes via the activation of AP-1 and NF-κB signaling in TNBC cells (14). Herein, NO2-OA significantly inhibited TNFα-induced TNBC cell migration and invasion (Fig. 5). Decreased expression of the pro-metastasis genes uPA and ICAM-1, via a decrease in NF-κB transcriptional activity, was also induced by NO2-OA (Fig. 6 (D and E) and Fig. S6 (A and B)). Consistent with this, electrophilic 15-deoxy-Δ12,14-prostaglandin J2, dithiolethione, and dimethyl fumarate also inhibit breast cancer cell migration (38, 64, 65). NO2-OA also limited the migration of MDA-MB-231 cells in the absence of TNFα induction (Fig. 5B). It is likely that NO2-OA inhibits cell mobility upon reaction with molecular targets in addition to NF-κB, because the electrophilic cyclopentenone 15-deoxy-Δ12,14-prostaglandin J2 also interferes with mammary cancer cell migration via inhibition of F-actin reorganization and focal adhesion disassembly (64). Additional studies are in progress to identify other metastasis-related protein targets and signaling pathways that could be impacted by NO2-OA–mediated alkylation reactions.
The proteolytic degradation of NF-κB subunits contribute to the termination of NF-κB activation. RelA protein is regulated by ubiquitin- and proteasome-dependent degradation signals that terminate NF-κB activation (42–44, 66). Thiol-alkylating and S-nitrosating agents also promote the degradation of the NF-κB subunit p50 via post-translational modification of Cys-62 in HT29 and HCT116 tumor cell lines (41). Thus, the NO2-OA alkylation of NF-κB RelA induces functional responses similar to other alkylating agents (41). Notably, the alkylation of RelA by NO2-OA induced an increase in RelA ubiquitination in TNBC cells, an effect not observed for non-electrophilic NO2-SA (Fig. 7D). PPARγ acts as an E3 ubiquitin ligase, inducing RelA protein ubiquitination and degradation via physically interacting with RelA protein. The PPARγ ligands troglitazone and pioglitazone increase PPARγ E3 ligase activity by promoting its interaction with RelA protein, in turn, decreasing RelA half-life (67). Because NO2-OA is a partial agonist of PPARγ (26), one can speculate that NO2-OA also activates PPARγ E3 ligase activity, thus further destabilizing RelA protein in TNBC.
The inhibition of NF-κB signaling represents a viable anticancer strategy, especially because the aberrant activation of NF-κB is closely linked with the development of diverse human cancers (68, 69). The immunomodulatory electrophile dimethyl fumarate, approved by the Food and Drug Administration as an oral drug for treating multiple sclerosis, also inhibits NF-κB activity in breast cancer cells and inhibits TNBC cell proliferation (65). The present results, in which NO2-OA inhibited multiple TNBC cell functions (proliferation, survival, mobility, and invasion), imply that electrophilic lipid nitroalkene species may display promising utility as pleiotropic chemotherapeutic agents.
In summary, we report that the lipid electrophile NO2-OA impacts NF-κB signaling in TNBC at multiple levels, including the suppression of IKKβ phosphorylation, inhibition of IκBα degradation, and enhanced ubiquitination and proteasomal degradation of RelA. These actions in turn contribute to the inhibition of TNBC cell migration and invasion in vitro. TNBC cells are in part more sensitive to NO2-OA due to lower GSH concentrations and suppression of NO2-OA export as the NO2-OA-SG adduct, a consequence of lower MRP1 expression. This GSH insufficiency-induced redox vulnerability of TNBC cells (70) in turn promotes more extensive protein thiol alkylation and oxidation reactions and instigates chemotherapeutic signaling responses at lower electrophile concentrations. The concentrations of endogenous free NO2-FAs, which are not protein-adducted or esterified to complex lipids, in healthy human plasma and urine are typically 1–5 nm (16, 18, 19). The oral administration of NO2-OA increased murine tumor NO2-OA levels to an extent sufficient to induce pharmacological responses, as evidenced by inhibition of MDA-MB-231 xenograft tumor growth. These results motivate more detailed future investigation of dose-response relationships and the impact of other lipid electrophiles on tumor growth and metastasis. At present, NO2-OA has cleared preclinical toxicology and pharmacokinetics testing in human Phase 1 safety trials of both oral and IV formulations (IV IND 122583; oral IND 124524) and is entering Phase 2 trials for treating chronic renal and pulmonary diseases. This present preclinical study provides the biochemical foundations for evaluating whether electrophilic NO2-FAs represent a useful new therapeutic candidate for treating breast cancer patients and possibly providing selectivity for treating TNBC, a cancer that currently lacks effective treatment options.
Experimental procedures
Cell culture and reagents
Cell lines were purchased from ATCC. MDA-MB-231 and MCF7 cells were cultured in Dulbecco's modified Eagle's medium, and MDA-MD-468 cells were cultured in improved minimum essential medium (Gibco), each supplemented with 5% fetal bovine serum (Hyclone, Logan, UT). MCF-10A cells were cultured in growth medium consisting of Dulbecco's modified Eagle's medium/F-12 (1:1) in 5% horse serum (Hyclone), and supplemented with 0.5 μg/ml hydrocortisone, 0.1 μg/ml cholera toxin, 20 ng/ml EGF, and 10 μg/ml insulin (Sigma-Aldrich). Cells were incubated at 37 °C in a 5% CO2 atmosphere. siRNAs directed against human RelA (L-003533-00-0005), human MRP1/ABCC1 siRNA (L-007308-00-0005), and non-targeting control siRNA (D-001810-10-05) were purchased from Dharmacon RNAi Technologies. Lipofectamine 2000 or 3000 (Life Technologies) was used for cell transfection. The MRP1 inhibitor Probenecid (4-[(dipropylamino)sulfonyl]benzoic acid) was purchased from Enzo Life Sciences and dissolved in 1 m sodium hydroxide. The NF-κB inhibitor JSH-23 (4-methyl-N1-(3-phenyl-propyl)-benzene-1,2-diamine) and proteasome inhibitor MG-132 (benzyloxycarbonyl-l-Leu-d-Leu-l-Leu-al) were purchased from Sigma-Aldrich. The IKKβ inhibitor BAY11-7082 (3-[(4-methylphenyl)sulfonyl]-(2E)-propenenitrile) was purchased from Calbiochem, and TNFα was from BD Biosciences.
Cell treatment for IKKβ phosphorylation, IκBα phosphorylation, and IκBα degradation
All studies used two TNBC cell lines, MDA-MB-231 and MDA-MB-468. To determine the effect of NO2-OA on IKKβ phosphorylation induced by TNFα in TNBC cells, cells were pretreated with NO2-OA (5 μm), NO2-SA (5 μm), or BAY11-7082 (10 μm) in serum-free medium (DMEM containing 0.1% fatty acid-free BSA) for 2 h before TNFα (20 ng/ml) stimulation for 5 min. For IκBα degradation, cells were treated as described above and stimulated for 10 min with 20 ng/ml TNFα. IκBα phosphorylation was measured in cells pretreated with MG-132 (10 μm) in combination with NO2-OA (5 μm), NO2-SA (5 μm), or BAY11-7082 (10 μm) in serum-free medium for 2 h before TNFα (20 ng/ml) stimulation for 10 min.
NO2-FA synthesis and use
OA was purchased from Nu-Chek Prep (Elysian, MN). Nitro-stearic acid (NO2-SA; 10-nitro-octadecanoic acid) was obtained by the reduction of 10-nitro-oleic acid. Specifically, NO2-OA was dissolved in tetrahydrofuran/methanol and cooled, and then sodium borohydride was added. The flask was stirred, and aliquots were monitored by UV analysis until there was full loss of the nitroalkene, and then the reactions were quenched with acetic acid. NO2-SA was purified by first adducting any remaining NO2-OA with added cysteine, and then NO2-SA was chromatographically fractionated on silica gel, using an ethyl acetate/hexane gradient. OA, NO2-OA, and NO2-SA were dissolved in absolute methanol and diluted in culture medium immediately before use in all experiments, at a maximum methanol concentration of 0.1% (v/v). Biotinylated NO2-FAs (Bt-NO2-OA, Bt-NO2-SA, and Bt-OA) were synthesized from corresponding free fatty acids and biotin-(polyethylene glycol)-amine (see Ref. 27 and supporting Methods).
Cell growth assay
Cells were plated at a cell density of 5000 cells/well in 96-well plates. After attachment overnight, the medium was replaced, and cells were treated with 0–15 μm NO2-OA, NO2-SA, or 0.1% methanol (vehicle) for 48 h. In an MRP inhibition study, MCF-10A cells were pretreated with 0.25 mm probenecid for 1 h, followed by 0–25 μm NO2-OA for 48 h. Cells were counted using the FluoReporter dsDNA quantitation kit (Molecular Probes) according to the manufacturer's instructions. Fluorescence was measured using a SpectraMax M2 plate reader (Molecular Devices). The half-maximal inhibitory concentration (IC50) of NO2-OA was determined by using CalcuSyn software from Biosoft. Three individual experiments were done (n = 5/each), and statistical comparison between two cell lines across doses was determined by two-way analysis of variance followed by Tukey post-test.
FACS
MCF-10A, MDA-MB-231, and MDA-MB-468 cells were plated at a cell density of 2.5 × 105 cells in 6-well plates for 24 h before treatment with 0.1% methanol (vehicle), 5 μm NO2-OA, NO2-SA, or OA for 24 h. Adherent and nonadherent cells were collected, centrifuged at 2000 rpm for 10 min, washed with ice-cold phosphate-buffered saline, fixed with cold 70% ethanol at 4 °C for 30 min, and stained with 50 μg/ml propidium iodide (Sigma-Aldrich). FACS analysis was performed at the University of Pittsburgh Department of Immunology Unified Flow Core Facility. Three individual experiments were done, and statistical comparisons among phases (G0/G1, S, and G2/M) were determined by one-way analysis of variance followed by Tukey post-test.
Cell migration analysis
MDA-MB-231 and MDA-MB-468 cells were subjected to cell migration analysis in Boyden chambers. The bottom of a 12-well membrane filter (BD Biosciences) was coated with 10 μg/ml fibronectin for 12 h before each experiment. Cells were pretreated with 5 μm NO2-OA or NO2-SA for 1 h and then in the absence or presence of TNFα (20 ng/ml) for an additional 2 h in culture medium containing 1% FBS. Cells were trypsinized and washed with migration medium (DMEM containing 0.1% fatty acid-free BSA) to remove serum. Cells at a density of 105/well were then placed in the upper chamber with migration medium containing the same pretreatment conditions. The cells were allowed to migrate toward the 5% FBS chemoattractant for 5 h. Non-migrated cells from the top surface were removed with cotton swabs. Migrated cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) and then stained with 0.5% crystal violet (Sigma-Aldrich) for 15 min. Migrated cell density on the filters was observed by microscopy. The crystal violet on migrated cells was destained with 10% acetic acid, and the absorbance in individual filters was determined at A573 nm. Images are representative of three individual experiments, and statistical comparison among treatments was determined by one-way analysis of variance followed by Tukey post-test.
Cell invasion assay
MDA-MB-468 cells were pretreated with NO2-OA (5 μm), NO2-SA (5 μm), or NF-κB inhibitor JSH-23 (10 μm) for 1 h and then in the absence or presence of TNFα (20 ng/ml) for an additional 2 h in culture medium containing 1% FBS. Cells were then suspended in migration medium and placed in the top well of invasion chambers (EMD Millipore). Chemoattractant (5% FBS) was placed in the lower chamber for 24 h at 37 °C to attract invasive cells. Cells were then harvested, and invasion rates were determined according to the manufacturer's protocol. Three individual experiments were done, and statistical comparison among treatments was determined by one-way analysis of variance followed by Tukey post-test.
Luciferase analysis of NF-κB activity
Luciferase chemiluminescence–based analysis of NF-κB transcriptional activity was performed as described previously (27) with minor modifications. MDA-MB-231 and MDA-MB-468 cells (∼70% confluence) in 12-well plates were transiently transfected with a NF-κB–luciferase reporter plasmid (Stratagene, La Jolla, CA) with Lipofectamine 3000. After transfection (24 h), cells were pretreated with NO2-OA (5 μm), NO2-SA (5 μm), OA (5 μm), or JSH-23 (20 μm) for 2 h, followed by 20 ng/ml TNFα for an additional 4 h. Each transfection was performed in triplicate. Luciferase activity was measured using the Dual-Luciferase assay kit (Promega). Relative light units (RLU) were measured using a 96-well plate luminometer, according to the manufacturer's instructions (Victor II, PerkinElmer Life Sciences). Protein concentration was determined using the BCA assay (Thermo Fisher Scientific). Data represent the ratio of treated samples to controls in the context of mean RLU/protein content ± S.D. Three individual experiments were done, and statistical significance was determined by Kruskal–Wallis test followed by Dunn's post-test with Bonferroni corrections for multiple comparisons.
NO2-FA protein alkylation reactions
To determine whether NO2-FAs bind to RelA (p65) or IKKβ in TNBC cells, MDA-MB-231 or MDA-MB-468 cells were treated with 5 μm Bt-NO2-OA, Bt-NO2-SA, or Bt-OA in DMEM containing 5% FBS. After 2 h, cells were harvested in lysis buffer containing 1% Triton X, 10% glycerol, 150 mm NaCl, 10 mm HEPES, 1 mm EDTA, 1 mm EGTA and supplemented with a mixture of protease and phosphatase inhibitors (Roche Applied Science) (26). Total cell lysates (0.5–1 mg) were mixed and incubated with streptavidin-agarose beads (Sigma-Aldrich) at 4 °C overnight. Beads were washed three times using lysis buffer. After SDS-PAGE, immunoblotting was performed using anti-RelA mouse monoclonal antibody (Santa Cruz Biotechnology) or anti-IKKβ rabbit polyclonal antibody (Cell Signaling). Proteomics analysis for the alkylation of RelA by NO2-OA was also conducted using recombinant RelA protein and LC-MS/MS analysis. See supporting Methods for more detail.
Immunoprecipitation and NO2-OA–induced RelA protein polyubiquitination
To determine the induction level of RelA protein polyubiquitination by NO2-FA, MDA-MB-231 and MDA-MB-468 cells were treated with 0.1% methanol (vehicle), NO2-OA (5 μm), or NO2-SA (5 μm) for 6 h, and then cell lysates were harvested in lysis buffer supplemented with a mixture of protease and phosphatase inhibitors. Lysates were clarified by centrifugation at 14,000 × g for 10 min. Protein lysates (1 mg) were incubated with anti-RelA antibody and Protein G/A-conjugated agarose beads (EMD Millipore, Bedford, MA) at 4 °C overnight. Immunoprecipitation fractions were obtained by centrifugation at 14,000 × g for 1 min at room temperature and washed with lysis buffer three times. The immunoprecipitated RelA was resolved by an 8% SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Bio-Rad) for immunoblotting probed with an anti-ubiquitin antibody (Santa Cruz Biotechnology). The blot was then stripped and probed with an anti-RelA antibody to assess amounts of RelA protein pulldown.
Western blotting
Western blotting was performed as described previously (26). 20–60 μg of total lysates per lane were loaded on 7, 10, or 12% SDS-PAGE and transferred onto nitrocellulose or polyvinylidene difluoride membranes (Bio-Rad). The membranes were probed with primary antibodies against caspase-3, MRP1, PARP-1, ubiquitin, or RelA; cyclin D1, p21, caspase-9, MRP4, IKKβ, pIKKβ, IκBα, or pIκBα (Cell Signaling); and caspase-8 (R&D Systems). Samples were normalized to β-actin (Sigma-Aldrich) or GAPDH (Trevigen). Protein bands were visualized, and digitized images were quantified using ImageLab software (Bio-Rad). Immunoblots are representative of at least three individual experiments. Quantitative results are an average of at least three individual experiments, and statistical significance was determined by one-way analysis of variance followed by Tukey post-test.
RNA extraction, quantitative PCR, and RT2 profiler PCR array
To determine the effect of NO2-OA on expression of NF-κB target genes in TNFα-induced MDA-MB-231 and MDA-MB-468 cells, cells were pretreated with NO2-OA (5 μm) for 2 h and then stimulated with TNFα (20 ng/ml) for 6 h. Total RNA samples of tissues or cells were extracted using TRIzol reagents according to the manufacturer's instructions (Invitrogen). Total RNA (1 μg) was reverse transcribed using the iScript cDNA kit (Bio-Rad) according to the manufacturer's instructions. cDNA (25 ng) was used for each subsequent real-time qPCR. All real-time qPCR was performed on the StepOne PLUS PCR system (Thermo Fisher Scientific) using TaqMan gene expression assays. -Fold change was calculated using the ΔΔCt method with 18S ribosomal RNA or human β-actin RNA serving as the internal control. Three individual experiments were done, and statistical significance was determined by one-way analysis of variance followed by Tukey post-test. For the RT2 profiler PCR array, MDA-MB-468 cells were treated or untreated with NO2-OA (5 μm) for 24 h. The expression of 84 human NF-κB target genes was analyzed with a 96-well plate format as instructed in the manufacturer's handbook (Qiagen). PCR amplification was conducted by the StepOne PLUS PCR system, and -fold change of gene expression was calculated according to the manufacturer's instructions.
Analysis of NO2-OA-SG and NO2-OA in cell medium
MCF-10A, MDA-MB-231, or MDA-MB-468 cells were cultured in 6-well plates (1 × 106 cells/well) for 24 h. Before treatments, cell medium was replaced with DMEM containing 5% FBS. NO2-OA (5 μm) was added to the medium, and cells were incubated at 37 °C for 60 min before the cell culture medium was collected. For MRP1 inhibition studies, MCF-10A cells were pretreated with 1 mm probenecid for 1 h and then co-treated with 5 μm NO2-OA for an additional 1 h. For MRP1 siRNA knockdown studies, MCF-10A cells were transiently transfected with non-target siRNA (scrambled) or MRP1 siRNA for 48 h before treatment with 5 μm NO2-OA for 1 h. Cells were washed with PBS and then gently scraped off of the plate in 1 ml of PBS. 100 μl of cell suspensions was lysed by sonication and used for protein concentration measurements via a BCA protein assay. The remaining 0.9 ml of cell suspension was used to determine the amount of intracellular NO2-OA-SG. NO2-OA-SG and free NO2-OA were extracted using a modified Bligh-Dyer method with NO2-OA-SG partitioning into the polar phase and NO2-OA into the organic. The cell culture medium was spiked with 15NO2-d4-OA (5 nm) as an internal standard for free NO2-OA before extraction. Samples were centrifuged at 2800 rpm at room temperature for 5 min. The bottom (organic) layer was transferred to a clean vial, dried, and reconstituted in methanol before MS analysis. The upper (aqueous) layer containing NO2-OA-SG was desalted and concentrated using 3 ml of C18 SPE columns (Thermo Fisher Scientific). Columns were preconditioned with 1 column volume of 100% methanol, followed by 2 column volumes of 5% methanol before sample addition. Samples were vortexed and equilibrated at 4 °C for 5 min before extraction. Samples were washed with 2 column volumes of 5% methanol, and the column was dried under vacuum for 30 min before elution with 3 ml of 100% methanol. Solvent was then evaporated under N2, and the samples were reconstituted in methanol for further analysis.
GSH and GSSG extraction and analysis
MCF-10A, MDA-MB-231, and MDA-MB-468 cells were seeded in 24-well plates at a density of 3 × 105 cells/well. Cells were cultured overnight before treatment with 5 μm NO2-OA for the indicated times. At each time point, cell medium was aspirated and washed two times with sterile PBS. Cells were then incubated with PBS containing 25 mm N-ethylmaleimide (NEM) for 15 min at 37 °C. Derivatizing solution (50 μl of 15% MeOH, 40 mm HEPES, 50 mm NaCl, 1 mm EDTA, 2 μm [13C215N]GSH, 2 μm [13C415N2]GSSG, and 25 mm NEM) was added to each well and incubated for 15 min at room temperature. Next, 50 μl of 10% (w/v) sulfosalicylic acid solution was immediately added to each well to stabilize GSH and GSSG. Supernatant was collected by centrifugation at 15,000 rpm for 10 min at 4 °C. Samples were diluted 1:5 in 5% sulfosalicylic acid, and 20 μl was injected for HPLC-MS/MS analysis. Cell numbers at time 0 were quantitated by a Hoechst 33258 DNA stain assay and used to normalize GSH or GSSG levels expressed as nmol/cells (× 106).
LC-MS/MS
NO2-OA, NO2-OA-SG, GSH, and GSSG were analyzed by high-performance LC-MS/MS using a Shimadzu/CTC PAL HPLC coupled to a Sciex 5000 triple quadrupole mass spectrometer (Sciex, San Jose, CA). NO2-OA, NO2-OA-SG gradient solvent systems consisted of water + 0.1% acetic acid (solvent A) and acetonitrile + 0.1% acetic acid (solvent B). NO2-OA and its metabolites were resolved using a Luna C18 reversed phase column (2 mm × 100 mm, Phenomenex, Torrence, CA) at a flow rate of 0.65 ml/min. Samples were applied to the column at 30% B and eluted with a linear increase in solvent B (30–100% in 9.7 min). The column was washed at 100% B for 3 min before returning to initial conditions for equilibration (2 min). NO2-OA-SG conjugates were resolved using a Luna C18 reversed phase column (2 mm × 150 mm; Phenomenex) at a 0.25 ml/min flow rate. Samples were applied to the column at 20% B, held for 5 min, and eluted with a linear increase in solvent B (20–98% solvent B in 20 min), followed by a wash step at 98% B for 4.5 min, and switched back to initial conditions for 4 min. MS analyses for NO2-FAs used electrospray ionization in the negative-ion mode with the collision gas set at 5 units, curtain gas at 40 units, ion source gas number 1 at 55 units and number 2 at 60 units, ion spray voltage at −4500 V, and temperature at 600 °C. The declustering potential was −80 eV, entrance potential −5, collision energy −35, and the collision exit potential −3. Multiple-reaction monitoring (MRM) was used for the analysis of lipids showing loss of a nitro group (m/z 46) upon collision-induced dissociation (MRM: 326.2/46 and 331/47 for NO2-OA and 15NO2-d4-OA, respectively) in negative-ion mode. The following parameters for the mass spectrometers were used for NO2-OA-SG conjugates in positive-ion mode: gas number 1, 50 units; gas number 2, 55 units; ion spray voltage, 5000 V; source temperature, 550 °C; declustering potential, 70 eV; entrance potential, 5; collision energy, 17; and collision exit potential, 5. The following MRM transitions were used: 635.2/506.2 and 640.2/511.2 for NO2-OA-SG and 15NO2-d4-OA-SG (Fig. S7), respectively.
The method for simultaneous determination of GSH and GSSG involved sample (20 μl) separation on a Phenomenex C18 (2.1 × 150 mm; 3.5-μm pore size) column. The solvent system employed aqueous 0.1% formic acid (A) and 0.1% formic acid in acetonitrile (B) with a net flow rate of 0.6 ml/min. A linear gradient of 2% B to 75% B from 0.1 to 6.2 min, followed by wash with 100% B for 2 min and re-equilibration with 2% B for 6 min, was employed for separation. Unlabeled and 13C415N2-labeled GSSG eluted at 2 min, whereas unlabeled and 13C215N-labeled GS-NEM eluted at ∼2.7 min. The Sciex 5000 mass spectrometer settings were as follows: CAD, 4 units; curtain gas, 40 units; GS1, 45 units; GS2, 50 units; ion spray voltage, 5500 V; source temperature, 550 °C; EP, 5 V; and CXP, 10 V. Multiple-reaction monitoring was performed in positive-ion mode. Transitions for respective species were as follows: GSH (Q1 308.3 → Q3 179.1; declustering potential (DP) 60 V, collision energy (CE) 18.5 V). 13C215N GSH (Q1 311.3 → Q3 182.1; DP 60 V, CE 18.5 V). GS-NEM (Q1 433.0 → Q3 304.2; DP 65 V, CE 38 V); [13C215N]GS-NEM (Q1 436.0 → Q3 307.2; DP 65 V, CE 38 V); GSSG (Q1 613.2 → Q3 355.2; DP 60 V, CE 24 V); [13C215N]GSSG (Q1 619.2 → Q3 361.2; DP 60 V, CE 24 V). Calibration curves were generated using known GSH and GSSG standards and isotopic internal standards and showed linearity over 5 orders of magnitude, and the limit of quantification (71) for both GS-NEM and GSSG was 1 nm. Sample [GSH] and [GSSG] were determined from analyte/internal standard area ratios, and intracellular GSH and GSSG were normalized to cell number (106), with results expressed as nmol of GSH or GSSG per 106 cells.
Statistical analysis
Data analyses were conducted using Prism version 6 software (GraphPad Software). Results are presented as mean ± S.D. tumor volumes except in Fig. 1E, where results are presented as mean ± S.E. Statistical analysis was performed using Student's t test, one-way or two-way analysis of variance as appropriate. Statistical significance was achieved with p < 0.05.
Author contributions
C.-S. C. W., B. A. F., N. E. D., C. N., S. G. W., and Y. H. conceived the project; C.-S. C. W., Y. H., S. R. W., S. R. S., B. S., F. G.-B., and S. G. W. performed experimental studies; and all authors contributed to the writing of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Chunyu Cao for technical assistance with xenograft tumor studies and Drs. Steffi Oesterreich and Abdolreza Zarnegar (University of Pittsburgh) for helpful discussions and comments on the manuscript.
This study was supported by United States Army Breast Cancer Research Breakthrough Awards W81XWH-14-1-0237 (to Y. H.) and W81XWH-14-1-0238 (to N. E. D.) and National Institutes of Health Grants R01-HL058115, R01-HL64937, P30-DK072506, and P01-HL103455 (to B. A. F.), R21AI122071-01A1 (to S. G. W.), and 5P30-CA047904 (to N. E. D.). B. A. F., S. G. W., S. R. W., and C. C. W. acknowledge interest in Complexa, Inc. No potential conflicts of interest were disclosed by other authors. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting Methods and Figs. S1–S10.
- TNBC
- triple-negative breast cancer
- ER
- estrogen receptor
- NO2-FA
- electrophilic fatty acid nitroalkene derivatives
- PTM
- post-translational modification
- IKKβ
- inhibitor of NF-κB subunit kinase β
- ICAM-1
- intercellular adhesion molecule 1
- uPA
- urokinase-type plasminogen activator
- OA
- oleic acid
- NO2-OA
- 10-nitro-octadec-9-enoic acid
- NO2-SA
- nitro-stearate
- CDDO
- 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- TNF
- tumor necrosis factor
- NF-κB
- nuclear factor-κB
- Bt
- biotinylated
- RLU
- relative light units
- NEM
- N-ethylmaleimide
- MRM
- multiple-reaction monitoring
- DP
- declustering potential
- CE
- collision energy
- qPCR
- quantitative PCR.
References
- 1. Alexander R. L., Bates D. J., Wright M. W., King S. B., and Morrow C. S. (2006) Modulation of nitrated lipid signaling by multidrug resistance protein 1 (MRP1): glutathione conjugation and MRP1-mediated efflux inhibit nitrolinoleic acid-induced, PPARγ-dependent transcription activation. Biochemistry 45, 7889–7896 10.1021/bi0605639 [DOI] [PubMed] [Google Scholar]
- 2. Brenton J. D., Carey L. A., Ahmed A. A., and Caldas C. (2005) Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J. Clin. Oncol. 23, 7350–7360 10.1200/JCO.2005.03.3845 [DOI] [PubMed] [Google Scholar]
- 3. Bauer K. R., Brown M., Cress R. D., Parise C. A., and Caggiano V. (2007) Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype. Cancer 109, 1721–1728 10.1002/cncr.22618 [DOI] [PubMed] [Google Scholar]
- 4. Smith C., Mitchinson M. J., Aruoma O. I., and Halliwell B. (1992) Stimulation of lipid peroxidation and hydroxyl-radical generation by the contents of human atherosclerotic lesions. Biochem. J. 286, 901–905 10.1042/bj2860901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dent R., Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., Lickley L. A., Rawlinson E., Sun P., and Narod S. A. (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 10.1158/1078-0432.CCR-06-3045 [DOI] [PubMed] [Google Scholar]
- 6. Kwan M. L., Kushi L. H., Weltzien E., Maring B., Kutner S. E., Fulton R. S., Lee M. M., Ambrosone C. B., and Caan B. J. (2009) Epidemiology of breast cancer subtypes in two prospective cohort studies of breast cancer survivors. Breast Cancer Res. 11, R31 10.1186/bcr2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heitz F., Harter P., Lueck H.-J., Fissler-Eckhoff A., Lorenz-Salehi F., Scheil-Bertram S., Traut A., and du Bois A. (2009) Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur. J. Cancer 45, 2792–2798 10.1016/j.ejca.2009.06.027 [DOI] [PubMed] [Google Scholar]
- 8. Dawood S., Broglio K., Esteva F. J., Yang W., Kau S.-W., Islam R., Albarracin C., Yu T. K., Green M., Hortobagyi G. N., and Gonzalez-Angulo A. M. (2009) Survival among women with triple receptor-negative breast cancer and brain metastases. Ann. Oncol. 20, 621–627 10.1093/annonc/mdn682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huber M. A., Azoitei N., Baumann B., Grünert S., Sommer A., Pehamberger H., Kraut N., Beug H., and Wirth T. (2004) NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 114, 569–581 10.1172/JCI200421358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakshatri H., Bhat-Nakshatri P., Martin D. A., Goulet R. J. Jr., and Sledge G. W. Jr. (1997) Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell Biol. 17, 3629–3639 10.1128/MCB.17.7.3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biswas D. K., Shi Q., Baily S., Strickland I., Ghosh S., Pardee A. B., and Iglehart J. D. (2004) NF-κB activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10137–10142 10.1073/pnas.0403621101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sovak M. A., Bellas R. E., Kim D. W., Zanieski G. J., Rogers A. E., Traish A. M., and Sonenshein G. E. (1997) Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 100, 2952–2960 10.1172/JCI119848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamaguchi N., Ito T., Azuma S., Ito E., Honma R., Yanagisawa Y., Nishikawa A., Kawamura M., Imai J., Watanabe S., Semba K., and Inoue J. (2009) Constitutive activation of nuclear factor-κB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer Science 100, 1668–1674 10.1111/j.1349-7006.2009.01228.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiao Y., He H., Jonsson P., Sinha I., Zhao C., and Dahlman-Wright K. (2016) AP-1 is a key regulator of proinflammatory cytokine TNFα-mediated triple-negative breast cancer progression. J. Biol. Chem. 291, 5068–5079 10.1074/jbc.M115.702571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H.-H., Zhu H., Liu L.-S., Huang Y., Guo J., Li J., Sun X.-P., Chang C.-X., Wang Z.-H., and Zhai K. (2015) Tumour necrosis factor-α gene polymorphism is associated with metastasis in patients with triple negative breast cancer. Sci. Rep. 5, 10244 10.1038/srep10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonacci G., Baker P. R. S., Salvatore S. R., Shores D., Khoo N. K. H., Koenitzer J. R., Vitturi D. A., Woodcock S. R., Golin-Bisello F., Cole M. P., Watkins S., St Croix C., Batthyany C. I., Freeman B. A., and Schopfer F. J. (2012) Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J. Biol. Chem. 287, 44071–44082 10.1074/jbc.M112.401356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fazzari M., Khoo N. K., Woodcock S. R., Jorkasky D. K., Li L., Schopfer F. J., and Freeman B. A. (2017) Nitro-fatty acid pharmacokinetics in the adipose tissue compartment. J. Lipid Res. 58, 375–385 10.1194/jlr.M072058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salvatore S. R., Vitturi D. A., Baker P. R., Bonacci G., Koenitzer J. R., Woodcock S. R., Freeman B. A., and Schopfer F. J. (2013) Characterization and quantification of endogenous fatty acid nitroalkene metabolites in human urine. J. Lipid Res. 54, 1998–2009 10.1194/jlr.M037804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delmastro-Greenwood M., Hughan K. S., Vitturi D. A., Salvatore S. R., Grimes G., Potti G., Shiva S., Schopfer F. J., Gladwin M. T., Freeman B. A., and Gelhaus Wendell S. (2015) Nitrite and nitrate-dependent generation of anti-inflammatory fatty acid nitroalkenes. Free Radic. Biol. Med. 89, 333–341 10.1016/j.freeradbiomed.2015.07.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hughan K. S., Wendell S. G., Delmastro-Greenwood M., Helbling N., Corey C., Bellavia L., Potti G., Grimes G., Goodpaster B., Kim-Shapiro D. B., Shiva S., Freeman B. A., and Gladwin M. T. (2017) Conjugated linoleic acid modulates clinical responses to oral nitrite and nitrate. Hypertension 70, 634–644 10.1161/HYPERTENSIONAHA.117.09016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schopfer F. J., Cipollina C., and Freeman B. A. (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021 10.1021/cr200131e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baker L. M. S., Baker P. R. S., Golin-Bisello F., Schopfer F. J., Fink M., Woodcock S. R., Branchaud B. P., Radi R., and Freeman B. A. (2007) Nitro-fatty acid reaction with glutathione and cysteine: kinetic analysis of thiol alkylation by a Michael addition reaction. J. Biol. Chem. 282, 31085–31093 10.1074/jbc.M704085200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Codreanu S. G., Ullery J. C., Zhu J., Tallman K. A., Beavers W. N., Porter N. A., Marnett L. J., Zhang B., and Liebler D. C. (2014) Alkylation damage by lipid electrophiles targets functional protein systems. Mol. Cell. Proteomics 13, 849–859 10.1074/mcp.M113.032953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levonen A. L., Hill B. G., Kansanen E., Zhang J., and Darley-Usmar V. M. (2014) Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics. Free Radic. Biol. Med. 71, 196–207 10.1016/j.freeradbiomed.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kansanen E., Bonacci G., Schopfer F. J., Kuosmanen S. M., Tong K. I., Leinonen H., Woodcock S. R., Yamamoto M., Carlberg C., Ylä-Herttuala S., Freeman B. A., and Levonen A.-L. (2011) Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 286, 14019–14027 10.1074/jbc.M110.190710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schopfer F. J., Cole M. P., Groeger A. L., Chen C.-S., Khoo N. K. H., Woodcock S. R., Golin-Bisello F., Motanya U. N., Li Y., Zhang J., Garcia-Barrio M. T., Rudolph T. K., Rudolph V., Bonacci G., Baker P. R. S., et al. (2010) Covalent peroxisome proliferator-activated receptor γ adduction by nitro-fatty acids: selective ligand activity and anti-diabetic signaling actions. J. Biol. Chem. 285, 12321–12333 10.1074/jbc.M109.091512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cui T., Schopfer F. J., Zhang J., Chen K., Ichikawa T., Baker P. R. S., Batthyany C., Chacko B. K., Feng X., Patel R. P., Agarwal A., Freeman B. A., and Chen Y. E. (2006) Nitrated fatty acids: endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 281, 35686–35698 10.1074/jbc.M603357200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Villacorta L., Chang L., Salvatore S. R., Ichikawa T., Zhang J., Petrovic-Djergovic D., Jia L., Carlsen H., Schopfer F. J., Freeman B. A., and Chen Y. E. (2013) Electrophilic nitro-fatty acids inhibit vascular inflammation by disrupting LPS-dependent TLR4 signalling in lipid rafts. Cardiovasc. Res. 98, 116–124 10.1093/cvr/cvt002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Snyder N. W., Golin-Bisello F., Gao Y., Blair I. A., Freeman B. A., and Wendell S. G. (2015) 15-Oxoeicosatetraenoic acid is a 15-hydroxyprostaglandin dehydrogenase-derived electrophilic mediator of inflammatory signaling pathways. Chem. Biol. Interact. 234, 144–153 10.1016/j.cbi.2014.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pledgie-Tracy A., Sobolewski M. D., and Davidson N. E. (2007) Sulforaphane induces cell type–specific apoptosis in human breast cancer cell lines. Mol. Cancer Ther. 6, 1013–1021 10.1158/1535-7163.MCT-06-0494 [DOI] [PubMed] [Google Scholar]
- 31. So J. Y., Lin J. J., Wahler J., Liby K. T., Sporn M. B., and Suh N. (2014) A synthetic triterpenoid CDDO-Im inhibits tumorsphere formation by regulating stem cell signaling pathways in triple-negative breast cancer. PLoS One 9, e107616 10.1371/journal.pone.0107616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rudnicki M., Faine L. A., Dehne N., Namgaladze D., Ferderbar S., Weinlich R., Amarante-Mendes G. P., Yan C. Y., Krieger J. E., Brüne B., and Abdalla D. S. (2011) Hypoxia inducible factor-dependent regulation of angiogenesis by nitro-fatty acids. Arterioscler. Thromb. Vasc. Biol. 31, 1360–1367 10.1161/ATVBAHA.111.224626 [DOI] [PubMed] [Google Scholar]
- 33. Lin D., Saleh S., and Liebler D. C. (2008) Reversibility of covalent electrophile-protein adducts and chemical toxicity. Chem. Res. Toxicol. 21, 2361–2369 10.1021/tx800248x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koenitzer J. R., Bonacci G., Woodcock S. R., Chen C. S., Cantu-Medellin N., Kelley E. E., and Schopfer F. J. (2016) Fatty acid nitroalkenes induce resistance to ischemic cardiac injury by modulating mitochondrial respiration at complex II. Redox Biol. 8, 1–10 10.1016/j.redox.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moody W. E., Ferro C. J., Edwards N. C., Chue C. D., Lin E. L., Taylor R. J., Cockwell P., Steeds R. P., Townend J. N., and CRIB-Donor Study Investigators (2016) Cardiovascular effects of unilateral nephrectomy in living kidney donors. Hypertension 67, 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin H.-M., Kim M.-H., Kim B. H., Jung S.-H., Kim Y. S., Park H. J., Hong J. T., Min K. R., and Kim Y. (2004) Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-κB without affecting IκB degradation. FEBS Lett. 571, 50–54 10.1016/j.febslet.2004.06.056 [DOI] [PubMed] [Google Scholar]
- 37. Evani S. J., Prabhu R. G., Gnanaruban V., Finol E. A., and Ramasubramanian A. K. (2013) Monocytes mediate metastatic breast tumor cell adhesion to endothelium under flow. FASEB J. 27, 3017–3029 10.1096/fj.12-224824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Switzer C. H., Cheng R. Y.-S., Ridnour L. A., Murray M. C., Tazzari V., Sparatore A., Del Soldato P., Hines H. B., Glynn S. A., Ambs S., and Wink D. A. (2012) Dithiolethiones inhibit NF-κB activity via covalent modification in human estrogen receptor–negative breast cancer. Cancer Res. 72, 2394–2404 10.1158/0008-5472.CAN-11-3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rossi A., Kapahi P., Natoli G., Takahashi T., Chen Y., Karin M., and Santoro M. G. (2000) Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IκB kinase. Nature 403, 103–108 10.1038/47520 [DOI] [PubMed] [Google Scholar]
- 40. Ahmad R., Raina D., Meyer C., Kharbanda S., and Kufe D. (2006) Triterpenoid CDDO-Me blocks the NF-κB pathway by direct inhibition of IKKβ on Cys-179. J. Biol. Chem. 281, 35764–35769 10.1074/jbc.M607160200 [DOI] [PubMed] [Google Scholar]
- 41. Paranjpe A., and Srivenugopal K. S. (2013) Degradation of NF-κB, p53 and other regulatory redox-sensitive proteins by thiol-conjugating and -nitrosylating drugs in human tumor cells. Carcinogenesis 34, 990–1000 10.1093/carcin/bgt032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Natoli G., and Chiocca S. (2008) Nuclear ubiquitin ligases, NF-κB degradation, and the control of inflammation. Sci. Signal. 1, pe1 [DOI] [PubMed] [Google Scholar]
- 43. Tanaka T., Grusby M. J., and Kaisho T. (2007) PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat. Immunol. 8, 584–591 10.1038/ni1464 [DOI] [PubMed] [Google Scholar]
- 44. Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y.-C., Wulf G., Rottapel R., Yamaoka S., and Lu K. P. (2003) Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12, 1413–1426 10.1016/S1097-2765(03)00490-8 [DOI] [PubMed] [Google Scholar]
- 45. Dent R., Hanna W. M., Trudeau M., Rawlinson E., Sun P., and Narod S. A. (2009) Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res. Treat. 115, 423–428 10.1007/s10549-008-0086-2 [DOI] [PubMed] [Google Scholar]
- 46. Wang W., Li C., and Yang T. (2015) Protection of nitro-fatty acid against kidney diseases. Am. J. Physiol. Renal Physiol. 310, F697–F704 10.1152/ajprenal.00321.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang H., Jia Z., Sun J., Xu L., Zhao B., Yu K., Yang M., Yang T., and Wang R. (2015) Nitrooleic acid protects against cisplatin nephropathy: role of COX-2/mPGES-1/PGE2 cascade. Mediators Inflamm. 2015, 293474 10.1155/2015/293474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu S., Jia Z., Zhou L., Liu Y., Ling H., Zhou S. F., Zhang A., Du Y., Guan G., and Yang T. (2013) Nitro-oleic acid protects against adriamycin-induced nephropathy in mice. Am. J. Physiol. Renal Physiol. 305, F1533–F1541 10.1152/ajprenal.00656.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Batthyany C., Schopfer F. J., Baker P. R., Duran R., Baker L. M., Huang Y., Cervenansky C., Branchaud B. P., and Freeman B. A. (2006) Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J. Biol. Chem. 281, 20450–20463 10.1074/jbc.M602814200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sibhatu M. B., Smitherman P. K., Townsend A. J., and Morrow C. S. (2008) Expression of MRP1 and GSTP1–1 modulate the acute cellular response to treatment with the chemopreventive isothiocyanate, sulforaphane. Carcinogenesis 29, 807–815 10.1093/carcin/bgn013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song N.-Y., Kim D.-H., Kim E.-H., Na H.-K., Kim N.-J., Suh Y.-G., and Surh Y.-J. (2011) Multidrug resistance-associated protein 1 mediates 15-deoxy-Δ12,14-prostaglandin J2-induced expression of glutamate cysteine ligase expression via Nrf2 signaling in human breast cancer cells. Chem. Res. Toxicol. 24, 1231–1241 10.1021/tx200090n [DOI] [PubMed] [Google Scholar]
- 52. Alli E., Sharma V. B., Sunderesakumar P., and Ford J. M. (2009) Defective repair of oxidative DNA damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 69, 3589–3596 10.1158/0008-5472.CAN-08-4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Samudio I., Konopleva M., Hail N., Shi Y.-X., McQueen T., Hsu T., Evans R., Honda T., Gribble G. W., Sporn M., Gilbert H. F., Safe S., and Andreeff M. (2005) 2-Cyano-3,12-dioxooleana-1,9-dien-28-imidazolide (CDDO-Im) directly targets mitochondrial glutathione to induce apoptosis in pancreatic cancer. J. Biol. Chem. 280, 36273–36282 10.1074/jbc.M507518200 [DOI] [PubMed] [Google Scholar]
- 54. Wang Y.-Y., Zhe H., and Zhao R. (2014) Preclinical evidences toward the use of triterpenoid CDDO-Me for solid cancer prevention and treatment. Mol. Cancer 13, 30 10.1186/1476-4598-13-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hagos F. T., Daood M. J., Ocque J. A., Nolin T. D., Bayir H., Poloyac S. M., Kochanek P. M., Clark R. S., and Empey P. E. (2017) Probenecid, an organic anion transporter 1 and 3 inhibitor, increases plasma and brain exposure of N-acetylcysteine. Xenobiotica 47, 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cole M. P., Rudolph T. K., Khoo N. K. H., Motanya U. N., Golin-Bisello F., Wertz J. W., Schopfer F. J., Rudolph V., Woodcock S. R., Bolisetty S., Ali M. S., Zhang J., Chen Y. E., Agarwal A., Freeman B. A., and Bauer P. M. (2009) Nitro-fatty acid inhibition of neointima formation after endoluminal vessel injury. Circulation Res. 105, 965–972 10.1161/CIRCRESAHA.109.199075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Klinke A., Möller A., Pekarova M., Ravekes T., Friedrichs K., Berlin M., Scheu K. M., Kubala L., Kolarova H., Ambrozova G., Schermuly R. T., Woodcock S. R., Freeman B. A., Rosenkranz S., Baldus S., et al. (2014) Protective effects of 10-nitro-oleic acid in a hypoxia-induced murine model of pulmonary hypertension. Am. J. Respir. Cell Mol. Biol. 51, 155–162 10.1165/rcmb.2013-0063OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kelley E. E., Baust J., Bonacci G., Golin-Bisello F., Devlin J. E., St Croix C. M., Watkins S. C., Gor S., Cantu-Medellin N., Weidert E. R., Frisbee J. C., Gladwin M. T., Champion H. C., Freeman B. A., and Khoo N. K. (2014) Fatty acid nitroalkenes ameliorate glucose intolerance and pulmonary hypertension in high-fat diet-induced obesity. Cardiovasc. Res. 101, 352–363 10.1093/cvr/cvt341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ambrozova G., Martiskova H., Koudelka A., Ravekes T., Rudolph T. K., Klinke A., Rudolph V., Freeman B. A., Woodcock S. R., Kubala L., and Pekarova M. (2016) Nitro-oleic acid modulates classical and regulatory activation of macrophages and their involvement in pro-fibrotic responses. Free Radic. Biol. Med. 90, 252–260 10.1016/j.freeradbiomed.2015.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rudolph T. K., Ravekes T., Klinke A., Friedrichs K., Mollenhauer M., Pekarova M., Ambrozova G., Martiskova H., Kaur J. J., Matthes B., Schwoerer A., Woodcock S. R., Kubala L., Freeman B. A., Baldus S., and Rudolph V. (2016) Nitrated fatty acids suppress angiotensin II-mediated fibrotic remodelling and atrial fibrillation. Cardiovasc. Res. 109, 174–184 10.1093/cvr/cvv254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verescakova H., Ambrozova G., Kubala L., Perecko T., Koudelka A., Vasicek O., Rudolph T. K., Klinke A., Woodcock S. R., Freeman B. A., and Pekarova M. (2017) Nitro-oleic acid regulates growth factor-induced differentiation of bone marrow-derived macrophages. Free Radic. Biol. Med. 104, 10–19 10.1016/j.freeradbiomed.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jaramillo M. C., and Zhang D. D. (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 10.1101/gad.225680.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalimutho M., Parsons K., Mittal D., López J. A., Srihari S., and Khanna K. K. (2015) Targeted therapies for triple-negative breast cancer: Combating a stubborn disease. Trends Pharmacol. Sci. 36, 822–846 10.1016/j.tips.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 64. Diers A. R., Dranka B. P., Ricart K. C., Oh J. Y., Johnson M. S., Zhou F., Pallero M. A., Bodenstine T. M., Murphy-Ullrich J. E., Welch D. R., and Landar A. (2010) Modulation of mammary cancer cell migration by 15-deoxy-Δ12,14-prostaglandin J2: implications for anti-metastatic therapy. Biochem. J. 430, 69–78 10.1042/BJ20091193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kastrati I., Siklos M. I., Calderon-Gierszal E. L., El-Shennawy L., Georgieva G., Thayer E. N., Thatcher G. R. J., and Frasor J. (2016) Dimethyl fumarate inhibits the nuclear factor κB pathway in breast cancer cells by covalent modification of p65. J. Biol. Chem. 291, 3639–3647 10.1074/jbc.M115.679704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maine G. N., Mao X., Komarck C. M., and Burstein E. (2007) COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 26, 436–447 10.1038/sj.emboj.7601489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hou Y., Moreau F., and Chadee K. (2012) PPARγ is an E3 ligase that induces the degradation of NF-κB/p65. Nat. Commun. 3, 1300 10.1038/ncomms2270 [DOI] [PubMed] [Google Scholar]
- 68. Rayet B., and Gélinas C. (1999) Aberrant rel/nfκb genes and activity in human cancer. Oncogene 18, 6938–6947 10.1038/sj.onc.1203221 [DOI] [PubMed] [Google Scholar]
- 69. Karin M., Cao Y., Greten F. R., and Li Z.-W. (2002) NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2, 301–310 10.1038/nrc780 [DOI] [PubMed] [Google Scholar]
- 70. Lien E. C., Lyssiotis C. A., Juvekar A., Hu H., Asara J. M., Cantley L. C., and Toker A. (2016) Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat. Cell Biol. 18, 572–578 10.1038/ncb3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zalba G., Fortuño A., Orbe J., San José G., Moreno M. U., Belzunce M., Rodríguez J. A., Beloqui O., Páramo J. A., and Díez J. (2007) Phagocytic NADPH oxidase-dependent superoxide production stimulates matrix metalloproteinase-9: implications for human atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27, 587–593 10.1161/01.ATV.0000256467.25384.c6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.