Abstract
The Hippo pathway plays important roles in controlling organ size and in suppressing tumorigenesis through large tumor suppressor kinase 1/2 (LATS1/2)–mediated phosphorylation of YAP/TAZ transcription co-activators. The kinase activity of LATS1/2 is regulated by phosphorylation in response to extracellular signals. Moreover, LATS2 protein levels are repressed by the ubiquitin–proteasome system in conditions such as hypoxia. However, the mechanism that removes the ubiquitin modification from LATS2 and thereby stabilizes the protein is not well understood. Here, using tandem affinity purification (TAP), we found that anaphase-promoting complex/cyclosome (APC/C), a ubiquitin ligase complex, and USP9X, a deubiquitylase, specifically interact with LATS2. We also found that although APC1 co-localizes with LATS2 to intracellular vesicle structures, it does not regulate LATS2 protein levels and activity. In contrast, USP9X ablation drastically diminished LATS2 protein levels. We further demonstrated that USP9X deubiquitinates LATS2 and thus prevents LATS2 degradation by the proteasome. Furthermore, in pancreatic cancer cells, USP9X loss activated YAP and enhanced the oncogenic potential of the cells. In addition, the tumorigenesis induced by the USP9X ablation depended not only on LATS2 repression, but also on YAP/TAZ activity. We conclude that USP9X is a deubiquitylase of the Hippo pathway kinase LATS2 and that the Hippo pathway functions as a downstream signaling cascade that mediates USP9X's tumor-suppressive activity.
Keywords: anaphase-promoting complex, LATS2, USP9X, ubiquitination, deubiquitination, protein stability, tumor suppressor gene, signal transduction, Hippo pathway
Introduction
The Hippo pathway is an evolutionarily conserved mechanism that plays a fundamental role in control of organ size (1, 2). It limits cell number through transcriptional regulation of cell proliferation, apoptosis, and stem cell self-renewal. Deregulation of the Hippo pathway leads to drastic enlargement of organs such as eyes and wings in Drosophila as well as livers, hearts, and stomachs in mice (3–9). Furthermore, sustained inactivation of the Hippo pathway potently induces tumorigenesis in mice. More importantly, accumulating evidence clearly indicates that deregulation of the Hippo pathway in various human cancers plays important roles in cancer initiation and progression (10). These findings highlight the importance of thoroughly understanding the molecular mechanisms regulating the Hippo pathway.
Central to the Hippo pathway is a kinase cascade formed by the MST1/2 kinases of the STE-20 family and their downstream kinases LATS1/2 of the AGC family. MST1/2 activate LATS1/2 through direct phosphorylation and also through phosphorylation of SAV1 (an adaptor protein of MST1/2) and MOB1A/B (adaptor proteins of LATS1/2) (2). The Hippo pathway regulates gene expression via direct phosphorylation and inhibition of transcription co-activators Yes-associated protein (YAP)2 and its paralog transcriptional coactivator with PDZ-binding motif (TAZ) (11–15). Phosphorylation of YAP by LATS1/2 leads to its cytoplasmic retention, mediated by the scaffold protein 14-3-3, and degradation, mediated by the E3 ligase SCFβ-TRCP (12, 16). Nevertheless, inactivation of the Hippo pathway leads to YAP nuclear translocation and interaction with transcription factors, such as the TEAD family proteins, which results in expression of pro-proliferative and anti-apoptotic genes (17–21). The Hippo pathway is tightly regulated by upstream signals, such as mechanical stress, G-protein–coupled receptor signaling, and cellular energy status, which result in change of LATS1/2 phosphorylation level and activity (2). Interestingly, the protein level of LATS2 is also regulated by hypoxia condition through ubiquitination by the E3 ubiquitin ligase SIAH2 and subsequent degradation by proteasomes (22). LATS2 is also ubiquitinated by other E3 enzymes, such as NEDD4 and CRL4DCAF1 (23, 24). Therefore, the turnover of LATS1/2 protein could be another mechanism playing important roles in regulation of Hippo pathway activity. However, little is known about the deubiquitination process of LATS2, the other side of the coin.
Protein ubiquitination is a reversible post-translational modification that could be removed by a family of enzymes called deubiquitylases (DUBs). USP9X is an evolutionarily conserved member of the largest DUB family, the ubiquitin-specific proteases (USPs) (25). Recent investigations revealed important functions of USP9X in development and in diseases such as neurodegeneration and cancer. Interestingly, depending on the type of cancer, USP9X could be either oncogenic or tumor-suppressive. For example, elevation of USP9X may stabilize MCL1, a pro-survival BCL2 family protein, thus contributing to the development of lymphoma and multiple myeloma (26). Conversely, in a genetic screen for tumor suppressors of pancreatic ductal adenocarcinoma (PDAC) carried out in mice, Usp9x was found to be the most commonly mutated gene in >50% of the tumors (27), indicating its strong tumor-suppressive activity. However, the mechanism by which USP9X works as a tumor suppressor in PDAC remains obscure.
In this study, we identified the APC/C E3 complex and USP9X as specific LATS2-interacting proteins. Whereas APC/C does not seem to have a regulatory effect on LATS2, USP9X potently regulates the protein level of LATS2. In pancreatic cancer cells, ablation of USP9X diminishes the protein level of LATS2 and thus leads to YAP activation and enhanced oncogenic potential. We thus identified USP9X as a DUB of LATS2 and propose that deregulation of USP9X in PDAC promotes tumorigenesis through silencing of the Hippo pathway.
Results
LATS2 interacts with APC/C complex and USP9X
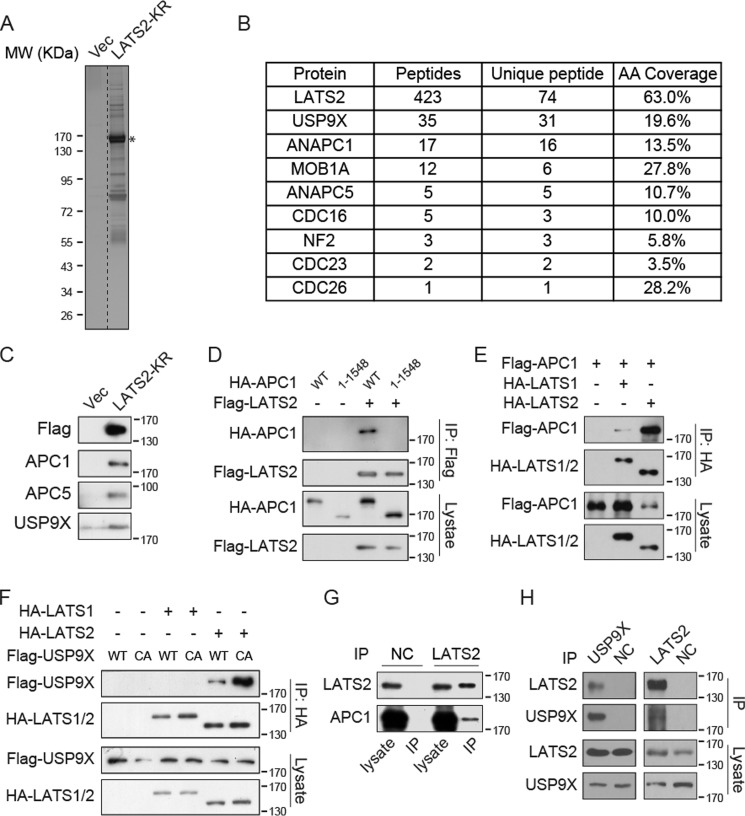
To further elucidate the regulation and function of LATS2 kinase, we carried out TAP of FLAG-streptavidin-binding peptide (SBP)–tagged LATS2 ectopically expressed in MCF10A cells. Due to the growth-suppressive activity of LATS2, we used the kinase-inactive KR mutant to avoid difficulty in stable cell propagation. In this purification, we recovered many proteins that specifically interact with LATS2 (Fig. 1A). Mass spectrometry identified known LATS2-interacting proteins, such as MOB1A and NF2 (Fig. 1B). Interestingly, several proteins regulating ubiquitination are also on top of the list, including USP9X deubiquitylase and several components of the APC/C ubiquitin ligase complex, including ANAPC1 (APC1), ANAPC5 (APC5), CDC16, CDC23, and CDC26 (Fig. 1B). The presence of APC1, APC5, and USP9X in the LATS2 TAP product was also confirmed by Western blotting with specific antibodies (Fig. 1C). We further determined the interaction between APC1 and LATS2 by their co-immunoprecipitation when co-expressed in HEK293T cells. Whereas LATS2 co-immunoprecipitated well with wild-type APC1, it lost interaction with a truncation mutant 1–1548, naturally occurring during subcloning (Fig. 1D). Interactions of LATS2 with other APC/C complex proteins, including APC5, APC2, CDC23, and APC7, have also been confirmed (Fig. S1, A and B). A couple of previous reports have investigated the Hippo pathway interactome using similar affinity purification approaches (28, 29). Interestingly, in their results, we also found USP9X and the APC/C complex as specific interacting proteins of LATS2 but not LATS1. We confirmed that in comparison with LATS1, LATS2 has much stronger interaction with APC1 (Fig. 1E). Similarly, USP9X only interacts with LATS2 and not LATS1, and the enzymatically inactive CA mutant of USP9X exhibits stronger interaction with LATS2 (Fig. 1F). Furthermore, we confirmed that LATS2 interacts with APC1 and USP9X at the endogenous level (Fig. 1, G and H). Angiomotin (AMOT) family proteins in the Hippo pathway have previously been shown to interact with and be deubiquitinated by USP9X (30, 31). We found that the interaction of USP9X with AMOT is much weaker than that with LATS2 (Fig. S1C), whereas the interaction of USP9X with AMOTL2 is similar to that with LATS2 (Fig. S1D). However, the USP9X-LATS2 interaction is not significantly affected by co-expression of AMOTL2 (Fig. S1D). The above results identified the APC/C complex and USP9X as ubiquitination-regulating enzymes that specifically interact with LATS2, which may thus play regulatory roles in the Hippo pathway.
Figure 1.
LATS2 interacts with APC/C complex and USP9X. A, TAP purification of LATS2-KR. MCF10A cells stably expressing FLAG-SBP–tagged LATS2-KR were established. Cell lysates were purified first with anti-FLAG resin and then with streptavidin resin. The final eluates were resolved on SDS-polyacrylamide gels and silver-stained. *, band of LATS2. MW, molecular weight. B, mass spectrometry analysis identified APC/C complex proteins and USP9X as LATS2-binding proteins. LATS2-KR TAP products were analyzed by MS-MS. The number of peptide hits for APC/C complex proteins, USP9X, and several known LATS2-binding proteins are shown. AA, amino acid. C, Western blotting of TAP samples confirmed that APC1, APC5, and USP9X interact with FLAG-LATS2-KR. TAP samples were examined by Western blotting using the indicated antibodies. D, APC1 co-immunoprecipitates with LATS2. FLAG-LATS2 and HA-APC1 were co-expressed in HEK293T cells, and immunoprecipitation (IP) was performed with anti-FLAG antibody. Samples were analyzed by Western blotting. E, APC1 preferentially binds to LATS2 rather than LATS1. Experiments were similar to those in D. F, USP9X specifically binds to LATS2. Experiments were similar to those in D. CA, C1566A inactive mutant of USP9X. G, LATS2 interacts with APC1 on the endogenous level. Endogenous LATS2 was immunoprecipitated from MCF10A cells with anti-LATS2 antibody, and samples were examined by Western blotting. H, LATS2 interacts with USP9X on the endogenous level. Experiments were similar to those in G. Data are representative of at least duplicate experiments.
LATS2 interacts with APC1 and USP9X through distinct regions
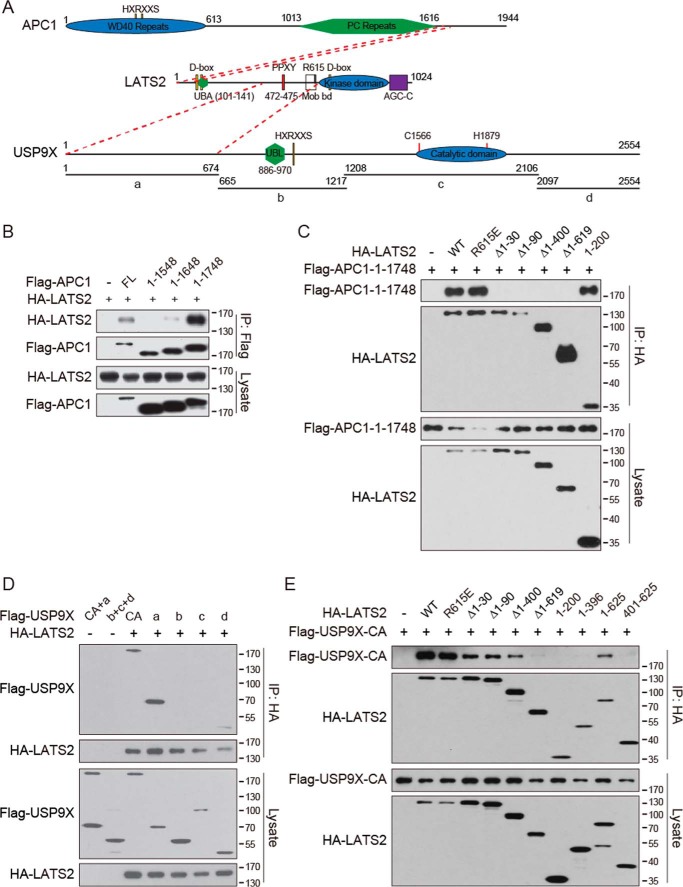
We next mapped the protein regions responsible for the interactions between LATS2 and APC1 or USP9X. Because the APC1 C-terminal truncation mutant lacking 400 amino acids (residues 1–1548) (Fig. 2A) lost interaction with LATS2, we added back 100 amino acids at a time to test restoration of the interaction. Clearly, adding back 200 amino acids (residues 1–1748) but not 100 amino acids (residues 1–1648) fully rescued the interaction with LATS2 (Fig. 2B), indicating that the region between APC1 amino acids 1648 and 1748 is critical for the interaction. For LATS2, deletion of the N-terminal 30 amino acids abolished the interaction with APC1 (Fig. 2C). Furthermore, deletion of amino acids 60–90, but not 30–60, also impairs LATS2 interaction with APC1 (Fig. S2A). Therefore, the 1–30 and 60–90 regions of LATS2 are both involved in the interaction with APC1. Consistently, the 1–200 region of LATS2 is sufficient to interact with APC1 (Fig. 2C). However, the interaction of LATS2 1–200 with the APC1 1549–1748 fragment is marginal (data not shown), suggesting that additional regions of APC1 are involved. The N terminus of LATS2 contains two D (destruction) boxes, targeting motifs recognized by the APC/C complex (32). However, they are not required for the interaction with APC1 (Fig. S2B). Similarly, the UBA domain of LATS2 is also dispensable for the interaction with APC1 (Fig. S2B).
Figure 2.
LATS2 interacts with APC1 and USP9X through distinct regions. A, LATS2, APC1, and USP9X protein domain organization. Domains and key residues of mouse LATS2, human APC1, and human USP9X are illustrated to scale. D box, binding motif recognized by APC/C; HXRXXS, LATS2 phosphorylation target consensus motif; PPXY, motif bound by WW domains; UBA, ubiquitin-associated domain; UBL, ubiquitin-like domain. Protein regions interacting with each other are indicated by dashed red lines. B, interaction of APC1 mutants with LATS2. The indicated plasmids were co-transfected into HEK293T cells. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody. Samples were examined by Western blotting. C, interaction of LATS2 mutants with APC1. Experiments were similar to those in B. D, interaction of USP9X mutants with LATS2. USP9X was truncated into four fragments as indicated in A. Experiments were similar to those in B. E, interaction of LATS2 mutants with USP9X. Experiments were similar to those in B. Data are representative of triplicate experiments.
To determine the LATS2-interacting region of USP9X, we truncated the USP9X CA protein into four fragments (Fig. 2A). Although the C-terminal fragment d showed weak interaction with LATS2, the N-terminal fragment a (residues 1–674) interacted with LATS2 similarly as the full-length protein (Fig. 2D), suggesting that a is the major LATS2-interacting region. Deletion of LATS2 from the N-terminal indicated that loss of the N-terminal 30 amino acids reduced but did not abolish interaction with USP9X (Fig. 2E). However, loss of a region from 400 to 619 almost abolished the interaction, whereas the fragment 1–625 interacted well with USP9X (Fig. 2E). These results suggest that LATS2 400–619 is playing a critical role in interaction with USP9X. However, this region alone is insufficient to interact with USP9X (Fig. 2E), suggesting that some other regions or an intact protein structure is required. It is noteworthy that mutation of Arg-615, a residue critical for LATS2 interaction with MOB1A/B (33), does not affect interaction with USP9X (Fig. 2E), indicating that the interaction of LATS2 with USP9X is independent of MOB1A/B. Taken together, LATS2 interacts with APC1 and USP9X through distinct regions of the protein (Fig. 2A), and loss of the N-terminal 30 amino acids of LATS2 inhibits the interaction with both APC1 and USP9X to various extents.
No evidence for LATS2 regulation by APC/C
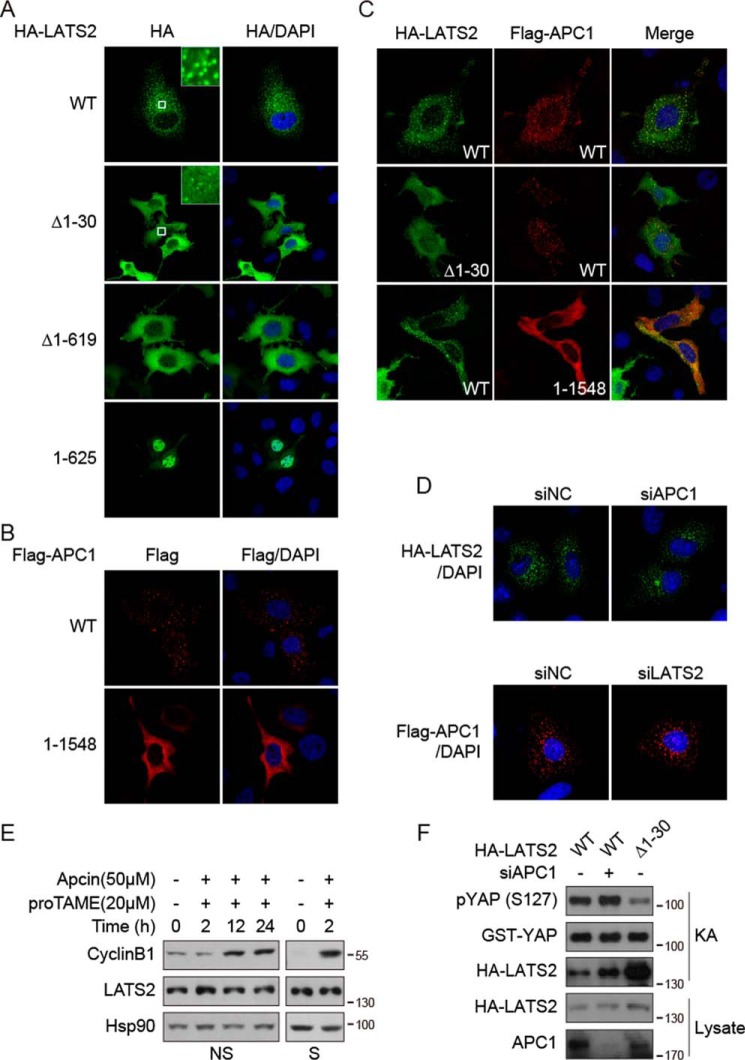
We next investigated whether APC/C has a regulatory effect on LATS2 by first examining the subcellular localization. When expressed in HeLa cells, wild-type LATS2 adopts vesicle-like localizations besides cytoplasmic distribution (Fig. 3A). However, deletion of the N-terminal 30 or 619 amino acids results in even distribution of LATS2 in the cytoplasm (Fig. 3A). Consistently, in biochemical fractionation, wild-type LATS2 was mainly recovered in the membrane fraction, which was dramatically reduced by deletion of 1–30 or 1–619 (Fig. S3A). The localization of LATS2 and its mutants has been confirmed in U2OS cells (data not shown). These results suggest that the 1–30 region of LATS2 might localize LATS2 to some intracellular vesicles. Interestingly, wild-type but not the LATS2 binding–deficient 1–1548 mutant of APC1 also adopts vesicle-like structures in HeLa cells (Fig. 3B). Fractionation experiments confirmed that APC1 is also mainly membrane-localized, which was reduced by deletion of the C-terminal after 1548 (Fig. S3B). To test whether LATS2 and APC1 promote the vesicle-like localization of each other, they were co-expressed for immunofluorescence staining. Indeed, vesicles positive for LATS2 were also co-occupied by APC1 (Fig. 3C). However, deletion of LATS2 1–30 or APC1 after 1548 only disrupts vesicle-like localization of the respective protein but not the other (Fig. 3C), suggesting that the vesicle-like localization of LATS2 and APC1 do not depend on each other. Indeed, knockdown of APC1 or LATS2 (Fig. S3C) did not affect the vesicle-like structures of LATS2 or APC1, respectively (Fig. 3D). These results demonstrate that both LATS2 and APC1 localize to internal vesicle structures in cells, which could be important for the interaction of these two proteins. However, this subcellular localization of LATS2 is not regulated by APC1. By co-staining, we found that these LATS2-positive vesicles are not Rab5- or Lamp1-positive endosomes (data not shown). Thus, the identity of these vesicle-like structures remains to be determined. During the study of LATS2 subcellular localization, we also noticed that the N-terminal fragment of LATS2 before the kinase domain adopts explicit nuclear localization (Fig. 3A), suggesting that it may mediate nuclear localization of LATS2. Indeed, although the nuclear localization of wild-type LATS2 is not significant, it was drastically enhanced by leptomycin B treatment, which blocks CRM1-dependent nuclear export (Fig. S3D). The nuclear localization of LATS1 is also enhanced by leptomycin B, although less potently (data not shown). These results suggest that the subcellular localization of LATS2 could be dynamically regulated, which might play an important role in the regulation and function of the Hippo pathway.
Figure 3.
APC1 does not regulate the localization, protein level, or activity of LATS2. A, subcellular localization of LATS2. The indicated proteins were expressed in HeLa cells and visualized by immunofluorescence staining. B, subcellular localization of APC1. Experiments were similar to those in A. C, co-localization of LATS2 and APC1. Experiments were similar to those in A. D, knockdown of APC1 or LATS2 does not affect the localization of the other. HeLa cells were co-transfected with indicated plasmids and siRNAs. HA-LATS2 and FLAG-APC1 were visualized by immunofluorescence staining. E, inhibition of APC/C does not affect the protein level of LATS2. Non-synchronized (NS) or thymidine-nocodazole–synchronized (S) HeLa cells were treated with APC/C inhibitors as indicated. Samples were examined by Western blotting. F, knockdown of APC1 does not affect the kinase activity of LATS2. LATS2 was immunoprecipitated from transfected HEK293T cells with anti-HA antibody and then subjected to in vitro kinase assay (KA) using recombinant GST-YAP as substrate. Data are representative of triplicate experiments.
The APC/C complex is a ubiquitin ligase–mediating protein degradation, which thus initiates the metaphase–anaphase transition and mitotic exit in mitosis (32). We thus examined whether inhibition of APC/C activity would increase LATS2 protein level. We treated non-synchronized or thymidine/nocodazole-synchronized HeLa cells with two inhibitors of APC/C, apcin and proTAME (34). The protein level of cyclin B1, a known substrate of APC/C, was clearly increased, but the protein level of LATS2 remained the same (Fig. 3E). Furthermore, knockdown of APC1 did not change the protein level or kinase activity of LATS2 (Fig. 3F). Therefore, LATS2 is not a substrate of APC/C. However, we noticed that deletion of amino acids 1–30 results in inhibition of LATS2 kinase activity (Fig. 3F), suggesting that LATS2 is probably activated on the membrane of intracellular vesicles. We have also noticed that APC1 has two consensus LATS2 target phosphorylation motifs, HXRXXS, at amino acids 304–309 and 338–343 (Fig. 2A). In addition, co-expression of APC1 with LATS2 could induce retarded migration of APC1 in electrophoresis on gels containing Phos-tag, which specifically retards phosphorylated proteins (Fig. S3E). Thus, LATS2 might phosphorylate and regulate APC/C, which is an interesting possibility beyond the scope of this study.
USP9X deubiquitinates and stabilizes LATS2
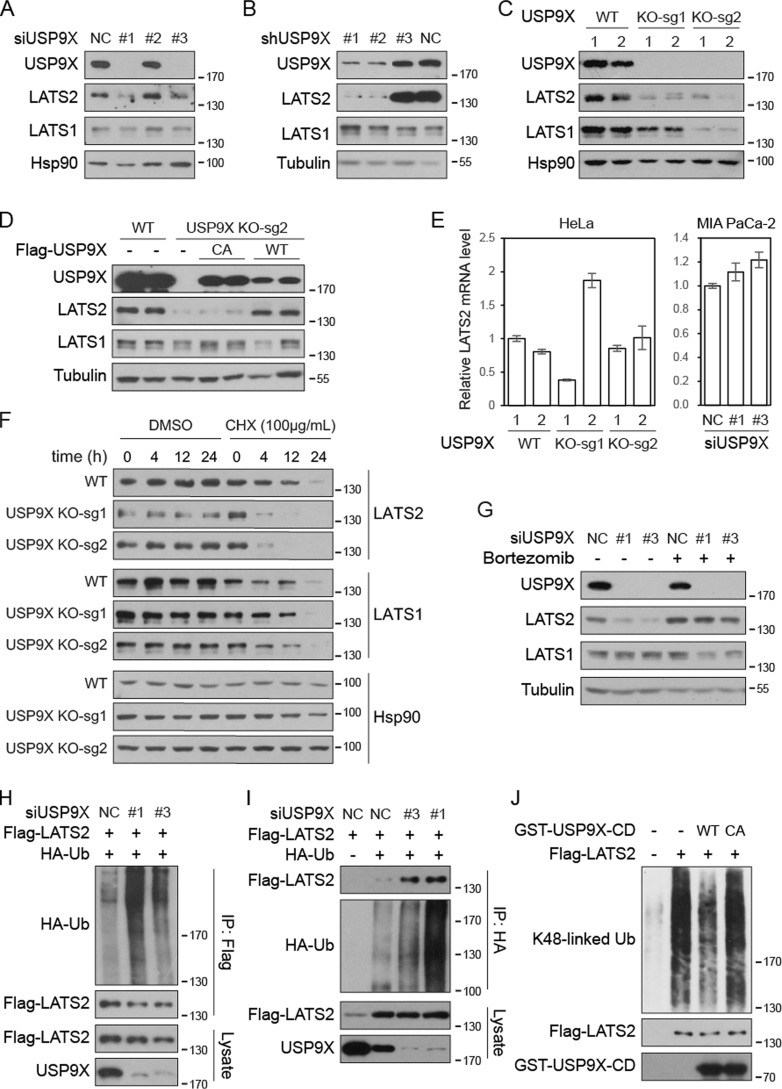
We further investigated whether USP9X regulates LATS2. siRNAs were used to knock down USP9X in pancreatic cancer cell line MIA PaCa-2. Interestingly, whereas siRNAs 1 and 3 efficiently repressed USP9X expression, the protein levels of LATS2 were also dramatically reduced (Fig. 4A). Similarly, siRNA-mediated knockdown of USP9X also diminished LATS2 protein levels in pancreatic cancer cell lines CFPAC-1, BxPC-3, and AsPC-1 and breast cancer cell line MDA-MB-468 as well as a transformed mammary epithelial cell line MCF10A (Fig. S4A). To further confirm these observations, shRNAs were used to knock down USP9X in BxPC-3 cells. Indeed, two shRNAs that efficiently repressed USP9X expression also diminished LATS2 protein expression (Fig. 4B). We next knocked out USP9X in HeLa and MIA PaCa-2 cells using CRISPR/Cas9 technology. Again, the LATS2 protein level was markedly reduced upon ablation of USP9X (Fig. 4, C and D). Importantly, the protein level of LATS2 could be rescued by re-expression of wild type but not the enzymatically inactive CA mutant of USP9X (Fig. 4D and Fig. S4B). Furthermore, the mRNA levels of LATS2 were not consistently repressed upon USP9X knockdown or knockout (Fig. 4E). These results highly suggest that USP9X maintains the protein level of LATS2 through deubiquitination and inhibition of LATS2 degradation. Whereas LATS1 protein levels largely remain unaffected by USP9X knockdown or knockout (Fig. 4, A, B, D, and G), they were sometimes found reduced in USP9X-knockout clones (Fig. 4C and Fig. S4 (C and D)). However, it is obvious that the reduction of LATS1 protein level is clone-specific and could not be rescued by proteasome inhibitor (Fig. S4C), suggesting that it is a nonspecific effect.
Figure 4.
USP9X deubiquitinates and stabilizes LATS2. A and B, knockdown of USP9X diminishes LATS2 protein level. USP9X was knocked down by siRNAs in MIA PaCa-2 cells (A) or by shRNAs in BxPC-3 cells (B). Cell lysates were examined by Western blotting. C, knockout of USP9X diminishes LATS2 protein level. USP9X was knocked out in HeLa cells by CRISPR-Cas9 technology using two independent sgRNAs. Two clones were examined for each genotype. D, enzymatically inactive USP9X fails to rescue LATS2 protein level in USP9X-knockout cells. FLAG-USP9X-WT and CA mutant were expressed in USP9X-knockout MIA PaCa-2 cells. Cell lysates were examined by Western blotting. E, loss of USP9X does not inhibit LATS2 mRNA expression. The mRNA levels of LATS2 in USP9X-knockout HeLa cells or USP9X knockdown MIA PaCa-2 cells were determined by quantitative RT-PCR. Values represent means ± S.D. (error bars) from three technical repeats. F, LATS2 is destabilized in USP9X-knockout cells. Wild-type and USP9X-knockout HeLa cells were treated with cycloheximide (CHX) as indicated before collection. Cell lysates were examined by Western blotting. G, proteasome inhibitor rescues LATS2 protein level in USP9X knockdown cells. Wild-type and USP9X knockdown MIA PaCa-2 cells were treated with 100 nm bortezomib for 24 h before collection. H and I, knockdown of USP9X promotes LATS2 ubiquitination. LATS2 and siRNAs were transfected into HeLa cells stably expressing HA-Ub or control HeLa cells, as indicated. FLAG-LATS2 (H) or HA-Ub (I) was immunoprecipitated (IP) with anti-FLAG or anti-HA antibodies, respectively. Samples were examined by Western blotting. J, USP9X deubiquitinates LATS2 in vitro. Wild-type or inactive catalytic domain of USP9X was purified from E. coli. FLAG-LATS2 was immunoprecipitated from bortezomib-treated transfected HeLa-Ub stable cells. LATS2 and USP9X catalytic domain were then incubated in deubiquitination assay buffer. Samples were analyzed by Western blotting. Data are representative of triplicate experiments.
We further examined the degradation of LATS2 after inhibition of protein synthesis by cycloheximide. The results indicate that the degradation of LATS2 but not LATS1 is much faster after ablation of USP9X (Fig. 4F). Furthermore, the diminished protein level of LATS2 caused by USP9X knockdown or knockout could be fully rescued by a proteasome inhibitor, bortezomib (Fig. 4G and Fig. S4C). However, inhibition of chaperone-mediated autophagy, another pathway for protein degradation, by chloroquine, could not rescue LATS2 protein level (Fig. S4D). Therefore, USP9X inhibits LATS2 degradation by proteasome. Consistently, knockdown of USP9X in HeLa cells stably expressing HA-tagged Ub clearly enhanced the level of LATS2 ubiquitination (Fig. 4H). In the same cells, immunoprecipitation of HA-Ub also revealed markedly increased pulldown of LATS2 after USP9X knockdown (Fig. 4I). These results indicate that USP9X inhibits polyubiquitination of LATS2. In addition, recombinant wild type but not the inactive USP9X catalytic domain purified from Escherichia coli could deubiquitinate LATS2 in vitro (Fig. 4J). Taken together, USP9X plays a key role in maintaining the protein level of LATS2 via prevention of proteasome-mediated degradation of the protein through direct deubiquitination.
YAP is activated by USP9X knockdown in pancreatic cancer cells
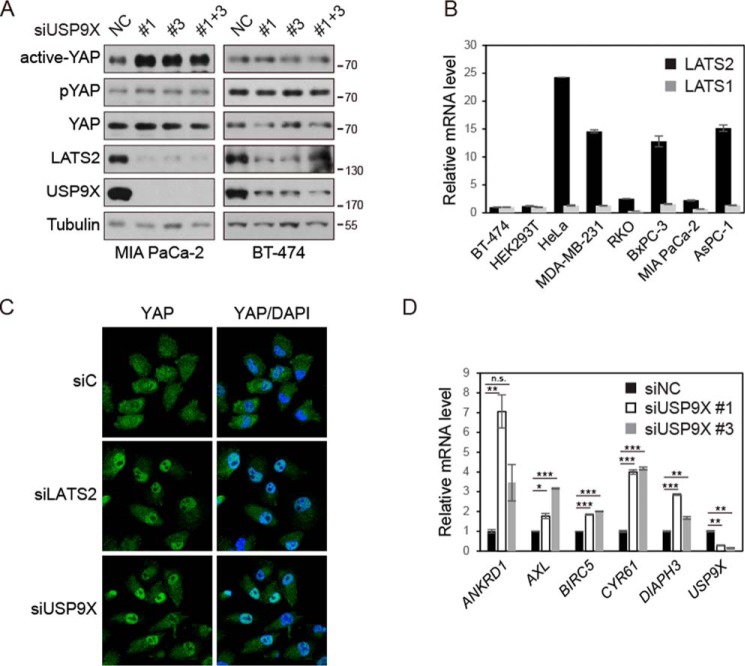
We hypothesized that USP9X may play a tumor-suppressive role by promoting the protein level of LATS2, thus inhibiting YAP in the Hippo pathway. Genetic evidence in mice indicates that USP9X is a tumor suppressor in PDAC, and in clinical samples, the expression level of USP9X is favorably associated with survival (27). We thus examined the effect of USP9X knockdown in pancreatic cancer cell line MIA PaCa-2. Surprisingly, using an antibody against YAP phosphorylated at Ser-127, we did not observe a significant decrease of YAP phosphorylation (Fig. 5A). Nevertheless, using an antibody that recognizes YAP only when it is dephosphorylated at Ser-127 (active YAP), the specificity of which has been previously validated (35, 36), we could demonstrate a marked increase of active YAP upon USP9X knockdown (Fig. 5A). These data also underscore the sensitivity and advantage of directly detecting active YAP, which is the fraction that executes downstream biological functions. However, in the BT-474 breast cancer cell line, knockdown of USP9X affected neither phospho-YAP nor active dephosphorylated YAP (Fig. 5A). This observation is consistent with the cancer type–specific function of USP9X as a tumor suppressor. Because YAP could be phosphorylated by both LATS1 and LATS2, we examined their mRNA expression levels in different cell lines. We found that the ratio between LATS2 and LATS1 is relatively low in BT-474 (Fig. 5B), which could be one of the reasons contributing to the insensitivity of YAP phosphorylation to USP9X in this cell line. Thereby, we examined additional cell lines with a high LATS2/LATS1 ratio, including AsPC-1, BxPC-3, and HeLa, and found that the phosphorylation of YAP is also regulated by USP9X in these cells (Fig. S5A). Consistent with the increase of active YAP level, USP9X knockdown led to increased nuclear localization of YAP in MIA PaCa-2 cells similarly to LATS2 knockdown (Fig. 5C). More importantly, knockdown of USP9X also enhanced the expression of many YAP target genes, such as CYR61, ANKRD1, AXL, BIRC5, and DIAPH3 (Fig. 5D), although the expression of CTGF, another well-known YAP target gene (20), was too low to be detected in this cell line. In contrast, the expression of YAP target genes was not stimulated by USP9X knockdown in BT-474 cells (Fig. S5B). These data demonstrate that USP9X suppresses YAP activity in pancreatic cancer cells probably through maintaining the protein level of LATS2.
Figure 5.
Knockdown of USP9X activates YAP in pancreatic cancer cells. A, knockdown of USP9X increases dephosphorylated active YAP in MIA PaCa-2 but not in BT-474 cells. Lysates of siRNA-transfected cells were examined by Western blotting. B, LATS1/2 mRNA levels in various cancer cell lines. Relative LATS1 and LATS2 mRNA levels in cancer cell lines were determined by quantitative RT-PCR. The levels of LATS1 and LATS2 in BT-474 cells were set to 1 arbitrary unit. C, knockdown of USP9X increases YAP in cell nuclei. siRNA-transfected MIA PaCa-2 cells were fixed and stained for YAP protein. D, knockdown of USP9X increases the expression of YAP target genes. mRNA levels of the indicated genes in siRNA-transfected MIA PaCa-2 cells were determined by quantitative RT-PCR. Values represent means ± S.D. (error bars) from three technical repeats. p value was calculated by Student's t test. n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data are representative of triplicate experiments.
USP9X suppresses tumorigenesis via the Hippo pathway
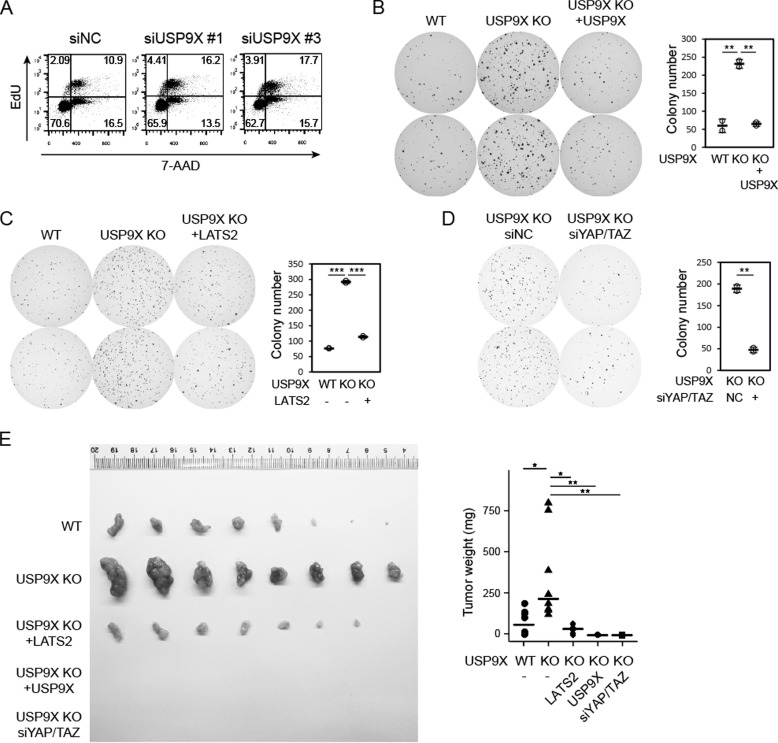
We next examined whether the Hippo pathway plays a role in tumorigenesis downstream of USP9X. Indeed, knockdown of USP9X increased proliferation of MIA PaCa-2 cells as determined by flow cytometry analysis of EdU incorporation (Fig. 6A). Furthermore, knockdown or knockout of USP9X promoted anchorage-independent growth, a well-known indicator of oncogenic potential (Fig. 6B and Fig. S6A). Importantly, this phenotype was normalized by re-expression of USP9X (Fig. 6B and Fig. S6B). These observations confirmed the tumor-suppressive activity of USP9X in pancreatic cancer cells. Remarkably, enhanced anchorage-independent growth induced by USP9X knockout was also inhibited by expression of LATS2 or knockdown of YAP/TAZ (Fig. 6 (C and D) and Fig. S6 (C and D)). Furthermore, USP9X knockout enhanced subcutaneous xenograft tumor growth of MIA PaCa-2 cells in nude mice, which was also dramatically inhibited by re-expression of USP9X or LATS2 or knockdown of YAP/TAZ (Fig. 6E). These data demonstrate that repression of LATS2 and the ensuing activation of the Hippo pathway effectors play a critical role in tumorigenesis induced by inhibition of USP9X, a tumor suppressor in pancreatic cancer.
Figure 6.
USP9X suppresses tumorigenesis via the Hippo pathway. A, USP9X knockdown increases the proportion of proliferating cells. S phase cells were labeled by EdU, and total DNA was labeled by 7-AAD. Samples were analyzed by flow cytometry. B, USP9X knockout promotes anchorage-independent growth. Wild-type, USP9X-knockout, and USP9X-restored MIA PaCa-2 cells were subjected to a soft agar colony formation assay. Colonies were stained by crystal violet and quantified (right). C, re-expression of LATS2 repressed colony formation induced by USP9X knockout. Experiments were similar to those in B. D, YAP/TAZ are required for colony formation induced by USP9X knockout. USP9X-knockout cells were transfected with siRNAs. Experiments were similar to those in B. E, tumorigenesis induced by USP9X knockout is dependent on LATS2 repression and YAP/TAZ activity. Nude mice were injected subcutaneously on the two flanks with cells the same as those in B–D. Tumors were dissected after 6 weeks. Values represent means ± S.D. (error bars). Individual data points are also shown. p value was calculated by Student's t test. n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data are representative of at least duplicate experiments.
Discussion
Biological functions of the Hippo pathway are mediated by YAP/TAZ transcription co-activators, which are tightly controlled via phosphorylation by LATS1/2 kinases (37). Therefore, regulation of LATS1/2 activity is central to mechanisms regulating the Hippo pathway. The activity of LATS1/2 is first regulated by phosphorylation on the activation loop and the hydrophobic motif by upstream kinases, such as MST1/2 in the canonical Hippo pathway and MAP4K family kinases as recently reported (38–41). It was also reported that phosphorylation of LATS2 by Aurora-A promotes its centrosomal localization (42). Recently, we have reported that Src inactivates LATS1 through direct phosphorylation on tyrosine residues (36). These investigations highlight the importance of phosphorylation in regulation of LATS1/2. However, the whole picture is more complicated. For instance, LATS1/2 are also regulated on the expression level by microRNAs, such as miR-372 and miR-373 (43). Furthermore, LATS1/2 protein levels are regulated by ubiquitination and proteasome-mediated degradation. LATS1 could be ubiquitinated by ubiquitin ligases Itch, WWP1, and NEDD4, and LATS2 could be ubiquitinated by CRL4DCAF1, NEDD4, and SIAH2 (22–24, 44–47). In this study, we found that USP9X is a DUB deubiquitinating LATS2 but not LATS1. LATS1 does not interact with USP9X, and repression of USP9X does not consistently decrease LATS1 protein level. It is interesting to note that despite functional redundancy of LATS1 and LATS2 in many circumstances, they could be individually regulated beyond what has been expected (e.g. LATS1 by Src (36) and LATS2 by USP9X or by YAP in the negative feedback loop (48–50)). Therefore, the impact of differential regulations of LATS1 and LATS2 in development and diseases should not be neglected.
In this study, we have also observed the differential contribution of LATS2 protein domains to its dynamic subcellular localization. Previous studies have proposed that interaction of LATS1/2 with MOB1A/B or NF2 could promote LATS1/2 recruitment to the plasma membrane, where they are activated by upstream kinases (51, 52). However, in this study, we found that the N-terminal 1–30 region of LATS2, which is different from mapped MOB1A/B- or NF2-interacting regions, mediates LATS2 distribution to the membrane fraction, especially intracellular vesicle structures. Importantly, deletion of this region not only inhibits interaction with APC1 and USP9X, but also impairs LATS2 kinase activity. Furthermore, we found that LATS2 is actively translocating between cytoplasm and the nucleus so that inhibition of nuclear export leads to nuclear accumulation of LATS2. In addition, the N-terminal domain of LATS2 (residues 1–625) exhibits clear nuclear localization. Interestingly, it was previously reported that LATS2 translocates to the nucleus following microtubule disruption (53). These observations raise the possibility of YAP phosphorylation by LATS2 in either the cytoplasm or the nucleus, which may have differential physiological consequences. The investigation of LATS2 subcellular localization in this work thus suggests the study of signals and mechanisms governing LATS2 subcellular localization to be an important direction in the future.
Our results also suggest potential regulation of APC/C by LATS2. In yeast, the LATS2 homolog Dbf2p is central to the mitotic exit network that regulates APC/C through Cdc14 phosphatase (54). However, no evidence supports the conservation of such a mechanism in mammals so far. Nevertheless, there is a report showing that LATS1/2 phosphorylates CDC26, which is also in our list of LATS2-interacting proteins identified by TAP, thus regulating APC/C assembly (55). The findings in this report suggest that LATS2 may further regulate APC/C through direct phosphorylation of APC1 or through binding to APC1 to phosphorylate other components of APC/C. Interestingly, there is evidence that activation of LATS2 is involved in cell cycle arrest induced by tetraploidy or expression of oncogenes (56, 57). The regulation of APC/C by LATS2 thus deserves further investigation.
The human genome encodes only around 95 DUBs. Therefore, it is certain that each DUB would have many substrates, which may mediate various biological functions. For USP9X, several substrates have been identified, such as MCL1, Smad4, and β-catenin, which are responsible for the oncogenic roles of USP9X in different types of cancer, such as lymphoma, multiple myeloma, melanoma, and breast cancer (26, 58, 59). However, a role of USP9X as a potent tumor suppressor in PDAC was shown by genetic screen in mice (27). Mutation of Usp9x was found in >50% of tumors, and it is the most mutated gene in this screen. Further experiments confirmed that loss of USP9X cooperated with KrasG12D to promote PDAC in vivo. However, the substrate of USP9X mediating the tumor-suppressive function of USP9X in PDAC remains obscure. In this study, we identified LATS2 as a critical substrate mediating the function of USP9X in pancreatic cancer cells through inhibition of YAP. Interestingly, YAP was found to be activated in relapsed PDAC in mouse models after cessation of KrasG12D expression and enables bypass of Kras addiction to sustain tumor growth (60). Many other reports also support the critical role of YAP in PDAC progression (61). Thus, the Hippo pathway probably mediates the tumor-suppressive function of USP9X in PDAC. The seemingly paradoxical roles of USP9X in different types of cancer might be explained by many reasons (e.g. differential oncogene addition and differential transcription level of various USP9X substrates in different cancers).
During the preparation of this manuscript, a study was published reporting a similar finding of USP9X as deubiquitylase of LATS1/2 (62). This study is consistent with our findings that USP9X stabilizes LATS2 and thus represses YAP to inhibit tumorigenesis of PDAC. Our study differs from this report in that we found USP9X is specific for LATS2 but not LATS1, suggesting that the ratio of LATS1 and LATS2 expression levels could also be an important factor in determining whether YAP activity could be regulated by USP9X. Our study provides a mechanism for the tumor-suppressive function of USP9X in pancreatic cancer and points to a new direction for therapy design.
Experimental procedures
Antibodies, plasmids, and other materials
We obtained anti-LATS2, anti-USP9X, anti-APC1, and anti-APC5 antibodies from Bethyl Laboratories; anti-K48-linked Ub, anti-LATS1, anti-phospho-YAP (Ser-127), and anti-cyclin B1 from Cell Signaling Technologies; anti-GST from Genscript; anti-α-tubulin and anti-FLAG from Sigma; anti-YAP from Santa Cruz Biotechnology, Inc.; anti-Hsp90 from BD Biosciences; anti-HA from Covance; anti-active-YAP from Abcam; Alexa Fluor 488– or 594–conjugated secondary antibodies from Thermo Fisher Scientific; and horseradish peroxidase–conjugated secondary antibodies from GE Healthcare.
pcDNA3-HA-LATS2 and pCDNA3-HA-LATS1 were described before (12), and then LATS2 was subcloned into pRK7-FLAG, pRK7-Myc, and pBABE-FLAG-SBP vectors. cDNA of USP9X was a gift from Dr. Dario R. Alessi (63) and was then subcloned into pRK7-FLAG and PB[Act-IRES-RFP]DS vectors. USP9X-catalytic domain was subcloned into pGEX-KG vector. APC1 was cloned into pDEST-HA vector from pDONR201 vector using gateway technology. Then APC1 was subcloned into pRK7-FLAG and pCDNA3-HA vectors. Human APC2, APC5, and CDC23 were cloned from the ORF LITE Clones library, and human APC7 was cloned from HeLa cDNA library and then subcloned into pRK7-FLAG and pCDNA3-HA vectors. Point mutations were generated by site-directed mutagenesis. Truncation mutants of LATS2, APC1, and USP9X were generated by subcloning.
Proteasome inhibitor bortezomib and DUB inhibitor PR-619 were purchased from Selleck, and APC/C inhibitors apcin and proTAME were from Boston Biochem. Leptomycin B was from Merck. All other chemicals were from Sigma or Sangon Biotech.
Cell culture, transfection, and viral infection
HEK293T, HeLa, HEK293P, MIA PaCa-2, BT-474, CFPAC-1, and MDA-MB-468 cells were cultured in DMEM (Thermo Fisher Scientific) containing 10% FBS (Thermo Fisher Scientific) and 50 μg/ml penicillin/streptomycin (P/S). AsPC-1 and BxPC-3 cells were cultured in RPMI1640 (Thermo Fisher Scientific) containing 10% FBS and 50 μg/ml P/S. MCF10A was cultured in DMEM/F-12 medium (Thermo Fisher Scientific) supplemented with 5% horse serum (Thermo Fisher Scientific), 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 100 ng/ml cholera toxin, and 50 μg/ml P/S. Plasmids were transfected with Lipofectamine (Thermo Fisher Scientific), which was performed according to the manufacturer's instructions. For siRNA transfection, we used Lipofectamine RNAiMax reagent (Thermo Fisher Scientific). For retroviral infection, HEK293P cells were used to package viral constructs that co-transfected with packaging plasmids for 48 h. Then viral supernatant was supplemented with 5 μg/ml Polybrene and applied for infection of target cells after filtering through a 0.45-μm filter. To generate USP9X re-expression cells from USP9X-knockout cells, we used the piggyBac transposon system. USP9X-knockout cells were transfected with PB[Act-FLAG-USP9X-IRES-RFP]DS and the Act-PB transposase plasmids. After 48 h, cells were collected and seeded at low density for colony formation. Individual clones were examined by Western blotting.
RNA interference
To generate shRNAs against human USP9X, oligonucleotides were cloned into pLKO.1 with the AgeI/EcoRI sites. Sequences of the sense oligonucleotides are as shown in Table 1.
Table 1.
Sense oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| USP9X#1 | GAGAGTTTATTCACTGTCTTA |
| USP9X#2 | CGCCTGATTCTTCCAATGAAA |
| USP9X#3 | CGATTCTTCAAAGCTGTGAAT |
siRNAs against human USP9X, APC1, LATS2, YAP, and TAZ were synthesized by RiboBio and are shown in Table 2.
Table 2.
siRNAs against human USP9X, APC1, LATS2, YAP, and TAZ
| siRNA | Sequence |
|---|---|
| USP9X#1 | AGAAATCGCTGGTATAAAT |
| USP9X#2 | TCTCAATAATGAAATTGAT |
| USP9X#3 | GAAATAACTTCCTACCGAA |
| LATS2#1 | GTTCGGACCTTATCAGAAA |
| LATS2#2 | GATCGGTGCCTTTGGAGAA |
| ANAPC1#1 | TCCTTGGATCATTGGACGA |
| ANAPC1#2 | CCAACGCTTGTCAGAACTA |
| YAP#1 | CCACCAAGCTAGATAAAGA |
| YAP#2 | GGTCAGAGATACTTCTTAA |
| YAP#3 | GCACCTATCACTCTCGAGA |
| TAZ#1 | CCAAATCTCGTGATGAATC |
| TAZ#2 | CCGCAGGGCTCATGAGTAT |
| TAZ#3 | AGGAACAAACGTTGACTTA |
CRISPR-Cas9–mediated knockout
We used CRISPR-Cas9 technology to generate USP9X-knockout cells. Two independent sgRNAs were designed by the Zhang laboratory CRISPR Design Tool. Annealed oligonucleotides were ligated into the PEP-KO vector digested with SapI. Cells were transfected with plasmids for 48 h and then selected with 2 μg/ml puromycin for another 48 h. Live cells were collected and seeded at low density for colony formation. Individual clones were examined by Western blotting. sgRNA hairpin sequences were as follows: TGCATTTCCACATACTGACT (USP9Xsg1) and GTGTCGAGTATGACAGCCAC (USP9Xsg2).
Immunoprecipitation and kinase assay
For LATS2 kinase assays, HEK293T cells were transfected with indicated plasmids. 48 h later, cells were lysed with mild lysis buffer (MLB) (10 mm Tris, pH 7.5, 100 mm NaCl, 10 mm EDTA, 0.5% Nonidet P-40, 50 mm NaF) with additive (1.5 mm Na3VO4, protease inhibitor mixture (Roche Applied Science), 1 mm DTT, and 1 mm PMSF) and immunoprecipitated with anti-HA antibody. The immunoprecipitates were washed three times with MLB and once with kinase assay buffer (30 mm HEPES, 50 mm potassium acetate, 5 mm MgCl2). The immunoprecipitated LATS2 was subjected to a kinase assay in the presence of 500 μm ATP and 1 μg of recombinant GST-YAP purified from E. coli as substrate. The reaction mixtures were incubated for 30 min at 30 °C. The reactions were terminated with SDS loading buffer.
Tandem affinity purification
TAP purification was performed as described (64). Briefly, MCF10A stable cells were lysed with buffer containing 0.3% CHAPS. Lysates were then incubated with anti-FLAG antibody–conjugated resin for 2 h at 4 °C and washed three times with lysis buffer. Then bound proteins were eluted with elution buffer containing 200 ng/μl 3× FLAG peptide. The eluates were further incubated with streptavidin-conjugated resin for 2 h at 4 °C. Washed resins were eluted with buffer containing 4 mm biotin. Samples were then processed for MS/MS analysis.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 for 5 min. After blocking in 3% BSA for 30 min, slides were incubated with first antibody diluted in 3% BSA overnight at 4 °C. Slides were washed with PBS and then incubated with Alexa Fluor 488– or 594–conjugated secondary antibodies for 45 min. Slides were then washed and mounted.
RNA isolation and real-time PCR
Total RNA was isolated from cells using TRIzol reagent (Thermo Fisher Scientific). cDNA was synthesized by reverse transcription using PrimeScrip RT Master Mix (Takara) and subjected to real-time PCR with gene-specific primers in the presence of SYBR Green (Applied Biosystems). Relative abundance of mRNA was calculated by normalization to GAPDH mRNA.
In vivo ubiquitination assay
HeLa HA-Ub stable cells were transfected with the indicated plasmids and then treated with bortezomib (100 nm) for 24 h and PR-619 (20 mm) for 1 h. Then cells were lysed with heated lysis buffer (10 mm Tris, pH 7.5, 100 mm NaCl, 10 mm EDTA, 1% SDS, 0.5% Nonidet P-40, 50 mm NaF, 1.5 mm Na3VO4, protease inhibitor mixture (Roche Applied Science), 1 mm DTT, 1 mm PMSF, 20 mm N-ethylmaleimide) and immunoprecipitated with anti-FLAG or anti-HA antibodies after 1:10 dilution with MLB. The immunoprecipitates were washed three times with MLB containing 0.1% SDS. Then samples were boiled before Western blot analysis.
In vitro deubiquitination assay
LATS2 was immunoprecipitated as that in the in vivo ubiquitination assay. The immunoprecipitated FLAG-LATS2 was then subjected to a deubiquitination assay in buffer containing 50 mm Tris, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 5 mm DTT in the presence of 2 μg of recombinant GST-USP9X-catalytic domain purified from E. coli as DUB. The reaction mixtures were incubated for 2 h at 37 °C. The reactions were terminated with SDS loading buffer and boiled before Western blotting analysis.
Flow cytometry
Cells were labeled with 50 μm EdU (RiboBio) for 45 min in the culture medium, and then cells were trypsinized and fixed with 4% paraformaldehyde for 30 min and then neutralized with 2 mg/ml glycine. After permeabilization with 0.1% Triton X-100 for 10 min, cells were stained according to instructions of the RiboBio EdU staining kit. Then DNA was stained with 7-AAD (BD Biosciences) for 20 min at room temperature. Cells were then resuspended in PBS for analysis on a Beckman Cytomic FC 500MCL. Data were analyzed using FlowJo software.
Soft agar colony formation assay
MIA PaCa-2 cells (1 × 103) in 1 ml of growth medium were mixed with 2 ml of 0.5% agarose and layered onto 2 ml of 0.6% agarose beds in 6-well plates. Cells were fed with 2 ml of growth medium every week for 4 weeks, after which colonies were stained, photographed, and counted with ImageJ.
Xenograft tumorigenesis model
All animal study protocols were approved by the Zhejiang University Animal Care and Use Committee. Nude mice (nu/nu, male 6–8-week-old) were injected subcutaneously with 7.5 × 105 MIA PaCa-2 cells. Around 5–6 weeks later, tumors were dissected, photographed, and weighted.
Subcellular fractionation
HeLa cells were lysed with low-salt buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA) with additive (2.5 mm NaF, protease inhibitor mixture (Roche Applied Science), 1 mm DTT, and 1 mm PMSF). After incubation on ice for 10 min, cells were passed through a 25-gauge needle for 10 strokes. After centrifugation at 1000 × g for 10 min at 4 °C, the pellet was washed and collected as nuclear fraction. The supernatant was processed for centrifugation at 1 × 104 × g for 5 min, and the pellet was washed and collected as mitochondrial fraction. The supernatant was further applied for another centrifugation at 1.5 × 105 × g for 40 min. Then the pellet was collected as membrane fraction, and the remaining supernatant was cytoplasmic fraction. Nuclear, membrane, and cytoplasmic fractions were assessed by Western blotting with lamin A/C, Na+/K+-ATPase, and tubulin as loading controls, respectively.
Author contributions
C.Z. and B.Z. conceptualization; C.Z., X.J., and B.Z. data curation; C.Z., X.J., H.Z., Q.Z., X.C., M.T., Y.S., and H.Y. investigation; C.Z., X.J., Q.Z., L.L., and T.L. methodology; C.Z. and B.Z. writing-original draft; C.Z., X.J., H.Z., Q.Z., X.C., M.T., L.L., T.L., X.-H.F., and B.Z. writing-review and editing; L.L., X.-H.F., and B.Z. supervision; B.Z. formal analysis; B.Z. funding acquisition; B.Z. project administration.
Supplementary Material
Acknowledgments
We thank Drs. Zongping Xia and Lin Hu for technical assistance, Dr. Dario R. Alessi for the USP9X cDNA, and Dr. Fangwei Wang for the HeLa cell line.
This work was supported by National Key R&D Program of China Grant 2017YFA0504502, National Natural Science Foundation of China Key Project Grant 81730069, Excellent Young Scholars Project Grant 31422036, General Project Grant 31471316, International Collaboration Project Grant 31661130150, 111 Project Grant B13026, the Qianjiang Scholar Plan of Hangzhou, the Thousand Young Talents Plan of China, and the Academy of Medical Sciences (United Kingdom) Newton Advanced Fellowship (to B. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- YAP
- Yes-associated protein
- EdU
- 5-ethynyl-2′-deoxyuridine
- P/S
- penicillin/streptomycin
- TAZ
- transcriptional coactivator with PDZ-binding motif
- DUB
- deubiquitylase
- USP
- ubiquitin-specific protease
- PDAC
- pancreatic ductal adenocarcinoma
- APC/C
- anaphase-promoting complex/cyclosome
- TAP
- tandem affinity purification
- SBP
- streptavidin-binding peptide
- AMOT
- angiomotin
- Ub
- ubiquitin
- 7-AAD
- 7-aminoactinomycin D
- MLB
- mild lysis buffer
- sgRNA
- single guide RNA.
References
- 1. Pan D. (2010) The Hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu F. X., Zhao B., and Guan K. L. (2015) Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu S., Huang J., Dong J., and Pan D. (2003) hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 10.1016/S0092-8674(03)00549-X [DOI] [PubMed] [Google Scholar]
- 4. Song H., Mak K. K., Topol L., Yun K., Hu J., Garrett L., Chen Y., Park O., Chang J., Simpson R. M., Wang C. Y., Gao B., Jiang J., and Yang Y. (2010) Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 107, 1431–1436 10.1073/pnas.0911409107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Udan R. S., Kango-Singh M., Nolo R., Tao C., and Halder G. (2003) Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 10.1038/ncb1050 [DOI] [PubMed] [Google Scholar]
- 6. Pantalacci S., Tapon N., and Léopold P. (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 10.1038/ncb1051 [DOI] [PubMed] [Google Scholar]
- 7. Harvey K. F., Pfleger C. M., and Hariharan I. K. (2003) The Drosophila Mst ortholog, Hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 10.1016/S0092-8674(03)00557-9 [DOI] [PubMed] [Google Scholar]
- 8. Jia J., Zhang W., Wang B., Trinko R., and Jiang J. (2003) The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519 10.1101/gad.1134003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D., and Hariharan I. K. (2002) salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- 10. Moroishi T., Hansen C. G., and Guan K. L. (2015) The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., and Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hao Y., Chun A., Cheung K., Rashidi B., and Yang X. (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 10.1074/jbc.M709037200 [DOI] [PubMed] [Google Scholar]
- 14. Lei Q. Y., Zhang H., Zhao B., Zha Z. Y., Bai F., Pei X. H., Zhao S., Xiong Y., and Guan K. L. (2008) TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol. Cell Biol. 28, 2426–2436 10.1128/MCB.01874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oka T., Mazack V., and Sudol M. (2008) Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J. Biol. Chem. 283, 27534–27546 10.1074/jbc.M804380200 [DOI] [PubMed] [Google Scholar]
- 16. Zhao B., Li L., Tumaneng K., Wang C. Y., and Guan K. L. (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J., and Zider A. (2008) SCALLOPED interacts with YORKIE, the nuclear effector of the Hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435–441 10.1016/j.cub.2008.02.034 [DOI] [PubMed] [Google Scholar]
- 18. Wu S., Liu Y., Zheng Y., Dong J., and Pan D. (2008) The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 19. Zhang L., Ren F., Zhang Q., Chen Y., Wang B., and Jiang J. (2008) The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., and Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang H., Liu C. Y., Zha Z. Y., Zhao B., Yao J., Zhao S., Xiong Y., Lei Q. Y., and Guan K. L. (2009) TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 284, 13355–13362 10.1074/jbc.M900843200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma B., Chen Y., Chen L., Cheng H., Mu C., Li J., Gao R., Zhou C., Cao L., Liu J., Zhu Y., Chen Q., and Wu S. (2015) Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 17, 95–103 [DOI] [PubMed] [Google Scholar]
- 23. Bae S. J., Kim M., Kim S. H., Kwon Y. E., Lee J. H., Kim J., Chung C. H., Lee W. J., and Seol J. H. (2015) NEDD4 controls intestinal stem cell homeostasis by regulating the Hippo signalling pathway. Nat. Commun. 6, 6314 10.1038/ncomms7314 [DOI] [PubMed] [Google Scholar]
- 24. Li W., Cooper J., Zhou L., Yang C., Erdjument-Bromage H., Zagzag D., Snuderl M., Ladanyi M., Hanemann C. O., Zhou P., Karajannis M. A., and Giancotti F. G. (2014) Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the Hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 26, 48–60 10.1016/j.ccr.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murtaza M., Jolly L. A., Gecz J., and Wood S. A. (2015) La FAM fatale: USP9X in development and disease. Cell Mol. Life Sci. 72, 2075–2089 10.1007/s00018-015-1851-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwickart M., Huang X., Lill J. R., Liu J., Ferrando R., French D. M., Maecker H., O'Rourke K., Bazan F., Eastham-Anderson J., Yue P., Dornan D., Huang D. C., and Dixit V. M. (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463, 103–107 10.1038/nature08646 [DOI] [PubMed] [Google Scholar]
- 27. Pérez-Mancera P. A., Rust A. G., van der Weyden L., Kristiansen G., Li A., Sarver A. L., Silverstein K. A., Grutzmann R., Aust D., Rümmele P., Knösel T., Herd C., Stemple D. L., Kettleborough R., Brosnan J. A., et al. (2012) The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486, 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Couzens A. L., Knight J. D., Kean M. J., Teo G., Weiss A., Dunham W. H., Lin Z. Y., Bagshaw R. D., Sicheri F., Pawson T., Wrana J. L., Choi H., and Gingras A. C. (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 6, rs15 10.1126/scisignal.2004712 [DOI] [PubMed] [Google Scholar]
- 29. Wang W., Li X., Huang J., Feng L., Dolinta K. G., and Chen J. (2014) Defining the protein-protein interaction network of the human Hippo pathway. Mol. Cell. Proteomics 13, 119–131 10.1074/mcp.M113.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim M., Kim M., Park S. J., Lee C., and Lim D. S. (2016) Role of angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 17, 64–78 10.15252/embr.201540809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thanh Nguyen H., Andrejeva D., Gupta R., Choudhary C., Hong X., Eichhorn P. J., Loya A. C., and Cohen S. M. (2016) Deubiquitylating enzyme USP9x regulates Hippo pathway activity by controlling angiomotin protein turnover. Cell Discov. 2, 16001 10.1038/celldisc.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Primorac I., and Musacchio A. (2013) Panta rhei: the APC/C at steady state. J. Cell Biol. 201, 177–189 10.1083/jcb.201301130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bothos J., Tuttle R. L., Ottey M., Luca F. C., and Halazonetis T. D. (2005) Human LATS1 is a mitotic exit network kinase. Cancer Res. 65, 6568–6575 10.1158/0008-5472.CAN-05-0862 [DOI] [PubMed] [Google Scholar]
- 34. Sackton K. L., Dimova N., Zeng X., Tian W., Zhang M., Sackton T. B., Meaders J., Pfaff K. L., Sigoillot F., Yu H., Luo X., and King R. W. (2014) Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514, 646–649 10.1038/nature13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo X., Zhao Y., Yan H., Yang Y., Shen S., Dai X., Ji X., Ji F., Gong X. G., Li L., Bai X., Feng X. H., Liang T., Ji J., Chen L., et al. (2017) Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 31, 247–259 10.1101/gad.294348.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Si Y., Ji X., Cao X., Dai X., Xu L., Zhao H., Guo X., Yan H., Zhang H., Zhu C., Zhou Q., Tang M., Xia Z., Li L., Cong Y. S., et al. (2017) Src inhibits the Hippo tumor suppressor pathway through tyrosine phosphorylation of Lats1. Cancer Res. 77, 4868–4880 10.1158/1538-7445.AM2017-4868 [DOI] [PubMed] [Google Scholar]
- 37. Zhu C., Li L., and Zhao B. (2015) The regulation and function of YAP transcription co-activator. Acta Biochim. Biophys. Sin. (Shanghai) 47, 16–28 10.1093/abbs/gmu110 [DOI] [PubMed] [Google Scholar]
- 38. Li Q., Li S., Mana-Capelli S., Roth Flach R. J., Danai L. V., Amcheslavsky A., Nie Y., Kaneko S., Yao X., Chen X., Cotton J. L., Mao J., McCollum D., Jiang J., Czech M. P., et al. (2014) The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev. Cell 31, 291–304 10.1016/j.devcel.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li S., Cho Y. S., Yue T., Ip Y. T., and Jiang J. (2015) Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discov. 1, 15038 10.1038/celldisc.2015.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meng Z., Moroishi T., Mottier-Pavie V., Plouffe S. W., Hansen C. G., Hong A. W., Park H. W., Mo J. S., Lu W., Lu S., Flores F., Yu F. X., Halder G., and Guan K. L. (2015) MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6, 8357 10.1038/ncomms9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng Y., Wang W., Liu B., Deng H., Uster E., and Pan D. (2015) Identification of Happyhour/MAP4K as alternative Hpo/Mst-like kinases in the Hippo kinase cascade. Dev. Cell 34, 642–655 10.1016/j.devcel.2015.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Toji S., Yabuta N., Hosomi T., Nishihara S., Kobayashi T., Suzuki S., Tamai K., and Nojima H. (2004) The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells 9, 383–397 10.1111/j.1356-9597.2004.00732.x [DOI] [PubMed] [Google Scholar]
- 43. Voorhoeve P. M., le Sage C., Schrier M., Gillis A. J., Stoop H., Nagel R., Liu Y. P., van Duijse J., Drost J., Griekspoor A., Zlotorynski E., Yabuta N., De Vita G., Nojima H., Looijenga L. H., and Agami R. (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 10.1016/j.cell.2006.02.037 [DOI] [PubMed] [Google Scholar]
- 44. Yeung B., Ho K. C., and Yang X. (2013) WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS One 8, e61027 10.1371/journal.pone.0061027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salah Z., Melino G., and Aqeilan R. I. (2011) Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 71, 2010–2020 10.1158/0008-5472.CAN-10-3516 [DOI] [PubMed] [Google Scholar]
- 46. Salah Z., Cohen S., Itzhaki E., and Aqeilan R. I. (2013) NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle 12, 3817–3823 10.4161/cc.26672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ho K. C., Zhou Z., She Y. M., Chun A., Cyr T. D., and Yang X. (2011) Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability [corrected]. Proc. Natl. Acad. Sci. U.S.A. 108, 4870–4875 10.1073/pnas.1101273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Q., Zhang N., Xie R., Wang W., Cai J., Choi K. S., David K. K., Huang B., Yabuta N., Nojima H., Anders R. A., and Pan D. (2015) Homeostatic control of Hippo signaling activity revealed by an endogenous activating mutation in YAP. Genes Dev. 29, 1285–1297 10.1101/gad.264234.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dai X., Liu H., Shen S., Guo X., Yan H., Ji X., Li L., Huang J., Feng X. H., and Zhao B. (2015) YAP activates the Hippo pathway in a negative feedback loop. Cell Res. 25, 1175–1178 10.1038/cr.2015.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moroishi T., Park H. W., Qin B., Chen Q., Meng Z., Plouffe S. W., Taniguchi K., Yu F. X., Karin M., Pan D., and Guan K. L. (2015) A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284 10.1101/gad.262816.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hergovich A., Schmitz D., and Hemmings B. A. (2006) The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 345, 50–58 10.1016/j.bbrc.2006.03.244 [DOI] [PubMed] [Google Scholar]
- 52. Yin F., Yu J., Zheng Y., Chen Q., Zhang N., and Pan D. (2013) Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355 10.1016/j.cell.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aylon Y., Michael D., Shmueli A., Yabuta N., Nojima H., and Oren M. (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 20, 2687–2700 10.1101/gad.1447006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hergovich A., Stegert M. R., Schmitz D., and Hemmings B. A. (2006) NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7, 253–264 10.1038/nrm1891 [DOI] [PubMed] [Google Scholar]
- 55. Masuda K., Chiyoda T., Sugiyama N., Segura-Cabrera A., Kabe Y., Ueki A., Banno K., Suematsu M., Aoki D., Ishihama Y., Saya H., and Kuninaka S. (2015) LATS1 and LATS2 phosphorylate CDC26 to modulate assembly of the tetratricopeptide repeat subcomplex of APC/C. PLoS One 10, e0118662 10.1371/journal.pone.0118662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aylon Y., Yabuta N., Besserglick H., Buganim Y., Rotter V., Nojima H., and Oren M. (2009) Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene 28, 4469–4479 10.1038/onc.2009.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ganem N. J., Cornils H., Chiu S., O'Rourke K. P., Arnaud J., Yimlamai D., Thery M., Camargo F., and Pellman D. (2014) Cytokinesis failure triggers Hippo tumor suppressor pathway activation. Cell 158, 833–848 10.1016/j.cell.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M. J., Soligo S., Morsut L., Inui M., Moro S., Modena N., Argenton F., Newfeld S. J., and Piccolo S. (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 10.1016/j.cell.2008.10.051 [DOI] [PubMed] [Google Scholar]
- 59. Taya S., Yamamoto T., Kanai-Azuma M., Wood S. A., and Kaibuchi K. (1999) The deubiquitinating enzyme Fam interacts with and stabilizes β-catenin. Genes Cells 4, 757–767 10.1046/j.1365-2443.1999.00297.x [DOI] [PubMed] [Google Scholar]
- 60. Kapoor A., Yao W., Ying H., Hua S., Liewen A., Wang Q., Zhong Y., Wu C. J., Sadanandam A., Hu B., Chang Q., Chu G. C., Al-Khalil R., Jiang S., Xia H., et al. (2014) Yap1 activation enables bypass of oncogenic kras addiction in pancreatic cancer. Cell 158, 185–197 10.1016/j.cell.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang W., Nandakumar N., Shi Y., Manzano M., Smith A., Graham G., Gupta S., Vietsch E. E., Laughlin S. Z., Wadhwa M., Chetram M., Joshi M., Wang F., Kallakury B., Toretsky J., et al. (2014) Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci. Signal. 7, ra42 10.1126/scisignal.2005049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Toloczko A., Guo F., Yuen H. F., Wen Q., Wood S. A., Ong Y. S., Chan P. Y., Shaik A. A., Gunaratne J., Dunne M. J., Hong W., and Chan S. W. (2017) Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the Hippo pathway. Cancer Res. 77, 4921–4933 10.1158/1538-7445.AM2017-4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., and Alessi D. R. (2008) Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 411, 249–260 10.1042/BJ20080067 [DOI] [PubMed] [Google Scholar]
- 64. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., and Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.