Abstract
Protein-disulfide isomerase-like protein of the testis (PDILT), a member of the protein-disulfide isomerase family, is a chaperone essential for the folding of spermatogenesis-specific proteins in male postmeiotic germ cells. However, the structural mechanisms that regulate the chaperone function of PDILTs are unknown. Here, we report the structures of human PDILT (hPDILT) determined by X-ray crystallography to 2.4 Å resolution and small-angle X-ray scattering (SAXS). Distinct from previously reported U-like structures of related PDI family proteins, our structures revealed that hPDILT folds into a compact L-like structure in crystals and into an extended chain-like structure in solution. The hydrophobic regions and the hydrophobic pockets in hPDILT, which are important for substrate recognition, were clearly delineated in the crystal structure. Moreover, our results of the SAXS analysis and of structure-based substitutions and truncations indicated that the C-terminal tail in hPDILT is required for suppression of aggregation of denatured proteins, suggesting that the tail is crucial for the chaperone activity of PDILT. Taken together, our findings have identified the critical regions and conformational changes of PDILT that enable and control its activity. These results advance our understanding of the structural mechanisms involved in the chaperone activity of PDILT.
Keywords: conformational change, molecular chaperone, protein structure, small-angle X-ray scattering (SAXS), X-ray crystallography
Introduction
Quality control of its proteins is essential for the survival of a cell, the proper function of organs, and the homeostasis of biological systems. Dysfunction of the cellular quality control system results in the accumulation of misfolded proteins, causing severe consequences such as Alzheimer's and Parkinson's diseases (1, 2). Molecular chaperones in the endoplasmic reticulum (ER)4 are important players to ensure protein quality control (3). Protein-disulfide isomerase (PDI) family protein is a type of molecular chaperone for secretory proteins. They assist protein folding by recognizing and binding to partially folded intermediates through hydrophobic interactions, thereby preventing their aggregation and promoting correct folding into their native structure (4). In addition, most of the PDI family proteins catalyze the formation of the disulfide bond in nascent polypeptides as oxidoreductase/isomerase, using canonical CXXC motifs as the redox-active catalytic sites (5, 6).
The PDI family members are involved in various cellular events, including ER-associated degradation (7), trafficking (8), calcium homeostasis (9), antigen presentation (10), and pathogen entry (11, 12). Among these family members, only expression of protein-disulfide isomerase-like protein of the testis (PDILT) has been shown to be controlled by developmental programs (13). PDILT lacks oxidoreductase activity due to mutation of the redox-active catalytic sites in CXXC motifs to SKQS and SKKC (14). Although recent studies identified PDILT as a novel, major gonadal auto-antigen in autoimmune polyendocrine syndrome type 1 (APS1) (15), the functional investigations of PDILT focused on its chaperone activity. It has been reported for a mouse system that PDILT resides exclusively in the ER of male postmeiotic germ cells (13), where PDILT cooperates with the testis-specific lectin chaperones, including calreticulin-3 and calmegin. These three proteins form a chaperone complex that assists the proper folding of spermatogenesis-specific proteins (16). For example, ADAM3 (a disintegrin and metalloproteinase 3), a sperm membrane protein critical for sperm migration and fertilization, is unable to fold properly in the absence of PDILT, preventing transportation to the sperm surface, resulting in mouse infertility (16). In addition, PDILT was shown to bind to unfolded substrates such as Δ-somatostatin peptide and unfolded bovine pancreatic trypsin inhibitor (13), supporting speculation that it acts as a molecular chaperone.
Similar to PDI, a prototype of the PDI family, PDILT consists of four tandem thioredoxin-like domains (a, b, b′, and a′), an x-linker between b′ and a′ domains, and a C-terminal tail. In the crystal structure of the b′–x-linker domains of PDILT, the b′ domain interacts with the aromatic residues in the x-linker of the adjacent molecule, which was considered as a structure showing the substrate recognition (17). However, the structure of full-length PDILT and the mechanisms of its chaperone function still need to be investigated. Here, we show both crystal structure and solution structure of full-length human PDILT (hPDILT), using X-ray diffraction and small-angle X-ray scattering (SAXS) methods. Based on the structural information, we analyzed the chaperone activity of hPDILT using in vitro biochemical assays. Our results provide the structural basis that should help to better understand the precise mechanisms of PDILT chaperone activity.
Results
Chaperone activity of hPDILT
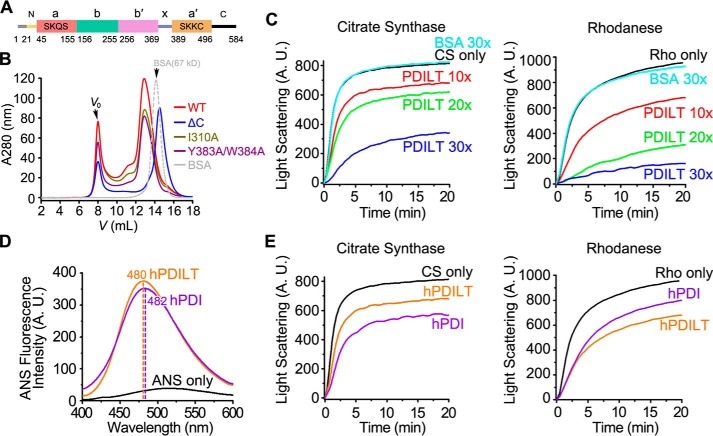
The full-length hPDILT consisting of 584 amino acids divided into 9 regions, including the signal peptide (Met-1–Ser-20), N-terminal loop (Ser-21–Ser-44), a domain (Leu-45–Lys-155), b domain (Ala-156–Thr-255), b′ domain (Asp-256–Lys-369), x-linker (Asn-370–Leu-388), a′ domain (Val-389–Lys-496), C-terminal tail (Ile-497–Val-580), and the ER retrieval peptide KEEL (Lys-581–Leu-584) (Fig. 1A). To study its structure and chaperone activity, we purified recombinant hPDILT proteins from Escherichia coli, which exhibited various assemble states as shown in size-exclusion chromatography (SEC) results (Fig. 1B). Protein samples (Ser-21–Leu-584) with predicted molecular mass at ∼80–140 kDa were used in the following chaperone activity studies.
Figure 1.
Chaperone activity of hPDILT. A, domain organization of hPDILT. The redox-inactive motifs are indicated in a and a′ domains. B, SEC profiles of WT, mutated, and C-terminally truncated hPDILT proteins. C, denatured and reduced citrate synthase (CS) (10 μm) or rhodanese (Rho) (22.5 μm) was 50-fold (for CS) or 100-fold (for Rho) diluted in the absence or presence of hPDILT or BSA as indicated in a molar ratio at 25 °C. A.U., arbitrary units. D, ANS fluorescence spectra of hPDILT and hPDI. 50 μm ANS was incubated without or with 5 μm hPDI and hPDILT for 20 min at 25 °C. ANS emission spectra were determined with excitation at 370 nm. E, chaperone activities of hPDILT and hPDI on CS or Rho were monitored as described in C. The molar ratio of hPDILT or hPDI to substrates is 10. A.U., arbitrary units.
The chaperone activity of hPDILT was first assessed by determining its ability to prevent the dithiothreitol (DTT)-induced aggregation of insulin B-chain (18, 19). At a molar ratio of 1:1, PDILT effectively protected the insulin B-chain from aggregation (Fig. S1). We also tested the chaperone activity of hPDILT by employing guanidine hydrochloride-denatured citrate synthase (CS) and rhodanese (Rho), which are widely used in chaperone activity assays (20–22), as substrates. The presence of hPDILT (Ser-21–Leu-584) in the refolding buffer prevents aggregation of denatured CS and Rho during the dilution-induced refolding (Fig. 1C). Increasing the ratio of hPDILT/substrate gradually enhanced the inhibition effects on substrate aggregation. In contrast, the negative control bovine serum albumin (BSA) failed to suppress substrate aggregation, even at a 30-fold concentration of substrate. As molecular chaperones utilize hydrophobic surfaces for the recognition of unfolded substrates, we measured the 1-anilino-8-naphthalene sulfonate (ANS) fluorescence of hPDILT. ANS has high affinity with the hydrophobic surface of protein, and higher hydrophobicity leads to stronger fluorescence and a blue-shift of the maximum emission peak. Compared with human PDI (hPDI), hPDILT showed similar ANS fluorescence intensity and maximum emission peak position, suggesting that hPDILT has similar exposed hydrophobic surface as hPDI (Fig. 1D). Consistent with this result, hPDILT showed chaperone activity comparable with that of hPDI on CS and Rho refolding (Fig. 1E). Together, these results indicated that hPDILT possesses chaperone activity in vitro.
Crystal structure of hPDILT
Our hPDILT crystal diffracted to 2.38 Å resolution, with space group of P32. The structure was solved using the molecular replacement method (23). Some unbiased areas of electron density after molecular replacement are shown in Fig. S2. The statistics for data collection and structure refinement are summarized in Table 1. Although we used recombinant hPDILT protein consisting of amino acids Ser-21–Val-580 for crystal growth (Fig. 2A), only amino acids Ser-32–Lys-495 are clearly distinguished in the final electron density map, covering parts of the N-terminal loop, four thioredoxin-like domains (a, b, b′, and a′), and the x-linker (Fig. 2B). This result suggests that the C-terminal tail of hPDILT is flexible in the crystal structure.
Table 1.
Statistics of crystallographic analysis
| Data collection | |
| Crystal | hPDILT |
| Wavelength (Å) | 0.979 |
| Space group | P32 |
| Cell dimensions | |
| a, b, c (Å) | 97.4, 97.4, 57.6 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 50.00–2.38 (2.42–2.38)a |
| Rsym | 0.066 (0.716) |
| Completeness (%) | 99.8 (100.0) |
| Redundancy | 3.8 (3.8) |
| I/σ(I) | 20.6 (2.0) |
| Refinement | |
| Resolution (Å) | 28.10–2.38 |
| No. of reflections | 24393 |
| Rwork /Rfree (%) | 19.7/23.5 |
| No. of atoms | |
| Protein/water | 3764/80 |
| B-Factors (Å2) | |
| Protein/water | 56.0/49.6 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.957 |
| Ramachandran statistics (%)b | |
| Most favored regions | 97.6 |
| Additional allowed regions | 2.4 |
| Disallowed regions | 0.0 |
| Side chain outliers | 0.0 |
| Clashscore | 0.0 |
a Highest resolution shell is shown in parentheses.
b Data were as defined by the validation suite MolProbity.
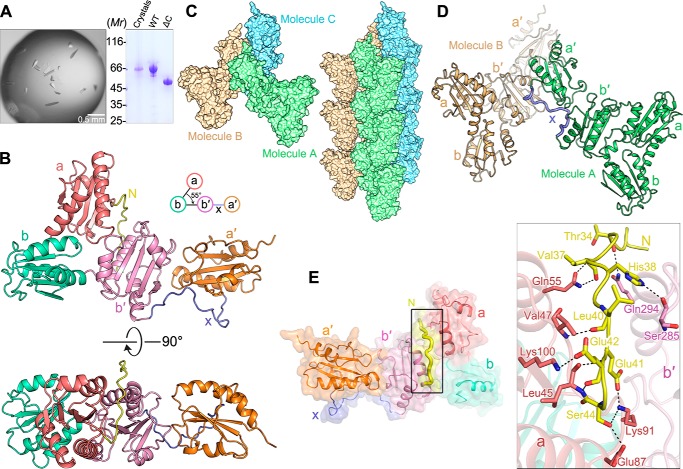
Figure 2.
Crystal structure of hPDILT. A, to test whether the C-terminal tail (Ile-497–Val-580) of hPDILT was degraded during crystallization, hPDILT crystals were washed with precipitant and dissolved in loading buffer for SDS-PAGE analysis. The band shown at 66 kDa suggests that the full-length hPDILT was present in the crystals. B, schematic diagrams of the crystal structure of hPDILT from the front view (upper panel) and the top view (lower panel). Color-coding of each domain is identical to that in Fig. 1A. C, crystal-packing reveals interactions between hPDILT molecules. D, x-linker of one hPDILT molecule A binds to the b′ domain of another symmetric molecule B. E, interactions between the N-terminal loop (Ser-32–Ser-44) and the thioredoxin-like a and b′ domains. Schematic and surface representation of hPDILT structure is colored as B. The N-terminal loop is highlighted in bold and yellow. Residues involved in the interaction are shown in stick representation on the right in the close-up view.
As seen in Fig. 2, C and D, the b′ domain of one hPDILT molecule interacts with the x-linker in another molecule. The interactions between b′ domain and x-linker are similar to those in the previously reported b′–x-linker structure (17) (Fig. S3). The hPDILT molecules regularly assemble into a fiber-like structure. In addition, the a domain of the hPDILT molecule in one fiber interacts with the b domain of hPDILT molecule in the adjacent fiber, using hydrogen bonds and electrostatic interactions (Fig. S4). Ser-72, Lys-73, Gln-74, Ser-75, Gln-110, Thr-116, and Lys-117 in the a domain of one molecule in the hPDILT fiber interact with Glu-185, Glu-186, Glu-189, and Asp-239 in the b domain of the other molecule in the adjacent fiber. These results provide the structural basis for the assembled states of recombinant hPDILT proteins (Fig. 1B).
Different from the canonical “U”-shaped crystal structures of other PDI family members so far reported, including bacterial DsbC (24), yeast Pdi1p (25), human ERp57 (26), and hPDI (Fig. S5) (27), our crystal structure of hPDILT resembles an “L”-shaped structure (Fig. 2B). The four thioredoxin-like domains are located in the same plane, whereas the b, b′, and a′ domains are arranged in a line, and the a domain is located above the line. In addition to the specific domain arrangement in hPDILT, the structures of the thioredoxin-like domains in hPDILT are different from the prototypical thioredoxin fold. The β-strands at the edges of the central β-sheet, β1 or β5, are shifted and twisted, leaving three or four-strands in the central β-sheets.
In the structure of hPDILT, the N-terminal loop (Ser-32–Ser-44) interacts with both a and b′ domains. Ten pairs of hydrogen bonds are observed in the structure, between residues Thr-34, Val-37, His-38, Leu-40, Glu-41, Glu-42, and Ser-44 in the N-terminal loop; Leu-45, Val-47, Gln-55, Glu-87, Lys-91, and Lys-100 in the a domain; and Ser-285 and Gln-294 in b′ domain (Fig. 2E). These interactions bring the a domain close to the b′ domain. Therefore, the first three thioredoxin-like domains form a compact triangular structure.
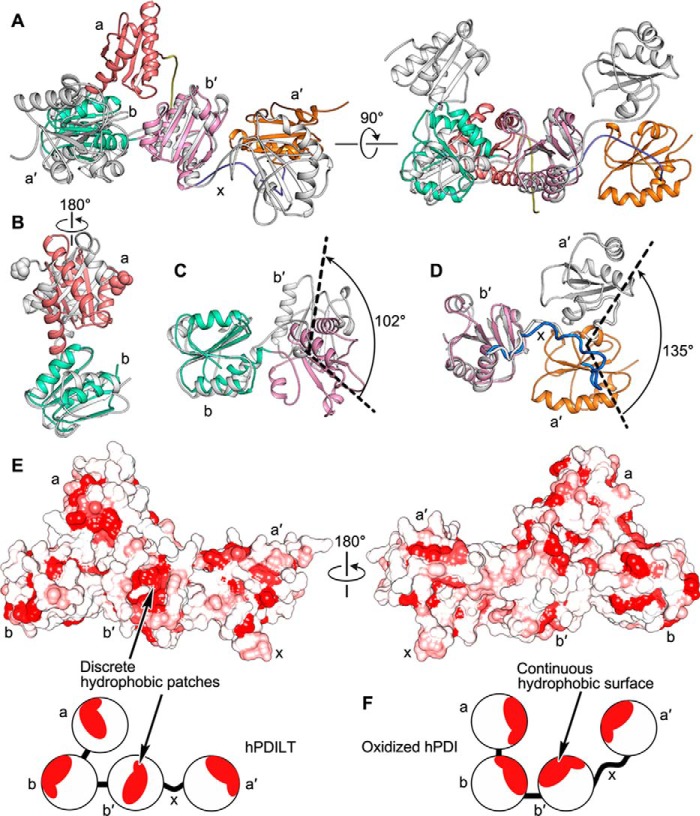
The arrangement of domains in the hPDILT structure was further compared with that in the oxidized hPDI structure (PDB code 4EL1) (27). When we superposed the crystal structures of hPDILT and the oxidized hPDI based on the b-b′ domains, we found that their a and a′ domains are located on different sides (Fig. 3A). When we superposed these two structures on the basis of the b domains, we found that the entire a domain of hPDILT rotates ∼180° counterclockwise compared with oxidized hPDI (Fig. 3B). In addition, the b′ domain of hPDILT rotates by ∼102° (Fig. 3C). Superposing the two structures using their b′ domains, we found that the a′ domain of hPDILT rotated ∼135° away from that of the oxidized hPDI form (Fig. 3D). In addition, we analyzed the hydrophobic regions of hPDILT as shown in Fig. 3E. The hPDILT structure possesses discrete hydrophobic patches, distinct from the structure of hPDI that displays a continuous hydrophobic surface in the inner face (Fig. 3F and Fig. S6) (27). These discrete hydrophobic patches probably result from the unique domain arrangement.
Figure 3.
Differences between the structures of hPDILT and hPDI. A, superposition of hPDILT (colored as Fig. 2B) and oxidized hPDI (PDB code 4EL1, in gray) structures based on the b-b′ domains. B and C, comparison of both a-b and b-b′ domains of hPDILT and oxidized hPDI based on the b domains. The start residues of two a domains are shown in sphere representation. D, superimposition of b′–x-a′ domains of hPDILT and oxidized hPDI based on the b′ domains. E, hydrophobic surface representation of hPDILT, colored from hydrophobic (red) to hydrophilic (white), according to the normalized consensus hydrophobicity scale of the surface-exposed residues by UCSF Chimera (45). The lower panel shows the schematic diagram. F, schematic diagram of oxidized hPDI (PDB code 4EL1).
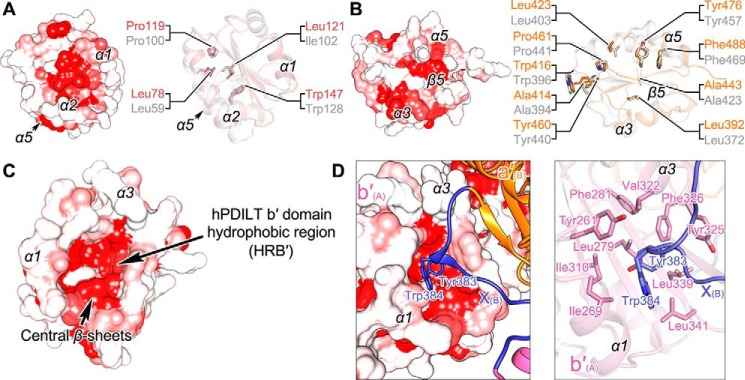
Because the hydrophobic regions in the a, a′, and b′ domains of hPDI are essential to assist the folding of unfolded substrates (28–31), we further analyzed the hydrophobic regions and residues in these domains of hPDILT. The hydrophobic regions of a and a′ domains (HRA and HRA′) of hPDILT scatter on the flank of the central β-sheets, which consist of α1, α2, and α5 for HRA and α3, α5, and β5 for HRA′ (Fig. 4, A and B). Hydrophobic residues in HRA and HRA′ are conserved between hPDI and hPDILT (Fig. 4, A and B). The hydrophobic region of the b′ domain (HRB′) locates in a pocket surrounded by α1, α3, and the central β-sheets (Fig. 4C). The hydrophobic pocket is composed of residues Tyr-261, Ile-269, Leu-279, Phe-281, Ile-310, Val-322, Tyr-325, Phe-326, Leu-339, and Leu-341 (Fig. 4D). Compared with other PDI family proteins, the hydrophobic pocket of the b′ domain in hPDILT is larger and deeper, which is helpful for interacting with the exposed aromatic residues in the unfolded proteins. Similar with those in the previously reported b′–x-linker structure (Fig. S3), the hydrophobic pocket is occupied by Tyr-383 and Trp-384 from the x-linker of another hPDILT molecule (Fig. 4D). Because the b′–x-linker structure was considered as a substrate-bound structure (17), we hypothesized that this crystal structure resembles the substrate-bound conformation of full-length hPDILT.
Figure 4.
Hydrophobic regions in the thioredoxin-like domains of hPDILT. A, HRA of hPDILT is shown on the left. The conserved hydrophobic residues in structures of the a domains of hPDILT and hPDI are shown on the right. hPDI is shown in gray (PDB code 4EKZ). B, HRA′ of hPDILT and the conserved hydrophobic residues in structures of a′ domains of hPDILT and hPDI. C, HRB′ of hPDILT. D, close-up views of the interactions between b′ domain and x-linker from another molecule. The residues involved in hydrophobic interactions are shown as sticks.
Solution structure of hPDILT
To detect the conformation of hPDILT in solution, we performed SAXS analysis. The overall structural parameters for SAXS analysis of hPDILT are summarized in Table 2. The experimental scattering profile, with the scattering intensity I(q) plotted versus momentum transfer q, is shown in Fig. 5A. The radii of gyration (Rg) derived from the SAXS data were determined as 48 Å, larger than that derived from the monomeric crystal structure of hPDILT, which is 29 Å. The molecular mass estimated from the SAXS data is ∼120 kDa, considerably larger than the molecular mass of hPDILT (66 kDa), similar to the size of a dimer (Table 2). These results indicate that hPDILT exists predominantly as a dimer in solution.
Table 2.
Statistics of SAXS analysis
| Data collection | |||
| Protein | WT | ΔN | ΔC |
| Wavelength (Å) | 1.033 | 1.033 | 1.033 |
| Exposure time (s) | 1 | 1 | 1 |
| Concentrations range (mg/ml) | 1.0–3.0 | 1.0–3.0 | 1.0–3.0 |
| No. collected images | 20 | 20 | 20 |
| Temperature (K) | 283 | 283 | 283 |
| Structural parameters | |||
| I(0) (from P(r)) | 321.4 | 311.9 | 106.7 |
| Rg (Å) (from P(r)) | 47.9 | 46.6 | 31.9 |
| I(0) (from Guinier) | 320.8 | 301.5 | 107.0 |
| Rg (Å) (from Guinier) | 45.9 | 42.9 | 31.8 |
| Dmax (Å) | 199 | 183 | 109 |
| Porod volume Vp (Å3) | 249916.4 | 225920.8 | 150638.1 |
| Correlation volume Vc (Å2) | 823.8 | 759.0 | 518.0 |
| Molecular mass | |||
| Mass (kDa)a | 127.4 | 116.2 | 75.6 |
| Mass (kDa)b | 120.2 | 102.6 | 68.4 |
| Mass (kDa)c | 122.2 | 100.8 | 66.1 |
| Mass (kDa)d | 132.5 (dimer) | 127.4 (dimer) | 56.5 (monomer) |
| Software employed | |||
| Primary data reduction | BioXTAS-RAW | ||
| Data processing | PRIMUS | ||
| Ab initio analysis | DAMMIN | ||
| Rigid body modeling | CORAL | ||
| Flexibility analysis | SCÅTTER | ||
| Validation and averaging | CRYSOL | ||
| SUPCOMB | |||
| DAMAVER | |||
a Mass was calculated from Porod volume Vp.
b Mass was calculated from correlation volume Vc.
c Mass was calculated from program SAXS MoW.
d Mass was calculated from protein sequence.
Figure 5.
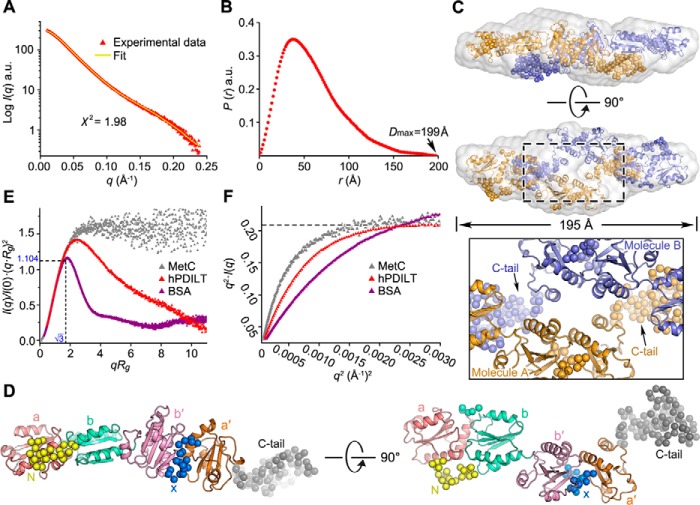
SAXS analysis of the hPDILT structure in solution. A, overlay of the experimental scattering profile with the back-calculated scattering profile from the CORAL model of hPDILT. B, pair distance distribution P(r) functions for hPDILT. C, superposition of low-resolution ab initio model and rigid body model. The ab initio model is shown as light-gray surface representation. Two molecules (A and B) are shown in different colors (orange and blue). The close-up view shows the dimeric interface. D, solution structure of hPDILT monomer. The thioredoxin-like domains in the rigid body model are colored as in Fig. 2B and the N-terminal loop, x-linker, and C-terminal tail are represented in spheres, shown in yellow, blue, and gray, respectively. E, superimposition of dimensionless Kratky plot representations of experimental data. The SAXS data of MetC was downloaded from Small Angle Scattering Biological Data Bank (code SASDBY5). F, Kratky-Debye plot of each data set is described in E. a.u., arbitrary units.
The pair distribution function P(r) of hPDILT, which describes the paired-set of all distances between all of the electrons within the hPDILT structure, is shown in Fig. 5B. The asymmetrical bell-shaped curve of this function indicates that hPDILT exists as an extended molecule in solution. We first generated three-dimensional reconstructions using program DAMMIN (32). Twenty ab initio models with P1 or P2 symmetry were generated independently (Fig. S7). Models with the same symmetry were aligned using SUPCOMB (33), averaged using DAMAVER (34), and filtered by DAMFILT (34) to generate the final ab initio model. The NSD value of the P1 models and the P2 models is 0.48 (Fig. S8), suggesting the structures resemble each other. Thus, P2 symmetry was applied in the modeling. The average normalized spatial discrepancy (NSDav) of models with P2 symmetry is 0.66, aligned and calculated by program SUPCOMB (33). The small NSDav indicates the ab initio models are similar and convincing. The most typical ab initio model with the lowest NSD (NSD = 0.61) displays an elongated shape with a maximum diameter of 195 Å (Fig. 5C), consistent with the result of the P(r) function. The back-calculated scattering profile of this model fits the experimental data with a discrepancy value χ2 of 1.64 (Table 2). Another set of ab initio models generated by GASBOR (35) was compared with those generated by DAMMIN (Fig. S8). The NSD values for model with P1 and P2 symmetry are 0.57 and 0.59, respectively. The low NSD values between DAMMIN and GASBOR models suggest that these models resemble each other.
To gain additional insights into the solution structure of hPDILT, we generated rigid models using the program CORAL (36), by translating and rotating the crystal structures. We first generated a model in P2 symmetry based on the crystal structure with four thioredoxin-like domains. However, the back-calculated scattering profile poorly fit the experimental data with χ2 at ∼10, indicating that the structure of hPDILT in solution is different from the crystal structure. Therefore, we employed the separated domains, a, b-b′, and a′ defined in a P2 symmetry-generated rigid model well fitted with the experimental data, with NSD at 1.51 and χ2 at 1.98 (Fig. 5A). Importantly, the NSD value of the most possible models generated by DAMMIN and CORAL is 1.74, suggesting that these two methods generated similar models, lending greater credibility to our results.
The arrangement of the thioredoxin-like domains in solution is different from that observed in the crystal. In the rigid model generated by CORAL, the C-terminal tails of hPDILT are involved in the dimer interface (Fig. 5C). The four thioredoxin-like domains in one molecule are arranged in a nearly linear manner and form a chain-like extended conformation (Fig. 5D). The extended structure exposes more hydrophobic regions of the thioredoxin-like domains, which is an important feature that allows hPDILT to interact with the unfolded protein substrates (Fig. S9). Compared with the crystal structure, we hypothesized that the structure hPDILT adopted in solution represents the substrate-free conformation of hPDILT.
We hypothesized that the solution structure is flexible, an important feature allowing hPDILT to fit substrates with different structures. To evaluate its flexibility in solution, we analyzed the dimensionless Kratky plot of hPDILT (37). As shown in Fig. 5E, the plot shows a qRg peak maximum at √3 for most globular compact proteins, obeying Guinier's approximation. By contrast, the plot shows a hyperbolic plateau for intrinsically disordered protein (37, 38). The curve of hPDILT differs from that of the well-folded globular BSA protein. The curve is slightly increased at higher scattering angles, with its peak shifting to 2.494 (Fig. 5E), indicating that hPDILT is flexible in solution. This feature was further supported by Flexibility Plot analysis using the program SCÅTTER (http://www.bioisis.net/scatter (51),5 where the plateau of hPDILT occurred in the Kratky-Debye plot but not Porod-Debye plot (Fig. 5F). Together, these results suggest that the solution structure of hPDILT represents the conformation prior to substrate binding.
Compared with the flexible SAXS structure, the L-shaped crystal structure is rigid and compact. Because the N-terminal loop is important to stabilize the crystal structure (Fig. 2E), we generated an N-terminal truncation, termed ΔN, and performed both SEC and SAXS analysis. The SEC profile, the Dmax, the calculated Rg, and the molecular mass derived from the SAXS for ΔN are similar with those for the wildtype hPDILT (Fig. S10 and Table 2). However, the dimensionless Kratky plot suggests that ΔN protein is more flexible than wildtype (Fig. S10D). This result is consistent with the crystal structure analysis (Fig. 2E).
The b′ domain, x-linker, and the C-terminal tail are important for the chaperone activity of hPDILT
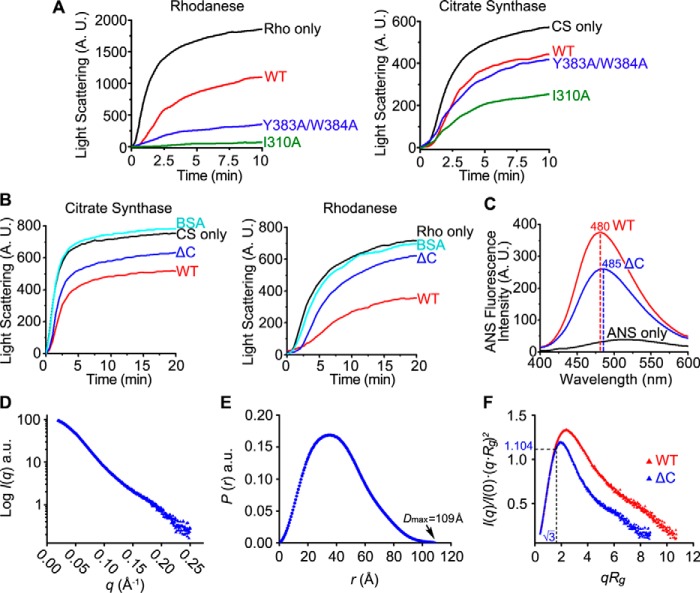
The chaperone activity of hPDILT was further studied using structure-based mutations. In the crystal structure, the aromatic residues (Tyr-383 and Trp-384) in the x-linker of hPDILT bind to the hydrophobic pocket of the b′ domain (Fig. 4D). Thus, the hydrophobic pocket of the b′ domain was considered as the principal binding element for substrates. Moreover, we hypothesized that the x-linker of hPDILT competes with the unfolded protein substrate for the binding site in the b′ domain and inhibits the chaperone activity. Consistent with this idea, chaperone activity of hPDILT improved greatly upon Y383A/W384A mutation (Fig. 6A). In addition, mutation of Ile-310 to Ala, which locates at the bottom of the hydrophobic pocket of b′ domain (Fig. 4D), greatly increased chaperone activity on unfolded CS and Rho (Fig. 6A). We attributed the increased chaperone activity of I310A mutation to the increased capacity for substrate recognition, as I310A enlarges the hydrophobic pocket of the b′ domain, which may destroy the interaction between b′ domain and x-linker.
Figure 6.
Chaperone activities and SAXS analysis of hPDILT C-tail truncation. A and B, chaperone activities of WT hPDILT, I310A, and Y383A/W384A mutants and the C-tail truncation (ΔC) on CS or Rho were monitored as described in Fig. 1C. The molar ratio of hPDILT proteins to substrates is 10. C, ANS fluorescence spectra of hPDILT and ΔC were monitored as described in Fig. 1D. A.U., arbitrary units. D, experimental scattering profile of ΔC hPDILT protein. E, pair distance distribution P(r) functions for ΔC hPDILT protein. F, superimposition of dimensionless Kratky plot representations of wildtype and ΔC hPDILT experimental data.
hPDILT possesses a functionally unknown C-terminal tail, which contains ∼90 residues rich in charged and hydrophobic amino acids (14, 17). Previous reports indicated that the flexible C-terminal tails of PDI family proteins were essential for substrate binding (25, 39, 40). In our SAXS data, the C-terminal tail of hPDILT locates in the dimer interface (Fig. 5C), which may contribute to the solution structure. We thus examined whether the C-terminal tail is involved in the chaperone activity. Using an hPDILT form truncated at the C-terminal (Ile-497–Val-580) tail (ΔC), we found that removal of this C-terminal tail impaired the ability of hPDILT to suppress the aggregation of the insulin B-chain (Fig. S1), as well as denatured CS and Rho (Fig. 6B). In addition, our ΔC truncation showed lower ANS fluorescence intensity than full-length hPDILT, with a red shift of the maximum emission peak from 480 to 485 nm (Fig. 6C), indicating that its hydrophobic surfaces were exposed to a smaller degree than those in the full-length form. Together, these results strongly indicate that the C-terminal tail is essential for the chaperone activity of hPDILT.
To study the importance of the C-terminal tail for the solution structure, we performed SAXS analysis for ΔC protein. The asymmetrical bell-shaped pair distribution function P(r) of the ΔC protein obtained by SAXS indicates that the Dmax of ΔC is 109 Å, which is ∼½ that of wildtype protein (Fig. 6, D and E, and Table 2). The calculated Rg derived from the SAXS data is ∼31 Å, comparable with 29 Å derived from the monomeric crystal structure of hPDILT. The molecular mass estimated from the SAXS data is ∼66 kDa, which is similar to the molecular mass of a monomer. The dimensionless Kratky plot reveals that ΔC protein is less flexible than wildtype (Fig. 6F). These results suggest that the C-terminal tail is important to stabilize the dimeric structure in solution, which is required for substrate binding.
Discussion
Here, we studied the structures and the chaperone activity of hPDILT, a PDI family protein specifically expressed in testis, using X-ray diffraction, SAXS, and biochemical methods. Our results show that the structure of hPDILT is dynamic. The N-terminal loop, the C-terminal tail, the x-linker, and the hydrophobic pockets in the thioredoxin-like domains are important for the chaperone activity. First, we provide an L-like crystal structure of hPDILT, which is distinct from the structures of its related proteins in the PDI family. The structural basis of chaperones during substrate recognition has been elusive because the substrates of chaperones, specifically unfolded or misfolded proteins, commonly exist in disordered or aggregate states, increasing the challenge of structural analysis. hPDILT includes both the b′ domain, the principal substrate-binding element of PDI family proteins (5), and the x-linker, the substrate of the b′ domain (41). We obtained the crystal structure of hPDILT benefit from its self-binding property. Compared with the reported b′–x-linker domain structure (17), our whole structure not only shows the interactions between residues in the hydrophobic pocket of the b′ domain from one molecule and residues in the x-linker from another molecule, but also reveals the cooperation among the four thioredoxin-like domains. In addition, we proposed that the x-linker of hPDILT competes with the unfolded protein substrate for the binding site in the b′ domain and inhibits the chaperone activity. Mutations that destroyed the interaction between the b′ domain and the x-linker, I310A and Y383A/W384A, increased the chaperone activity of hPDILT. The possible self-inhibition function of hPDILT in vivo still needs further studies.
Second, our crystal and SAXS structures results strongly suggest that hPDILT adopts distinct conformations to fulfill the chaperone activity. It has been reported that a structural dynamic of PDI based on the rearrangement of the thioredoxin-like domains is required during the substrate-binding-release cycles (4). The domain rearrangement of hPDI is regulated by its redox states. The oxidation of the hPDI active site in the a′ domain results in the conversion from the compact conformation to the open conformation with the substrate-binding surface more exposed, exhibiting higher chaperone activity to prevent substrate aggregation (21). Different from hPDI, hPDILT lacks an intact oxidoreductase active site (14). We propose that the dynamic structure of hPDILT is induced by substrate binding. hPDILT folds into an extended substrate-free conformation with large and continuous hydrophobic regions, which is accessible for substrate binding, similar to that found in the SAXS structure. Once it has recognized its substrate, hPDILT undergoes domain rearrangement, and it subsequently folds into a compacted L-shaped substrate-bound conformation as found in our crystal structure. Our results show that the N-terminal loop and the C-terminal tail are important to stabilize the crystal structure and the SAXS structure, respectively. Thus, the N-terminal loop and the C-terminal tails are important for the chaperone activity. Residues in the N-terminal loop and C-terminal tail are mainly conserved (Fig. S11), suggesting that the structures and functions of the N- and C-terminal tails are conserved in PDILT from different species.
On the basis of these insights, we propose that both substrate-bound and substrate-free states exist in solution (Fig. S12). The secondary state is predominant in solution as shown in the SAXS results. In high concentrations, both states tend to aggregate due to hydrophobic exposure. However, the first-state PDILT molecules are able to regularly assemble using the b′–x-linker interaction, the hydrogen bonds, and electrostatic interactions (Fig. S4). Under the crystallization condition, the transferring of the first-state PDILT from solution to crystal leads to a rapid decrease of its concentration in solution, which drives the shift of the dynamic equilibrium and the transformation of PDILT from the secondary state to the first state.
Taken together, we report here the mechanistic details of hPDILT chaperone activity based on our structural data. Our findings provide a framework to better understand the functions of hPDILT involved in biological processes and shed fresh light on the mechanism of other PDI family proteins.
Materials and methods
Protein expression and purification
The hPDILT gene lacking both the N-terminal signal peptide and ER retrieval peptide KEEL (Ser-21–Val-580) was amplified by PCR and subcloned into the pRSFDuetTM-1 vector (Novagen) using BamHI and XhoI restriction sites. The sequences of the recombinant vectors were verified by sequencing (Beijing Genomics Institute) and transformed into E. coli BL21 (DE3) cells. Transformed E. coli cells were cultured at 37 °C in Luria-Bertani medium (LB medium) containing 100 mg/liter kanamycin and induced at 16 °C by adding 1 mm isopropyl β-d-thiogalactopyranoside at an absorbance at 600 nm (A600) of 0.8.
Cells were harvested after 20 h and lysed by sonication with ice-cold lysis buffer containing 20 mm HEPES (pH 8.0), 1.3 m NaCl, 30 mm imidazole, 10% glycerol, and 0.5 mm phenylmethanesulfonyl fluoride (PMSF). The insoluble fraction was precipitated by ultracentrifugation (20,000 × g) for 30 min at 4 °C, and the supernatant was loaded onto a nickel-nitrilotriacetic acid superflow affinity column (Qiagen) and eluted with buffer containing 20 mm HEPES (pH 8.0), 150 mm NaCl, 400 mm imidazole, and 10% glycerol. The N-terminal His6-tagged hPDILT proteins were further purified by heparin-affinity chromatography, followed by SEC (Superdex-200; GE Healthcare) in 20 mm HEPES (pH 8.0), 150 mm NaCl, 10% glycerol buffer, and then concentrated to ∼15 mg/ml using a concentration tube (Millipore) with the 50-kDa cutoff. The strategies of molecular cloning and protein purification for mutants and truncations are the same as the wildtype hPDILT mentioned above.
Crystallization, data collection, and structure determination
Crystallization was performed using the hanging-drop vapor-diffusion method at 16 °C by mixing equal volumes (1 μl) of protein (15 mg/ml) and reservoir solution. Good-quality crystals appeared in drops containing 21% (w/v) PEG 3350 and 100 mm succinic acid, pH 7.0, after a week. The crystals were then flash-frozen in liquid nitrogen and cryoprotected by 25% glycerol.
Diffraction data were collected on an ADSC Q315 CCD detector at beamline BL17U of Shanghai Synchrotron Radiation Facility (SSRF, China). 360 frames were collected at λ = 0.979 Å, 100 K, with an oscillation step of 1 per frame for the crystal.
Structural data were processed using the HKL2000 program suite (42), and structures were solved by molecular replacement using PHASER (23). The b′ domain of hPDILT (PDB code 4NWY) and the three isolated thioredoxin-like domains (domains a, b, and a′) of hPDI (PDB code 4EKZ) without flexible loops, including the N-terminal loop, x-linker, and C-tail, were employed as search models. Employing the four search models in the order of the b′ domain of hPDILT, a domain, b domain, and a′ domain of hPDI, the molecular replacement was performed using program PHASER in the CCP4 software package with default parameters. A definite solution with high LLG and TFZ values (LLG, 553.313; RFZ = 7.5 and TFZ = 12.1; LLG = 158, RFZ = 4.5, and TFZ = 12.8; LLG = 266, RFZ = 3.9, and TFZ = 10.8; LLG = 346, RFZ = 3.8, and TFZ = 18.5) was obtained. Only one hPDILT molecule was found per asymmetric unit. The structural model was built by PHENIX.AUTOBUILD (43) and manually modified using COOT (44) followed by several cycles of refinement with PHENIX.REFINE (43). The Rwork and Rfree values of the final model are 19.7 and 23.5%, respectively. Data collection and structure refinement statistics are summarized in Table 1. All figures representing structures were prepared with UCSF Chimera (45) and PyMOL (51). The coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB entry 5XF7).
Chaperone activity assays
Denaturation and reduction of CS (Sigma) or Rho (Sigma) were carried out by incubation of 10 μm CS or 22.5 μm Rho in 6 m guanidine hydrochloride with 20 mm DTT in 50 mm Tris-HCl buffer (pH 8.0) for 2 or 1 h at 25 °C. Refolding was initiated by 50-fold (for CS, resulting in a final concentration at 0.2 μm) or 100-fold (for Rho, resulting in a final concentration at 0.225 μm) dilution of the denatured and reduced CS and Rho into 100 mm potassium phosphate buffer (pH 7.4) containing 2.5 mm EDTA with the absence or presence of a 10-fold molar ratio concentration of hPDI or hPDILT proteins (or 20- and 30-fold in Fig. 1C) relative to the final concentration of CS and Rho at 25 °C. Aggregation produced during the refolding was monitored by recording the 90° light scattering at 360 nm for CS or 350 nm for Rho using a spectrofluorometer.
ANS fluorescence measurements
ANS (Sigma) was added to a final concentration of 50 μm to solutions of 5 μm protein, followed by incubation in 50 mm Tris-HCl buffer (pH 7.6) containing 150 mm NaCl for 20 min at 25 °C in the dark. ANS fluorescence emission spectra (400–600 nm) were determined using a spectrofluorometer at an excitation wavelength of 370 nm. The concentration of ANS was determined with an extinction coefficient at 350 nm of 5000 m−1 cm−1.
SAXS data collection and analysis
The peak fractions of full-length hPDILT purified by SEC were collected and prepared at three concentrations (1, 2, and 3 mg/ml) in buffer containing 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 5% glycerol, and 2 mm DTT as background control. SAXS data were collected on beamline BL19U2 at Shanghai Synchrotron Radiation Facility (SSRF, China). The wavelength of X-ray radiation was set to 1.033 Å. Scattered X-ray intensities were measured using a Pilatus 1M detector (DECTRIS Ltd.). The sample-to-detector distance was adjusted to set the detecting range of momentum transfer q to 0.01–0.5 Å−1. The exposure time was set to 1 s, and to obtain optimal signal-to-noise ratios, 20 images were taken for each protein sample as well as buffer-only samples.
All 2D scattering images were converted to 1D SAXS curves using the software package BioXTAS RAW (46). Program PRIMUS (47) and the ATSAS package (48) were used for subsequent data processing and three-dimensional modeling. The molecular mass was calculated using Vp, Vc, and program SAXS MoW, which determines the molecular mass of proteins with an error smaller than 10%, using experimental data of a single SAXS curve measured on a relative scale (49). The 20 individual ab initio DAMMIN (32) or GASBOR (35) models were aligned using SUPCOMB (33), averaged using DAMAVER (34), and filtered by DAMFILT (34) to generate the final ab initio model. For rigid body modeling, hundreds of rigid body models were calculated by the program CORAL (36) employing the separated domains (a, b–b′, and a′) defined in P2 symmetry. Several models that fitted the experimental data with χ2 <2 were selected. Among these models, the one with the lowest NSD value aligned with the final ab initio model was chose to generate the final rigid body model. The fitting of the theoretical scattering curves between models and the experimental data were calculated using CRYSOL (50). Flexibility analysis was performed using SCÅTTER (http://www.bioisis.net/scatter) (52)5. Scattering curves were generated using ORIGINPRO (http://originlab.com) and structural figures were generated by PyMOL. Final statistics for data collection and scattering-derived parameters are summarized in Table 2.
Author contributions
H. Li and K. Y. performed most of the experiments. H. Li and W. W. analyzed the SAXS data and Y. D. supervised. K. Y. and Y. N. performed the chaperone assays. J. L. collected the X-ray diffraction data. Y. L., C. W., L. W., and H. Liang designed the experiment and analyzed the results. H. Li, K. Y., L. W., and H. Liang wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the staff at the BL17U and BL19U2 beamlines of the Shanghai Synchrotron Radiation Facility (SSRF, China) for assistance during data collection. We thank Dr. Xiaoxia Yu and Tao Li for technical support, Dr. Torsten Juelich for linguistic assistance, and Dr. Xi Wang for helpful discussion.
This work was supported by National Natural Science Foundation of China Grants 31530015, 31571163, and 31370775, National Key R&D Program of China Grants 2016YFA0500200 and 2017YFA0504000, the Strategic Priority Research Program, CAS Grant XDB08000000, Frontier Sciences Project, CAS Grant QYZDYSSW-SMC018, and Youth Innovation Promotion Association CAS (to L. W). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12 and supporting Refs. 1–2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ER
- endoplasmic reticulum
- PDI
- protein-disulfide isomerase
- PDILT
- protein-disulfide isomerase-like protein of testis
- hPDILT
- human PDILT
- SAXS
- small-angle X-ray scattering
- ANS
- 1-anilino-8-naphthalene sulfonate
- SEC
- size-exclusion chromatography
- NSD
- normalized spatial discrepancy
- CS
- citrate synthase
- Rho
- rhodanese.
References
- 1. Lipton S. A., Gu Z., and Nakamura T. (2007) Inflammatory mediators leading to protein misfolding and uncompetitive/fast off-rate drug therapy for neurodegenerative disorders. Int. Rev. Neurobiol. 82, 1–27 10.1016/S0074-7742(07)82001-0 [DOI] [PubMed] [Google Scholar]
- 2. Labbadia J., and Morimoto R. I. (2015) The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sitia R., and Braakman I. (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426, 891–894 10.1038/nature02262 [DOI] [PubMed] [Google Scholar]
- 4. Wang L., Wang X., and Wang C.-C. (2015) Protein disulfide–isomerase, a folding catalyst and a redox-regulated chaperone. Free Radic. Biol. Med. 83, 305–313 10.1016/j.freeradbiomed.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 5. Kozlov G., Määttänen P., Thomas D. Y., and Gehring K. (2010) A structural overview of the PDI family of proteins. FEBS J. 277, 3924–3936 10.1111/j.1742-4658.2010.07793.x [DOI] [PubMed] [Google Scholar]
- 6. Hatahet F., and Ruddock L. W. (2009) Protein-disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 11, 2807–2850 10.1089/ars.2009.2466 [DOI] [PubMed] [Google Scholar]
- 7. Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., and Nagata K. (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572 10.1126/science.1159293 [DOI] [PubMed] [Google Scholar]
- 8. Fraldi A., Zito E., Annunziata F., Lombardi A., Cozzolino M., Monti M., Spampanato C., Ballabio A., Pucci P., Sitia R., and Cosma M. P. (2008) Multistep, sequential control of the trafficking and function of the multiple sulfatase deficiency gene product, SUMF1 by PDI, ERGIC-53 and ERp44. Hum. Mol. Genet. 17, 2610–2621 10.1093/hmg/ddn161 [DOI] [PubMed] [Google Scholar]
- 9. Higo T., Hattori M., Nakamura T., Natsume T., Michikawa T., and Mikoshiba K. (2005) Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell 120, 85–98 10.1016/j.cell.2004.11.048 [DOI] [PubMed] [Google Scholar]
- 10. Park B., Lee S., Kim E., Cho K., Riddell S. R., Cho S., and Ahn K. (2006) Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell 127, 369–382 10.1016/j.cell.2006.08.041 [DOI] [PubMed] [Google Scholar]
- 11. Diwaker D., Mishra K. P., Ganju L., and Singh S. B. (2015) Protein-disulfide isomerase mediates dengue virus entry in association with lipid rafts. Viral Immunol. 28, 153–160 10.1089/vim.2014.0095 [DOI] [PubMed] [Google Scholar]
- 12. Tsai B., Rodighiero C., Lencer W. I., and Rapoport T. A. (2001) Protein-disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104, 937–948 10.1016/S0092-8674(01)00289-6 [DOI] [PubMed] [Google Scholar]
- 13. van Lith M., Karala A. R., Bown D., Gatehouse J. A., Ruddock L. W., Saunders P. T., and Benham A. M. (2007) A developmentally regulated chaperone complex for the endoplasmic reticulum of male haploid germ cells. Mol. Biol. Cell 18, 2795–2804 10.1091/mbc.E07-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Lith M., Hartigan N., Hatch J., and Benham A. M. (2005) PDILT, a divergent testis-specific protein-disulfide isomerase with a non-classical SXXC motif that engages in disulfide-dependent interactions in the endoplasmic reticulum. J. Biol. Chem. 280, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 15. Landegren N., Sharon D., Freyhult E., Hallgren Å., Eriksson D., Edqvist P. H., Bensing S., Wahlberg J., Nelson L. M., Gustafsson J., Husebye E. S., Anderson M. S., Snyder M., and Kämpe O. (2016) Proteome-wide survey of the autoimmune target repertoire in autoimmune polyendocrine syndrome type 1. Sci. Rep. 6, 20104 10.1038/srep20104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tokuhiro K., Ikawa M., Benham A. M., and Okabe M. (2012) Protein-disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility [corrected]. Proc. Natl. Acad. Sci. U.S.A. 109, 3850–3855 10.1073/pnas.1117963109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bastos-Aristizabal S., Kozlov G., and Gehring K. (2014) Structure of the substrate-binding b′ domain of the protein-disulfide isomerase-like protein of the testis. Sci. Rep. 4, 4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smulders R. H., Carver J. A., Lindner R. A., van Boekel M. A., Bloemendal H., and de Jong W. W. (1996) Immobilization of the C-terminal extension of bovine αA-crystallin reduces chaperone-like activity. J. Biol. Chem. 271, 29060–29066 10.1074/jbc.271.46.29060 [DOI] [PubMed] [Google Scholar]
- 19. Farahbakhsh Z. T., Huang Q. L., Ding L. L., Altenbach C., Steinhoff H. J., Horwitz J., and Hubbell W. L. (1995) Interaction of α-crystallin with spin-labeled peptides. Biochemistry 34, 509–516 10.1021/bi00002a015 [DOI] [PubMed] [Google Scholar]
- 20. Jakob U., Gaestel M., Engel K., and Buchner J. (1993) Small heat shock proteins are molecular chaperones. J. Biol. Chem. 268, 1517–1520 [PubMed] [Google Scholar]
- 21. Wang C., Yu J., Huo L., Wang L., Feng W., and Wang C. C. (2012) Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a′. J. Biol. Chem. 287, 1139–1149 10.1074/jbc.M111.303149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uehara T., Nakamura T., Yao D., Shi Z.-Q., Gu Z., Ma Y., Masliah E., Nomura Y., and Lipton S. A. (2006) S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517 10.1038/nature04782 [DOI] [PubMed] [Google Scholar]
- 23. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCarthy A. A., Haebel P. W., Torronen A., Rybin V., Baker E. N., and Metcalf P. (2000) Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat. Struct. Biol. 7, 196–199 10.1038/73295 [DOI] [PubMed] [Google Scholar]
- 25. Tian G., Xiang S., Noiva R., Lennarz W. J., and Schindelin H. (2006) The crystal structure of yeast protein-disulfide isomerase suggests cooperativity between its active sites. Cell 124, 61–73 10.1016/j.cell.2005.10.044 [DOI] [PubMed] [Google Scholar]
- 26. Dong G., Wearsch P. A., Peaper D. R., Cresswell P., and Reinisch K. M. (2009) Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity 30, 21–32 10.1016/j.immuni.2008.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang C., Li W., Ren J., Fang J., Ke H., Gong W., Feng W., and Wang C.-C. (2013) Structural insights into the redox-regulated dynamic conformations of human protein-disulfide isomerase. Antioxid. Redox Signal. 19, 36–45 10.1089/ars.2012.4630 [DOI] [PubMed] [Google Scholar]
- 28. Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., and Ruddock L. W. (2004) Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein-disulfide isomerase. J. Biol. Chem. 279, 10374–10381 10.1074/jbc.M312193200 [DOI] [PubMed] [Google Scholar]
- 29. Koivunen P., Salo K. E., Myllyharju J., and Ruddock L. W. (2005) Three binding sites in protein-disulfide isomerase cooperate in collagen prolyl 4-hydroxylase tetramer assembly. J. Biol. Chem. 280, 5227–5235 10.1074/jbc.M412480200 [DOI] [PubMed] [Google Scholar]
- 30. Klappa P., Ruddock L. W., Darby N. J., and Freedman R. B. (1998) The b′ domain provides the principal peptide-binding site of protein-disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 17, 927–935 10.1093/emboj/17.4.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Denisov A. Y., Määttänen P., Dabrowski C., Kozlov G., Thomas D. Y., and Gehring K. (2009) Solution structure of the bb′ domains of human protein-disulfide isomerase. FEBS J. 276, 1440–1449 10.1111/j.1742-4658.2009.06884.x [DOI] [PubMed] [Google Scholar]
- 32. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 10.1016/S0006-3495(99)77443-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 10.1107/S0021889800014126 [DOI] [Google Scholar]
- 34. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 10.1107/S0021889803000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 10.1016/S0006-3495(01)76260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petoukhov M. V., and Svergun D. I. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 10.1529/biophysj.105.064154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Receveur-Brechot V., and Durand D. (2012) How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 10.2174/138920312799277901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Durand D., Vivès C., Cannella D., Pérez J., Pebay-Peyroula E., Vachette P., and Fieschi F. (2010) NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 10.1016/j.jsb.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 39. Wang L., Wang L., Vavassori S., Li S., Ke H., Anelli T., Degano M., Ronzoni R., Sitia R., Sun F., and Wang C. C. (2008) Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 9, 642–647 10.1038/embor.2008.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watanabe S., Harayama M., Kanemura S., Sitia R., and Inaba K. (2017) Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface. Proc. Natl. Acad. Sci. U.S.A. 114, E3224–E3232 10.1073/pnas.1621426114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nguyen V. D., Wallis K., Howard M. J., Haapalainen A. M., Salo K. E., Saaranen M. J., Sidhu A., Wierenga R. K., Freedman R. B., Ruddock L. W., and Williamson R. A. (2008) Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b′ domain. J. Mol. Biol. 383, 1144–1155 10.1016/j.jmb.2008.08.085 [DOI] [PubMed] [Google Scholar]
- 42. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 43. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 45. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 46. Nielsen S. S., Møller M., and Gillilan R. E. (2012) High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J. Appl. Crystallogr. 45, 213–223 10.1107/S0021889812000957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 10.1107/S0021889803012779 [DOI] [Google Scholar]
- 48. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fischer H., de Oliveira Neto M., Napolitano H. B., Polikarpov I., and Craievich A. F. (2010) Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43, 101–109 10.1107/S0021889809043076 [DOI] [Google Scholar]
- 50. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 10.1107/S0021889895007047 [DOI] [Google Scholar]
- 51. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5, Schrödinger, LLC, New York [Google Scholar]
- 52. Rambo R. P., and Tainer J. A. (2013) Accurate assessment of mass, models, and resolution by small-angle scattering. Nature 496, 477–481 10.1038/nature12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.