Abstract
Notch signaling is a ubiquitous signal transduction pathway found in most if not all metazoan cell types characterized to date. It is indispensable for cell differentiation as well as tissue growth, tissue remodeling, and apoptosis. Although the canonical Notch signaling pathway is well characterized, accumulating evidence points to the existence of multiple, less well-defined layers of regulation. In this study, we investigated the function of the intracellular domain (ICD) of the Notch ligand Delta-like 4 (DLL4). We provide evidence that the DLL4 ICD is required for normal DLL4 subcellular localization. We further show that it is cleaved and interacts with the JUN proto-oncogene, which forms part of the activator protein 1 (AP-1) transcription factor complex. Mechanistically, the DLL4 ICD inhibited JUN binding to DNA and thereby controlled the expression of JUN target genes, including DLL4. Our work further demonstrated that JUN strongly stimulates endothelial cell tube formation and that DLL4 constrains this process. These results raise the possibility that Notch/DLL4 signaling is bidirectional and suggest that the DLL4 ICD could represent a point of cross-talk between Notch and receptor tyrosine kinase (RTK) signaling.
Keywords: angiogenesis, AP1 transcription factor (AP-1), gene regulation, Notch pathway, signaling
Introduction
The generic Notch signaling network is a central regulator of cell fate (1, 2). This pathway is absolutely necessary for normal development and tissue homeostasis, and corruption of it has been implicated in numerous diseases, including the majority of solid tumors where it plays diverse oncogenic and tumor-suppressive roles (3–5). In vertebrates, the Notch signaling system is composed of four single-pass cell-surface receptors (Notch1–4) and five type 1 transmembrane ligands: Jagged (JAG)4 1, JAG2, Delta-like (DLL) 1, DLL3, and DLL4 (1, 6, 7). Operationally, the canonical Notch signaling pathway is relatively well characterized (7–9). It is activated by a trans interaction between a Notch receptor on one cell and a ligand expressed by a neighboring cell. This triggers a cascade of proteolytic events terminating in the γ-secretase–mediated cleavage of the Notch intracellular domain, which translocates to the nucleus whereupon it regulates expression of Notch target genes (1, 8). By these means, the molecular and cellular asymmetries required for tissue maintenance and development are established across populations of cells. In recent years, studies have identified manifold, unique facets of Notch signaling (9). These include cis receptor-ligand interactions (because each are expressed in the same cell) (10), ligand-independent signaling (11), endocytosis of Notch and Notch ligands as an essential mediator of signaling (12, 13), and in the case of DLL4 signaling at a distance through the incorporation of DLL4 into exosomes (14, 15). Adding further to the complexity is the extensive cross-talk between the Notch pathway and other major signaling networks such as receptor tyrosine kinase signaling (16); WNT, hedgehog, and transforming growth factor (TGF)-β signaling (17); Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling (18); and hypoxia signaling (19). To understand these mechanisms in greater depth, new studies are beginning to elucidate the role of the Notch ligands in this process. The five ligands share a similar overall architecture: module at the N terminus of Notch ligands (MNNL) domains, a Delta/Serrate/Lag-2 (DSL) domain, between six and 14 EGF-like repeats, a transmembrane segment, and an intracellular domain (ICD) 100–150 amino acids in length (20). The extracellular moiety is essential for establishing the direct contacts with the Notch receptor necessary for eliciting Notch signaling (21). Biochemical and genetic evidence has shown that the intracellular domain is clearly essential for normal functioning of the full-length protein (22–25). Ligand ICDs harbor putative PDZ domains that couple them to membrane-bound proteins required for the maintenance of cell-cell junctions and likely play a central role in assembling those complexes necessary for ligand trafficking (26, 27). Consistent with this, in Drosophila (28) and in vertebrates (29), Delta (and Serrate) ligands lacking an intracellular domain behave as dominant-negative mutants such that the phenotypes resemble Notch or Delta loss-of-function mutants. Likewise, corruption of the DLL1 C terminus has been shown to provoke mislocalization of the ligand (22).
In common with the Notch receptor, the DLL1 and JAG ligands can be sequentially processed by proteases. Both a disintegrin and metalloprotease (ADAM) metalloendopeptidases (30, 31) and matrix metalloproteinase 14 (in the case of DLL1) (32) mediate ectodomain shedding by cleavage of the ligand close to the transmembrane domain on the extracellular side. Subsequent intramembrane processing by γ-secretase liberates the ICD (33, 34). A growing body of evidence suggests that these ICDs might participate in signaling and downstream transcription, and ectopically expressed ICDs have been localized to the nucleus (35). Moreover, it has been reported that both DLL1 (36) and JAG1 ICDs (37) are able to bind to and disrupt the function of the Notch ICD by mediating its degradation in the case of the JAG ICD. The DLL1 ICD has also been pinpointed as a modulator of TGF-β/activin signaling through binding to SMAD proteins (38). A number of studies have described the effects of ligand ICDs at the cellular level. Inhibition of Notch1 signaling by the JAG1 ICD was shown to regulate cardiac homeostasis in the postnatal heart (39). When ectopically expressed in mesenchymal stromal cells, the JAG1 ICD regulated hematopoietic stem and progenitor cell proliferation (40). Finally, overexpression of the DLL1 ICD brought about the growth arrest of primary endothelial cells (41).
Collectively, the mechanisms described above determine the strength, direction, specificity, and nature of the output of the Notch signaling pathway. The emerging evidence that ICDs are biologically active is thus important for fully understanding Notch signaling in both normal and disease contexts, not least because the Notch pathway in general (42) and the DLL4 ligand in particular (43, 44) have emerged as a potential target for novel therapeutic interventions in cancer. The DLL4 ligand is integral to the development and homeostasis of numerous tissues, most notably the endothelium where it plays a central role in angiogenesis (45–49). It also fundamentally involved in hematopoiesis (50) and the maintenance of the stem cell compartment of intestinal crypts (51). To date, little is known about the function of the DLL4 intracellular domain. Here, we provide evidence that the DLL4 ICD is needed for appropriate DLL4 subcellular localization. We show that it encodes a functional PDZ-binding domain that is necessary for associating to the MUPP1 scaffold protein. We further show that the DLL4 ICD is cleaved. The liberated DLL4 (and DLL1) ICD could interact with the bZIP domain of the JUN proto-oncogene, thereby blocking its binding to the consensus AP-1 DNA-binding site. We found that JUN strongly stimulated endothelial cell sprouting, which was inhibited by the ICDs of both DLL4 and DLL1. These data highlight a previously unreported role for JUN as a potent pro-angiogenic signal and the DLL4 ICD as a potential regulator of Notch signaling. Our evidence suggests that the ICDs of DLL4 (and DLL1) could control endothelial cell sprouting by linking Notch signaling to the action of the AP-1 transcription factor complex.
Results
The intracellular domain of DLL4 is cleaved
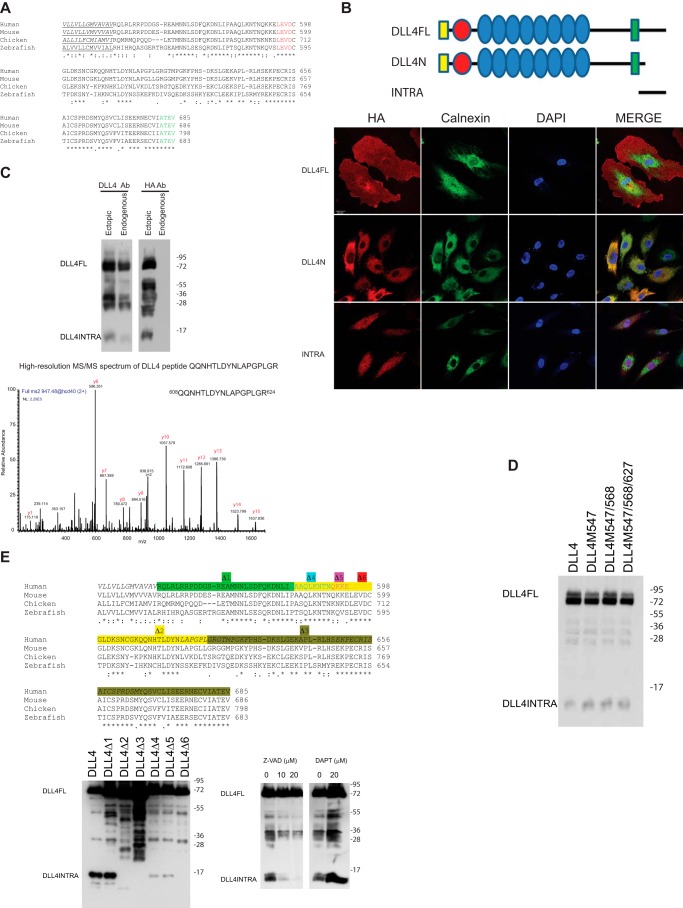
The amino acid sequence of the intracellular domain of DLL4 (hereafter referred to as DLL4INTRA) has been highly conserved throughout vertebrate evolution (Fig. 1A). The four C-terminal amino acids (ATEV) encode a putative PDZ-binding domain (52). PDZ-binding domains have been shown to mediate protein-protein interactions, cell adhesion, tight junction integrity, and trafficking (27), and loss of DLL4INTRA led to a significant disruption of normal DLL4 localization in primary human endothelial cells (Fig. 1B). Costaining a specific calnexin antibody showed that DLL4 lacking the intracellular domain (DLL4N) is entrapped in the endoplasmic reticulum/Golgi apparatus. DLL4INTRA harbors a number of additional motifs that suggest it is functionally important, including GSK consensus phosphorylation sites (which are utilized),5 a sumoylation motif, and putative ubiquitination sites. Moreover, ectopically expressed DLL4INTRA is able to accumulate in the nucleus (Fig. 1B), indicating that it may play a role independently of ensuring appropriate DLL4 subcellular distribution. To explore the function of this domain, we raised custom-made antibodies directed against epitopes unique to the C terminus of human DLL4. Fig. 1C shows that these antibodies recognized full-length endogenous DLL4 as well as a number of smaller DLL4 species (predominant forms with masses between ∼25 and 35 kDa and a lower molecular mass form of ∼12 kDa). Similarly sized fragments could be detected by Western blotting of lysates prepared from cells ectopically expressing full-length DLL4 fused to a C-terminal HA epitope tag (Fig. 1C). Through mass spectrometry, we determined that these fragments constitute the C terminus of DLL4 (Fig. 1C). The 25–35-kDa fragments are predicted to encompass the entire ICD, the transmembrane domain, and sequences of the extracellular domain EGF repeats. The size of the shorter fragment indicates that it is composed uniquely of ICD sequences. Our evidence supports the view that this fragment is generated by post-translational proteolysis of the ICD distal to the transmembrane domain on the intracellular side. First, the molecular mass of the observed endogenous fragment precludes the possibility of it being generated by known mechanisms of alternative splicing alone. Second, mutation of the intracellular domain methionine residues failed to eliminate detection of the band in cells, suggesting that it is not produced through use of internal initiation codons (Fig. 1D). Third, a comprehensive deletion analysis mapped a putative cleavage site to a highly conserved LEVD motif (amino acids 594–597) (Fig. 1E). LEVD closely resembles a consensus caspase cleavage site (53), and the pan-caspase inhibitor Z-VAD-fmk blocked the production of the DLL4ICD (Fig. 1E). It has been reported that the closely related DLL1 ligand undergoes intramembrane processing mediated by γ-secretase to yield an intracellular fragment (31, 33). Although we cannot formally rule out γ-secretase–dependent cleavage of DLL4, we found that release of the observed DLL4INTRA was not blocked by DAPT (Fig. 1E). Together, these observations reveal two unique features of DLL4INTRA: it is necessary for accurate ligand trafficking, and it is proteolytically cleaved (and this proteolysis is caspase-dependent).
Figure 1.
A, the ICD of DLL4 is evolutionarily conserved. Highlighted are the transmembrane domain (underlined), a putative caspase cleavage site (red), and a PDZ-binding motif (green). *, conserved amino acids; :, partly conserved amino acids (different amino acids are similar); ., partly conserved amino acids (different amino acids are not similar). B, the DLL4 ICD is necessary for appropriate DLL4 subcellular localization, and untethered DLL4INTRA is enriched in the nucleus. HUVECs were infected with lentiviruses for stably expressing the indicated HA epitope–tagged DLL4 constructs. DLL4N lacks the ICD and is otherwise identical to wildtype DLL4. DLL4INTRA encompasses the ICD alone. The subcellular distribution of DLL4 was visualized by immunofluorescence using an HA antibody. Scale bar, 20 μm. C–E, the ICD of DLL4 is cleaved. C, mass spectrometry of cleaved DLL4 fragments. Endogenous DLL4 or DLL4 expressing a C-terminal HA epitope tag was immunopurified from HUVECs. Samples were separated by SDS-PAGE, and DLL4 ICDs were subjected to in-gel digestion with trypsin followed by on-line nanoflow LC-MS/MS analysis of the peptide mixture on an LTQ Orbitrap Velos mass spectrometer. The MS/MS spectrum of a recovered peptide corresponding to amino acids 608–624 of the DLL4 ICD is shown. D, the indicated ICD methionine residues of HA epitope–tagged full-length DLL4 were mutated to alanine. HA antibody Western blotting was performed on lysates prepared from HUVECS stably expressing the indicated DLL4 ligands. E, a conserved LEVD motif is required for cleavage of the DLL4 ICD. Left panel, the indicated nested deletions of HA epitope–tagged full-length DLL4 were generated by site-directed mutagenesis. HA antibody Western blotting was performed on lysates prepared from HUVECs stably expressing the indicated DLL4 ligands. Corresponding deletions are highlighted above. Right panel, DLL4 ICD cleavage is blocked by the pan-caspase inhibitor Z-VAD but not DAPT. Cells expressing wildtype HA epitope–tagged full-length DLL4 were incubated overnight with or without the indicated concentrations of Z-VAD or DAPT. DLL4 protein species were detected by HA antibody Western blotting. Ab, antibody; DLL4FL, full-length DLL4.
DLL4 interacts with the PDZ domain of MUPP1 and the bZIP domain of JUN
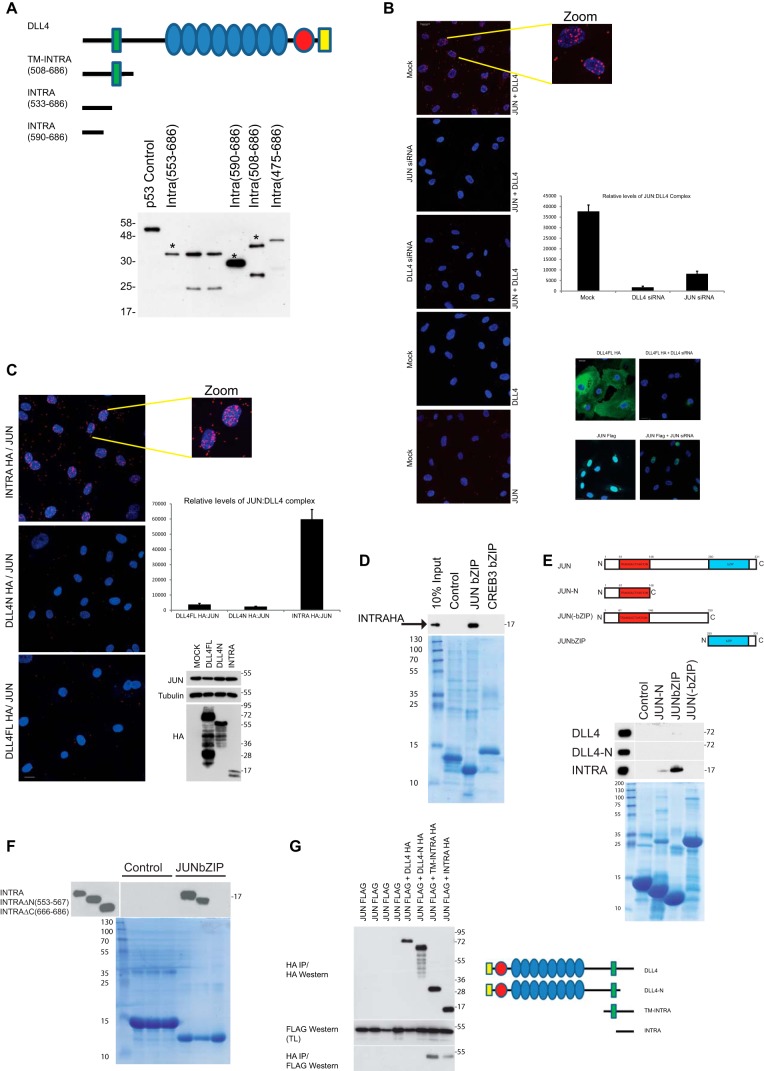
To investigate the potential molecular function(s) of the DLL4 intracellular domain, we performed yeast two-hybrid screens to identify partner proteins of DLL4INTRA. Three different DLL4 C terminus constructs were used as baits to independently screen a library prepared from primary HUVECs (Fig. 2A). The PDZ domain–containing scaffold protein MUPP1 was obtained in all screens. It is involved in cell-cell junction integrity, including endothelial cell junctions, and protein trafficking (26, 54). Immunoprecipitation studies in tissues culture cells corroborated the interaction between DLL4 and MUPP1 and its dependence on the extreme C terminus (ATEV) of DLL4, lending substance to the view that DLL4INTRA has a bona fide PDZ-binding domain (Fig. S1 and see Fig. 1A). Less expected was the interaction with the bZIP-containing transcription factor JUN. We have previously established that JUN levels are dynamically balanced by VEGF receptor signaling in primary endothelial cells (55), and several lines of evidence confirmed that DLL4INTRA and JUN can interact both in vitro and in cells. First, we used the sensitive and quantitative proximity ligation assay to visualize the endogenous DLL4-JUN complex. Using custom-made antibodies directed against the DLL4 C terminus and an endogenous JUN antibody, we found that endogenous JUN and DLL4 associated (predominantly in the nucleus) in HUVECs (Fig. 2B). By stably expressing engineered DLL4 mutants in HUVECs, we showed that DLL4INTRA was necessary and sufficient for this interaction (Fig. 2C). Second, using purified recombinant JUN protein domains and in vitro translated DLL4 proteins, we demonstrated that DLL4INTRA bound efficiently to the bZIP domain of JUN but not to the bZIP domain of CREB (Fig. 2D). Third, Fig. 2, E and G, substantiate our proximity ligation assay (PLA) findings (see Fig. 2, B and C) by showing that DLL4INTRA, but not DLL4N, is necessary for binding to JUN in vitro (Fig. 2E) and in tissue culture cells (Fig. 2G). Furthermore, the association between DLL4INTRA and JUN requires the bZIP domain of JUN (Fig. 2E) as well as the highly conserved C-terminal 20 amino acids of DLL4, which are shared with the ICD of DLL1 (Fig. 2F and see Fig. S4). The PDZ-binding domain of DLL4INTRA was dispensable for this interaction (see Fig. 3C). In these experiments, JUN did not appear to interact with full-length DLL4, which does contain the intracellular domain, suggesting that DLL4 is folded in such a way as to preclude this interaction. It is also possible that DLL4 monomers oligomerize/dimerize, which could render the C terminus inaccessible for JUN interactions.5 Collectively, these data highlight potentially a dual role for the DLL4ICD: it can interact with the bZIP domain of JUN, and in addition it encodes a PDZ-binding domain that mediates a functional interaction with MUPP1.
Figure 2.
DLL4INTRA interacts with the bZIP domain of JUN. A, yeast two-hybrid baits were constructed by fusing DLL4 ICD fragments in-frame with a LexA DNA-binding domain either at the N or the C terminus of the ICD. Constructs were expressed in yeast to verify expression by Western blotting of lysates using a LexA antibody. A p53-LexA fusion is included as a control (Dualsystems Biotech). Predicted full-length proteins of constructs used to screen a library prepared from HUVECs are marked with an asterisk. B, a PLA revealed an endogenous DLL4-JUN complex. The graph shows relative complex formation per cell (right upper panel) that was abolished by ablation of either DLL4 or JUN (siRNA efficacies were demonstrated using cells ectopically expressing either DLL4 or JUN; see right lower panels). The endogenous DLL4-JUN complex was undetectable using single antibodies alone. Quantification was performed as described (see “Experimental procedures”) with an average of 100 cells scored. Scale bar, 20 μm. C, JUN specifically associated with the ICD of DLL4. PLA was performed on HUVECs ectopically expressing HA epitope–tagged versions of either full-length DLL4 (DLL4FL); DLL4N, which lacks the ICD but retains the transmembrane domain; or the non-membrane–tethered DLL4INTRA. Relative protein levels of DLL4 and endogenous JUN were determined by Western blotting with the indicated antibodies (right lower panel). The DLL4-JUN complex was detected using a combination of HA (for DLL4) and endogenous JUN antibodies. The graph shows relative complex formation per cell (right upper panel). Quantification was performed as described (see “Experimental procedures”) with an average of 100 cells scored. Scale bar, 20 μm. D and E, the ICD of DLL4 interacts biochemically with the bZIP domain of JUN. D, recombinant bZIP domains of JUN and CREB3 (and a control protein consisting of the DNA-binding domain of the TEL/ETV6 ETS transcription factor) were incubated with in vitro translated HA epitope–tagged DLL4INTRA. Recombinant proteins were visualized by Coomassie staining. Bound DLL4INTRA was detected by Western blotting. E, recombinant JUN domains (and a control protein) were incubated with in vitro translated HA epitope–tagged full-length DLL4, DLL4N, or DLL4INTRA. Recombinant proteins were visualized by Coomassie staining. Bound DLL4 was detected by Western blotting. F, the C terminus of DLL4INTRA is necessary for binding of DLL4INTRA to the JUN bZIP domain. Experiments were performed as in D and E. DLL4INTRAΔN lacks the N-terminal amino acids 553–567. DLL4INTRAΔC lacks the C-terminal amino acids 666–686. G, the indicated constructs were stably expressed in HUVECs. JUN-DLL4 complexes were immunopurified from cell lysates using a rabbit HA polyclonal antibody and visualized by Western blotting with a FLAG mouse monoclonal antibody. TM, transmembrane domain; IP, immunoprecipitation; TL, total lysate.
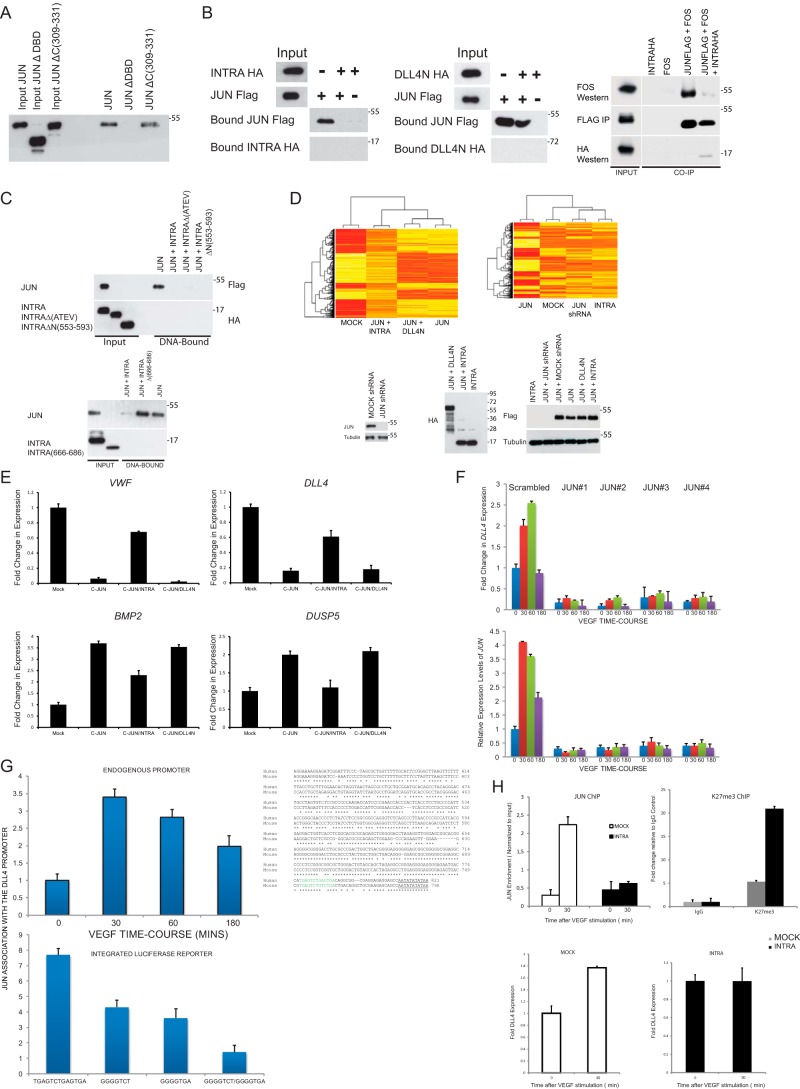
Figure 3.
DLL4INTRA antagonizes JUN DNA binding. A, JUN binds to a consensus AP-1 DNA-binding site in vitro. A biotinylated double-stranded oligonucleotide harboring three consensus AP-1–binding sites was incubated with the indicated FLAG epitope–tagged in vitro translated proteins. JUNΔDBD lacks the DNA-binding domain. JUNΔC(309–331) lacks the C-terminal amino acids abutting the DNA-binding domain. DNA-bound JUN was detected by Western blotting. B, DLL4INTRA impedes JUN binding to DNA. As in A, AP-1 DNA–binding sites were incubated with the indicated in vitro translated proteins. DNA-bound JUN was detected by Western blotting. Right panel, in vitro translated FLAG epitope–tagged JUN, untagged FOS, and HA epitope–tagged INTRA were incubated as shown in the absence of the AP-1 site. FLAG epitope–tagged JUN was immunopurified on FLAG beads, and associated complexes were detected by Western blotting. C, the putative PDZ-binding motif of DLL4INTRA is dispensable for DLL4INTRA binding to JUN. As in A and B, AP-1 DNA-binding sites were incubated with the indicated in vitro translated proteins. INTRAΔ(ATEV) lacks the putative C-terminal PDZ-binding motif. INTRAΔN(553–593) lacks the highlighted N-terminal amino acids. INTRAΔ(666–686) lacks the C-terminal 20 amino acids. DNA-bound JUN was detected by Western blotting. D, DLL4INTRA alters the JUN-controlled transcriptome. A microarray analysis was performed on HUVECs stably expressing the indicated constructs. Protein levels were determined by Western blotting with the indicated antibodies (lower panel). Heat maps show a comparison of global gene expression profiles (upper panels). E, expression levels in HUVECs of the indicated transcripts were determined by real-time qPCR. All values were averaged relative to TBP, SRPR, and CAPNS1. Values were normalized against mock-treated cells. Values represent ±S.D. (n = 3). F, JUN controls DLL4 expression. Real-time qPCR was performed on cDNA prepared from cells stably infected with one of four different JUN shRNA constructs and stimulated with or without VEGF for the shown time course (minutes). All values were averaged relative to TBP, SRPR, and CAPNS1. Values were normalized against the time 0 time point of mock-infected cells. Values represent ±S.D. (n = 3). G and H, JUN associates with the proximal promoter of human DLL4. G, upper panel, a ChIP analysis was performed on HUVECs incubated with or without 50 ng/ml VEGF for the indicated times. Three different primer sets centered on the illustrated promoter region were used, and a single representative is shown (all three gave very similar results). Equivalent amounts of rabbit IgG were used as a control, and results are presented as -fold changes in recovery (as a fraction of input) relative to the zero time point. Lower panel, expression of a stably integrated luciferase reporter was placed under the control of the depicted wildtype DLL4 proximal promoter or the same promoter in which the putative AP-1 sites have been singly or doubly mutated. The alignment of the human and mouse DLL4 promoter regions highlights the presumed transcription start site (underlined). Conserved consensus AP-1 DNA-binding sites are shown in green. A ChIP analysis was performed on reporter-expressing HUVECs incubated for 30 min with 50 ng/ml VEGF. Equivalent amounts of rabbit IgG were used as a control, and results are presented as -fold changes in recovery (as a fraction of input) relative to the control IgG antibody. Two different primer sets were centered on the integrated luciferase gene. A single representative is shown (both gave comparable results). H, upper graphs, ChIP analyses were performed with the indicated antibodies (as described in G) on control HUVECs and HUVECs stably expressing DLL4INTRA. Lower graphs, DLL4 expression levels in control HUVECs and HUVECs stably expressing DLL4INTRA were determined by real-time qPCR. All values were averaged relative to TBP, SRPR, and CAPNS1. Values were normalized against mock-treated cells. Values represent ±S.D. (n = 3). IP, immunoprecipitation; VWF, von Willebrand factor. Error bars represent S.D.
DLL4INTRA blocks JUN binding to DNA
To determine the mechanistic consequences of DLL4INTRA-JUN binding, we established an in vitro assay to recapitulate JUN binding to DNA. For this, we utilized a biotinylated consensus AP-1 DNA-binding site and in vitro translated proteins. Fig. 3A shows that JUN efficiently bound to the AP-1 site. As expected, JUN binding to the AP-1 site required the bZIP domain of JUN because a mutant lacking this domain, but not a mutant lacking amino acids C-terminal of this domain, was unable to bind the consensus sites, thus authenticating the specificity of the assay (Fig. 3A). Compellingly, DLL4INTRA, but not DLL4N, inhibited JUN association to the AP-1 site (Fig. 3B) and could impede AP-1 dimerization (Fig. 3B). Both the PDZ-binding motif of DLL4INTRA and sequences N-terminal of the LEVD motif were dispensable for this inhibition (Fig. 3C), whereas the highly conserved C-terminal 20 amino acids of DLL4 appear to be necessary for inhibiting in vitro JUN DNA binding (in agreement with our observation that these sequences are needed for binding to the JUN bZIP domain; see Fig. 2F). JUN represses or activates gene expression in a context-dependent fashion by docking on target promoters and through the differential recruitment of corepressors and coactivators. To ascertain a more global view of regulation of JUN activity by DLL4INTRA, we performed transcriptome profiling of HUVECs ectopically expressing JUN in the presence or absence of DLL4INTRA or DLL4N. Ectopic expression of JUN strongly stimulates endothelial cell sprouting (see Fig. 4) and leads to a dramatic alteration in the set of expressed genes (Fig. 3D, compare mock and JUN). Whereas coexpression of DLL4N had relatively more modest effects on the JUN-controlled transcriptome, DLL4INTRA significantly altered the expression pattern of JUN-controlled genes (Fig. 3D). These findings were verified by real-time quantitative PCR (qPCR) of a subset of the genes, including DLL4 (Fig. 3E). To test whether these effects could have resulted from DLL4INTRA directly occluding JUN DNA binding, we investigated JUN recruitment to the proximal promoter of DLL4. We have previously shown that DLL4 expression (and JUN expression) is highly attuned to VEGF receptor signaling in HUVECs (55). Fig. 3F shows that short hairpin (sh) RNA-mediated loss of JUN in HUVECs abrogated DLL4 levels and abolished the VEGF-stimulated wave of DLL4 expression. In support of the view that DLL4 expression is governed by JUN, we found that endogenous JUN associated with the proximal promoter of DLL4 and, using an integrated luciferase reporter, that loss of a consensus AP-1 site upstream of the DLL4 TATA box significantly diminished the detectable levels of promoter-bound JUN (Fig. 3G). Accordantly, ectopic expression of DLL4INTRA inhibited VEGF-dependent association of JUN with the DLL4 promoter (Fig. 3H, upper graph) and led to a concomitant inhibition of VEGF-stimulated DLL4 expression (Fig. 3H, lower graphs). Consistent with these findings, stable expression of DLL4INTRA augmented the levels of the repressive H3K27me chromatin methylation mark found at the DLL4 proximal promoter (Fig. 3H, upper graph). These data indicate that DLL4INTRA can negatively regulate JUN binding to DNA and, in consequence, control expression of genes required for endothelial sprouting.
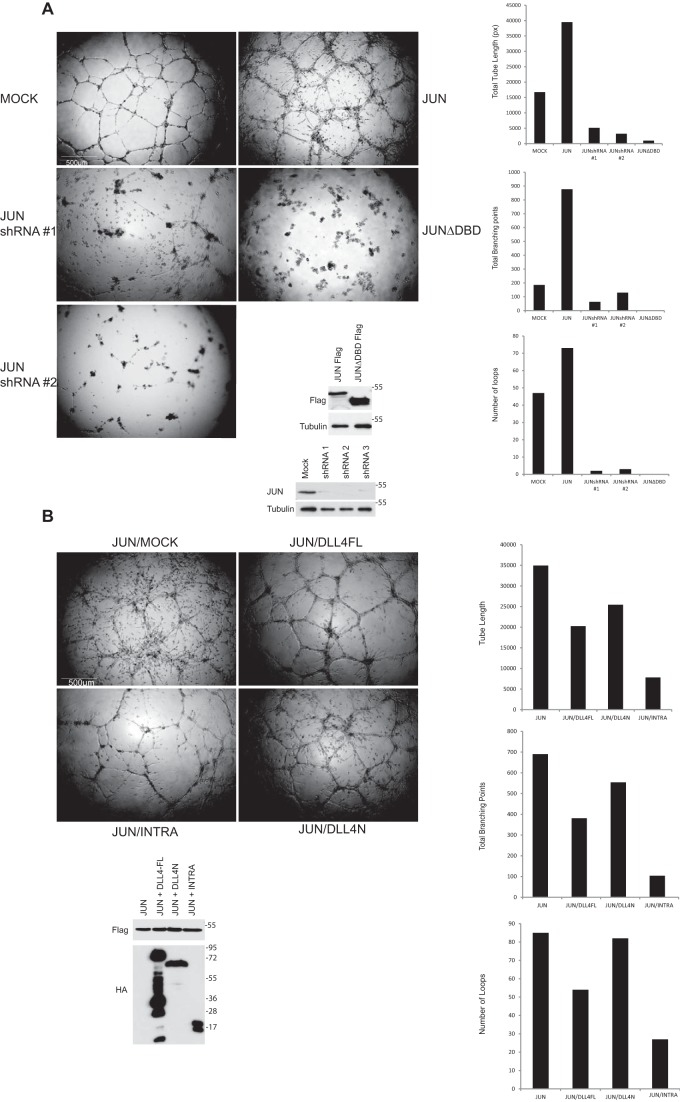
Figure 4.
A, JUN strongly stimulates endothelial cell tube formation/sprouting. HUVECs lacking endogenous JUN or ectopically expressing the indicated JUN proteins were cultured in Matrigel in the presence of 50 ng/ml VEGF. A representative of several independent experiments is shown. After 24 h, in-house computer software was used to quantify the total length of the sprouts, the number of branch points, and the number of loops. Protein levels were determined by Western blotting with the indicated antibodies. Scale bar, 500 μm. B, DLL4INTRA attenuates JUN-mediated sprouting. HUVECs ectopically expressing the indicated proteins were cultured in Matrigel in the presence of 50 ng/ml VEGF. Experiments were conducted and quantified as in A. Scale bar, 500 μm. DBD, DNA-binding domain; DLL4FL, full-length DLL4.
DLL4INTRA attenuates JUN-mediated endothelial cell tube formation/sprouting
To elucidate the biological consequences of DLL4INTRA binding to JUN, we used a 3D Matrigel sprouting assay. Fig. 4A shows that enhanced JUN levels hugely increased endothelial sprouting (including filopodia formation), whereas loss of endogenous JUN abolished the ability of HUVECs to sprout. JUN-stimulated sprouting depended on the bZIP domain of JUN and thus presumably relies on functional JUN binding to DNA because a JUN mutant lacking this domain acted as a dominant negative, resulting in strong inhibition of sprouting (Fig. 4A). In this assay, wildtype sprouting networks are relatively short-lived and collapse after ∼24 h. By contrast, endothelial cells ectopically expressing JUN were robustly sustained and continued to sprout for several weeks. Additionally, JUN-expressing cells could sprout efficiently in the absence of exogenous VEGF.5 In agreement with our prior findings that DLL4INTRA obstructs JUN DNA binding (see Fig. 3), Fig. 4B shows that coexpression of DLL4INTRA, but not DLL4N, restricted JUN-driven endothelial sprouting.
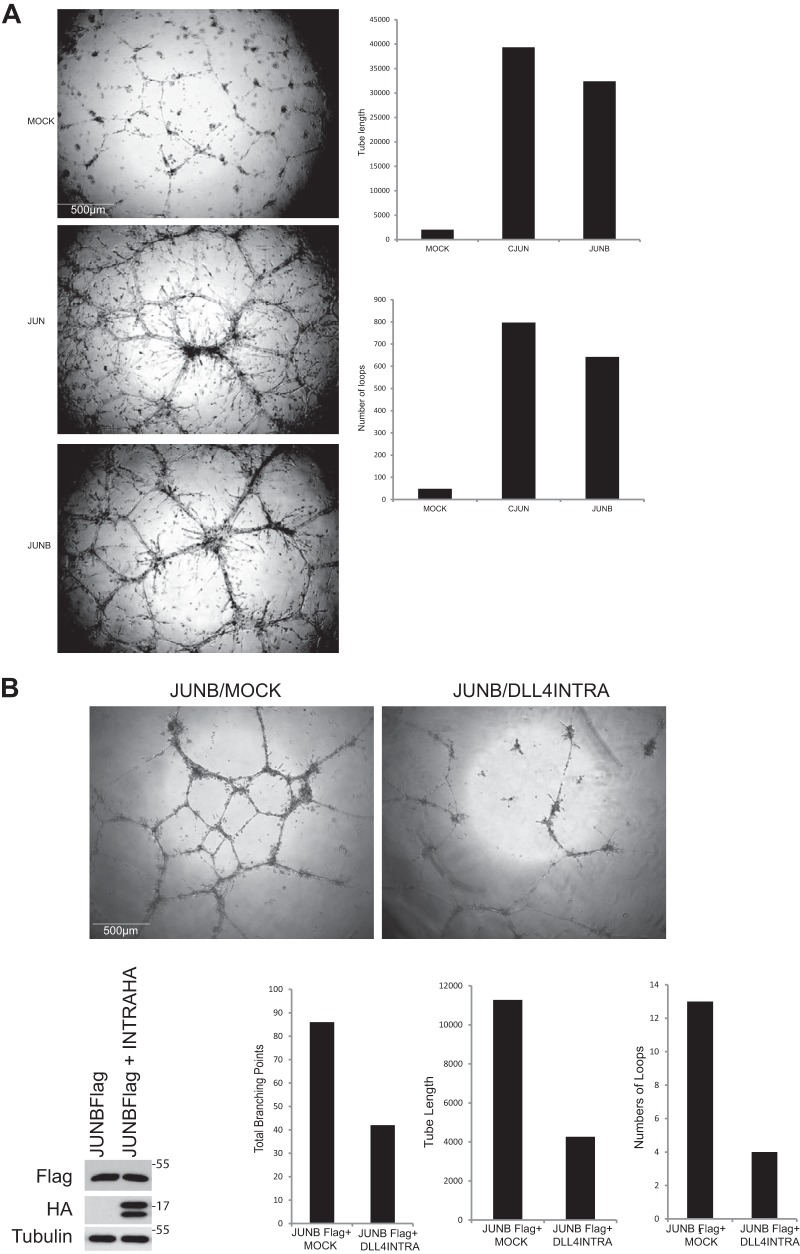
AP-1 transcription factors are characterized by the conserved bZIP domain, which mediates homo- and heterodimerization of AP-1 family members and DNA binding. This begs the question as to whether DLL4INTRA might also influence the activity of other AP-1 family members such as JUNB, which has an established role in vascular morphogenesis (56–58). Our experiments show that JUNB, like JUN, strongly stimulated endothelial tube formation (Fig. 5A), and DLL4INTRA could inhibit this process (Fig. 5B).
Figure 5.
A, JUNB strongly stimulates endothelial cell tube formation/sprouting. HUVECs ectopically expressing JUN or JUNB were cultured in Matrigel in the presence of 50 ng/ml VEGF. A representative of several independent experiments is shown. After 48 h, in-house computer software was used to quantify the total length of the sprouts, the number of branch points, and the number of loops. Protein levels were determined by Western blotting with the indicated antibodies. Scale bar, 500 μm. B, DLL4INTRA attenuates JUNB-mediated sprouting. HUVECs ectopically expressing the indicated proteins were cultured in Matrigel in the presence of 50 ng/ml VEGF. A representative of several independent experiments is shown. After 24 h, in-house computer software was used to quantify the total length of the sprouts, the number of branch points, and the number of loops. Protein levels were determined by Western blotting with the indicated antibodies. Scale bar, 500 μm.
Discussion
In this study, we have presented evidence that DLL4INTRA could perform a dual function: it is essential for establishing normal DLL4 subcellular localization, and strikingly, the untethered ICD interacted with and inhibited the activity of the JUN transcription factor. By means of chemical inhibitors and targeted mutagenesis, we demonstrated that one mechanism of ICD cleavage is caspase-dependent (see Fig. 1). The pattern of cellular DLL4 protein species indicates that multiple DLL4 cleavage events could occur, but their precise nature is yet to be delineated. What triggers the cleavage event(s) or whether proteolysis is constitutive is also currently unclear and will be important to establish. Certainly, DLL4INTRA is highly conserved and harbors a number of motifs that might control its function (see Introduction) and could influence its fate. We found that endogenous DLL4INTRA accumulated following incubation of cells with VEGF (Fig. S2). Future work will determine whether this simply reflects increased levels of DLL4 and/or specific signaling events. Related to this, unraveling the mechanism(s) of DLL4INTRA turnover could shed further light on its function. The DLL4 ICD contains an intrinsically disordered region terminating in a structured C-terminal PDZ-binding domain. This conformation resembles the proto-oncogene FOS (59, 60) and other signaling proteins and transcription factors (61). We recently found that the C terminus of FOS is composed of a comparable intrinsically disordered region/structured extreme C terminus arrangement that controls intrinsic, rapid proteasomal degradation of FOS, and a similar mechanism might control the activity of DLL4INTRA. Indeed, we found that DLL4INTRA protein levels are strongly stabilized when the proteasome is blocked by MG132, indicating that DLL4INTRA is rapidly turned over.5
JUN is a member of the AP-1 family of transcription factors that plays a pivotal role in cell growth, differentiation, and survival as well as the DNA damage response (62, 63). Our results suggest that DLL4INTRA can block the activity of both JUN and JUNB (see Figs. 4 and 5). A related question is whether other Notch ligand ICDs can function similarly to DLL4INTRA. The DLL1 ligand shares a high degree of amino acid identity with DLL4, including ∼40% of the ICD (see Fig. S3). Our results suggest that DLL1INTRA, like DLL4INTRA, can bind to the bZIP domain of JUN (Fig. 3A) and block JUN binding to a consensus AP-1 DNA site (Fig. 3B). Interestingly, unlike DLL4INTRA, DLL1INTRA could also interact with the bZIP domain of CREB3 in vitro (see Fig. S3A), raising the possibility that they have specific as well as overlapping activities. At the cellular level, DLL1INTRA, in common with DLL4INTRA, inhibited JUN-driven endothelial cell sprouting (Fig. S4C). Collectively, these results highlight a potential link between untethered Notch ligand ICDs and the immediate-early gene AP-1 transcription factor complex. Genome-wide screens have established the importance of ETS and AP-1 transcription factor cooperation by their association to neighboring AP-1 and ETS DNA-binding sites (64). Because ETS transcription factors are essential for angiogenesis (55, 65, 66) and have been reported to control DLL4 expression (55, 67), it is noteworthy that JUN potently stimulates endothelial sprouting and is required for normal expression of DLL4 (see Fig. 3). JUN-DLL4INTRA interactions, therefore, could form part of a feedback loop whereby VEGF-dependent changes in DLL4 expression (as well as other angiogenesis-regulating genes) depend on ETS/AP-1 factors and are regulated by DLL ICDs.
In summary, our data lend support to the idea that biologically active Notch ligand ICDs could help establish appropriate endothelial cell responses through cross-talk with AP-1–dependent signal transduction pathways. They further highlight the possibility that corruption of this mechanism might play a role in disease processes such as tumor angiogenesis and developmental disorders. This is important because the Notch pathway has emerged as a primary target for the design of novel therapeutic interventions in cancer (42). Also, it has recently been reported that heterozygous loss-of-function DLL4 mutations are a potential cause of Adams-Oliver syndrome, a rare congenital disorder characterized by multiple defects, including vascular and cardiac anomalies (68). Two of the identified DLL4 mutations are nonsense mutations that are predicted to lead to complete loss of the DLL4 ICD.
Experimental procedures
Yeast two-hybrid screens
Screens were performed according to the manufacturer's protocol (Dualsystems Biotech). Bait and prey were rescued from yeast colonies, and genuine interactions were confirmed through retransfection of yeast. Protein-protein interactions were tested quantitatively using a LacZ reporter assay (Dualsystems Biotech).
Proximity ligation assay
For the PLA, cells were seeded into eight-chamber cell culture slides (Lab-Tek II). The next day, cells were rinsed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min at room temperature followed by permeabilization for 5 min with PBS + 0.2% Triton X-100. Following extensive washing, the proximity ligation assay was performed using a Duolink in situ PLA kit (Sigma) according to the manufacturer's instructions. Briefly, blocking solution was added to each sample, and the slide was incubated for 30 min. All the incubations were performed in a preheated humidity chamber at +37 °C. Subsequently, primary antibodies were used at optimized concentrations in 1× antibody diluent for 1 h. Sequentially, two different PLA probes (anti-mouse MINUS and anti-rabbit PLUS) were mixed and incubated on the slides for 1 h. Ligation and amplification reagents were diluted in water and incubated on the slides for 30 and 100 min, respectively. Mounting medium was used for preservation of the PLA signals. Images were recorded on a Leica SP8 confocal microscope system using wavelengths of 488 and 561 nm. Statistical analysis of fluorescence signals was performed using BlobFinder software (http://www.cb.uu.se/∼amin/BlobFinder/).6 For each condition, an average of 100 cells was scored. Error bars show the standard error of the mean.
Mass spectrometry
Endogenous DLL4 or HA epitope–tagged DLL4 was immunopurified from duplicate lysates, each prepared by lysing HUVECS (20 × 15- and 10 × 15-cm dishes, respectively) in ice-cold lysis buffer (50 mm Tris (pH 7.5), 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mm NaCl). Protein bands were excised from SDS-polyacrylamide gels and subjected to in-gel tryptic digestion as described previously (69, 70).
Cell culture, biochemistry, and molecular biology
Primary HUVECs (Lonza) were cultured in EGM2 medium (Lonza). Human embryonic kidney 293T cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco). 293T cells were typed using short-tandem-repeat analysis of the DNA, and all cell lines were checked for Mycoplasma with the MycoAlert kit (Lonza). Transfections, lentivirus production and cell infections, Western blotting, and coimmunoprecipitations have been described previously (71). All lysis buffers contained a mixture of protease inhibitors (phenylmethylsulfonyl fluoride, trypsin inhibitor, pepstatin A, leupeptin, and aprotinin).
Recombinant protein production/in vitro protein-protein interaction
Domains for recombinant protein production were cloned into the pET 28a vector in-frame to an N-terminal His6 epitope. His epitope–tagged proteins were produced in Escherichia coli BL21(DE3). Following sonication (Misonix Sonicator 3000) in 3 ml of ice-cold buffer/50 ml of bacterial culture (150 mm NaCl, 2.7 mm KCl, Na2HPO4, KH2PO4, 20 mm imidazole, 10 mm β-mercaptoethanol), proteins were purified onto 50 μl of nickel-agarose beads (Qiagen) by 3 h of rolling at 4 °C. Beads were washed 10 times with 1 ml of the same buffer. Protein yields were determined by Bradford assay (Bio-Rad), and relative protein integrity and purity were determined by SDS-PAGE and colloidal blue staining (Invitrogen). 5–10 μl of His beads (purified recombinant protein) in 1 ml of buffer were incubated for 2 h at 4 °C with in vitro translated DLL4 proteins made using the TnT coupled reticulocyte in vitro translation system (Promega). Beads were washed 10 times with 1 ml of buffer. Proteins were separated by SDS-PAGE, and associated proteins were detected by Western blotting.
Plasmid and shRNA construction
Unless otherwise stated, all cDNAs were fused in-frame with a FLAG or an HA epitope tag and cloned into the pLV lentiviral vector and pCS2 expression plasmid. The luciferase reporter was constructed by cloning 1 kb of the DLL4 proximal promoter downstream of the luciferase gene in the pLV lentiviral vector. All mutants were engineered by site-directed mutagenesis using Phusion high-fidelity DNA polymerase (Thermo Fisher). All constructs were verified by Sanger sequencing (Macrogen). The following Mission shRNA library clones (Sigma) were used: shRNA TRCN0000010366 JUN, shRNA TRCN0000039589 JUN, shRNA TRCN0000009845 JUN, shRNA TRCN0000039588 JUN, shRNA TRCN0000039590 JUN, shRNA TRCN0000039591 JUN, and shRNA TRCN0000039592 JUN. The following siRNA duplexes (Thermo Scientific) were used (sense strands are shown): DLL4 siRNA 1, GCUCCACUGCGAGAAGAAU; DLL4 siRNA 2, GCAUGGUGGCAGUGGCUGUUU; JUN siRNA 1, GGAUCAAGGCGGAGAGGAAUU; and scrambled, ON-TARGETplus Non-targeting siRNA (D-001810-01-05).
HUVECs cells were transfected with duplexes using DharmaFECT transfection reagent according to the manufacturer's recommendations (Thermo Scientific). Transfected cells were incubated for 72 h prior to PLAs. Efficient delivery of siRNA and subsequent silencing were tested by immunostaining and Western blotting.
Transcriptome profiling
Total RNA was isolated using TRIzol (Invitrogen) and further purified using an RNeasy minikit (Qiagen). Total RNA was labeled using an Illumina RNA amplification kit (Ambion). Duplicate samples from two independent experiments were tested. Gene profiles were determined by hybridization to Sentrix HumanHT-12 Expression Beadchips (ServiceXS).
Analysis of mRNA expression
RNA isolation, first-strand cDNA synthesis, and analysis of expression of transcripts by quantitative PCR were performed as described previously (55). The following primer sets were used (5′ to 3′ orientation): DLL4FOR, ccctggcaatgtacttgtgat; DLL4REV, tggtgggtgcagtagttgag; JUNFOR, cgcctgataatccagtcca; JUNREV, ttcttggggcacaggaact; DUSP5FOR, caaatggatccctgtggaa; DUSP5REV, cccttttccctgacacagtc; VWFFOR, gtgcagacccaacttcacct; VWFREV, gggtggggacactcttttg; BMP2FOR, cagaccaccggttggaga; and BMP2REV, ccactcgtttctggtagttcttc.
All qPCR values were averaged relative to the control gene, TATA-binding protein (TBP), signal recognition particle receptor (SRPR), and calcium-activated neutral proteinase 1 (CAPNS1). For each data point, PCRs were performed in triplicate, and error bars show standard deviations from the mean.
HUVEC tube formation/sprouting assay
96-well plates were coated with 60 μl of Matrigel/well 30 min prior to seeding 25000 HUVECs/well. EGM2 medium was supplemented with 50 ng/ml recombinant human VEGF 165 (R&D Systems). Images were taken at multiple time points. Analysis of the sprouting was performed with Stacks (in-house software; Department of Molecular Cell Biology, Leiden University Medical Center). Each individual space bounded by connected tubes constitutes a loop. Branches are the intersections made by connected tubes, and total length is the combined length of all tubes.
Immunofluorescence
Immunostaining was performed as previously described (72) using Alexa Fluor 488 goat anti-mouse secondary antibodies (Thermo Fisher scientific). Imaging was performed with a Leica SP8 confocal microscope.
Protein-DNA interaction assays
In vitro translated protein was made using the TnT coupled reticulocyte in vitro translation system (Promega). 50 pmol of biotinylated double-stranded oligonucleotides harboring three contiguous AP-1 DNA-binding sites were coupled to MyOne streptavidin C1 beads (Invitrogen). Reactions were incubated at 4 °C with vigorous shaking for 30 min in the presence of 1 μg of poly(dI/dC), 4 mm spermidine, 50 mm KCl, 10 mm HEPES (pH 7.6), 5 mm MgCl2, 10 mm Tris (pH 8), 0.05 mm EDTA (pH 8), 0.05 mm, 0.1% Triton X-100, 20% glycerol. Beads were successively washed three times with the aforementioned buffer. Associated proteins were eluted in Laemmli buffer, and protein-DNA interactions were determined by Western blotting.
ChIP
Confluent 10-cm tissue culture dishes of HUVECs were cross-linked for 10 min with formaldehyde (final concentration, 1%). Glycine was added to a final concentration of 0.125 m and incubated for 5 min. Cells were washed two times with PBS and subsequently lysed in ChIP lysis buffer (1%SDS,10 mm EDTA, 50 mm Tris-HCl (pH 8.1)) supplemented with protease inhibitors. Chromatin was sheared by sonication (Bioruptor UCD-200 ultrasound sonicator, Diagenode), resulting in DNA fragments between 500 and 1000 bp in size. After centrifugation, 10% of the sample was kept as input. Chromatin was diluted 10× in dilution buffer (1%Triton X-100, 2 mm EDTA (pH 8.0), 150 mm NaCl, 20 mm Tris-HCl (pH 8.1) supplemented with protease inhibitors). For immunoprecipitation, 2 μg of test or control antibody were added to the diluted chromatin and incubated overnight at 4 °C. Blocked protein A-Sepharose beads (10 μg of sonicated herring sperm DNA) were added for 2 h. Beads were extensively washed (0.1% SDS, 0.1% sodium deoxycholate, 1% TritonX-100, 0.15 m NaCl, 1 mm EDTA, 0.5 mm EGTA, 20 mm HEPES), and complexes were eluted with elution buffer (1% SDS, 0.1 m NaH2CO3, 0.2 m NaCl) at 65 °C overnight to reverse cross-linking. Associated DNA was purified by phenol/chloroform extraction and ethanol precipitation (in the presence of 15 μg/ml glycogen (Roche Applied Science)). Real-time qPCR was used to determine recovery of specific DNA fragments. The following primers were used (5′ to 3′ orientation): DLL4promFOR A, ttctttttacctgctttggaaca; DLL4promREV A, agtccctgtaggctgtgcat; DLL4promFOR B, aatgaccatgagtctgagtgaca; DLL4promREV B, cgccgctactgaaacctg; DLL4promFOR C, gggtgggcactcataggtt; DLL4promREV C, aaaccagcgctagggaaatc; DLL4promFOR D, tcaggagagttcctccttgc; DLL4promREV D, tgagtccagcttcagttcctg; DLL4promFOR E, acgctcccaacctcttgtt; DLL4promREV E, ccgagcatggtctgattttt; DLL4promFRO F, tcatgaatgtcttttgatgctga; DLL4promREV F, tcccagagatctagaaaggctct; DLL4promFOR G, gaacacgaggccaagagc; DLL4promREV G, cgcgtcttctgtctaatcctg; LuciferaseFOR 1, catgaccgagaaggagatcg; LuciferaseREV 1, cagcttcttggcggttgta; LuciferaseFOR 2, tgagtacttcgaaatgtccgttc; and LuciferaseREV 2, gtattcagcccatatcgtttcat.
Antibodies and drugs
The following antibodies were obtained from the indicated sources: custom-made rabbit and goat polyclonal antibodies were generated by Eurogentec; FLAG mouse M2 monoclonal (Sigma-Aldrich), anti-HA.11 mouse monoclonal (Covance), anti-HA rabbit polyclonal (Abcam), anti-JUN rabbit (Cell Signaling Technology), anti-JUN mouse (Santa Cruz Biotechnology), anti-FLAG rabbit (Sigma), anti-γ-tubulin (Sigma), H3K27me3 (Bethyl Laboratories), calnexin rabbit polyclonal (Abcam), and FOS rabbit polyclonal (Cell Signaling Technology). Z-VAD was purchased from Sigma.
Author contributions
Z. F. and F. R., assisted by D. A. B., performed the majority of experiments. A. L. and J. V. O. performed mass spectrometry analyses. D. A. B. supervised the study and wrote the paper. All authors read and approved the paper.
Supplementary Material
Acknowledgments
We thank members of the Department of Molecular Cell Biology for helpful discussions, technical advice, and support, in particular Professor Peter ten Dijke. We are very grateful to Dr. Jan Oosting for the analyses of the microarray data and Annelies van der Laan for help with the confocal microscopy. We are indebted to Dr. Hans Vrolijk for designing computer software for quantifying sprouting assays.
This work was supported by Dutch Cancer Society Grant 30861 (to D. A. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
Z. Forghany and D. A. Baker, unpublished data.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- JAG
- Jagged
- ICD
- intracellular domain
- DLL
- Delta-like
- AP-1
- activator protein 1
- DLL4INTRA
- intracellular domain of DLL4
- DLL4N
- DLL4 lacking the intracellular domain
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- bZIP
- basic leucine zipper
- DAPT
- N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester
- HUVEC
- human umbilical vein endothelial cell
- CREB
- cAMP-response element–binding protein
- PLA
- proximity ligation assay
- qPCR
- quantitative PCR
- H3K27me
- histone H3 Lys-27 methylation
- TBP
- TATA-binding protein
- SRPR
- signal recognition particle receptor
- CAPNS1
- calcium-activated neutral proteinase 1.
References
- 1. Kopan R., and Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation. Cell 137, 216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guruharsha K. G., Kankel M. W., and Artavanis-Tsakonas S. (2012) The Notch signaling system: recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 13, 654–666 10.1038/nrg3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aster J. C., Pear W. S., and Blacklow S. C. (2017) The varied roles of Notch in cancer. Annu. Rev. Pathol. 12, 245–275 10.1146/annurev-pathol-052016-100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braune E. B., and Lendahl U. (2016) Notch—a Goldilocks signaling pathway in disease and cancer therapy. Discov. Med. 21, 189–196 [PubMed] [Google Scholar]
- 5. Ranganathan P., Weaver K. L., and Capobianco A. J. (2011) Notch signaling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11, 338–351 10.1038/nrc3035 [DOI] [PubMed] [Google Scholar]
- 6. Bray S. J. (2016) Notch signaling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735 10.1038/nrm.2016.94 [DOI] [PubMed] [Google Scholar]
- 7. Fiúza U.-M., and Arias A. M. (2007) Cell and molecular biology of Notch. J. Endocrinol. 194, 459–474 10.1677/JOE-07-0242 [DOI] [PubMed] [Google Scholar]
- 8. Kitagawa M. (2016) Notch signaling in the nucleus: roles of Mastermind-like (MAML) transcriptional coactivators. J. Biochem. 159, 287–294 10.1093/jb/mvv123 [DOI] [PubMed] [Google Scholar]
- 9. Bray S. J. (2006) Notch signaling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- 10. del Álamo D., Rouault H., and Schweisguth F. (2011) Mechanisms and significance of cis-inhibition in Notch signaling. Curr. Biol. 21, R40–R47 10.1016/j.cub.2010.10.034 [DOI] [PubMed] [Google Scholar]
- 11. Palmer W. H., and Deng W.-M. (2015) Ligand independent mechanisms of Notch activity. Trends Cell Biol. 25, 697–707 10.1016/j.tcb.2015.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le Borgne R., Bardin A., and Schweisguth F. (2005) The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132, 1751–1762 10.1242/dev.01789 [DOI] [PubMed] [Google Scholar]
- 13. Nichols J. T., Miyamoto A., and Weinmaster G. (2007) Notch signaling—constantly on the move. Traffic 8, 959–969 10.1111/j.1600-0854.2007.00592.x [DOI] [PubMed] [Google Scholar]
- 14. Sheldon H., Heikamp E., Turley H., Dragovic R., Thomas P., Oon C. E., Leek R., Edelmann M., Kessler B., Sainson R. C., Sargent I., Li J.-L., and Harris A. L. (2010) New mechanism of Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood 116, 2385–2394 10.1182/blood-2009-08-239228 [DOI] [PubMed] [Google Scholar]
- 15. Sharghi-Namini S., Tan E., Sharon Ong L.-L., Ge R., and Asada H. H. (2014) DLL4-containing exosomes induce capillary sprout retraction in a 3D microenvironment. Sci. Rep. 4, 4031 10.1038/srep04031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doroquez D. B., and Rebay I. (2006) Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit. Rev. Biochem. Mol. Biol. 41, 339–385 10.1080/10409230600914344 [DOI] [PubMed] [Google Scholar]
- 17. Borggrefe T., Lauth M., Zwijsen A., Huylebroeck D., Oswald F., and Giaimo B. D. (2016) The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFβ/BMP and hypoxia pathways. Biochim. Biophys. Acta 1863, 303–313 10.1016/j.bbamcr.2015.11.020 [DOI] [PubMed] [Google Scholar]
- 18. Josten F., Fuss B., Feix M., Meissner T., and Hoch M. (2004) Cooperation of JAK/STAT and Notch signaling in the Drosophila foregut. Dev. Biol. 267, 181–189 10.1016/j.ydbio.2003.11.016 [DOI] [PubMed] [Google Scholar]
- 19. Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U., and Bondesson M. (2005) Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617–628 10.1016/j.devcel.2005.09.010 [DOI] [PubMed] [Google Scholar]
- 20. D'Souza B., Miyamoto A., and Weinmaster G. (2008) The many facets of Notch ligands. Oncogene 27, 5148–5167 10.1038/onc.2008.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luca V. C., Jude K. M., Pierce N. W., Nachury M. V., Fischer S., and Garcia K. C. (2015) Structural basis for Notch1 engagement of Delta-like 4. Science 347, 847–853 10.1126/science.1261093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heuss S. F., Ndiaye-Lobry D., Six E. M., Israël A., and Logeat F. (2008) The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc. Natl. Acad. Sci. U.S.A. 105, 11212–11217 10.1073/pnas.0800695105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pintar A., De Biasio A., Popovic M., Ivanova N., and Pongor S. (2007) The intracellular region of Notch ligands: does the tail make the difference? Biol. Direct 2, 19 10.1186/1745-6150-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Six E. M., Ndiaye D., Sauer G., Laâbi Y., Athman R., Cumano A., Brou C., Israël A., and Logeat F. (2004) The notch ligand Delta1 recruits Dlg1 at cell-cell contacts and regulates cell migration. J. Biol. Chem. 279, 55818–55826 10.1074/jbc.M408022200 [DOI] [PubMed] [Google Scholar]
- 25. Ascano J. M., Beverly L. J., and Capobianco A. J. (2003) The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J. Biol. Chem. 278, 8771–8779 10.1074/jbc.M211427200 [DOI] [PubMed] [Google Scholar]
- 26. Dejana E. (2004) Endothelial cell-cell junctions: happy together. Nat. Rev. Mol. Cell Biol. 5, 261–270 10.1038/nrm1357 [DOI] [PubMed] [Google Scholar]
- 27. Lee H.-J., and Zheng J. J. (2010) PDZ domains and their binding partners: structure, specificity and modification. Cell Commun. Signal. 8, 8 10.1186/1478-811X-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun X., and Artavanis-Tsakonas S. (1996) The intracellular deletions of DELTA and SERRATE define dominant negative forms of the Drosophila Notch ligands. Development 122, 2465–2474 [DOI] [PubMed] [Google Scholar]
- 29. Chitnis A., Henrique D., Lewis J., Ish-Horowicz D., and Kintner C. (1995) Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 375, 761–766 10.1038/375761a0 [DOI] [PubMed] [Google Scholar]
- 30. Dyczynska E., Sun D., Yi H., Sehara-Fujisawa A., Blobel C. P., and Zolkiewska A. (2007) Proteolytic processing of Delta-like 1 by ADAM proteases. J. Biol. Chem. 282, 436–444 10.1074/jbc.M605451200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Six E., Ndiaye D., Laabi Y., Brou C., Gupta-Rossi N., Israel A., and Logeat F. (2003) The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and γ-secretase. Proc. Natl. Acad. Sci. U.S.A. 100, 7638–7643 10.1073/pnas.1230693100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin G., Zhang F., Chan K. M., Xavier Wong H. L., Liu B., Cheah K. S., Liu X., Mauch C., Liu D., and Zhou Z. (2011) MT1-MMP cleaves Dll1 to negatively regulate Notch signaling to maintain normal B-cell development. EMBO J. 30, 2281–2293 10.1038/emboj.2011.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ikeuchi T., and Sisodia S. S. (2003) The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent γ-secretase cleavage. J. Biol. Chem. 278, 7751–7754 10.1074/jbc.C200711200 [DOI] [PubMed] [Google Scholar]
- 34. LaVoie M. J., and Selkoe D. J. (2003) The Notch ligands, Jagged and Delta, are sequentially processed by α-secretase and presenilin/γ-secretase and release signaling fragments. J. Biol. Chem. 278, 34427–34437 10.1074/jbc.M302659200 [DOI] [PubMed] [Google Scholar]
- 35. Liebler S. S., Feldner A., Adam M. G., Korff T., Augustin H. G., and Fischer A. (2012) No evidence for a functional role of bi-directional Notch signaling during angiogenesis. PLoS One 7, e53074 10.1371/journal.pone.0053074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jung J., Mo J.-S., Kim M.-Y., Ann E.-J., Yoon J.-H., and Park H.-S. (2011) Regulation of Notch1 signaling by Delta-like ligand 1 intracellular domain through physical interaction. Mol. Cells 32, 161–1665 10.1007/s10059-011-1046-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim M.-Y., Jung J., Mo J.-S., Ann E.-J., Ahn J.-H., Yoon J.-H., and Park H.-S. (2011) The intracellular domain of Jagged-1 interacts with Notch1 intracellular domain and promotes its degradation through Fbw7 E3 ligase. Exp. Cell Res. 317, 2438–2446 10.1016/j.yexcr.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 38. Hiratochi M., Nagase H., Kuramochi Y., Koh C.-S., Ohkawara T., and Nakayama K. (2007) The Delta intracellular domain mediates TGF-β/Activin signaling through binding to Smads and has an important bi-directional function in the Notch-Delta signaling pathway. Nucleic Acids Res. 35, 912–922 10.1093/nar/gkl1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Metrich M., Bezdek Pomey A., Berthonneche C., Sarre A., Nemir M., and Pedrazzini T. (2015) Jagged1 intracellular domain-mediated inhibition of Notch1 signalling regulates cardiac homeostasis in the postnatal heart. Cardiovasc. Res. 108, 74–86 10.1093/cvr/cvv209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duryagina R., Thieme S., Anastassiadis K., Werner C., Schneider S., Wobus M., Brenner S., and Bornhäuser M. (2013) Overexpression of Jagged-1 and its intracellular domain in human mesenchymal stromal cells differentially affect the interaction with hematopoietic stem and progenitor cells. Stem Cells Dev. 22, 2736–2750 10.1089/scd.2012.0638 [DOI] [PubMed] [Google Scholar]
- 41. Kolev V., Kacer D., Trifonova R., Small D., Duarte M., Soldi R., Graziani I., Sideleva O., Larman B., Maciag T., and Prudovsky I. (2005) The intracellular domain of Notch ligand Delta1 induces cell growth arrest. FEBS Lett. 579, 5798–5802 10.1016/j.febslet.2005.09.042 [DOI] [PubMed] [Google Scholar]
- 42. Andersson E. R., and Lendahl U. (2014) Therapeutic modulation of Notch signaling—are we there yet? Nat. Rev. Drug Discov. 13, 357–378 10.1038/nrd4252 [DOI] [PubMed] [Google Scholar]
- 43. Kuhnert F., Kirshner J. R., and Thurston G. (2011) Dll4-Notch signaling as a therapeutic target in tumor angiogenesis. Vasc. Cell 3, 20 10.1186/2045-824X-3-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sainson R. C., and Harris A. L. (2007) Anti-Dll4 therapy: can we block tumour growth by increasing angiogenesis. Trends Mol. Med. 13, 389–395 10.1016/j.molmed.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 45. Adams R. H., and Alitalo K. (2007) Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 8, 464–478 10.1038/nrm2183 [DOI] [PubMed] [Google Scholar]
- 46. Herbert S. P., and Stainier D. Y. (2011) Molecular control of endothelial cell behavior during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 12, 551–564 10.1038/nrm3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carmeliet P., and Jain R. K. (2011) Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 10.1038/nature10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chung A. S., and Ferrara N. (2011) Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 27, 563–584 10.1146/annurev-cellbio-092910-154002 [DOI] [PubMed] [Google Scholar]
- 49. Geudens I., and Gerhardt H. (2011) Coordinating cell behavior during blood vessel formation. Development 138, 4569–4583 10.1242/dev.062323 [DOI] [PubMed] [Google Scholar]
- 50. Ayllón V., Bueno C., Ramos-Mejía V., Navarro-Montero O., Prieto C., Real P. J., Romero T., García-León M. J., Toribio M. L., Bigas A., and Menendez P. (2015) The Notch ligand DLL4 specifically marks human hematoendothelial progenitors and regulates their hematopoietic fate. Leukemia 29, 1741–1753 10.1038/leu.2015.74 [DOI] [PubMed] [Google Scholar]
- 51. Clevers H. (2013) The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 10.1016/j.cell.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 52. De Biasio A., Guarnaccia C., Popovic M., Uversky V. N., Pintar A., and Pongor S. (2008) Prevalence of intrinsic disorder in the intracellular region of human single-pass type I proteins: the case of the Notch ligand Delta-4. J. Proteome Res. 7, 2496–2506 10.1021/pr800063u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Timmer J. C., and Salvesen G. S. (2007) Caspase substrates. Cell Death Differ. 14, 66–72 10.1038/sj.cdd.4402059 [DOI] [PubMed] [Google Scholar]
- 54. Balasubramanian S., Fam S. R., and Hall R. A. (2007) GABAB receptor association with the PDZ scaffold Mupp1 alters receptor stability and function. J. Biol. Chem. 282, 4162–4171 10.1074/jbc.M607695200 [DOI] [PubMed] [Google Scholar]
- 55. Roukens M. G., Alloul-Ramdhani M., Baan B., Kobayashi K., Peterson-Maduro J., van Dam H., Schulte-Merker S., and Baker D. A. (2010) Control of endothelial sprouting by a Tel:CtBP complex. Nat. Cell Biol. 12, 933–942 10.1038/ncb2096 [DOI] [PubMed] [Google Scholar]
- 56. Licht A. H., Pein O. T., Florin L., Hartenstein B., Reuter H., Arnold B., Lichter P., Angel P., and Schorpp-Kistner M. (2006) JunB is required for endothelial cell morphogenesis by regulating core-binding factor β. J. Cell Biol. 175, 981–991 10.1083/jcb.200605149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kanno T., Kamba T., Yamasaki T., Shibasaki N., Saito R., Terada N., Toda Y., Mikami Y., Inoue T., Kanematsu A., Nishiyama H., Ogawa O., and Nakamura E. (2012) JunB promotes cell invasion and angiogenesis in VHL-defective renal cell carcinoma. Oncogene 31, 3098–3110 10.1038/onc.2011.475 [DOI] [PubMed] [Google Scholar]
- 58. Schorpp-Kistner M., Wang Z. Q., Angel P., and Wagner E. F. (1999) JunB is essential for mammalian placentation. EMBO J. 18, 934–948 10.1093/emboj/18.4.934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Campbell K. M., Terrell A. R., Laybourn P. J., and Lumb K. J. (2000) Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry 39, 2708–2713 10.1021/bi9923555 [DOI] [PubMed] [Google Scholar]
- 60. Gomard T., Jariel-Encontre I., Basbous J., Bossis G., Moquet-Torcy G., and Piechaczyk M. (2008) Fos family protein degradation by the proteasome. Biochemical Society Transactions 36, 858–863 10.1042/BST0360858 [DOI] [PubMed] [Google Scholar]
- 61. Erales J., and Coffino P. (2014) Ubiquitin-independent proteasomal degradation. Biochim. Biophys. Acta 1843, 216–221 10.1016/j.bbamcr.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shaulian E., and Karin M. (2001) AP-1 in cell proliferation and survival. Oncogene 20, 2390–2400 10.1038/sj.onc.1204383 [DOI] [PubMed] [Google Scholar]
- 63. Shaulian E., and Karin M. (2002) AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- 64. Plotnik J. P., Budka J. A., Ferris M. W., and Hollenhorst P. C. (2014) ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res. 42, 11928–11940 10.1093/nar/gku929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Craig M. P., and Sumanas S. (2016) ETS transcription factors in embryonic vascular development. Angiogenesis 19, 275–285 10.1007/s10456-016-9511-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Randi A. M., Sperone A., Dryden N. H., and Birdsey G. M. (2009) Regulation of angiogenesis by ETS transcription factors. Biochem. Soc. Trans. 37, 1248–1253 10.1042/BST0371248 [DOI] [PubMed] [Google Scholar]
- 67. Shah A. V., Birdsey G. M., Peghaire C., Pitulescu M. E., Dufton N. P., Yang Y., Weinberg I., Osuna Almagro L., Payne L., Mason J. C., Gerhardt H., Adams R. H., and Randi A. M. (2017) The endothelial transcription factor ERG mediates Angiopoietin-1-dependent control of Notch signaling and vascular stability. Nat. Commun. 8, 16002 10.1038/ncomms16002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meester J. A., Southgate L., Stittrich A.-B., Venselaar H., Beekmans S. J., den Hollander N., Bijlsma E. K., Helderman-van den Enden A., Verheij J. B., Glusman G., Roach J. C., Lehman A., Patel M. S., de Vries B. B., Ruivenkamp C., et al. (2015) Heterozygous loss-of-function nutations in DLL4 cause Adams-Oliver syndrome. Am. J. Hum. Genet. 97, 475–482 10.1016/j.ajhg.2015.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lundby A., and Olsen J. V. (2011) GeLCMS for in-depth protein characterization and advanced analysis of proteomes. Methods Mol. Biol. 753, 143–155 10.1007/978-1-61779-148-2_10 [DOI] [PubMed] [Google Scholar]
- 70. Lundby A., Rossin E. J., Steffensen A. B., Acha M. R., Newton-Cheh C., Pfeufer A., Lynch S. N., QT Interval International GWAS Consortium (QT-IGC), Olesen S. P., Brunak S., Ellinor P. T., Jukema J. W., Trompet S., Ford I., Macfarlane P. W., et al. (2014) Annotation of loci from genome-wide association studies using tissue-specific quantitative interaction proteomics. Nat. Methods 11, 868–874 10.1038/nmeth.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roukens M. G., Alloul-Ramdhani M., Moghadasi S., Op den Brouw M., and Baker D. A. (2008) Downregulation of vertebrate Tel (ETV6) and Drosophila Yan is facilitated by an evolutionarily conserved mechanism of F-box-mediated ubiquitination. Mol. Cell. Biol. 28, 4394–4406 10.1128/MCB.01914-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Roukens M. G., Alloul-Ramdhani M., Vertegaal A. C., Anvarian Z., Balog C. I., Deelder A. M., Hensbergen P. J., and Baker D. A. (2008) Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol. Cell. Biol. 28, 2342–2357 10.1128/MCB.01159-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.