Abstract
Aberrant expression of O-glycans is a hallmark of epithelial cancers. Mucin-type O-glycosylation is initiated by a large family of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (GalNAc-Ts) that target different proteins and are differentially expressed in cells and organs. Here, we investigated the expression patterns of all of the GalNAc-Ts in colon cancer by analyzing transcriptomic data. We found that GalNAc-T6 was highly up-regulated in colon adenocarcinomas but absent in normal-appearing adjacent colon tissue. These results were verified by immunohistochemistry, suggesting that GalNAc-T6 plays a role in colon carcinogenesis. To investigate the function of GalNAc-T6 in colon cancer, we used precise gene targeting to produce isogenic colon cancer cell lines with a knockout/rescue system for GALNT6. GalNAc-T6 expression was associated with a cancer-like, dysplastic growth pattern, whereas GALNT6 knockout cells showed a more normal differentiation pattern, reduced proliferation, normalized cell–cell adhesion, and formation of crypts in tissue cultures. O-Glycoproteomic analysis of the engineered cell lines identified a small set of GalNAc-T6–specific targets, suggesting that this isoform has unique cellular functions. In support of this notion, the genetically and functionally closely related GalNAc-T3 homolog did not show compensatory functionality for effects observed for GalNAc-T6. Taken together, these data strongly suggest that aberrant GalNAc-T6 expression and site-specific glycosylation is involved in oncogenic transformation.
Keywords: cancer biology, cell adhesion, colon cancer, differentiation, glycobiology, GalNAc, GalNAc-Ts, genetic engineering, glycoproteomics, tissue model
Introduction
Malignant transformation is closely associated with changes in the glycosylation of proteins and lipids (1, 2). One well-documented example, which is observed in the majority of epithelial cancers and premalignant lesions, is cancer-associated changes in GalNAc O-glycosylation (1, 3–5). GalNAc-type O-glycosylation is an abundant and diverse form of post-translational modification (6). It is initiated by a family of up to 20 polypeptides, termed GalNAc-transferases (GalNAc-Ts),2 that catalyzes the addition of GalNAc residues to the hydroxyl groups of selected serine and threonine residues in proteins (6–19). In healthy cells, the initiating GalNAc residue (GalNAcα1, also known as the Tn antigen) is elongated, branched, and capped with different carbohydrate structures in sequential processing steps. In contrast, cancer cells are often characterized by the expression of immature and truncated O-glycan structures, such as Tn and sialylated Tn (STn) (20). The expression of these short and truncated O-glycans strongly correlates with poor prognosis (21–24). Although the association between the expression of truncated O-glycans and cancer prognosis per se is well-established, the importance of GalNAc-T-mediated site-specific glycosylation in cancer is unclear, due mainly to technical limitations. Until recently, it was not possible to conduct a global analysis of GalNAc-T function to identify which proteins are glycosylated with this moiety and the glycosylation sites within the proteins. We recently developed a differential global glycoproteomic strategy, which, in combination with genetic engineering, enables us to investigate the function of specific GalNAc-Ts in cell line models and to begin to investigate the site-specific functions of GalNAc glycosylation in cancer (25–27).
GalNAc-Ts control the initiation of O-glycan biosynthesis and are differentially expressed in cells and tissues. They have distinct, partly overlapping acceptor substrate specificities (6, 28, 29). GalNAc-T glycosylation has been implicated in numerous important biological functions, including pro-protein processing, ecto-domain shedding, cell signaling, and cell adhesion (30–33). Furthermore, GalNAc-Ts are reported to influence several key processes that are important for tumor formation, including growth (34–37), immune evasion (38, 39), and invasion and metastasis (36, 37, 40–44). Notably, the exact relationship of site-specific O-glycosylation and tumor formation is unknown, and we do not yet understand how up-regulation of selective GalNAc-Ts affects carcinogenesis.
A homologous pair of glycosyltransferases, GalNAc-T3 and GalNAc-T6, is the most prominent example of cancer-associated GalNAc-Ts (6, 10, 28, 29, 45, 46). GalNAc-T3 and GalNAc-T6 display high similarity at both DNA and amino acid levels, having similar genomic organization with nine identically positioned intron/exon boundaries in the coding regions (10). GalNAc-T3 is expressed in almost all normal epithelia, and dysregulated expression of GalNAc-T3 has been observed in various carcinomas, including oral (41, 47), colon (44), pancreatic (48–50), breast (51), gallbladder (52), gastric (53, 54), prostate (55), renal (56), lung (57), esophageal (58), thyroid (59), and extrahepatic bile duct (42) carcinomas. GalNAc-T6 is also reported to be aberrantly expressed in various types of cancer, including breast (60–64), gastric (40), renal (56), and pancreatic (49) carcinomas. However, in contrast to GalNAc-T3, GalNAc-T6 is absent in most healthy tissues. Furthermore, the correlation of GalNAc-T6 expression with early stage cancer tissue (65) implies that GalNAc-T6 plays a role in early carcinogenesis. This is supported by reports suggesting that GalNAc-T6 is involved in the induction of the epithelial-to-mesenchymal transition (EMT) and cadherin-switching, which are followed by morphological changes and increased metastatic potential (66–68).
In this study, we examined the expression patterns of all GalNAc-Ts in colon cancer using transcriptomic data analysis, and we observed selective up-regulation of GalNAc-T6 but not GalNAc-T3. To shed light on the function of GalNAc-T6 in colon cancer development, we developed a cell model system in which specific ablation of the gene encoding GalNAc-T6 (GALNT6) was followed by detailed polyomic analysis. Precise genome-targeted knockout of GALNT6, GALNT3, or a combination of the two in the LS174T colon cancer cell line demonstrated that GalNAc-T6 expression was essential for the acquisition of oncogenic features such as hyperproliferation, loss of normal colonic epithelial architecture, and the disruption of cell–cell adhesion. Thus, LS174T GALNT6 knockout cells showed terminal differentiation traits and formed crypt-like structures that resembled the tissue architecture of a healthy colon, features that were reverted upon reintroduction of exogenous GalNAc-T6. Differential transcriptomic analysis confirmed that the expression profile of the GalNAc-T6-expressing LS174T cells resembled that of colon cancer cells, whereas LS174T GALNT6 knockout cells had an expression profile that was more similar to that of normal colon tissue. Furthermore, differential O-glycoproteomic analysis identified unique GalNAc-T6 targets, including several important cellular adhesion proteins. These results support the notion that aberrantly expressed GalNAc-T6 plays an important role in colorectal carcinogenesis.
Results
Selective up-regulation of GalNAc-T6 in colon cancer tissue
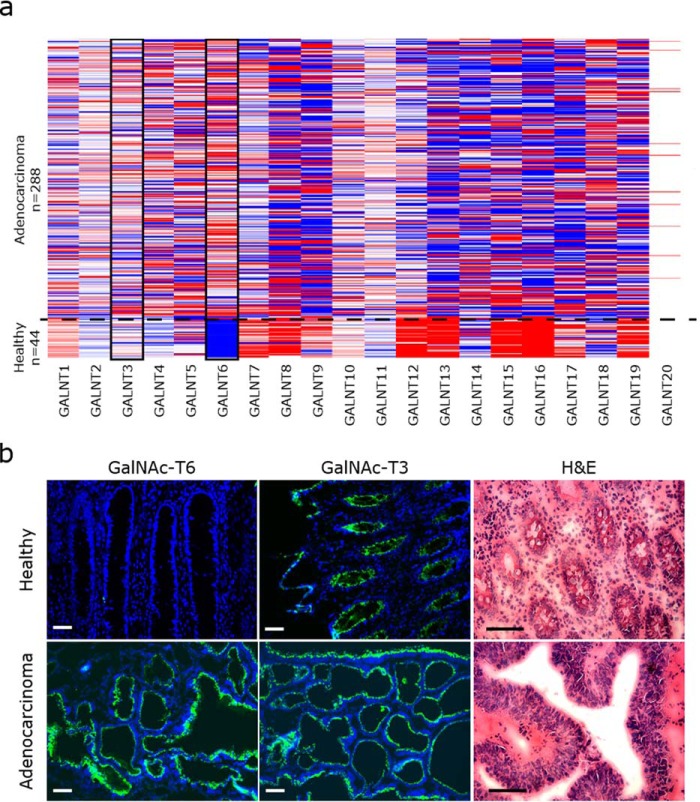
We first used The Cancer Genome Atlas (TCGA) on colon adenocarcinomas to identify potential cancer-associated changes in the expression of GalNAc-Ts. Expression profiles for all 20 GalNAc-T isoforms were analyzed in 288 colon adenocarcinomas and in 41 healthy colon tissue samples using RNAseq transcriptome data (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/)3 (Fig. 1a and Fig. S1). Of the 20 GalNAc-T isoforms, GalNAc-T6 was the only GalNAc-T that was expressed de novo in colon cancer, i.e. was absent from healthy colon tissue. In contrast, the majority of GalNAc-Ts was either unregulated or down-regulated in colon cancer (Fig. 1a and Fig. S1). To confirm the cancer-specific up-regulation of GalNAc-T6 at the protein level, we evaluated the expression of GalNAc-T6 in 39 cases of colorectal carcinomas and in healthy colorectal mucosa by immunostaining. The expression pattern of GalNAc-T6 was compared with the expression of its close homolog GalNAc-T3 (Fig. 1b; Table 1). GalNAc-T6 expression was detected in 34 of 39 carcinomas with antibody labeling restricted to the perinuclear area, suggesting its localization to the Golgi apparatus. GalNAc-T6 expression was not detected in normal colorectal mucosa in four of four cases. High levels of GalNAc-T3 were detected in all 39 colorectal carcinomas, and GalNAc-T3 was almost homogeneously expressed in all layers of the normal-appearing crypts as well as in the tumor tissues. Thus, in contrast to GalNAc-T3, GalNAc-T6 was overexpressed in tumor tissue but not in normal colon tissue. This establishes that GalNAc-T6 is up-regulated at both RNA and protein levels in a cancer-specific manner and suggests that aberrantly expressed GalNAc-T6 has a unique function in colon cancer progression.
Figure 1.
Expression pattern of GalNAc-T6 and GalNAc-T3 in colon tissue. a, TCGA IlluminaHiSeq RNAseq data obtained from https://genome-cancer.ucsc.edu/proj/site/hgHeatmap3 show the expression of GalNAc-Ts in 288 colon adenocarcinomas and 44 healthy colon tissue samples. Red >0, white = 0, blue <0, gray = no data. The data are normalized by subtracting the mean of the RNAseq values from each sample value for each of the 20 GalNAc-T and shown in red or blue color. GalNAc-T6 is specifically up-regulated in colon adenocarcinoma, whereas GalNAc-T3 expression is unchanged. b, immunofluorescence staining of GalNAc-T6 (mAb 2F3) and GalNAc-T3 (mAb 2D10) (green) in colorectal adenocarcinoma and healthy colon mucosa (blue, DAPI). GalNAc-T6 is strongly expressed in tumor tissue and absent in normal tissue, whereas GalNAc-T3 is expressed in both types of tissue. Hematoxylin and eosin (H&E) staining shows the morphology of tumor tissue compared with normal tissue in the present sample. Scale bar, 50 μm.
Table 1.
GalNAc-T6 and GalNAc-T3 expression in colon adenocarcinoma
Tissues were evaluated as positive when more than 25% of the cells were labeled. Labeling intensities were scored from 0 (negative) to 3 (high intensity staining).
| GalNAc-T6 |
GalNAc-T3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Labeling intensities |
Total positive (>1) | Labeling intensities |
Total positive (>1) | |||||||
| 0 | 1 | 2 | 3 | 0 | 1 | 2 | 3 | |||
| Colorectal carcinoma | ||||||||||
| Well-differentiated (n = 22) | 1 | 2 | 3 | 16 | 95% (21/22) | 0 | 0 | 0 | 22 | 100% (22/22) |
| Moderately differentiated (n = 10) | 2 | 0 | 2 | 6 | 80% (8/10) | 0 | 0 | 0 | 9 | 100% (9/9) |
| Poorly differentiated (n = 1) | 1 | 0 | 0 | 0 | 0% (0/1) | 0 | 0 | 0 | 1 | 100% (1/1) |
| No information (n = 6) | 1 | 1 | 3 | 1 | 83% (5/6) | 0 | 0 | 0 | 6 | 100% (6/6) |
| Total | 5 | 3 | 8 | 23 | 87% (34/39) | 0 | 0 | 0 | 39 | 100% (39/39) |
| Healthy | 4 | 0 | 0 | 0 | 0% (0/4) | 0 | 0 | 0 | 2 | 100% (2/2) |
GalNAc-T6 disrupts the formation of actin-lined lumens and is associated with the expression of cancer-associated genes in vitro
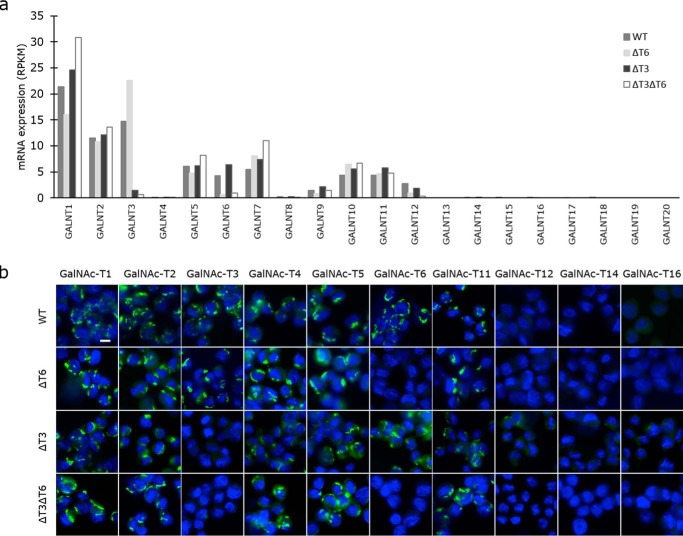
We next used the well-differentiated human LS174T colon adenocarcinoma cell line as a cell model to evaluate colon cell growth in the presence and absence of GalNAc-T6. LS174T cells exhibit unrestricted growth and grow as separate clusters of cells, supposedly due to inhibited p21WAF1 expression (69). GALNT6 and GALNT3 were knocked out in LS174T cells, individually or combined, using zinc finger nuclease (ZFN)-based genome editing to produce ΔT6 and ΔT3 cells. Successful out-of-frame mutagenesis was confirmed in individual single-cell clones (Table S1). RNAseq verified that non-sense–mediated RNA decay had removed the targeted transcripts (Fig. 2a), which was also shown by immunofluorescence staining (Fig. 2b). Knockout of GALNT6 was accompanied by an increase in GALNT3 transcripts, and similarly, the knockout of GALNT3 was associated with an increase in GALNT6 transcripts, which suggests that these two enzymes can compensate for each other (Fig. 2a). There were no notable changes in the expression of other GalNAc-Ts after select knockout of either GALNT3 or GALNT6. Knockout of both GalNAc-T3 and -T6, however, resulted in an increase in the expression of GALNT1.
Figure 2.
Expression of various GalNAc-T isoforms in LS174T WT and GALNT6 and/or GALNT3 knockout cells at the RNA level in the transcriptomic analysis (a) and at the protein level as analyzed by immunofluorescence staining (b). Green, GalNAc-T; blue, DAPI. Scale bar, 10 μm in all images.
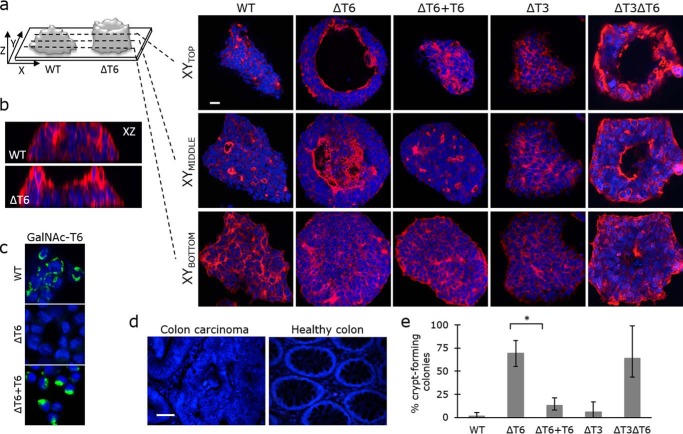
In accordance with previous reports (69), wildtype (WT) LS174T cells formed multilayered colonies, thereby replicating colon cancer growth. Phalloidin staining, to detect F-actin cytoskeletal protein, showed that WT LS174T colon cancer cells, expressing high levels of GalNAc-T6, grew as clusters of cells with dense tubular structures and multiple small, actin-lined lumens, which could resemble the disordered crypts seen in colon cancer tissue (Fig. 3, a, b, and d). Intriguingly, knockout of GALNT6 resulted in cells that grew as colonies with one large actin-lined lumen surrounded by a wall of cells of varying thickness. Staining of healthy colon tissue revealed similarity of these luminal structures with healthy colonic crypts (Fig. 3d).
Figure 3.
GALNT6 knockout induces crypt-like morphology in the LS174T colon cancer cell line. a, actin-lined lumens were detected by phalloidin staining (red) that was visualized by confocal microscopy. XY micrographs from three z-sections are presented. Scale bar, 20 μm. b, XZ micrographs of WT and ΔT6 colonies generated by confocal z-stacks. c, expression of GalNAc-T6 after re-introduction of GALNT6 (ΔT6+T6) in GALNT6 knockout cells (ΔT6). Green, GalNAc-T6; blue, DAPI. Scale bar, 10 μm. d, colon carcinoma and healthy tissue section stained with DAPI (blue). e, more than 300 colonies of each cell type were investigated, and the percentage of crypt-forming colonies was determined in a blinded manner. *, t test, p value = 0.008853.
To confirm that the phenotypic change observed in LS174TΔT6 cells was the result of GALNT6 knockout rather than a clonal effect, we re-introduced functional, constitutively expressed GALNT6 into ΔT6 cells to create ΔT6+T6 cells. This was accomplished using a recently published site-specific ZFN-mediated knockin strategy (Fig. 3c and Table S1) (70, 71). GALNT6 re-introduction rescued the phenotype, and the ΔT6+T6 cells formed disorganized clusters of cells with multiple small actin-lined lumens (Fig. 3a). Interestingly, no major phenotypic changes were observed after knocking out the close homolog GALNT3 in a WT or ΔT6 cell background (ΔT3 and ΔT3ΔT6) (Fig. 3a). When we observed more than 300 colonies of ΔT6 and ΔT3ΔT6 cells, we found that 69 and 64% formed crypt-like structures, respectively, compared with 1.5% of WT cells, 1% of ΔT3 cells, and 13% of ΔT6+T6 cells (Fig. 3e). Taken together, these results indicate that the specific up-regulation of GalNAc-T6 expression during malignant transformation disrupts colon crypt formation.
GalNAc-T6 influences the proliferation and differentiation of colon cancer cells
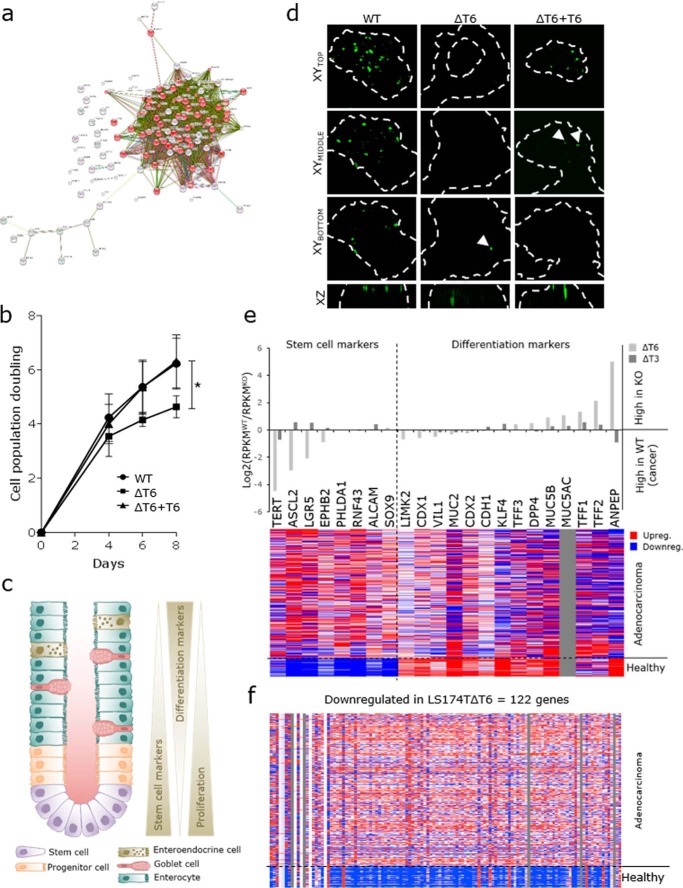
To investigate the functional role of GalNAc-T6 in colon carcinogenesis, we performed RNAseq analysis of LS174T cells that did or did not express GalNAc-T6 and GalNAc-T3 (Table S2). We defined 122 genes that were significantly down-regulated in LS174TΔT6 cells (RPKM(WT) ≥10, log2(ΔT6/WT) ≤ −2) (Table S3). String (http://string-db.org)3 and functional enrichment analysis (Gene Ontology terms) revealed that half of these genes (62/122) are involved in the cell cycle (Fig. 4a). Investigation of the proliferative potential of LS174T cells showed lower cell population doubling times for cells lacking GalNAc-T6 compared with wildtype cells (Fig. 4b). When GalNAc-T6 expression was restored in LS174T cells, the doubling curves phenocopied those of the WT cells. No changes in proliferation were observed in ΔT3 cells. These findings suggest that GalNAc-T6 promotes cellular proliferation in colon cancer.
Figure 4.
GalNAc-T6 expression stimulates proliferation and suppresses differentiation in LS174T colon cancer cells. a, Gene Ontology term analysis of genes that are down-regulated in GALNT6 knockout cells (string-db.org).3 The genes involved in cell cycle are depicted in red. b, cell population doubling curves for LS174T cells with the indicated genotypes. *, t test, p value = 0.0149. c, morphology and cell types in a normal colon crypt. Stem cells at the crypt base differentiate into progenitor cells, which further differentiate into enterocytes, goblet cells, and enteroendocrine cells. Stem cell markers and proliferating cells are found at the bottom part of the crypt, and differentiated cells are found toward the mucosal surface of the colon. d, LGR5 expression (green) in WT and ΔT6 colonies shown in XY and XZ micrographs generated by confocal z-stacks. Colony borders were visualized by actin staining and are depicted as white dashed lines. Scale bar, 20 μm. e, upper panel, regulation of stem cell markers and differentiation markers in the transcriptomes of ΔT6 and ΔT3 cells compared with WT cells. Data are shown by the relative level of RPKM on a log2 scale. e, lower panel, expression of the same stem cell and differentiation markers in adenocarcinomas and in healthy colon tissue using the TCGA IlluminaHiSeq RNAseq data obtained from https://genome-cancer.ucsc.edu/proj/site/hgHeatmap3 showing gene expression in 292 colon adenocarcinomas and 37 healthy colon tissue samples. Red, >0; white = 0; blue, <0; gray = no data. f, TCGA IlluminaHiSeq RNAseq data obtained from https://genome-cancer.ucsc.edu/proj/site/hgHeatmap3 showing gene expression in 292 colon adenocarcinomas and 37 healthy colon tissue samples of genes that were down-regulated at least 4-fold (log2 ≤ −2) in the transcriptome of ΔT6 cells compared with WT cells. Only genes with RPKM values greater than 10 are included. Red, >0; white = 0; blue, <0; gray = no data.
In normal colon crypts, cells arise from stem cells located in the crypt base. As the cells differentiate, they lose their ability to proliferate (Fig. 4c). In cancer, however, a lack of differentiation allows sustained proliferation. The formation of organized crypts in LS174TΔT6 cells suggests a shift toward a more differentiated state and might explain the decrease in proliferative potential. To test this hypothesis, we stained LS174T cells for the leucine-rich repeat-containing G-protein–coupled receptor 5 (LGR5). LGR5 is considered one of the most selective stem cell markers in the intestine (72), and its expression is restricted to the crypt base of normal colon mucosa. LS174TΔT6 cells showed positive LGR5 staining only at the crypt base of the colonies (Fig. 4d), mimicking the LGR5 expression pattern in healthy colon tissue. In contrast, LS174T cells that expressed GalNAc-T6 had a high number of LGR5-positive cells in the upper layers of the colonies, with increasing staining intensity at the luminal surface, similar to the expression pattern of LGR5 in human colon cancer (73). This may indicate that GalNAc-T6 expression induces a cancer-like LGR5 expression pattern in these colonies (Fig. 4d). The number of LGR5-positive cells was higher in ΔT6+T6 cultures than in ΔT6 cultures, with increased staining toward the top of the colonies, although reintroduction of GalNAc-T6 expression did not completely revert the ΔT6 phenotype.
Next, we assessed the expression of markers that define various differentiation stages in the colon in the transcriptome of LS174T cells, and we found a correlation between the loss of GalNAc-T6 and increased transcription of differentiation marker genes. Conversely, the transcriptome of LS174T cells that expressed GalNAc-T6 had an expression signature that was characteristic of non-differentiated colon cells (Fig. 4e, upper panel, and Table S4). We used the colon adenocarcinoma TCGA transcriptome database to assess whether this GalNAc-T6-dependent gene expression pattern resembled the expression pattern in human colon cancer tissue (Fig. 4e, lower panel). Remarkably, the general low expression of differentiation markers in LS174T WT cancer cells mimics the low expression of these markers in human colon adenocarcinomas (Fig. 4e). Conversely, the expression profile of LS174T cells lacking GalNAc-T6 resembled the expression profile of healthy colon tissue in that both had decreased expression of stem cell markers and increased expression of genes associated with the differentiated state. Knockout of GALNT3 did not affect the expression of differentiation markers.
The ability to form crypts, keep a low proliferation rate, and retain a highly differentiated state are all properties of healthy colon cells that are lost during carcinogenesis. In LS174T cancer cells, elimination of GalNAc-T6 slows down proliferation and allows the cells to differentiate and form crypt-like structures, which suggests a shift toward a less dysplastic phenotype. To investigate whether the change in expression detected upon knockout of GALNT6 resembles the shift in gene expression from healthy colon tissue to colon adenocarcinoma, we used the dataset from the TCGA transcriptome database. The vast majority of the 122 genes down-regulated in LS174TΔT6 cells was also expressed at lower levels in healthy colon tissue when compared with colon adenocarcinomas (Fig. 4f). The trend was less clear for the 43 genes that were significantly up-regulated in ΔT6 cells (RPKM(ΔT6) ≥10, log2(ΔT6/WT) ≥2) (Table S3). In this case, we found only a portion of the genes to be up-regulated in healthy colon tissue compared with colon adenocarcinomas (data not shown). In contrast, the expression of only a few genes was changed in ΔT3 cells (Tables S2 and S3 and data not shown).
GalNAc-T6 controls cell–cell adhesion but does not induce the EMT
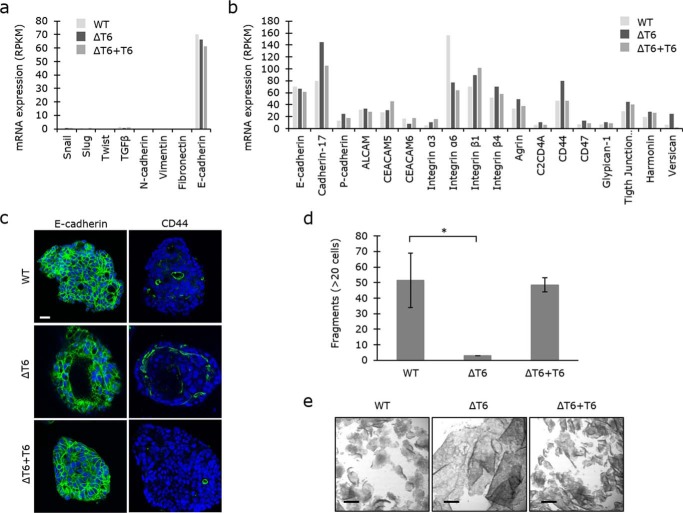
GalNAc-T6 expression has previously been associated with induction of the EMT (68). We therefore investigated whether the LS174T transcriptome expressed markers that are associated with the epithelial phenotype (E-cadherin/CDH1) or mesenchymal markers (N-cadherin/CDH2; vimentin/VIM; fibronectin/FN1) (Fig. 5a). We did not detect any notable change in any of these markers or in inducers of EMT (SNAI1/Snail; SNAI2/Slug; TWIST1/Twist; TGFB1/transforming growth factor β), which implies that GalNAc-T6 expression alone does not induce EMT. Knockdown of GalNAc-T6 was recently shown to induce a switch from P-cadherin to E-cadherin expression in pancreatic cancer cells (67). However, our colon transcriptome data indicated rather an opposite shift from E-cadherin to P-cadherin expression (Fig. 5b). In addition to P-cadherin, several other proteins involved in adhesion, predominantly cell–cell adhesion, were up-regulated upon GALNT knockout, including cadherin-17, CD44, and versican (Fig. 5, b and c). This indicates that expression of GalNAc-T6 disrupts the intercellular adhesive potential. To test this hypothesis, we performed a cellular dissociation assay on confluent sheets of LS147T cells that did or did not express GalNAc-T6. Cell–cell adhesion was significantly stronger in LS174TΔT6 cells than in LS174T WT cells, where the confluent cell sheet was easily disrupted (Fig. 5, d and e). Reintroduction of GalNAc-T6 expression in LS174TΔT6 cells rescued the WT phenotype, indicating that GalNAc-T6 decreases intercellular adhesion in LS174T cell cultures (Fig. 5, d and e). However, whether the change in expression of adhesion molecules is responsible for the changes in cell–cell adhesion that we observed in LS174TΔT6 cells remains to be determined.
Figure 5.
GALNT6 knockout increases cell–cell adhesion. a, mRNA expression of markers of epithelial-to-mesenchymal transition in LS174T WT, ΔT6, and ΔT6+T6 cells. b, mRNA expression of proteins involved in adhesion. c, fluorescence staining of E-cadherin and CD44 proteins in colonies of LS174T WT, ΔT6, and ΔT6+T6 cells (green, DAPI in blue). Scale bar, 20 μm. d and e, cells grown to confluency were dissociated from the plastic surface as complete cell layers by dispase treatment. After pipetting the cells up and down three times, cell fragments were counted as a measure of cell–cell adhesion. Fragments with at least 20 cells were counted. e, micrographs of the cell layers/fragments. Scale bars, 500 μm. *, t test, p value = 0.0436.
Differential O-glycoproteomic analysis identifies substrates glycosylated by GalNAc-T6
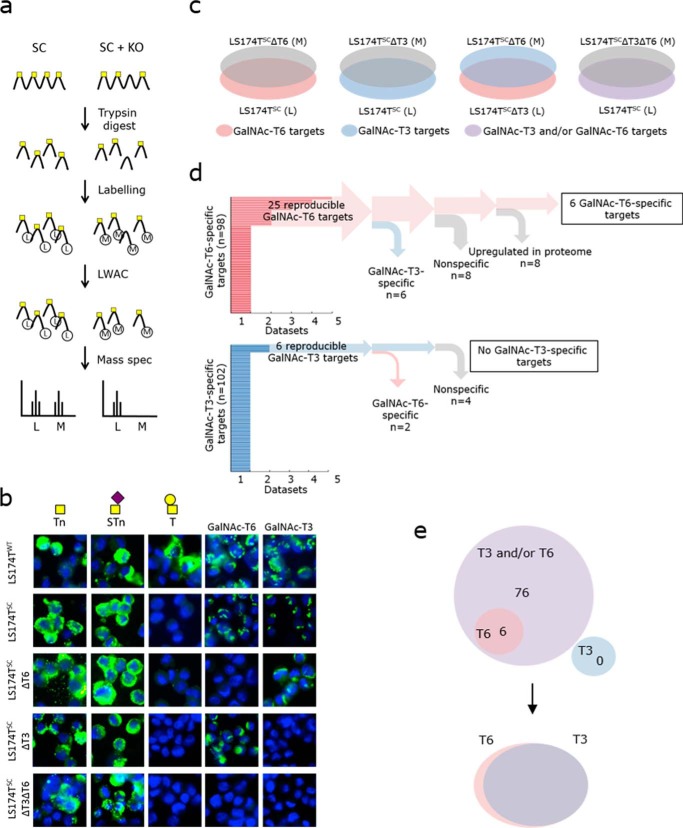
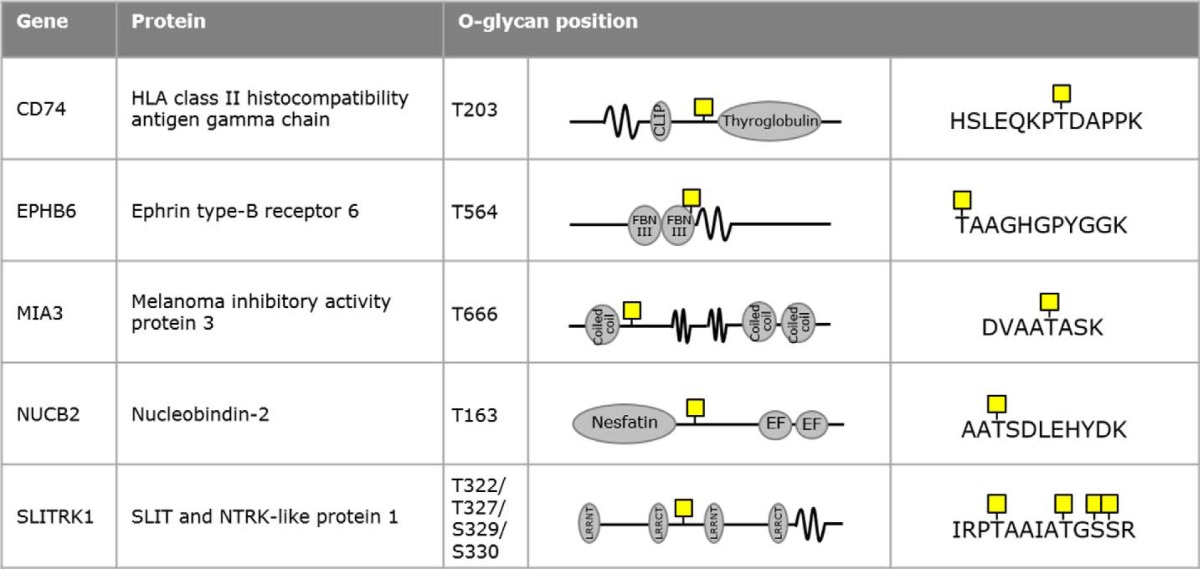
To further investigate the role of GalNAc-T6 in colon cancer development, we analyzed GalNAc-T6 target sites using the SimpleCell strategy. The SimpleCell strategy (Fig. 6a) uses homogeneous truncation of O-glycosylation to produce short glycan structures (Tn and STn) that allow enrichment of glycopeptides and determination of glycosites by nanoflow liquid chromatography-tandem mass spectrometry with electron transfer dissociation (ETD). This strategy can be combined with targeted knockout and knockin of individual GALNTs for broad ex vivo discovery of GalNAc-T isoform-specific functions (27, 29). SimpleCell versions of LS174T (LS174TSC) cells that do or do not express GalNAc-T6 and/or GalNAc-T3 were developed by ZFN-mediated knockout of COSMC (Fig. 6b and Table S1) (29, 32). Stable isotype dimethyl labeling (74) of total tryptic peptide digests from isogenic cell pairs with either light (L) or medium (M) reagent allowed quantitative profiling of GalNAc O-glycopeptides using sensitive O-glycoproteomic mass spectrometry (Fig. 6a) (75). The strategy and the light/medium pairs are shown in Fig. 6c. Elimination of specific GalNAc-T isoforms in LS174TSC cells is thus expected to reveal glycosylation sites specific to those isoforms. Specifically, we looked for down-regulation or loss of M-labeled glycopeptides compared with L-labeled glycopeptides by analysis of the complete data sets from all of the differential O-glycoproteomes (Table S5). Based on earlier observations (27, 29), we considered 10-fold down-regulated M-labeled glycopeptides (log10(M/L) ≤ −1) to be GalNAc-T6–specific and/or GalNAc-T3-specific sites. When comparing GalNAc-T3 and GalNAc-T6 directly (Fig. 6c), specific GalNAc-T3 targets were the result of a loss of L-labeled LS174TSCΔT3 glycopeptides, producing log10(M/L) values that were equal to or higher than 1. The analysis was performed on total cell lysates (TCL) as well as on secretomes (SEC) with two or three replicates for each set. 43 potential GalNAc-T6–specific glycosylation sites were identified in LS174TSCΔT6 cells, whereas 67 sites were identified from the LS174TSCΔT3 versus LS174TSCΔT6 comparison; 12 sites were found using both approaches leaving a total of 98 potential GalNAc-T6–specific sites. In addition, 102 potential GalNAc-T3 targets were identified (35 sites from LS174TSC versus LS174TSCΔT3 comparison and 72 sites from LS174TSCΔT3 versus LS174TSCΔT6 comparison, and five sites were found using both approaches). Finally, 130 potential GalNAc-T3- and/or GalNAc-T6–specific targets were identified by the comparison of LS174TSC and LS174TSCΔT3ΔT6 cells (Fig. 6d and Table S5). To refine the results, we only considered targets found in at least two datasets. Furthermore, we rejected all glycosites that were targets of both GalNAc-T6 and GalNAc-T3. Finally, we performed focused proteome analysis (data not shown) to ensure that the observed loss of glycosite was not due to down-regulation of acceptor substrates. After this rigorous selection, six glycopeptides in six different proteins were found to be selectively glycosylated by GalNAc-T6 (Fig. 6d and Table 2). Another 82 glycopeptides in 68 proteins were glycosylated by GalNAc-T3 and/or GalNAc-T6 (Fig. 6e and Table S6). The six GalNAc-T6–specific targets that we identified were as follows: melanoma inhibitory activity protein 3 (Thr-666); ephrin type-B receptor 6 (Thr-564); coiled-coil domain-containing protein 14 (Thr-807/Ser-819)4; HLA class II histocompatibility antigen γ chain (Thr-203); nucleobindin-2 (Thr-163); and SLIT and NTRK-like protein 1 (Thr-322/Thr-327/Ser-329/Ser-330). In contrast, we did not find any reproducible, selective GalNAc-T3 targets that were not shared with GalNAc-T6 (Fig. 6, d and e). These results suggest that the majority of the glycosites in LS174T cells can be targeted by the two homologs, GalNAc-T3 and GalNAc-T6. In addition, GalNAc-T6 can glycosylate a small number of unique receptors or secreted matrix proteins that are involved in cell–cell adhesion (Fig. 6e).
Figure 6.
Differential O-glycoproteomic analysis of GalNAc-T6 and GalNAc-T3 using the SimpleCell strategy. a, digests of cell pairs were labeled with either light (L) or medium (M) isotopes and mixed 1:1. The glycopeptides were submitted to VVA LWAC for GalNAc-glycopeptide enrichment and then analyzed further by mass spectrometry. b, production of SimpleCells (LS174TSC). Knockout of COSMC generates cells with short glycans (Tn and sTn). The SimpleCells strategy was described by Steentoft et al. (26). Additional knockouts of GALNT6 (ΔT6), GALNT3 (ΔT3), or both (ΔT3ΔT6) in LS174TSC cells are shown. Green, GalNAc-T/O-glycans; blue, DAPI. Scale bar, 10 μm in all images. c, differential O-glycoproteomes were analyzed for the indicated pairs: LS174TSC (L) versus LS174TSCΔT6 (M); LS174TSC (L) versus LS174TSCΔT3 (M); LS174TSC (L) versus LS174TSCΔT3ΔT6 (M); and LS174TSCΔT3 (L) versus LS174TSCΔT6 (M). Red, potential GalNAc-T6 O-glycosylation targets; blue, potential GalNAc-T3 targets; purple, potential GalNAc-T3 and/or GalNAc-T6 targets. d, 98 GalNAc-T6–specific glycosylation sites were identified. Of 25 reproducible targets, 19 targets were also detected as GalNAc-T3 specific, non-specific, or false positives due to up-regulated protein levels, leaving six specific targets for GalNAc-T6. Of 102 GalNAc-T3 targets, no specific targets were detected. e, 82 GalNAc-T3/GalNAc-6 targets were detected, which included six GalNAc-T6–specific targets. There were no GalNAc-T3-specific targets, suggesting that GalNAc-T3 and GalNAc-T6 have overlapping functions, with just a few GalNAc-T6–specific O-glycosylation targets.
Table 2.
GalNAc-T6-specific O-glycosylation sites
Six GalNAc-T6-specific O-glycosylation sites were identified as described in Fig. 6. CCDC14 was excluded from the table due to its presumed cytoplasmic localization, which could question the nature of the HexNAc residues. When several O-glycans are depicted, the precise position of the GalNAc-T6 specific O-glycosylation could not be determined.
Discussion
Aberrant expression of O-glycans is a hallmark of epithelial cancers (76–80) and is associated with poor prognosis and survival (20). Although many studies have investigated how changes in O-glycan elongation and branching affect cellular behavior in cancer, only a few studies have investigated the impact of changes in the initiating step of O-glycan biosynthesis, which is governed by GalNAc-Ts. Here, we demonstrated that the de novo expression of GalNAc-T6 suppresses human colon cancer cell differentiation. This suggests that the glycosylation pattern governed by GalNAc-T6 has a unique role in colon carcinogenesis. Furthermore, this implies that the GalNAc-T6 near-identical homolog, GalNAc-T3, does not functionally compensate for GalNAc-T6 and suggests that the high similarity sub-family members, which most GalNAc-Ts belong to, can serve unique functions.
Up-regulated GalNAc-T6 levels were previously reported in various types of cancer, including breast (60–64), gastric (40), renal (56), and pancreatic (49) carcinomas. Here, we report a de novo expression of GalNAc-T6 in colon adenocarcinoma. Approximately 95% of colon cancers are of the adenocarcinoma type. The remaining 5% includes sarcomas and squamous cell and carcinoid tumors. However, the regulation of GalNAc-T6 in these less common carcinomas remains to be evaluated.
To assess the importance of GalNAc-T6 expression in differentiation of colonic epithelia, we used the LS174T tumor cell line, which is derived from a human adenocarcinoma (81). LS174T cells are well-differentiated and exhibit unrestricted growth as separate clusters of cells; this is thought to be due to inhibited p21WAF1 expression (69). We demonstrated that the knockout of GALNT6 in LS174T cells reverted their cancer-like growth characteristics and promoted defined tissue organization with the formation of crypt-like structures. In contrast, knockout of the close homolog GALNT3, which is expressed in both healthy colon tissue and in colon cancer, did not induce any changes in growth.
The increased expression of GalNAc-T6 has previously been suggested to promote morphological changes in several human cancers (66, 82). In breast cancer, overexpression of GalNAc-T6 was reported to decrease cellular adhesion and disrupt mammary acinar morphogenesis (66, 67, 82). Furthermore, it has been shown that GalNAc-T6 expression in pancreatic cancer causes a switch from E-cadherin expression to P-cadherin expression affecting cellular adhesion to the underlying matrix (67). The cancer-associated expression of GalNAc-T6 has also been proposed to induce EMT (66, 68, 83) as evidenced by decreased E-cadherin expression and enhanced expression of mesenchymal markers (68). Whereas our results support the idea that GalNAc-T6 expression negatively regulates adhesion of epithelial cells, we did not observe any changes in E-cadherin expression and hence could not confirm the potential effect of GalNAc-T6 on EMT previously observed in prostate cancer cell lines (68).
Because a large proportion of the proteins that pass through the secretory apparatus are potential substrates for GalNAc-T6 (84, 85), it is challenging to determine the specific molecular mechanisms underlying the observed oncogenic effects of GalNAc-T6. Using the isogenic LS174T cell model system, we began to characterize the effects associated with the loss of the GALNT6 gene both at the global transcriptomic and global O-glycoproteomic level.
We confirmed that the transcriptomic profile of LS174T cancer cells resembled the expression profile of human colon cancer found in the CancerBrowser UCSC database. Both data sets showed low expression of colon cancer differentiation markers, such as pyruvate dehydrogenase kinase 1 (PDK1), trefoil factor 2 (TFF2), and alanyl aminopeptidase (ANPEP), and high expression of stem cell markers, such as olfactomedin 4 (OLFM4), achaete-scute family of bHLH transcription factor 2 (ASCL2), and SPARC-related modular calcium-binding 2 (SMOC2). In contrast, the expression profile of GALNT6 knockout cells had many features that are found in the transcriptomic profile of healthy colon tissue, e.g. higher expression of differentiation markers and down-regulated expression of stem cell markers. Thus, the transcriptomic data confirmed that loss of GalNAc-T6 causes the LS174T cells to resemble cells found in normal human colon tissue. Furthermore, the transcriptomic analysis revealed a much larger impact by the loss of GalNAc-T6 than by the loss of GalNAc-T3. This implies that inactivation of GALNT3 in LS174T cells has wider consequences than inactivation of GALNT3 supporting the phenotypic characterization of cells with and without GalNAc-T3 and GalNAc-T6. These findings are intriguing, because GalNAc-T3 and GalNAc-T6 have previously been suggested to perform very similar functions.
Our global differential O-glycoproteomic analysis of cells with and without GalNAc-T3 and GalNAc-T6 further confirmed specific functions related to GalNAc-T6 but not GalNAc-T3 in LS174T cells. We used our previously developed SimpleCell strategy (26) and performed proteome-wide analysis to identify O-glycoproteins with O-glycan attachment sites directly comparing the O-glycoproteomes of isogenic cell lines with and without GalNAc-T6 and/or GalNAc-T3. We identified 81 shared O-glycosylation targets for GalNAc-T6 and GalNAc-T3. Surprisingly, of these targets, only six were GalNAc-T6–specific, whereas none of the targets were specific for GalNAc-T3. Interestingly, several of the GalNAc-T6–specific target proteins play important roles in cell–cell and cell–matrix adhesion. For example, MIA3 (melanoma inhibitory activity family, member 3) has an ortholog, TANGO1, that is important for transportation and polarized secretion of collagen 7 in Drosophila and mice (86–88). Another interesting GalNAc-T6 target is the ephrin B6 receptor (EphB6) (89). Ephrin receptors are a family of transmembrane proteins involved in cell adhesion and migration (90). Interestingly, the GalNAc-T6 glycosylation site in EphB6 is located in close proximity to the proposed binding pocket, which opens for the possibility that glycosylation interferes with ephrin B ligand binding. A few other GalNAc-T6–specific targets were found, including SLITRK1, associated with Tourette's syndrome (91) and homologous to the Slit proteins. Slit proteins are important for regulating axonal guidance, cell migration, and axonal branching by altering cellular adhesiveness and cytoskeletal organization (92, 93). The effect of SLITRK1 on synaptic adhesion is mediated through two extracellular leucine-rich repeats (LRR1 and LRR2) that interact with presynaptic leukocyte common antigen-related receptor protein-tyrosine phosphatases (LAR-RPTPs) (94). Interestingly, the GalNAc-T6–specific site in SLITRK1 is localized in close proximity to the binding interface with LAR-RPTPs. It is, however, an open question whether the GalNAc-T6-mediated glycosylation influences SLITRK1 functions, such as lateral assembly of LAR-RPTPs–Slitrks complexes (94), and whether the interaction between SLITRK1 and LAR-RPTPs is important for adhesion between colonic cells. Notably, the molecular mechanisms by which these individual site-specific glycosylation events affect the function of proteins are not easy to elucidate and will require an in-depth analysis of each individual protein in future studies. We and others have demonstrated that site-specific O-glycosylation of proteins affects protein function in several ways, including proprotein processing, modulation of the ligand-binding properties of receptors, and regulation of ectodomain shedding and cell signaling (30–33). One prominent example of the function of single O-glycan sites in receptor dimerization comes from the demonstration that site-specific O-glycosylation of the granulocyte-CSF receptor regulates receptor homodimerization and signaling. Moreover, a common somatic mutation in a single O-glycosite is a cancer driver in a large percentage of patients with chronic neutrophilic leukemia (95). It is thus very likely that GalNAc-T6 can influence the function of its protein targets in several ways, but further studies are needed to identify the molecular mechanisms underlying GalNAc-T6-mediated regulation of protein function.
A potential role for GalNAc-T6 in modulation of the interaction with the immune system was suggested by the sit-specific glycosylation of HLA class II. The GalNAc-T6–specific site was localized in the HLA class II histocompatibility antigen γ chain, also known as CD74 or invariant chain, which is involved in the stabilization and transport of MHC class II proteins (96, 97). Disrupting the interaction between CD74 and MHC class II molecules, which could be the result of GalNAc-T6–specific O-glycosylation, might decrease the presence of MHC class II molecules on the cell surface, leading to immune evasion of cancer cells. Although validation in future studies is required, it would be intriguing if a site-specific cancer-associated glycosylation event directly interfered with the expression of MHCII on the cancer surface. The cancer-specific expression of GalNAc-T6 might also interact with the immune system indirectly through induction of cancer-associated autoantibodies targeting novel cancer-specific glycopeptide epitopes (98–102). Such site-specific glycosylation events may also represent potential targets for monoclonal antibody therapy (98, 103), and our findings may provide support for the development of immunotherapeutic strategies that target aberrant O-glycoproteins or glycopeptide epitopes created by GalNAc-T6. Furthermore, the cancer-specific expression of GalNAc-T6 in colon cancer also suggests that isoform-specific inhibitors could prove useful in the future treatment of human cancers.
In conclusion, we here present evidence that overexpression of GalNAc-T6, as observed in several types of epithelial cancer, intrinsically promotes an oncogenic phenotype in the LS174T colon cancer cell model. This phenotype is characterized by increased proliferation and dysplasia, compromised adhesion, and the loss of normal differentiation, all of which are characteristics of colon cancer (104). Our study demonstrates that GalNAc-T6 is a potential key regulator of the malignant phenotype in colon cancer and suggests that overexpression of GalNAc-T6 is an early event during cancer development that provides a permissive environment for malignant evolution. In this context it will be of interest to test the effect of overexpression of GalNAc-T6 in healthy colon cancer cells in future studies.
Experimental procedures
Tissues
Tissue microarrays were purchased from U.S. Biomax, Inc. The tissue microarray (FCO401b) frozen tissue samples were from colo-rectal adenocarcinoma patients. Healthy control samples were evaluated from frozen multiple organ normal tissue array. The sections were fixed in cold 10% buffered neutral formalin for 15 min or in cold acetone for 10 min. Immunohistochemistry was performed as described under “Immunofluorescence” below.
Cell culture
The human colon adenocarcinoma LS174T cell line was chosen as a model system based on its phenotypic characteristics and stable genome (70, 81). Cells were grown in 50% DMEM 1965 and 50% Ham's F12 supplemented with 10% FBS and 1% l-glutamine. Prior to staining, cells were trypsinized and seeded on diagnostic slides, dried overnight, and fixed in ice-cold acetone for 5–10 min. Alternatively, cells were cultured directly on coverslips in 24-well plates (5 × 104 cells/well) for 4 days and fixed directly in the wells with 4% paraformaldehyde and 1% Triton X-100 for at least 2 h at room temperature.
Immunofluorescence
Slides or coverslips with cultured cells were incubated for 24 h at 4 °C with undiluted hybridoma supernatant (anti-GalNAc-T6, mouse mAb 2F3; anti-GalNAc-T3, mouse mAb 2D10; anti-GalNAc-T1 (Mab UH3 4D8), -T2 (Mab UH4 4C4), -T4 (UH6 4G4), -T5 (Mab 5F11), -T11 (Mab UH8 1B2), -T12 (Mab 1F9), -T14 (Mab 3D2, -T16 (Mab 4C6) (112) anti-Tn, mouse mAb 5F4; anti-STn, mouse mAb 3F1; and anti-T, mouse mAb 3C9) or with anti-LGR5 rabbit mAb (1:100, Novus Biologicals). Bound mAbs were detected with FITC-conjugated rabbit anti-mouse immunoglobulin (1:100; Dako, Denmark), anti-mouse Alexa 488 (1:500; Invitrogen), or swine anti-rabbit FITC (1:100; DAKO). Actin was detected with Alexa 594-conjugated phalloidin (1:500; Invitrogen). Slides were mounted with Prolong Gold antifade reagent with DAPI (Invitrogen). Fluorescence micrographs were obtained on a Leica wide-field fluorescence microscope or a Zeiss LSM710 confocal microscope. Image processing was performed in ImageJ.
ZFN knockout gene targeting
LS174T ΔT3 and/or ΔT6 in a WT and SimpleCell (COSMC knockout) background were generated as described previously (26). Briefly, ZFN constructs targeting COSMC, GALNT6, and GALNT3 were custom produced (Sigma), and LS174T cells were transfected with 2 μg of GFP- or DsRed2-tagged ZFN plasmids (105) using nucleofection with Amaxa Nucleofector (Lonza). GFP+/DsRed2+ cells were enriched by fluorescence-activated cell sorting (FACS) and single-cell cloned by limited dilution. Indels at the respective target sites were characterized by Indel Detection by Amplicon Analysis (IDAA) (106), and indels identified in individual cell clones that were selected were confirmed by Sanger sequencing.
Precise GALNT6-targeted integration
Precise targeted integration of GALNT6 into GALNT6−/− cells for stable expression of GALNT6 was performed using the ObLiGaRe knockin strategy (107) targeting the AAVS1 locus (also known as the PPP1R12C locus) on human chromosome 19. Briefly, an ObLiGaRe donor scaffolding vector was constructed encompassing the left and right inverted AAVS1 ZFN-binding sites flanking a CMV-GALNT6-Bgh-UTR expression cassette surrounded by insulator sequences. LS174TΔT6 cells were transfected with 5 μg of ObLiGaRe-GALNT6 vector plasmid and 2.5 μg of each AAVS1 ZFN pair (Sigma) using nucleofection (Amaxa Nucleofector; Lonza). Nucleofected cells were single-cell cloned by limiting dilution, and GALNT6 knockin clones were screened for GalNAc-T6 expression by immunohistochemistry using our well-characterized anti-GalNAc-T6 monoclonal antibody (2F3) (108). Precise targeted integration was verified by junction PCR across the target integration sites.
RNA transcriptomic analysis
Total RNA was extracted from exponentially growing cells using the RNeasy® kit (Qiagen). RNA integrity and quality were assessed using Bioanalyzer instrumentation (Agilent Technologies). The analysis was performed on total RNA from one clone of each of the following types of cells: LS174T WT, ΔT6, ΔT6+T6,ΔT3, and ΔT3ΔT6. Transcriptome analysis of the extracted total RNA samples was performed by the Beijing Genomics Institute (BGI) as described previously (29). Briefly, a library was constructed using the Illumina Truseq RNA Sample Preparation Kit and subjected to PCR amplification and quality control before undergoing next generation sequencing with the Illumina HiSeq 2000 System (Illumina, San Diego).
Bioinformatics analysis
Bioinformatics analysis was performed as described previously (29). Briefly, aligned reads from the RNAseq analysis were analyzed using the DESeq (109) and EdgeR (110) packages for R and Bioconductor to identify differentially expressed transcripts. DESeq and EdgeR analyses were run using default parameters following previously described protocols (111).
Proliferation assay
A total of 5 × 104 cells/well were plated in duplicate in 24-well dishes on day 0 for each time point. Cells were trypsinized and counted on days 4, 6, and 8.
LWAC isolation of Tn-O-glycopeptides
Vicia villosa agglutinin (VVA) LWAC isolation of Tn-O-glycopeptides was performed as described previously (29). Briefly, peptides obtained from trypsin digestion of TCL or culture supernatant (SEC) from LS174TSC cells were neuraminidase-treated to remove sialic acid residues, followed by labeling with light or medium isotopomeric dimethyl labels (74). The labeled digestion reactions were mixed in 1:1 ratios as follows: LS174TSC (light)/LS174TSCΔT6 (medium); LS174TSC (light)/LS174TSCΔT3 (medium); LS174TSC (light)/LS174TSCΔT3ΔT6 (medium); and LS174TSCΔT3 (light)/LS174TSCΔT6 (medium). GalNAc-binding VVA lectin was used for LWAC to separate the digests into glycopeptide (eluate) and peptide fractions (flow-through) (26). Isoelectric focusing was performed on the peptides using a 3100 OFFGEL fractionator (Agilent) using pH 3–10 strips (GE Healthcare) (75). All peptide and glycopeptide samples were desalted by self-made Stage Tips (C18 sorbent from 3 M Empore) and subjected to LC-MS and HCD/ETD-MS/MS analysis.
Mass spectrometry and data analysis
EASY-nLC 1000 UHPLC (Thermo Fisher Scientific) interfaced via nanoSpray Flex ion source to an LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific) was used for analysis. The nLC was operated in a single analytical column set up using PicoFrit Emitters (New Objectives, 75 μm inner diameter) packed in-house with Reprosil-Pure-AQ C18 phase (Dr. Maisch, 1.9-μm particle size, 19–21 cm column length). Each sample dissolved in 0.1% formic acid was injected onto the column and eluted in a gradient from 2 to 20% B in 95 min, from 20% to 80% B in 10 min, and 80% B for 15 min at 200 nl/min (solvent A, 100% H2O; solvent B, 100% acetonitrile; both containing 0.1% (v/v) formic acid).
A precursor MS1 scan (m/z 350–1,700) of intact peptides was acquired in the Orbitrap at a nominal resolution setting of 60,000, followed by Orbitrap HCD-MS2 and ETD-MS2 (m/z of 120–2,000) of the five most abundant multiply charged precursors in the MS1 spectrum; a minimum MS1 signal threshold of 50,000 was used for triggering data-dependent fragmentation events; MS2 spectra were acquired at a resolution of 15,000 for HCD MS2 and 30,000 for ETD MS2. Isolation width was 3 mass units, and usually one microscan was collected for each spectrum. Automatic gain control targets were 500,000 ions for Orbitrap MS1 and 100,000 for MS2 scans. Supplemental activation (25%) of the charge-reduced species was used in the ETD analysis to improve fragmentation. Dynamic exclusion for 60 s was used to prevent repeated analysis of the same components. Polysiloxane ions at m/z 445.12003 were used as a lock mass in all runs.
Data processing was performed using Proteome Discoverer 1.4 software (Thermo Fisher Scientific) using Sequest HT node as a search engine. In all cases the precursor mass tolerance was set to 10 ppm and fragment ion mass tolerance to 20 milli-mass units. All spectra were initially searched at the full cleavage specificity, filtered according to the confidence level (medium, low, and unassigned), and further searched with the semi-specific enzymatic cleavage. Up to two missed cleavages were allowed. Carbamidomethylation on cysteine residues was used as a fixed modification. Methionine oxidation and HexNAc and HexHexNAc attachment to serine, threonine, and tyrosine were used as variable modifications for ETD-MS2. All HCD-MS2s were pre-processed as described (75) and searched under the same conditions mentioned above using only methionine oxidation as variable modification. All spectra were searched against a concatenated forward/reverse human-specific database (UniProt, January 2013, containing 20,232 canonical entries. In addition, another 251 common contaminants and 3187 entries of viruses known to infect humans were included in the search) using a target false discovery rate of 1%.
Cell–cell adhesion assay
Cells that were grown to 100% confluence in 24-well plates were washed in HBSS and treated with 2.4 mg/ml dispase in HBSS for 20 min, leaving intact cell layers that were disassociated from the plastic wells. PBS (1 ml) was added carefully, and after the cell solution was pipetted up and down three times, fractions with more than 20 cells were counted.
Author contributions
K. L., S. D., and H. H. W. designed and performed experiments, analyzed data, and wrote the paper. E. P. B. and M. F. provided essential reagents and revised the paper. L. H., A. M. R. L., A. D., A. D. H., U. M., and S. Y. V. designed and/or performed experiments.
Supplementary Material
This work was supported in part by Danish Research Councils Grant 1331-00133B (to K. L., S. D., and H. H. W.), Programme of Excellence 2016 Copenhagen as the Next Leader in Precise Genetic Engineering Grant CDO2016 from the University of Copenhagen (to K. L. and H. H. W.), Danish National Research Foundation Grant DNRF107, and The Lundbeck Foundation (to K. L. and H. H. W.). H. H. W. is a consultant and owns stock in GO therapeutics.
This article was selected as one of our Editors' Picks.
This article contains Fig. S1 and Tables S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
CCDC14 was excluded from Table 2 due to its presumed cytoplasmic localization, which could question the nature of the HexNAc residues.
- GalNAc-T
- N-acetylgalactosaminyltransferase
- EMT
- epithelial-to-mesenchymal transition
- ZFN
- zinc finger nuclease
- nLC/MS/MS
- lectin chromatography and nanoflow liquid chromatography tandem mass spectrometry
- ETD
- electron transfer dissociation
- VVA
- Vicia villosa agglutinin
- LWAC
- lectin weak affinity chromatography
- SEC
- secretome
- TCL
- total cell lysate
- LAR-RPTP
- leukocyte common antigen-related receptor protein tyrosine phosphatase
- HBSS
- Hanks' balanced salt solution
- seq
- sequence
- L
- light
- M
- medium
- TCGA
- The Cancer Genome Atlas
- RPKM
- reads per kilobase of transcript per million mapped reads.
References
- 1. Brockhausen I. (2006) Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 7, 599–604 10.1038/sj.embor.7400705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daniotti J. L., Vilcaes A. A., Torres Demichelis V., Ruggiero F. M., and Rodriguez-Walker M. (2013) Glycosylation of glycolipids in cancer: basis for development of novel therapeutic approaches. Front. Oncol. 3, 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hakomori S. (2002) Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 10.1073/pnas.172380699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hollingsworth M. A., and Swanson B. J. (2004) Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 4, 45–60 10.1038/nrc1251 [DOI] [PubMed] [Google Scholar]
- 5. Tarp M. A., and Clausen H. (2008) Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 1780, 546–563 10.1016/j.bbagen.2007.09.010 [DOI] [PubMed] [Google Scholar]
- 6. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 10.1093/glycob/cwr182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennett E. P., Hassan H., and Clausen H. (1996) cDNA cloning and expression of a novel human UDP-N-acetyl-α-d-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-t3. J. Biol. Chem. 271, 17006–17012 10.1074/jbc.271.29.17006 [DOI] [PubMed] [Google Scholar]
- 8. Bennett E. P., Hassan H., Mandel U., Mirgorodskaya E., Roepstorff P., Burchell J., Taylor-Papadimitriou J., Hollingsworth M. A., Merkx G., van Kessel A. G., Eiberg H., Steffensen R., and Clausen H. (1998) Cloning of a human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J. Biol. Chem. 273, 30472–30481 10.1074/jbc.273.46.30472 [DOI] [PubMed] [Google Scholar]
- 9. Bennett E. P., Hassan H., Hollingsworth M. A., and Clausen H. (1999) A novel human UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 460, 226–230 10.1016/S0014-5793(99)01268-5 [DOI] [PubMed] [Google Scholar]
- 10. Bennett E. P., Hassan H., Mandel U., Hollingsworth M. A., Akisawa N., Ikematsu Y., Merkx G., van Kessel A. G., Olofsson S., and Clausen H. (1999) Cloning and characterization of a close homolog of human UDP-N-acetyl-α-d-galactosamine:Polypeptide N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. J. Biol. Chem. 274, 25362–25370 10.1074/jbc.274.36.25362 [DOI] [PubMed] [Google Scholar]
- 11. Hagen F. K., Van Wuyckhuyse B., and Tabak L. A. (1993) Purification, cloning, and expression of a bovine UDP-GalNAc: polypeptide N-acetyl-galactosaminyltransferase. J. Biol. Chem. 268, 18960–18965 [PubMed] [Google Scholar]
- 12. Hagen F. K., Ten Hagen K. G., Beres T. M., Balys M. M., VanWuyckhuyse B. C., and Tabak L. A. (1997) cDNA cloning and expression of a novel UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 272, 13843–13848 10.1074/jbc.272.21.13843 [DOI] [PubMed] [Google Scholar]
- 13. Homa F. L., Hollander T., Lehman D. J., Thomsen D. R., and Elhammer A. P. (1993) Isolation and expression of a cDNA clone encoding a bovine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 268, 12609–12616 [PubMed] [Google Scholar]
- 14. Ten Hagen K. G., Bedi G. S., Tetaert D., Kingsley P. D., Hagen F. K., Balys M. M., Beres T. M., Degand P., and Tabak L. A. (2001) Cloning and characterization of a ninth member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, ppGaNTase-T9. J. Biol. Chem. 276, 17395–17404 10.1074/jbc.M009638200 [DOI] [PubMed] [Google Scholar]
- 15. Ten Hagen K. G., Hagen F. K., Balys M. M., Beres T. M., Van Wuyckhuyse B., and Tabak L. A. (1998) Cloning and expression of a novel, tissue specifically expressed member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family. J. Biol. Chem. 273, 27749–27754 10.1074/jbc.273.42.27749 [DOI] [PubMed] [Google Scholar]
- 16. Ten Hagen K. G., Tetaert D., Hagen F. K., Richet C., Beres T. M., Gagnon J., Balys M. M., VanWuyckhuyse B., Bedi G. S., Degand P., and Tabak L. A. (1999) Characterization of a UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase that displays glycopeptide N-acetylgalactosaminyltransferase activity. J. Biol. Chem. 274, 27867–27874 10.1074/jbc.274.39.27867 [DOI] [PubMed] [Google Scholar]
- 17. White K. E., Lorenz B., Evans W. E., Meitinger T., Strom T. M., and Econs M. J. (2000) Molecular cloning of a novel human UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T8, and analysis as a candidate autosomal dominant hypophosphatemic rickets (ADHR) gene. Gene 246, 347–356 10.1016/S0378-1119(00)00050-0 [DOI] [PubMed] [Google Scholar]
- 18. White T., Bennett E. P., Takio K., Sørensen T., Bonding N., and Clausen H. (1995) Purification and cDNA cloning of a human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 270, 24156–24165 10.1074/jbc.270.41.24156 [DOI] [PubMed] [Google Scholar]
- 19. Clausen H., and Bennett E. P. (1996) A family of UDP-GalNAc: polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology 6, 635–646 10.1093/glycob/6.6.635 [DOI] [PubMed] [Google Scholar]
- 20. Pinho S. S., and Reis C. A. (2015) Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 10.1038/nrc3982 [DOI] [PubMed] [Google Scholar]
- 21. Baldus S. E., Zirbes T. K., Hanisch F. G., Kunze D., Shafizadeh S. T., Nolden S., Mönig S. P., Schneider P. M., Karsten U., Thiele J., Hölscher A. H., and Dienes H. P. (2000) Thomsen-Friedenreich antigen presents as a prognostic factor in colorectal carcinoma: A clinicopathologic study of 264 patients. Cancer 88, 1536–1543 10.1002/(SICI)1097-0142(20000401)88:7%3C1536::AID-CNCR6%3E3.0.CO%3B2-W [DOI] [PubMed] [Google Scholar]
- 22. David L., Nesland J. M., Clausen H., Carneiro F., and Sobrinho-Simões M. (1992) Simple mucin-type carbohydrate antigens (Tn, sialosyl-Tn and T) in gastric mucosa, carcinomas and metastases. APMIS Suppl. 27, 162–172 [PubMed] [Google Scholar]
- 23. Ferreira J. A., Videira P. A., Lima L., Pereira S., Silva M., Carrascal M., Severino P. F., Fernandes E., Almeida A., Costa C., Vitorino R., Amaro T., Oliveira M. J., Reis C. A., Dall'Olio F., Amado F., and Santos L. L. (2013) Overexpression of tumour-associated carbohydrate antigen sialyl-Tn in advanced bladder tumours. Mol. Oncol. 7, 719–731 10.1016/j.molonc.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ozaki H., Matsuzaki H., Ando H., Kaji H., Nakanishi H., Ikehara Y., and Narimatsu H. (2012) Enhancement of metastatic ability by ectopic expression of ST6GalNAcI on a gastric cancer cell line in a mouse model. Clin. Exp. Metastasis 29, 229–238 10.1007/s10585-011-9445-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steentoft C., Bennett E. P., and Clausen H. (2013) Glycoengineering of human cell lines using zinc finger nuclease gene targeting: SimpleCells with homogeneous GalNAc O-glycosylation allow isolation of the O-glycoproteome by one-step lectin affinity chromatography. Methods Mol. Biol. 1022, 387–402 10.1007/978-1-62703-465-4_29 [DOI] [PubMed] [Google Scholar]
- 26. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., and Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 10.1038/nmeth.1731 [DOI] [PubMed] [Google Scholar]
- 27. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., and Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 10.1073/pnas.1203563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kong Y., Joshi H. J., Schjoldager K. T., Madsen T. D., Gerken T. A., Vester-Christensen M. B., Wandall H. H., Bennett E. P., Levery S. B., Vakhrushev S. Y., and Clausen H. (2015) Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology 25, 55–65 10.1093/glycob/cwu089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schjoldager K. T., Joshi H. J., Kong Y., Goth C. K., King S. L., Wandall H. H., Bennett E. P., Vakhrushev S. Y., and Clausen H. (2015) Deconstruction of O-glycosylation-GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 16, 1713–1722 10.15252/embr.201540796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boskovski M. T., Yuan S., Pedersen N. B., Goth C. K., Makova S., Clausen H., Brueckner M., and Khokha M. K. (2013) The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504, 456–459 10.1038/nature12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kariya Y., Kanno M., Matsumoto-Morita K., Konno M., Yamaguchi Y., and Hashimoto Y. (2014) Osteopontin O-glycosylation contributes to its phosphorylation and cell-adhesion properties. Biochem. J. 463, 93–102 10.1042/BJ20140060 [DOI] [PubMed] [Google Scholar]
- 32. Schjoldager K. T., and Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing- deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 10.1016/j.bbagen.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 33. Goth C. K., Halim A., Khetarpal S. A., Rader D. J., Clausen H., and Schjoldager K. T. (2015) A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 112, 14623–14628 10.1073/pnas.1511175112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng R. Q., Wan H. Y., Li H. F., Liu M., Li X., and Tang H. (2012) MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. J. Biol. Chem. 287, 14301–14309 10.1074/jbc.M111.337642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X., Wang J., Li W., Xu Y., Shao D., Xie Y., Xie W., Kubota T., Narimatsu H., and Zhang Y. (2012) Characterization of ppGalNAc-T18, a member of the vertebrate-specific Y subfamily of UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 22, 602–615 10.1093/glycob/cwr179 [DOI] [PubMed] [Google Scholar]
- 36. Ding M. X., Wang H. F., Wang J. S., Zhan H., Zuo Y. G., Yang D. L., Liu J. Y., Wang W., Ke C. X., and Yan R. P. (2012) ppGalNAc T1 as a potential novel marker for human bladder cancer. Asian Pac. J. Cancer Prev. 13, 5653–5657 10.7314/APJCP.2012.13.11.5653 [DOI] [PubMed] [Google Scholar]
- 37. Wang Z. Q., Bachvarova M., Morin C., Plante M., Gregoire J., Renaud M. C., Sebastianelli A., and Bachvarov D. (2014) Role of the polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin O-glycosylation. Oncotarget 5, 544–560 10.18632/oncotarget.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adachi T., Hinoda Y., Nishimori I., Adachi M., and Imai K. (1997) Increased sensitivity of gastric cancer cells to natural killer and lymphokine-activated killer cells by antisense suppression of N-acetylgalactosaminyltransferase. J. Immunol. 159, 2645–2651 [PubMed] [Google Scholar]
- 39. Gaziel-Sovran A., Segura M. F., Di Micco R., Collins M. K., Hanniford D., Vega-Saenz de Miera E., Rakus J. F., Dankert J. F., Shang S., Kerbel R. S., Bhardwaj N., Shao Y., Darvishian F., Zavadil J., et al. (2011) miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20, 104–118 10.1016/j.ccr.2011.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gomes J., Marcos N. T., Berois N., Osinaga E., Magalhães A., Pinto-de-Sousa J., Almeida R., Gärtner F., and Reis C. A. (2009) Expression of UDP-N-acetyl-d-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 in gastric mucosa, intestinal metaplasia, and gastric carcinoma. J. Histochem. Cytochem. 57, 79–86 10.1369/jhc.2008.952283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harada Y., Izumi H., Noguchi H., Kuma A., Kawatsu Y., Kimura T., Kitada S., Uramoto H., Wang K. Y., Sasaguri Y., Hijioka H., Miyawaki A., Oya R., Nakayama T., Kohno K., and Yamada S. (2015) Strong expression of polypeptide N-acetylgalactosaminyltransferase 3 independently predicts shortened disease-free survival in patients with early stage oral squamous cell carcinoma. Tumour Biol. 36, 10003–10004 10.1007/s13277-015-4211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inoue T., Eguchi T., Oda Y., Nishiyama K., Fujii K., Izumi H., Kohno K., Yamaguchi K., Tanaka M., and Tsuneyoshi M. (2007) Expression of GalNAc-T3 and its relationships with clinicopathological factors in 61 extrahepatic bile duct carcinomas analyzed using stepwise sections–special reference to its association with lymph node metastases. Mod. Pathol. 20, 267–276 [DOI] [PubMed] [Google Scholar]
- 43. Matsumoto Y., Zhang Q., Akita K., Nakada H., Hamamura K., Tokuda N., Tsuchida A., Matsubara T., Hori T., Okajima T., Furukawa K., Urano T., and Furukawa K. (2012) pp-GalNAc-T13 induces high metastatic potential of murine Lewis lung cancer by generating trimeric Tn antigen. Biochem. Biophys. Res. Commun. 419, 7–13 10.1016/j.bbrc.2012.01.086 [DOI] [PubMed] [Google Scholar]
- 44. Shibao K., Izumi H., Nakayama Y., Ohta R., Nagata N., Nomoto M., Matsuo K., Yamada Y., Kitazato K., Itoh H., and Kohno K. (2002) Expression of UDP-N-acetyl-α-d-galactosamine-polypeptide galNAc N-acetylgalactosaminyl transferase-3 in relation to differentiation and prognosis in patients with colorectal carcinoma. Cancer 94, 1939–1946 10.1002/cncr.10423 [DOI] [PubMed] [Google Scholar]
- 45. Wandall H. H., Hassan H., Mirgorodskaya E., Kristensen A. K., Roepstorff P., Bennett E. P., Nielsen P. A., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., and Clausen H. (1997) Substrate specificities of three members of the human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 272, 23503–23514 10.1074/jbc.272.38.23503 [DOI] [PubMed] [Google Scholar]
- 46. Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., et al. (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J. Biol. Chem. 277, 22623–22638 10.1074/jbc.M202684200 [DOI] [PubMed] [Google Scholar]
- 47. Mandel U., Hassan H., Therkildsen M. H., Rygaard J., Jakobsen M. H., Juhl B. R., Dabelsteen E., and Clausen H. (1999) Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology 9, 43–52 10.1093/glycob/9.1.43 [DOI] [PubMed] [Google Scholar]
- 48. Yamamoto S., Nakamori S., Tsujie M., Takahashi Y., Nagano H., Dono K., Umeshita K., Sakon M., Tomita Y., Hoshida Y., Aozasa K., Kohno K., and Monden M. (2004) Expression of uridine diphosphate N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 in adenocarcinoma of the pancreas. Pathobiology 71, 12–18 10.1159/000072957 [DOI] [PubMed] [Google Scholar]
- 49. Li Z., Yamada S., Inenaga S., Imamura T., Wu Y., Wang K. Y., Shimajiri S., Nakano R., Izumi H., Kohno K., and Sasaguri Y. (2011) Polypeptide N-acetylgalactosaminyltransferase 6 expression in pancreatic cancer is an independent prognostic factor indicating better overall survival. Br. J. Cancer 104, 1882–1889 10.1038/bjc.2011.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taniuchi K., Cerny R. L., Tanouchi A., Kohno K., Kotani N., Honke K., Saibara T., and Hollingsworth M. A. (2011) Overexpression of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell growth. Oncogene 30, 4843–4854 10.1038/onc.2011.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sutherlin M. E., Nishimori I., Caffrey T., Bennett E. P., Hassan H., Mandel U., Mack D., Iwamura T., Clausen H., and Hollingsworth M. A. (1997) Expression of three UDP-N-acetyl-α-d-galactosamine:polypeptide GalNAc N-acetylgalactosaminyltransferases in adenocarcinoma cell lines. Cancer Res. 57, 4744–4748 [PubMed] [Google Scholar]
- 52. Miyahara N., Shoda J., Kawamoto T., Furukawa M., Ueda T., Todoroki T., Tanaka N., Matsuo K., Yamada Y., Kohno K., and Irimura T. (2004) Expression of UDP-N-acetyl-α-d-galactosamine-polypeptide N-acetylgalactosaminyltransferase isozyme 3 in the subserosal layer correlates with postsurgical survival of pathological tumor stage 2 carcinoma of the gallbladder. Clin. Cancer Res. 10, 2090–2099 10.1158/1078-0432.CCR-1024-03 [DOI] [PubMed] [Google Scholar]
- 53. Ishikawa M., Kitayama J., Nariko H., Kohno K., and Nagawa H. (2004) The expression pattern of UDP-N-acetyl-α-d-galactosamine: polypeptide N-acetylgalactosaminyl transferase-3 in early gastric carcinoma. J. Surg. Oncol. 86, 28–33 10.1002/jso.20042 [DOI] [PubMed] [Google Scholar]
- 54. Onitsuka K., Shibao K., Nakayama Y., Minagawa N., Hirata K., Izumi H., Matsuo K., Nagata N., Kitazato K., Kohno K., and Itoh H. (2003) Prognostic significance of UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-3 (GalNAc-T3) expression in patients with gastric carcinoma. Cancer Sci. 94, 32–36 10.1111/j.1349-7006.2003.tb01348.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Landers K. A., Burger M. J., Tebay M. A., Purdie D. M., Scells B., Samaratunga H., Lavin M. F., and Gardiner R. A. (2005) Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int. J. Cancer 114, 950–956 10.1002/ijc.20760 [DOI] [PubMed] [Google Scholar]
- 56. Kitada S., Yamada S., Kuma A., Ouchi S., Tasaki T., Nabeshima A., Noguchi H., Wang K. Y., Shimajiri S., Nakano R., Izumi H., Kohno K., Matsumoto T., and Sasaguri Y. (2013) Polypeptide N-acetylgalactosaminyl transferase 3 independently predicts high-grade tumours and poor prognosis in patients with renal cell carcinomas. Br. J. Cancer 109, 472–481 10.1038/bjc.2013.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gu C., Oyama T., Osaki T., Li J., Takenoyama M., Izumi H., Sugio K., Kohno K., and Yasumoto K. (2004) Low expression of polypeptide GalNAc N-acetylgalactosaminyl transferase-3 in lung adenocarcinoma: impact on poor prognosis and early recurrence. Br. J. Cancer 90, 436–442 10.1038/sj.bjc.6601531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ishikawa M., Kitayama J., Kohno K., and Nagawa H. (2005) The expression pattern of UDP-N-acetyl-α-d-galactosamine-polypeptide N-acetyl-galactosaminyl transferase-3 in squamous cell carcinoma of the esophagus. Pathobiology 72, 139–145 10.1159/000084117 [DOI] [PubMed] [Google Scholar]
- 59. Mochizuki Y., Ito K., Izumi H., Kohno K., and Amano J. (2013) Expression of polypeptide N-acetylgalactosaminyl transferase-3 and its association with clinicopathological factors in thyroid carcinomas. Thyroid 23, 1553–1560 10.1089/thy.2012.0613 [DOI] [PubMed] [Google Scholar]
- 60. Andergassen U., Liesche F., Kölbl A. C., Ilmer M., Hutter S., Friese K., and Jeschke U. (2015) Glycosyltransferases as markers for early tumorigenesis. BioMed Res. Int. 2015, 792672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berois N., Mazal D., Ubillos L., Trajtenberg F., Nicolas A., Sastre-Garau X., Magdelenat H., and Osinaga E. (2006) UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J. Histochem. Cytochem. 54, 317–328 10.1369/jhc.5A6783.2005 [DOI] [PubMed] [Google Scholar]
- 62. Freire T., Berois N., Sóñora C., Varangot M., Barrios E., and Osinaga E. (2006) UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a potential new marker for detection of bone marrow-disseminated breast cancer cells. Int. J. Cancer 119, 1383–1388 10.1002/ijc.21959 [DOI] [PubMed] [Google Scholar]
- 63. Patani N., Jiang W., and Mokbel K. (2008) Prognostic utility of glycosyltransferase expression in breast cancer. Cancer Genomics Proteomics 5, 333–340 [PubMed] [Google Scholar]
- 64. Potapenko I. O., Lüders T., Russnes H. G., Helland Å., Sørlie T., Kristensen V. N., Nord S., Lingjærde O. C., Børresen-Dale A. L., and Haakensen V. D. (2015) Glycan-related gene expression signatures in breast cancer subtypes; relation to survival. Mol. Oncol. 9, 861–876 10.1016/j.molonc.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liesche F., Kölbl A. C., Ilmer M., Hutter S., Jeschke U., and Andergassen U. (2016) Role of N-acetylgalactosaminyltransferase 6 in early tumorigenesis and formation of metastasis. Mol. Med. Rep. 13, 4309–4314 10.3892/mmr.2016.5044 [DOI] [PubMed] [Google Scholar]
- 66. Park J. H., Katagiri T., Chung S., Kijima K., and Nakamura Y. (2011) Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia 13, 320–326 10.1593/neo.101440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tarhan Y. E., Kato T., Jang M., Haga Y., Ueda K., Nakamura Y., and Park J. H. (2016) Morphological changes, cadherin switching, and growth suppression in pancreatic cancer by GALNT6 knockdown. Neoplasia 18, 265–272 10.1016/j.neo.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Freire-de-Lima L., Gelfenbeyn K., Ding Y., Mandel U., Clausen H., Handa K., and Hakomori S. I. (2011) Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process. Proc. Natl. Acad. Sci. U.S.A. 108, 17690–17695 10.1073/pnas.1115191108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. de Carné Trécesson S., Guillemin Y., Bélanger A., Bernard A. C., Preisser L., Ravon E., Gamelin E., Juin P., Barré B., and Coqueret O. (2011) Escape from p21-mediated oncogene-induced senescence leads to cell dedifferentiation and dependence on anti-apoptotic Bcl-xL and MCL1 proteins. J. Biol. Chem. 286, 12825–12838 10.1074/jbc.M110.186437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pinto R., Hansen L., Hintze J., Almeida R., Larsen S., Coskun M., Davidsen J., Mitchelmore C., David L., Troelsen J. T., and Bennett E. P. (2017) Precise integration of inducible transcriptional elements (PrIITE) enables absolute control of gene expression. Nucleic Acids Res. 45, e123 10.1093/nar/gkx371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang Z., Wang S., Halim A., Schulz M. A., Frodin M., Rahman S. H., Vester-Christensen M. B., Behrens C., Kristensen C., Vakhrushev S. Y., Bennett E. P., Wandall H. H., and Clausen H. (2015) Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol. 33, 842–844 10.1038/nbt.3280 [DOI] [PubMed] [Google Scholar]
- 72. Walker F., Zhang H. H., Odorizzi A., and Burgess A. W. (2011) LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS One 6, e22733 10.1371/journal.pone.0022733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Takeda K., Kinoshita I., Shimizu Y., Matsuno Y., Shichinohe T., and Dosaka-Akita H. (2011) Expression of LGR5, an intestinal stem cell marker, during each stage of colorectal tumorigenesis. Anticancer Res. 31, 263–270 [PubMed] [Google Scholar]
- 74. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 10.1038/nprot.2009.21 [DOI] [PubMed] [Google Scholar]
- 75. Vakhrushev S. Y., Steentoft C., Vester-Christensen M. B., Bennett E. P., Clausen H., and Levery S. B. (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 12, 932–944 10.1074/mcp.O112.021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ching C. K., Holmes S. W., Holmes G. K., and Long R. G. (1993) Comparison of two sialosyl-Tn binding monoclonal antibodies (MLS102 and B72.3) in detecting pancreatic cancer. Gut 34, 1722–1725 10.1136/gut.34.12.1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lyubsky S., Madariaga J., Lozowski M., Mishriki Y., Schuss A., Chao S., and Lundy J. (1988) A tumor-associated antigen in carcinoma of the pancreas defined by monoclonal antibody B72.3. Am. J. Clin. Pathol. 89, 160–167 10.1093/ajcp/89.2.160 [DOI] [PubMed] [Google Scholar]
- 78. Kim G. E., Bae H. I., Park H. U., Kuan S. F., Crawley S. C., Ho J. J., and Kim Y. S. (2002) Aberrant expression of MUC5AC and MUC6 gastric mucins and sialyl Tn antigen in intraepithelial neoplasms of the pancreas. Gastroenterology 123, 1052–1060 10.1053/gast.2002.36018 [DOI] [PubMed] [Google Scholar]
- 79. Itzkowitz S., Kjeldsen T., Friera A., Hakomori S., Yang U. S., and Kim Y. S. (1991) Expression of Tn, sialosyl Tn, and T antigens in human pancreas. Gastroenterology 100, 1691–1700 10.1016/0016-5085(91)90671-7 [DOI] [PubMed] [Google Scholar]
- 80. Hruban R. H., Goggins M., Parsons J., and Kern S. E. (2000) Progression model for pancreatic cancer. Clin. Cancer Res. 6, 2969–2972 [PubMed] [Google Scholar]
- 81. Tom B. H., Rutzky L. P., Jakstys M. M., Oyasu R., Kaye C. I., and Kahan B. D. (1976) Human colonic adenocarcinoma cells. I. Establishment and description of a new line. In Vitro 12, 180–191 10.1007/BF02796440 [DOI] [PubMed] [Google Scholar]
- 82. Park J. H., Nishidate T., Kijima K., Ohashi T., Takegawa K., Fujikane T., Hirata K., Nakamura Y., and Katagiri T. (2010) Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 70, 2759–2769 10.1158/0008-5472.CAN-09-3911 [DOI] [PubMed] [Google Scholar]
- 83. Zhang L., Gallup M., Zlock L., Chen Y. T., Finkbeiner W. E., and McNamara N. A. (2014) Pivotal role of MUC1 glycosylation by cigarette smoke in modulating disruption of airway adherens junctions in vitro. J. Pathol. 234, 60–73 10.1002/path.4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Apweiler R., Hermjakob H., and Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 10.1016/S0304-4165(99)00165-8 [DOI] [PubMed] [Google Scholar]
- 85. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Saito K., Chen M., Bard F., Chen S., Zhou H., Woodley D., Polischuk R., Schekman R., and Malhotra V. (2009) TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell 136, 891–902 10.1016/j.cell.2008.12.025 [DOI] [PubMed] [Google Scholar]
- 87. Wilson D. G., Phamluong K., Li L., Sun M., Cao T. C., Liu P. S., Modrusan Z., Sandoval W. N., Rangell L., Carano R. A., Peterson A. S., and Solloway M. J. (2011) Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J. Cell Biol. 193, 935–951 10.1083/jcb.201007162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang L., Syed Z. A., van Dijk Härd I., Lim J. M., Wells L., and Ten Hagen K. G. (2014) O-Glycosylation regulates polarized secretion by modulating Tango1 stability. Proc. Natl. Acad. Sci. U.S.A. 111, 7296–7301 10.1073/pnas.1322264111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Peng L., Tu P., Wang X., Shi S., Zhou X., and Wang J. (2014) Loss of EphB6 protein expression in human colorectal cancer correlates with poor prognosis. J. Mol. Histol. 45, 555–563 10.1007/s10735-014-9577-0 [DOI] [PubMed] [Google Scholar]
- 90. Herath N. I., and Boyd A. W. (2010) The role of Eph receptors and ephrin ligands in colorectal cancer. Int. J. Cancer 126, 2003–2011 [DOI] [PubMed] [Google Scholar]
- 91. Abelson J. F., Kwan K. Y., O'Roak B. J., Baek D. Y., Stillman A. A., Morgan T. M., Mathews C. A., Pauls D. L., Rasin M. R., Gunel M., Davis N. R., Ercan-Sencicek A. G., Guez D. H., Spertus J. A., Leckman J. F., et al. (2005) Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science 310, 317–320 10.1126/science.1116502 [DOI] [PubMed] [Google Scholar]
- 92. Brose K., and Tessier-Lavigne M. (2000) Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr. Opin. Neurobiol. 10, 95–102 10.1016/S0959-4388(99)00066-5 [DOI] [PubMed] [Google Scholar]
- 93. Aruga J., and Mikoshiba K. (2003) Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol. Cell. Neurosci. 24, 117–129 10.1016/S1044-7431(03)00129-5 [DOI] [PubMed] [Google Scholar]
- 94. Um J. W., Kim K. H., Park B. S., Choi Y., Kim D., Kim C. Y., Kim S. J., Kim M., Ko J. S., Lee S. G., Choii G., Nam J., Heo W. D., Kim E., Lee J. O., Ko J., and Kim H. M. (2014) Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion. Nat. Commun. 5, 5423 10.1038/ncomms6423 [DOI] [PubMed] [Google Scholar]
- 95. Maxson J. E., Luty S. B., MacManiman J. D., Abel M. L., Druker B. J., and Tyner J. W. (2014) Ligand independence of the T618I mutation in the colony-stimulating factor 3 receptor (CSF3R) protein results from loss of O-linked glycosylation and increased receptor dimerization. J. Biol. Chem. 289, 5820–5827 10.1074/jbc.M113.508440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Stumptner-Cuvelette P., and Benaroch P. (2002) Multiple roles of the invariant chain in MHC class II function. Biochim. Biophys. Acta 1542, 1–13 10.1016/S0167-4889(01)00166-5 [DOI] [PubMed] [Google Scholar]
- 97. Bergmann H. (2012) Business as usual: the p35 isoform of human CD74 retains function in antigen presentation. Immunol. Cell Biol. 90, 839–840 10.1038/icb.2012.51 [DOI] [PubMed] [Google Scholar]
- 98. Lavrsen K., Madsen C. B., Rasch M. G., Woetmann A., Ødum N., Mandel U., Clausen H., Pedersen A. E., and Wandall H. H. (2013) Aberrantly glycosylated MUC1 is expressed on the surface of breast cancer cells and a target for antibody-dependent cell-mediated cytotoxicity. Glycoconj. J. 30, 227–236 10.1007/s10719-012-9437-7 [DOI] [PubMed] [Google Scholar]
- 99. Madsen C. B., Petersen C., Lavrsen K., Harndahl M., Buus S., Clausen H., Pedersen A. E., and Wandall H. H. (2012) Cancer associated aberrant protein O-glycosylation can modify antigen processing and immune response. PLoS One 7, e50139 10.1371/journal.pone.0050139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pedersen J. W., Blixt O., Bennett E. P., Tarp M. A., Dar I., Mandel U., Poulsen S. S., Pedersen A. E., Rasmussen S., Jess P., Clausen H., and Wandall H. H. (2011) Seromic profiling of colorectal cancer patients with novel glycopeptide microarray. Int. J. Cancer 128, 1860–1871 10.1002/ijc.25778 [DOI] [PubMed] [Google Scholar]
- 101. Pedersen J. W., Gentry-Maharaj A., Fourkala E. O., Dawnay A., Burnell M., Zaikin A., Pedersen A. E., Jacobs I., Menon U., and Wandall H. H. (2013) Early detection of cancer in the general population: a blinded case-control study of p53 autoantibodies in colorectal cancer. Br. J. Cancer 108, 107–114 10.1038/bjc.2012.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Pedersen J. W., Gentry-Maharaj A., Nøstdal A., Fourkala E. O., Dawnay A., Burnell M., Zaikin A., Burchell J., Papadimitriou J. T., Clausen H., Jacobs I., Menon U., and Wandall H. H. (2014) Cancer-associated autoantibodies to MUC1 and MUC4–a blinded case-control study of colorectal cancer in UK collaborative trial of ovarian cancer screening. Int. J. Cancer 134, 2180–2188 10.1002/ijc.28538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Danielczyk A., Stahn R., Faulstich D., Löffler A., Märten A., Karsten U., and Goletz S. (2006) PankoMab: a potent new generation anti-tumour MUC1 antibody. Cancer Immunol. Immunother. 55, 1337–1347 10.1007/s00262-006-0135-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ashley N., Yeung T. M., and Bodmer W. F. (2013) Stem cell differentiation and lumen formation in colorectal cancer cell lines and primary tumors. Cancer Res. 73, 5798–5809 10.1158/0008-5472.CAN-13-0454 [DOI] [PubMed] [Google Scholar]
- 105. Duda K., Lonowski L. A., Kofoed-Nielsen M., Ibarra A., Delay C. M., Kang Q., Yang Z., Pruett-Miller S. M., Bennett E. P., Wandall H. H., Davis G. D., Hansen S. H., and Frödin M. (2014) High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res. 42, e84 10.1093/nar/gku251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yang Z., Steentoft C., Hauge C., Hansen L., Thomsen A. L., Niola F., Vester-Christensen M. B., Frödin M., Clausen H., Wandall H. H., and Bennett E. P. (2015) Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 43, e59 10.1093/nar/gkv126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Maresca M., Lin V. G., Guo N., and Yang Y. (2013) Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546 10.1101/gr.145441.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wandall H. H., Dabelsteen S., Sørensen J. A., Krogdahl A., Mandel U., and Dabelsteen E. (2007) Molecular basis for the presence of glycosylated onco-foetal fibronectin in oral carcinomas: the production of glycosylated onco-foetal fibronectin by carcinoma cells. Oral Oncol. 43, 301–309 10.1016/j.oraloncology.2006.03.005 [DOI] [PubMed] [Google Scholar]
- 109. Anders S., and Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. McCarthy D. J., Chen Y., and Smyth G. K. (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Anders S., McCarthy D. J., Chen Y., Okoniewski M., Smyth G. K., Huber W., and Robinson M. D. (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- 112. Vester-Christensen M. B., Bennett E. P., Clausen H., and Mandel U. (2013) Generation of monoclonal antibodies to native active human glycosyltransferases. Methods Mol. Biol. 1022, 403–420 10.1007/978-1-62703-465-4_30 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.