Abstract
It has been long assumed that post-mitotic neurons only utilize the error-prone non-homologous end–joining pathway to repair double-strand breaks (DSBs) associated with oxidative damage to DNA, given the inability of non-replicating neuronal DNA to utilize a sister chromatid template in the less error-prone homologous recombination (HR) repair pathway. However, we and others have found recently that active transcription triggers a replication-independent recombinational repair mechanism in G0/G1 phase of the cell cycle. Here we observed that the HR repair protein RAD52 is recruited to sites of DNA DSBs in terminally differentiated, post-mitotic neurons. This recruitment is dependent on the presence of a nascent mRNA generated during active transcription, providing evidence that an RNA-templated HR repair mechanism exists in non-dividing, terminally differentiated neurons. This recruitment of RAD52 in neurons is decreased by transcription inhibition. Importantly, we found that high concentrations of amyloid β, a toxic protein associated with Alzheimer's disease, inhibits the expression and DNA damage response of RAD52, potentially leading to a defect in the error-free, RNA-templated HR repair mechanism. This study shows a novel RNA-dependent repair mechanism of DSBs in post-mitotic neurons and demonstrates that defects in this pathway may contribute to neuronal genomic instability and consequent neurodegenerative phenotypes such as those seen in Alzheimer's disease.
Keywords: Alzheimer's disease, amyloid β (Aβ), neuron, recombination, RNA, oxidative damage
Introduction
The most deleterious form of DNA damage, double-strand breaks (DSBs),2 can arise from endogenous metabolic processes or exogenous environmental factors such as radiation or chemicals (1). Oxidative damage caused by reactive oxygen species (ROS) and consequent DSBs has been implicated in neurodegenerative disorders such as Alzheimer's disease (AD). ROS are generated endogenously by cellular metabolism and a variety of exogenous agents, such as ionizing radiation (IR). In studies conducted on replicating cells, it has been found that metabolically generated ROS can cause around 10,000 oxidative lesions per day (2).
ROS-induced damage predominantly leads to base or deoxyribose modifications that lead to single-strand breaks (SSBs). DSBs can arise because of replication past ROS-induced lesions or when SSBs occur in close proximity (3). These lesions are repaired either via non-homologous end joining (NHEJ) or homologous recombination (HR). In NHEJ, the broken DNA ends are processed/digested and then directly ligated, potentially leading to nucleotide deletions and consequent frameshift mutations. Thus, NHEJ is considered to be error-prone (4). HR utilizes undamaged DNA templates to direct error-free repair of the damaged strands. It has long been assumed that HR can only take place during the late S/G2 phases of the cell cycle, where sister chromatids are present as templates (5). However, recent studies in terminally differentiated cells have revealed an RNA-templated HR repair mechanism of DSBs at active transcription sites during the G0/G1 phase (transcription-coupled homologous recombination (TC-HR)). For this mechanism to occur, Cockayne syndrome B protein is recruited to an actively transcribed damage site, followed by recruitment of replication protein A (RPA), RAD51, RAD51C, and RAD52; then, repair occurs utilizing the nascent RNA template produced by active transcription (6). An RNA-templated, RAD52-directed mechanism of HR has also been reported in yeast (7). In addition, a recent study identifies yeast and human RAD52 inverse strand exchange, in which RAD52 forms a complex with dsDNA and promotes strand exchange with homologous ssRNA or ssDNA, indicating that RAD52 plays an important role in RNA-templated DSB repair in vivo (8). However, it is not known whether the transcription-dependent HR pathway exists in neurons. Except in the hippocampus and striatum, most neurons in the adult human brain are terminally differentiated and non-dividing and therefore are assumed to be incapable of utilizing HR for DSB repair (9, 10).
In this study, utilizing our site-specific damage induction systems, we measured the recruitment of the TC-HR–associated repair protein RAD52 to sites of DNA damage with and without transcription inhibition. We found that post-mitotic neurons employ this RNA-based recombinational mechanism for the DNA damage response. We also discovered preferential binding of RAD52 protein to R-loops, DNA–RNA hybrid structures present during active transcription (11), as further evidence of the role of RAD52 in TC-HR. Given our finding of the novel TC-HR DNA repair mechanism in post-mitotic cells, we wondered whether this mechanism might be affected in neurodegenerative disorders such as AD. We utilized Aβ1–42, neurotoxic oligomers 42 amino acids in length that are heavily implicated in AD pathology (12). We found that Aβ1–42 oligomers down-regulate the expression and damage response of the essential TC-HR repair protein RAD52. How this dysregulation of DNA repair may significantly contribute to the development of neurodegenerative diseases such as AD is discussed.
Results
DNA repair factors are recruited to laser-induced damage in neurons
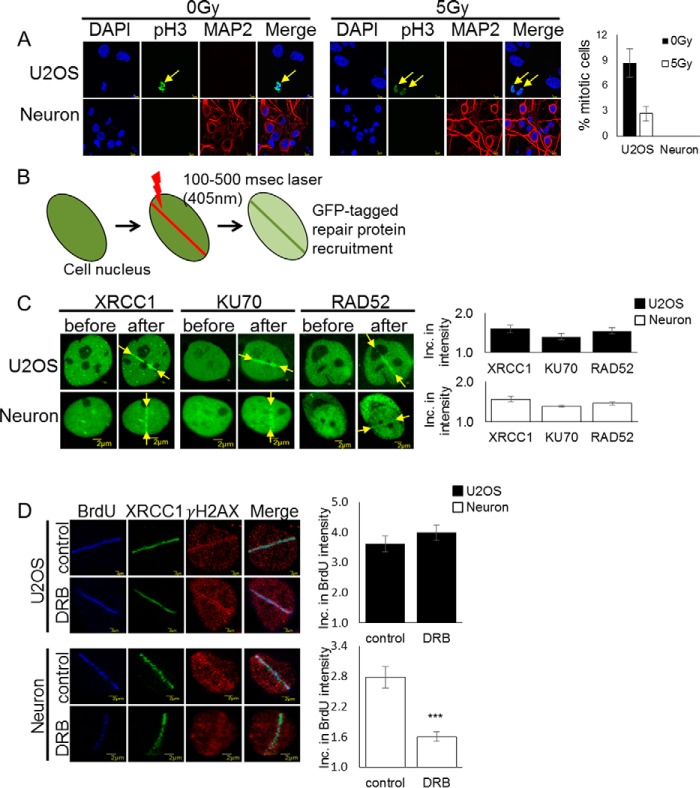
To understand how non-dividing neurons repair oxidative DNA damage, we first verified that the primary rat cortical neurons utilized for experimentation were post-mitotic. Some findings have shown a link between cell cycle activation in post-mitotic neurons and a DNA damage response leading to apoptosis (13). Therefore, we probed day in vitro 12 (DIV12) neuronal cell cultures with anti-phosphorylated histone H3 (Ser-10) before and after DSB-inducing γ irradiation (Fig. 1A). Studies have shown a correlation between this phosphorylated histone H3 (Ser-10) and mitotic chromosome condensation during early prophase, suggesting that anti-phosphohistone H3 can be used as a mitosis-specific marker (14). Here we observed no evidence of mitosis in primary cortical neurons compared with replicating U2OS cells before or 24 h after 5-gray γ irradiation.
Figure 1.
Post-mitotic neurons recruit the TC-HR protein RAD52. A, rat DIV12 cortical neurons and U2OS cells were treated with and without 5-gray (Gy) irradiation and probed with anti-phospho-Histone H3 (Ser-10) antibody. Error bars indicate the standard error of three separate experiments (n = 100), and the p values were determined by using Student's two-tailed t test. Cortical neurons do not show expression of phosphorylated H3 (Ser-10). B, schematic of the 405-nm scan laser system for induction of DSBs in single cells, where single cell nuclei are targeted with 100–500 ms of laser microirradiation to visualize and quantify GFP-tagged repair protein recruitment at damage sites. C, recruitment of the GFP-tagged DNA repair proteins XRCC1 (SSBR), KU70 (NHEJ), and RAD52 (HR) at sites of DNA damage in U2OS and DIV12 cortical neurons before and 1 min after 100- to 500-ms laser microirradiation. Error bars indicate the standard error of two separate experiments, and the p values were determined by using Student's two-tailed t test. Inc., increase. D, BrdU incorporation after 100-ms laser microirradiation with and without pretreatment with DRB (40 μm) for 24 h in U2OS and DIV12 cortical neurons. Error bars indicate the standard error of three separate experiments (n = 10), and the p values were determined by using Student's two-tailed t test (***, p ≪ 0.01). BrdU incorporation into repaired DNA was significantly reduced after transcription inhibition in post-mitotic neurons.
Prior studies in our laboratory have shown that terminally differentiated cells utilize an RNA-templated homologous recombination mechanism for repair of DSBs in G0/G1 phase of the cell cycle. This mechanism of repair can only occur with active transcription (6). We utilized 405-nm laser microirradiation to induce DSBs at localized sites in single cells, which makes it possible to observe repair protein recruitment in non-dividing cells (15) (Fig. 1B). We then investigated recruitment of GFP-tagged repair proteins specific to particular repair pathways, including XRCC1 (DNA single-strand break repair (SSBR)), KU70 (NHEJ), and RAD52 (HR) (Fig. 1C). In this experiment, we were surprised to find recruitment of the TC-HR repair protein RAD52 to sites of damage in post-mitotic neurons despite the lack of a sister chromatid of the neuron to utilize as a template for homologous repair.
DNA incorporation at sites of damage is affected by transcription inhibition
We next investigated whether transcription inhibition would affect the repair synthesis process of HR in post-mitotic neurons. We pretreated the cells with the RNA polymerase II inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), whichinhibits transcription elongation, and analyzed the incorporation of bromodeoxyuridine (BrdU), a synthetic analog of thymidine, into newly repaired DNA after laser damage. Importantly, the replication-mediated incorporation of BrdU into genomic DNA of post-mitotic neurons does not occur. Therefore, BrdU staining in neurons should indicate the repair-triggered BrdU incorporation. This incorporation was significantly decreased after DRB treatment (Fig. 1D), indicating that active transcription and a nascent RNA template are necessary for HR repair in post-mitotic neurons.
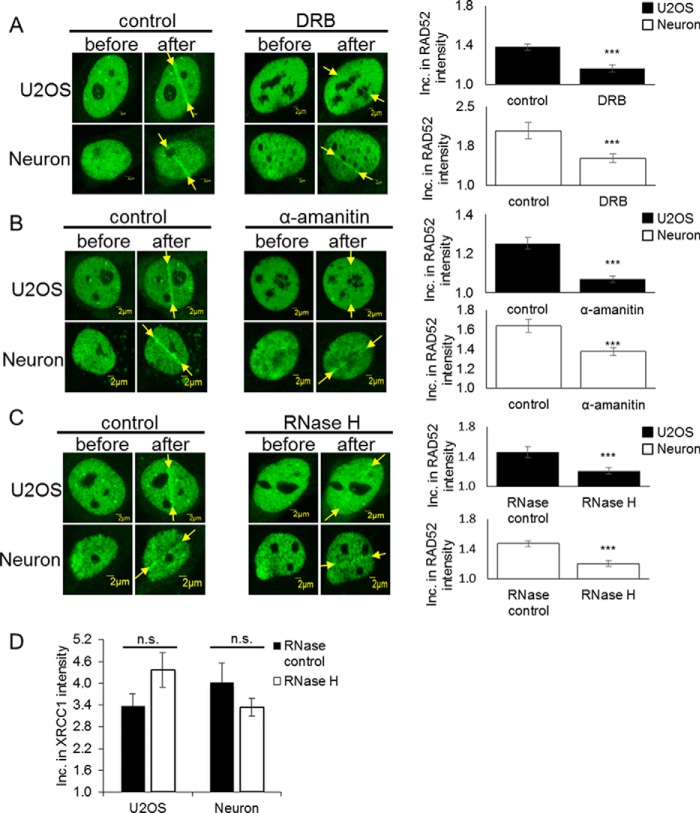
Transcription inhibition or RNase H treatment reduces the recruitment of RAD52 in neurons
RAD52 can be utilized in HR or a mutagenic repair process known as single-strand annealing. Single-strand annealing, which results in a deletion rearrangement between homologous repeating sequences, is independent of active transcription (16). Because our prior study indicated that the RAD52 dependent, RNA-templated recombination only occurs at sites of active transcription (6), we then tried to determine whether post-mitotic neurons utilize this particular recombinational mechanism for error-free repair of DSBs. To determine whether active transcription is necessary for the damage response of RAD52 in post-mitotic neurons, we pretreated cells with the RNA polymerase II inhibitor DRB, which inhibits transcription elongation, or α-amanitin, which inhibits transcription initiation and elongation (Fig. 2, A and B). Both inhibitors block transcription, thereby preventing the production of an mRNA template for use in RNA-templated HR. We found that, after inhibiting RNA polymerase II, recruitment of RAD52, an essential factor for RNA-templated recombinational repair, was significantly reduced at sites of laser damage in post-mitotic neurons. This supports our findings that RNA-templated HR in post-mitotic neurons requires active transcription to take place.
Figure 2.
Transcription inhibition reduces template-driven repair in post-mitotic neurons. A, recruitment of RAD52 before and 1 min after 200- to 500-ms laser microirradiation treatment with and without pretreatment with the RNA polymerase II inhibitor (40 μm) DRB for 24 h in U2OS cells and DIV12 ventral neurons. Inc., increase. Error bars indicate the standard error of two separate experiments (n = 10), and the p values were determined by using Student's two-tailed t test (***, p < 0.01). B, recruitment of RAD52 before and 1 min after 200- to 500-ms laser microirradiation treatment with and without pretreatment with the RNA polymerase II inhibitor (100 μg/ml) α-amanitin for 0.5 h in U2OS cells and DIV12 ventral neurons. Error bars indicate the standard error of two separate experiments (n = 10), and the p values were determined by using Student's two-tailed t test (***, p ≪ 0.01). RAD52 recruitment to damage sites was significantly reduced in post-mitotic neurons after DRB and α-amanitin treatment. C, recruitment of RAD52 before and 1 min after 500-ms laser microirradiation treatment with and without pretreatment with 15 units of RNase H for 15 min in U2OS cells and DIV12 ventral neurons. Error bars indicate the standard error of two different experiments (n = 10), and the p values were determined by using Student's two-tailed t test (***, p ≪ 0.01). RAD52 recruitment was significantly reduced after RNase H treatment in post-mitotic neurons. D, recruitment of XRCC1 before and 1 min after 500-ms laser microirradiation treatment with and without pretreatment with 15 units of RNase H for 15 min in U2OS cells and DIV12 ventral neurons. Error bars indicate the standard error of two different experiments (n = 10), and the p values were determined by using Student's two-tailed t test. XRCC1 recruitment was not significantly reduced after RNase H treatment in post-mitotic neurons. n.s., not significant.
To further establish that the recombination in post-mitotic neurons is associated with RNA-templated repair, we examined the effect of RNase H, an endoribonuclease that specifically degrades the RNA strand in an RNA–DNA hybrid structure, on the recruitment of RAD52 (Fig. 2C). After treatment with RNase H, RAD52 recruitment was significantly inhibited at sites of laser damage in post-mitotic neurons. To test whether the effects of the RNase H assay were due to the enzyme activity or the treatment itself, we measured recruitment of the SSBR protein XRCC1 after RNase H treatment. Because XRCC1 does not rely on an RNA strand, recruitment of XRCC1 should not be affected by RNase H activity. As evidenced in Fig. 2D, we did not see a significant reduction in XRCC1 activity after RNase H treatment. Combined, these results confirm that a nascent RNA template is necessary for HR repair to occur in post-mitotic neurons.
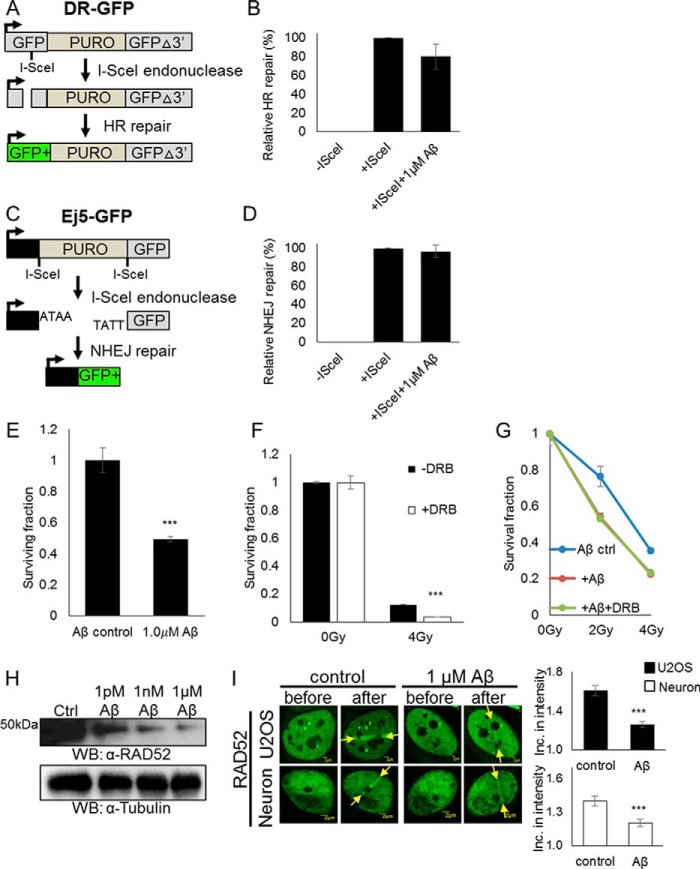
Aβ1–42 sensitizes cells to IR in the absence of active transcription
The accumulation of Aβ in AD may be associated with DNA damage induced by oxidative stress. A recent study has shown that high concentrations of Aβ1–42 (1 μm) in mitotic mouse hippocampal neuron cultures and in brain samples from AD patients contributed to reduced levels of BRCA1, leading to increased DSBs and reduced repair (17). However, it is not known whether Aβ1–42 affects the major DSB repair pathways: the error-prone NHEJ pathway and the less error-prone HR pathway.
We investigated the effects of Aβ1–42 on overall HR and NHEJ in dividing U2OS cells using the HR (DR-GFP) and NHEJ (Ej5-GFP) reporter assays (Fig. 3, A and C). For the HR reporter assay, the DR-GFP cell line utilizes an I-SceI recognition site inserted into a GFP coding sequence to create a nonfunctional GFP transgene. A wildtype GFP fragment has been inserted downstream of the transgene. After transfection of an I-SceI–expressing plasmid, a DSB is induced at the I-SceI cleavage site, and after HR occurs, fully functional GFP is restored, which can be detected by FACS. For the NHEJ assay, the Ej5-GFP cell line utilizes a GFP coding sequence interrupted by a puromycin gene that is flanked by I-SceI recognition sites. After transfection of the I-SceI–expressing plasmid, a DSB is induced at both sites, and, after NHEJ occurs, the fully functioning GFP is restored, which will be detectable by FACS (18, 19). Both assays were treated with 1 μm Aβ1–42 to determine its effect on each pathway. Aβ1–42 samples were investigated via Western blotting and transmission electron microscopy to verify that they contained oligomers (Fig. S2, A and B). We found that Aβ1–42 negatively affects classical HR in dividing cells (Fig. 3, B and D) to a slight extent.
Figure 3.
1 μm Aβ1–42 reduces TC-HR in dividing U2OS cells and post-mitotic neurons. A, schematic of the DR-GFP reporter used to measure HR efficiency in U2OS cells. B, DR-GFP reporter cells were pretreated with 1 μm Aβ1–42 for 5 h, and expression of GFP was measured by flow cytometry. Error bars indicate the standard error of three separate experiments, and the p values were determined by using Student's two-tailed t test. 1 μm Aβ1–42 treatment did not significantly decrease the HR efficiency. C, schematic of the Ej5-GFP reporter used to measure NHEJ efficiency in U2OS cells. D, Ej5-GFP reporter cells were pretreated with 1 μm Aβ1–42 for 5 h, and expression of GFP was measured by flow cytometry. Error bars indicate the standard error of three separate experiments, and the p values were determined by using Student's two-tailed t test. 1 μm Aβ1–42 treatment had no effect on NHEJ efficiency. E, U2OS cells were pretreated with 1 μm Aβ1–42 for 24 h. A colony-forming assay was performed, and the surviving fraction is shown. Error bars indicate the standard error of three separate experiments, and the p values were determined by using Student's two-tailed t test (***, p≪0.01). 1 μm Aβ1–42 negatively affected cell survival. F, U2OS cells were pretreated for 24 h with DRB (40 μm) or left untreated and then irradiated with IR at the indicated dose. A colony-forming assay was performed, and the surviving fraction is shown. Error bars indicate the standard error of three separate experiments, and the p values were determined by using Student's two-tailed t test (***, p ≪ 0.01). DRB treatment rendered cells more sensitive to IR. G, U2OS cells were pretreated for 24 h with DRB (40 μm) or left untreated and then treated with 1 μm Aβ1–42 for 5 h or left untreated and irradiated with IR at the indicated dose. A colony-forming assay was performed, and the surviving fraction is shown. Error bars indicate the standard error of three separate experiments, and the p values were determined by using Student's two-tailed t test. 1 μm Aβ1–42 in addition to DRB treatment did not render cells more sensitive to IR. Gy, gray. H, Western blot of RAD52 in DIV12 cortical neurons with and without 1 pm, 1 nm, and 1 μm Aβ1–42 treatment for 5 h. An increasing dose of Aβ1–42 reduced endogenous RAD52 protein levels in post-mitotic neurons. Ctrl, control; WB, Western blot. I, recruitment of RAD52 before and 1 min after 500-ms laser microirradiation treatment with and without 1 μm Aβ1–42 for 24 h in U2OS and DIV12 cortical neurons. Error bars indicate the standard error of two separate experiments (n = 10), and the p values were determined by using Student's two-tailed t test (***, p ≪ 0.01). Recruitment of RAD52 to damage sites was reduced in post-mitotic neurons after 24-h treatment of 1 μm Aβ1–42.
To determine whether the slightly negative effect of Aβ1–42 on classical HR was through the TC-HR pathway, we utilized the RNA polymerase II inhibitor DRB to disrupt transcription in U2OS cells and measured clonogenic survival against IR. 1 μm Aβ1–42 alone negatively affects overall U2OS survival (Fig. 3E), and, as shown in Fig. 3F, DRB treatment rendered cells more sensitive to IR. However, addition of 1 μm Aβ1–42 to the DRB treatment did not further increase the sensitivity of the cells to IR (Fig. 3G). This indicates that the effect of Aβ1–42 on classical HR is dependent on active transcription.
High concentrations of Aβ1–42 down-regulate the damage response of RAD52 in neurons
After determining that the RNA-templated recombination in neurons is active, we tested how Aβ1–42 might compromise repair efficiency in non-dividing neurons. We treated primary rat cortical neuron cell cultures with 1 μm Aβ1–42 (Fig. 3, H and I). Western blot analysis showed that 1 μm Aβ1–42 reduced overall RAD52 protein levels in post-mitotic neurons (Fig. 3H) and led to increased γH2AX expression (Fig. S1). After 24 h of treatment, it significantly reduced RAD52 recruitment to damage sites in both post-mitotic neurons and U2OS cancer cells (Fig. 3I). This indicates that Aβ1–42 affects the same repair pathway in both dividing and non-dividing cells, compromising genomic stability in a similar manner by contributing to the reduction of TC-HR repair efficiency.
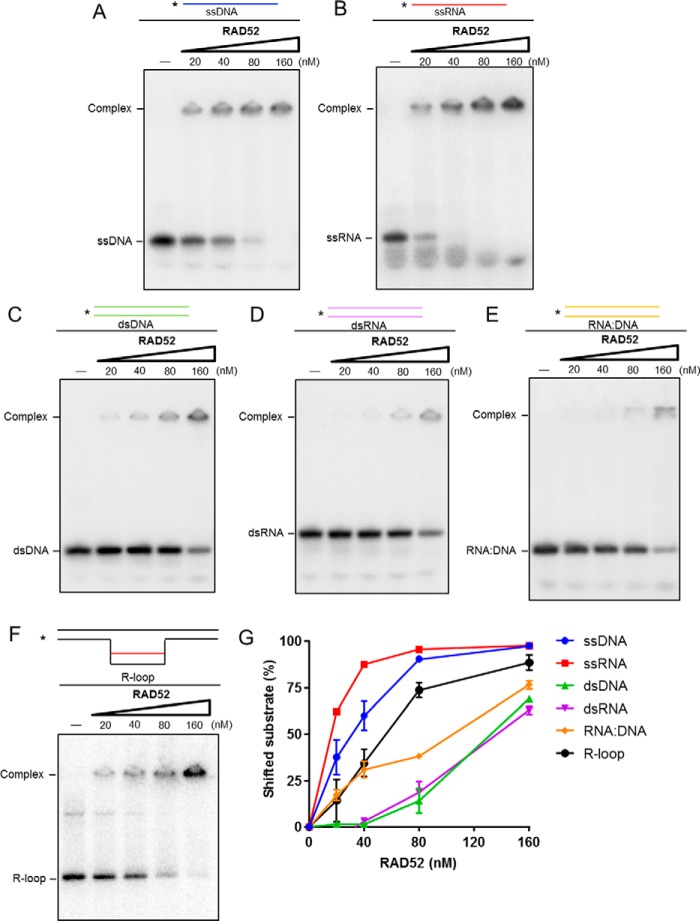
RAD52 binds to an R-loop substrate and preferentially binds to ssRNA
RAD52 protein has been demonstrated to bind DNA structures with DSBs (20, 21). Because previous studies from others and our study both indicate that RAD52 is involved in RNA-templated recombination repair, we tested the affinity of RAD52 to RNA-related substrates by electrophoretic mobility shift assay. The results show that purified human RAD52 protein efficiently binds both ssDNA and RNA (Fig. 4, A and B), although its binding affinity to ssRNA is higher than that of ssDNA. The binding affinities of RAD52 to dsDNA and RNA (Fig. 4, C and D) are significantly reduced compared with single-stranded substrates (Fig. 4G). Interestingly, RAD52 has a slightly higher affinity to an RNA–DNA hybrid duplex than dsDNA and dsRNA (Fig. 4E). Furthermore, RAD52 has a higher binding affinity to R-loop substrates (Fig. 4F), the damage-prone structures at transcriptionally active sites in the genome (11), than hybrid structures. These in vitro results are consistent with the possibility that RAD52 may bind ssRNA and the co-transcriptional R-loop upon transcriptional stress in post-mitotic neurons.
Figure 4.
RAD52 preferentially binds ssRNA and R-loop substrate. A–F, electrophoretic mobility shift assay to test binding of the RAD52 protein to ssDNA (A), ssRNA (B), dsDNA (C), dsRNA (D), the RNA–DNA hybrid (E), and the R-loop substrate (10 nm, F). G, representative graph of A–F. RAD52 preferentially binds ssRNA and the R-loop.
Discussion
In this study, we first discovered that recruitment of the HR factor RAD52 to sites of DNA damage is dependent on the presence of a nascent mRNA template for HR, indicating the existence of a novel RNA-templated recombinational repair pathway in post-mitotic neurons. We also discovered that RAD52 preferentially binds ssRNA and has direct affinity for R-loops, reinforcing the involvement of RAD52 in TC-HR repair of double-strand breaks. Utilization of this less error-prone pathway is essential for non-dividing neurons to maintain their genomic integrity when faced with environmental pressures such as metabolically induced oxidative damage and other endogenous and exogenous stressors.
Considering the fact that terminally differentiated, post-mitotic neurons must survive for a human lifetime, utilizing an error-prone pathway such as NHEJ would be vastly insufficient. In a study on cell cycle–arrested cells, it was found that NHEJ activity was responsible for approximately half of all replication-independent frameshift mutations, primarily because of deletions. Mutants deficient for HR (RAD54 deletion) contributed no increase in the amount of replication-independent mutations in the cell cycle–arrested cells but RAD52-deletion mutants did (22). This study confirms both the mutagenic nature of NHEJ and the possibility of a RAD52-dependent pathway that counteracts the frameshift-producing NHEJ pathway, such as TC-HR, which we have elucidated here and in our previous work.
Although post-mitotic neurons do not face the acquisition of mutations after replication past DNA lesions, DNA DSBs still pose massive risks to essential genes when not repaired. Failure to completely repair DNA may result in cellular senescence or apoptosis as a means for the cell to cope with an excess of damage and avoid malignant transformation (23, 24). However, unrepaired cells may also accumulate mutations that affect genes involved in regulating apoptosis, cell division, and DNA repair. If proliferating stem and neural progenitor cells incur mutations because of unrepaired DNA during development, then disease may later develop as a result of disrupted essential genes passed down through cell division or when tumor suppressing or oncogenes are no longer properly regulated (25). In post-mitotic neurons, mutations may contribute to neuronal dysfunction and apoptosis, as seen in the pathology of AD, where the cerebral cortex and hippocampus suffer extensive neuronal loss (26). Many neurodegenerative pathologies have also been linked to genetic deficiencies in DNA repair genes. For example, in ataxia telangiectasia, a mutation in the PI3K ataxia telangiectasia–mutated (ATM) protein, which is activated by DNA DSBs and is responsible for phosphorylating numerous downstream targets involved in DNA repair, cell cycle arrest, and apoptosis, causes individuals to experience higher rates of cancer and undergo neurodegeneration before the age of five (27–29). New research has suggested that the neuronal loss observed in AD may also result from partial loss of ATM function as AD pathology progresses (30).
In this study, we also found that high concentrations (micromolar) of Aβ1–42 oligomers lead to a reduction in the key RNA-templated HR repair protein RAD52. Given the critical role of RAD52 in the RNA-templated recombination process but not classic recombination in S/G2 phase, this reduction in overall function may result in increased DSBs because of a loss in high-fidelity repair capacity. The question remains, however, of the direct cause of increased concentrations of Aβ1–42 found in AD patients and how they directly or indirectly down-regulate essential repair proteins. Whether deficiencies in DNA repair, genetic abnormalities, or exogenous factors are the source of this pathology is still under investigation.
Study of repair mechanisms in post-mitotic neurons is essential if we are to understand the complex nature of neurodegenerative diseases. Although certain genetic aberrations may constitute a significant role in contributing to physiological aspects of AD, such as the APOEϵ4 allele and its association with increased AD risk, other contributing factors remain unknown (31). These factors may include deficiencies in DNA repair directly related to environmental exposure or internal processes such as the production of Aβ1–42.
Experimental procedures
Plasmids
GFP-tagged plasmids containing full-length KU70, RAD52, and XRCC1 were constructed as described previously (32).
Cell cultures and transfection
U2OS cells were cultured in high-glucose Dulbecco's modified Eagle's medium with 10% (v/v) fetal bovine serum (FBS) at 37 °C, 5% CO2. DR-GFP and Ej5-GFP stable cells were cultured in Dulbecco's modified Eagle's medium with 4.5 g/liter glucose and l-glutamine without sodium pyruvate with 10% (v/v) FBS and 0.1× GlutaMAX (Gibco) at 37 °C, 5% CO2. Primary neuronal cultures were prepared as described previously (33) with minor modifications. Rat cortical tissues were dissected from embryonic day 17 Sprague-Dawley rat brains. For the first 48 h, brain cultures were maintained in minimum Eagle's medium (Life Technologies) containing 2% heat-inactivated fetal bovine serum (Cellgro), 2% heat-inactivated horse serum (Life Technologies), 1 g/liter glucose (Sigma), 2 mm GlutaMAX (Life Technologies), 1 mm sodium pyruvate (Cellgro), 100 μm non-essential amino acids (Cellgro), 50 units/ml penicillin, and 50 μg/ml streptomycin (Corning). The culture medium was then changed to 0.5 ml/well of fresh serum-free Neurobasal medium containing 2% B27 supplement (Life Technologies), 2 mm GlutaMAX I (Life Technologies), 0.5 mg/ml AlbuMAX I (Life Technologies), and penicillin–streptomycin (Corning). Primary neurons were plated at 1.0 × 105 on 35-mm glass-bottom culture dished coated with poly-d-lysine (0.1 mg/ml, MatTek) or on 12-mm glass coverslips coated with poly-d-lysine (Millipore) in 6-well plates. Neuronal cells were treated or transfected at DIV12–14. For transfection, Neurobasal medium was removed and saved in 37 °C and 5% CO2, and 1 ml of prewarmed transfection medium was added (1× minimum Eagle's medium without bicarb (11430-030, Invitrogen), 2× GlutaMAX (35050-061, Invitrogen), 0.02 m Hepes (H30237.01, Thermo Fisher), 1 mm sodium pyruvate (11360-070, Invitrogen), and 0.033 m filter-sterilized glucose (G5400, Sigma)). Transfected neurons were incubated at 37 °C, 0% CO2 for 1.5 h, and then transfection medium was removed and replaced with warmed saved Neurobasal medium. All cells were transfected using Lipofectamine 2000 (Invitrogen) and Opti-MEM (Gibco). All neurons were transfected using 1 ml of transfection medium, 500 μl of Opti-MEM, 5 μl of Lipofectamine 2000, and 4 μg of total DNA per 3.5-cm MatTek glass-bottom dish.
Aβ1–42 oligomer preparation
Recombinant Aβ1–42 oligomers were prepared from lyophilized monomers treated with hexafluoro-2-propanol (HFIP) (A-1163-2, rPeptide) as described previously (17, 34). Aβ was resuspended in DMSO at 1 mg/ml and further diluted with ice-cold Neurobasal medium to 100–200 μm. Protein concentration was measured using reverse-phase HPLC and the Pierce BCA protein assay kit (Thermo Scientific). The ratio of monomers versus oligomers was determined using size exclusion chromatography on a sample incubated for 24 h. We also investigated the pelleted and supernatant fractions and observed that the supernatant fraction was the toxic fraction and that DMSO did not induce the resultant effects. Additionally, we investigated Aβ1–42 oligomers via Western blotting by running diluted fractions with monomeric controls on a 4–20% gradient gel (Mini-Protean TGX, 4561096) and probing with anti-6E10 antibody (BioLegend, 803004) (Fig. S2A). Aβ control medium was generated using equivalent amounts of DMSO and Neurobasal medium.
Transmission electron microscopy
Transmission electronic microscopy was used to detect the presence of Aβ1–42 oligomers (Fig. S2B). The Aβ1–42 sample (5 μl) was placed on a formvar grid and allowed to partly dry at room temperature. It was washed by touching it three times to the surface of a drop of distilled water, and excess water was removed by touching the grid to filter paper. Then a small drop of 1–3% uranyl acetate in distilled deionized water was added to the grid. After 10 s, excess stain was removed by touching the edge to filter paper. The grid was allowed to air-dry at room temperature before examination using a transmission electron microscope (Jeol JEM 1400, Tokyo, Japan) and imaging at 80 kV with the MegaView III Soft Imaging System.
Polymerase II inhibitors
The RNA polymerase II inhibitor DRB (D1916, Sigma) was added with a final concentration of 40 μm for 24 h, or α-amanitin (A2263, Sigma) was used at 100 μg/ml for 15 min before laser microirradiation.
RNase H treatment
U2OS cells and neurons were rinsed once with PBS and incubated with 1× RNase H buffer with or without RNase H (EN0201, Thermo Scientific) at room temperature for 15 min. Cells were treated with 0.1 mm 8-MOP (Methoxsalen) and incubated for 10 min at 37 °C, followed by confocal microscopy.
Immunostaining
For endogenous pH3, γH2AX, and MAP2 staining, after damage, neurons and U2OS cells were washed with PBS three times and fixed with 3.7% paraformaldehyde for 15 min at room temperature. The fixed cells were rinsed three times with PBS, treated with 0.2% Triton for 10 min at room temperature, and then rinsed three times with PBS. For staining of pH3, γH2AX, MAP2, and BrdU, cells were incubated with primary antibody overnight at 4 °C. Cells were then washed three times with PBS and incubated with Alexa Fluor 488 goat anti-mouse immunoglobulin G and Alexa Fluor 594 goat anti-rabbit immunoglobulin G conjugate or Alexa Fluor 488 goat anti-rabbit immunoglobulin G conjugate, Alexa Fluor 594 goat anti-mouse immunoglobulin G conjugate, and Alexa Fluor 405 goat anti-mouse immunoglobulin G conjugate (Invitrogen). Cells were washed three times with PBS. For 4′,6-diamidino-2-phenylindole staining, cells were treated with 1:1000 4′,6-diamidino-2-phenylindole for 5 min, washed three times in PBS and once with water, dried, mounted with DABCO-Tris-Glycerol (DTG) mounting buffer, and imaged using an Olympus FV1000 confocal microscopy system (catalog no. F10PRDMYR-1, Olympus). FV1000 software was used for acquisition of images. Primary antibodies used in this research were anti-γH2AX (1:2000, JBW301, Millipore), anti-MAP2 (1:1000, MAB378, Millipore), anti-BrdU (IIB5, I1212, Santa Cruz Biotechnology), and anti-phospho-histone H3 (Ser-10, 1:100, PA5-17869, Invitrogen).
Microscopy and laser light irradiation
Fluorescence images were obtained and processed with an FV-500 confocal scanning laser microscopy system (Olympus). A 405-nm scan laser system (Olympus) for irradiation of cells in the epifluorescence path of the microscope system was used. One scan of the laser light at full power delivers ∼1600 nanowatts. We scanned cells 100–500 times with the 405-nm laser at full power focused through a ×40 objective lens, which has been shown to induce SSBs and DSBs. Live cells were treated with 0.1 mm 8-MOP in Opti-MEM (Gibco) in glass-bottom dishes, incubated for 10 min at 37 °C, and then placed on a temperature-controlled (37 °C) plate. “Before” images were taken prior to laser microirradiation. The fluorescence intensity at an irradiated site was initially measured with a laser power/energy monitor (Orion, Ophir Optronics). The -fold increase in fluorescence intensity of each site was quantified by measuring fluorescent intensity after damage induction/background nuclear intensity using ImageJ software (15, 32, 35). Each experiment was done at least twice, and we observed 10 or more cells to obtain one result. Standard deviations and standard errors were derived from at least two independent experiments, and statistical significance (p values) was assessed using an unpaired Student's t test: *, p < 0.05; **, p < 0.01; ***, p ≪ 0.01.
Protein expression and purification
His6-tagged human RAD52 was expressed in Escherichia coli Rosetta cells and purified as described previously (36).
Electrophoretic mobility shift assay
RAD52 protein was serially diluted to the indicated concentration and mixed with 10 nm 32P-labeled ssDNA, ssRNA, dsDNA, dsRNA, RNA–DNA, or R-loop substrate in electrophoretic mobility shift assay buffer (15 mm HEPES (pH 7.5), 1 mm MgCl2, 2% glycerol, 0.2 μg/μl BSA, 100 mm KCl, and 0.16 units/μl RNaseOut). The reaction mixtures were incubated at 37 °C for 10 min. After addition of gel loading buffer (50% glycerol, 20 mm Tris-HCl (pH 7.4), 0.5 mm EDTA, and 0.05% orange G), the reaction mixtures were resolved by 5% native polyacrylamide gel electrophoresis in 1× TBE buffer (90 mm Tris borate (pH 8.0) and 2 mm EDTA) at 4 °C. The gels were dried, and the products were visualized by PhosphorImager.
HR and NHEJ assays
DR-GFP and Ej5-GFP stable cells were incubated in Dulbecco's modified Eagle's medium with 10% FBS. Cells were transfected with the pCMV-I-SceI plasmid using Lipofectamine 2000 (Life Technologies) for 24 h and kept in the dark. 1 μm Aβ1–42 or Aβ control medium was added for 5 h. Then cells were spun at 600 rpm, washed with 1× PBS three times, and analyzed by FACS.
BrdU incorporation
U2OS or DIV12 rat cortical neurons previously transfected with GFP-XRCC1 were treated with 40 μm DRB for 24 h or left untreated. Cells were treated with laser microirradiation, and then 0.01 mm BrdU was added for 24 h. Cells were fixed with 3.7% paraformaldehyde for 15 min at room temperature, rinsed three times with PBS, and then treated with 2.5 n HCl for 30 min at 37 °C. Cells were rinsed three times with PBS, permeabilized with 0.2% Triton for 5 min at room temperature, and then rinsed three times with PBS. Immunofluorescence staining proceeded as above.
Cell survival
To determine the effect of 1 μm Aβ1–42 on U2OS cell survival, U2OS cells were treated with 1 μm Aβ1–42 or Aβ control medium for 24 h or left untreated, seeded onto 6-cm dishes, and incubated at 37 °C for 10 days. Colonies were fixed and stained with 0.3% crystal violet, and then survival (number of colonies) was expressed as a percentage of non-treated colonies. To determine the effect of 1 μm Aβ1–42 on U2OS IR sensitivity, U2OS cells were treated with 40 μm DRB for 24 h and/or 1 μm Aβ1–42 or Aβ control medium for 5 h or left untreated and then seeded onto 6-cm dishes. After seeding, cells were exposed to radiation as indicated and incubated for 10 days at 37 °C. Colonies were fixed and stained with 0.3% crystal violet, and then survival (number of colonies) was expressed as a percentage of non-irradiated colonies.
Lysates and Western blotting
Primary DIV12 cortical neurons seeded on a 24-well plate (3 wells/treatment) or 6-well plate (1 well/treatment) and U2OS cells seeded on 6-cm dishes were treated as indicated. Cell lysates were prepared with 100–150 μl of lysis buffer/well (62.5 mm Tris (pH 6.8), 5% glycerol, 2% SDS, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride). Samples were run on 8–12% SDS-polyacrylamide gels or a 4–20% gradient gel (Mini-Protean TGX, 4561096) and immunoblotted using a monoclonal antibody specific for the anti-RAD52 antibody (K1512, Santa Cruz Biotechnology) or anti-γH2AX (ab12267, Abcam).
Author contributions
S. W. conducted the major experiments and wrote the manuscript. Y. T., Z. L., W. Z., L. H. S., M. E. R., R. K., and A. T. helped with the assays. J. T. G., A. T., and R. W. helped analyze the results. A. S. L. and L. L. provided overall experimental guidance. All authors edited their specific contributions and reviewed the manuscript.
Supplementary Material
Acknowledgment
This project used the University of Pittsburgh Cancer Institute Cytometry Facility, supported in part by National Institutes of Health Grant P30CA047904.
This work was supported in part by National Institutes of Health Grants GM118833 (to L. L.) and DC013048 (to M. E. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
- DSB
- double-strand break
- ROS
- reactive oxygen species
- AD
- Alzheimer's disease
- IR
- ionizing radiation
- SSB
- single-strand break
- NHEJ
- non-homologous end joining
- HR
- homologous recombination
- TC
- transcription-coupled
- ssRNA
- single-stranded RNA
- ssDNA
- single-stranded DNA
- DIV
- days in vitro
- SSBR
- single-strand break repair
- DRB
- 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- BrdU
- bromodeoxyuridine
- ATM
- ataxia telangiectasia–mutated
- FBS
- fetal bovine serum
- DR
- direct repeats.
References
- 1. Fang Y. Z., Yang S., and Wu G. (2002) Free radicals, antioxidants, and nutrition. Nutrition 18, 872–879 10.1016/S0899-9007(02)00916-4 [DOI] [PubMed] [Google Scholar]
- 2. Ames B. N., Shigenaga M. K., and Hagen T. M. (1993) Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. U.S.A. 90, 7915–7922 10.1073/pnas.90.17.7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- 4. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sonoda E., Hochegger H., Saberi A., Taniguchi Y., and Takeda S. (2006) Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5, 1021–1029 10.1016/j.dnarep.2006.05.022 [DOI] [PubMed] [Google Scholar]
- 6. Wei L., Nakajima S., Böhm S., Bernstein K. A., Shen Z., Tsang M., Levine A. S., and Lan L. (2015) DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 112, E3495–E3504 10.1073/pnas.1507105112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keskin H., Shen Y., Huang F., Patel M., Yang T., Ashley K., Mazin A. V., and Storici F. (2014) Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439 10.1038/nature13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazina O. M., Keskin H., Hanamshet K., Storici F., and Mazin A. V. (2017) Rad52 inverse strand exchange drives RNA-templated DNA double-strand break repair. Mol. Cell 67, 19–29.e3 10.1016/j.molcel.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eriksson P. S., Perfilieva E., Björk-Eriksson T., Alborn A. M., Nordborg C., Peterson D. A., and Gage F. H. (1998) Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317 10.1038/3305 [DOI] [PubMed] [Google Scholar]
- 10. Ernst A., Alkass K., Bernard S., Salehpour M., Perl S., Tisdale J., Possnert G., Druid H., and Frisén J. (2014) Neurogenesis in the striatum of the adult human brain. Cell 156, 1072–1083 10.1016/j.cell.2014.01.044 [DOI] [PubMed] [Google Scholar]
- 11. Skourti-Stathaki K., and Proudfoot N. J. (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396 10.1101/gad.242990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yankner B. A., Dawes L. R., Fisher S., Villa-Komaroff L., Oster-Granite M. L., and Neve R. L. (1989) Neurotoxicity of a fragment of the amyloid precursor associated with Alzheimer's disease. Science 245, 417–420 10.1126/science.2474201 [DOI] [PubMed] [Google Scholar]
- 13. Kruman I. I., Wersto R. P., Cardozo-Pelaez F., Smilenov L., Chan S. L., Chrest F. J., Emokpae R. Jr., Gorospe M., and Mattson M. P. (2004) Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 41, 549–561 10.1016/S0896-6273(04)00017-0 [DOI] [PubMed] [Google Scholar]
- 14. Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., BazettJones D. P., and Allis C. D. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- 15. Lan L., Nakajima S., Komatsu K., Nussenzweig A., Shimamoto A., Oshima J., and Yasui A. (2005) Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 118, 4153–4162 10.1242/jcs.02544 [DOI] [PubMed] [Google Scholar]
- 16. Bhargava R., Onyango D. O., and Stark J. M. (2016) Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suberbielle E., Djukic B., Evans M., Kim D. H., Taneja P., Wang X., Finucane M., Knox J., Ho K., Devidze N., Masliah E., and Mucke L. (2015) DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat. Commun. 6, 8897 10.1038/ncomms9897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakanishi K., Cavallo F., Brunet E., and Jasin M. (2011) Homologous recombination assay for interstrand cross-link repair. Methods Mol. Biol. 745, 283–291 10.1007/978-1-61779-129-1_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helfricht A., Wiegant W. W., Thijssen P. E., Vertegaal A. C., Luijsterburg M. S., and van Attikum H. (2013) Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle 12, 3070–3082 10.4161/cc.26033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haber J. E. (1999) DNA repair: gatekeepers of recombination. Nature 398, 665–667 10.1038/19423 [DOI] [PubMed] [Google Scholar]
- 21. Ristic D., Modesti M., Kanaar R., and Wyman C. (2003) Rad52 and Ku bind to different DNA structures produced early in double-strand break repair. Nucleic Acids Res. 31, 5229–5237 10.1093/nar/gkg729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heidenreich E., Novotny R., Kneidinger B., Holzmann V., and Wintersberger U. (2003) Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. EMBO J. 22, 2274–2283 10.1093/emboj/cdg203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Campisi J. (2001) From cells to organisms: can we learn about aging from cells in culture?. Exp. Gerontol. 36, 607–618 10.1016/S0531-5565(00)00230-8 [DOI] [PubMed] [Google Scholar]
- 24. Collado M., and Serrano M. (2010) Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer 10, 51–57 10.1038/nrc2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawson A. R., Hindley G. F., Forshew T., Tatevossian R. G., Jamie G. A., Kelly G. P., Neale G. A., Ma J., Jones T. A., Ellison D. W., and Sheer D. (2011) RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology. Genome Res. 21, 505–514 10.1101/gr.115782.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wenk G. L. (2003) Neuropathologic changes in Alzheimer's disease. J. Clin. Psychiatry 64, Suppl. 9, 7–10 [PubMed] [Google Scholar]
- 27. Shiloh Y., and Rotman G. (1996) Ataxia-telangiectasia and the ATM gene: linking neurodegeneration, immunodeficiency, and cancer to cell cycle checkpoints. J. Clin. Immunol. 16, 254–260 10.1007/BF01541389 [DOI] [PubMed] [Google Scholar]
- 28. Barlow C., Brown K. D., Deng C. X., Tagle D. A., and Wynshaw-Boris A. (1997) ATM selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat. Genet. 17, 453–456 10.1038/ng1297-453 [DOI] [PubMed] [Google Scholar]
- 29. Khanna K. K. (2000) Cancer risk and the ATM gene: a continuing debate. J. Natl. Cancer Inst. 92, 795–802 10.1093/jnci/92.10.795 [DOI] [PubMed] [Google Scholar]
- 30. Shen X., Chen J., Li J., Kofler J., and Herrup K. (2016) Neurons in vulnerable regions of the Alzheimer's disease brain display reduced ATM signaling. eNeuro 3, ENEURO.0124–0115.2016 10.1523/ENEURO.0124-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., and Pericak-Vance M. A. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261, 921–923 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- 32. Lan L., Hayashi T., Rabeya R. M., Nakajima S., Kanno S., Takao M., Matsunaga T., Yoshino M., Ichikawa M., Riele H., Tsuchiya S., Tanaka K., and Yasui A. (2004) Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to cis-diamminedichloroplatinum(II) in mammalian cells. DNA Repair 3, 135–143 10.1016/j.dnarep.2003.10.005 [DOI] [PubMed] [Google Scholar]
- 33. Sanders L. H., McCoy J., Hu X., Mastroberardino P. G., Dickinson B. C., Chang C. J., Chu C. T., Van Houten B., and Greenamyre J. T. (2014) Mitochondrial DNA damage: molecular marker of vulnerable nigral neurons in Parkinson's disease. Neurobiol. Dis. 70, 214–223 10.1016/j.nbd.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cissé M., Halabisky B., Harris J., Devidze N., Dubal D. B., Sun B., Orr A., Lotz G., Kim D. H., Hamto P., Ho K., Yu G. Q., and Mucke L. (2011) Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature 469, 47–52 10.1038/nature09635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei L., Nakajima S., Hsieh C. L., Kanno S., Masutani M., Levine A. S., Yasui A., and Lan L. (2013) Damage response of XRCC1 at sites of DNA single strand breaks is regulated by phosphorylation and ubiquitylation after degradation of poly(ADP-ribose). J. Cell Sci. 126, 4414–4423 10.1242/jcs.128272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma C. J., Kwon Y., Sung P., and Greene E. C. (2017) Human RAD52 interactions with replication protein A and the RAD51 presynaptic complex. J. Biol. Chem. 292, 11702–11713 10.1074/jbc.M117.794545 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.