Abstract
The protein C receptor (PROCR) has emerged as a stem cell marker in several normal tissues and has also been implicated in tumor progression. However, the functional role of PROCR and the signaling mechanisms downstream of PROCR remain poorly understood. Here, we dissected the PROCR signaling pathways in breast cancer cells. Combining protein array, knockdown, and overexpression methods, we found that PROCR concomitantly activates multiple pathways. We also noted that PROCR-dependent ERK and PI3k–Akt–mTOR signaling pathways proceed through Src kinase and transactivation of insulin-like growth factor 1 receptor (IGF-1R). These pathway activities led to the accumulation of c-Myc and cyclin D1. On the other hand, PROCR-dependent RhoA–ROCK–p38 signaling relied on coagulation factor II thrombin receptor (F2R). We confirmed these findings in primary cells isolated from triple-negative breast cancer–derived xenografts (PDX) that have high expression of PROCR. To the best our knowledge, this is the first comprehensive study of PROCR signaling in breast cancer cells, and its findings also shed light on the molecular mechanisms of PROCR in stem cells in normal tissue.
Keywords: breast cancer, cell signaling, mammary gland, signal transduction, stem cells, PROCR, breast cancer, TNBC, ERK, Akt, RhoA
Introduction
The protein C receptor (PROCR)3 has emerged as a stem cell marker in several tissues, including the mammary gland (1), hematopoietic system (2–5), and vascular endothelial cells (6). Besides being a surface marker, its signaling mechanisms in the stem cells are unknown. PROCR has also been implicated in tumor progression. However, there is conflicting evidence on the mechanisms of action of PROCR. It has been reported that PROCR promotes tumor growth (7–9), but it has also been suggested that PROCR inhibits tumor progression (10). Hence, the functional roles as well as the signaling pathways of PROCR in cancer cells are also poorly understood.
PROCR is a single-pass transmembrane receptor, and is best known for its expression on vascular cells and its anticoagulation activity (11). PROCR activates its ligand, a protease precursor protein C (PROC), to become active PROC, which is then dissociated from PROCR and exerts anticoagulation effect directly via inactivation of Factor Va and Factor VIIIa (reviewed in Refs. 12, 13).
There is evidence that PROCR activates intracellular signaling, resulting in cytoprotective effects in endothelial cells, monocyte, keratinocyte, and intestinal epithelial cells (14–18). It is widely accepted that the central event of PROCR intracellular signaling is the activation of a G protein–coupled receptor (GPCR), coagulation factor II thrombin receptor (F2R; also called protease-activated receptor-1, PAR-1) (19). active PROC uses PROCR as a co-receptor for the cleavage of F2R, enabling F2R to activate downstream signaling events (reviewed in Refs. 12, 13). The PROCR–F2R axis has been shown to increase endothelial cell barrier function, survival, proliferation or migration through activation of the mitogen-activated protein kinase (MAPK), phosphatidylinositol-3 kinase (PI3K), or endothelial nitric oxide synthase (eNOS) pathways or through inhibition of p53 (20–23). There are limited data on PROCR-induced signal transduction in non-endothelial cells. In lymphocytes, epithermal keratinocytes, and breast cancer epithelial cells, it has been reported that active PROC–PROCR–F2R can stimulate the MAPK pathway via activation of epidermal growth factor receptor (EGFR) (17, 24, 25).
In this study, we utilized breast cancer cell lines and patient-derived xenograft (PDX) tumor cells to investigate the signaling pathways of PROCR in breast cancer cells. The results described here provide evidence that PROCR induces the activation of ERK, PI3K–Akt, and RhoA signaling. Distinct from previous reports, the PROCR-dependent ERK and PI3K–Akt activities in breast cancer cells are not via F2R and EGFR, instead they are through Src and IGF-1R activation.
Results
PROCR activates ERK, PI3K–Akt–mTOR, and RhoA–ROCK pathways in TNBC cells
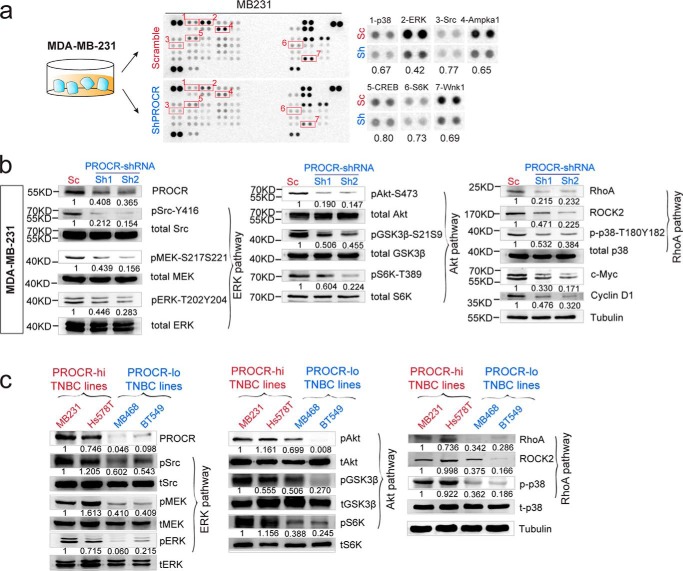
We found that PROCR is differentially expressed in breast cancer cell lines. PROCR expression is more prevalent in triple-negative breast cancer (TNBC) cells compared with ER+/PR+ (estrogen receptor–positive, progesterone receptor–positive) and HER2+ cells that we examined (Fig. S1). Within TNBC, MDA-MB-231, Hs 578T, HCC38, CAL51, and HCC1806 cells exhibited higher PROCR expression level compared with other lines (MDA-MB-468, BT549, MDA-MB-436, HCC1937, HCC1599, HCC2157), ER+/PR+ lines, and HER+ lines (Fig. S1). To dissect the intracellular pathways that PROCR activates, we performed a phosphokinase antibody array using lysates of MDA-MB-231 cells (a PROCR–high TNBC line) harvested at 48 h post lentiviral infection. PROCR silencing with shRNA (shPROCR) led to inhibition of the phosphorylation of several kinases, including p38α (Thr-180/Tyr-182), ERK (Thr-202/Tyr-204, Thr-221/Tyr-223), Src (Tyr-419), Ampka1 (Thr-183), CREB (Ser-133), S6K (Thr-389), and Wnk1 (Thr-60) (Fig. 1a). We investigated whether inhibition of PROCR affects MAPK signaling (in view of the down-regulation of pSrc and pERK), PI3K–Akt–mTOR signaling (given the observed down-regulation of pCREB and pS6K), and RhoA–ROCK signaling cascades (given the down-regulation of p38α), which are key signaling pathways in breast cancer (26). Western blot analysis with two distinct shRNAs targeting PROCR confirmed the down-regulation of pSrc (Tyr-416) and pERK (Thr-202/Tyr-204), and further revealed reduced levels of pMEK (Ser-217/Ser-221), indicating a reduced ERK signaling activity (Fig. 1b). In addition, Western blot analysis confirmed that pAkt (Ser-473) level and the indicator of Akt activity such as pGSK3β (Ser-21/Ser-9) were also decreased upon PROCR silencing (Fig. 1b). Further downstream, the activity of mTOR1 signaling was also suppressed, as seen by reduced levels of pS6K (Thr-389), and decreased protein levels of c-Myc and cyclin D1 (Fig. 1b). These results indicated that PI3K–Akt–mTOR signaling activity is reduced upon PROCR inhibition. Moreover, the decreased RhoA, ROCK2, and p-p38α (Thr-180/Tyr-182) levels were detected when PROCR was inhibited (Fig. 1b). These data suggest that PROCR induces the activation of ERK, PI3K–Akt–mTOR, and RhoA–ROCK signal cascades in MDA-MB-231 cells.
Figure 1.
PROCR activates ERK, PI3K–Akt–mTOR, and RhoA–ROCK signaling pathways in breast cancer cells. a, illustration and representing image of phosphokinase antibody array using lysates of MDA-MB-231 cells with scramble control and PROCR-shRNA (Sh2). Seven proteins with most evident down-regulation following PROCR knockdown are indicated. b, Western blot showing down-regulation of ERK, PI3K–Akt–mTOR, and RhoA–ROCK pathway activities in MDA-MB-231 cells with PROCR-shRNA (Sh1 and Sh2) knockdown. Tubulin was used as loading controls. c, Western blot showing differential activities of ERK, PI3K–Akt–mTOR, and RhoA–ROCK pathway in PROCR–high TNBC cell lines (MDA-MB-231, Hs 578T) and PROCR–low TNBC cell lines (MDA-MB-468, BT549). Tubulin was used as loading control. Western blots in the same panel are from the same batch of cells using the same loadings, thus only one loading control is shown at the end of the panel. For a better illustration, they are shown as three separated columns representing ERK, Akt, and RhoA pathway, respectively. Each Western blot analyses was repeated for three times or more.
Next, we chose another PROCR–high TNBC line, Hs 578T, and confirmed the down-regulation of these pathway activities upon PROCR knockdown (Fig. S1b). The association of PROCR expression with the activities of the three pathways was further compared between the PROCR–high TNBC lines (MDA-MB-231 and Hs 578T) and PROCR–low TNBC lines (MDA-MB-468 and BT549). Western blot analysis validated their PROCR level status (Fig. 1c). Indeed, all three pathways were activated in both PROCR–high TNBC lines, MDA-MB-231, and Hs 578T (Fig. 1c). In contrast, in both PROCR–low TNBC lines, no concomitant activation of the three pathways was observed (Fig. 1c). Of note, PI3K–Akt–mTOR pathway activation in MDA-MB-468 is likely because of the known EGFR amplification in this line (27). In BT549, all three pathways were in low activities (Fig. 1c). Overall, these results support our model that PROCR activates ERK, PI3K–Akt–mTOR and RhoA–ROCK signaling cascades in PROCR–high TNBC cells.
Validating ERK, PI3K–Akt–mTOR, and RhoA–ROCK signaling activities in PDX cells
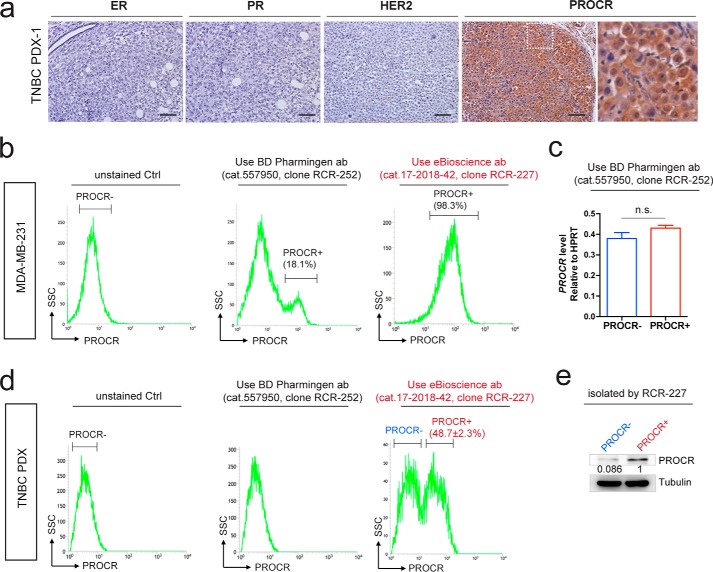
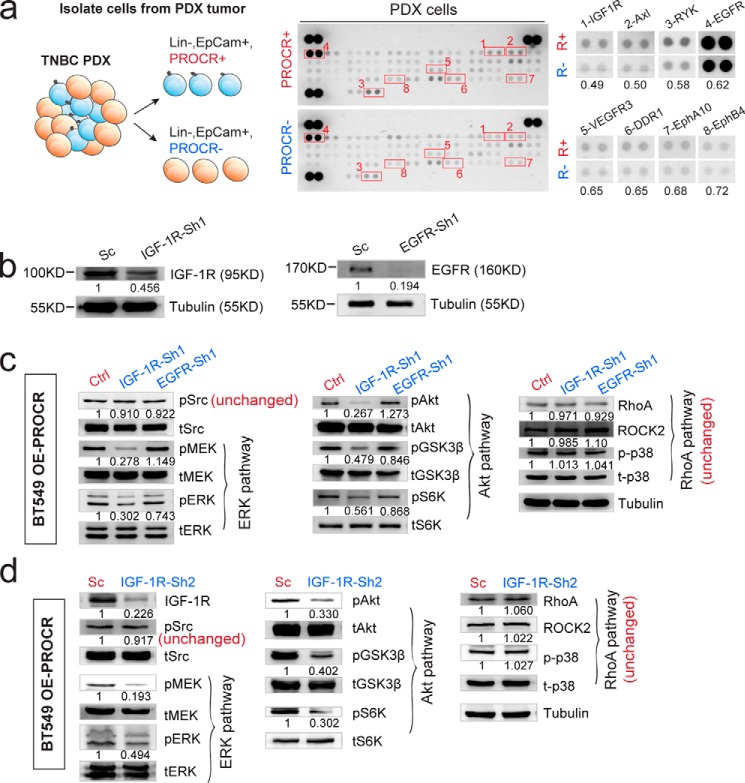
Next we investigated the signaling activities in PROCR+ cells in tumors. We used patient-derived xenograft TNBC cells with high expression of PROCR (Fig. 2a). To properly isolate PROCR+ and PROCR− cells, we looked for an antibody effective for fluorescence-associated cell sorter (FACS). We found that clone RCR-227 can accurately distinguish PROCR+ and PROCR− cells by FACS, whereas clone RCR-252 cannot. The comparisons were using both MDA-MB-231 cells and TNBC PDX-1 tumor cells. First, in MDA-MB-231 cells, consistent with previous studies (9, 28), RCR-252 antibody detected a small portion (18.1%) of PROCR+ cells by FACS (Fig. 2b). However, the isolated cells displayed no differential expression of PROCR by quantitative polymerase chain reaction (qPCR) analysis (Fig. 2c), suggesting an inaccurate separation of PROCR+ and PROCR− cells using this antibody. In contrast, FACS analysis using RCR-227 showed that almost all MDA-MB-231 cells (98.3%) are PROCR+ (Fig. 2b), suggesting RCR-227 is a more potent antibody in this assay compared with RCR-252. The comparison of the two antibodies was further carried out using freshly dissociated cells from PROCR–high TNBC PDX-1. RCR-252 was ineffective in recognizing PROCR+ cells by FACS (Fig. 2d), whereas FCAS analysis using RCR-227 established that 48.7% of the PDX-1 cells are PROCR+ (Figs. 2d and 3a). Western blot analysis of PROCR protein levels confirmed the correct isolation using RCR-227 (Fig. 2e).
Figure 2.
Validate PROCR antibody for flow cytometry. a, immunohistochemistry (IHC) indicating that the breast cancer PDX-1 sample is ER-, PR-, HER2-, and PROCR-high. Scale bars represent 100 μm. b, FACS analysis of MDA-MB-231 cells using two distinct monoclonal antibodies of PROCR. RCR-252 (BD Pharmingen, cat. no. 557950) only detects 18.1% of cells are positive for PROCR, whereas RCR-227 (eBioscience, cat. no. 17-2018-42) indicates 98.3% of cells are positive for PROCR. c, qPCR analysis indicating no significant difference of PROCR expression levels between isolated PROCR+/− cells using RCR-252. Data are presented as mean ± S.E. d, FACS analysis of PDX cells using the two PROCR antibodies. Analysis with RCR-252 was not able to detect cells that are positive for PROCR, whereas analysis with RCR-227 indicates 48.7% of cells are positive for PROCR. e, Western analysis indicating obvious higher level of PROCR in isolated PROCR+ cells compared with PROCR− cells, confirming the correct isolation using RCR-227 antibody. Each experiments was repeated for three times or more.
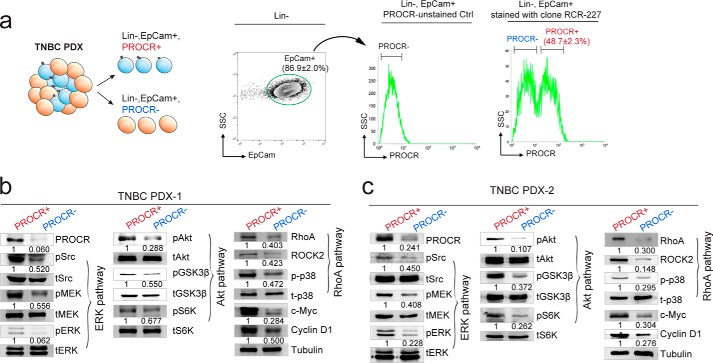
Figure 3.
Validate PROCR-dependent signal activities in human breast cancer PDX. a, illustration of isolation of PROCR+ and PROCR− tumor epithelial cells in TNBC PDX. FACS analysis of PDX cells using PROCR antibody (clone RCR-227) indicating that 86.9% of cells are EpCam+ epithelial cells. Within EpCam+ cells, 48.7% of cells are positive for PROCR. b and c, Western blot showing differential activities of ERK, PI3K–Akt–mTOR, and RhoA–ROCK pathway in PROCR+ and PROCR− cells isolated from PROCR+ TNBC PDX-1 (b) and PDX-2 (c) tumor. Tubulin was used as loading control. Western blots in the same panel are from the same batch of cells using the same loadings, thus only one loading control is shown at the end of the panel. For a better illustration, they are shown as three separated columns representing ERK, Akt, and RhoA pathway, respectively. Each experiment was repeated three times or more.
Upon proper isolation of PROCR+ and PROCR− cells, the signaling activities of the three pathways (ERK, PI3K–Akt, and RhoA) were examined. Western blot analyses showed that PROCR+ tumor cells exhibit markedly more robust signaling activities in all three pathways compared with PROCR− tumor cells (Fig. 3b). PROCR+ cells also had distinctly higher expression of c-Myc and cyclin D1 compared with PROCR− cells (Fig. 3b). Consistent results were observed using an additional TNBC PDX sample (PDX-2) (Fig. 3c). These data reinforce that ERK, PI3K–Akt–mTOR, and RhoA–ROCK–p38 signal cascades are intracellular effectors of PROCR in breast cancer cells.
PROCR activates RhoA–ROCK–p38 signaling via F2R
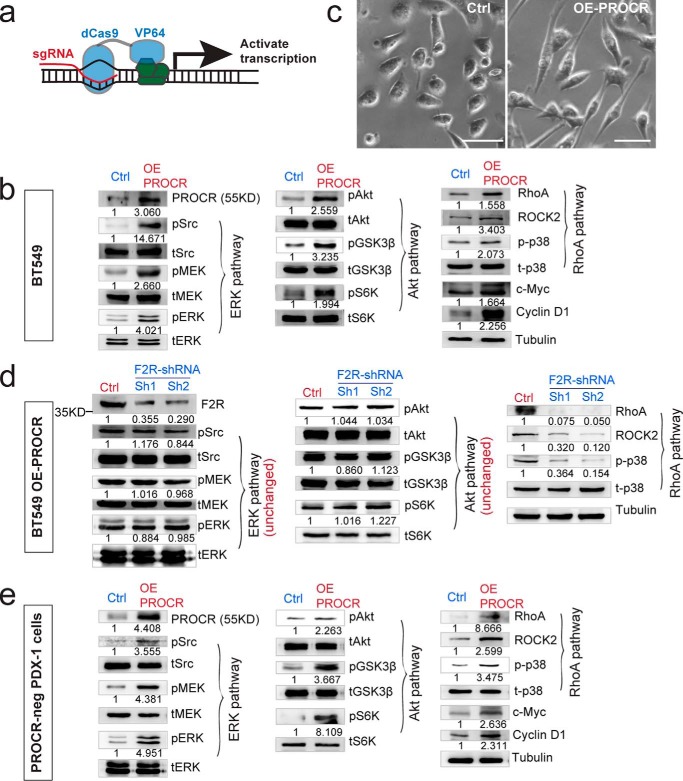
Next, we investigated the cell surface components through which PROCR activates downstream signaling. We established a PROCR overexpression system that can activate its downstream signaling. In a PROCR–low TNBC cell line BT549, which has low baseline activities of the three above signaling pathways, we employed the CRISPR interference system to activate endogenous PROCR expression (29). BT549 cells were virally infected with dCas9-VP64 and sgRNA (sgPROCR) (Fig. 4a). Enhancement of PROCR expression is confirmed using this system (Fig. 4b). ERK, PI3K–Akt–mTOR signaling, and RhoA–ROCK signal cascades were all up-regulated upon PROCR overexpression, including accumulation of c-Myc and cyclin D1 (Fig. 4b). Change of cell shape was also associated with PROCR overexpression: BT549 cells with enhanced PROCR expression became more elongated compared with the control (Fig. 4c). In this overexpression system, we interrogated which surface effectors are required for PROCR-dependent signaling. Previous studies have reported that the G protein–coupled receptor, F2R, is central for the cytoprotective activity of PROCR in various cell types. Thus, we examined whether F2R is required for PROCR signaling in breast cancer cells. Two F2R shRNAs were generated and their knockdown efficacies were confirmed (Fig. 4d). Interestingly, knockdown of F2R only attenuated RhoA–ROCK and p38 signaling induced by PROCR overexpression, leaving the other two pathways (ERK and PI3K–Akt–mTOR) unaffected (Fig. 4d). These results suggest that RhoA–ROCK–p38 signaling induced by PROCR is dependent on F2R, whereas ERK and PI3K–Akt–mTOR activation is dependent on other surface effectors, not F2R.
Figure 4.
F2R mediates the activation of RhoA pathway by PROCR, not the activation of ERK and PI3K–Akt–mTOR pathways. a, illustration of activating endogenous PROCR expression using CRISPR interference system, through viral infection with dCas9-VP64 and sgPROCR. b, Western blot indicating that ERK, PI3K–Akt–mTOR, and RhoA–ROCK signaling activities are all up-regulated as a consequence of PROCR overexpression in BT549 cells using the system in (a), including increased c-Myc and cyclin D1 levels. c, overexpression of PROCR in BT549 cells induces change of cell shape as visualized by phase contrast. Scale bars represent 20 μm. d, Western blot showing that in BT549 cells with PROCR overexpression, knockdown of F2R by two independent shRNAs (Sh1 or Sh2) attenuates F2R level and RhoA–ROCK signaling, whereas it is ineffective to ERK and PI3K–Akt–mTOR pathways. e, Western blot indicating that ERK, PI3K–Akt–mTOR, and RhoA–ROCK signaling activities are all up-regulated in isolated PROCR-neg PDX-1 cells as a consequence of PROCR overexpression, including increased c-Myc and cyclin D1 levels. Western blots in the same panel are from the same batch of cells using the same loadings, thus only one loading control is shown at the end of the panel. For a better illustration, they are shown as three separated columns representing ERK, Akt, and RhoA pathways, respectively. Each experiment was repeated three times or more.
It is noteworthy that using the CRISPR interference system to activate endogenous PROCR expression is a powerful means to activate its downstream signaling, not only in BT549 cell line, but also in PDX tumor cells. We found that restoration of PROCR in PROCR-negative PDX-1 cells potently enhances the activities of the three signaling pathways (Fig. 4e).
PROCR engages IGF-1R for the activation of ERK and PI3K–Akt–mTOR pathways
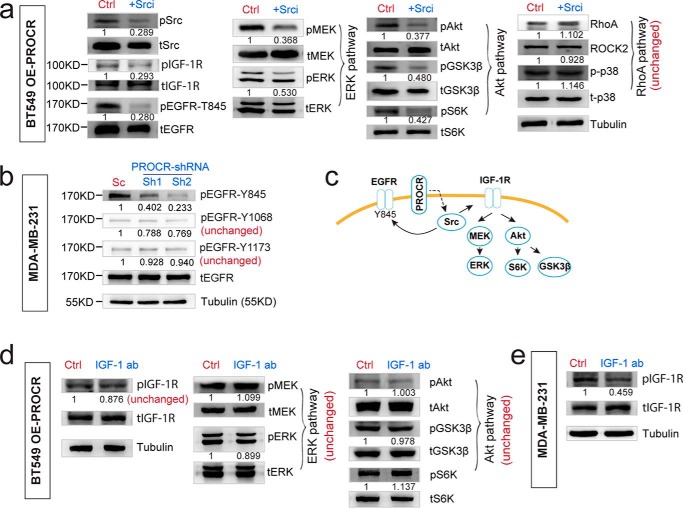
Because PROCR-dependent ERK and PI3K–Akt–mTOR signal activation does not involve F2R, we asked whether they rely on receptor tyrosine kinases (RTKs). To identify potential RTK candidates, we performed a RTK antibody array using lysates of PROCR+ cells and PROCR− cells isolated from PROCR–high TNBC PDX tumor. Higher activities of IGF-1R, Axl, RYK, and EGFR were apparent in PROCR+ cells (Fig. 5a). Therefore, we tested their requirement for ERK and PI3K–Akt–mTOR signals induced by PROCR overexpression. Successful knockdown of IGF-1R or EGFR expression by shRNA was validated by Western blot analysis (Fig. 5b). Strikingly, knockdown of IGF-1R exhibited significantly reduced ERK and PI3K–Akt–mTOR signaling, whereas RhoA–ROCK signaling was unchanged (Fig. 5c). These results were further confirmed using an additional shRNA targeting IGF-1R (Fig. 5d). In contrast, knockdown of EGFR by shRNA affected none of the three signal cascades induced by PROCR (Fig. 5c). Similarly, knockdown of Axl and RYK had no effect on the three signal cascades induced by PROCR (data not shown). We reasoned that the increased activity of EGFR, Axl, and RYK in PROCR+ cells is likely a correlation or consequence, and they do not mediate the actions of PROCR in activating the three intracellular signals. These data suggest that IGF-1R is the key RTK for the stimulation of ERK and PI3K–Akt–mTOR signals in these cells.
Figure 5.
IGF-1R, not EGFR, mediates the activation of ERK and PI3K–Akt–mTOR signals induced by PROCR. a, illustration and representing image of phospho-RTK antibody array using PROCR+ (Lin−, EpCam+, PROCR+) and PROCR− (Lin−, EpCam+, PROCR−) cells isolated from PDX tumor. Eight proteins with most evident differential activities are indicated. b, Western blot showing the knockdown efficacy of IGF-1R and EGFR by shRNAs. c, Western blot showing that in BT549 cells with PROCR overexpression, knockdown of IGF-1R by shRNA does not affect Src activity, whereas it diminishes both ERK and PI3K–Akt–mTOR pathways, and it is ineffective to RhoA–ROCK signaling; knockdown of EGFR by shRNA affects none of Src and the three PROCR-dependent signaling. d, Western blot showing that in BT549 cells with PROCR overexpression, knockdown of IGF-1R by shRNA (Sh2) does not affect Src activity, whereas it diminishes both ERK and PI3K–Akt–mTOR pathways, and it is ineffective to RhoA–ROCK signaling. Western blots in the same panel are from the same batch of cells using the same loadings, thus only one loading control is shown at the end of the panel. For a better illustration, they are shown as three separated columns representing ERK, Akt, and RhoA pathways, respectively. Each Western blot analysis was repeated three times or more.
PROCR engages Src kinase to transactivate IGF-1R
Interestingly, in PROCR overexpression background, knockdown of IGF-1R did not affect the level of pSrc, implying that Src activation is upstream of IGF-1R (Fig. 5, c and d). To further investigate this, we inhibited Src using KX2–391. Inhibition of Src resulted in attenuation of IGF-1R (Fig. 6a), supporting the idea that Src is upstream of IGF-1R for its transactivation. Consistently, Src inhibition led to reduced activities of ERK and PI3k–Akt–mTOR pathways, but did not affect the RhoA–ROCK pathway (Fig. 6a), in line with the notion that Src is upstream of IGF-1R. In addition, inhibition of Src also attenuated EGFR–Tyr-845 activity (Fig. 6a), consistent with the idea that increased EGFR activity in PROCR+ cells is a consequence of Src activation. It has been established that Src can phosphorylate EGFR at Tyr-845 (30). Indeed, we found that inhibition of PROCR only attenuates EGFR activity at Tyr-845, but not other sites (Tyr-1068, Tyr-1173) (Fig. 6b). These results further supported that increased EGFR activity in PROCR+ cells is a consequence of Src activation, and EGFR itself is not the key RTK involved in the PROCR signaling relay (illustrated in Fig. 6c).
Figure 6.
PROCR engages Src kinase to transactivate IGF-1R and other RTKs. a, Western blot showing that in BT549 cells with PROCR overexpression, inhibition of Src using KX2–391 diminishes the activities of Src, IGF-1R, EGFR-T845, and both ERK and PI3K–Akt–mTOR pathways, whereas it is ineffective to RhoA–ROCK signaling. b, Western blot analysis indicating that in MDA-MB-231 cells, knockdown of PROCR with two independent shRNAs attenuates the activity of EGFR at Tyr-845, but does not affect EGFR Tyr-1068 or Tyr-1173 phosphorylation. c, illustration of PROCR-dependent intracellular signaling pathways. Impact on EGFR-T845 activity is a subsequence of activation of Src by PROCR. d, Western blot showing that in BT549 cells with PROCR overexpression, incubation with IGF-1 neutralizing antibody (12 μg/ml, 2 h) could not inhibit IGF-1R, ERK, and Akt pathways induced by etopic PROCR. e, Western blot showing that in MDA-MB-231 cells, incubation with IGF-1 neutralizing antibody (12 μg/ml, 2 h) is sufficient to inhibit endogenous IGF-1R activity. Western blots in the same panel are from the same batch of cells using the same loadings, thus only one loading control is shown. For a better illustration, they are shown as three or four separated columns representing ERK, Akt, and RhoA pathways, respectively. Each experiment was repeated three times or more.
Next, we address whether IGF-1R activation (Tyr-1135, Tyr-1136) induced by PROCR-Src axis depends on IGF-1. To this end, an IGF-1 neutralizing antibody that inhibits its interaction with IGF-1R was incubated with BT549 cells in PROCR overexpression background. We found that the antibody inhibits neither the increased IGF-1R activity nor the increased MEK–ERK and PI3K–Akt activities induced by ectopic PROCR expression (BT549 OE-PROCR) (Fig. 6d). The efficacy of the antibody was demonstrated in another experiment with MDA-MB-231 cells, in which the antibody in the same concentration (12 μg/ml) and incubation duration (2 h) was sufficient to inhibit the endogenous IGF-1R activity (Fig. 6e). It is likely that in the in vitro setting, endogenous IGF-1R activity in MDA-MB-231 cells is jointly regulated by the endogenous PROCR-Src axis and serum IGF-1. Together, these data suggest that PROCR engages Src to transactivate IGF-1R and other RTKs, and this PROCR–Src–IGF-1R axis is independent of IGF-1.
Protein C serves as the ligand for the activation of PROCR intracellular signaling in breast cancer cells
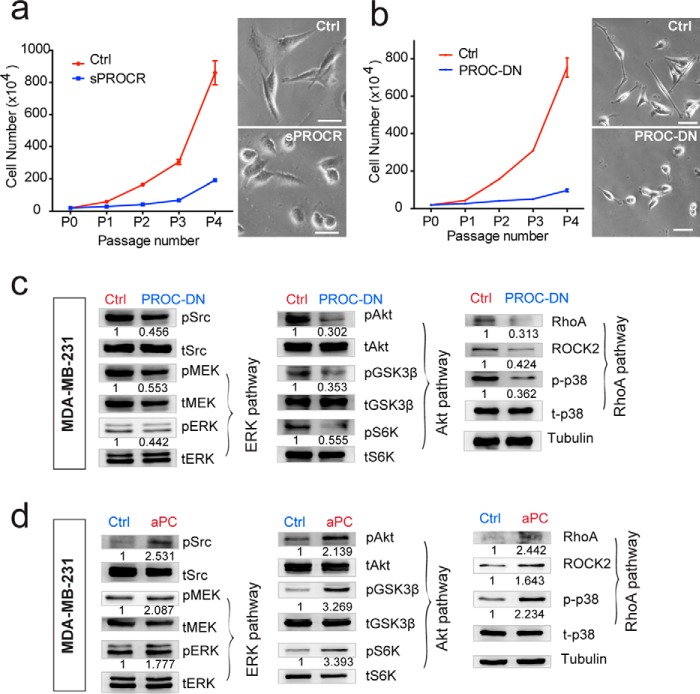
To investigate whether the activation of PROCR intracellular signaling in breast cancer cells requires a ligand, we utilized soluble PROCR (sPROCR, extracellular domain of PROCR) that can compete with the membrane form of PROCR (31). Addition of sPROCR in MDA-MB-231 culture resulted in decreased proliferation, accompanied by cell shape changes. The spindle-shaped morphology of MDA-MB-231 was altered to become more spherical (Fig. 7a). Similar effects on cell proliferation and morphology were observed when PROCR was knocked down by shRNA (data not shown). These results suggest that the extracellular domain of PROCR that facilitates ligand binding is important for its function in breast cancer cells. Protein C, a coagulation protease, is a well-established ligand in endothelial cells for anticoagulation, anti-inflammation, and cytoprotective activities of PROCR (14, 15, 19, 32–34). To address the possibility that the same ligand binds to PROCR in breast cancer cells, we generated the protease dead form of PROC (PROC-DN, dominant negative form). Addition of PROC-DN led to decreased proliferation and similar morphological changes in MDA-MB-231 cells (Fig. 7b). Importantly, the activities of the three intracellular signals of PROCR were blocked in the presence of PROC-DN (Fig. 7c). Moreover, incubation with active protein C provided direct evidence. Addition of active PROC potently enhanced the three PROCR-dependent intracellular signals (Fig. 7d). These data suggest that PROC serves as the ligand for PROCR in breast cancer cells.
Figure 7.
Protein C serves as the ligand for the activation of PROCR intracellular signaling in breast cancer cells. a, MDA-MB-231 cells were cultured in the presence of Ctrl or sPROCR (6 μg/ml) for four passages in complete media. Cell numbers that are counted in each passage showing that sPROCR markedly inhibited cell proliferation. The spindle-shaped morphology of MDA-MB-231 (Ctrl, upper right panel) was altered to become more spherical in the presence of sPROCR (bottom right panel). One of three similar experiments is shown. Data are presented as mean ± S.E. b, MDA-MB-231 cells were cultured in the presence of Ctrl or PROC-DN (2 μg/ml) for four passages in complete media. Cell numbers that are counted in each passage show that PROC-DN markedly inhibited cell proliferation. The spindle-shaped morphology of MDA-MB-231 (Ctrl, upper right panel) was altered to become more spherical in the presence of PROC-DN (bottom right panel). One of three similar experiments is shown. Data are presented as mean ± S.E. c, Western blot showing that addition of PROC-DN in MDA-MB-231 cells (24 h) diminishes the activities of Src and all three PROCR intracellular signaling pathways (ERK, PI3K–Akt–mTOR, and RhoA–ROCK signaling). d, Western blot showing that addition of active PROC (10 μg/ml) in MDA-MB-231 cells (8 h) enhances the activities of Src and all three PROCR intracellular signaling pathways. Western blots in the same panel are from the same batch of cells using the same loadings, thus only one loading control is shown at the end of the panel. For a better illustration, they are shown as three separated columns representing ERK, Akt, and RhoA pathways, respectively. Each experiments was repeated three times or more.
Blockage of PROCR intracellular signal impedes clonogenicity of breast cancer cells
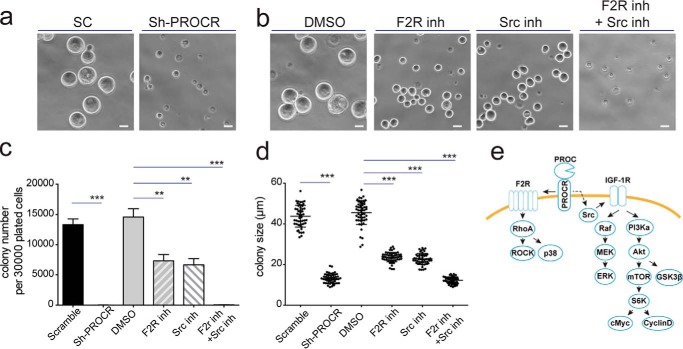
To investigate whether PROCR function through these intracellular signals to regulate stemness, we performed in vitro colony formation assays as described previously (35). Cells isolated from PDX-1 sample were plated in 3D Matrigel culture and their colony-forming abilities were examined upon inhibition of PROCR or its downstream signals studied. We found that the colonies occurred at a ratio of one colony per two PDX epithelial cells plated, in line with the notion that about 50% of PDX cells are PROCR+ (Fig. 3a). Knockdown of PROCR completely blocked the colony formation (Fig. 8a, c, and d). Inhibition of F2R by sch79797, or inhibition of Src by KX2–391 partially blocked the colony formation shown by decreased colony formation rate and colony sizes, whereas joint inhibition of F2R and Src delivered strongest effects and completely blocked the colony formation (Fig. 8, b–d). These data suggest that the three downstream signals of PROCR are functionally important for the stem cell activities in breast cancer cells.
Figure 8.
Blockage of PROCR intracellular signaling impedes clonogenicity of breast cancer cells. a and b, colonies formed from PDX-1 single cells were dissolved from Matrigel for imaging. Representative pictures are shown. Knockdown of PROCR abolished colony formation (a). F2R inhibitor (sch79797) or Src inhibitor (KX2–391) attenuated colony formation, and the combined treatment completely blocked colony formation (b). Scale bars, 20 μm. c and d, quantification indicating that colony formation efficiency (c) and colony sizes (d) are significantly reduced when PROCR or its downstream signaling component is inhibited. The combined treatment of F2R inhibitor and IGF-1R inhibitor completely blocked colony formation, similar to the effect of PROCR knockdown. **, p < 0.002; ***, p < 0.0001. Data are pooled from three independent experiments. Data are presented as mean ± S.E. e, the model of PROCR signaling mechanisms in breast cancer cells.
Discussion
PROCR has been implicated in tumor progression and is an important surface marker for normal stem cells in several tissues. However, the signaling mechanism of PROCR had remained elusive. In the present study, we investigated PROCR signaling mechanism in breast cancer cells. We revealed that PROCR induces the activation of ERK and PI3K–Akt–mTOR signal through transactivation of IGF-1R by Src and concomitantly stimulates RhoA–ROCK–p38 signals through F2R (illustrated in Fig. 8e). These findings were further validated in PROCR+ cells and PROCR− cells isolated from PDX tumors.
In this study we found that F2R does not account for all PROCR activities in breast cancer cells, which is in contrast to previously described PROCR intracellular signaling mechanisms, in which F2R is an essential mediator of PROCR in cell survival, anti-inflammation, and migration (15, 17–19, 24, 25). In breast cancer cells, only the RhoA–ROCK–p38 signal is dependent on F2R, whereas ERK and PI3k–Akt–mTOR signals are F2R-independent, and instead are dependent on Src and subsequent activation of IGF-1R. This is the first report that IGF-1R mediates the signaling function of PROCR. Our results agree with previous reports that EGFR is activated by PROCR (17, 24, 25). However our study indicates that EGFR is not required for PROCR-dependent ERK and PI3k–Akt–mTOR signals, and that EGFR activation is sequential to PROCR-dependent Src activation. It has been studied how F2R is activated by PROCR–active PROC axis. F2R is activated when the N terminus is cleaved by active PROC, creating a new N terminus that acts as a tethered ligand that binds intramolecularly to the receptor to initiate transmembrane signals (36). There remains a gap in knowing how Src is activated by PROCR–active PROC. Our current findings cannot rule out that some of the effects on signal pathways are indirect.
The activities of PROCR-dependent signals are attenuated in the presence of dominant negative form of PROC, and are increased in the presence of active PROC. These results provide direct evidence that PROC is a ligand of PROCR in breast cancer cells. Potentially, either increased expression of the receptor or increased exposure to the ligand would promote the activities of these pathways. Increased PROCR expression has been observed in a subset of TNBC cell lines (MDA-MB-231, Hs 578T, HCC38, CAL51, and HCC1806). Robust PROCR expression has also been seen in primary TNBC samples.4 Regarding the source of the ligand, one possibility is the circulation system, where the plasma protein C concentration is 70 nm (4 μg/ml) (37). It is also plausible that protein C is locally produced, considering that autocrine protein C has been reported in skin keratinocytes for promoting their survival, growth, and migration (38). Further experiments are needed to investigate the source of protein C in breast cancers.
Considering that PROCR promotes the activities of ERK, PI3K–Akt–mTOR, and RhoA pathways, and leads to accumulation of c-Myc and cyclin D1, which are key signal events in breast cancer (26), our findings support the tumor-promoting role of PROCR. Previous studies in normal mammary gland and in breast cancer cells have suggested that PROCR+ cells have increased epithelial and mesenchymal transition (EMT) characteristics (1, 28). In this study, observations on the cell shape changes upon modulation of PROCR expression may also be because of alteration of EMT program. EMT could be another channel through which PROCR signaling promotes tumor progression. In the current study, the effective RTK, IGF-1R was identified in a screen using phospho antibody array. We are aware that our antibody array approach could have missed some other RTK candidates. Considering the versatile roles of PROCR in the transactivation of various RTKs in different cell types (17, 24, 25), it would be no surprise if PROCR triggers a more complex signal cascade. Nevertheless, selectively blocking single kinases involved in ERK or PI3K pathways has been associated with limited or sporadic responses in clinical studies (39). Simultaneously attenuating multiple pathways, e.g. with a reagent that inhibits PROCR, can potentially be a more effective means to attenuate the complex signaling network in breast cancer cells.
In conclusion, our study illustrated the signaling mechanisms of PROCR in the breast cancer cells, elucidating the potential functional role of PROCR in TNBC. These findings may guide the development of anti-PROCR therapeutic agents for breast cancer treatment.
Experimental procedures
Cell lines and cell culture
The MCF7, SK-BR-3, MDA-MB-231, Hs 578T, T-47D, ZR-75–1, MDA-MB-415, MDA-MB-453, BT474, MDA-MB-436, BT549, HCC38, CAL51, HCC1806, MDA-MB-468, HCC1937, HCC1599, and HCC2157 human breast cancer cell lines were obtained from the Shanghai Cell Bank Type Culture Collection Committee or American Type Culture Collection (ATCC) and maintained in complete growth medium as recommended by the distributor.
Generation of patient-derived xenografts from human breast cancers
PDX lines were originally initiated by implantation of a fresh patient tumor fragment into the mammary fat pad of recipient SCID/Beige mice and were maintained by serial passage in vivo at intervals characteristic for each line, and in accordance with Institutional Animal Care and Use Committee requirements. This study was approved by the Institutional Review Board (IRB) of Fudan University Shanghai Cancer Center (FDSCC).
Antibodies
Antibodies used in immunohistochemistry were mouse anti-human PROCR (1:300, Abcam), mouse anti-ER (1:50, Dako), mouse anti-PR (1:50, Dako), rabbit anti-HER2 (1:50, Proteintech).
Antibodies used in Western blotting were rabbit anti-human PROCR (1:200, Novus), rabbit anti-human phospho-Src (1:1000, Cell Signaling Technology), rabbit anti-human total Src (1:1000, Cell Signaling Technology), rabbit anti-human phospho-MEK (1:1000, Cell Signaling Technology), mouse anti-human total MEK (1:1000, Cell Signaling Technology), rabbit anti-human phospho-ERK (1:1000, Cell Signaling Technology), rabbit anti-human total ERK (1:100, Santa Cruz Biotechnology), rabbit anti-human phospho-Raf (1:100, Santa Cruz Biotechnology), rabbit anti-human total Raf (1:100, Santa Cruz Biotechnology), rabbit anti-human phospho-Akt (1:1000, Cell Signaling Technology), rabbit anti-human total Akt (1:1000, Cell Signaling Technology), rabbit anti-human phospho-GSK3β(1:1000, Cell Signaling Technology), rabbit anti-human total GSK3β(1:1000, Cell Signaling Technology), rabbit anti-human phospho-CREB (1:1000, Cell Signaling Technology), rabbit anti-human total CREB (1:1000, Cell Signaling Technology), rabbit anti-human phospho-S6K (1:1000, Cell Signaling Technology), rabbit anti-human total S6K (1:1000, Cell Signaling Technology), mouse anti-human c-Myc (1:100, Santa Cruz Biotechnology), mouse anti-human cyclin D1 (1:100, Santa Cruz Biotechnology), mouse anti-human RhoA (1:100, Santa Cruz Biotechnology), rabbit anti-human ROCK2 (1:1000, Cell Signaling Technology), rabbit anti-human phospho-p38 (1:1000, Cell Signaling Technology), rabbit anti-human p38 (1:1000, Cell Signaling Technology), rabbit anti-human phospho-IGF1R (1:1000, Cell Signaling Technology), rabbit anti-human IGF-1R (1:1000, Cell Signaling Technology), rabbit anti-human phospho-EGFR (Tyr-1068, Tyr-1173, Tyr-845) (1:1000, Cell Signaling Technology), rabbit anti-human EGFR (1:1000, Cell Signaling Technology), mouse anti tubulin (1:5000, Sigma), and mouse anti beta-Actin (1:2000, Sigma).
Antibody for neutralizing IGF-1 in culture cells was goat anti IGF-1 (12 μg/ml, R&D Systems). Antibodies for FACS were used in 1:200 dilutions: PE/cy7-anti-human EpCam, FITC-anti-human CD31, FITC-anti-human CD45, FITC-anti-human CD235a (BioLegend), APC-anti-human PROCR (eBioscience), and PE-anti-human PROCR (BD Pharmingen).
Phospho protein array
The Human Phospho-Kinase Array (R&D Systems, ARY003B) was performed as the procedure attached in the kit. 106 of MDA-MB-231 cells were used. The Human Phospho-RTK Array (R&D Systems, ARY001B) was performed as the procedure attached in the kit. 106 of freshly isolated PROCR+ and PROCR− cells from PDX tumors were used. Protein samples are normalized by tubulin level through Western blotting before use in array analysis.
Primary cell preparation
The minced primary tumor was placed in culture medium (RPMI 1640 with 25 mm HEPES, 5% fetal bovine serum, 1% penicillin-streptomycin-glutamine (PSQ), 300 units ml−1 collagenase III (Worthington)) and digested for up to 3 h at 37 °C. After lysis of the red blood cells in NH4Cl, a single-cell suspension was obtained by sequential incubation with 0.25% trypsin-EDTA at 37 °C for 5 min and 0.1 mg/ml DNase I (Sigma) for 5 min with gentle pipetting, followed by filtration through 70-μm cell strainers.
Overexpression, shRNA, and sgRNA constructs
Expression constructs for sPROCR (1–214 amino acids, extracellular domain) and protein C (1–252 amino acids, a truncation of the protease domain) were made using pCMV-Fc vector (Addgene).
The shRNAs targeting PROCR sequences were constructed in lentivirus-based pLKO.1-EGFP constructs (Addgene). The efficiency of individual shRNA was validated by Western blotting or qPCR. The shRNA sequences were as follows: PROCR-sh1, TGGCCTCCAAAGACTTCATAT; PROCR-sh2, GCAGCAGCTCAATGCCTACAA; F2R-sh1, GCATTACTCATTCCTTTCTCA; F2R-sh2, CCCGGTCATTTCTTCTCAGGA; IGF-1R-sh1, GCGGTGTCCAATAACTACATT; IGF-1R-sh2, GCCTTTCACATTGTACCGCAT; EGFR-sh, CGCAAAGTGTGTAACGGAATA.
The dCas9-VP64 plasmid was from Addgene. The sgRNAs targeting PROCR genome sequence were constructed in lentivirus-based plasmid (MP177, Addgene). The efficiency of individual sgRNA was validated by Western blotting. The sgRNA sequence for PROCR activation was TCCTGCCGGCGCTGACTCAG.
In vitro MDA-MB-231 and BT549 morphology assay
MDA-MB-231 cells infected with scramble or PROCR shRNA or BT549 cells infected with control or PROCR sgRNA were plated at a low density (5 × 104) onto coverslips in 12-well plate using complete culture medium. After 12 h when cells were adhered to the coverslip, the plates were washed with PBS followed by fixation with 4% PFA for 10 min. Cells on coverslips were stained with vimentin and DAPI counterstain. To examine the effect of various proteins on MDA-MB-231 cell morphology, purified sPROCR (6 μg/ml) or protein kinase C dead (2 μg/ml) were used when cells were plated.
In vitro colony formation assay
FACS-sorted cells were resuspended at a density of 6 × 105 cells ml−1 in chilled 100% growth factor reduced Matrigel (Corning), and the mixture was allowed to polymerize before covering with culture medium (DMEM/F12), ITS (1:100, Sigma), 50 ng ml−1 EGF, and 10 ng ml−1 bFGF, with or without F2R inhibitor (sch79797, 500 nm, Abcam) or Src inhibitor (KX2–391, 100 nm, Selleck). Culture medium was changed every 2 days. Colony numbers and sizes were scored after 6 days in culture. The colonies were mostly spherical. In cases in which colonies were oval, the long axis was measured.
Quantification and statistical analysis
In all Western blot analyses, when quantifying non-phosphoproteins, i.e. PROCR, c-Myc, cyclin D1, RhoA, ROCK2, they were normalized to tubulin; when quantifying the levels of phosphoproteins, they were normalized to the total protein counterparts. In the latter case, all total proteins should be 1. Student's t test was performed and the p value was calculated in Prism on data represented by bar charts, which consisted of results from three independent experiments unless specified otherwise. For all experiments with error bars, the standard error of the mean (S.E.M.) was calculated to indicate the variation within each experiment.
Author contributions
D. W. and Y. A. Z. designed the experiments; D. W., C. L., J. W., and Y. J. performed all in vitro experiments; X. H. and Z. S. provided PDX sample; D. W., H. J., and Y. A. Z. analyzed the data and wrote the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Dr. Esther Verheyen and Dr. Roel Nusse for critical reading of the manuscript. We thank Dr. Weiguo Zou for helpful discussion.
This work was supported by National Natural Science Foundation of China Grants 31530045 and 31371500 (to Y. A. Z.), Ministry of Science and Technology of China Grant 2014CB964800 (to Y. A. Z.), and Chinese Academy of Sciences Grants XDB19000000 and XDA12020349 (to Y. A. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
D. Wang and Y. A. Zeng, unpublished data.
- PROCR
- protein C receptor
- mTOR
- mechanistic target of rapamycin
- F2R
- coagulation factor II thrombin receptor
- PDX
- patient-derived xenograft
- EGFR
- epidermal growth factor receptor
- TNBC
- triple-negative breast cancer
- CRISPR
- clustered regularly interspaced short palindromic repeats
- RTK
- receptor tyrosine kinase
- EMT
- epithelial and mesenchymal transition
- qPCR
- quantitative polymerase chain reaction.
References
- 1. Wang D., Cai C., Dong X., Yu Q. C., Zhang X. O., Yang L., and Zeng Y. A. (2015) Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81–84 10.1038/nature13851 [DOI] [PubMed] [Google Scholar]
- 2. Balazs A. B., Fabian A. J., Esmon C. T., and Mulligan R. C. (2006) Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107, 2317–2321 10.1182/blood-2005-06-2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwasaki H., Arai F., Kubota Y., Dahl M., and Suda T. (2010) Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood 116, 544–553 10.1182/blood-2009-08-240903 [DOI] [PubMed] [Google Scholar]
- 4. Fares I., Chagraoui J., Lehnertz B., MacRae T., Mayotte N., Tomellini E., Aubert L., Roux P. P., and Sauvageau G. (2017) EPCR expression marks UM171-expanded CD34+ cord blood stem cells. Blood 129, 3344–3351 10.1182/blood-2016-11-750729 [DOI] [PubMed] [Google Scholar]
- 5. Zhou F., Li X., Wang W., Zhu P., Zhou J., He W., Ding M., Xiong F., Zheng X., Li Z., Ni Y., Mu X., Wen L., Cheng T., Lan Y., Yuan W., Tang F., and Liu B. (2016) Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487–492 10.1038/nature17997 [DOI] [PubMed] [Google Scholar]
- 6. Yu Q. C., Song W., Wang D., and Zeng Y. A. (2016) Identification of blood vascular endothelial stem cells by the expression of protein C receptor. Cell Res. 26, 1079–1098 10.1038/cr.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beaulieu L. M., and Church F. C. (2007) Activated protein C promotes breast cancer cell migration through interactions with EPCR and PAR-1. Exp. Cell Res. 313, 677–687 10.1016/j.yexcr.2006.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antón I., Molina E., Luis-Ravelo D., Zandueta C., Valencia K., Ormazabal C., Martínez-Canarias S., Perurena N., Pajares M. J., Agorreta J., Montuenga L. M., Segura V., Wistuba I. I., De Las Rivas J., Hermida J., and Lecanda F. (2012) Receptor of activated protein C promotes metastasis and correlates with clinical outcome in lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 186, 96–105 10.1164/rccm.201110-1826OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schaffner F., Yokota N., Carneiro-Lobo T., Kitano M., Schaffer M., Anderson G. M., Mueller B. M., Esmon C. T., and Ruf W. (2013) Endothelial protein C receptor function in murine and human breast cancer development. PLoS One 8, e61071 10.1371/journal.pone.0061071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keshava S., Sahoo S., Tucker T. A., Idell S., Rao L. V., and Pendurthi U. R. (2013) Endothelial cell protein C receptor opposes mesothelioma growth driven by tissue factor. Cancer Res. 73, 3963–3973 10.1158/0008-5472.CAN-12-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukudome K., and Esmon C. T. (1994) Identification, cloning, and regulation of a novel endothelial cell protein C/activated protein C receptor. J. Biol. Chem. 269, 26486–26491 [PubMed] [Google Scholar]
- 12. Griffin J. H., Zlokovic B. V., and Mosnier L. O. (2012) Protein C anticoagulant and cytoprotective pathways. Int. J. Hematol. 95, 333–345 10.1007/s12185-012-1059-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mohan Rao L. V., Esmon C. T., and Pendurthi U. R. (2014) Endothelial cell protein C receptor: A multiliganded and multifunctional receptor. Blood 124, 1553–1562 10.1182/blood-2014-05-578328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bae J. S., Yang L., Manithody C., and Rezaie A. R. (2007) The ligand occupancy of endothelial protein C receptor switches the protease-activated receptor 1-dependent signal specificity of thrombin from a permeability-enhancing to a barrier-protective response in endothelial cells. Blood 110, 3909–3916 10.1182/blood-2007-06-096651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng T., Liu D., Griffin J. H., Fernández J. A., Castellino F., Rosen E. D., Fukudome K., and Zlokovic B. V. (2003) Activated protein C blocks p53-mediated apoptosis in ischemic human brain endothelium and is neuroprotective. Nat. Med. 9, 338–342 10.1038/nm826 [DOI] [PubMed] [Google Scholar]
- 16. Vetrano S., Ploplis V. A., Sala E., Sandoval-Cooper M., Donahue D. L., Correale C., Arena V., Spinelli A., Repici A., Malesci A., Castellino F. J., and Danese S. (2011) Unexpected role of anticoagulant protein C in controlling epithelial barrier integrity and intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 108, 19830–19835 10.1073/pnas.1107140108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue M., Chow S. O., Dervish S., Chan Y. K., Julovi S. M., and Jackson C. J. (2011) Activated protein C enhances human keratinocyte barrier integrity via sequential activation of epidermal growth factor receptor and Tie2. J. Biol. Chem. 286, 6742–6750 10.1074/jbc.M110.181388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang X. V., Banerjee Y., Fernández J. A., Deguchi H., Xu X., Mosnier L. O., Urbanus R. T., de Groot P. G., White-Adams T. C., McCarty O. J., and Griffin J. H. (2009) Activated protein C ligation of ApoER2 (LRP8) causes Dab1-dependent signaling in U937 cells. Proc. Natl. Acad. Sci. U.S.A. 106, 274–279 10.1073/pnas.0807594106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riewald M., Petrovan R. J., Donner A., Mueller B. M., and Ruf W. (2002) Activation of endothelial cell protease activated receptor 1 by the protein C pathway. Science 296, 1880–1882 10.1126/science.1071699 [DOI] [PubMed] [Google Scholar]
- 20. Russo A., Soh U. J., Paing M. M., Arora P., and Trejo J. (2009) Caveolae are required for protease-selective signaling by protease-activated receptor-1. Proc. Natl. Acad. Sci. U.S.A. 106, 6393–6397 10.1073/pnas.0810687106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uchiba M., Okajima K., Oike Y., Ito Y., Fukudome K., Isobe H., and Suda T. (2004) Activated protein C induces endothelial cell proliferation by mitogen-activated protein kinase activation in vitro and angiogenesis in vivo. Circ. Res. 95, 34–41 10.1161/01.RES.0000133680.87668.FA [DOI] [PubMed] [Google Scholar]
- 22. Minhas N., Xue M., Fukudome K., and Jackson C. J. (2010) Activated protein C utilizes the angiopoietin/Tie2 axis to promote endothelial barrier function. FASEB J. 24, 873–881 10.1096/fj.09-134445 [DOI] [PubMed] [Google Scholar]
- 23. Finigan J. H., Dudek S. M., Singleton P. A., Chiang E. T., Jacobson J. R., Camp S. M., Ye S. Q., and Garcia J. G. (2005) Activated protein C mediates novel lung endothelial barrier enhancement: Role of sphingosine 1-phosphate receptor transactivation. J. Biol. Chem. 280, 17286–17293 10.1074/jbc.M412427200 [DOI] [PubMed] [Google Scholar]
- 24. Feistritzer C., Mosheimer B. A., Sturn D. H., Riewald M., Patsch J. R., and Wiedermann C. J. (2006) Endothelial protein C receptor-dependent inhibition of migration of human lymphocytes by protein C involves epidermal growth factor receptor. J. Immunol. 176, 1019–1025 10.4049/jimmunol.176.2.1019 [DOI] [PubMed] [Google Scholar]
- 25. Gramling M. W., Beaulieu L. M., and Church F. C. (2010) Activated protein C enhances cell motility of endothelial cells and MDA-MB-231 breast cancer cells by intracellular signal transduction. Exp. Cell Res. 316, 314–328 10.1016/j.yexcr.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polyak K., and Metzger Filho O. (2012) SnapShot: Breast cancer. Cancer Cell 22, 562.e1 10.1016/j.ccr.2012.06.021 [DOI] [PubMed] [Google Scholar]
- 27. Yunokawa M., Koizumi F., Kitamura Y., Katanasaka Y., Okamoto N., Kodaira M., Yonemori K., Shimizu C., Ando M., Masutomi K., Yoshida T., Fujiwara Y., and Tamura K. (2012) Efficacy of everolimus, a novel mTOR inhibitor, against basal-like triple-negative breast cancer cells. Cancer Sci. 103, 1665–1671 10.1111/j.1349-7006.2012.02359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hwang-Verslues W. W., Kuo W. H., Chang P. H., Pan C. C., Wang H. H., Tsai S. T., Jeng Y. M., Shew J. Y., Kung J. T., Chen C. H., Lee E. Y., Chang K. J., and Lee W. H. (2009) Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS One 4, e8377 10.1371/journal.pone.0008377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A., Lim W. A., Weissman J. S., and Qi L. S. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sato K., Nagao T., Iwasaki T., Nishihira Y., and Fukami Y. (2003) Src-dependent phosphorylation of the EGF receptor Tyr-845 mediates Stat-p21waf1 pathway in A431 cells. Genes Cells 8, 995–1003 10.1046/j.1356-9597.2003.00691.x [DOI] [PubMed] [Google Scholar]
- 31. Kurosawa S., Stearns-Kurosawa D. J., Hidari N., and Esmon C. T. (1997) Identification of functional endothelial protein C receptor in human plasma. J. Clin. Invest. 100, 411–418 10.1172/JCI119548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor F. B. Jr., Peer G. T., Lockhart M. S., Ferrell G., and Esmon C. T. (2001) Endothelial cell protein C receptor plays an important role in protein C activation in vivo. Blood 97, 1685–1688 10.1182/blood.V97.6.1685 [DOI] [PubMed] [Google Scholar]
- 33. Taylor F. B. Jr., Stearns-Kurosawa D. J., Kurosawa S., Ferrell G., Chang A. C., Laszik Z., Kosanke S., Peer G., and Esmon C. T. (2000) The endothelial cell protein C receptor aids in host defense against Escherichia coli sepsis. Blood 95, 1680–1686 [PubMed] [Google Scholar]
- 34. Esmon C. T. (2003) The protein C pathway. Chest 124, 26S–32S 10.1378/chest.124.3_suppl.26S [DOI] [PubMed] [Google Scholar]
- 35. Zeng Y. A., and Nusse R. (2010) Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 6, 568–577 10.1016/j.stem.2010.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schuepbach R. A., Madon J., Ender M., Galli P., and Riewald M. (2012) Protease-activated receptor-1 cleaved at R46 mediates cytoprotective effects. J. Thromb. Haemost. 10, 1675–1684 10.1111/j.1538-7836.2012.04825.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Griffin J. H., Evatt B., Zimmerman T. S., Kleiss A. J., and Wideman C. (1981) Deficiency of protein C in congenital thrombotic disease. J. Clin. Invest. 68, 1370–1373 10.1172/JCI110385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xue M., Campbell D., and Jackson C. J. (2007) Protein C is an autocrine growth factor for human skin keratinocytes. J. Biol. Chem. 282, 13610–13616 10.1074/jbc.M610740200 [DOI] [PubMed] [Google Scholar]
- 39. Hiscox S., and Nicholson R. I. (2008) Src inhibitors in breast cancer therapy. Expert Opin. Ther. Targets 12, 757–767 10.1517/14728222.12.6.757 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.