Abstract
Binding of receptor activator of NF-κB ligand (RANKL) to its receptor RANK on osteoclast (OC) precursors up-regulates c-Fos and CCAAT/enhancer–binding protein-α (C/EBPα), two critical OC transcription factors. However, the effects of c-Fos and C/EBPα on osteoclastogenesis have not been compared. Herein, we demonstrate that overexpression of c-Fos or C/EBPα in OC precursors up-regulates OC genes and initiates osteoclastogenesis independently of RANKL. However, although C/EBPα up-regulated c-Fos, c-Fos failed to up-regulate C/EBPα in OC precursors. Consistently, C/EBPα overexpression more strongly promoted OC differentiation than did c-Fos overexpression. RANK has a cytoplasmic 535IVVY538 (IVVY) motif that is essential for osteoclastogenesis, and we found that mutation of the IVVY motif blocked OC differentiation by partly inhibiting expression of C/EBPα but not expression of c-Fos. We therefore hypothesized that C/EBPα overexpression might rescue osteoclastogenesis in cells expressing the mutated IVVY motif. However, overexpression of C/EBPα or c-Fos failed to stimulate osteoclastogenesis in the mutant cells. Notably, the IVVY motif mutation abrogated OC gene expression compared with a vector control, suggesting that the IVVY motif might counteract OC inhibitors during osteoclastogenesis. Consistently, the IVVY motif mutant triggered up-regulation of recombinant recognition sequence–binding protein at the Jκ site (RBP-J) protein, a potent OC inhibitor. Mechanistically, C/EBPα or c-Fos overexpression in the mutant cells failed to control the up-regulated RBP-J expression, leading to suppression of OC genes. Accordingly, RBP-J silencing in the mutant cells rescued osteoclastogenesis with C/EBPα or c-Fos overexpression with C/EBPα exhibiting a stronger osteoclastogenic effect. Collectively, our findings indicate that C/EBPα is a stronger inducer of OC differentiation than c-Fos, partly via C/EBPα regulation by the RANK 535IVVY538 motif.
Keywords: bone, c-Fos, CCAAT/enhancer-binding protein (C/EBP), differentiation, NFAT transcription factor, osteoclast, transcription factor
Introduction
Osteoclasts (OCs)3 are multinucleated giant cells that are responsible for bone resorption (1–3). Through their bone-resorbing functions, these polykaryons carry essential roles in skeletal development and bone homeostasis. As such, deregulated OC differentiation has been regarded among the main causes of many osteolytic bone disorders (4, 5). OCs originate from the macrophage lineage upon stimulation by the macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB (RANK) ligand (RANKL) (5, 6). M-CSF acts through its receptor, colony-stimulating factor 1 receptor, to promote the proliferation of OC precursors, and RANKL activates its receptor RANK on OC precursors to mediate OC lineage commitment and differentiation. Importantly, RANK contains a cytoplasmic 535IVVY538 (IVVY) motif that is critical for OC differentiation both in vivo and in vitro (7–10). Targeting of the RANK IVVY motif has been shown to prevent bone destruction in mouse models of inflammatory and ovariectomy-induced bone loss (10). These findings are underscored by a genetic study reporting that a truncating mutation causing the loss of a RANK region containing the IVVY motif results in osteopetrosis in humans (11).
RANKL mediates osteoclastogenesis by inducing the expressions of key transcription factors, including FBJ osteosarcoma oncogene (c-Fos) and nuclear factor of activated T cells, C1 (NFATc1) (12–15). NFATc1 is regarded as a master regulator of OC differentiation that plays essential roles in inducing the expressions of many OC genes, including cathepsin K (Ctsk) and tartrate-resistant acid phosphatase (TRAP) (16–18). Mice deficient in the NFATc1 gene (NFATc1−/− mice) exhibit a severe osteopetrotic phenotype due to defective OC development (14). Likewise, mice deficient in the c-Fos gene (c-Fos−/− mice) also exhibit osteopetrosis from impaired OC development (13). However, unlike the NFATc1−/− mice, most c-Fos−/− mice show almost a normal lifespan despite other issues associated with delayed or absent gametogenesis, lymphocythemia, and altered behavior, indicating that c-Fos may not be required for the growth of most cell types during development. With regard to the molecular basis of its role during osteoclastogenesis, c-Fos can induce the expression of NFATc1, establishing NFATc1 as a target gene of c-Fos in OC (19).
CCAAT/enhancer–binding protein-α (C/EBPα) is a transcription factor of the C/EBP family of transcription factors that is crucial for hematopoiesis through its ability to bind on gene promoters to regulate gene expression in a lineage-specific manner (20–22). Global deletion of the C/EBPα gene in mice (C/EBPα−/− mice) results in early death from defective granulocyte development and impaired homeostasis (23, 24). Recently, our group has reported that C/EBPα is strongly up-regulated by RANKL during osteoclastogenesis and carries a critical function in osteoclastogenesis by appointing OC precursor cells to the OC lineage (25). We showed that C/EBPα overexpression in OC precursors can induce the expressions of various OC markers, including NFATc1, independently of RANKL and thereby promotes lineage commitment. Moreover, we have recently reported that C/EBPα is also important for OC differentiation and function (26). Consistent with this posture, we revealed that newborn C/EBPα−/− mice are osteopetrotic due to impaired OC development (25).
Although the transcription factors C/EBPα and c-Fos are both critical for osteoclastogenesis in vitro and in vivo and are essential for the expressions of OC markers, including NFATc1, Ctsk, and TRAP, during OC differentiation (12, 13, 25, 27), the effects of their roles in osteoclastogenesis have not yet been compared. Moreover, although C/EBPα is critical for OC lineage commitment, a process that was shown to be regulated by the RANK IVVY motif (8), it remains unknown whether the RANK IVVY motif can also regulate C/EBPα expression during osteoclastogenesis. In the current study, we compared the roles of c-Fos and C/EBPα in OC differentiation by using a gain-of-function strategy in a RANK IVVY motif–dependent manner. This study not only enhances our understanding of the roles of c-Fos and C/EBPα in OC differentiation but also provides important insight into the role of RANK signaling and transcription factors in OC differentiation.
Results
C/EBPα overexpression shows a stronger osteoclastogenic effect than c-Fos overexpression
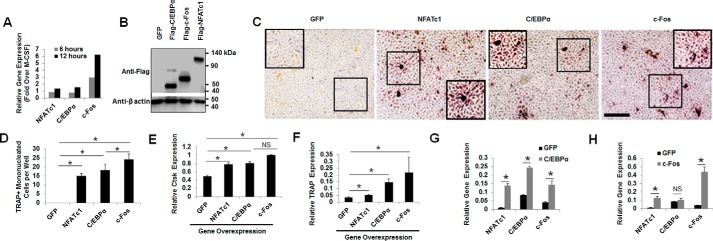
c-Fos is a critical transcription factor for osteoclastogenesis (12, 13). Likewise, C/EBPα is also an essential transcription factor for osteoclastogenesis (25, 26). In comparing the roles of C/EBPα and c-Fos in osteoclastogenesis, we first compared their requirements for OC lineage commitment by using their target gene, NFATc1, as a positive control (Fig. 1). Transient stimulation of mouse bone marrow (MBM) cells, widely used as primary OC precursors, with M-CSF and RANKL was shown to up-regulate NFATc1, c-Fos, and C/EBPα during osteoclastogenesis (14–16, 25). We recapitulated this finding and confirmed that stimulation of MBM cells by M-CSF and RANKL for 12 h could up-regulate the expressions of NFATc1, C/EBPα, and c-Fos as compared with M-CSF alone (Fig. 1A), supporting their involvement in early stages of osteoclastogenesis. In line with this notion, NFATc1 (14), c-Fos (12), and C/EBPα (25) were each shown to be able to appoint OC precursors to the OC lineage. Hence, we examined the effect of overexpressing C/EBPα or c-Fos in inducing OC lineage priming as compared with NFATc1 overexpression (Fig. 1, B–D). In overexpressing the genes in OC precursor cells, we used the 293GPG retroviral system, which has been extensively used to study various genes in OC biology through gene overexpression by using the pMX retroviral plasmid (28, 29). Using a pMX-puro-GFP control vector, we generated a retrovirus encoding the GFP cDNA to infect MBM cells. We showed that GFP was highly expressed on days 2, 3, and 4 postinfection (Fig. S1), confirming that this retroviral system can sustain high gene expression for our standard osteoclastogenesis assays (6, 25, 26). We showed that overexpression of C/EBPα, c-Fos, or NFATc1 in MBM cells, as confirmed by Western blot analysis (Fig. 1B), could initiate osteoclastogenesis by generating TRAP-positive mononucleated cells without RANKL stimulation as compared with a GFP control (Fig. 1, C and D). This finding recapitulates the previous reports that C/EBPα (25), c-Fos (12), and NFATc1 (14) can all induce lineage priming. In addressing the molecular basis of this finding, we showed that overexpression of C/EBPα, c-Fos, or NFATc1 could significantly up-regulate the OC genes encoding Ctsk (Fig. 1E) and TRAP (Fig. 1F) as compared with a GFP control in the absence of RANKL (27). These results confirmed the previous reports that C/EBPα and c-Fos could induce the expressions of OC marker genes and thereby promote OC lineage commitment.
Figure 1.
C/EBPα or c-Fos can initiate osteoclastogenesis independently of RANKL. A, MBM cells were transiently stimulated with M-CSF (10 ng/ml) or M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 6 or 12 h. Gene expression was examined by qPCR. B–D, MBM cells expressing a GFP control, FLAG-NFATc1, FLAG-C/EBPα, or FLAG-c-Fos were treated with M-CSF (10 ng/ml) for 4 days for Western blot analysis using β-actin as a loading control from at least three independent experiments (B) or TRAP staining for osteoclastogenesis (C). Scale bar, 250 μm. Quantification for C is shown as mean number of mononucleated TRAP-positive cells per well from at least three independent experiments (D). E and F, MBM cells expressing GFP, NFATc1, C/EBPα, or c-Fos were cultured with M-CSF (10 ng/ml) for 4 days for analysis of the expressions of Ctsk (E) and TRAP (F) by qPCR. G, MBM cells expressing GFP or C/EBPα were treated with M-CSF (10 ng/ml) for 4 days. H, MBM cells expressing GFP or c-Fos were treated with M-CSF (10 ng/ml) for 4 days. Gene expressions in A, D, E, F, G, and H were assessed by qPCR using Hprt as a loading control in three independent experiments. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
Next, in comparing the abilities of C/EBPα and c-Fos to mediate OC lineage commitment, we examined their abilities to up-regulate each other independently of RANKL. Whereas C/EBPα overexpression could induce the expressions of c-Fos and its target gene NFATc1 in MBM cells without RANKL stimulation (Fig. 1G), c-Fos overexpression could only induce the expression of its target gene NFATc1, but not that of C/EBPα, in the absence of RANKL (Fig. 1H). This finding indicated that c-Fos is a target gene of C/EBPα during osteoclastogenesis. As a control, NFATc1 overexpression in MBM cells was unable to up-regulate C/EBPα and c-Fos in the absence of RANKL (data not shown), confirming that NFATc1 is a target of C/EBPα and c-Fos during OC differentiation and was thus unable to up-regulate these genes.
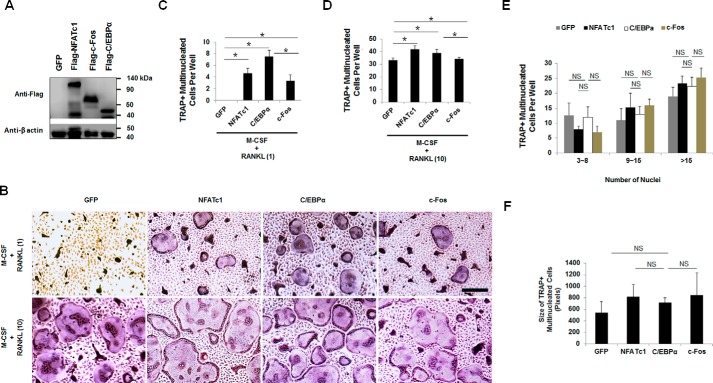
Subsequently, we compared the roles of C/EBPα and c-Fos in OC differentiation by using a gain-of-function strategy (Fig. 2). Treatment of MBM cells with permissive levels (one-tenth of the amount required for normal osteoclastogenesis) of RANKL was shown to mediate OC lineage commitment but was insufficient to promote OC differentiation (30, 31). We utilized this approach to examine the roles of C/EBPα and c-Fos overexpression in OC differentiation as compared with NFATc1 overexpression. Our standard osteoclastogenesis assay requires the treatment of MBM cells with M-CSF and 10 ng/ml RANKL for 4 days (25, 26). Therefore, we overexpressed C/EBPα or c-Fos in the presence of M-CSF and RANKL (1 ng/ml) to promote OC differentiation with RANKL-evoked lineage priming. The data showed that overexpression of C/EBPα in MBM cells, as confirmed by Western blotting (Fig. 2A), could promote OC differentiation with permissive RANKL dosages as compared with a GFP control (Fig. 2B), confirming our recent report that C/EBPα is not only involved in OC lineage priming but can also promote OC differentiation (26). Comparatively, we noted that although overexpression of C/EBPα, c-Fos, or NFATc1 in MBM cells could each promote OC differentiation with 1 ng/ml RANKL (Fig. 2B), the C/EBPα overexpressers generated significantly more OCs than c-Fos overexpression or the GFP control (Fig. 2C). Consistently, the C/EBPα overexpressers also formed significantly more OCs with stimulation by 10 ng/ml RANKL than the cells overexpressing c-Fos or expressing GFP control (Fig. 2, B and D). These results indicated that C/EBPα exhibited a stronger role than c-Fos in mediating OC differentiation from the RANKL-precommitted cells. However, we found that C/EBPα overexpression did not influence OC size as compared with the GFP control as well as c-Fos or NFATc1 overexpression (Fig. 2, E and F).
Figure 2.
C/EBPα is a stronger inducer of OC differentiation than c-Fos. A, MBM cells expressing a GFP control, FLAG-NFATc1, FLAG-C/EBPα, or FLAG-c-Fos were cultured with M-CSF (10 ng/ml) for 4 days for Western blot analysis using β-actin as a loading control from at least three independent assays. B, MBM cells expressing GFP, FLAG-NFATc1, FLAG-C/EBPα, or FLAG-c-Fos were cultured with M-CSF (10 ng/ml) and RANKL (1 ng/ml) or M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 4 days and then stained for TRAP activity. Scale bar, 250 μm. C and D, quantifications for B are shown for M-CSF and RANKL (1 ng/ml) (C) and M-CSF and RANKL (10 ng/ml) (D) as mean number of multinucleated TRAP-positive cells per well from at least three independent assays. E and F, quantifications of OC size for M-CSF and RANKL (10 ng/ml) shown in B via the number of nuclei (E) or area (F) of multinucleated TRAP-positive cells. The numbers in parentheses show concentrations in ng/ml. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
Mutational inactivation of the RANK IVVY motif blocks OC differentiation and inhibits C/EBPα, but not c-Fos, expression
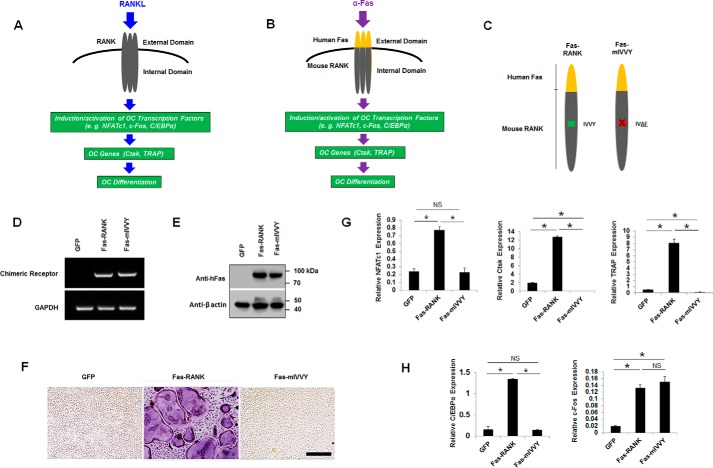
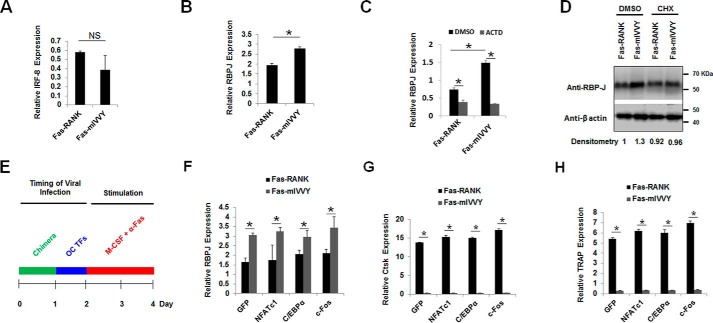
Activation of the RANK IVVY motif, through RANKL-induced RANK activation, induces the expressions of Ctsk and TRAP during OC differentiation (8–10). In further comparing the roles of C/EBPα and c-Fos in OC differentiation, we investigated whether the RANK IVVY motif could regulate their expressions during OC differentiation (Fig. 3). RANK and Fas are members of the tumor necrosis factor receptor superfamily, which are activated by ligand-induced receptor trimerization (32). Upon binding to RANK, RANKL triggers RANK trimerization, leading to signal transduction for induction of OC transcription factors and genes to promote OC differentiation (Fig. 3A) (7, 32). Xu et al. (33) developed a chimeric receptor system that consists of the human Fas extracellular domain linked to the transmembrane and cytoplasmic domains of mouse RANK to study RANK signaling during osteoclastogenesis. The chimeric receptor system can be exclusively activated by a human Fas-activating antibody (α-Fas) in a similar fashion as RANKL to induce OC transcription factors and genes for OC differentiation (Fig. 3B). In this study, we utilized two chimeras (Fas-RANK and Fas-mIVVY) that were previously generated and validated by Xu et al. (8). Fas-RANK is constructed with the human Fas extracellular domain linked to the transmembrane and intracellular domains of normal mouse RANK, and Fas-mIVVY has the human Fas extracellular domain linked to the transmembrane and intracellular domains of mouse RANK containing an inactivating mutation in the IVVY motif (Fig. 3C). Following retrovirally induced gene expression, the chimeras were shown to be highly expressed on the cell surface of OC precursors and respond successfully to stimulation by α-Fas and M-CSF to promote OC differentiation in a similar fashion as the RANKL/RANK system (8, 33–37). MBM cells expressing a GFP control, Fas-RANK, or Fas-mIVVY, as confirmed by reverse transcription (RT)-PCR (Fig. 3D) and Western blotting (Fig. 3E), were stimulated with M-CSF and α-Fas to promote OC differentiation (Fig. 3F). We recapitulated the findings of the previous studies and showed that only the cells expressing Fas-RANK, but not the Fas-mIVVY expressers, could generate OCs with stimulation by M-CSF and α-Fas (Fig. 3F) (8–10). Furthermore, we used the macrophage RAW264.7 cell line and confirmed that RAW264.7 cells expressing Fas-RANK, but not the Fas-mIVVY expressers, could mediate osteoclastogenesis with α-Fas stimulation (Fig. S2). At the molecular level, our quantitative real-time PCR (qPCR) analysis confirmed that mutational inactivation of the RANK IVVY motif significantly abrogated the expressions NFATc1, Ctsk, and TRAP during OC differentiation (34–36) (Fig. 3G). Importantly, we also found that inactivation of the RANK IVVY motif significantly attenuated the expression of C/EBPα, but exhibited no overt effect on c-Fos expression, during OC differentiation (Fig. 3H). These results indicated that the RANK IVVY motif promoted OC differentiation in part by regulating C/EBPα expression but was dispensable to c-Fos expression.
Figure 3.
Mutational inactivation of the RANK cytoplasmic IVVY motif blocks osteoclastogenesis and inhibits C/EBPα, but not c-Fos, expression. A, a schematic of RANK activation by RANKL to induce OC differentiation. B, a schematic of the chimeric receptor system, which can be specifically activated by α-Fas to stimulate OC differentiation. C, schematics of Fas-RANK and Fas-mIVVY. D and E, MBM cells expressing a GFP control, Fas-RANK, or Fas-mIVVY were treated with M-CSF (10 ng/ml) for 4 days for gene expression analysis by RT-PCR (D) and Western blotting (E) using GAPDH and β-actin as loading controls, respectively. F, MBM cells expressing GFP, Fas-RANK, or Fas-mIVVY were treated with M-CSF (10 ng/ml) and α-Fas (100 ng/ml) for 4 days. Cultures from at least three independent assays were then stained for TRAP activity. Scale bar, 250 μm. G and H, MBM cells expressing GFP, Fas-RANK, or Fas-mIVVY were treated with M-CSF (10 ng/ml) and α-Fas (100 ng/ml) for 2 days before being submitted to qPCR analysis using Hprt as a loading control in three independent repeats. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
Overexpression of C/EBPα or c-Fos in cells expressing mutated RANK IVVY motif fails to mediate OC differentiation
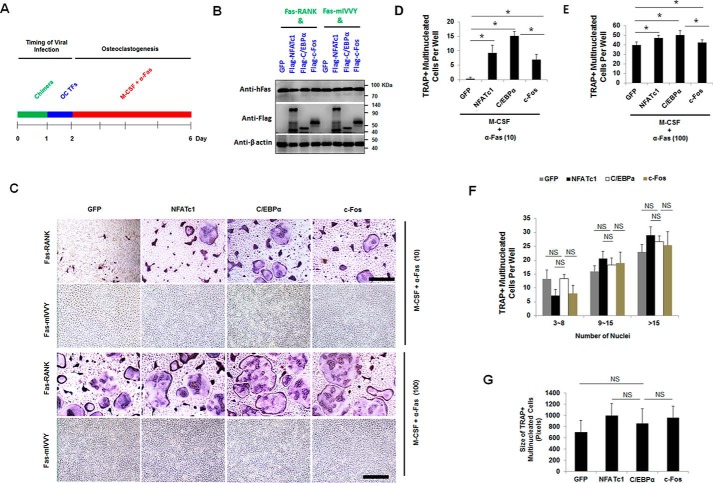
Given that inactivation of the RANK IVVY motif blocked OC differentiation and inhibited NFATc1 and C/EBPα, but not c-Fos, expression (Fig. 3), we reasoned that overexpression of C/EBPα or NFATc1 might rescue OC differentiation in MBM cells expressing the mutated RANK IVVY motif. In addressing this issue, MBM cells doubly expressing Fas-RANK or Fas-mIVVY and a GFP control, NFATc1, C/EBPα, or c-Fos (Fig. 4A), as confirmed by Western blotting (Fig. 4B), were stimulated with permissive (10 ng/ml) or optimum (100 ng/ml) levels of α-Fas in the presence of M-CSF for 4 days as validated in previous studies to promote OC differentiation (34–36) (Fig. 4C). As expected, MBM cells doubly expressing Fas-RANK and C/EBPα, c-Fos, or NFATc1 generated numerous OCs with either 10 (Fig. 4, C and D) or 100 ng/ml (Fig. 4, C and E) α-Fas as compared with the GFP control. Notably, overexpression of C/EBPα in the Fas-RANK expressers generated significantly more OCs than the c-Fos and Fas-RANK double expressers (Fig. 4, D and E), further confirming that C/EBPα exhibited a stronger osteoclastogenic effect than c-Fos. However, we found that MBM cells doubly expressing Fas-mIVVY and C/EBPα, c-Fos, or NFATc1 failed to generate OCs with 10 (Fig. 4, C and D) or even 100 ng/ml (Fig. 4, C and E) α-Fas as compared with cells expressing the chimeric receptor with normal RANK, indicating that gene overexpression could not rescue osteoclastogenesis from inactivation of the RANK IVVY motif. Moreover, we found that, similarly to the RANKL-induced osteoclastogenesis assays (Fig. 2, E and F), C/EBPα or c-Fos overexpression showed no overt effect on OC size from the α-Fas-mediated osteoclastogenesis assays (Fig. 4, F and G). In confirming this finding, we demonstrated that RAW264.7 cells doubly expressing Fas-RANK and C/EBPα, c-Fos, or NFATc1 could promote OC differentiation with both permissive (10 ng/ml) and optimum (100 ng/ml) α-Fas stimulation as compared with the GFP control (Fig. S3, A–E). Nevertheless, forced expression of C/EBPα, c-Fos, or NFATc1 in RAW264.7 cells expressing the mutated RANK IVVY motif failed to promote OC differentiation (Fig. S3, A–E). Moreover, we demonstrated that C/EBPα overexpression showed no overt effect on OC size as compared with cells overexpressing c-Fos or expressing the GFP control (Fig. S3F).
Figure 4.
C/EBPα or c-Fos fails to mediate osteoclastogenesis with mutational inactivation of the RANK cytoplasmic IVVY motif. A, experimental strategy. B, MBM cells doubly expressing Fas-RANK or Fas-mIVVY and a GFP control, FLAG-NFATc1, FLAG-C/EBPα, or FLAG-c-Fos were cultured with M-CSF (10 ng/ml) for 4 days for Western blotting using β-actin as a loading control. C, MBM cells were cultured as in B but treated with M-CSF (10 ng/ml) and α-Fas (10 ng/ml) or M-CSF (10 ng/ml) and α-Fas (100 ng/ml) for 4 days to stimulate OC differentiation. The cultures were then stained for TRAP activity. Scale bars, 250 μm. D and E, quantifications for C are shown for M-CSF and α-Fas (10 ng/ml) (D) or M-CSF and α-Fas (100 ng/ml) (E) as mean number of multinucleated TRAP-positive cells per well from at least three independent assays. F and G, quantification of OC size for M-CSF and α-Fas (100 ng/ml) for the Fas-RANK expressers shown in C via the number of nuclei (F) or area (G) of multinucleated TRAP-positive cells per well. The numbers in parentheses show concentrations in ng/ml. Error bars show averages ± S.D. *, p < 0.05; NS, not significant. TFs, transcription factors.
Mutational inactivation of RANK IVVY motif up-regulates recombinant recognition sequence–binding protein at the Jκ site (RBP-J)
Our data thus far demonstrated that although the RANK IVVY motif could regulate the expressions of C/EBPα and NFATc1, but not that of c-Fos, during OC differentiation (Fig. 3) overexpression of C/EBPα, NFATc1, or c-Fos in cells expressing the mutated RANK IVVY motif could not mediate OC differentiation (Fig. 4). We were surprised that although overexpression of C/EBPα or c-Fos in MBM cells could initiate osteoclastogenesis independently of RANKL (Fig. 1) and promote OC differentiation (Fig. 2) overexpression of C/EBPα or c-Fos in cells expressing the mutated RANK IVVY motif was unable to promote osteoclastogenesis. Notably, a closer look at the role of the RANK IVVY motif in regulating the expressions of Ctsk and TRAP during osteoclastogenesis revealed that the inactivation of the RANK IVVY motif led to a significantly lower Ctsk or TRAP expression as compared with the GFP control (Fig. 3G). The result suggested that the RANK IVVY motif, besides positively regulating the expressions of OC genes, might also counteract the expressions of OC inhibitors during OC differentiation. This is consistent with the notion that deregulated expressions of potent negative regulators of osteoclastogenesis in the context of inactivation of the RANK IVVY motif may negatively affect the abilities of NFATc1, C/EBPα, or c-Fos to promote OC differentiation (38). Among the known factors that can negatively regulate OC differentiation, RBP-J (39–41) and interferon-regulatory factor 8 (IRF-8) (42, 43) have been regarded as the leading factors. Hence, we investigated the ability of the RANK IVVY motif to regulate the expressions of RBP-J and IRF-8 during OC differentiation. The data showed that mutational inactivation of the RANK IVVY motif displayed no significant effect on IRF-8 expression (Fig. 5A) but triggered a significant increase in RBP-J expression (Fig. 5B), indicating that the RANK IVVY motif could negatively regulate RBP-J expression during OC differentiation. For additional insight into the role of the RANK IVVY motif in regulating RBP-J expression, we used the RNA synthesis inhibitor actinomycin D (ACTD) and the protein synthesis inhibitor cycloheximide (CHX) to examine the effects of IVVY inactivation on RBP-J expression at the transcriptional and translation levels. As expected, treatment of MBM cells expressing Fas-mIVVY with DMSO, used as a control, triggered a significant increase in RBP-J mRNA levels as compared with the Fas-RANK expressers upon stimulation by M-CSF and α-Fas (Fig. 5C). However, we noted that the Fas-RANK and Fas-mIVVY expressers showed comparable RBP-J mRNA levels upon treatment with ACTD to inhibit RNA synthesis, indicating that the RANK IVVY motif induced RBP-J expression at the transcriptional level (Fig. 5C). Furthermore, treatment of the Fas-mIVVY expressers with DMSO triggered a 30% increase in RBP-J protein levels as compared with the Fas-RANK expressers upon stimulation by M-CSF and α-Fas (Fig. 5D). However, treatment of Fas-mIVVY expressers with CHX to inhibit protein synthesis showed similar RBP-J protein levels as the Fas-RANK expressers upon activation by M-CSF and α-Fas. These results indicated that mutational inactivation of the RANK IVVY motif triggered a drastic increase in RBP-J mRNA levels, which translated into increased protein levels.
Figure 5.
Mutational inactivation of the RANK cytoplasmic IVVY motif represses OC genes but up-regulates RBP-J. A and B, MBM cells expressing a GFP control, Fas-RANK, or Fas-mIVVY were treated with M-CSF (10 ng/ml) and α-Fas (100 ng/ml) for 2 days. The expressions of IRF-8 (A) and RBP-J (B) were examined by qPCR. C, MBM cells expressing Fas-RANK or Fas-mIVVY were cultured with M-CSF (10 ng/ml) for 3 days before being treated with ACTD (1 μg/ml) for 12 h while undergoing stimulation by M-CSF (10 ng/ml) and α-Fas (100 ng/ml). The expression of RBP-J was then examined by qPCR. D, MBM cells expressing Fas-RANK or Fas-mIVVY were cultured with M-CSF (10 ng/ml) for 3 days before being treated with CHX (5 μg/ml) for 12 h while undergoing stimulation by M-CSF (10 ng/ml) and α-Fas (100 ng/ml). The expression of RBP-J was then examined by Western blotting using β-actin as a loading control in three independent experiments. E, experimental strategy for F–H. F–H, MBM cells doubly expressing Fas-RANK or Fas-mIVVY and GFP, FLAG-NFATc1, FLAG-C/EBPα, or FLAG-c-Fos were treated with M-CSF (10 ng/ml) and α-Fas (100 ng/ml) as indicated in E. The expressions of RBP-J (F), Ctsk (G), and TRAP (H) were examined by qPCR. The qPCR analyses in A, B, C, F, G, and H were performed using Hprt as a loading control in three independent experiments. Error bars show averages ± S.D. *, p < 0.05; NS, not significant. TFs, transcription factors.
RBP-J was shown to suppress OC differentiation by inhibiting the immunoreceptor tyrosine-based activation motif (ITAM)–associated receptor costimulatory signaling, which is required for osteoclastogenesis (39–41, 44). Accordingly, we found that overexpression of C/EBPα, c-Fos, or NFATc1 failed to control the up-regulated RBP-J expression from the inactivation of the RANK IVVY motif as compared with cells expressing the normal RANK cytoplasmic domain (Fig. 5, E and F). Additionally, consistent with the inability of gene overexpression to rescue OC differentiation in cells expressing the mutated RANK IVVY motif (Fig. 4), overexpression of C/EBPα, c-Fos, or NFATc1 in MBM cells expressing the mutated RANK IVVY motif failed to induce the expressions of Ctsk (Fig. 5, E and G) and TRAP (Fig. 5, E and H), mimicking the reported role of RBP-J in inhibiting gene expression during OC differentiation (39). Collectively, these results indicated that the inability of NFATc1, C/EBPα, or c-Fos to mediate OC differentiation in the context of inactivation of the RANK IVVY motif stemmed in part from transcriptionally up-regulated RBP-J expression, which negatively affected the expressions of OC marker genes.
Ctsk is a direct target of repression by RBP-J
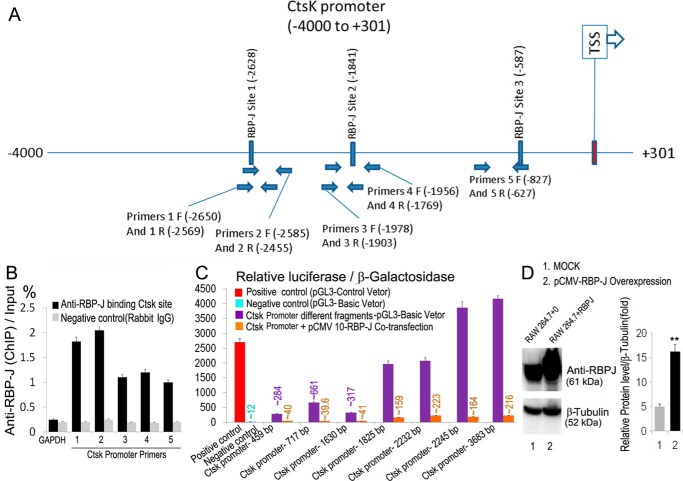
We next examined whether the inhibition of OC genes, such as Ctsk, was due to a direct transcriptional repression by RBP-J (Fig. 6). Using an online transcription factor–binding site predictive tool (alggen.lsi.upc.es),4 we identified three binding sites for RBP-J on the 4-kb promoter region upstream of the murine Ctsk gene (Fig. 6A). Two of these binding regions were approximated 2628 (Site I) and 1841bp (Site 2) upstream of the Ctsk transcriptional start site (TSS) (−1841 to +1 bp relative to TSS and −2628 to +1 bp relative to TSS, respectively) (Fig. 6A). The third binding region was ∼587 bp (Site 3) upstream of the Ctsk TSS (−587 to +1 bp relative to TSS) (Fig. 6A). Consistently, chromatin immunoprecipitation (ChIP) sequencing analysis demonstrated that Site 1 is a major binding site for RBP-J on the Ctsk promoter, whereas Site 2 and Site 3 were minor binding sites (Fig. 6B). Next, we performed luciferase assays using a reporter carrying different fragments of the Ctsk promoter (Fig. 6, C and D). RAW cells expressing RBP-J, as confirmed by Western blotting (Fig. 6D), were cotransfected with different Ctsk promoter fragments carrying the luciferase gene (Fig. 6C). The data showed that coexpression of two Ctsk promoter regions (−2445 to +1 bp relative to TSS and −2232 to +1 bp relative to TSS) containing Site 1 and Site 2 resulted in a drastic increase in luciferase activity as compared with three other regions (−1825 to +1 bp relative to TSS, −1630 to +1 bp relative to TSS, and −717 to +1 bp relative to TSS) containing only Site 1 (Fig. 6C). As controls, transfection of RAW cells with a pGL3-Control vector (positive control) showed a drastic increase in luciferase activity, whereas transfection of RAW cells with pGL3-Basic vector (negative control) failed to induce luciferase activity (Fig. 6C). Collectively, the data indicated that RBP-J could directly bind on Ctsk promoter to repress its expression in OC precursors.
Figure 6.
Ctsk is a direct target of repression by RBP-J. A, scheme of a 4-kb region of the murine Ctsk promoter showing the conserved RBP-J–binding sites and the location of the primers used for ChIP. B, ChIP qPCR assay using 293T cells, anti-RBP-J antibody, rabbit IgG (negative control), and primers as indicated in A. The data are shown as percent input from three independent experiments. C, luciferase activity from the Ctsk promoter with the indicated deletions in the presence or absence of RBP-J protein in RAW cells is shown from three independent repeats. RAW cells expressing pGL3-Control vector or pGL3-Basic vector served as positive and negative controls, respectively. D, Western blot analysis of RAW cells expressing pCMV-RBP-J or empty vector (MOCK) with anti-RBP-J antibody using anti-tubulin as a loading control. Error bars show averages ± S.D. **, p < 0.01.
RBP-J silencing rescues osteoclastogenesis in cells expressing mutated RANK IVVY motif
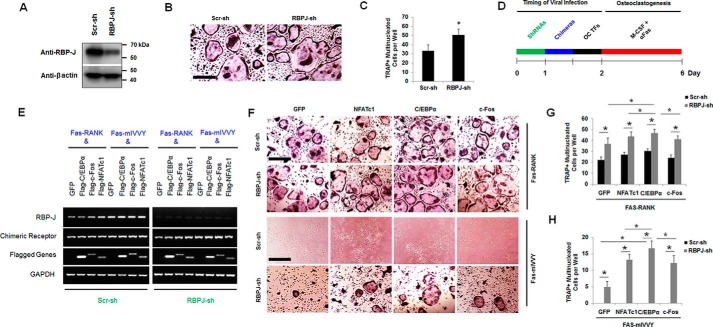
Next, we investigated the effect of silencing the RBP-J gene on rescuing osteoclastogenesis in the Fas-mIVVY expressers. We were able to silence the RBP-J gene using an shRNA construct that was purchased from Sigma-Aldrich as assessed by Western blot analysis (Fig. 7A). We found that, consistent with the reported role of RBP-J in suppressing osteoclastogenesis (39, 41), RBP-J silencing in MBM cells drastically enhanced RANKL-induced OC differentiation (Fig. 7, B and C). Accordingly, RBP-J silencing in MBM cells doubly expressing Fas-RANK and C/EBPα, c-Fos, or NFATc1 (Fig. 7, D and E) formed more OCs than RBPJ-depleted cells expressing the GFP control and Fas-RANK as well as MBM cells triply expressing the scrambled shRNA control, Fas-RANK, and C/EBPα, c-Fos, or NFATc1 (Fig. 7F, top two rows). Interestingly, RBP-J silencing in MBM cells doubly expressing Fas-mIVVY and C/EBPα, c-Fos, or NFATc1 could partially rescue osteoclastogenesis as compared with the scrambled shRNA control doubly expressing Fas-mIVVY and C/EBPα, c-Fos, or NFATc1 (Fig. 7F, last two rows). Consistently, the C/EBPα overexpressers doubly expressing the RBP-J shRNA construct and Fas-RANK or Fas-mIVVY generated significantly more OCs than the c-Fos overexpressers doubly expressing the RBP-J shRNA construct and Fas-RANK or Fas-mIVVY (Fig. 7, G and H), further confirming the stronger osteoclastogenic effect of C/EBPα than of c-Fos. Notably, RBP-J silencing in cells doubly expressing Fas-mIVVY and the GFP control could also generate a few TRAP-positive multinucleated cells as compared with cells triply expressing the Fas-mIVVY, the GFP control, and the scrambled shRNA control (Fig. 7, F and H), indicating that RBP-J silencing could initiate osteoclastogenesis in the cells expressing the mutated RANK IVVY motif. We further confirmed this finding by silencing the RBP-J gene in the RAW cells (Fig. S4). First, we showed that RBP-J depletion in RAW cells led to significantly more OCs than the scrambled shRNA control upon RANKL stimulation (Fig. S4, A–D). Moreover, RBP-J silencing in RAW cells doubly expressing Fas-RANK and C/EBPα, c-Fos, or NFATc1 formed more OCs than RBPJ-depleted cells expressing the GFP control and Fas-RANK as well as RAW cells triply expressing the scrambled shRNA control, Fas-RANK, and C/EBPα, c-Fos, or NFATc1 (Fig. S4, E–G). RBP-J silencing in the RAW cells doubly expressing Fas-mIVVY and C/EBPα, c-Fos, or NFATc1 could also partially rescue OC differentiation (Fig. S4, E and F). Moreover, the C/EBPα overexpressers doubly expressing the RBP-J shRNA construct and Fas-RANK or Fas-mIVVY generated significantly more OCs than the c-Fos overexpressers doubly expressing the RBP-J shRNA construct and Fas-RANK or Fas-mIVVY (Fig. S4, G and H). Consistently, RBP-J silencing in RAW cells doubly expressing Fas-mIVVY and the GFP control could also generate a few TRAP-positive multinucleated cells as compared with cells triply expressing Fas-mIVVY, the GFP control, and the scrambled shRNA control (Fig. S4, F and H). Taken together, the results indicated that RBP-J silencing could initiate osteoclastogenesis in cells expressing the mutated RANK IVVY motif, and overexpression of C/EBPα, c-Fos, or NFATc1 in the mutant cells could further enhance OC differentiation.
Figure 7.
RBP-J silencing rescues osteoclastogenesis from the mutational inactivation of the RANK cytoplasmic IVVY motif. A, MBM cells expressing an RBP-J shRNA construct (RBPJ-sh) or a scrambled shRNA control (Scr-sh) were cultured with M-CSF (10 ng/ml) for 4 days. Gene silencing was examined by Western blotting using β-actin as a loading control in three independent experiments. B, MBM cells expressing RBP-J shRNA construct or scrambled shRNA control were treated with M-CSF and RANKL (10 ng/ml) for 4 days to stimulate OC differentiation. Scale bar, 250 μm. C, quantification for B is shown as mean number of multinucleated TRAP-positive cells per well in three independent assays. D, experimental strategy for E–H. E, MBM cells triply expressing an shRNA construct (RBP-J shRNA or scrambled shRNA control), a chimera (Fas-RANK or Fas-mIVVY), and a GFP control, FLAG-NFATc1, FLAG-C/EBPα, or FLAG-c-Fos were treated with M-CSF for 4 days for gene expression analysis by RT-PCR using GAPDH as a loading control. F, the triple expressers as in E were stimulated with M-CSF (10 ng/ml) and α-Fas (100 ng/ml) to promote osteoclastogenesis. The cultures were then stained by TRAP. Scale bars, 250 μm. G and H, quantifications for F are shown for the Fas-RANK (G) and Fas-mIVVY (H) expressers as mean number of multinucleated TRAP-positive cells per well from three independent assays, respectively. Error bars show averages ± S.D. *, p < 0.05; NS, not significant. TFs, transcription factors.
Discussion
In this study, we compared the roles of C/EBPα and c-Fos in OC differentiation by using a gain-of-function strategy.
Although overexpression of C/EBPα or c-Fos can initiate osteoclastogenesis independently of RANKL, C/EBPα overexpression shows a stronger osteoclastogenic effect than c-Fos
C/EBPα and c-Fos are induced early during osteoclastogenesis, suggesting their involvement in OC lineage priming. Accordingly, we showed that overexpression of C/EBPα or c-Fos in MBM cells could up-regulate OC genes and thereby initiate osteoclastogenesis independently of RANKL. This finding is in agreement with other studies showing that C/EBPα or NFATc1 overexpression can initiate osteoclastogenesis independently of RANKL (14, 25). However, to our knowledge, this is the first report on the role of c-Fos overexpression in initiating osteoclastogenesis independently of RANKL. In the absence of RANKL, we found that although C/EBPα could up-regulate c-Fos, c-Fos failed to up-regulate C/EBPα in OC precursors. This finding confirms our previous report that c-Fos is a target gene of C/EBPα during osteoclastogenesis (25), which is further supported by the fact that C/EBPα overexpression displays a stronger osteoclastogenic effect than does c-Fos overexpression. We believe that the ability of C/EBPα to up-regulate c-Fos may in part account for its stronger effect in mediating OC differentiation than c-Fos.
RANK IVVY motif mediates osteoclastogenesis in part by regulating C/EBPα, but not c-Fos, expression
We confirmed that the RANK IVVY motif is essential for osteoclastogenesis through regulation of OC genes, including NFATc1, Ctsk, and TRAP (9, 10, 34, 35), during OC differentiation. We demonstrated that inactivation of the RANK cytoplasmic IVVY motif inhibited the expressions of the aforementioned OC genes and thereby blocked OC differentiation. Importantly, we revealed that inactivation of the RANK IVVY motif repressed C/EBPα but was dispensable for c-Fos expression during OC differentiation, suggesting that the stronger ability of C/EBPα than of c-Fos to promote OC differentiation may be mediated through regulation by the RANK IVVY motif. The fact that NFATc1, a target gene of C/EBPα and c-Fos, is also repressed with inactivation of the RANK IVVY motif despite normal c-Fos expression indicates that C/EBPα may be a major factor responsible for NFATc1 up-regulation during OC differentiation. It will be important to further investigate this issue in future studies. The RANK IVVY motif was initially identified due to its essential role in OC lineage commitment (8) and was later shown to promote TNF-α– and IL-1–mediated OC differentiation (34, 35). The fact that c-Fos can promote OC lineage priming although it is not regulated by the RANK IVVY motif suggests that c-Fos overexpression may be unable to fully activate the osteoclastogenic machinery during the early stages of osteoclastogenesis, which is critical for cell differentiation. Nevertheless, it is also possible that the RANK IVVY motif by itself may be insufficient to up-regulate C/EBPα during osteoclastogenesis because it was reported that the IVVY motif functions by cooperating with two other RANK cytoplasmic motifs (559PVQEET564 and 604PVQEQG609) to induce OC genes during OC differentiation (36). We therefore anticipate that the IVVY motif may function with other RANK motifs to up-regulate C/EBPα during OC differentiation.
Overexpression of NFATc1, C/EBPα, or c-Fos cannot mediate osteoclastogenesis in cells bearing inactivated RANK IVVY motif
Given the role of the RANK IVVY motif in inducing the expressions of NFATc1 and C/EBPα during OC differentiation, we reasoned that overexpression of C/EBPα or NFATc1, unlike c-Fos, might rescue OC differentiation in cells expressing the mutated RANK IVVY motif. However, we later found that gene overexpression failed to induce OC differentiation in the mutant cells despite the ability of C/EBPα, c-Fos, or NFATc1 to initiate osteoclastogenesis independently of RANKL and promote OC differentiation. Notably, we confirmed this finding in both primary cells and the RAW264.7 cell line. Moreover, in deciphering the molecular mechanism for this finding, we showed that C/EBPα, NFATc1, or c-Fos overexpression was unable to induce OC genes with inactivation of the RANK IVVY motif. This finding prompted the assumption that the RANK IVVY motif might also down-regulate OC inhibitors, which might counteract the effect of NFATc1, c-Fos, or C/EBPα in promoting OC differentiation with inactivation of the RANK IVVY motif (38, 44). This is consistent with the idea that inactivation of the RANK IVVY motif might trigger dysregulation of potent OC inhibitors (e.g. RBP-J and IRF-8) in inhibiting OC differentiation. We revealed that RBP-J, but not IRF-8, was significantly up-regulated in cells expressing the mutated RANK IVVY motif, and overexpression of NFATc1, C/EBPα, or c-Fos was unable to control the up-regulated RBP-J expression in the mutant cells, which prevented C/EBPα, c-Fos, and NFATc1 from inducing OC genes. Our finding is in agreement with other studies showing that RBP-J can directly repress gene expression in various cell types. Boggs et al. (45) showed that RBP-J binds to the p53 promoter to repress p53 gene expression in fibroblasts. Moreover, RBP-J binds to Sox9, a critical transcription factor for chondrogenesis, to repress Sox9 expression and thereby inhibits chondrocyte differentiation (46). Furthermore, Rozenberg et al. (47) demonstrated that methylation-dependent binding of RBP-J to a GC repressor element negatively regulates smooth muscle myosin heavy chain promoter activity and inhibits smooth muscle cell marker genes. Consistent with these studies, we showed that the mechanism by which RBP-J inhibits OC differentiation involves its direct binding on an OC gene promoter to repress gene expression during osteoclastogenesis. Remarkably, whereas NFATc1 can bind on Ctsk promoter to induce its expression (48), NFATc1, C/EBPα, or c-Fos overexpression was unable to induce Ctsk expression with RBP-J up-regulation from inactivation of the RANK IVVY motif. This suggests that binding of RBP-J on Ctsk promoter may prevent the osteoclastic transcription factors from accessing the promoter to promote gene expression. Future study is warranted to further address the mechanistic basis of the inhibitory effect of RBP-J on OC genes.
Notably, we demonstrated that inactivation of the RANK IVVY motif led to increased RBP-J mRNA levels, which translated into increased protein levels. Although this finding indicates that the RANK IVVY motif regulates RBP-J at mainly a transcriptional level, it does not exclude the possibility that the RANK IVVY motif may also regulate RBP-J by other means, including protein stability. Hence, the mechanism by which the RANK IVVY motif regulates RBP-J remains incomplete. With regard to the transcriptional regulation, we anticipate that the RANK IVVY motif may activate some unknown factors that can bind on the RBP-J promoter to down-regulate its expression and that inactivation of this motif may release this inhibitory brake on RBP-J expression. Importantly, RBP-J can suppress ITAM-mediated costimulatory signaling and limits the cross-talk between ITAM and RANK signaling, which ultimately inhibit OC differentiation through inhibition of the Ca2+/calmodulin signaling pathway (39). Consistently, we showed that RBP-J silencing could partially rescue osteoclastogenesis in both primary cells and RAW6264.7 cells doubly expressing the mutated RANK IVVY motif and C/EBPα or c-Fos with C/EBPα exhibiting a stronger osteoclastogenic effect.
The role of RANK IVVY motif in osteoclastogenesis remains complex
Whereas the RANK IVVY motif can up-regulate NFATc1 and C/EBPα, but not c-Fos, and down-regulate RBP-J during OC differentiation, overexpression of NFATc1, C/EBPα, or c-Fos can only partially rescue OC differentiation in RBPJ-depleted cells expressing the mutated RANK IVVY motif. These results indicated that the main mechanism(s) by which the RANK IVVY motif mediates osteoclastogenesis remains incomplete. Furthermore, despite its role in OC formation, the RANK IVVY motif is dispensable for the activation of the RANKL-induced signaling pathways (ERK, p38, JNK, and NF-κB) (8, 9). Our results indicate that the RANK IVVY motif can mediate OC differentiation by orchestrating a cascade of critical functions, which include induction of transcription factors, up-regulation of OC genes, and down-regulation of OC suppressors. Further studies on the role of the RANK IVVY motif in osteoclastogenesis should generate important insights into this issue. A key long-standing question is to unambiguously identify the protein(s) that is recruited by the RANK IVVY motif to mediate osteoclastogenesis. We anticipate that the RANK IVVY motif may recruit different factors at different stages of osteoclastogenesis and thereby exhibits distinct roles in different stages of osteoclastogenesis.
In conclusion, we demonstrated that C/EBPα exhibits a stronger osteoclastogenic effect than c-Fos in part through regulation by the RANK cytoplasmic IVVY motif. Moreover, the IVVY motif promotes osteoclastogenesis by positively regulating OC markers and negatively regulating RBP-J. It is unknown whether the stronger effect of C/EBPα than of c-Fos in mediating OC differentiation in vitro also applies in vivo. It will be important to validate this finding in vivo through comparative analyses of bone phenotypes in C/EBPα−/− mice and c-Fos−/− mice in conjunction with gene rescuing strategies. Nevertheless, our study provides important comparative insight into the roles of transcription factors and RANK signaling in OC differentiation.
Experimental procedures
Chemical and biological reagents
All chemicals were obtained from Sigma-Aldrich. Anti-FLAG antibody (catalogue number F1804–1 mg) was purchased from Sigma-Aldrich. Recombinant mouse RANKL (catalogue number 462-TEC) and M-CSF (catalogue number 416-ML) were purchased from R&D Systems. Anti-β-actin (catalogue number SC-81178) antibody was from Santa Cruz Biotechnology, and anti-human Fas-activating antibody (α-Fas; catalogue number 05–201) was purchased from Millipore. ACTD and CHX were purchased from Sigma-Aldrich.
Construct generation and retroviral transduction
We previously constructed the pMX-puro-3xFLAG vector by cloning a synthesized 3xFLAG oligonucleotide into the pMX-puro vector (26, 28). The pMX-puro-3xFLAG-C/EBPα (FLAG-C/EBPα), pMX-puro-3xFLAG-c-Fos (FLAG-c-Fos), and pMX-puro-3xFLAG-NFATc1 (FLAG-NFATc1) constructs were prepared by first amplifying the mouse C/EBPα, c-Fos, and NFATc1 cDNAs from the pSport6-C/EBPα (Addgene), pSport6-c-Fos (Addgene), and pSport6-NFATc1 (Addgene) vectors, respectively. The amplified cDNAs were then subcloned in-frame with the 3xFLAG sequence into the pMx-puro-3xFLAG vector. The constructs were confirmed by sequencing. The pMX-puro-GFP (GFP), pMX-puro-hFas-RANK (Fas-RANK), and pMX-puro-hFas-mIVVY (Fas-mIVVY) were generated in previous studies (8, 33). For retrovirus generation, the 293GPG retroviral packaging cell line was cultured in DMEM with 10% heat-inactivated FBS, G418, tetracycline, penicillin/streptomycin, and puromycin (29) before transient transfection with pMX retroviral constructs using the calcium phosphate precipitation method. The retroviral supernatant was harvested at 48, 72, and 96 h post-transfection and then utilized to infect cells for osteoclastogenesis assays (35).
Lentiviral transduction
RBP-J shRNA or scrambled shRNA lentiviral construct (Sigma-Aldrich) and packaging constructs were co-transfected into HEK-293T cells using the calcium phosphate precipitation method (49). The lentiviral supernatant was collected at 60 h post-transfection and then utilized to infect cells for osteoclastogenesis.
In vitro osteoclastogenesis assays from MBM cells
MBM cells were isolated from long bones of 4–6-week-old C57BL/6 mice and cultured as described previously (50–52). Briefly, MBM cells (5 × 104 cells/well) were cultured in 24-well culture dishes in α-minimal essential medium with 10% heat-inactivated FBS and M-CSF (20 ng/ml) for 24 h. Some cells were directly differentiated into OCs as indicated in individual experiments. Other cells were infected with a retrovirus/lentivirus in the presence of M-CSF (10 ng/ml) and Polybrene (8 μg/ml) for 24 h before being cultured as indicated in related experiments for OC formation. The cultures were stained for TRAP activity using a leukocyte acid phosphatase kit (catalogue number 387-A, Sigma) according to the manufacturer's instructions at the end of the assays to examine OC differentiation. The osteoclastogenesis assays were quantified by counting and/or assessing the size of multinucleated TRAP-positive cells (more than three nuclei) in a representative area under a microscope. The experiments involving mice were approved by the University of Alabama at Birmingham institutional animal care and use committee.
In vitro osteoclastogenesis assays from RAW264.7 cells
The macrophage RAW264.7 cell line (1.5 × 104cells/well) was cultured in 24-well culture dishes in DMEM with 10% heat-inactivated FBS for 24 h before infection with a retrovirus/lentivirus in the presence of Polybrene (8 μg/ml) for 24 h followed by osteoclastogenesis assays as indicated in individual experiments. At the end of the assays, the cultures were stained for TRAP activity as indicated above. The assays were quantified by counting and/or assessing the size of multinucleated TRAP-positive cells (more than three nuclei) in a representative area under a microscope.
Western blot analysis
Western blot analysis was carried out as described previously (53). Briefly, cells were cultured as indicated in individual experiments before protein collection for gel electrophoresis. The membranes were washed, and enhanced chemiluminescence detection was carried using Luminata Forte HRP substrate from Millipore. The membranes were visualized using a C-DiGit® blot scanner and Image Studio software from LI-COR Biosciences.
qPCR analysis
qPCR analysis was performed as described previously (54). Cells were cultured as indicated in individual experiments, and total RNA was collected using TRIzol reagent (Life Technologies). 1 μg of total RNA was transcribed into cDNA using the ProtoScript® first strand cDNA synthesis kit (New England Biolabs) according to the manufacturer's instructions. qPCRs were carried using Fast SYBR® Green Master Mix reagent (Life Technologies) using hypoxanthine-guanine phosphoribosyltransferase (Hprt) as an endogenous control for normalization. PCR conditions and primer sequences are available upon request.
RT-PCR analysis
Cells were cultured as indicated in individual experiments, and total RNA was collected for cDNA synthesis as indicated above. Gene amplification analysis was carried using Taq DNA polymerase (catalogue number E001, Novo Protein) as indicated previously (34). RT-PCR primers used to detect the chimeric receptors (Fas-RANK and Fas-mIVVY) were 5′-ATGCTGGGCATCTGGACCCTCCTA-3′ for the human Fas extracellular domain (forward) and 5′-GAAGTCACAGCCCTCAGAATC-3′ for the mouse RANK intracellular domain (reverse). RT-PCR primers used to examine the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), used as an endogenous control, were 5′-TCATTGAGAGCAATGCCAGC-3′ (forward) and 5′-ACATCATCCCTGCATCCACTG-3′ (reverse). 20 μl of the PCR was loaded on a 2% agarose gel for electrophoretic analysis.
ChIP
ChIP was performed as described in a previous study (55). After immunoprecipitation using anti-RBP-J antibody (catalogue number 5313, Cell Signaling Technology) and DNA extraction, quantitative PCR was performed using the primers in the promoter region of the murine Ctsk gene. PCR conditions and primer sequences are available upon request.
Promoter-luciferase assay
The luciferase assay was carried as described in a previous study (55). Briefly, different fragments of the promoter region of the murine Ctsk gene were amplified by PCR using a Ctsk bacterial artificial chromosome clone. The promoter regions were then inserted into the pGL3-Basic vector to construct the pGL3-Ctsk promoter regions. Subsequently, RAW cells were transiently transfected with a DNA mixture containing the pGL3-Ctsk construct (0.3 μg) and β-gal-expressing plasmids (0.03 μg) with or without RBP-J–expressing vector (pCMV-RBP-J; 0.3 μg) using FuGENE 6. Luciferase activity was examined using the Dual-Glo luciferase assay system (Promega) 48 h post-transfection. The β-gal activity of the cell lysates was analyzed using the β-galactosidase Enzyme Assay System (Promega). The level of luciferase activity was normalized to the level of β-gal activity.
Statistical analysis
Data are reported as averages ± S.D. Statistical significance was assessed using Student's t test. p values less than 0.05 were considered significant.
Author contributions
Y.-P. L. designed the study. J. J. and W. C. carried out experiments. J. J., W. C., X. F., and Y.-P. L. analyzed data and prepared the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Jue Wang for extensive reading and discussion of the manuscript. We appreciate the assistance of the Center for Metabolic Bone Disease at the University of Alabama at Birmingham (UAB) (supported by National Institutes of Health Grant P30 AR046031). We are also grateful for the assistance of the Small Animal Phenotyping Core, Metabolism Core, and Neuroscience Molecular Detection Core Laboratory at UAB (supported by National Institutes of Health GrantP30 NS0474666).
This work was supported by National Institutes of Health Grants R01-AR-44741 and R01-DE-023813 (to Y.-P. L.) and R01-AR-070135 (to W. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- OC
- osteoclast
- ACTD
- actinomycin D
- C/EBP
- CCAAT/enhancer–binding protein
- CHX
- cycloheximide
- c-Fos
- FBJ osteosarcoma oncogene
- Ctsk
- cathepsin K
- Fas-RANK
- a chimeric receptor system with the human Fas extracellular domain linked to the normal mouse RANK transmembrane and intracellular domains
- Fas-mIVVY
- a chimeric receptor system with the human Fas extracellular domain linked to the mouse RANK transmembrane and intracellular domains bearing an inactivating mutation in the cytoplasmic 535IVVY538 motif
- Hprt
- hypoxanthine-guanine phosphoribosyltransferase
- IRF-8
- interferon-regulatory factor 8
- ITAM
- immunoreceptor tyrosine-based activation motif
- MBM
- mouse bone marrow
- M-CSF
- macrophage colony-stimulating factor
- NFATc1
- nuclear factor of activated T cells, C1
- RANK
- receptor activator of NF-κB
- RANKL
- receptor activator of NF-κB ligand
- RBP-J
- recombinant recognition sequence–binding protein at the Jκ site
- TRAP
- tartrate-resistant acid phosphatase
- qPCR
- quantitative real-time PCR
- TSS
- transcriptional start site
- hFas
- human Fas.
References
- 1. Boyle W. J., Simonet W. S., and Lacey D. L. (2003) Osteoclast differentiation and activation. Nature 423, 337–342 10.1038/nature01658 [DOI] [PubMed] [Google Scholar]
- 2. Teitelbaum S. L. (2000) Bone resorption by osteoclasts. Science 289, 1504–1508 10.1126/science.289.5484.1504 [DOI] [PubMed] [Google Scholar]
- 3. Li Y. P., Chen W., and Stashenko P. (1996) Molecular cloning and characterization of a putative novel human osteoclast-specific 116-kDa vacuolar proton pump subunit. Biochem. Biophys. Res. Commun. 218, 813–821 10.1006/bbrc.1996.0145 [DOI] [PubMed] [Google Scholar]
- 4. Feng X., and McDonald J. M. (2011) Disorders of bone remodeling. Annu. Rev. Pathol. 6, 121–145 10.1146/annurev-pathol-011110-130203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang S., Hao L., McConnell M., Zhou X., Wang M., Zhang Y., Mountz J. D., Reddy M., Eleazer P. D., Li Y. P., and Chen W. (2013) Inhibition of Rgs10 expression prevents immune cell infiltration in bacteria-induced inflammatory lesions and osteoclast-mediated bone destruction. Bone Res. 1, 267–281 10.4248/BR201303005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Y. P., Chen W., Liang Y., Li E., and Stashenko P. (1999) Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat. Genet. 23, 447–451 10.1038/70563 [DOI] [PubMed] [Google Scholar]
- 7. Das S., Sepahi I., Duthie A., Clark S., and Crockett J. C. (2014) RANK receptor oligomerisation in the regulation of NFκB signalling. J. Mol. Endocrinol. 53, 81–91 10.1530/JME-14-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu D., Wang S., Liu W., Liu J., and Feng X. (2006) A novel receptor activator of NF-κB (RANK) cytoplasmic motif plays an essential role in osteoclastogenesis by committing macrophages to the osteoclast lineage. J. Biol. Chem. 281, 4678–4690 10.1074/jbc.M510383200 [DOI] [PubMed] [Google Scholar]
- 9. Taguchi Y., Gohda J., Koga T., Takayanagi H., and Inoue J. (2009) A unique domain in RANK is required for Gab2 and PLCγ2 binding to establish osteoclastogenic signals. Genes Cells 14, 1331–1345 10.1111/j.1365-2443.2009.01351.x [DOI] [PubMed] [Google Scholar]
- 10. Kim H., Choi H. K., Shin J. H., Kim K. H., Huh J. Y., Lee S. A., Ko C. Y., Kim H. S., Shin H. I., Lee H. J., Jeong D., Kim N., Choi Y., and Lee S. Y. (2009) Selective inhibition of RANK blocks osteoclast maturation and function and prevents bone loss in mice. J. Clin. Investig. 119, 813–825 10.1172/JCI36809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guerrini M. M., Sobacchi C., Cassani B., Abinun M., Kilic S. S., Pangrazio A., Moratto D., Mazzolari E., Clayton-Smith J., Orchard P., Coxon F. P., Helfrich M. H., Crockett J. C., Mellis D., Vellodi A., et al. (2008) Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am. J. Hum. Genet. 83, 64–76 10.1016/j.ajhg.2008.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grigoriadis A. E., Wang Z. Q., Cecchini M. G., Hofstetter W., Felix R., Fleisch H. A., and Wagner E. F. (1994) c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 266, 443–448 10.1126/science.7939685 [DOI] [PubMed] [Google Scholar]
- 13. Johnson R. S., Spiegelman B. M., and Papaioannou V. (1992) Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell 71, 577–586 10.1016/0092-8674(92)90592-Z [DOI] [PubMed] [Google Scholar]
- 14. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., and Taniguchi T. (2002) Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 10.1016/S1534-5807(02)00369-6 [DOI] [PubMed] [Google Scholar]
- 15. Huang H., Chang E. J., Ryu J., Lee Z. H., Lee Y., and Kim H. H. (2006) Induction of c-Fos and NFATc1 during RANKL-stimulated osteoclast differentiation is mediated by the p38 signaling pathway. Biochem. Biophys. Res. Commun. 351, 99–105 10.1016/j.bbrc.2006.10.011 [DOI] [PubMed] [Google Scholar]
- 16. Asagiri M., and Takayanagi H. (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 10.1016/j.bone.2006.09.023 [DOI] [PubMed] [Google Scholar]
- 17. Li Y. P., Alexander M., Wucherpfennig A. L., Yelick P., Chen W., and Stashenko P. (1995) Cloning and complete coding sequence of a novel human cathepsin expressed in giant cells of osteoclastomas. J. Bone Miner. Res. 10, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 18. Chen W., Yang S., Abe Y., Li M., Wang Y., Shao J., Li E., and Li Y. P. (2007) Novel pycnodysostosis mouse model uncovers cathepsin K function as a potential regulator of osteoclast apoptosis and senescence. Hum. Mol. Genet. 16, 410–423 10.1093/hmg/ddl474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsuo K., Galson D. L., Zhao C., Peng L., Laplace C., Wang K. Z., Bachler M. A., Amano H., Aburatani H., Ishikawa H., and Wagner E. F. (2004) Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 279, 26475–26480 10.1074/jbc.M313973200 [DOI] [PubMed] [Google Scholar]
- 20. Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., and McKnight S. L. (1988) Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 2, 786–800 10.1101/gad.2.7.786 [DOI] [PubMed] [Google Scholar]
- 21. Nerlov C. (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 17, 318–324 10.1016/j.tcb.2007.07.004 [DOI] [PubMed] [Google Scholar]
- 22. Ye M., Zhang H., Amabile G., Yang H., Staber P. B., Zhang P., Levantini E., Alberich-Jordà M., Zhang J., Kawasaki A., and Tenen D. G. (2013) C/EBPα controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol. 15, 385–394 10.1038/ncb2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang D. E., Zhang P., Wang N. D., Hetherington C. J., Darlington G. J., and Tenen D. G. (1997) Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 569–574 10.1073/pnas.94.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang N. D., Finegold M. J., Bradley A., Ou C. N., Abdelsayed S. V., Wilde M. D., Taylor L. R., Wilson D. R., and Darlington G. J. (1995) Impaired energy homeostasis in C/EBPα knockout mice. Science 269, 1108–1112 10.1126/science.7652557 [DOI] [PubMed] [Google Scholar]
- 25. Chen W., Zhu G., Hao L., Wu M., Ci H., and Li Y. P. (2013) C/EBPα regulates osteoclast lineage commitment. Proc. Natl. Acad. Sci. U.S.A. 110, 7294–7299 10.1073/pnas.1211383110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jules J., Chen W., Feng X., and Li Y. P. (2016) CCAAT/enhancer binding protein α (C/EBPα) is important for osteoclast differentiation and activity. J. Biol. Chem. 291, 16390–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao Q., Shao J., Chen W., and Li Y. P. (2007) Osteoclast differentiation and gene regulation. Front. Biosci. 12, 2519–2529 10.2741/2252 [DOI] [PubMed] [Google Scholar]
- 28. Onishi M., Nosaka T., Misawa K., Mui A. L., Gorman D., McMahon M., Miyajima A., and Kitamura T. (1998) Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18, 3871–3879 10.1128/MCB.18.7.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ory D. S., Neugeboren B. A., and Mulligan R. C. (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 10.1073/pnas.93.21.11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lam J., Takeshita S., Barker J. E., Kanagawa O., Ross F. P., and Teitelbaum S. L. (2000) TNF-α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Investig. 106, 1481–1488 10.1172/JCI11176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei S., Kitaura H., Zhou P., Ross F. P., and Teitelbaum S. L. (2005) IL-1 mediates TNF-induced osteoclastogenesis. J. Clin. Investig. 115, 282–290 10.1172/JCI200523394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horowitz M. C., Xi Y., Wilson K., and Kacena M. A. (2001) Control of osteoclastogenesis and bone resorption by members of the TNF family of receptors and ligands. Cytokine Growth Factor Rev. 12, 9–18 10.1016/S1359-6101(00)00030-7 [DOI] [PubMed] [Google Scholar]
- 33. Xu D., Shi Z., McDonald J., Pan G., Cao X., Yu X., and Feng X. (2004) Development of a chimaeric receptor approach to study signalling by tumour necrosis factor receptor family members. Biochem. J. 383, 219–225 10.1042/BJ20040961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jules J., Shi Z., Liu J., Xu D., Wang S., and Feng X. (2010) Receptor activator of NF-κB (RANK) cytoplasmic IVVY535–538 motif plays an essential role in tumor necrosis factor-α (TNF)-mediated osteoclastogenesis. J. Biol. Chem. 285, 37427–37435 10.1074/jbc.M110.149484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jules J., Zhang P., Ashley J. W., Wei S., Shi Z., Liu J., Michalek S. M., and Feng X. (2012) Molecular basis of requirement of receptor activator of nuclear factor κB signaling for interleukin 1-mediated osteoclastogenesis. J. Biol. Chem. 287, 15728–15738 10.1074/jbc.M111.296228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jules J., Wang S., Shi Z., Liu J., Wei S., and Feng X. (2015) The IVVY motif and tumor necrosis factor receptor-associated factor (TRAF) sites in the cytoplasmic domain of the receptor activator of nuclear factor κB (RANK) cooperate to induce osteoclastogenesis. J. Biol. Chem. 290, 23738–23750 10.1074/jbc.M115.667535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ashley J. W., McCoy E. M., Clements D. A., Shi Z., Chen T., and Feng X. (2011) Development of cell-based high-throughput assays for the identification of inhibitors of receptor activator of nuclear factor-κB signaling. Assay Drug Dev. Technol. 9, 40–49 10.1089/adt.2010.0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao B., and Ivashkiv L. B. (2011) Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res. Ther. 13, 234 10.1186/ar3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li S., Miller C. H., Giannopoulou E., Hu X., Ivashkiv L. B., and Zhao B. (2014) RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. J. Clin. Investig. 124, 5057–5073 10.1172/JCI71882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swarnkar G., Karuppaiah K., Mbalaviele G., Chen T. H., and Abu-Amer Y. (2015) Osteopetrosis in TAK1-deficient mice owing to defective NF-κB and NOTCH signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 154–159 10.1073/pnas.1415213112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao B., Grimes S. N., Li S., Hu X., and Ivashkiv L. B. (2012) TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J. Exp. Med. 209, 319–334 10.1084/jem.20111566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao B., Takami M., Yamada A., Wang X., Koga T., Hu X., Tamura T., Ozato K., Choi Y., Ivashkiv L. B., Takayanagi H., and Kamijo R. (2009) Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15, 1066–1071 10.1038/nm.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wagner H. (2010) Bone diseases: Interferon regulatory factor-8 suppresses osteoclastogenesis. Nat. Rev. Rheumatol. 6, 73–74 10.1038/nrrheum.2009.274 [DOI] [PubMed] [Google Scholar]
- 44. Ma J., Liu Y. L., Hu Y. Y., Wei Y. N., Zhao X. C., Dong G. Y., Qin H. Y., Ding Y., and Han H. (2013) Disruption of the transcription factor RBP-J results in osteopenia attributable to attenuated osteoclast differentiation. Mol. Biol. Rep. 40, 2097–2105 10.1007/s11033-012-2268-6 [DOI] [PubMed] [Google Scholar]
- 45. Boggs K., Henderson B., and Reisman D. (2009) RBP-Jκ binds to and represses transcription of the p53 tumor suppressor gene. Cell Biol. Int. 33, 318–324 10.1016/j.cellbi.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 46. Chen S., Tao J., Bae Y., Jiang M. M., Bertin T., Chen Y., Yang T., and Lee B. (2013) Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. J. Bone Miner. Res. 28, 649–659 10.1002/jbmr.1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rozenberg J. M., Tesfu D. B., Musunuri S., Taylor J. M., and Mack C. P. (2014) DNA methylation of a GC repressor element in the smooth muscle myosin heavy chain promoter facilitates binding of the Notch-associated transcription factor, RBPJ/CSL1. Arterioscler. Thromb. Vasc. Biol. 34, 2624–2631 10.1161/ATVBAHA.114.304634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharma S. M., Bronisz A., Hu R., Patel K., Mansky K. C., Sif S., and Ostrowski M. C. (2007) MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J. Biol. Chem. 282, 15921–15929 10.1074/jbc.M609723200 [DOI] [PubMed] [Google Scholar]
- 49. Yang D. Q., Feng S., Chen W., Zhao H., Paulson C., and Li Y. P. (2012) V-ATPase subunit ATP6AP1 (Ac45) regulates osteoclast differentiation, extracellular acidification, lysosomal trafficking, and protease exocytosis in osteoclast-mediated bone resorption. J. Bone Miner. Res. 27, 1695–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu H., Xu G., and Li Y. P. (2009) Atp6v0d2 is an essential component of the osteoclast-specific proton pump that mediates extracellular acidification in bone resorption. J. Bone Miner. Res 24, 871–885 10.1359/jbmr.081239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Feng S., Deng L., Chen W., Shao J., Xu G., and Li Y. P. (2009) Atp6v1c1 is an essential component of the osteoclast proton pump and in F-actin ring formation in osteoclasts. Biochem. J. 417, 195–203 10.1042/BJ20081073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang S., and Li Y. P. (2007) RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev. 21, 1803–1816 10.1101/gad.1544107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen W., Ma J., Zhu G., Jules J., Wu M., McConnell M., Tian F., Paulson C., Zhou X., Wang L., and Li Y. P. (2014) Cbfβ deletion in mice recapitulates cleidocranial dysplasia and reveals multiple functions of Cbfβ required for skeletal development. Proc. Natl. Acad. Sci. U.S.A. 111, 8482–8487 10.1073/pnas.1310617111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tian F., Wu M., Deng L., Zhu G., Ma J., Gao B., Wang L., Li Y. P., and Chen W. (2014) Core binding factor β (Cbfβ) controls the balance of chondrocyte proliferation and differentiation by up-regulating Indian hedgehog (Ihh) expression and inhibiting parathyroid hormone-related protein receptor (PPR) expression in postnatal cartilage and bone formation. J. Bone Miner. Res. 29, 1564–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu M., Wang Y., Shao J. Z., Wang J., Chen W., and Li Y. P. (2017) Cbfβ governs osteoblast-adipocyte lineage commitment through enhancing β-catenin signaling and suppressing adipogenesis gene expression. Proc. Natl. Acad. Sci. U.S.A. 114, 10119–10124 10.1073/pnas.1619294114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.