Summary
Growth is characterized by the interplay between cell division and cell expansion, two processes that occur separated along the growth zone at the maize leaf. To gain further insight into the transition between cell division and cell expansion, conditions were investigated in which the position of this transition zone was positively or negatively affected. High levels of gibberellic acid (GA) in plants overexpressing the GA biosynthesis gene GA20‐OXIDASE (GA20OX‐1 OE) shifted the transition zone more distally, whereas mild drought, which is associated with lowered GA biosynthesis, resulted in a more basal positioning. However, the increased levels of GA in the GA20OX‐1 OE line were insufficient to convey tolerance to the mild drought treatment, indicating that another mechanism in addition to lowered GA levels is restricting growth during drought. Transcriptome analysis with high spatial resolution indicated that mild drought specifically induces a reprogramming of transcriptional regulation in the division zone. ‘Leaf Growth Viewer’ was developed as an online searchable tool containing the high‐resolution data.
Keywords: maize, mild drought, gibberellic acid, cell division
Introduction
Plants are continuously producing organs that grow to fulfil specific roles during plant development, and the sessile nature of plants urges them to adjust their growth when the environment changes. Therefore, it is pivotal to gain better insights into the coordination of the processes of cell division and cell expansion and to understand how intrinsic signals and environmental cues impinge on these. Depending on the organ, the processes of cell division and cell expansion are mainly viewed as spatially and/or temporally regulated (Gonzalez et al., 2012; Nelissen et al., 2012; Sozzani and Iyer‐Pascuzzi, 2014), and it becomes increasingly clear that the regulation of the growth mechanisms in dicot and monocot leaves is to a great extend conserved (Nelissen et al., 2016).
One of these conserved mechanisms that also represents an important developmental switch is the transition from cell division to cell expansion, for which already a large number of genes and molecular pathways have been identified (Breuninger and Lenhard, 2010; Gonzalez et al., 2012; Nelissen et al., 2012; Sozzani and Iyer‐Pascuzzi, 2014). These genes function in transcriptional regulation (Gonzalez et al., 2010; Horiguchi et al., 2005; Mizukami and Fischer, 2000; Nath et al., 2003; Vercruyssen et al., 2014), protein degradation (Disch et al., 2006; Li et al., 2008), hormone metabolism and signalling (Achard et al., 2009; Hu et al., 2003) and the production of a non‐cell‐autonomous growth‐promoting signal (Czesnick and Lenhard, 2015; Kazama et al., 2010). In Arabidopsis, the cell cycle arrest front, visualized by a CYCLINB1;1 reporter gene (Donnelly et al., 1999), is often used together with the analysis of growth over time (kinematic analysis; Andriankaja et al., 2012; Nelissen et al., 2013) to gain insights into perturbations in the transition from cell division to cell expansion. Differences in the cell cycle arrest front have been observed in genetic (Mizukami and Fischer, 2000; Nath et al., 2003; Vercruyssen et al., 2014) and environmental (Skirycz et al., 2011) perturbations.
During steady‐state growth of the maize leaf, the growth‐promoting processes are spatially separated. At the leaf base, cells are dividing, and as the cells move upwards in the leaf, they transition to expanding and later to mature cells (Nelissen et al., 2013). The regions in the maize growth zone in which these processes occur, are quite large, encompassing more than 10 mm and are referred to as division zone (DZ), expansion zone (EZ) and mature zone (MZ). The respective transitions are defined here as transition zone 1 (TZ1) and transition zone 2 (TZ2). TZ1 has been shown to be characterized by a local accumulation of bioactive gibberellic acid (GA; Nelissen et al., 2012). This GA peak is instrumental to determine the position of TZ1, and thus in defining the position where cells decide to exit cell division and to enter cell expansion (Nelissen et al., 2012). Water deficit has been shown to affect the growth‐promoting processes differently depending on the position along the growth zone (Tardieu et al., 2011, 2000).
The aim of this study was to gain further insights into the molecular processes that dynamically regulate the transition from cell division to cell expansion in growing maize leaves. Therefore, we examined the effects of perturbations that positively (overexpression of GA20OX‐1; Nelissen et al., 2012) or negatively (mild drought stress) affect the position of TZ1. Strikingly, a reduced level of bioactive GA (GA1 and GA4) and its biosynthetic precursors was observed at TZ1 under mild drought conditions. However, merely increasing GA levels was insufficient to overcome this mild drought phenotype, because the GA20OX‐1 OE line was not more drought tolerant, compared with its nontransgenic siblings. We analysed the high‐resolution transcriptomics data of mild drought‐treated and GA20OX‐1 OE plants by aligning the samples according to the relative position of TZ1. In this manner, growth zone‐specific, mild drought‐induced changes in the expression of transcription factors, E2F/DP target genes, aquaporins and photosynthesis‐related genes were shown. To visualize and provide access to these high‐resolution data, we developed ‘Leaf Growth Viewer’ (LGV).
Results
Mild drought affects the position of transition zone 1
Overexpression of the rate‐limiting GA biosynthesis gene GA20‐OXIDASE (GA20OX‐1 OE) has an enhancing effect on the size of the DZ and results in a more distal position of TZ1 (Nelissen et al., 2012). Here, we investigated the effect of mild drought on this transition zone between cell division and cell expansion in B104, an inbred that is closely related to B73 (Liu et al., 2003) and that can be routinely transformed (Coussens et al., 2012; Frame et al., 2006).
Our mild drought conditions reduced the leaf elongation rate (LER) by 28% compared with plants grown in well‐watered conditions (Table 1). A kinematic analysis revealed that this growth reduction was partially the result of a reduced cell production (16%), which was in turn caused by a reduction in DZ size (22%, P‐value = 0.04), bringing about a significantly reduced cell number (14%, P‐value = 0.005), and thus a more basal position of TZ1. The duration of one cell cycle and the rate of cell division were not significantly altered (P‐value = 0.5 and 0.4, respectively, Table 1). In addition to this effect on cell division, also a reduction in cell expansion was observed, because the mature cell size was significantly lowered by 15% (P‐value = 0.04). Whereas both cell division and cell expansion were negatively affected by mild drought, the final reduction in leaf length was remarkably less pronounced (10%, P‐value = 0.002), because the duration of growth, also referred to as leaf elongation duration (LED), was significantly increased by 14% (P‐value = 0.0025), indicating that although under mild drought the LER was reduced, growth was maintained for a longer period of time. The same mild drought treatment resulted in a similar repositioning of TZ1 and an increased LED in B73 (Table 1).
Table 1.
Growth‐related parameters of mild drought‐treated and well‐watered B104 and B73 plants
| Parameters | B104 | B73 | ||||||
|---|---|---|---|---|---|---|---|---|
| Control* | Drought* | % Change | P‐value# | Control* | Drought* | % Change | P‐value# | |
| LER (mm/h) | 2.9 ± 0.1 | 2.1 ± 0.1 | −29 | 8.10−5 | 3.1 ± 0.1 | 2.2 ± 0.2 | −30 | 0.008 |
| FLL (mm) | 557 ± 8.6 | 499.2 ± 9.8 | −10 | 0.002 | na | na | na | na |
| Lma (μm) | 123 ± 1 | 104 ± 1 | −15 | 0.04 | 136 ± 7 | 110 ± 4 | −15 | 0.07 |
| Lez (mm) | 28.6 ± 2.3 | 26.7 ± 2.4 | −7 | 0.6 | 50.0 ± 0.8 | 47.0 ± 2.6 | −7 | 0.3 |
| Nez | 393 ± 32 | 469 ± 56 | 19 | 0.3 | 657 ± 71 | 882 ± 142 | 34 | 0.2 |
| P (cells/h) | 24 ± 0.23 | 20 ± 0.2 | −16 | 7.10−4 | 23 ± 1 | 19 ± 1 | −18 | 0.05 |
| Ldz (mm) | 11.6 ± 0.6 | 9 ± 0.2 | −22 | 0.04 | 14.9 ± 1.5 | 9.8 ± 0.4 | −34 | 0.03 |
| Ndz (cells) | 613 ± 11 | 528 ± 7 | −14 | 0.005 | 747 ± 78 | 619 ± 9 | −17 | 0.2 |
| D (cells/cells.h) | 0.039 ± 0.0001 | 0.038 ± 0.001 | −2 | 0.4 | 0.032 ± 0.005 | 0.030 ± 0.002 | −4 | 0.8 |
| Tc (h) | 18 ± 0.4 | 18 ± 0.1 | 2 | 0.5 | 23 ± 3 | 23 ± 1 | 2 | 0.9 |
*Mean ± standard error. # P‐values as obtained by Student's t‐test (n = 3).
na, not analysed; LER, leaf elongation rate; FLL, final leaf length; Lma, mature cell length; Lez, expansion zone size; Nez, number of cells in expansion zone; P, cell proliferation rate; Ldz, division zone size; Ndz, number of dividing cells; D, cell division rate; Tc, cell cycle duration.
Mild drought lowers auxin and cytokinin levels in the division zone and gibberellic acid levels at the transition zone
The effect of the repositioning of TZ1 by elevated GA levels or mild drought was assessed at the hormone level. The growth zone was sampled every 0.5 cm, while the remainder of the leaf until 10 cm was sampled in 1‐cm pieces (Figure 1). Because TZ1 in B104 is positioned around 1.3 cm, the most basal half cm reflects the basal DZ, while the second half cm represents the distal DZ. The third half cm coincides with the location of TZ1 and thus contains cells that are both dividing and expanding. The fourth half cm contains cells in early cell expansion, while later phases of cell expansion and cell maturity are present in the successive 1‐cm‐long leaf samples taken along the leaf gradient (Figure 1). TZ2 is located between 4 and 5 cm in B104 plants.
Figure 1.
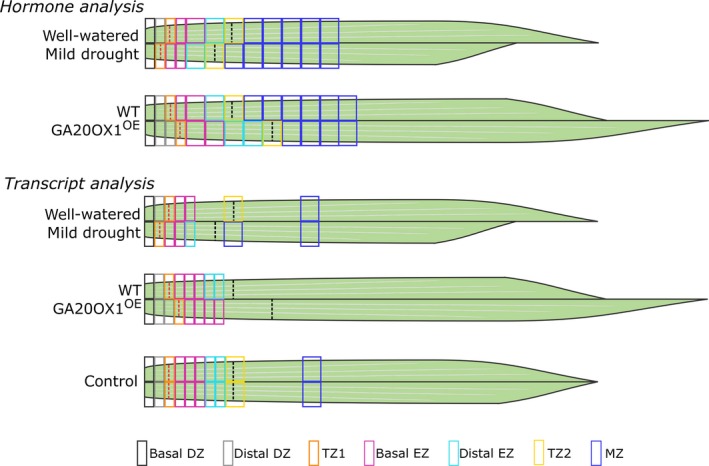
Schematic overview of the sampling strategy of leaf four samples for hormone and transcriptome analysis. The transition between cell division and cell expansion (TZ1) is indicated by a red dotted line, and the transition between cell expansion and mature cells (TZ2) is indicated by a black dotted line. Although the boundaries between the different zones look fixed, the transition between different zones is a gradual process. To investigate the robust transcriptional changes with high resolution in the growth zone, the control samples from the mild drought (well‐watered) and GA20OX‐1 OE (WT) experiment were analysed together (bottom panel). WT, wild type; DZ, division zone; TZ, transition zone; EZ, expansion zone; MZ, mature zone.
There was a large increase in bioactive GAs (GA1 and GA4) in GA20OX‐1 OE plants (Figure 2a), confirming our previous findings (Nelissen et al., 2012). The higher GA levels also resulted in elevated levels of the auxin indole‐3‐acetic acid (IAA; Figure 2b), abscisic acid (ABA; Figure 2c) and jasmonoyl‐isoleucine (JA‐Ile; Figure 2d). Cytokinin levels [N6‐(Δ2‐isopentenyl)adenine (iP) and trans‐zeatin (tZ)] were unaffected in the DZ of the GA20OX‐1 OE line, but showed a delayed rise towards the end of the EZ (Figure 2e). This rise in iP corresponds to the position of TZ2, which is shifted to a more distal position as compared with the nontransgenic siblings (Figure 1), suggesting that iP plays a role at the transition between cell expansion and cell maturity. Salicylic acid (SA) levels were not significantly affected in the GA20OX‐1 OE plants (Figure 2f).
Figure 2.
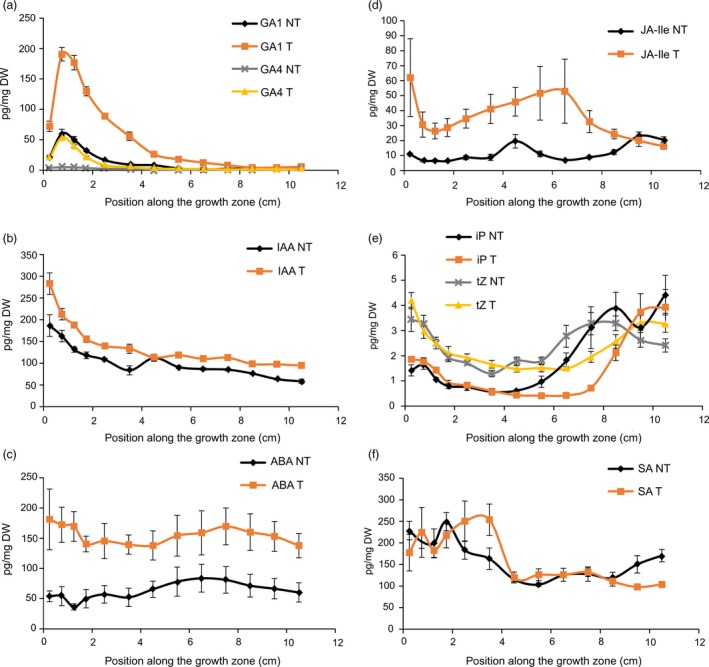
Hormone levels [pg/mg dry weight (DW)] along the growth zone of transgenic GA20OX‐1 OE plants (T) and segregating nontransgenic siblings (NT): (a) gibberellic acids (GA); (b) auxin indole‐3‐acetic acid (IAA); (c) abscisic acid (ABA); (d) jasmonoyl‐isoleucine (Ja‐Ile); (e) cytokinins N6‐(Δ2‐isopentenyl)adenine (iP) and trans‐zeatin (tZ); (f) salicylic acid (SA). Values are average ± standard error (n = 5).
Under mild drought, the levels of ABA and SA (Figure 3c,f) showed a strong increase. Strikingly, the levels of SA under mild drought significantly increased between the basal and distal DZ to stay maximal throughout the EZ and MZ (Figure 3f). No significant differences between mild drought and well‐watered plants were found for JA and JA‐Ile levels (Figure 3d). Under mild drought, both IAA and tZ levels were found to be significantly decreased at those positions in the growth zone where their levels were the highest in well‐watered conditions, that is at the leaf base (Figure 3b,e). In contrast to the more distal shift of the position of TZ2 in the GA20OX‐1 OE plants, its position (Figure 1) and a concomitant rise in iP (Figure 3e) were shifted to a more basal position under mild drought as compared with the well‐watered B104. The levels of the bioactive GA1 and GA4 (Figure 3a) were down‐regulated by mild drought and maximal levels were reached approximately 5 mm closer to the leaf basis as compared with control leaves. This shift in GA maxima towards the leaf base corresponds to the shift in TZ1 at the cellular level (Table 1).
Figure 3.

Hormone levels (pg/mg dry weight (DW)) along the growth zone under mild drought and well‐watered (control) conditions: (a) gibberellic acids (GA); (b) auxin indole‐3‐acetic acid (IAA); (c) abscisic acid (ABA); (d) jasmonic acid (JA) and jasmonoyl‐isoleucine (Ja‐Ile); (e) cytokinins N6‐(Δ2‐isopentenyl)adenine (iP) and trans‐zeatin (tZ); (f) salicylic acid (SA). Values are average ± standard error (n = 5).
GA biosynthesis is lowered by mild drought stress
The reason for the shift and reduction in the GA profile was examined by measuring all GA biosynthetic intermediates from GA12 onwards and two major GA inactivation products (GA29 and GA8; Figure 4). The levels of the first‐formed GA (GA12) were increased by the drought treatment, but the levels of the subsequent metabolites (GA15 and GA24) were significantly lowered. The GA biosynthetic intermediates GA9, GA51 and GA34 were hardly detectable, and differences in their levels were not significant. For most metabolite precursors of GA4 and GA1 (GA15, GA44, GA24, GA20), a significant interaction between the drought treatment and the position along the growth zone was shown, indicating that the accumulation pattern of the GA metabolites was shifted towards the leaf base. The lower levels of bioactive GAs also resulted in a significant reduction in the levels of the GA2‐OXIDASE (GA2‐OX)‐mediated degradation products (GA29 and GA8; Figure 4).
Figure 4.
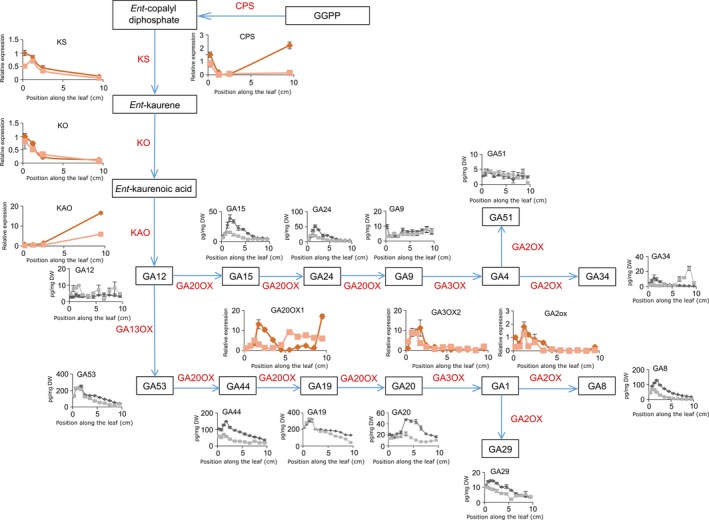
Gibberellic acid (GA) biosynthesis and catabolism along the growth zone of B104 leaves. GA metabolites along the growth zone under mild drought and well‐watered conditions (grey and black, respectively). Values are average ± standard error (n = 5). The expression levels of enzymes under mild drought and well‐watered conditions are indicated in dark and light red, respectively. Expression values are represented as values relative to the expression value at the leaf base in well‐watered plants. Values are average ± standard error (n = 3). CPS, ent‐copalyl diphosphate synthase; KS, ent‐kaurene synthase; KO, ent‐kaurene oxidase; KAO, ent‐kaurenoic acid oxidase.
The expression levels of several GA biosynthetic genes, as well as of the GA inactivation genes, GA2‐ox, were analysed by qRT‐PCR (Figure 4). No significant down‐regulation of the expression levels of CPS (ent‐copalyl diphosphate synthase), KS (ent‐kaurene synthase), KO (ent‐kaurene oxidase) or KAO (ent‐kaurenoic acid oxidase) was observed in the growth zone, whereas all tested genes encoding GA metabolic enzymes from the GA13 oxidation step onwards were significantly down‐regulated by the drought treatment, with the gene encoding the rate‐limiting biosynthetic enzyme GA20‐OXIDASE (Nelissen et al., 2012) being the earliest GA biosynthetic enzyme to show a lowered and shifted expression pattern.
High GA levels in GA20OX‐overexpressing plants do not affect the response to mild drought
Because mild drought lowered the expression level of GA20‐OXIDASE, boosting GA levels by overexpressing the GA20‐OXIDASE could render plants more tolerant to drought. To test this hypothesis, we grew hemizygous GA20OX‐1 OE plants under mild drought conditions. Although under well‐watered conditions, leaf growth of the GA20OX‐1 OE plants was enhanced compared with the nontransgenic siblings (Nelissen et al., 2012), the relative growth reduction caused by mild drought was comparable (26% and 23% in the hemizygous GA20OX‐1 OE plants and the nontransgenic siblings, respectively; Figure 5a). The mild drought assay was repeated using homozygous GA20OX‐1 OE plants and the nontransgenic siblings, showing similar results (23% and 24%, respectively). Kinematic analysis revealed that mild drought caused a significant reduction in the DZ size (24%) in the homozygous GA20OX‐1 OE plants, which was similar to that observed for the nontransgenic siblings (22%), indicating that high levels of GA were not sufficient to overcome the reduction in the DZ size caused by mild drought (Table 2). The differences in the GA1 levels in the GA20OX‐1 OE plants and their nontransgenic siblings under well‐watered and water‐deficit conditions were concomitant with the changes in the growth curve (Figure 5a). The water deficit caused a comparable reduction in GA1 levels for both transgenic and nontransgenic siblings, but the GA1 levels in the transgenic plants grown under drought were comparable or higher than those in the nontransgenic siblings grown in well‐watered conditions (Figure 5b).
Figure 5.
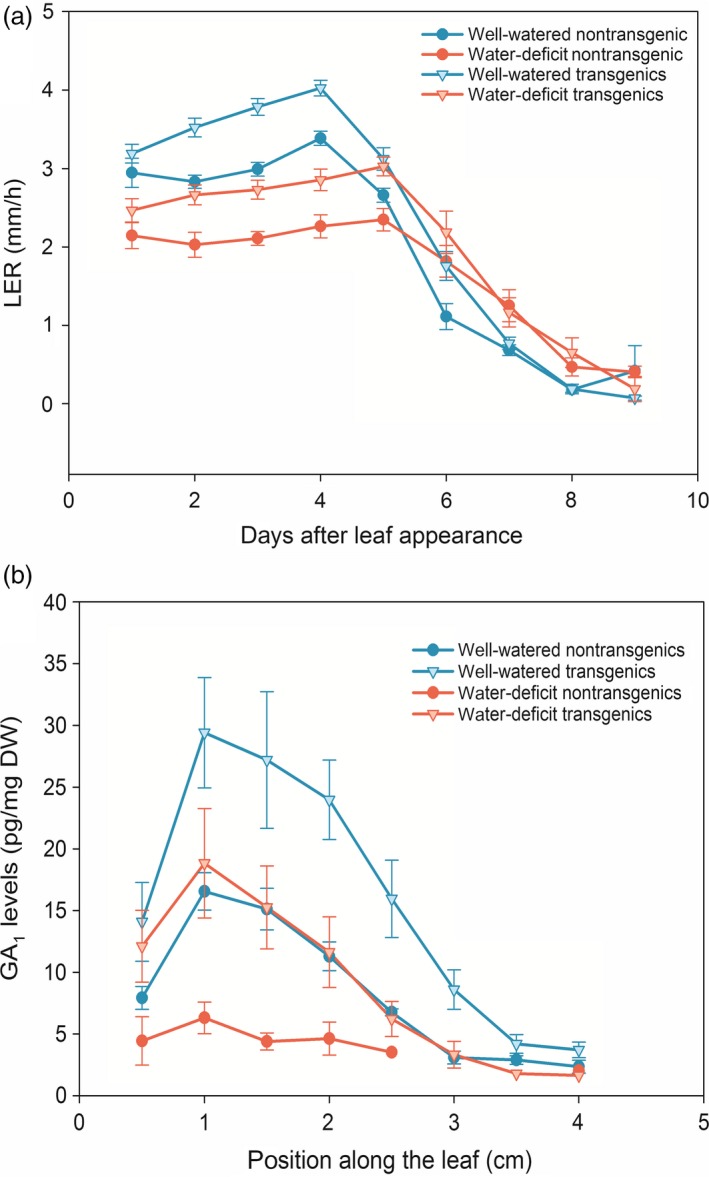
Growth curve (a) and GA1 levels (b) of hemizygous GA20OX‐1 OE transgenic and nontransgenic siblings grown under well‐watered and water‐deficit conditions. (a) n ≥ 12, error bars represent standard error; (b) n = 3, error bars represent standard error.
Table 2.
Growth‐related parameters of mild drought‐treated and well‐watered plants for the homozygous GA20OX‐1 OE transgenic and nontransgenic siblings
| Parameters | GA20OX‐1 OE nontransgenic siblings | GA20OX‐1 OE transgenic siblings | ||||||
|---|---|---|---|---|---|---|---|---|
| Control* | Drought* | % Change | P‐value# | Control* | Drought* | % Change | P‐value# | |
| LER (mm/h) | 3 ± 0.1 | 2.2 ± 0.1 | −24 | 8.10−6 | 3.8 ± 0.3 | 2.9 ± 0.2 | −23 | 0.001 |
| FLL (mm) | 560 ± 6.4 | 468.8 ± 13.2 | −16 | 0.001 | 816.5 ± 14.6 | 669.5 ± 32.5 | −18 | 0.09 |
| Lma (μm) | 119 ± 2 | 97 ± 2 | −18 | 8.10−4 | 127 ± 2 | 98 ±4 | −23 | 0.008 |
| Lez (mm) | 30.5 ± 3.1 | 25.5 ± 0.6 | −16 | 0.2 | 44.3 ± 8.1 | 30.9 ± 5.3 | −22 | 0.004 |
| Nez | 412 ± 41 | 403 ± 28 | −2 | 0.9 | 504 ± 83 | 460 ± 81 | −9 | 0.7 |
| P (cells/h) | 25 ± 0.3 | 23 ± 0.4 | −7 | 0.03 | 30 ± 1 | 30 ± 1 | −0.4 | 0.9 |
| Ldz (mm) | 14.8 ± 0.5 | 11.5 ± 0.3 | −22 | 0.006 | 22.9 ± 0.5 | 17.5 ± 0.5 | −24 | 0.005 |
| Ndz (cells) | 694 ± 32 | 540 ± 16 | −22 | 0.025 | 1026 ± 67 | 795 ± 63 | −22 | 0.048 |
| D (cells/cells.h) | 0.036 ± 0.002 | 0.043 ± 0.002 | 19 | 0.04 | 0.029 ± 0.003 | 0.038 ± 0.005 | 31 | 0.2 |
| Tc (h) | 19 ± 1 | 16 ± 1 | −16 | 0.05 | 24 ± 2 | 19 ± 2 | −22 | 0.1 |
*Mean ± standard error. # P‐values as obtained by Student's t‐test (n = 3).
LER, leaf elongation rate; FLL, final leaf length; Lma, mature cell length; Lez, expansion zone size; Nez, number of cells in expansion zone; P, cell proliferation rate; Ldz, division zone size; Ndz, number of dividing cells; D, cell division rate; Tc, cell cycle duration.
Leaf Growth Viewer (LGV) as a tool to query high‐resolution transcriptomics data
Because mild drought might act through additional mechanisms besides GA biosynthesis to affect the position of TZ1, we compared high‐resolution, genome‐wide transcriptomics data obtained by sampling the growth zone with 0.5‐cm intervals till TZ2 of mild drought‐treated and GA20OX‐1 OE plants (up to five and eight cm from the leaf base, respectively; Figure 1). The mild drought experiment was complemented with a sample representative of the late expansion zone (between 4 and 5 cm) and one of the MZ (between 8 and 9 cm; Figure 1). We developed an online search engine, called Leaf Growth Viewer (LGV; accessible through the following link: https://psblgv01.psb.ugent.be) that allows to query the data set starting from gene identifiers to obtain their leaf growth zone‐specific expression profile, or to search for genes that follow a specific expression profile.
The obtained expression profiles can be exported as image files (heat maps) or data list files. Because all gene identifiers are linked to PLAZA3.0 (Proost et al., 2015), each query results automatically and in parallel with the expression profiles in a gene ontology (GO) enrichment and shows the Arabidopsis homologues for the selected genes, which can also be easily exported as tabular data.
Transcriptional changes within the division and expansion zone discriminate each zone into a basal and distal part
Because the two controls (well‐watered and nontransgenic GA20OX‐1 OE plants) were both in the B104 background and all 0.5‐cm samples contained the same zones (Figure 1), their data sets were merged and statistically analysed together. 1020 genes were significantly up‐regulated along the leaf gradient (FC > 2; FDR < 0.05), of which 611 between the EZ and the MZ, and 1055 genes were significantly down‐regulated (FC < −2; FDR < 0.05), of which 661 between the EZ and the MZ (Table S1).
Transcriptional changes were observed between the basal part and the more distal part of the DZ. The genes that were more highly expressed in the basal part were enriched in nucleotide and amino acid biosynthesis and transcriptional regulation (Figure 6; Table S1c). Ten transcription factors were specifically up‐regulated at the base of the DZ, among which four Dof (DNA‐binding with one finger) zinc finger transcription factors, GROWTH REGULATING FACTOR15, three TGACG SEQUENCE_SPECIFIC BINDING PROTEIN (TGA) transcription factors (among which FASCIATED EAR4 (FEA4) and LIGULELESS2), BEL1‐like homeodomain transcription factor and ETHYLENE REGULATED FACTOR1 (Table S1c). Conversely, the genes that were less expressed in the basal half cm compared with the more distal half cm of the DZ were involved in GA‐mediated signalling (gibberellin 3‐beta‐dioxygenase 2‐2 and GATA transcription factor 22) and translation (Figure 6; Table S1b).
Figure 6.
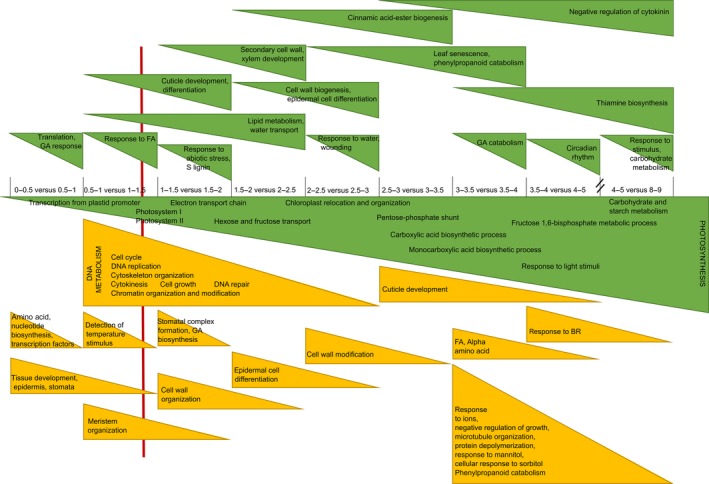
Schematic overview of the main categories of transcriptional changes between the sequential samples (Figure 1; 0–9 cm from the leaf base) in a growing maize leaf in well‐watered conditions. The GO categories of transcripts that were up‐regulated and therefore associated with the maturity of the tissue are indicated in green, and the GO categories of transcripts that were down‐regulated are indicated in yellow. The red vertical line indicates the transition zone between cell division and cell expansion (TZ1). GO enrichment is characterized by PLAZA.
Between the DZ and TZ1, genes with a function in ‘tissue development’, ‘epidermis’, ‘stomata’, ‘cell fate’, ‘growth’ and ‘maintenance of meristematic identity’ were down‐regulated. Towards the TZ1, the processes of ‘DNA replication’, ‘chromatin assembly’, ‘chromosome organization’, ‘cytokinesis’ and ‘cell cycle’ were enriched among the down‐regulated genes relative to the DZ samples (Figure 6; Table S1b). The down‐regulation of genes involved in ‘DNA metabolism’, ‘cell cycle’ and ‘cytokinesis’ persisted till after TZ1, whereas the process of ‘meristem organization’ was specifically down‐regulated at TZ1, and ‘GA biosynthesis’ (gibberellin 3‐beta‐dioxygenase 2‐2) dropped directly after TZ1. The sample containing TZ1 was also characterized by the up‐regulation of distinct transport pathways (oligopeptide, peptides, amide and water) and the GO categories ‘response to abiotic stimulus’, ‘light intensity’, ‘(far) red’, ‘UV‐A’, ‘fatty acids’, ‘blue light’ and ‘radiation’. In the early EZ samples, relative to the TZ1 sample, genes involved in ‘lignin biosynthesis’, ‘biogenesis of cinnamic esters’ and ‘secondary metabolism’ were up‐regulated, indicating that lignification starts soon after TZ1. In the later EZ, many genes involved in ‘phenylpropanoid metabolism’ and ‘cell wall organization’ were up‐regulated as compared with the early EZ samples. Towards the distal part of the EZ and TZ2, genes involved in ‘leaf senescence’ and ‘negative regulation of cytokinin’ and ‘GA catabolism’ were up‐regulated relative to the early EZ samples (Figure 6; Table S1c). Towards the distal EZ and the MZ, compared with the early EZ samples, genes involved in response to ions (magnesium, calcium and potassium) and mannitol and sorbitol were significantly down‐regulated, while genes involved in ‘thiamine biosynthesis’ were up‐regulated. In the MZ, ‘growth’, ‘protein polymerization’, ‘microtubule organization’ and ‘response to brassinosteroids’ were down‐regulated relative to the EZ.
Remarkably, the category ‘photosynthesis’ was significantly up‐regulated at all comparisons between successive samples from the leaf base to the MZ, starting within the DZ. A distinction could be made between different subprocesses of photosynthesis as cells progressed through the different zones. At the leaf base, genes involved in the biosynthesis of precursors of photosynthetic pigments were already expressed, and ‘transcription from plastid promoters’ was up‐regulated at the basal half cm compared with the next half cm. Around TZ1, an enrichment in the genes involved in light harvesting through photosystem I (PS I) and subsequently PS II was observed, relative to the DZ samples. The GO categories ‘chloroplast ribulose bisphosphate carboxylase complex biogenesis’ and ‘electron transport chain’ were up‐regulated from TZ1 to EZ, followed by ‘chloroplast relocation and organization’ between TZ1 and more basal EZ. Towards the end of the distal EZ, the GO categories ‘pentose‐phosphate shunt’ and ‘carboxylic acid biosynthetic process’ were up‐regulated relative to the early EZ. At TZ2, genes involved in the ‘circadian rhythm’ were significantly up‐regulated, while the genes involved in the actual production of sugars and starch were up‐regulated towards the MZ as compared with earlier samples (Figure 6; Table S1b and c).
Mild drought stimulates a transcriptional reprogramming and lowers photosynthesis in the division zone and extends cell division at the transition zone
A principle component analysis (PCA) using all samples normalized relative to the wild‐type samples showed that the majority of the transcriptional changes could be explained by six principle components (Figure 7a). The genes that positively contributed to the first principle component (PC1), which separates the samples according to their position along the leaf, were enriched for the GO categories ‘photosynthesis’, ‘response to abiotic stimulus’, ‘very long‐chain fatty acid metabolic process’ and ‘secondary cell wall biogenesis’. The genes that negatively affected PC1 were enriched in ‘DNA replication’, ‘cell cycle’ and ‘nucleosome assembly and organization’. PC2 also divided the samples according to their position along the leaf, but was the highest for the samples in the basal EZ compared with the samples in the DZ and the MZ. The genes that positively contributed to PC2 were enriched for ‘lipid metabolic process’, ‘response to osmotic stress’, whereas the cells that negatively contributed were mainly involved in ‘leaf senescence’, ‘response to jasmonic acid’ and ‘response to oxidative stress’. Both PC1 and PC2 caused a shift in the mild drought, control and GA20OX‐1 OE samples, a difference that was opposite between mild drought and GA20OX‐1 OE (Figure 7b) and that corresponded to the microscopically determined position of the TZs.
Figure 7.
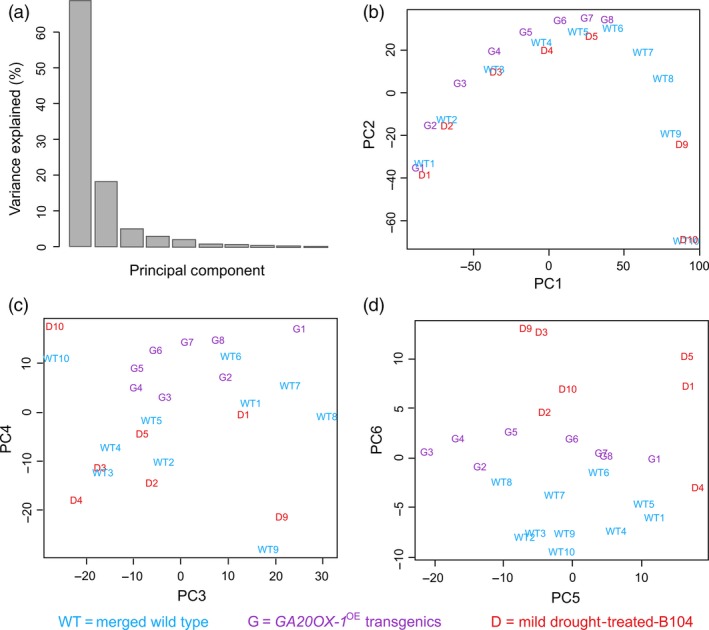
Principal component analysis (PCA) of the transcriptomics data. (a) Bar chart showing the explained variance for each component of PCA. (b–d) PCA 1, 2, 3, 4, 5 and 6 representing the classifications of the transcriptomics data. The numbers indicate the leaf samples: 1–10 as indicated in Figure 1 with mild drought (D), GA20OX‐1 OE (G) and the merged well‐watered and wild‐type control samples (WT).
The genes that contributed to PC4 that discriminated between the DZ, EZ and MZ samples were enriched for ‘proline transport’, ‘lipid’ and ‘phenylpropanoid metabolic process’ (Figure 7c). Remarkably, it was mainly PC6 that discriminated the drought samples from all well‐watered samples, indicating that the transcriptional changes over the gradient were much more prominent than the ones due to drought or GA20OX‐1 OE. The genes that contributed to PC6 in the same direction as the mild drought classification were enriched in a plethora of ‘responses to abiotic stimulus’ (including heat and reactive oxygen species), whereas the oppositely regulated genes were mainly involved in the ‘carbohydrate catabolic process’ (Figure 7d).
LGV can also be used to narrow down the number of genes identified, because of chained queries (a query within a query). In this way, we searched for genes that have opposite expression profiles in TZ1 in plants grown under drought conditions and in the GA20OX‐1 OE line. However, because the position of TZ1 changes in opposite direction in the mild drought and GA20OX‐1 OE samples (Figure 1), above comparison is likely to reflect mainly developmentally regulated genes. To resolve this, we performed a shift in both experiments, based on the cellular analyses that were used to determine the position of TZ1 (Table 1). As a consequence of the alignment of TZ1, all other zones shifted as well.
Few genes were specifically differentially expressed in GA20OX‐1 OE plants (and not under mild drought) as compared with the wild type (16 or 1 genes at the DZ, 4 or 7 genes at TZ1 and 5 or 10 genes at the EZ, respectively; Table S2a and b).
In contrast, more genes, but still a modest set of genes, were specifically differentially expressed by mild drought (Table S2c and e): 140 and 76 at the DZ, 180 and 206 at TZ1 and 103 and 84 at the EZ, respectively. The genes that were up‐regulated by mild drought in the DZ were enriched for ‘regulation of gene expression’, ‘response to light intensity’, ‘response to reactive oxygen species’, ‘leaf morphogenesis’ and ‘stomatal complex formation’ (Table S2f). The genes involved in the ‘regulation of gene expression’ were several Dof transcription factors, two TGAs, ethylene response factor1 (ERF1), NAC67, SPEECHLESS, KNOTTED1 and a B3 domain‐containing transcription factor, of which some were also identified to be specific in the basal DZ (Table S1). Remarkably, at TZ1 and EZ, the GO categories ‘DNA replication’ and ‘cell cycle’ were significantly enriched among the genes that were up‐regulated by drought. These genes were mainly targets of E2F/DP because a significant enrichment was obtained of the TZ1 mild drought up‐regulated genes (31 of the 180) that showed homology to Arabidopsis genes that were computationally and experimentally shown to be E2F/DP targets (Table S3; Vandepoele et al., 2005; Verkest et al., 2014). At the EZ, several histones (H4, H2A, H2B) were significantly up‐regulated under mild drought, together with genes involved in DNA replication and cell cycle (Table S2e).
Among the drought‐mediated down‐regulated genes at the DZ are GA3‐oxidase (consistent with our Q‐PCR data, Figure 4), proline dehydrogenase involved in proline catabolism, and two transcription factors that were already associated with abiotic stress responses (Table S2c). Both at the DZ and TZ1, genes involved in PS II, RuBisCO (ribulose bisphosphate carboxylase) complex biogenesis and light harvesting complex were down‐regulated upon mild drought (Table S2d) and at TZ1 and EZ several aquaporins (PIP2‐3, PIP2‐4 and PIP2‐5) were significantly down‐regulated.
Discussion
High‐resolution sampling reveals the complex regulation of maize leaf growth
Kinematic analysis allows to quantify the contribution of cell division or cell expansion to a given leaf growth phenotype and to delineate zones in which cells are dividing (DZ) or expanding (EZ), until they reach their mature size (MZ; Nelissen et al., 2013, 2012). However, our data show that this rather simplistic growth model that merely discriminates between cell division, cell expansion and cell maturity is more complex. High‐resolution transcriptome analysis and quantification of hormones showed that within the DZ and EZ, there are substantial differences between the basal and distal parts of these zones. Indeed, the levels of auxin and tZ declined throughout the division zone, to reach their basal level around TZ1 (this work; Nelissen et al., 2012), reinforcing the concept that cells in the basal and distal parts of the DZ are molecularly distinct, whereas the presence of mitotic figures in the kinematic analysis (Nelissen et al., 2013) classified them all as dividing cells. These data suggest that there are gradients of molecular processes within the growth zone that govern cell cycle progression. It is likely that such gradients are at least partially mediated by diffusible factors, such as auxin, originating from the leaf base and gradually diminishing in concentration when cells are getting displaced further away from the leaf base.
This concept is also exemplified by a number of transcription factors that were specifically expressed in the basal half cm of the growing maize leaf, of which some were previously described to have a role in leaf development. The BEL1 domain‐containing protein, which is an orthologue of the Arabidopsis BELLRINGER, and FEA4, which is orthologous to Arabidopsis PERANTHIA, were both shown to regulate AGAMOUS (Bao et al., 2004; Pautler et al., 2015). The expression of FEA4 during leaf growth coincides with the position where the highest levels of auxin were observed and FEA4 was shown to regulate many different transcription factors involved in leaf differentiation and polarity (Pautler et al., 2015). Other genes that are specifically highly expressed in the basal half cm are LIGULELESS‐2, which is involved in leaf patterning along the proximo‐distal axis (Walsh et al., 1998) and GROWTH REGULATING FACTOR15 (GRF15). Previously, we have shown that GRF15 was more highly expressed in the DZ compared with the EZ and that that GRF15 protein was identified as a significantly enriched part of the AN3‐mediated chromatin remodelling SWI/SNF complex in the DZ (Nelissen et al., 2015).
The fine sampling strategy also underlines the importance of TZ1 as an integration point of molecular cues because both at the transcript and at the hormone level, numerous changes occur exactly at TZ1. Previously, the local accumulation of GA at TZ1 resulted in a more distal shift of TZ1 and hence in more cells in the DZ (Nelissen et al., 2012). Here, we show that the more proximal shift of TZ1 under mild drought was associated with lowered levels of bioactive GA, which was also observed when analysing the biosynthetic intermediates and transcript levels of GA biosynthetic enzymes.
Some processes, such as photosynthesis, were found to be transcriptionally regulated in a gradient throughout the entire growth zone, although the high‐resolution sampling allows for discriminating subprocesses that show spatial specificity. At the leaf base, in the DZ, an up‐regulation was observed of genes enriched in the transcription from plastid promoters. Genes involved in plastid gene expression and the regulation of photosynthesis typically anticorrelate with final leaf size and timing of growth‐related parameters (Baute et al., 2015, 2016). Already at the leaf base, which is shielded from light by the surrounding older leaves, cells are preparing for their final role in photosynthesis, showing that the basal part of the growing leaf is instrumental to determine the final size and shape, as well as its role as energy source of the plant.
Perturbations of the organization of the growth zone allow to identify novel mechanisms
Positively and negatively disturbing the organization of the growth zone allowed the identification of novel mechanisms. Remarkably, although the final level of the cytokinin iP was the same in all examined conditions, its level transiently increased in a zone corresponding to the TZ2, which shifted, together with TZ1, more basally or more distally, under mild drought and in GA20OX‐1 OE, respectively. As cytokinins have been shown to promote shoot development, to inhibit leaf senescence and to delay differentiation (Kieber and Schaller, 2014), it is unlikely that the increase in cytokinin is causing the transition between cell expansion and maturation. On the other hand, it is known that cytokinin influences chloroplast ultrastructure, number and biosynthesis (Cortleven and Schmülling, 2015). Because the increase in cytokinin occurs at a position that is close to the point of leaf emergence from the sheet and that thus might already perceive incident light, we hypothesize that towards the end of the EZ, cytokinin biosynthesis is activated to steer the development and activity of chloroplasts. This increase in cytokinin levels is concomitant with an increased expression in multiple successive samples in the EZ and MZ of a gene encoding an SCF‐type F‐box protein that is the orthologue of KISS ME DEADLY‐LIKE2 (KMD2). Elevated KMD2 expression targets the B‐type Arabidopsis response regulators (ARRs), key transcription factors in the cytokinin response for degradation (Kim et al., 2013).
SA was found to gradually accumulate within the DZ, a phenomenon that became more apparent when the levels of SA were elevated under mild drought. An increase in endogenous levels of SA promotes stomatal closure and drought tolerance (Miura et al., 2013; Miura and Tada, 2014) and SA can influence both plant growth and drought tolerance in a dose‐dependent manner (Kang et al., 2012, 2013; Miura and Tada, 2014). Because stomata start to form within the DZ, the up‐regulation of SA in the DZ might reflect a role of SA in stomatal closure. The extended high levels of SA in the expanding and the mature part of the leaf might be more related to the antioxidant defence system of SA to protect against oxidative damage caused by drought (Miura and Tada, 2014).
Although ABA and GA have been reported to act antagonistically during germination, growth and flower development (Weiss and Ori, 2007; White et al., 2000), ABA was significantly up‐regulated in the GA20OX‐1 OE transgenic plants, albeit to the same level as under mild drought. Because high levels of ABA inhibit GA biosynthesis (Toh et al., 2008), the elevated ABA levels in the GA20OX‐1 OE plants might serve as a negative feedback loop to limit the maximal GA levels that can be obtained in the plants. Conversely, the drought‐evoked increase in ABA levels might result in an inhibition of GA biosynthesis, resulting in the lower levels of bioactive GA and its precursors under mild drought.
Growth reduction by mild drought stress cannot be reversed by elevated GA levels
Because overexpression of the rate‐limiting enzyme GA20‐OXIDASE cannot render plants relatively more tolerant to mild drought, a comparative analysis was performed to identify transcripts that were differentially expressed specifically under mild drought and not in GA20OX‐1 OE transgenic plants. Remarkably, five of the ten transcription factors that were highly expressed at the leaf base in normal conditions were significantly up‐regulated under mild drought. One of the up‐regulated Dof (Dof22) transcription factor genes has previously been shown to be up‐regulated in maize seedlings under salt treatment (Chen and Cao, 2015). The ERF1 protein is orthologous to RELATED TO APETALA2‐2 (RAP2.2), for which it has been shown that elevated levels sustain ABA‐mediated activation of stress response genes (Papdi et al., 2015). Also, the expression of KNOTTED1 (KN1) is specifically up‐regulated by mild drought in the DZ. Interestingly, KN1 has been shown to negatively modulate the accumulation of GA through the control of GA2‐OXIDASE and overexpression of KN1 results in smaller plants (Bolduc and Hake, 2009), raising the possibility that the up‐regulated KN1 expression might be involved in the drought‐mediated growth reduction.
Genes involved in proline accumulation, photosynthesis and aquaporins were specifically down‐regulated by drought stress. The down‐regulation of PROLINE DEHYDROGENASE (ProDH) during stress is widely accepted to promote proline accumulation (Verslues and Sharma, 2010). Our results are consistent with the down‐regulation of ProDH at the transition from cell division to cell expansion under mild drought in Arabidopsis (Clauw et al., 2015). Also, genes involved in PS II and the light harvesting complex were down‐regulated by mild drought at the DZ and TZ1, which is in line with the observation that drought stress results in damage to PS II and photochemistry in mature leaves (Lu and Zhang, 1999; Sperdouli and Moustakas, 2012). Transcriptomics data also revealed that several genes encoding PLASMA MEMBRANE INTRINSIC PROTEINS (PIPs), belonging to aquaporins that facilitate the water diffusion across cell membranes, were significantly down‐regulated at TZ1 and EZ by drought stress. Under water stress, the expression of several PIPs in leaves is down‐regulated, and ABA represses aquaporin activity (Alexandersson et al., 2005; Shatil‐Cohen et al., 2011). The down‐regulation of aquaporins might be a way for plants to minimize water flow through cell membranes and to maintain leaf turgor, which is required for both cell division and cell expansion (Chaumont and Tyerman, 2014).
Although increased GA levels could not compensate the mild drought‐induced growth reduction, the growth advantage of leaves from GA20OX‐1 OE plants was maintained. The leaf length of GA20OX‐1 OE plants under mild drought still exceeded that of the well‐watered nontransgenic siblings. These data indicate that the effects of growth‐promoting genes can still persist under mild abiotic stress conditions, although the genes are not necessarily involved in stress tolerance or survival.
Plants grown under mild drought stress have adapted their growth, molecularly and cellularly
Our kinematic analysis showed that the cell division rate and the cell cycle duration were not significantly affected by mild drought, and also at the transcript level, no differential expression of cell cycle‐related genes was observed at the DZ. However, effects on cell division rate and down‐regulation of cell cycle genes have been observed in other studies that examined the effect of drought in the maize leaf (Avramova et al., 2017, 2015), indicating that the significance of differential expression of cell cycle genes might depend on the applied drought stress or additional environmental changes between different growth chambers. Alternatively, our high‐resolution analysis and the alignment of the samples according to the position of the TZ1 might explain some of the differences. By aligning the samples according to the TZ1, transcripts that are known downstream targets of E2F/DP transcription factors, which are well‐known negative regulators of the G1/S transition whose ectopic expression results in an increased cell division duration (De Veylder et al., 2002), were significantly up‐regulated at TZ1 and the basal EZ under mild drought during steady‐state growth. These data suggest that the mild drought‐induced expression of E2F/DP targets around TZ1 might provide the possibility to maintain the capacity to resume growth upon water availability.
During osmotic stress in Arabidopsis leaves, the initial reduction in cell division is in part compensated by meristemoid divisions, generating extra pavement cells while forming stomata (Bergmann and Sack, 2007; Geisler et al., 2000; Skirycz et al., 2011). Unlike Arabidopsis, where the meristemoid divisions contribute substantially to the final leaf size (Gonzalez et al., 2012), the polarized stomatal divisions in maize barely affect it (Larkin et al., 1997). Instead, our data show that the prolonged duration of leaf growth (or LED), at least partly, compensates for the mild drought‐induced growth reduction. More studies will be needed to examine the biological relevance of the prolonged LED as a compensation for the drought‐induced growth reduction, and to analyse whether a prolonged LED allows to resume growth upon re‐watering. Recently, we identified PLA1, a cytochrome P450 78A, as one of the key players involved in the prolonged LED under mild drought and cold stress (Sun et al., 2017).
At the molecular level, the effects of the mild drought treatment only explained approximately 1 % of the variation in the transcriptomics data, indicating that the changes over the leaf gradient explained the majority of the transcriptional variation rather than the drought treatment. Alternatively, the plants that were subjected to mild drought throughout their entire life most likely adapted their metabolism, resulting in limited differences at the molecular level to be observed as opposed to acute drought stress responses. The imposed stress most likely does not exceed the tolerance limitations but is mild enough to not cause permanent damage, but to rather evoke a new physiological homeostasis (Gaspar et al., 2002; Pinheiro and Chaves, 2011; Verslues et al., 2006).
Leaf Growth Viewer allows querying transcriptome changes over the leaf growth gradient under mild drought stress and at elevated GA levels
To date, many different research groups use the maize leaf as a model, albeit with a different biological question (Bonhomme et al., 2012; Facette et al., 2013; Jaskiewicz et al., 2011; Li et al., 2010; Nelissen et al., 2012; Ponnala et al., 2014; Tausta et al., 2014; van Wijk et al., 2014; Zhang et al., 2014). We developed a user‐friendly and searchable tool, LGV, to visualize our transcriptomics data. LGV provides a scaffold that can be further developed to integrate multiple data sets and analytical tools. Recently, the AIM database in Arabidopsis (Wang et al., 2014) exemplified the need and power of integrating big data sets. Genes that are transcriptionally coordinated are often functionally related, indicating the importance to use different transcriptome studies to identify co‐expressed genes (De Bodt et al., 2010; Hansen et al., 2014). It has been shown that a comparative analysis of co‐expression networks over different species offers additional value to remove false positives and to increase the power of predictions (Hansen et al., 2014). In addition, proteins can interact with many different interactors and thereby often affect distinct processes (Tucker et al., 2001). Because more and more experimental data about proteome (Facette et al., 2013), metabolome (Wang et al., 2014) and protein–protein interactions (Bommert et al., 2013; Nelissen et al., 2015) become available in the maize leaf, together with the wealth of transcriptomics data (Li et al., 2010; Mattiello et al., 2014; Tausta et al., 2014; Wang et al., 2014), it would be a great opportunity to boost the maize leaf as a model system to understand organ growth in monocots by integrating these different levels of information. Such a multilevel integration, ultimately even complemented with phenotypes, could use LGV as a starting point.
Experimental procedures
Growth conditions
All experiments were executed in growth chambers with controlled relative humidity (55%), temperature (24 °C) and light intensity (170 mmol/m2/s photosynthetically active radiation at plant level) in a 16‐h/8‐h (day/night) cycle. For mild drought treatment, the water content was allowed to drop to a soil water content of 70% of the well‐watered condition, corresponding to a soil water potential of −0.518 and −0.023 MPa, respectively.
Hormone profiling and microarray analysis
For hormone profiling, sampling, extraction, purification and hormone metabolic profiling were performed as described in Nelissen et al. (2012). The details of the microarray analysis are explained in Data S1.
Leaf growth viewer
The back‐end of LGV is written in Python using the Django framework (https://www.djangoproject.com/), which connects to a MySQL database (https://www.mysql.com/). The dynamic HTML front‐end are JavaScript‐based, interactive charts using HighCharts (http://www.highcharts.com/).
Conflicts of interest
No conflicts of interest are to be declared.
Supporting information
Table S1 The differentially expressed transcripts along the leaf gradient in the merged well‐watered and non‐transgenic GA20OX‐1 OE control data set.
Table S2 The specifically differentially expressed transcripts in GA20OX‐1 OE transgenic plants under drought conditions.
Table S3 List of E2F/DP targets that were specifically up‐regulated at the TZ1 by mild drought stress.
Data S1 Details of the microarray analysis.
Acknowledgements
The research was funded by the European Research Council under the European Community's Seventh Framework Programme [FP7/2007‐2013] under ERC Grant Agreement N° [339341‐AMAIZE]11, Ghent University (‘Bijzonder Onderzoeksfonds Methusalem Project’ No. BOF08/01M00408 and Multidisciplinary Research Partnership ‘Biotechnology for a Sustainable Economy’ Grant 01MR0510W), the Interuniversity Attraction Poles Programme (IUAP P7/29 ‘MARS’) initiated by the Belgian Science Policy Office. X.S. is funded by a PhD grant from the Chinese Scholarship Council (CSC). We thank Dr. Annick Bleys for the critical reading of the manuscript.
References
- Achard, P. , Gusti, A. , Cheminant, S. , Alioua, M. , Dhondt, S. , Coppens, F. , Beemster, G.T.S. et al (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis . Curr. Biol. 19, 1188–1193. [DOI] [PubMed] [Google Scholar]
- Alexandersson, E. , Fraysse, L. , Sjövall‐Larsen, S. , Gustavsson, S. , Fellert, M. , Karlsson, M. , Johanson, U. et al (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484. [DOI] [PubMed] [Google Scholar]
- Andriankaja, M. , Dhondt, S. , De Bodt, S. , Vanhaeren, H. , Coppens, F. , De Milde, L. , Mühlenbock, P. et al (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not‐so‐gradual process. Dev. Cell, 22, 64–78. [DOI] [PubMed] [Google Scholar]
- Avramova, V. , AbdElgawad, H. , Zhang, Z. , Fotschki, B. , Casadevall, R. , Vergauwen, L. , Knapen, D. et al (2015) Drought induces distinct growth response, protection, and recovery mechanisms in the maize leaf growth zone. Plant Physiol. 169, 1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, V. , AbdElgawad, H. , Vasileva, I. , Petrova, A.S. , Holek, A. , Mariën, J. , Asard, H. et al (2017) High antioxidant activity facilitates maintenance of cell division in leaves of drought tolerant maize hybrids. Front. Plant Sci. 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, X. , Franks, R.G. , Levin, J.Z. and Liu, Z. (2004) Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell, 16, 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baute, J. , Herman, D. , Coppens, F. , De Block, J. , Slabbinck, B. , Dell'Acqua, M. , Pè, M.E. et al (2015) Correlation analysis of the transcriptome of growing leaves with mature leaf parameters in a maize RIL population. Genome Biol. 16, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baute, J. , Herman, D. , Coppens, F. , De Block, J. , Slabbinck, B. , Dell'Acqua, M. , Pè, M.E. et al (2016) Combined large‐scale phenotyping and transcriptomics in maize reveals a robust growth regulatory network. Plant Physiol. 170, 1848–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, D.C. and Sack, F.D. (2007) Stomatal development. Annu. Rev. Plant Biol. 58, 163–181. [DOI] [PubMed] [Google Scholar]
- Bolduc, N. and Hake, S. (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1 . Plant Cell, 21, 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert, P. , Je, B.I. , Goldshmidt, A. and Jackson, D. (2013) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature, 502, 555–558. [DOI] [PubMed] [Google Scholar]
- Bonhomme, L. , Valot, B. , Tardieu, F. and Zivy, M. (2012) Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol. Cell Proteomics, 11, 957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger, H. and Lenhard, M. (2010) Control of tissue and organ growth in plants. Curr. Top. Dev. Biol. 91, 185–220. [DOI] [PubMed] [Google Scholar]
- Chaumont, F. and Tyerman, S.D. (2014) Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 164, 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. and Cao, J. (2015) Comparative analysis of Dof transcription factor family in maize. Plant Mol. Biol. Rep. 33, 1245–1258. [Google Scholar]
- Clauw, P. , Coppens, F. , De Beuf, K. , Dhondt, S. , Van Daele, T. , Maleux, K. , Storme, V. et al (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 167, 800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven, A. and Schmülling, T. (2015) Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66, 4999–5013. [DOI] [PubMed] [Google Scholar]
- Coussens, G. , Aesaert, G. , Verelst, W. , Demeulenaere, M. , De Buck, S. , Njuguna, E. , Inzé, D. et al (2012) Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J. Exp. Bot. 63, 4263–4273. [DOI] [PubMed] [Google Scholar]
- Czesnick, H. and Lenhard, M. (2015) Size control in plants—lessons from leaves and flowers. Cold Spring Harb. Perspect. Biol. 7, a019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt, S. , Carvajal, D. , Hollunder, J. , Van den Cruyce, J. , Movahedi, S. and Inzé, D. (2010) CORNET: a user‐friendly tool for data mining and integration. Plant Physiol. 152, 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L. , Beeckman, T. , Beemster, G.T.S. , de Almeida Engler, J. , Ormenese, S. , Maes, S. , Naudts, M. et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa‐DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch, S. , Anastasiou, E. , Sharma, V.K. , Laux, T. , Fletcher, J.C. and Lenhard, M. (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage‐dependent manner. Curr. Biol. 16, 272–279. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M. , Bonetta, D. , Tsukaya, H. , Dengler, R.E. and Dengler, N.G. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis . Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Facette, M.R. , Shen, Z. , Björnsdóttir, F.R. , Briggs, S.P. and Smith, L.G. (2013) Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell, 25, 2798–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, B.R. , McMurray, J.M. , Fonger, T.M. , Main, M.L. , Taylor, K.W. , Torney, F.J. , Paz, M.M. et al (2006) Improved Agrobacterium‐mediated transformation of three maize inbred lines using MS salts. Plant Cell Rep. 25, 1024–1034. [DOI] [PubMed] [Google Scholar]
- Gaspar, T. , Franck, T. , Bisbis, B. , Kevers, C. , Jouve, L. , Hausman, J.F. and Dommes, J. (2002) Concepts in plant stress physiology. Application to plant tissue cultures. Plant Growth Regul. 37, 263–285. [Google Scholar]
- Geisler, M. , Nadeau, J. and Sack, F.D. (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell, 12, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, N. , De Bodt, S. , Sulpice, R. , Jikumaru, Y. , Chae, E. , Dhondt, S. , Van Daele, T. et al (2010) Increased leaf size: different means to an end. Plant Physiol. 153, 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, N. , Vanhaeren, H. and Inzé, D. (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. [DOI] [PubMed] [Google Scholar]
- Hansen, B.O. , Vaid, N. , Musialak‐Lange, M. , Janowski, M. and Mutwil, M. (2014) Elucidating gene function and function evolution through comparison of co‐expression networks in plants. Front. Plant Sci. 5, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi, G. , Kim, G.‐T. and Tsukaya, H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana . Plant J. 43, 68–78. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Xie, Q. and Chua, N.‐H. (2003) The Arabidopsis auxin‐inducible gene ARGOS controls lateral organ size. Plant Cell, 15, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz, M. , Peterhansel, C. and Conrath, U. (2011) Detection of histone modifications in plant leaves. J. Vis. Exp. 55, e3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, G. , Li, G. , Xu, W. , Peng, X. , Han, Q. , Zhu, Y. and Guo, T. (2012) Proteomics reveals the effects of salicylic acid on growth and tolerance to subsequent drought stress in wheat. J. Proteome Res. 11, 6066–6079. [DOI] [PubMed] [Google Scholar]
- Kang, G.Z. , Li, G.Z. , Liu, G.Q. , Xu, W. , Peng, X.Q. , Wang, C.Y. , Zhu, Y.J. et al (2013) Exogenous salicylic acid enhances wheat drought tolerance by influence on the expression of genes related to ascorbate‐glutathione cycle. Biol. Plant. 57, 718–724. [Google Scholar]
- Kazama, T. , Ichihashi, Y. , Murata, S. and Tsukaya, H. (2010) The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5‐dependent mobile growth factor in developing leaves of Arabidopsis thaliana . Plant Cell Physiol. 51, 1046–1054. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J. and Schaller, G.E. (2014) Cytokinins. Arabidopsis Book, 12, e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J. , Chiang, Y.‐H. , Kieber, J.J. and Schaller, G.E. (2013) SCFKMD controls cytokinin signaling by regulating the degradation of type‐B response regulators. Proc. Natl Acad. Sci. USA, 110, 10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C. , Marks, M.D. , Nadeau, J. and Sack, F. (1997) Epidermal cell fate and patterning in leaves. Plant Cell, 9, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zheng, L. , Corke, F. , Smith, C. and Bevan, M.W. (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana . Genes Dev. 22, 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Ponnala, L. , Gandotra, N. , Wang, L. , Si, Y. , Tausta, S.L. , Kebrom, T.H. et al (2010) The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Liu, K. , Goodman, M. , Muse, S. , Smith, J.S. , Buckler, E. and Doebley, J. (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics, 165, 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C. and Zhang, J. (1999) Effects of water stress on photosystem II photochemistry and its thermostability in wheat plants. J. Exp. Bot. 50, 1199–1206. [Google Scholar]
- Mattiello, L. , Begcy, K. , da Silva, F.R. , Jorge, R.A. and Menossi, M. (2014) Transcriptome analysis highlights changes in the leaves of maize plants cultivated in acidic soil containing toxic levels of Al3+ . Mol. Biol. Rep. 41, 8107–8116. [DOI] [PubMed] [Google Scholar]
- Miura, K. and Tada, Y. (2014) Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Okamoto, H. , Okuma, E. , Shiba, H. , Kamada, H. , Hasegawa, P.M. and Murata, Y. (2013) SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid‐induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 73, 91–104. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y. and Fischer, R.L. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl Acad. Sci. USA, 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, U. , Crawford, B.C.W. , Carpenter, R. and Coen, E. (2003) Genetic control of surface curvature. Science, 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Nelissen, H. , Rymen, B. , Jikumaru, Y. , Demuynck, K. , Van Lijsebettens, M. , Kamiya, Y. , Inzé, D. et al (2012) A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 22, 1183–1187. [DOI] [PubMed] [Google Scholar]
- Nelissen, H. , Rymen, B. , Coppens, F. , Dhondt, S. , Fiorani, F. and Beemster, G.T.S. (2013) Kinematic analysis of cell division in leaves of mono‐ and dicotyledonous species: a basis for understanding growth and developing refined molecular sampling strategies. Methods Mol. Biol. 959, 247–264. [DOI] [PubMed] [Google Scholar]
- Nelissen, H. , Eeckhout, D. , Demuynck, K. , Persiau, G. , Walton, A. , van Bel, M. , Vervoort, M. et al (2015) Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell, 27, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen, H. , Gonzalez, N. and Inzé, D. (2016) Leaf growth in dicots and monocots: so different yet so alike. Curr. Opin. Plant Biol. 33, 72–76. [DOI] [PubMed] [Google Scholar]
- Papdi, C. , Pérez‐Salamó, I. , Joseph, M.P. , Giuntoli, B. , Bögre, L. , Koncz, C. and Szabados, L. (2015) The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3 . Plant J. 82, 772–784. [DOI] [PubMed] [Google Scholar]
- Pautler, M. , Eveland, A.L. , LaRue, T. , Yang, F. , Weeks, R. , Lunde, C. , Je, B.I. et al (2015) FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell, 27, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, C. and Chaves, M.M. (2011) Photosynthesis and drought: can we make metabolic connections from available data? J. Exp. Bot. 62, 869–882. [DOI] [PubMed] [Google Scholar]
- Ponnala, L. , Wang, Y. , Sun, Q. and van Wijk, K.J. (2014) Correlation of mRNA and protein abundance in the developing maize leaf. Plant J. 78, 424–440. [DOI] [PubMed] [Google Scholar]
- Proost, S. , Van Bel, M. , Vaneechoutte, D. , Van de Peer, Y. , Inzé, D. , Mueller‐Roeber, B. and Vandepoele, K. (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43, D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatil‐Cohen, A. , Attia, Z. and Moshelion, M. (2011) Bundle‐sheath cell regulation of xylem‐mesophyll water transport via aquaporins under drought stress: a target of xylem‐borne ABA? Plant J. 67, 72–80. [DOI] [PubMed] [Google Scholar]
- Skirycz, A. , Claeys, H. , De Bodt, S. , Oikawa, A. , Shinoda, S. , Andriankaja, M. , Maleux, K. et al (2011) Pause‐and‐stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell, 23, 1876–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani, R. and Iyer‐Pascuzzi, A. (2014) Postembryonic control of root meristem growth and development. Curr. Opin. Plant Biol. 17, 7–12. [DOI] [PubMed] [Google Scholar]
- Sperdouli, I. and Moustakas, M. (2012) Differential response of photosystem II photochemistry in young and mature leaves of Arabidopsis thaliana to the onset of drought stress. Acta Physiol. Plant. 34, 1267–1276. [Google Scholar]
- Sun, X. , Cahill, J. , Van Hautegem, T. , Feys, K. , Whipple, C. , Novák, O. , Delbare, S. et al (2017) Altered expression of maize PLASTOCHRON1 enhances biomass and seed yield by extending cell division duration. Nat. Commun. 8, 14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu, F. , Reymond, M. , Hamard, P. , Granier, C. and Muller, B. (2000) Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J. Exp. Bot. 51, 1505–1514. [DOI] [PubMed] [Google Scholar]
- Tardieu, F. , Granier, C. and Muller, B. (2011) Water deficit and growth. Co‐ordinating processes without an orchestrator? Curr. Opin. Plant Biol. 14, 283–289. [DOI] [PubMed] [Google Scholar]
- Tausta, S.L. , Li, P. , Si, Y. , Gandotra, N. , Liu, P. , Sun, Q. , Brutnell, T.P. et al (2014) Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4‐related processes. J. Exp. Bot. 65, 3543–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, S. , Imamura, A. , Watanabe, A. , Nakabayashi, K. , Okamoto, M. , Jikumaru, Y. , Hanada, A. et al (2008) High temperature‐induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146, 1368–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, C.L. , Gera, J.F. and Uetz, P. (2001) Towards an understanding of complex protein networks. Trends Cell Biol. 11, 102–106. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K. , Vlieghe, K. , Florquin, K. , Hennig, L. , Beemster, G.T.S. , Gruissem, W. , Van de Peer, Y. et al (2005) Genome‐wide identification of potential plant E2F target genes. Plant Physiol. 139, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen, L. , Verkest, A. , Gonzalez, N. , Heyndrickx, K.S. , Eeckhout, D. , Han, S.‐K. , Jégu, T. et al (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell, 26, 210–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest, A. , Abeel, T. , Heyndrickx, K.S. , Van Leene, J. , Lanz, C. , Van De Slijke, E. , De Winne, N. et al (2014) A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol. 164, 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues, P.E. and Sharma, S. (2010) Proline metabolism and its implications for plant‐environment interaction. Arabidopsis Book, 8, e0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues, P.E. , Agarwal, M. , Katiyar‐Agarwal, S. , Zhu, J. and Zhu, J.‐K. (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. [DOI] [PubMed] [Google Scholar]
- Walsh, J. , Waters, C.A. and Freeling, M. (1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade–sheath boundary. Genes Dev. 12, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Czedik‐Eysenberg, A. , Mertz, R.A. , Si, Y. , Tohge, T. , Nunes‐Nesi, A. , Arrivault, S. et al (2014) Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 32, 1158–1165. [DOI] [PubMed] [Google Scholar]
- Weiss, D. and Ori, N. (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144, 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, C.N. , Proebsting, W.M. , Hedden, P. and Rivin, C.J. (2000) Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 122, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk, K.J. , Friso, G. , Walther, D. and Schulze, W.X. (2014) Meta‐analysis of Arabidopsis thaliana phospho‐proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell, 26, 2367–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Liu, H. , Tao, P. and Chen, H. (2014) Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PLoS One, 9, e98215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The differentially expressed transcripts along the leaf gradient in the merged well‐watered and non‐transgenic GA20OX‐1 OE control data set.
Table S2 The specifically differentially expressed transcripts in GA20OX‐1 OE transgenic plants under drought conditions.
Table S3 List of E2F/DP targets that were specifically up‐regulated at the TZ1 by mild drought stress.
Data S1 Details of the microarray analysis.


