Summary
Potato is one of the four most important food crop plants worldwide and is strongly affected by drought. The following two pairs of potato cultivars, which are related in ancestry but show different drought tolerances, were chosen for comparative gene expression studies: Gwiazda/Oberon and Tajfun/Owacja. Comparative RNA‐seq analyses of gene expression differences in the transcriptomes obtained from drought‐tolerant versus drought‐sensitive plants during water shortage conditions were performed. The 23 top‐ranking genes were selected, 22 of which are described here as novel potato drought‐responsive genes. Moreover, all but one of the potato genes selected have homologues in the Arabidopsis genome. Of the seven tested A. thaliana mutants with altered expression of the selected homologous genes, compared to the wild‐type Arabidopsis plants, six showed an improved tolerance to drought. These genes encode carbohydrate transporter, mitogen‐activated protein kinase kinase kinase 15 (MAPKKK15), serine carboxypeptidase‐like 19 protein (SCPL19), armadillo/beta‐catenin‐like repeat‐containing protein, high‐affinity nitrate transporter 2.7 and nonspecific lipid transfer protein type 2 (nsLPT). The evolutionary conservation of the functions of the selected genes in the plant response to drought confirms the importance of these identified potato genes in the ability of plants to cope with water shortage conditions. Knowledge regarding these gene functions can be used to generate potato cultivars that are resistant to unfavourable conditions. The approach used in this work and the obtained results allowed for the identification of new players in the plant response to drought.
Keywords: potato drought‐responsive genes, transcriptomics, RNA‐seq, Arabidopsis homologues, functional confirmation of the selected gene importance to drought, Solanum tuberosum
Introduction
Potato (Solanum tuberosum L.) is a crop plant cultivated worldwide that is of great economic importance. There are more than 4500 potato varieties in the World Catalogue of Potato Varieties 2009/10 (www.euroseeds.eu/potatoes). These potato cultivars differ in their bulking time, growth rate and sensitivity to pathogens and various abiotic stresses. Many classical and molecular studies have been performed to identify the genetic loci responsible for agricultural traits using diploid potato plants (Anithakumari et al., 2011, 2012; Gebhardt, 2016; Khan et al., 2015; Mani and Hannachi, 2015). Recently, due to the development of high‐throughput RNA sequencing techniques, which enable qualitative and quantitative global analyses of gene expression, novel approaches have been proposed to understand many physiological and agronomical traits in crop plants (Chen et al., 2015; Gramazio et al., 2016; Prince et al., 2015; Shankar et al., 2016; Wang et al., 2016; Zhang et al., 2014a,b). This methodology enables the study of global gene expression in tetraploid plants, such as potato plants. The identification of gene expression profiles in potato cultivars that are closely related but differ in a particular trait appears to be a promising approach for understanding the mechanisms involved in plant stress tolerance regulation.
Drought is one of the main climate threats limiting crop plant production. Plants are sessile organisms that can tolerate and survive even severe drought by developing escape, avoidance and tolerance strategies that are not mutually exclusive and may act synergistically (Blum, 1996; Levitt, 1980 1980). The strategy employed by plants strongly depends on the plant species, phase of plant development, and intensity and duration of the drought progression (Pinheiro and Chaves, 2011). Potato belongs to a group of crop plants that are considered sensitive to water shortage (Hijmans, 2003; Obidiegwu et al., 2015). To diminish the effect of the forecasted potato harvest losses, it is crucial to identify the strategies used by potato plants to withstand long drought periods during the vegetative season. Therefore, we decided to analyse transcriptome differences in the following two selected pairs of potato cultivars: Gwiazda/Oberon and Tajfun/Owacja. The cultivars in each pair are closely related to each other but differ in their sensitivity to drought conditions (Nowacki, 2012).
In this study, by comparing closely related cultivars, we identified 23 potato genes with significantly different expression profiles during drought. From the list of homologous Arabidopsis genes, we selected seven genes and obtained a homozygous mutant of each gene. The A. thaliana mutants with altered expression of six of the seven genes tested showed a significant improvement in tolerance to drought, demonstrating the evolutionary conservation of the functions of the genes selected between potato and Arabidopsis. Thus, the approach applied and results obtained allowed us to identify novel important and conserved players in the plant response to drought.
Results
Selection of closely related potato cultivars that differ in water shortage tolerance
Previous studies on various potato cultivars led us to select four cultivars: Gwiazda, Tajfun, Oberon and Owacja, which differ in their tolerance to drought (Nowacki, 2012). In addition, they can be grouped as pairs of cultivars based on their origins as follows: Gwiazda and Oberon have one parent in common and, thus, a similar genetic background, while Tajfun and Owacja originated from the clone PS646, which is a parent of the Tajfun cultivar and a grandparent of the Owacja cultivar (Figure S1 and Data S1). The potato cultivars within each pair differ in their ability to withstand drought conditions. Figure 1 (also Figure S2) shows a comparison of the growth of the Gwiazda/Oberon and Tajfun/Owacja cultivar plants under 13 days of drought conditions and after rewatering. As expected, Gwiazda and Tajfun withstood drought better than Oberon and Owacja. A strong wilting phenotype was observed in the Oberon and Owacja cultivars by the sixth day of drought, while on the 13th day of the experiment, all cultivars suffered from the water scarcity. On the 10th day without watering, the lower leaves of the Owacja plants began to yellow intensely, while on day 13, approximately half of the Owacja leaves were yellow or dry. This finding was not observed in the plants of the other three cultivars. Rewatering allowed the plants from the three cultivars to recover completely; however, the Owacja plants were unable to fully regenerate because some of the leaves were irreversibly damaged by the drought stress (Figure 1). After 6, 9 and 13 days of drought stress, the transpirational water loss in the drought‐tolerant cultivars (Gwiazda and Tajfun) was significantly lower than that observed in the drought‐sensitive cultivars (Oberon and Owacja) (Figure 2). The higher RWC levels in the Gwiazda and Tajfun plants indicate that their drought tolerance is superior to that of the Oberon and Owacja plants. Gwiazda developed better mechanisms to reduce transpirational water loss than Tajfun. This finding is also supported by the phenotypic observations in the Gwiazda and Tajfun plants under the drought conditions (Figure 1; the 6th, 10th and 13th days of drought stress).
Figure 1.

Potato cultivars Gwiazda and Tajfun show an improved tolerance to drought. Two‐month‐old well‐watered plants of the four studied cultivars were subjected to water stress. Plants before the application of water stress (upper panel) and after 6, 10 and 13 days without watering are shown (lower panels). After 13 days of drought, the plants were rewatered. The plants are shown three days after rewatering (the lowest panel).
Figure 2.
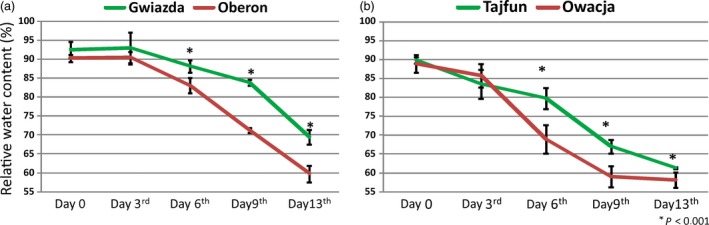
Potato plants from selected cultivars show differences in the RWC in the subsequent days of drought. RWC was measured in leaves from the four studied cultivars after drought stress: (a) cultivars Gwiazda and Oberon and (b) cultivars Tajfun and Owacja. RWC was measured on day 0, 3, 6, 9 and 13 of drought stress. The measurements taken at various drought time points are presented as a percentage of the RWC of the plant at day 0. The leaves were detached and weighed for the initial FW (fresh weight), SW (saturated weight) and DW (dry weight) values. Calculations were performed as described in the Experimental Procedures section, and values are shown as mean ± SD (n = 3) of three independent experiments. *P < 0.001, Mann–Whitney U‐test.
Establishing water deficit conditions: Molecular monitoring during the drought experiment
The main aim of these studies was to identify new genes that are involved in the potato plant response to drought. Therefore, we searched for differentially expressed genes during a drought period in potato plants representing closely related pairs of cultivars that differ in their sensitivity to water shortage. To perform this analysis, plants of the four cultivars studied were subjected to drought three weeks after tuberization began. At this stage of development, potato plants are extremely sensitive to a lack of water (Głuska, 2004). The plants were subjected to drought, and their leaves were collected before the experiment and on the 4th, 6th, 8th, 10th, 12th and 13th days of the experiment (Figure 3a).
Figure 3.
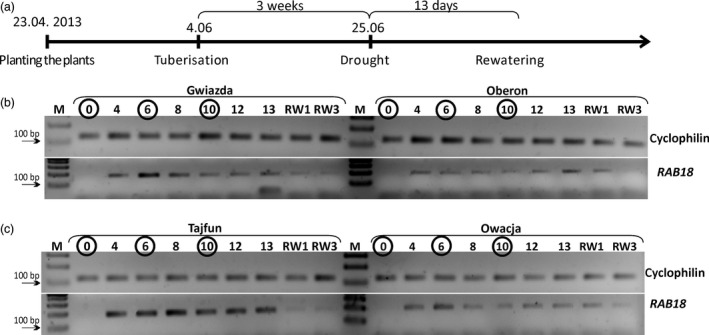
The scheme and molecular marker monitoring of the drought experiment. (a) Time course of the drought experiment. Drought was applied three weeks after the initiation of the tuberization process and was carried out for 13 days. On the 13th day, the plants were rewatered. (b and c) Expression profile of the RAB18 gene. Gel electrophoresis of RAB18 (PGSC0003DMG400003531) and cyclophilin (PGSC0003DMG400001630) cDNA RT‐PCR products on selected days of drought is shown. The results of the cultivar pairs Gwiazda/Oberon and Tajfun/Owacja are shown in panels b and c, respectively. The numbers above the gels indicate the subsequent days of drought; the numbers in circles show the time points when the plant RNA was isolated and subjected to the RNA‐seq analysis. RW—days after plants were rewatered; M—DNA molecular weight marker.
The induction of RAB18 gene expression (Responsive to ABA 18; PGSC0003DMG400003531) at the mRNA level has been shown to occur in plants exposed to various abiotic stresses or exogenous ABA treatment (Hong et al., 2008; Lang and Palva, 1992). RAB18 expression is strongly induced in potato leaves upon drought and dehydration stress (Pieczynski et al., 2013). We performed a semiquantitative RT‐PCR analysis to determine the RAB18 gene expression during our drought experiment using RNA isolated from potato leaves derived from plants representing the four cultivars studied (Figure 3b and c). RAB18 mRNA accumulation was detectable starting on the fourth day of water deficit in all cultivars studied. Generally, RAB18 mRNA accumulated in the drought‐tolerant cultivars to a much higher extent than that in the drought‐sensitive cultivars (Figure 3b and c). In the Gwiazda plants, the highest level of RAB18 expression was observed on the sixth day of drought. During the subsequent days of water deficit, the level of RAB18 mRNA in the Gwiazda plants continuously decreased (Figure 3b). However, in the Oberon cultivar, the level of RAB18 accumulation was relatively stable during all days of drought (Figure 3b). In the Tajfun plants, RAB18 showed the highest expression level on days 6 and 8 and then slightly decreased and remained at the same level until the end of the drought experiment (Figure 3c). The Owacja plants exhibited the highest RAB18 mRNA expression level on the sixth day, and then, the RAB18 expression remained relatively stable until the last day of our drought experiment (Figure 3c).
On the first and third days after the plants were rewatered (Figure 3b and c, RW1 and RW3), a strong reduction in the RAB18 mRNA level was observed in the drought‐tolerant cultivars (Gwiazda and Tajfun), while in the drought‐sensitive cultivars (Oberon and Owacja), the down‐regulation of RAB18 mRNA was slower and was observed on the third day after rewatering (RW3). RT‐PCR experiments were carried out using two additional biological replicates on days 0, 6 and 10, and the results were the same as those obtained in the RT‐PCR experiment using the first biological replicate. Moreover, RT‐qPCR was performed in all time points (days 0, 4, 8, 10, 12, 13, RW1 and RW3) to confirm the semiquantitative RT‐PCR results obtained in the Gwiazda/Oberon cultivars (Figure S3). This RT‐qPCR experiment was performed using one biological replicate with three technical repeats. The results mirrored those obtained using RT‐PCR (Figures 3 and S3). The analysis of the RAB18 mRNA level during the drought experiment allowed us to select the days of drought stress to be used for the RNA‐seq. We performed the RNA‐seq on days 0 (no RAB18 mRNA; control conditions), 6 (the maximum level of RAB18 mRNA expression; strong plant response to drought) and 10 (weaker RAB18 mRNA expression; plant response to extended drought).
Identification of differentially expressed genes during drought stress in the Gwiazda/Oberon and Tajfun/Owacja cultivars
Total RNA was isolated from plants leaves before being subjected to the drought conditions and on the 6th and 10th days of the experiment. Differential analyses of the whole transcriptome sequencing data obtained were carried out by comparing the expression levels of the same transcripts in the Gwiazda/Oberon and Tajfun/Owacja pairs. Normalized reads for all splicing isoforms found for a particular transcript were summed and treated as the final expression of a particular gene as described in detail in the Experimental Procedures section.
Table S2 presents the number of clean and mapped reads obtained for all RNA libraries sequenced, the distribution and percentage of RNA‐seq reads mapped onto genomic loci, and the number and percentage of reads mapped onto multiple loci for the two pairs of drought‐tolerant and drought‐sensitive cultivars under control (day 0) and drought conditions (days 6 and 10). After the sequencing and removing the low‐quality reads, the total number of reads that varied between the samples ranged from 39 to 43 million. The obtained reads were mapped onto the reference S. tuberosum group Phureja potato genome. Depending on the RNA library, the percentage of reads that mapped onto the potato genome varied from 67.9 to 83.8. The percentage of reads that mapped onto a single position within the genome varied from 80.2 to 94.8, while the percentage of reads that mapped onto more than one locus varied from 5.2 to 19.8.
To select the differentially expressed genes upon drought conditions in both pairs of drought‐tolerant/drought‐sensitive cultivars, we designed a pipeline that allowed us to limit the number of genes selected to the best candidate genes. First, of the differentially expressed genes in the pairs of cultivars studied, only those that exhibited the same level of expression on day 0 in either the Gwiazda/Oberon or Tajfun/Owacja cultivars (the differences in the expression of these genes were statistically insignificant) were selected for further study (Figure S4A, B; horizontal two‐headed arrows on day 0). Then, from this selected starting pool of genes, only the genes that displayed statistically significant differences in their expression pattern between days 0 and 6 or days 0 and 10, independently in each drought‐tolerant cultivar (Gwiazda or Tajfun) (Figure S4A, B; vertical arrows), passed the filtering step. In parallel, the selected genes had to display statistically significant differences between the drought‐tolerant and drought‐sensitive cultivars on any given day of drought (Figure S4A, B; horizontal two‐headed arrows on days 6 and 10). These analyses resulted in the selection of genes that significantly differed in their expression levels under drought conditions. In the Gwiazda/Oberon comparison, 134 genes were selected (66 genes were up‐regulated, and 68 genes were down‐regulated in the Gwiazda versus Oberon cultivars), and in the Tajfun/Owacja comparison, 460 genes were selected (261 genes were up‐regulated, and 199 genes were down‐regulated in the Tajfun versus Owacja cultivars) (Figure S5A).
We also selected genes that exhibited a low fold change (0.8 < fold change < 1.2) in their expression patterns during the time course of our drought experiment in plants representing all four cultivars (Figure S4C). We identified eight genes with stable expression under the control and drought conditions. These genes represent a pool of genes that can potentially serve as gene expression controls in drought experiments. The stable expression of these genes was confirmed by RT‐PCR (Figure S6).
The genes that showed statistically significant changes in their expression levels in the drought‐tolerant versus drought‐sensitive cultivars were subjected to further selection steps. The second round of filtering consisted of the selection of those genes showing the highest differences in their expression profiles obtained in the RNA‐seq data during the time course of the drought experiment (days 6 and 10). The comparison of the normalized number of reads for transcripts derived from all genes identified in the first round of selection (594 genes) during the time course of the drought experiment is presented in Table S3. In addition to this tabular data presentation, the expression profile of each gene is shown in a separate chart containing the normalized number of reads from the RNA‐seq data for the Gwiazda/Oberon or Tajfun/Owacja pair of cultivars (Tables S1, S3).
Of the 33 drought‐related genes obtained in second round of selection for Gwiazda/Oberon, the expression profiles of 18 genes were up‐regulated, while the expression profiles of 15 genes were down‐regulated in the comparison of the Gwiazda and Oberon cultivars (Figure S4A). In the Tajfun/Owacja cultivars (53 selected genes in second round), the expression profiles of 28 genes were up‐regulated, and 25 genes were down‐regulated when the Tajfun cultivar was compared to the Owacja cultivar (Figure S5A). All genes from the second round of selection were further assessed by confirming the gene expression profile differences in each gene in the drought‐tolerant versus drought‐sensitive cultivars using RT‐PCR (third round of selection). Only genes showing consistency in their expression profiles between the RNA‐seq and RT‐PCR results were used for further analysis.
The third selection stage allowed us to reduce the number of selected genes to the nine top‐ranking genes in the Gwiazda/Oberon pair and 15 genes in the Tajfun/Owacja pair (Figure S5A). The expression profiles of six genes were up‐regulated, while the expression profiles of three genes were down‐regulated when the Gwiazda plants were compared to the Oberon plants, and the expression of nine genes was up‐regulated, while the expression of six genes was down‐regulated when the Tajfun plants were compared to the Owacja plants (Figure S5A). Notably, one of the selected genes (PGSC0003DMG400014293) was up‐regulated by drought in both drought‐tolerant cultivars (Gwiazda and Tajfun), and thus, the overall number of selected genes was 23 (the accession numbers of these genes are presented in Fig. S5B). The results of the third round of selection are shown in Figure S7. RT‐PCRs were carried out to test the expression profiles of the 23 selected genes in three biological replicates. The RNA‐seq data and RT‐PCR results of seven potato genes (two in Gwiazda/Oberon and five in Tajfun/Owacja) displaying the highest differences in gene expression during the water shortage conditions in the drought‐tolerant and drought‐sensitive cultivars are shown in Figure 4, and the expression data of the remaining 17 genes are shown in Figure S7.
Figure 4.
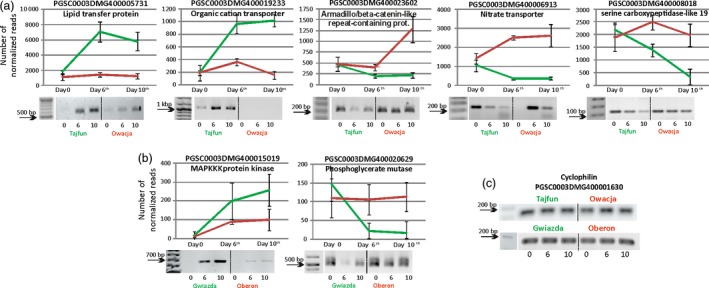
Seven selected potato genes showing the highest differences in gene expression between the studied cultivars during the drought experiment. Above each graph, the accession number of a given gene and its function are displayed. (a) Graphs showing the normalized number of reads for genes from Tajfun/Owacja cultivars (green/red lines) and (b) Gwiazda/Oberon cultivars (green/red lines). Values are shown as mean ± SD (n = 3), P < 0.05, of three independent RNA‐seq experiments. Statistical methods for the differential gene expression analysis are described in the Experimental Procedures section. The homologues of these genes were further studied in Arabidopsis plants. Gel electrophoresis analyses of the RT‐PCR products for all seven genes are presented below each graph, confirming the RNA‐seq results. (c) Gel electrophoresis of the stably expressed cyclophilin gene during the drought experiment in all cultivars studied. The numbers at the bottom of each gel indicate the days of the drought experiment.
A. thaliana genes homologous to the selected drought‐related potato genes also affect the Arabidopsis response to water deficit conditions
Twenty‐two A. thaliana genes that are homologous to the best potato drought‐related candidate genes were identified (one potato gene—PGSC0003DMG400024093—has no homologue in the Arabidopsis genome; Figure 5a). Of these 22 genes, seven arbitrarily chosen Arabidopsis homozygous mutants were obtained from the SALK and GABI‐Kat mutant collections. These mutants were genotyped to confirm that they were homozygous lines (Alonso et al., 2003 ; Kleinboelting et al., 2012). The RT‐PCR analyses revealed that in the case of three genes, T‐DNA insertions resulted in the complete abolishment of gene expression (AT5G14570, AT1G22170 and AT5G09640). A down‐regulation of gene expression was observed only in one mutant (AT3G18280), and in the case of the other three genes, T‐DNA insertions resulted in the up‐regulation of the expression of each gene analysed (AT3G20660, AT4G16490 and AT5G55090) (Figure 5b).
Figure 5.
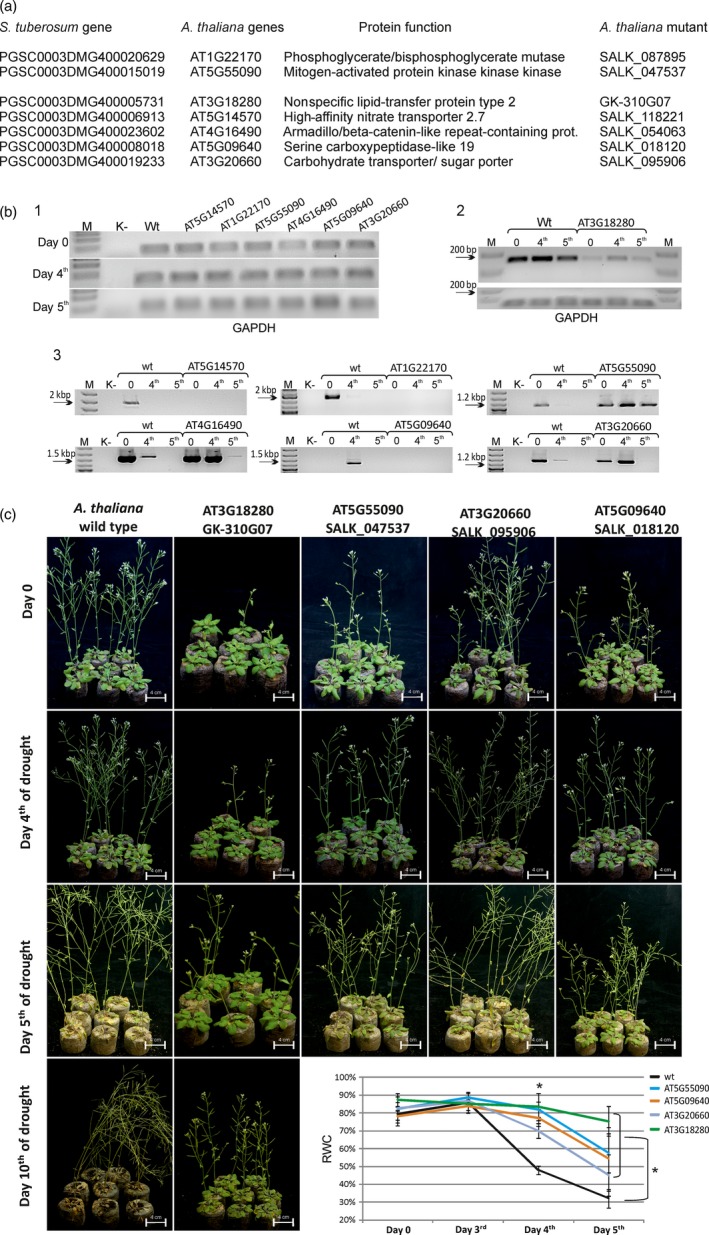
Analysis of Arabidopsis mutant plants with altered expression of genes homologous to the selected drought‐related potato genes. (a) Accession numbers of the seven top‐ranking drought‐related potato genes (left column) and the accession numbers and protein functions of their Arabidopsis homologues (middle columns) were obtained from the Spud DB and TAIR database (Hirsch et al., 2014; Huala et al., 2001). The right column presents the accession numbers of the selected Arabidopsis SALK or GABI‐Kat mutants of each gene (Alonso et al., 2003; Kleinboelting et al., 2012). (b) RT‐PCR analysis of the expression of selected Arabidopsis genes under control (day 0) and drought conditions (days 4 and 5). (1) Gel electrophoresis of the RT‐PCR products of the GAPDH gene in wild‐type and selected Arabidopsis SALK line plants on days 0, 4 and 5 of the drought experiment; (2 and 3) gel electrophoresis of the RT‐PCR products of selected Arabidopsis genes in wild‐type and mutant plants grown under control and drought conditions, respectively. SALK accession numbers are presented above each electrophoretic line. M—DNA marker; K—RT‐PCR nontemplate control; wt—wild‐type plant. (c) Arabidopsis plants subjected to water deficit stress. Plants are shown on days 0, 4, 5 and 10 of the drought experiment. Only wild‐type and four mutant plants are shown (the other three are presented in Fig. S8). Lower bottom right panel of (c)—RWC analysis in the leaves of wild‐type and selected mutant Arabidopsis plants. Mutant plants show higher water content than wild‐type plants. RWC was measured 0, 4 and 5 days after the introduction of drought stress. Values are shown as mean ± SD (n = 3) of three independent experiments. *P < 0.001, Mann–Whitney U‐test.
Four‐week‐old Arabidopsis wild‐type and mutant plants were subjected to drought, which was continued until plant death (up to 10 days). Comparative RT‐PCR analyses of the gene expression in wild‐type and mutant plants under drought conditions showed that in the null mutants analysed, there was no expression of the genes tested (AT5G14570, AT1G22170 and AT5G09640), although the expression was detectable in the wild‐type plants (Figure 5b). Interestingly, the expression in the control wild‐type plants was detectable only on day 0 in the case of two genes (AT5G14570 and AT1G22170), but during drought stress, the expression was abolished, while in the case of the AT5G09640 gene, its expression was observed only on day 4 of drought stress. In the case of the AT3G18280 gene, we observed a slight up‐regulation of its expression in the wild‐type plants upon drought, and in the SALK mutant of this gene, the expression was also up‐regulated, although at a much lower level than that in the wild‐type plants (Figure 5B). The expression of the AT3G20660, AT4G16490 and AT5G55090 genes was strongly down‐regulated by drought in the wild‐type plants, while in the mutant plants, up‐regulated gene expression was observed, although for the AT4G16490 and AT3G20660 genes, there was no detectable expression on the fifth day of drought (Figure 5B). Compared with the wild‐type plants, all mutant plants, except for one (AT1G22170), showed an improved tolerance to drought starting on day 4 of drought; additionally, the wild‐type plants presented a strong wilting phenotype on the same day (Figures 5C and S8B). In almost all mutant plants, the wilting phenotype was visible on the fifth day of the stress conditions; however, in the case of the AT3G18280 gene mutant, the plants still showed a strong resistance to the lack of water on the 10th day of the stress experiment.
The best drought‐tolerant Arabidopsis mutant plants are shown in Figure 5C, while the remaining three mutants are presented in (Figure S8). The drought‐tolerant phenotypic observations of the Arabidopsis mutants were confirmed by the RWC measurements. Figure 5C (the bottom right panel) and Figure S8C indicate a strong transpirational water loss reduction in six of the seven Arabidopsis mutants tested compared to that in the wild‐type Arabidopsis plants. The AT1G22170 mutant exhibited higher RWC values than the wild‐type plants on the fourth day of drought; however, on the fifth day of drought stress, the leaf RWC in the mutant was the same as that in the wild‐type plants.
These experiments demonstrated that the selected genes exhibit evolutionary conservation in their functions in the plant response to drought, at least in potato and Arabidopsis plants.
Discussion
Comparative transcriptomic studies identified novel genes involved in the potato response to drought
Next‐generation sequencing (NGS) of transcriptomes provides a useful tool for exploring differential gene expression between closely related plant species, cultivars or even individuals to identify genes that are responsive to different environmental stresses (Prince et al., 2015; Chen et al., 2015; Wang et al., 2016; Gramazio et al., 2016; Shankar et al., 2016). The studies presented herein provide in‐depth insight into the responses of potato plants of closely related cultivars with contrasting tolerances to drought stress during tuberization. A large number of genes were found to be differentially expressed in the cultivars studied. This number was higher in the Tajfun/Owacja pair than in the Gwiazda/Oberon pair. We cannot exclude the possibility that this difference is due to the more distant relationship between Tajfun and Owacja than that between the Gwiazda and Oberon cultivars, resulting in a higher variability of allele content (see Figure S1).
We limited the number of compared genes to those related to water shortage conditions by selecting only those genes that displayed a similar level of expression on day 0 and for which the induction or down‐regulation of gene expression was observed in the drought‐tolerant cultivar compared to the level in the drought‐sensitive cultivar within a given pair on days 6 and 10. This selection process allowed us to avoid any ambiguities in whether the differential gene expression was caused by reasons other than drought. Using this strategy, we identified the 23 top‐ranking drought‐related genes that successfully passed the designed filtering pipeline. One gene was found to be selected in both pairs of analysed cultivars (PGSC0003DMG400014293); this gene encodes the CAP160 protein, which is already known to be stress‐related. Interestingly, only this single gene of the 23 identified here was also found by the Wang group (Zhang et al., 2014a,2014b,c) in their transcriptome‐wide studies on drought‐responsive genes in the drought‐tolerant potato cultivar Longshu3 (Cheng et al., 2013; Zhang et al., 2014a,2014b,c). Their results on the CAP160 gene are consistent with our data showing a strong up‐regulation of this gene in drought‐tolerant cultivars under water deficit conditions. Although we also found that the potato CAP160 mRNA level was up‐regulated when water was scarce in the drought‐sensitive cultivars, the induction of CAP160 expression was much lower in the drought‐sensitive cultivars than in the drought‐tolerant cultivars (see Figure S7). Interestingly, CAP160 expression is also up‐regulated in spinach and pepper by both water and low‐temperature stresses (Kaye et al., 1998; Park et al., 2016). These results suggest that the up‐regulation of CAP160 expression during water deficit in drought‐tolerant potato cultivars may be related to the improved response of these cultivars to drought stress.
Transcriptomic analyses show that the identified potato genes were also related to drought in other plant species
We compared the expression patterns of the 23 selected potato genes with the expression of their homologues in Arabidopsis and rice under drought conditions using available high‐throughput data (Table 1). Surprisingly, the analysis of these data revealed that 17 of the genes show expression changes in either all or two of the species compared. Six selected genes show the same direction of expression change in potato, Arabidopsis and rice; nine genes show the same direction of expression change only in rice and potato; and one gene shows similar expression changes in potato and Arabidopsis. Two genes show changes that are opposite to the potato homologue expression: one in rice and one in Arabidopsis (Table 1; Winter et al., 2007; Kapushesky et al., 2012; Petryszak et al., 2016; Moumeni et al., 2015; Petryszak et al., 2016; Zielezinski et al., 2017). To the best of our knowledge, the identified common drought‐responsive genes have not been studied in rice, Arabidopsis or potato regarding their roles in the plant response to a water deficit.
Table 1.
Comparison of the 23 top‐ranking drought‐related genes in S. tuberosum and their homologues in Arabidopsis thaliana and Oryza sativa and the direction of the expression change during drought stress
| No. | Names of the cultivars | Potato gene | Arabidopsis gene | Rice gene | Common drought‐related genes | |
|---|---|---|---|---|---|---|
| Potato vs. Arabidopsis | Potato vs. rice | |||||
| 1 | Gwiazda/Oberon | PGSC0003DMG400001621 | AT4G38690 | LOC_Os09g36520 | ↗ | ↗ |
| 2 | PGSC0003DMG400012174 | AT1G64110 | LOC_Os01g12660 | – | ↗ | |
| 3 | PGSC0003DMG400014293* | AT5G52300 | LOC_Os10g36180 | – | ↗ | |
| 4 | PGSC0003DMG400015019 | AT5G55090 | LOC_Os02g21700 | – | nd | |
| 5 | PGSC0003DMG400024849 | AT1G77120 | LOC_Os11g10480 | – | ↗ | |
| 6 | PGSC0003DMG401015935 | AT4G33150 | LOC_Os02g54254 | – | ↗ | |
| 7 | PGSC0003DMG400000723 | AT4G34490 | LOC_Os03g51250 | – | nd | |
| 8 | PGSC0003DMG400002943 | AT3G11170 | No homologue | – | nd | |
| 9 | PGSC0003DMG400020629 | AT1G22170 | LOC_Os02g51590 | – | nd | |
| 10 | Tajfun/Owacja | PGSC0003DMG400001771 | AT3G09720 | LOC_Os07g45360 | – | ≠ |
| 11 | PGSC0003DMG400003688 | AT1G19530 | LOC_Os03g61150 | ↗ | ↗ | |
| 12 | PGSC0003DMG400005731 | AT3G18280 | LOC_Os05g47700 | ↗ | ↗ | |
| 13 | PGSC0003DMG400005917 | AT3G49400 | LOC_Os01g50690 | ↗ | nd | |
| 14 | PGSC0003DMG400008497 | AT3G51810 | LOC_Os05g28210 | – | ↗ | |
| 15 | PGSC0003DMG400019233 | AT3G20660 | LOC_Os04g53930 | ↗ | ↗ | |
| 16 | PGSC0003DMG400024093 | No homologue | LOC_Os02g33820 | nd | ↗ | |
| 17 | PGSC0003DMG400039484 | AT1G59860 | LOC_Os03g16030 | – | ↗ | |
| 18 | PGSC0003DMG400006913 | AT5G14570 | LOC_Os01g50820 | – | ↘ | |
| 19 | PGSC0003DMG400007427 | AT3G05410 | LOC_Os01g70820 | ↘ | ↘ | |
| 20 | PGSC0003DMG400008018 | AT5G09640 | LOC_Os11g27264 | – | nd | |
| 21 | PGSC0003DMG400014954 | AT4G31940 | No homologue | – | nd | |
| 22 | PGSC0003DMG400022225 | AT4G27030 | LOC_Os08g08850 | ↘ | ↘ | |
| 23 | PGSC0003DMG400023602 | AT4G16490 | LOC_Os07g39590 | ≠ | ↘ | |
Homologues were identified using Orcan software in the ComBio platform for Arabidopsis and the UniProt database for rice (http://bar.utoronto.ca/efp2/Arabidopsis/Arabidopsis_eFPBrowser2.html; http://www.combio.pl/orcan; http://www.ebi.ac.uk/gxa/home), with the potato protein as a blastp query (The UniProt Consortium, 2017; Zielezinski et al., 2017). *—transcripts found to be affected in both pairs of potato cultivars; ↗—genes up‐regulated in the three compared plant species; ↘—genes down‐regulated in the three compared plant species; nd—no data available;–—no significant changes in Arabidopsis; ≠—different directions of change in the compared plant species.
Altogether, 70% of the selected potato genes show evolutionary conservation with rice genes, and 35% of the selected potato genes show evolutionary conservation with Arabidopsis genes. This finding suggests that the identified genes play a pivotal role in shaping the plant cell metabolism during the drought stress response.
The GO database annotation in the biological processes category shows that among the 22 top‐ranking Arabidopsis homologues, eight are linked to various abiotic stresses (Table S4). Five homologues are associated with water deprivation, heat stress, salt stress, osmotic stress and response to ABA (AT5G52300, AT1G77120, AT3G51810, AT3G20660 and AT1G59860). Drought stress is accompanied by salinity, by osmotic stresses and often by heat in natural conditions of plant growth. Plant metabolic pathways responding to different environmental cues often overlap (Barciszewska‐Pacak et al., 2015; Xiong et al., 1999). Consequently, genes with induced expression under these stresses were selected in the potato cultivars in the drought experiment performed here (Krasensky and Jonak, 2012; Zhang et al., 2014a,2014b,c). ABA signalling is one of the main pathways that integrate various stress signals and control downstream stress responses (Tuteja, 2007). Therefore, it is not surprising that three of the 23 genes analysed were found to be responsive to ABA or even participate in ABA‐activated signalling pathways (AT1G77120, AT5G52300 and AT3G51810).
Four of the 23 selected genes were involved in oxidation–reduction processes (AT1G77120, AT3G11170, AT4G31940 and AT4G27030), and it might be connected with accumulation of reactive oxygen species (ROS) that play a central role in coordinating defence signalling in plants in response to many abiotic stresses (Iyer et al., 2012; Mittler, 2002). Cell membrane composition changes, such as changes in phospholipids and proteins, help to maintain membrane integrity, preserve cell compartmentalization and activate phospholipid signalling pathways in response to drought (Jarzyniak and Jasinski, 2014; Liu et al., 2013; Ruelland et al., 2014; Schulz, 2011). Five of the 23 genes studied are involved in unsaturated fatty acid and fatty acid biosynthetic processes and transmembrane and lipid transport (AT3G11170, AT3G18280, AT3G20660, AT5G14570 and AT4G27030).
Functional analysis of selected potato homologous genes in Arabidopsis confirms their importance in plant tolerance to drought
Six of the seven studied Arabidopsis mutants exhibited an improved tolerance to drought compared to the wild‐type Arabidopsis plants that were grown under the same conditions. The only mutant that did not show an improved response to drought in Arabidopsis plants contained a T‐DNA insertion within the phosphoglycerate/bisphosphoglycerate mutase gene (AT1G22170). For five of the homologous genes (AT3G20660/PGSC0003DMG400019233, AT5G09640/PGSC0003DMG400008018, AT5G55090/PGSC0003DMG400015019, AT4G16490/PGSC0003DMG400023602 and AT5G14570/PGSC0003DMG400006913), the gene expression changes in the drought‐tolerant cultivar were in the same direction as those in the Arabidopsis mutant plants tested. These genes encode carbohydrate transporter, mitogen‐activated protein kinase kinase kinase 15 (MAPKKK15), serine carboxypeptidase‐like 19 protein (SCPL19), armadillo/beta‐catenin‐like repeat‐containing protein and high‐affinity nitrate transporter 2.7 (transmembrane transport, nitrate assimilation/transport), and three of these genes have already been reported in high‐throughput studies as genes responsive to drought in other plants (carbohydrate transporter, MAPKKK15 and NRT2.7).
Carbohydrate transporters are reduced in maize under drought stress, leading to a decreased carbohydrate supply in developing reproductive plant parts. Moreover, the phosphorylation level of the maize carbohydrate transporter/sugar porter transporter (B6U8S7) was up‐regulated under heat stress (Aslam et al., 2015; Hu et al., 2015).
Mitogen‐activated protein kinases (MAPKs) are involved in signal transduction. The functions of many of these kinases in Arabidopsis are unknown. The MAPKKKs form the largest family of MAP kinases. Very little is known about MAPKKK15. However, MAPKKK15 has been already shown to be activated at the transcriptional level under drought conditions (Menges et al., 2008).
Serine carboxypeptidase‐like protein 49 (SCPL49), which is involved in protein degradation (proteolysis), was identified among the drought‐adaptive genes in a cross‐species meta‐analysis comparing Arabidopsis, rice, wheat and barley plants (Shaar‐ Moshe et al., 2015). To the best of our knowledge, SCPL19 has not previously been reported as a drought‐responsive gene. However, our studies clearly show that the down‐regulated expression of this gene in drought‐tolerant potato cultivars and the knockout of this gene activity in the A. thaliana mutant correlate with the enhanced tolerance to water deficit stress (see Figures 5 and S8).
Armadillo/beta‐catenin‐like repeat‐containing proteins are involved in developmental processes and stress signalling. In rice, several members of this family were found to be up‐regulated by drought, salt and/or cold stress (Sharma et al., 2014). In addition, several members of this family in rice and Arabidopsis contain a U‐Box motif and are thought to be implicated in protein degradation and response to drought stress (Cho et al., 2008; Seo et al., 2012; Sharma et al., 2014). However, proteins encoded by the Arabidopsis AT4G16490 and potato PGSC0003DMG400023602 genes do not contain the U‐Box motif. Expression of this gene was down‐regulated in the drought‐tolerant Tajfun cultivar and was also strongly down‐regulated on the fifth day of drought conditions in the corresponding Arabidopsis mutant (see Figures 5 and S8). The exact function of this gene product remains to be elucidated.
The expression of the homologous genes AT5G14570/PGSC0003DMG400006913 in Arabidopsis and potato is strongly down‐regulated in the Arabidopsis mutant (no detectable expression) and the drought‐tolerant potato cultivar Tajfun. These genes encode NTR2.7, which is known to control nitrate content in dry seeds. One of the high‐affinity nitrate transporters (MdNRT2.4) has been found to be involved in the drought response in apple roots. The expression of this gene was elevated in plant roots likely due to the reduction of the nitrate concentration in response to drought (Bassett et al., 2014). However, AtNTR2.7 mRNA was absent in the drought‐tolerant Arabidopsis mutant, and in the drought‐tolerant potato cultivar Tajfun, the NTR2.7 transcript level was down‐regulated. The exact role of NTR2.7 in the plant response to drought has yet to be elucidated.
In the case of the AT3G18280/PGSC0003DMG400005731 homologous genes, in the drought‐tolerant Tajfun cultivar, the expression of the potato gene was up‐regulated compared to that in the drought‐sensitive Owacja plants. However, in the Arabidopsis mutant of this gene, the expression was strongly down‐regulated, although the mutant was extremely resistant to drought stress. This gene encodes nonspecific lipid transfer protein type 2 (nsLPT). This gene has been previously identified in microarray studies as a marker for drought stress in Arabidopsis plants. Its expression was up‐regulated twofold by drought but was not affected by salt, cold or heat stress, suggesting that nsLPT may play a very specific role in the plant response to water shortage (El Ouakfaoui and Miki, 2005). nsLPTs play a role in phospholipid and fatty acid transfer between membranes (Kader, 1996). Some members of this family in plants have been shown to participate in wax and/or cutin monomer transport (Kim et al., 2013; Sterk et al., 1991). Detailed studies on the nsLPT gene family in maize showed induced but also down‐regulated expression of particular family members in response to drought. Our studies on potato and Arabidopsis support the importance of the nsLPT gene family in the plant response to drought. To explain the observed discrepancies in the gene expression profiles between the drought‐tolerant potato cultivar and the relevant Arabidopsis mutant, more detailed studies are required to reveal the functions of particular genes in this large family in both species studied.
The data presented in this study show a comparison of the gene expression profiles between closely related cultivars, which represents a successful approach for identifying genes responsive to drought with interspecies conserved functions.
Experimental procedures
Potato plant material and growth conditions
The following four potato cultivars were used: Gwiazda and Oberon (bred by Potato Breeding Zamarte Ltd., Co. IHAR‐PIB Group, Poland) and Tajfun and Owacja (bred by the Pomeranian‐Mazurian Potato Breeding Company, Strzekecino, Poland). Plants were grown as described in Boguszewska et al. (2010) (Data S1). All experiments were performed in three biological replicates. During the drought experiment, leaf samples were taken every day from three different plants for each cultivar at a given time point and frozen in liquid nitrogen. The third, fourth and fifth whirls from the top of the compound leaves were collected from each plant.
Arabidopsis plant material and growth conditions
The following A. thaliana plants were used in this study: ecotype Columbia‐0 (wild‐type plants) and the homozygous T‐DNA insertion mutants GK‐310G07, SALK_118221, SALK_087895, SALK_047537, SALK_054063, SALK_018120 and SALK_095906 (Alonso et al., 2003; Kleinboelting et al., 2012). The plants were grown in soil (Jiffy‐7 42 mm; Jiffy Products International AS, Norway) in a growth chamber as described in Szarzynska et al. (2009). Four‐week‐old Arabidopsis plants were subjected to drought conditions by the cessation of watering for ten days without further rewatering.
DNA and RNA isolation, cDNA synthesis and PCR and RT‐PCR amplification
Genomic DNA and total RNA were isolated from plant leaves as described in Szarzynska et al. (2009) (Data S1). All primer sequences are shown in Figure S2. The PCR and RT‐qPCR were performed as previously described (Pieczynski et al., 2013; Szarzynska et al., 2009).
Relative water content (RWC) measurements
The RWC was calculated as described in Boguszewska et al. (2010) (Data S1).
RNA‐seq
Total RNA was extracted from the plant tissues using TRIzol reagent as described in Pant et al. (2009) and Szarzynska et al. (2009) (Data S1). The strand‐specific library construction and RNA sequencing (RNA‐seq) (PE100) using an Illumina HiSeq 2500 were conducted by BGI Tech Solutions Co., Ltd (Hong Kong). Basic information regarding the RNA‐seq data is provided in Table S2. All row data have been deposited in the GEO database (GSE97776) under the link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97776.
Bioinformatic analyses of RNA deep sequencing data
Analysis of bioinformatic data is described in Data S1.
Primer design
All PCR primers used for the expression analysis of selected genes in potato were designed as described in Data S1.
Supporting information
Figure S1 Genetic background of the four studied potato cultivars Gwiazda and Oberon (A) and Tajfun and Owacja (B).
Figure S2 Gwiazda/Oberon and Tajfun/Owacja cultivar plants grown in the half‐open glasshouse on day 13 of drought.
Figure S3 RT‐qPCR analysis of RAB18 gene expression in leaves detached from Gwiazda and Oberon cultivar plants during the drought experiment.
Figure S4 Schematic representation of drought‐related gene selection using RNA‐seq data from two pairs of potato cultivars, Gwiazda/Oberon and Tajfun/Owacja.
Figure S5 Drought‐related gene selection pipeline.
Figure S6 Eight stably expressed genes in all potato cultivars studied during drought stress.
Figure S7 Seventeen of 23 potato genes selected after the third round of selection showing the highest differences in gene expression between the studied drought‐tolerant and drought‐sensitive cultivars during the drought experiment.
Figure S8 Phenotypic and RWC analyses of three Arabidopsis mutant plants with altered expression of genes homologous to the selected drought‐related potato genes.
Table S1 Comparison of the normalized number of reads for transcripts derived from genes identified in the first round of selection (594 genes) during the time course of the drought experiment.
Table S2 GO database annotation of the biological process, molecular function and cellular component categories of 8 genes selected from among the 22 top‐ranking Arabidopsis genes.
Table S3 Primer sequences.
Table S4 Summary of RNA‐seq Data.
Data S1 Experimental procedures ancillary information.
Acknowledgements
This work was supported by the project NSC UMO‐2012/05/B/NZ9/03383 and the KNOW RNA Research Centre in Poznan (01/KNOW2/2014). KM was supported by the Foundation for Polish Science (FNP) in the International PhD Program cofinanced by the European Union Regional Development Fund (MPD 2010/3). We would like to thank Prof. Ewa Zimnoch‐Guzowska and Prof. Waldemar Marczewski for discussions and critical reading of the manuscript. The authors declare no conflict of interest.
References
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. et al (2003) Genome‐wide Insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Anithakumari, A.M. , Dolstra, O. , Vosman, B. , Visser, R.G.F. and van der Linden, C.G. (2011) In vitro screening and QTL analysis for drought tolerance in diploid potato. Euphytica, 181, 357–369. [Google Scholar]
- Anithakumari, A.M. , Nataraja, K.N. , Visser, R.G.F. and van der Linden, C.G. (2012) Genetic dissection of drought tolerance and recovery potential by quantitative trait locus mapping of a diploid potato population. Mol. Breeding, 30, 1413–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam, M. , Maqbool, M. A. and Cengiz, R. (2015) Drought stress in maize (Zea mays L.). Effects, resistance, mechanisms, global achievements and biological strategies for improvement. Springer Briefs in Agriculture: Heidelberg New York London:Springer Cham. [Google Scholar]
- Barciszewska‐Pacak, M. , Milanowska, K. , Knop, K. , Bielewicz, D. , Nuc, P. , Plewka, P. , Pacak, A.M. et al (2015) Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 6, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett, C.L. , Baldo, A.M. , Moore, J.T. , Jenkins, R.M. , Soffe, D.S. , Wisniewski, M.E. , Norelli, J.L. et al (2014) Genes responding to water deficit in apple (Malus × domestica Borkh.) roots. BMC Plant Biol. 14, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, A. (1996) Crop responses to drought and the interpretation of adaptation. Plant Growth Regul. 20, 135–148. [Google Scholar]
- Boguszewska, D. , Grudkowska, M. and Zagdańska, B. (2010) Drought‐responsive antioxidant enzymes in potato (Solanum tuberosum L.). Potato Res. 53, 373–382. [Google Scholar]
- Chen, H. , Chen, X. , Chen, D. , Li, J. , Zhang, Y. and Wang, A. (2015) A comparison of the low temperature transcriptomes of two tomato genotypes that differ in freezing tolerance: Solanum lycopersicum and Solanum habrochaites. BMC Plant Biol. 15, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.‐J. , Deng, X.‐P. , Kwak, S.‐S. , Chen, W. and Eneji, A.E. (2013) Tolerant potato cultivar selection under multiple abiotic stresses. JFAE, 11, 760–766. [Google Scholar]
- Cho, S.K. , Ryu, M.Y. , Song, C. , Kwak, J.M. and Kim, W.T. (2008) Arabidopsis PUB22 and PUB23 are homologous U‐Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell, 20, 1899–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ouakfaoui, S. and Miki, B. (2005) The stability of the Arabidopsis transcriptome in transgenic plants expressing the marker genes nptII and uidA. Plant J. 41, 791–800. [DOI] [PubMed] [Google Scholar]
- Gebhardt, K. (2016) The historical role of species from the Solanaceae plant family in genetic research. Theor. Appl. Genet. 129, 2281–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głuska, A. (2004) Wpływ zmiennego rozkładu opadów na cechy bulw ziemniaka (Solanum tuberosum L) w warunkach polowych oraz wyznaczenie okresu krytycznego wrażliwości na niedobór wody u odmian o różnej długości okresu wegetacji. Zeszyty Prob. Post. Nauk Roln. 496, 217–222. [Google Scholar]
- Gramazio, P. , Blanca, J. , Ziarsolo, P. , Herraiz, F.J. , Plazas, M. , Prohens, J. and Vilanova, S. (2016) Transcriptome analysis and molecular marker discovery in Solanum incanum and S. aethiopicum, two close relatives of the common eggplant (Solanum melongena) with interest for breeding. BMC Genom. 17, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans, R.J. (2003) The effect of climate change on global potato production. Am. J. Potato Res. 80, 271–280. [Google Scholar]
- Hirsch, C.D. , Hamilton, J.P. , Childs, K.L. , Cepela, J. , Crisovan, E. , Vaillancourt, B. , Hirsch, C.N. et al (2014) Spud DB: a resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. Plant Genome, 7, 1–12. [Google Scholar]
- Hong, Y. , Zheng, S. and Wang, X. (2008) Dual functions of phospholipase Dalpha1 in plant response to drought. Mol. Plant, 1, 262–269. [DOI] [PubMed] [Google Scholar]
- Hu, X. , Wu, L. , Zhao, F. , Zhang, D. , Li, N. , Zhu, G. , Li, C. et al (2015) Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala, E. , Dickerman, A. , Garcia‐Hernandez, M. , Weems, D. , Reiser, L. , LaFond, F. , Hanley, D. et al (2001) The Arabidopsis Information Resource (TAIR): A comprehensive database and web‐based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, N.J. , Jia, X. , Sunkar, R. , Tang, G. and Mahalingam, R. (2012) MicroRNAs responsive to ozone‐induced oxidative stress in Arabidopsis thaliana . Plant Signal. Behav. 7, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzyniak, K.M. and Jasinski, M. (2014) Membrane transporters and drought resistance – a complex issue. Front. Plant Sci. 5, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader, J. C. (1996) Lipid‐transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 47, 627–654. [DOI] [PubMed] [Google Scholar]
- Kapushesky, M. , Adamusiak, T. , Burdett, T. , Culhane, A. , Farne, A. , Filippov, A. , Holloway, E. et al (2012) Gene Expression atlas update‐ a value‐added database of microarray and sequencing‐based functional genomics experiments. Nucleic Acids Res. 40, D1077–D1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye, C. , Neven, L. , Hofig, A. , Li, Q.B. , Haskell, D. and Guy, C. (1998) Characterization of a gene for spinach CAP160 and expression of two spinach cold‐acclimation proteins in tobacco. Plant Physiol. 116, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.A. , Saravia, D. , Munive, S. , Lozano, F. , Farfan, E. , Eyzaguirre, R. and Bonierbale, M. (2015) Multiple QTLs linked to agro‐morphological and physiological traits related to drought tolerance in potato. Plant Mol. Biol. Rep. 3, 1286–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. , Lee, S.B. , Kim, H.J. , Min, M.K. , Hwang, I. and Suh, M.C. (2012) Characterization of glycosylphosphatidylinositol‐anchored lipid transfer Protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana . Plant Cell Physiol. 53, 1391–1403. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting, N. , Huep, G. , Kloetgen, A. , Viehoever, P. and Weisshaar, B. (2012) GABI‐Kat SimpleSearch: new features of the Arabidopsis thaliana T‐DNA mutant database. Nucleic Acids Res. 40, D1211–D1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky, J. and Jonak, C. (2012) Drought, salt, and temperature stress‐induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, V. and Palva, E.T. (1992) The expression of a rab‐related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mo. Biol. 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Levitt, J. (1980) Responses of Plants to Environmental Stresses. vol. II. Water, Radiation, Salt and Other Stresses, 2nd edn Academic Press, New York: 1980. pp. 607 ISBN 0124455026. [Google Scholar]
- Liu, X. , Zhai, S. , Zhao, Y. , Sun, B. , Liu, C. , Yang, A. and Zhang, J. (2013) Overexpression of the phosphatidylinositol synthase gene (ZmPIS) conferring drought stress tolerance by altering membrane lipid composition and increasing ABA synthesis in maize. Plant, Cell Environ. 36, 1037–1055. [DOI] [PubMed] [Google Scholar]
- Mani, F. and Hannachi, C. (2015) Genomic advances in potato drought tolerance. J. Chem. Bio. Phy. Sci. Sec. B 5, 1677–1699. [Google Scholar]
- Menges, M. , Dóczi, R. , Ökrész, L. , Morandini, P. , Mizzi, L. , Soloviev, M. , Murray, J.A.H. et al (2008) Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 179, 643–662. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Moumeni, A. , Satoh, K. , Venuprasad, R. , Serraj, R. , Kumar, A. , Leung, H. and Kikuchi, S. (2015) Transcriptional profiling of the leaves of near‐isogenic rice lines with contrasting drought tolerance at the reproductive stage in response to water deficit. BMC Genom. 16, 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki, W. (2012). Charakterystyka Krajowego Rejestru Odmian Ziemniaka. Wydanie XV. ISBN: 83‐891172‐55‐0.
- Obidiegwu, J.E. , Bryan, G.J. , Jones, H.G. and Prashar, A. (2015) Coping with drought: stress and adaptive responses in potato and perspectives for improvement. Front. Plant Sci. 6, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, B.D. , Musialak‐ Lange, M. , Nuc, P. , May, P. , Buhtz, A. , Kehr, J. , Walther, D. et al (2009) Identification of nutrient‐responsive Arabidopsis and rapeseed microRNAs by comprehensive real‐time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C. , Woo Lim, C.W. and Lee, S.C. (2016) The pepper CaOSR1 protein regulates the osmotic stress response via abscisic acid signaling. Front. Plant Sci. 7, 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak, R. , Keays, M. , Tang, Y. A. , Fonseca, N. A. , Barrera, E. , Burdett, T. , Füllgrabe, A. et al (2016) Expression atlas update‐ an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Research 44, D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak, R. , Burdett, T. , Fiorelli, B. , Fonseca, N.A. , Gonzalez‐Porta, M. , Hastings, E. , Huber, W. et al (2016) Expression atlas update‐ a database of gene and transcript expression from microarray and sequencing‐based functional genomics experiments. Nucleic Acids Res. 42, D926–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczynski, M. , Marczewski, W. , Hennig, J. , Dolata, J. , Bielewicz, D. , Piontek, P. , Wyrzykowska, A. et al (2013) Down‐regulation of CBP80 gene expression as a strategy to engineer a drought‐tolerant potato. Plant Biotechnol. J. 11, 459–469. [DOI] [PubMed] [Google Scholar]
- Pinheiro, C. and Chaves, M.M. (2011) Photosynthesis and drought: can we make metabolic connections from available data? J. Exp. Bot. 62, 869–882. [DOI] [PubMed] [Google Scholar]
- Prince, S.J. , Joshi, T. , Mutava, R.N. , Syed, N. , Vitor, J. , Mdos, S. , Patil, G. et al (2015) Comparative analysis of the drought‐responsive transcriptome in soybean lines contrasting for canopy wilting. Plant Science 240, 65–78. [DOI] [PubMed] [Google Scholar]
- Ruelland, E. , Kravets, V. , Derevyanchuk, M. , Martinec, J. , Zachowski, A. and Pokotylo, I. (2014) Role of phospholipid signalling in plant environmental responses. Env. Exp. Bot. 114, 129–143. [Google Scholar]
- Schulz, B. (2011) Functional classification of plant plasma membrane transporters In The Plant Plasma Membrane, (Murphy A. S., Schulz B. and Peer W., eds), pp. 131–176. Berlin: Springer. [Google Scholar]
- Seo, D.H. , Ryu, M.Y. , Jammes, F. , Hwang, J.H. , Turek, M. , Kang, B.G. , Kwak, J.M. et al (2012) Roles of four Arabidopsis U‐Box E3 ubiquitin ligases in negative regulation of abscisic acid‐mediated drought stress responses. Plant Physiol. 160, 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaar‐ Moshe, L. , Hubner, S. and Peleg, Z. (2015) Identification of conserved drought‐adaptive genes using a cross‐species meta‐analysis approach. BMC Plant Biol. 15, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar, R. , Bhattacharjee, A. and Jain, M. (2016) Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 6, 23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, M. , Singh, A. , Shankar, A. , Pandey, A. , Baranwal, V. , Kapoor, S. , Tyagi, A.K. et al (2014) Comprehensive expression analysis of rice armadillo gene family during abiotic stress and development. DNA Res. 21, 267–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, P. , Booij, H. , Schellekens, G.A. , Van Kammen, A. and De Vries, S.C. (1991) Cell‐specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell, 3, 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarzynska, B. , Sobkowiak, L. , Pant, B.D. , Balazadeh, S. , Scheible, W.R. , Mueller‐ Roeber, B. , Jarmolowski, A. et al (2009) Gene structures and processing of Arabidopsis thaliana HYL1‐dependent pri‐miRNAs. Nucleic Acids Res. 37, 3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja, N. (2007) Abscisic acid and abiotic stress signalling. Plant Signalling Behavior, 2, 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.M. , Zhang, L.D. , Chen, J.B. , Huang, D.F. and Zhang, Y.D. (2016) Physiological analysis and transcriptome comparison of two muskmelon (Cucumis melo L.) cultivars in response to salt stress. Genet. Mol. Res. 15, 1–18. [DOI] [PubMed] [Google Scholar]
- Winter, D. , Vinegar, B. , Nahal, H. , Ammar, R. , Wilson, G.V. and Provart, N.J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS ONE, 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L. , Ishitani, M. and Zhu, J.‐K. (1999) Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis . Plant Physiol., 119, 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Lu, G. , Long, W. , Zou, X. , Li, F. and Nishio, T. (2014a) Recent progress in drought and salt tolerance studies in Brassica crops. Breed. Sci. 64, 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N. , Lu, B. , Ma, C. , Zhang, G. , Chang, J. , Si, H. and Wang, D. (2014b) Transcriptome characterization and sequencing‐based identification of drought‐responsive genes in potato. Mol. Biol. Rep. 41, 505–517. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Yang, J. , Wang, Z. , Wen, Y. , Wang, J. , He, W. , Liu, B. et al (2014c) Identification of novel and conserved MicroRNAs related to drought stress in potato by deep sequencing. PLoS ONE, 9, e95489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielezinski, A. , Dziubek, M. , Sliski, J. and Karlowski, W.M. (2017) ORCAN‐a web‐based meta‐server for real‐time detection and functional annotation of orthologs. Bioinformatics, 33, 1224–1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Genetic background of the four studied potato cultivars Gwiazda and Oberon (A) and Tajfun and Owacja (B).
Figure S2 Gwiazda/Oberon and Tajfun/Owacja cultivar plants grown in the half‐open glasshouse on day 13 of drought.
Figure S3 RT‐qPCR analysis of RAB18 gene expression in leaves detached from Gwiazda and Oberon cultivar plants during the drought experiment.
Figure S4 Schematic representation of drought‐related gene selection using RNA‐seq data from two pairs of potato cultivars, Gwiazda/Oberon and Tajfun/Owacja.
Figure S5 Drought‐related gene selection pipeline.
Figure S6 Eight stably expressed genes in all potato cultivars studied during drought stress.
Figure S7 Seventeen of 23 potato genes selected after the third round of selection showing the highest differences in gene expression between the studied drought‐tolerant and drought‐sensitive cultivars during the drought experiment.
Figure S8 Phenotypic and RWC analyses of three Arabidopsis mutant plants with altered expression of genes homologous to the selected drought‐related potato genes.
Table S1 Comparison of the normalized number of reads for transcripts derived from genes identified in the first round of selection (594 genes) during the time course of the drought experiment.
Table S2 GO database annotation of the biological process, molecular function and cellular component categories of 8 genes selected from among the 22 top‐ranking Arabidopsis genes.
Table S3 Primer sequences.
Table S4 Summary of RNA‐seq Data.
Data S1 Experimental procedures ancillary information.


