Summary
The regulation of plant carotenogenesis is an active research area for both biological discovery and practical implementation. In tomato (Solanum lycopersicum), we demonstrate additional bottlenecks exist in the poly‐cis‐transformation of phytoene to lycopene in the context of ripening‐induced PSY1 expression and activity and reveal phytoene desaturase (PDS), as a target for manipulation towards elevated lycopene content in maturing tomato fruit. Overexpression of Arabidopsis PDS, AtPDS, elevated PDS transcript abundance in all aerial tissues resulting in both altered carotenoid accumulation and associated pathway gene expression in a tissue‐specific manner. Significant increases in downstream carotenoids (all‐trans‐lycopene and β‐carotene) and minimal changes in carotenogenic gene expression (carotenoid isomerase‐like 1, CRTIL1) suggest overexpression of heterologous AtPDS in tomato circumvents endogenous regulatory mechanism observed with previous strategies. In transgenic leaves, depletion of the PDS substrate, phytoene, was accompanied by minor, but significant increases in xanthophyll production. Alterations in the leaf carotenogenic transcript profile, including the upstream MEP pathway, were observed revealing unique feedback and feedforward regulatory mechanisms in response to AtPDS overexpression. AtPDS overexpression in the background of the tangerine (carotenoid isomerase, CRTISO) mutant exposes its potential in elevating downstream cis‐lycopene accumulation in ripe tomato fruit, as cis‐lycopene is more bioavailable yet less abundant than all‐trans‐lycopene in the wild‐type control. In summary, we demonstrate the limitation of PDS in ripening fruit, its utility in modifying carotenoid profiles towards improved quality, and reveal novel carotenoid pathway feedback regulation.
Keywords: Phytoene desaturase, carotenoid isomerization, bioavailability, fruit ripening, tomato (Solanum lycopersicum)
Introduction
Carotenoids (e.g. lutein, β‐carotene and lycopene) and their catabolites (i.e. vitamin A and other apocarotenoids) are integral in the human diet as they ensure proper development and aid in the prevention of numerous age‐related diseases (Ip et al., 2013; Lian and Wang, 2008; Rhinn and Dollé, 2012; Sharoni et al., 2012). Humans’ inability to synthesize carotenoids de novo makes them dependent on plants as their primary source of dietary carotenoids, the acquisition of which is dependent upon both content in specific plant tissues and their ability to be absorbed in the digestive tract (bioavailability). The capacity of many fruits to accumulate elevated levels of carotenoids as they ripen makes fruits an important source of dietary carotenoids in many diets. Lycopene, the red carotenoid predominant in ripe tomatoes, has received increased attention as a target for manipulation due to evidence that lycopene‐derived apocarotenoid signals aid in cancer prevention (Ip et al., 2013; Lian and Wang, 2008).
In plants, carotenoids are essential for light absorption in photosynthetic tissues and as provisional colorants in flowers and fruit facilitating the attraction of pollinators and seed dispersing organisms (reviewed in Nisar et al., 2015). The enhanced accumulation of carotenoids in ripening fruit is dependent on the switch from a photosynthetic to a nonphotosynthetic‐state, which embodies a remarkable transition of chloroplast to chromoplast‐rich cells, concurrent with strategic adjustments to the carotenoid profile within (Barry et al., 2008, 2012). Ripening‐associated carotenogenic gene expression in tomato is regulated by a complex symphony of transcription factors, the gaseous hormone ethylene and epigenome dynamics driving the flux of the pathway to all‐trans‐lycopene (and inhibition of its subsequent metabolism) conferring the typical red colour of tomato (Barry et al., 2005; Eriksson et al., 2004; Gallusci et al., 2016; Giovannoni et al., 2017; Martel et al., 2011; Vrebalov et al., 2002, 2009; Zhong et al., 2013). In the mature fruit, phytoene synthase (the first committed step in carotenogenesis) is encoded by the highly regulated PSY1 gene and is the major limiting activity for carotenoid flux (Figure 1a) (Fraser et al., 2002). Once synthesized by PSY1, phytoene undergoes a poly‐cis‐transformation conferred by four desaturation reactions catalysed by phytoene desaturase (PDS) and ζ‐carotene desaturase (ZDS) and two isomerizations facilitated by ζ‐carotene isomerase (ZISO) and carotene isomerase (CRTISO) ultimately producing all‐trans‐lycopene (Figure 1a) (Alba et al., 2005; Fantini et al., 2013; Isaacson et al., 2002; Yu et al., 2011). Subsequent processing of all‐trans‐lycopene is prevented through downstream repression of lycopene ε‐ and β‐cyclases involved in synthesizing α‐carotene (ε,β‐rings), the precursor to lutein and the provitamin A, β‐carotene (β,β‐rings) (Figure 1a) (Ronen et al., 1999, 2000, respectively). These combined activities, tightly regulated to push carotenoid flux towards specific products in the ripe fruit, demonstrate finely tuned pathway regulation.
Figure 1.
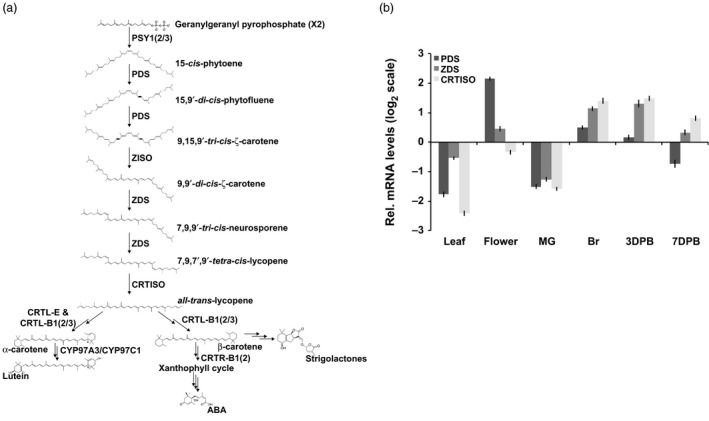
PDS is limiting in carotenogenesis during tomato fruit ripening. (a) Carotenoid biosynthetic pathway with enzymes abbreviated in bold: PSY1, 2 and 3; phytoene synthase 1, 2 and 3; PDS, phytoene desaturase; ZISO, ζ‐carotene isomerase; ZDS, ζ‐carotene desaturase; CRTISO, carotene isomerase; CRTL‐E (LCY‐E), lycopene ε‐cyclase; CRTL‐B1 and 2 (LCY‐B and BCYC, respectively), lycopene β‐cyclase; CYP97A3, carotene ε‐hydroxylase; CRTR‐B1 and 2, carotene β‐hydroxylase 1 and 2; ABA is abscisic acid. (b) Transcript levels of PDS,ZDS and CRTISO relative to a reference sample (combination of equal amounts of RNA from each tissue analysed) (n = 3, in triplicate; error bars, ±SEM). Abbreviated tissues represent different stages of fruit ripening, mature green fruit, MG; breaker or onset of ripening, Br; and 3 and 7 days post breaker, 3DPB and 7DPB, respectively.
Multiple strategies have been pursued to enhance lycopene content in tomato deploying either plant or bacterial transgenes (reviewed in Fraser et al., 2009). In carotenoid deficient staple crops (e.g. rice and potato), the use of the bacterial carotenoid desaturase, CRTI, has proven effective due to its ability to catalyse all six reactions required to convert phytoene to all‐trans‐lycopene (Diretto et al., 2007; Misawa et al., 1990; Ye et al., 2000). Due to mechanisms yet to be defined, CRTI introduction in planta drives carotenogenesis beyond all‐trans‐lycopene to β‐carotene via induced lycopene β‐cyclase expression (Enfissi et al., 2017; Nogueira et al., 2013; Römer et al., 2000; Ye et al., 2000). In contrast to the successful applications in low carotenoid tissues, ectopic expression of CRTI was counterproductive for the enhancement of lycopene content in tomato fruit.
CRTI's structure has been solved facilitating characterization of its function as an FAD‐dependent oxidase/isomerase (Schaub et al., 2012). Two subsequent studies provided insight on the causal processes leading to β‐carotene enhancement in the tomato including CRTI protein localization (Nogueira et al., 2013) in the plastid and lycopene‐mediated pathway regulation (Enfissi et al., 2017). Remaining unclear is the potential for an uncontrolled elevation of a foreign carotenogenic protein (i.e. CRTI) to disrupt one or more of the many protein interactions involved in plant carotenogenesis, thus negatively affecting carotenoid gene expression (reviewed in Shumskaya and Wurtzel, 2013). Targeted manipulation of one or more of the four native plant enzymes carrying out limiting CRTI reactions in ripening fruit may facilitate enhanced all‐trans‐lycopene accumulation in an already high carotenoid tissue and provide insight into aspects of endogenous pathway regulation.
Here, we (i) identify PDS within the poly‐cis‐transformation of phytoene to lycopene as a useful target for manipulation towards carotenoid enhancement in ripening tomato fruit, (ii) fully characterize the effect of PDS overexpression on carotenoid content and carotenoid gene expression in aerial organs and (iii) demonstrate the utility of PDS overexpression combined with CRTISO repression via the tangerine (t 3183) mutation as a means to both elevate carotenoid levels and modify carotenoid accumulation profiles towards more bioavailable forms compared to wild‐type control tomatoes (Cooperstone et al., 2015).
Results
PDS becomes limiting once PSY1 is elevated during ripening
There is overwhelming evidence that PSY1 represents the major rate‐limiting step in ripening‐associated carotenogenesis (McQuinn et al., 2015; Nisar et al., 2015). The prevalence of intermediates in the poly‐cis‐transformation of 15‐cis‐phytoene to all‐trans‐lycopene as the fruit ripens suggests additional points of pathway limitation providing targets to develop alternate strategies for enhancing flux to desired downstream carotenoids. In terms of amino acid sequence identity and conserved domains, PDS and ZDS, enzymes are predicted to have evolved from CRTI, with 41% and 30% identity, respectively, and both contain the same functional domains, NAD‐binding domain and amine oxidase domain (NCBI Conserved Domains: http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Figure S1). In contrast, ZISO is considerably more distant and unrelated lacking both the NAD‐binding and amine oxidase domains and instead contains a nitric acid domain (NCBI Conserved Domains: http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Figure S1). Furthermore, analysis of protein structure (TOPCONS.net) indicates ZISO is the only enzyme in this part of the pathway predicted as a transmembrane protein with multiple transmembrane domains, consistent with previous reports (Beltrán et al., 2015). For these reasons and to gain a better understanding of the previously demonstrated metabolic constraints associated with CRTI overexpression, both PDS and ZDS were the focus of the efforts reported here, while ZISO, a more distant and distinct protein at the amino acid sequence and structural level (Beltrán et al., 2015), was left for later examination.
In an effort to identify the optimal target for increasing carotenoid pathway flux post‐PSY1 activity, expression patterns of PDS and ZDS were measured through four stages of fruit ripening (Mature Green, MG; breaker, Br; and 3 and 7 days postbreaker, 3DPB and 7DPB) using Quantitative Real‐Time PCR. PDS was the lowest expressed gene in the fruit as compared to ZDS and CRTISO (Figure 1b). While transcription does not determine enzyme activity, the elevated prevalence of PDS substrates (phytoene and phytofluene) compared to that of ZDS (9,9′di‐cis‐ζ‐carotene and neurosporene) in ripe fruit, as well as trace levels of PDS substrates observed in leaves (Table 1) suggest PDS activity, may be limiting during carotenogenesis. PDS shares the substrate, phytoene, with CRTI allowing for the additional assessment of potential feedback or feedforward effects from depletion of a specific substrate (i.e. phytoene) or over accumulation of a specific downstream product (i.e. all‐trans‐lycopene) as speculated in previous reports (Enfissi et al., 2017; Fantini et al., 2013; Kachanovsky et al., 2012). Together these observations suggested PDS as a high value target within the poly‐cis‐transformation for manipulation with the goal of elevating carotenoid accumulation.
Table 1.
Available carotenoid content (μg/g FW) in the poly‐cis‐transformation of phytoene to all‐trans‐lycopene in leaves, flowers, and ripe fruit from wild type (AC) tomato plants. (n ≥ 5)
| PDS | ZDS | CRTISO | |||
|---|---|---|---|---|---|
| 15′ cis‐phytoene | cis‐phytofluene | 9,9′ di cis‐ζ‐carotene | cis‐neurosporene | Prolycopene | |
| Leaf | 0.07 ± 0.004 | n.d. | n.d. | n.d. | n.d. |
| Flower | n.d. | n.d. | n.d. | n.d. | n.d. |
| Ripe fruit | 1.72 ± 0.11 | 0.95 ± 0.06 | 0.23 ± 0.03 | n.d. | n.d. |
Carotenoid content is presented as μg/g fresh weight (FW). Values represent the mean of a minimum of five biological replicates ± standard error. n.d. denotes not detected.
Heterologous overexpression of Arabidopsis PDS in tomato enhances downstream carotenoid content in ripe fruit
The potential for activating an endogenous targeted gene repression system (e.g. siRNA‐mediated gene silencing) in response to the introduction of a transgene can be exacerbated due to high nucleotide sequence similarity between the endogenous gene and the transgene. The coding sequence of AtPDS shares only 72% of sequence identity with that of the endogenous SlPDS, reducing the potential for activating DNA sequence homology‐based repression systems. For this reason, the Arabidopsis homolog of PDS, AtPDS (GenBank accession no. NM_202816) was deployed for overexpression rather than SlPDS, under the control of the constitutive CaMV 35S promoter in the wild‐type tomato background cv. Ailsa Craig (AC++). Four independent homozygous transgenic lines (i.e. AtPDS1A.1; AtPDS3.2; AtPDS4.2; AtPDS6.2) displaying elevated levels of AtPDS transcripts in the three aerial plant organs analysed (i.e. leaves, flowers and ripe fruits) were propagated to the T2 generation (Figure 2a). The observed minimal effect on endogenous PDS transcript levels (Figure 2a) indicated that any operating suppression systems had been bypassed. Ripe fruit from all four AtPDS overexpressing lines presented noticeably deeper red pigmentation compared to their wild‐type counterparts (Figure 2b).
Figure 2.
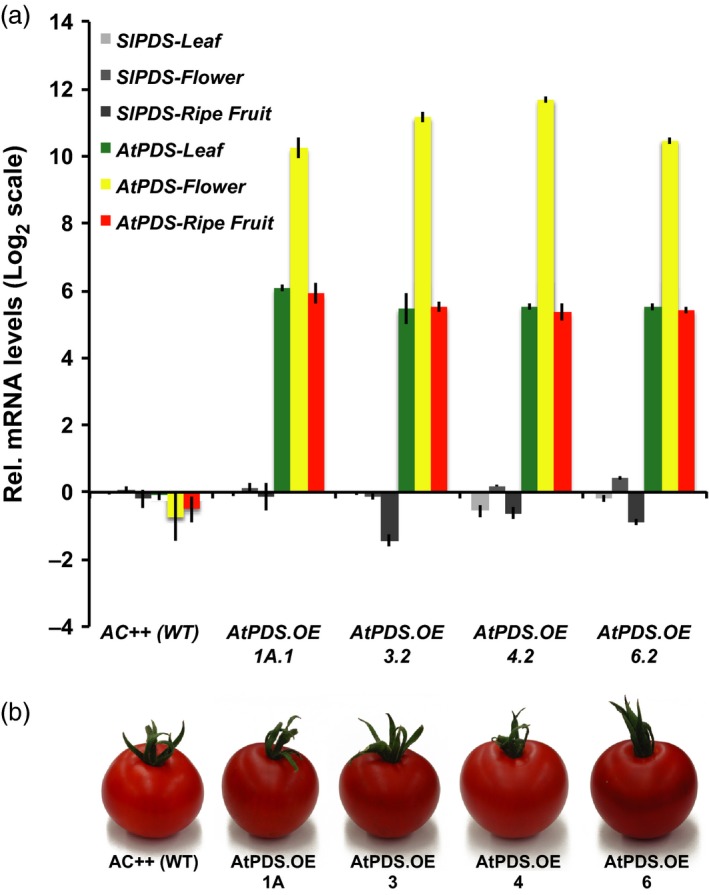
Overexpression of AtPDS enhances tomato fruit colour. (a) Transcript levels of tomato and Arabidopsis PDS (SlPDS and AtPDS, respectively) in stable, T2 generation AtPDS overexpression lines relative to the wild‐type control (cv. Ailsa Craig). Ripe fruit represents 7DPB (n = 5, in triplicate; error bars, ±SEM). (b) Visual phenotypes of the chromoplast‐rich 7DPB fruit of the stable, T2 generation AtPDS.OE lines compared to the wild‐type control (cv. Ailsa Craig).
Ripe fruit displayed the greatest increase in carotenoids downstream of PDS. Flowers and leaves had reduced and elevated lutein levels, respectively (Table 2). The metabolic effectiveness of AtPDS overexpression was evident in the reduction of steady‐state levels of phytoene and phytofluene, the substrate and intermediate of the PDS catalysed reaction, respectively, in both ripe tomato fruit (Table 2 and Figure 3a) and leaves (Table 2 and Figure S3). Total lycopene and β‐carotene were increased up to 31.1% and 42.8%, respectively (Table 2 and Figure 3b,c). With regard to the elevated total lycopene content, 31.1% equals an increase of 21.3 μg/g fresh weight in this already high lycopene tissue (Figure 3b). The observed increases in total lycopene and β‐carotene content in AtPDS overexpressing ripe are not a consequence of reduced fruit size (Figure S4). The largest metabolic shifts were observed in the immediate products of the PDS enzymatic reaction where total ζ‐carotene levels were >160% higher than in wild‐type fruit (Table 2 and Figure 3a).
Table 2.
Carotenoid composition in ripe fruit (7DPB), flower petals, and leaves of stable T2 generatio AtPDS.OE tomato plants compared to wild type (AC) tomato plants
| Genotype | Carotenoid composition (% of control ±SEM, n > 5) | |||||||
|---|---|---|---|---|---|---|---|---|
| Phytoene | Phytofluene | ζ‐carotene | Lycopene | ζ‐carotene | Lutein | Xanthophylls | Total carotenoids/xanthophylls | |
| Fruit (7DPB) | ||||||||
| Ailsa Craig (WT) | 100.0 ± 6.4 | 100.0 ± 6.5 | 100.0 ± 13.9 | 100.0 ± 3.9 | 100.0 ± 2.6 | 100.0 ± 7.2 | 100.0 ± 3.7 | |
| AtPDS.OE.1A.1 | 28.5 ± 1.9 | 24.9 ± 1.7 | 234.6 ± 23.8 | 110.1 ± 6.4 | 123.4 ± 9.7 | 90.9 ± 6.4 | 108.3 ± 6.3 | |
| AtPDS.OE.3.2 | 30.0 ± 3.0 | 26.1 ± 2.2 | 228.6 ± 24.1 | 121.9 ± 7.0 | 131.2 ± 11.5 | 102.9 ± 3.2 | 119.4 ± 6.3 | |
| AtPDS.OE.4.2 | 32.1 ± 3.2 | 28.5 ± 2.4 | 262.6 ± 24.3 | 131.1 ± 3.5 | 142.8 ± 6.9 | 101.6 ± 0.9 | 128.5 ± 3.0 | |
| AtPDS.OE.6.2 | 35.3 ± 4.7 | 30.7 ± 3.8 | 213.2 ± 28.3 | 100.8 ± 5.8 | 108.4 ± 5.7 | 86.9 ± 7.8 | 99.2 ± 5.5 | |
| Flower petals | ||||||||
| Ailsa Craig (WT) | 100.0 ± 6.2 | 100.0 ± 3.5 | 100.0 ± 4.9 | 100.0 ± 4.5 | ||||
| AtPDS.OE.1A.1 | 110.9 ± 5.8 | 92.4 ± 3.3 | 106.3 ± 4.6 | 105.7 ± 4.2 | ||||
| AtPDS.OE.3.2 | 100.7 ± 6.5 | 92.3 ± 5.4 | 101.8 ± 4.0 | 101.2 ± 3.5 | ||||
| AtPDS.OE.4.2 | 95.5 ± 5.1 | 89.1 ± 3.1 | 103.8 ± 5.1 | 102.2 ± 4.7 | ||||
| AtPDS.OE.6.2 | 109.6 ± 4.2 | 95.4 ± 3.2 | 108.5 ± 2.6 | 107.8 ± 2.4 | ||||
| Leaves | ||||||||
| Ailsa Craig (WT) | Trace | 100.0 ± 6.4 | 100.0 ± 2.9 | 100.0 ± 3.0 | 100.0 ± 3.3 | |||
| AtPDS.OE.1A.1 | n.d. | 102.3 ± 6.4 | 118.2 ± 6.6 | 115.0 ± 4.5 | 113.3 ± 4.7 | |||
| AtPDS.OE.3.2 | n.d. | 101.8 ± 4.6 | 110.9 ± 4.2 | 103.6 ± 3.5 | 104.0 ± 3.6 | |||
| AtPDS.OE.4.2 | n.d. | 108.3 ± 2.8 | 117.6 ± 2.8 | 104.1 ± 2.7 | 106.6 ± 2.5 | |||
| AtPDS.OE.6.2 | n.d. | 102.4 ± 4.1 | 106.4 ± 5.9 | 92.4 ± 5.0 | 96.1 ± 4.9 | |||
Carotenoid contents are presented as percent of control [Ailsa Craig(WT)] unless otherwise indicated. Values represent a mean of a minimum of 5 biological replicates ±SEM. Trace denotes detected at very low levels, n.d. denotes not detected. (Representative chromatograph and UV spectrums of carotenoids detected in the wildtype ripe tomato shown in Figure S2).
Figure 3.
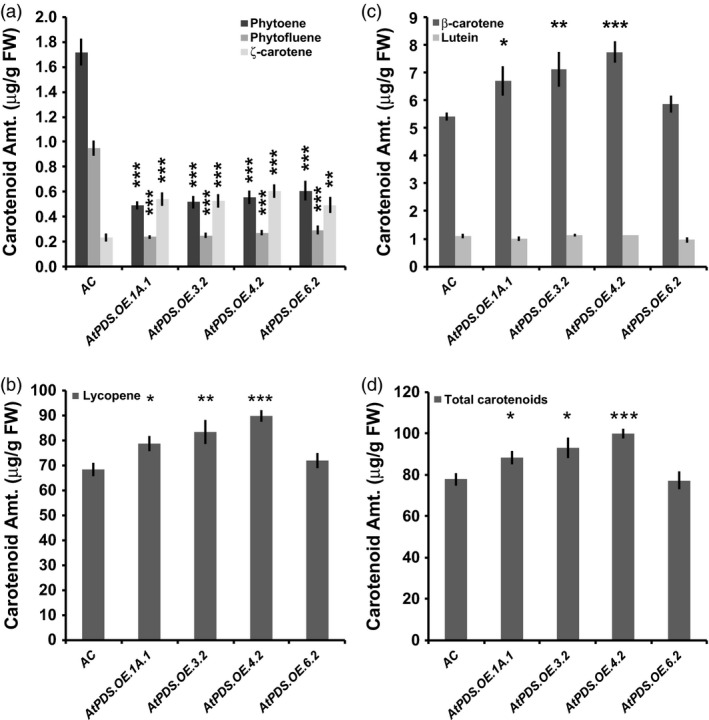
AtPDS overexpression pushes residual phytoene towards synthesis of downstream carotenoids in ripe tomato fruit. (a) Phytoene; phytofluene; and ζ‐carotene content (μg/g FW) in ripe fruit of stable, T2 generation AtPDS.OE lines compared to wild type (n = 5, error bars ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (b) Lycopene content (μg/g FW) in ripe fruit of stable, T2 generation AtPDS.OE lines compared to wild type (n = 5, error bars ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (c) β‐Carotene content (μg/g FW) in ripe fruit of stable, T2 generation AtPDS.OE lines compared to wild type (n = 5, error bars ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (d) Total carotenoid content (μg/g FW) in ripe fruit of stable, T2 generation AtPDS.OE lines compared to wild type (n = 5, error bars ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001).
AtPDS overexpression in ripening fruit resulted in altered transcription of CRTIL1
Considering the previously observed deregulation of transcripts associated with carotenoid biosynthesis in CRTI overexpressing fruit (Römer et al., 2000), it was necessary to quantify transcript abundance of genes throughout the biosynthetic pathway in ripe fruit of AtPDS.OE lines. A comprehensive analysis of the resulting carotenogenic transcript profile in AtPDS.OE ripe fruit was carried out via qRT–PCR with gene‐specific primers (Table S2) and compared to transcript abundance observed in the WT ripe fruit. Contrary to the observed off‐target changes in carotenogenic gene expression in CRTI overexpressing fruit first described in Römer et al. (2000), no consistent or statistically significant changes in PSY1, CRTL‐B1 and CRTL‐B2 gene expression were identified (Figure 4a). Analysis of relative transcript levels for the remaining carotenoid biosynthetic genes (DXS; DXR; IDI1; PSY2, ZISO; ZDS; CRTISO; CRTIL1; CRTR‐B1; CRTR‐B2; and CRTL‐E) further shows limited impact of elevated PDS on carotenoid pathway self‐regulation. A solitary significant change was observed in the reduction of CRTIL1 transcript levels, a gene suggested to be involved in the isomerization of ζ‐carotenes (Fantini et al., 2013) (Figure 4b). These results indicate that PDS overexpression can provide a means for elevating carotenoid content in ripe fruit with limited feedback or feedforward effects on carotenoid gene expression and furthermore that CRTIL1 responds to changes in flux through early steps in the pathway during tomato ripening.
Figure 4.
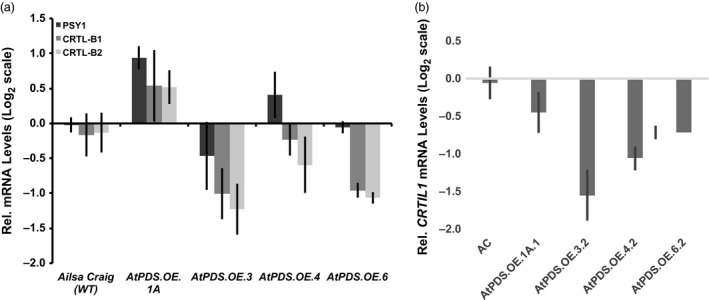
Carotenoid gene expression changes limited to CRTIL1 in AtPDS.OE ripe fruit. (a) Transcript levels of PSY1,CRTL‐B1 and CRTL‐B2 in the ripe fruit of stable, T2 generation AtPDS.OE lines relative to the wild‐type control (cv. Ailsa Craig) (n = 3, in triplicate; error bars ±SEM). (b) Transcript levels of CRTIL1 in the ripe fruit of stable, T2 generation AtPDS.OE lines relative to the wild‐type control (cv. Ailsa Craig) (n = 3, in triplicate; error bars ±SEM).
AtPDS overexpression in tomato leaves and flowers alters gene expression throughout the pathway
AtPDS overexpression in leaves enhanced carotenoid production in downstream xanthophylls, lutein, β‐cryptoxanthin and zeaxanthin to small but measurable degrees (Figure 5a,b). Analysis of transcript levels of carotenoid biosynthetic genes by qRT–PCR revealed that AtPDS overexpression in tomato leaves had broad regulatory consequences and influenced multiple steps throughout the carotenogenic pathway. The gene representing the rate‐limiting step of the MEP pathway (Rodríguez‐Concepción and Boronat, 2002), deoxyxylulose 5‐phosphate (DXP) synthase (DXS) was induced approximately twofold in the AtPDS.OE lines compared to the wild‐type control (Figure 5c). PSY1 was also elevated in leaves, but to a lesser amount than DXS in AtPDS.OE leaves (Figure 5c). Lycopene β‐cyclase 1 (CRTL‐B1) and β‐carotene‐hydroxylase 1 (CRTR‐B1) were both expressed at higher levels in the AtPDS.OE leaves (Figure 5c). CRTR‐B1 increased the most, (threefold to fourfold), and in combination with other changes in carotenoid gene expression provides insight into the mechanism underlying the elevated lutein, β‐cryptoxanthin and zeaxanthin content (Figure 5). β‐Carotene content remained unchanged (Figure 5).
Figure 5.
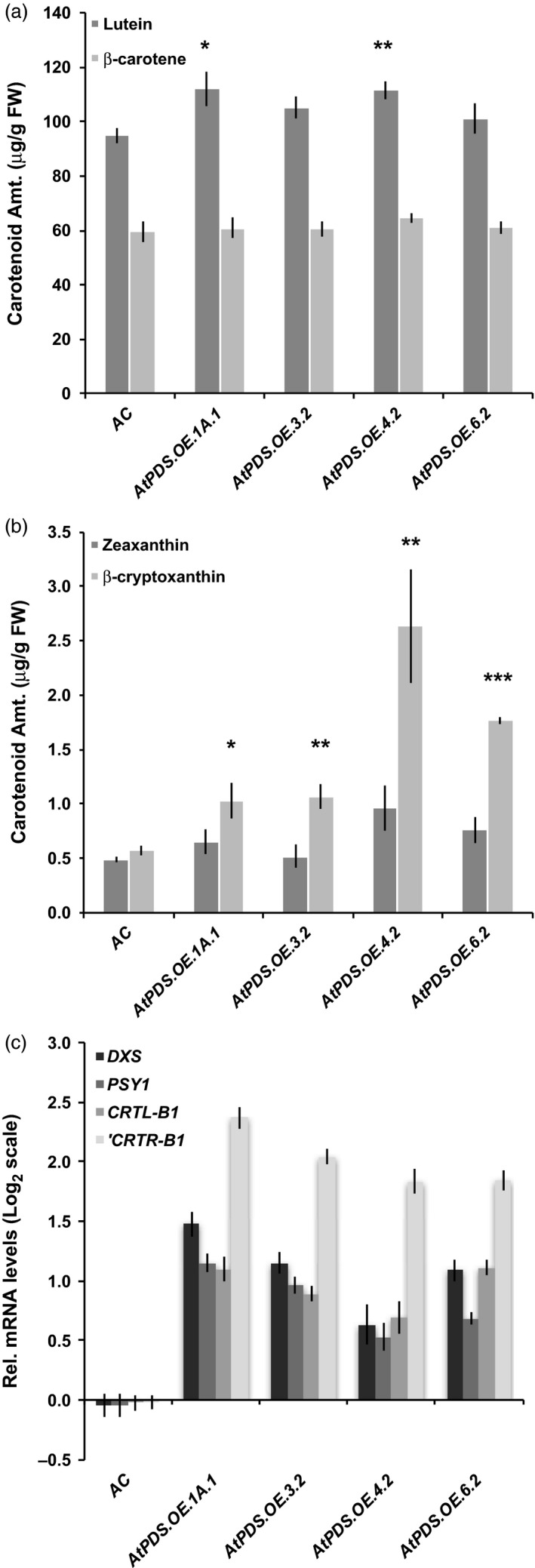
Enhanced photoprotective xanthophyll content in young leaves of AtPDS.OE lines. (a) Lutein and β‐carotene contents in young leaves of stable, T2 generation AtPDS.OE lines compared to wild type (n = 5, error bars ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (b) Zeaxanthin and β‐cryptoxanthin contents in young leaves of stable, T2 generation AtPDS.OE lines compared to wild type (n = 5, error bars ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (c) Endogenous regulation of carotenoid biosynthetic genes, deoxyxylulose 5‐phosphate (DXP) synthase, DXS; phytoene synthase 1, PSY1; chloroplast‐specific lycopene β‐cyclase CRTL‐B1; and chloroplast‐specific carotene β‐hydroxylase, CRTR‐B1 (n = 5, in triplicate; error bars ±SEM).
Carotenoid biosynthetic gene expression analysis of the chromoplast‐rich flowers displayed limited alterations in response to heterologous PDS expression, and this was again even more limited in the maturing fruit as noted above. The gene encoding the step immediately following PDS, ζ‐carotene isomerase (ZISO), was elevated approximately twofold compared to wild‐type flowers (Figure 6). CRTR‐B1 was reduced slightly in the AtPDS.OE flowers, potentially limiting the ability to increase xanthophyll content of the petals and anthers as confirmed by HPLC (Figure 6).
Figure 6.
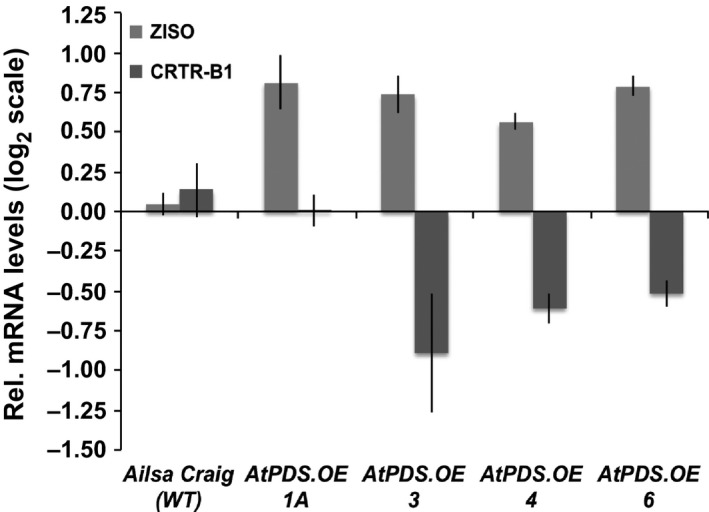
Deregulation of ZISO and CRTR‐B1 in AtPDS.OE flowers. Transcript levels of ZISO and CRTR‐B1 in the anthesis flowers of stable, T2 generation AtPDS.OE lines relative to the wild‐type control (cv. Ailsa Craig) (n = 3, in triplicate; error bars ±SEM).
AtPDS overexpression enhances cis‐lycopene accumulation in tangerine (CRTISO) mutant ripe fruit
In an effort to further amplify the value of the metabolic shift towards cis‐carotenoids in tangerine (t 3183) mutant fruit, both AtPDS3.2 and the high pigment 1 (hp1) mutations were crossed independently into the t 3183 background. The hp1 mutation in the tomato DAMAGED DNA BINDING PROTEIN1 (DDB1) gene was selected as it confers an increase in plastid number resulting in elevated accumulation of carotenoids in ripe fruit (Liu et al., 2004), a means of carotenoid elevation distinct from the direct increase of pathway flux conferred by AtPDS overexpression in tomato. hp1;t 3183 double‐mutant lines were also developed as a standard for comparison of metabolic pathway shift.
Plants homozygous for the tangerine (t 3183 ) mutation and positive for the AtPDS transgene or hp1 were selected in F2 populations. All mutations and transgenes were nearly isogenic in the inbred Ailsa Craig background to minimize additional genetic variation in double‐mutant lines. Phenotypic and HPLC analyses were carried out on the ripe fruit of the AtPDS3.2;t 3183 double mutant, the t 3183 parent line and the hp1;t 3183 double mutant. Compared to both the parental t 3183 and hp1;t 3183 ripe fruit, AtPDS3.2;t 3183 fruit were darker in colour (Figure 7a). The enhanced pigmentation associated with AtPDS3.2;t 3813 was observed throughout the pericarp of cut fruit (Figure 7b).
Figure 7.
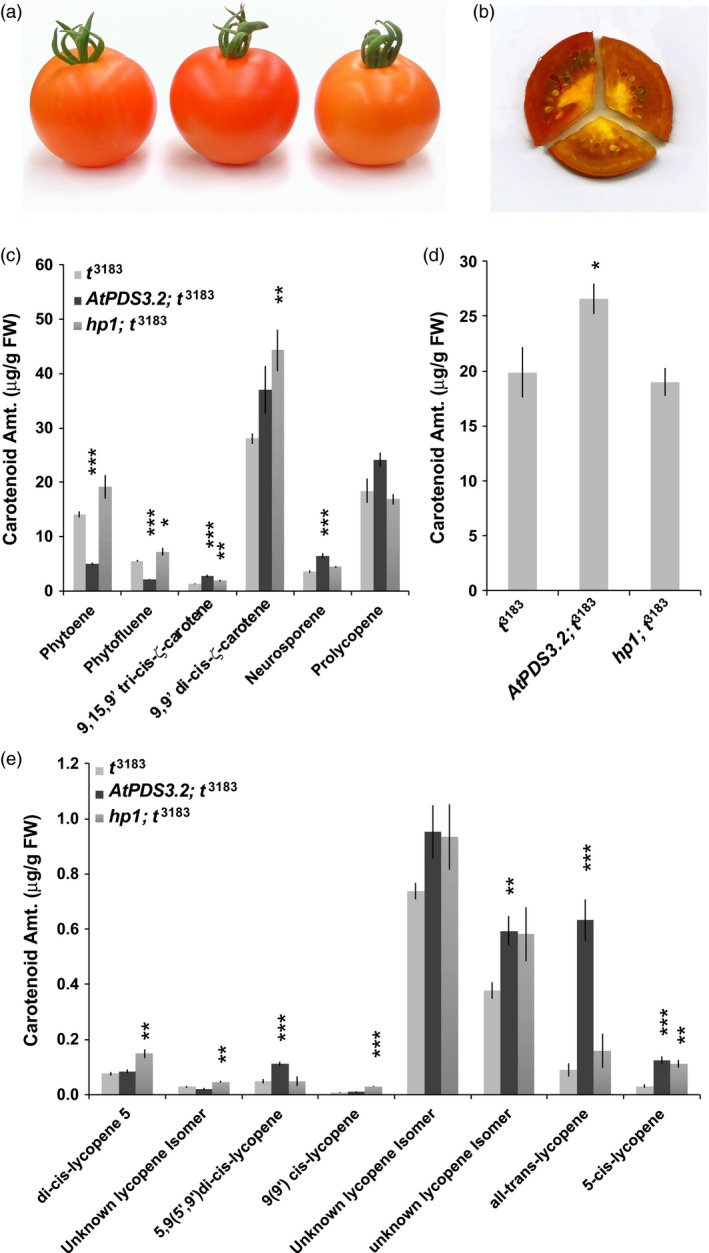
Overexpression of AtPDS enhances downstream cis‐carotenoid accumulation in the tangerine mutant. (a) External visual phenotype of tangerine (t 3183) compared to AtPDS3.2; t 3183 and hp1; t 3183 double mutants. (b) Cross sections of ripe fruit display internal pericarp pigmentation in t 3183, AtPDS3.2; t 3183 and hp1; t 3183. (c) Cis‐carotenoid content (μg/g FW) from the poly‐cis‐transformation of phytoene to all‐trans‐lycopene in t 3183, AtPDS3.2; t 3183 and hp1; t 3183 7DPB fruit (n = 5; error bars, ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (d) Total lycopene content in 7DPB fruit of t 3183, AtPDS3.2; t 3183 and hp1; t 3183 (n = 5; error bars, ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). (e) Cis‐ and all‐trans‐lycopene content (μg/g FW) accumulating in 7DPB fruit of t 3183, AtPDS3.2; t 3183 and hp1; t 3183 (n = 5; error bars, ±SEM) (*P < 0.05; **P < 0.01; ***P < 0.001). All carotenoid names identify peaks with similar characteristics and retention timing as those identified in Melendez‐Martinez et al. (2013).
HPLC analysis revealed that AtPDS overexpression reduced steady‐state levels of phytoene and phytofluene by 64.8% and 61.8%, respectively, consistent with reductions observed when overexpressed in the wild‐type background (Figure 7c and Table 2; Figure S5). AtPDS efficiency appears to be influenced by tissue type as AtPDS3.2;t 3183 anthesis flowers only displayed a 30% reduction relative to t 3183 phytoene and phytofluene levels (Figures 7c and S6). Both AtPDS overexpression and the hp1 mutation led to a similar increase in 9,9′‐di‐cis‐ζ‐carotene content when combined with t 3183 (Figures 7c and S5), while AtPDS3.2;t 3183 alone displayed significant increases in products downstream of 9,9′‐di‐cis‐ζ‐carotene when compared to both t 3183 and hp1;t 3183 (Figures 7c and S5). Total neurosporene accumulates up to 84.5% higher in AtPDS3.2;t 3183 compared to t 3183 and 44.5% higher than hp1;t 3183 (Figures 7c and S5). Additionally, tetra‐cis‐lycopene (prolycopene) levels are 30.3% and 42.2% higher than in t 3183 and hp1;t 3183, respectively (Figures 7c and S5).
Total lycopene levels followed the same trend as prolycopene in the three genotypes with AtPDS3.2;t 3183 displaying the highest levels (Figures 7d and S5). Subsequent lycopene isomer identification and quantification in the ripe fruit demonstrates genotype‐specific effects on particular lycopene isomers (Figures 7e and S7; Table S2). Introduction of AtPDS overexpression and the hp1 mutation in t 3183 conferred elevations in two unidentified lycopene isomers and 5‐cis‐lycopene relative to the t 3183 parental line (Figure 7e). AtPDS3.2;t 3183 tomatoes accumulate 2 and 7 times more 5,9(5′,9′) di‐cis‐lycopene and all‐trans‐lycopene, respectively, compared to levels found in t 3183 tomatoes, while levels remain relatively unchanged in hp1;t 3183 (Figure 7e). The presence of genotype‐specific changes in cis‐lycopene isomers and elevated total lycopene content observed in AtPDS3.2;t 3183 demonstrates the potential for targeting PDS in a means that bypasses endogenous regulation in concert with CRTISO to enhance bioavailable cis‐lycopene isomers in tomato.
Discussion
AtPDS overexpression effectively enhanced carotenoid accumulation in fruit and significantly alters the carotenoid gene expression pattern in a tissue‐specific manner
Carotenogenesis is heavily regulated regardless of plant organ, with PSY1 activity the major rate‐limiting step in ripening‐associated carotenoid biosynthesis (McQuinn et al., 2015; Nisar et al., 2015). This study demonstrates that in the context of normal ripening induction and function of PSY1, PDS activity becomes limiting and when elevated through deployment of a heterologous transgene, can significantly increase downstream carotenoid (i.e. ζ‐carotene, lycopene and β‐carotene) accumulation and total carotenoid output (Table 2 and Figure 3). Moreover, when overexpressed in the ripening tomato, PDS metabolized 70% of the remaining phytoene in the plastid ultimately contributing to the elevated downstream carotenoids including the desired all‐trans‐lycopene (Table 2). The largest relative increase in carotenoid content was in ζ‐carotene suggesting that as PDS activity was elevated, ZISO and ZDS now became limiting. These two genes thus present interesting targets for further carotenoid enhancement in ripening fruit with elevated PDS.
Importantly and in contrast to previous observations with transgenic tomatoes expressing the bacterial CRTI (Enfissi et al., 2017; Nogueira et al., 2013; Römer et al., 2000), total carotenoid content was significantly elevated in AtPDS.OE ripe fruit (Table 2 and Figure 3d). Furthermore, PSY1 transcripts did not decrease, nor did lycopene β‐cyclase transcripts (CRTL‐B1 and CRTL‐B2) increase across the four independent transgenic lines (Figure 4). The lack of feedback regulation within the carotenogenic pathway may rely on the fact that an enzyme native to plants, PDS, was introduced rather than a more distant bacterial enzyme, CRTI. Given that both PDS and CRTI considerably deplete the phytoene pool in the plastids (based on observed steady‐state levels), the observed feedback regulation does not appear directly attributable to a response to lower substrate availability. It has recently been suggested that this response may be due to an elevation in lycopene content (Enfissi et al., 2017). PDS increases the accumulation of all‐trans‐lycopene in ripe tomato plastids more than with CRTI overexpression (Table 2 and Figure 3b)( Enfissi et al., 2017; Nogueira et al., 2013; Römer et al., 2000). While the lack of elevated lycopene in response to CRTI is due to enhanced lycopene β‐cyclase activity, additional studies with CRTB (bacterial PSY) overexpressed in ripening tomatoes increased lycopene, absent increases in lycopene β‐cyclase (CRTL‐B1 and CRTL‐B2) expression (Fraser et al., 2002; Nogueira et al., 2013). It is possible that misregulation throughout the pathway in response to CRTI may be through means other than or in addition to elevated lycopene.
While PDS evolved from CRTI, PDS and CRTI function in contrast to each other with regard to maintenance of the FAD redox state and may differentially influence carotenoid pathway regulation in the ripening tomato fruit. It has been established that the plastid terminal oxidase, PTOX and the plastoquinone (PQ) pool controls the redox state and function of PDS in plants (Carol et al., 1999; Joët et al., 2002; Josse et al., 2003; Norris et al., 1995; Yu et al., 2014). Moreover, while in photosynthetic chloroplasts, alternative mechanisms appear to aid in the maintenance of PDS's redox state, in nonphotosynthetic chromoplasts, PDS is solely dependent on PTOX and the PQ pool (Shahbazi et al., 2007). Norflurazon inhibits PDS activity by maintaining the FAD within PDS in a reduced state once it is lodged in the active site (Gemmecker et al., 2015). In contrast, CRTI cannot be inhibited by norflurazon (Sandmann and Fraser, 1993), and as suggested by Schaub et al. (2012), it remains unclear whether CRTI can be oxidized similarly to PDS by PQ in planta. Therefore, in contrast to PDS overexpression, ripe fruit expressed CRTI protein may have the potential to disrupt the redox state of the PQ pool providing an alternative explanation for the observed carotenoid pathway misregulation.
AtPDS overexpression successfully circumvents the negative feedback mechanisms observed in response to bacterial CRTI. Indeed, AtPDS overexpression associated elevation of lycopene and β‐carotene in the T2 generation ripe fruit is improved compared to values observed with other strategies (Enfissi et al., 2004; Fraser et al., 2007). Lycopene content resulting from AtPDS overexpression is higher than reported in DXS overexpression lines in the T1 generation, while total carotenoid content stays comparable (Enfissi et al., 2004). Observations by Fraser et al. (2007) demonstrated that PSY1 overexpression increases phytoene content in ripe transgenic fruit, but has no overall effect of the steady‐state levels of lycopene when compared to wild‐type fruit. In contrast, AtPDS overexpression overcomes the apparent limitation in the pathway at the PDS step and increases the steady‐state levels of lycopene in the ripe transgenic tomato fruit.
Many examples of pathway self‐regulation have been reported including, but not limited to, the feedback regulation observed when CRTI is overexpressed in tomato fruit (Enfissi et al., 2017; Nogueira et al., 2013; Römer et al., 2000) and epistatic regulation of PSY1 in the spontaneous tangerine (CRTISO) mutation (Kachanovsky et al., 2012; and reviewed in Cazzonelli and Pogson, 2010). Therefore, it was not surprising to observe tissue‐specific transcriptional perturbations of carotenogenic genes in response to AtPDS overexpression (Figure 8). Interestingly, artificially increasing flux via elevated PDS triggered major alterations in gene expression throughout the carotenoid biosynthetic pathway in leaves (Figures 5 and 8), while modifications in transgenic ripe fruit across all four independent AtPDS.OE lines were limited to a small change in CRTIL1 (Figures 4b and 8). Additionally, minor unique changes in pathway gene expression were also detected in the flowers but with no significant alterations in carotenoid content (Figures 6 and 8).
Figure 8.
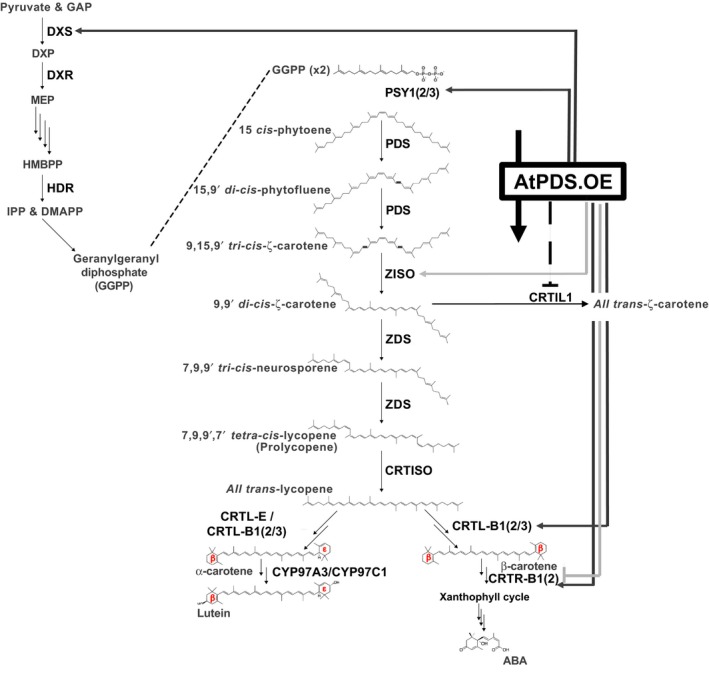
Model of feedback and feedforward regulation induced by AtPDS overexpression in ripe fruit, leaves and flowers. Enzymes within the simplified MEP pathway are abbreviated in bold. DXS, deoxyxylulose 5‐phosphate (DXP) synthase; DXR, DXP reductase; HDR, 4‐hydroxy‐3‐methylbut‐2‐enyl diphosphate reductase. Enzyme products are abbreviated as follows: GAP, glyceraldehyde 3‐phosphate; DXP, deoxyxylulose 5‐phosphate; MEP, methyl‐d‐erythritol 4‐phosphate; HMBPP, 4‐hydroxy‐3‐methylbut‐2‐enyl diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; and GGPP, geranylgeranyl diphosphate. Carotenoid biosynthetic enzymes are abbreviated in bold: PSY1, 2 and 3, phytoene synthase 1, 2 and 3; PDS, phytoene desaturase; ZISO, ζ‐carotene isomerase; ZDS, ζ‐carotene desaturase; CRTISO, carotene isomerase; CRTIL1, CRTISO‐like 1; CRTL‐E (LCY‐E), lycopene ε‐cyclase; CRTL‐B1 and 2 (LCY‐B and BCYC, respectively), lycopene β‐cyclase; CYP97A3, carotene ε‐hydroxylase; CRTR‐B1 and 2, carotene β‐hydroxylase 1 and 2; and ABA is abscisic acid. Black dotted bar represents negative impacts on gene expression in response to AtPDS overexpression in ripe fruit. Dark grey arrows represent positive impacts by AtPDS overexpression in leaves. Light grey arrows/bars represent positive/negative impacts by AtPDS overexpression in anthesis flowers.
Considering the important protective roles of carotenoids in chloroplast stress tolerance, it is understandable that the carotenoid pathway would be more responsive to perturbations within leaf chloroplasts. The transcript induction resulting from AtPDS overexpression was similar, yet distinct from gene expression changes in tomato leaves expressing CRTI reported in Nogueira et al. (2013). In the AtPDS.OE leaves, induced DXS, PSY1, CRTL‐B1 and CRTR‐B1 transcripts and subsequent elevation of xanthophylls (i.e. lutein, β‐cryptoxanthin and zeaxanthin) are consistent with previous reports of elevated zeaxanthin levels in cyanobacteria from elevated PDS protein (Chamovitz et al., 1993). However, the minimal changes observed in xanthophyll accumulation in response to the elevation in gene expression throughout the pathway are intriguing. Similar observations have been observed when manipulating PSY in photosynthetic tissues of Arabidopsis (Lätari et al., 2015; Maass et al., 2009; Zhou et al., 2015). Whether via overexpression of PSY or the PSY activating ORANGE protein, carotenoid enrichment was limited to nonphotosynthetic tissues (Maass et al., 2009; Zhou et al., 2015). Lätari et al. (2015) demonstrate that overexpression of PSY in the ccd4 mutant background results in xanthophyll accumulation to toxic levels, suggesting a role for CCD4 in controlling xanthophyll accumulation. PDS‐associated enrichment of xanthophylls in tomato leaves may be similarly restricted via CCD4.
A strategy for enhancing lycopene content and bioavailability in tomato fruit
Bioavailability as it is most commonly defined (Macrae et al., 1993) equally depends on both the accessibility of the nutrient to the human body and its absorption properties therein. Impaired accessibility of carotenoids can negatively influence the overall effectiveness of carotenoid enhanced crops in addressing food security and nutritional concerns. Recent reports have suggested chromoplast ultrastructure and carotenoid sequestration effect the overall bioavailability of carotenoids, where bioavailability is improved via sequestration of carotenoids in a lipid‐dissolved liquid–crystalline state (Schweiggert et al., 2012, 2014). However, in tomatoes and other all‐trans‐lycopene‐rich fruits, all‐trans‐lycopene forms long crystalline aggregates of approximately 15 μm in length in the chromoplasts inhibiting its accessibility (Cooperstone et al., 2015; Schweiggert et al., 2012, 2014).
Chromoplast ultrastructure has been observed to be dependent on carotenoid composition (Harris and Spurr, 1969). Furthermore, an abundance of cis‐carotenoids in chromoplast‐rich tissues has been suggested to promote lipid‐associated sequestration of carotenoids (Ben‐Amotz et al., 1988). Microscopic analysis of chromoplasts in tangerine mutant (t 3183) fruit demonstrates that the abundance of cis‐lycopene isomers promotes carotenoid sequestration in globular lipid‐rich structures improving the accessibility of all lycopene isomers (Ben‐Amotz et al., 1988; Cooperstone et al., 2015). Therefore, overexpression of AtPDS in the tangerine mutant (t 3183) of tomato provides a unique and effective strategy capable of elevating total lycopene content without complementing the tangerine mutation, thereby accumulating higher levels of the bioavailable cis‐lycopene isomers (e.g. tetra‐cis‐lycopene; 5,9‐di‐cis‐lycopene; 5‐cis‐lycopene, etc.) and maintaining enhanced accessibility of carotenoids, including all‐ trans‐lycopene, sequestered in a lipid‐dissolved liquid crystalline state (Ben‐Amotz et al., 1988; Cooperstone et al., 2015; Schweiggert et al., 2012, 2014). The strategy overexpressing AtPDS in the t 3183 background reported here provides an initial step towards the enhancement of bioavailable carotenoids in fruit crops enabled in part by regulatory insights revealed through heterologous expression of PDS in tomato fruit.
Experimental procedures
Plant materials and growth conditions
Wild‐type control, tangerine single mutant (LA3183) and tangerine; high‐pigment1 double‐mutant (LA3367) seed (Solanum lycopersicum cv Ailsa Craig) were obtained from the Tomato Genetics Resource Center, UC Davis (http://tgrc.ucdavis.edu/). Plants were grown in glasshouses at the Guterman Bioclimate Laboratory and Greenhouse Complex, Cornell University, Ithaca, NY. All plants were grown under natural light conditions consisting of 16‐h day/8‐h nights. AtPDS overexpression transgenic lines were carried on to the T2 generation. T2 generation plants showing the strongest consistent phenotypes were used for crosses to make double mutants/transgenics with tangerine. Leaf tissue was harvested from the 4th, 5th and 6th leaves from the meristem of a two‐month‐old plant. Whole petals and anthers were harvested from flowers at the point of anthesis/pollination. Ripe fruits were collected 7 days after tagging fruit at the first sign of colour change at the blossom end (i.e. breaker stage). The tomato fruit ripening time course is represented by fruit collected at four stages of fruit ripening (Mature Green, MG; breaker or onset of ripening, Br; and 3 [light red] and 7 days [full red] postbreaker, 3DPB and 7DPB, respectively).
Development of overexpression constructs and plant transformation
The AtPDS overexpression (AtPDS.OE) construct was generated as described previously in Gleave (1992) using the pART7 and pART27 binary vector strategy. RNA from Arabidopsis thaliana (accession Columbia‐0) leaf tissue was converted to cDNA via iScript™ cDNA synthesis kit (Cat. No. 170‐8891; Bio‐Rad; Hercules, CA). Resulting cDNA was used to amplify the full‐length AtPDS ORF via FastStart High‐fidelity PCR system (Cat No. 04‐738‐292‐001; Roche Applied Sciences; Indianapolis, IN) with AtPDS‐OE‐KpnI.for and AtPDS‐OE‐XbaI.rev primers (Table S1). The resulting AtPDS.OE construct was sequence‐verified and transformed into S. lycopersicum cv Ailsa Craig by Agrobacterium tumafaciens (strain LBA‐4404) as previously described (Van Eck et al., 2006).
DNA isolation and zygosity and copy number analysis
Genomic DNA was isolated from fresh meristematic leaf tissue as previously described (Barry et al., 2005). Verification of insertion events in T0 plants was confirmed via PCR using primers specific for the 35S promoter in the pHELLSGATE 2 vector, 35S‐for and 35S‐rev (Table S1). Zygosity and copy number were determined in the T1 generation via Quantitative PCR relative to the single copy polygalacturonase 2a gene (PG2a, Accession No. X04583) using a modified protocol described in Haurogné et al. (2007) Quantitative PCR was performed in 5 μL reactions containing 2.5 μL SYBR® Green PCR Master Mix (Cat No. 4309155; Applied Biosystems, Foster City, CA); 0.75 μL of 10 μm of each primer; and 1 μL of gDNA using an ABI PRISM™ 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) under the following reaction conditions: 48 °C for 2 min; 95 °C for 10 min; and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Amplification was followed by a dissociation curve to verify specificity of the 35S promoter‐specific and PG gDNA‐specific primers used (Table S1).
Identification of positive double mutants/transgenics was performed using a forward and reverse primers in the interior of the 35S promoter and AtPDS ORF (35S‐internal‐FOR and AtPDS‐internal‐REV, respectively) and primers spanning the deletion in the tangerine mutant (i.e. tangerine.mut‐for and tangerine.mut‐rev) (Table S1). All positive double mutants/transgenics were further verified by gene expression via qRT–PCR and identification of appropriate flower and fruit phenotypes (Isaacson et al., 2002).
RNA isolation and quantitative RT–PCR analysis
Total RNA was isolated from all previously described tissue using a modified protocol from the RNeasy Minikit (Cat. No 74106, Qiagen Sciences, Germantown, MD). RNA was extracted from 200 to 300 mg of frozen powdered tissue via the addition of 800 μL modified RLT buffer (4 m guanidine isothiocyanate; 0.2 m sodium acetate, pH 5.2; 25 mm EDTA; 2.5% (w/v) PVP‐40) containing 1% (v/v) β‐mercaptoethanol and 2% sarcosine and separated upon the addition of 800 μL chloroform. Total RNA in the upper aqueous phase was collected on the Econospin™ minispin column (Cat. No. 1920250; Epoch Life Sciences, Missouri City, TX). The column was washed twice with 500 μL of RPE buffer (Cat. No. 1018013; Qiagen Sciences; MD), and RNA was eluted with 360 μL of nuclease‐free water. DNase treatment of total RNA was achieved by adding 40 μL DNase reaction buffer (60 mm MgCl2; 400 mm Tris, pH 7.5) and 3 μL of RQ1 DNase enzyme (Cat. No. M6101; Promega, Fitchburg, WI) and allowed to incubate for 45 min at 37 °C. Finally, RNA samples were washed with an equal volume of phenol: chloroform solution (500 g phenol crystals; 500 mL chloroform; 20 mL isoamyl alcohol; 0.5 g 8‐hydroxy‐quinoline; equilibrated with 100 mm Tris, pH 8.0) and lastly with an equal volume of chloroform. Total RNA was then precipitated overnight at −20 °C and resuspended in 50–100 μL of DEPC‐treated water.
Quantitative Real‐time PCR was performed using the Power SYBR® Green RNA‐to‐CT™ 1‐Step Kit (Cat No. 4309169; Applied Biosystems, Foster City, CA) in a 5 μL reaction volume (2.5 μL 2X Master Mix; 1 μm forward and reverse primers; 1 μL of total RNA; 0.46 μL DEPC‐treated water; 0.04 μL RT enzyme mix). All tissue samples were represented by three biological replicates, each being carried out in triplicate. Gene‐specific primers were checked for efficiency using wild‐type RNA (for primer sequence see Table S1). To be able to apply the standard curve method described in Applied Biosystems User Bulletin # 2 (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf), a standard curve was included on each plate for the specific gene being analysed using wild‐type RNA (serial dilutions: 50 ng; 5 ng; 0.5 ng; 0.05 ng) in triplicates. For each gene analysis, template‐free and negative‐RT controls were included. Real‐time PCRs were carried out using an ABI PRISM™ 7900HT Sequence Detection System (Applied Biosystems, CA) under the following reaction conditions: reverse transcription at 48 °C for 30 min; enzyme activation at 95 °C for 10 min; followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. A dissociation curve was added at the end of the run for verification of primer specificity.
ABI PRISM™ SDS version 2.3 software (Applied Biosystems, CA) was used to determine gene‐specific threshold cycles (CT) using the endogenous reference (18S rRNA) for every sample. CTs were extracted, and the standard curve method (Applied Biosystems, 1997) was applied to calculate relative mRNA levels in comparison with the wild‐type control or the reference sample (equal volume and concentration of RNA from each tissue combined), which was employed for the wild‐type differential expression profile.
Carotenoid extraction and analysis
Carotenoids were extracted from 200 mg of frozen tomato pericarp using a modified protocol from Alba et al. (2005). Carotenoid content of anthesis flowers was extracted from frozen petals and anthers of a single flower. The frozen tissue was homogenized in a Savant FP120 Fast Prep machine first with 15 mg Mg‐carbonate and 450 μL of tetrahydrofuran (THF) twice and then a third time with 450 μL of methanol containing 2,6‐Di‐tert‐butyl‐4‐methylphenol (MeOH/BHT). The homogenate was then transferred and filtered through Spin‐X centrifuge filters (0.45‐mm nylon filter; Corning/Costar #8170; Corning Inc., Corning, NY). Tissue debris was re‐extracted with an additional 500 μL of THF to ensure complete extraction of carotenoids. The carotenoid/nonpolar phase was separated from the aqueous phase through two separation steps, first with 375 μL of petroleum ether and 150 μL of 25% NaCl and next with 500 μL of petroleum ether. The two upper phase aliquots were combined and dried down in a vacufuge (Eppendorf). Prior to drying the flower extracts, xanthophylls went through a saponification as described in Galpaz et al. (2006). The dried extract from fruit was resuspended in 250 μL of ethyl acetate, while dried flower extracts were resuspended in 500 μL. All solvents used were HPLC grade.
Carotenoid analysis was carried out using a Dionex HPLC machine (ThermoFisher, Waltham, MA) (P680 HPLC pump, ASI‐100 automated sample injector, PDA‐100 photodiode array detector) and Chromeleon (v6.40) software package (ThermoFisher, Waltham, MA). Separation of carotenoids was achieved under a polar to nonpolar gradient (0–5 min 100% methanol: 0.1% ammonium acetate; 6‐ to 25‐min ramp to 4% methanol: ammonium acetate and 96% methyl t‐butyl ether; 26‐ to 30‐min ramp to 100% methanol: ammonium acetate; 31–35 min 100% methanol: ammonium acetate) through a guard cartridge (YMC carotenoid S‐5, 4.0 × 20 mm DC guard; Waters, Milford, MA), C30 column (YMC carotenoid S‐5, 4.6 × 250 mm; Waters, Milford, MA) assembly. Seven channels were used for data acquisition, channel 1 (286 nm); channel 2 (348 nm); channel 3 (398 nm); channel 4 (428 nm); channel 5 (437 nm) and channel 6 (450 nm); channel 7 (464 nm). Peak identification was performed as described in Alba et al. (2005). Lycopene isomers identification was determined relevant to results in Melendez‐Martinez et al. (2013) (Figure S6 and Table S2).
Supporting information
Figure S1 Assessment of protein structure of tomato PDS; ZDS; and ZISO compared to CRTI from bacteria.
Figure S2 Representative chromatograph (285 nm—best wavelength to show all peaks) of carotenoids detected in the wild type ripe tomato.
Figure S3 Absence of phytoene (μg/g FW) accumulation in young leaves of AtPDS.OE.1A, AtPDS.OE.3, AtPDS.OE.4, and AtPDS.OE.6 (n = 5, Error bars ±SEM).
Figure S4 Fruit size in terms of mass (g) of AtPDS.OE.1A, AtPDS.OE.3, AtPDS.OE.4, and AtPDS.OE.6 ripe fruit compared to wild type ripe fruit (n > 10, Error bars ±SEM).
Figure S5 Chromatographs with carotenoids detected in the tangerine (t 3183); AtPDS3.2;t 3183; and hp1;t 3183 ripe tomatoes.
Figure S6 Tissue type negatively affects the extent of AtPDS over‐expression in tangerine mutant flower.
Figure S7 Representative chromatograph (454 nm) of stereoisomers of lycopene red, all‐trans‐lycopene standard (4 °C) and blue, stereomutated all‐trans‐lycopene (80 °C for 1 h).
Table S1 Primer sequences
Table S2 Identification of lycopene stereoisomers detected in the tangerine (t 3183) ripe fruit based on artificial stereomutation of All‐trans‐lycopene standard according to Melendez et al. (2013)
Acknowledgements
The authors thank Patricia R. Keen and Joyce Van Eck for their assistance in tomato transformation and access to the tissue culture facility. The authors also thank Nigel E. Gapper for access to the vectors used in the study and valuable communication regarding construct design. The authors declare no conflict of interest. This work was supported by grants IOS‐1539831and IOS‐1339287 from the National Science Foundation to JG and by the United Stated Department of Agriculture ‐ Agricultural Research Service.
Accession number: Arabidopsis thaliana: PDS (AT4G14210.1); Solanum lycopersicum: DXS (Solyc01 g067890) PSY1, 2 and 3 (Soly03 g031860, Solyc02 g081330, Solyc01 g005940, respectively); PDS (Solyc03 g123760); ZISO (Solyc12 g098710); ZDS (Solyc01 g097810); CRTISO (Solyc10 g081650); CRTIL1 (Solyc05 g010180); CRTL‐E (Solyc12 g008980); CRTL‐B1 and 2 (Solyc04 g040190, Solyc06 g074240, respectively); CRTR‐B1 and 2 (Solyc06 g036260, Solyc03 g007960, respectively).
Seed stocks: Wild‐type cv. Ailsa Craig (LA2838A) tangerine single mutant (LA3183) tangerine; high‐pigment1 double mutant (LA3367).
References
- Alba, R. , Payton, P. , Fei, Z. , McQuinn, R. , Debbie, P. , Martin, G.B. , Tanksley, S.D. et al (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell, 17, 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S. , Aldridge, G.M. , Herzog, G. , Ma, Q. , McQuinn, R.P. , Hirschberg, J. and Giovannoni, J.J. (2012) Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol. 159, 1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S. , McQuinn, R.P. , Chung, M.‐Y. , Besuden, A. and Giovannoni, J.J. (2008) Amino acid substitutions in homologs of the Stay‐Green protein are responsible for the green‐flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S. , McQuinn, R.P. , Thompson, A.J. , Seymour, G.B. , Grierson, D. and Giovannoni, J.J. (2005) Ethylene insensitivity conferred by the Green‐ripe and Never‐ripe 2 ripening mutants of tomato. Plant Physiol. 138, 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán, J. , Kloss, B. , Hosler, J.P. , Geng, J. , Liu, A. , Modi, A. , Dawson, J.H. et al (2015) Control of carotenoid biosynthesis through a heme‐based cis‐trans isomerase. Nat. Chem. Biol. 11, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Amotz, A. , Lers, A. and Avron, M. (1988) Stereoisomers of ‐carotene and phytoene in the alga dunaliella bardawil. Plant Physiol. 86, 1286–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol, P. , Stevenson, D. , Bisanz, C. , Breitenbach, J. , Sandmann, G. , Mache, R. , Coupland, G. et al (1999) Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell, 11, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli, C.I. and Pogson, B.J. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15, 266–274. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D. , Sandmann, G. and Hirschberg, J. (1993) Molecular and biochemical characterization of herbicide‐resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate‐limiting step in carotenoid biosynthesis. J. Biol. Chem. 268, 17348–17353. [PubMed] [Google Scholar]
- Cooperstone, J.L. , Ralston, R.A. , Riedl, K.M. , Haufe, T.C. , Schweiggert, R.W. , King, S.A. , Timmers, C.D. et al (2015) Enhanced bioavailability of lycopene when consumed as cis ‐isomers from tangerine compared to red tomato juice, a randomized, cross‐over clinical trial. Mol. Nutr. Food Res. 59, 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto, G. , Al‐Babili, S. , Tavazza, R. , Papacchioli, V. , Beyer, P. and Giuliano, G. (2007) Metabolic engineering of potato carotenoid content through tuber‐specific overexpression of a bacterial mini‐pathway. PLoS ONE, 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi, E.M.A. , Fraser, P.D. , Lois, L.‐M. , Boronat, A. , Schuch, W. and Bramley, P.M. (2004) Metabolic engineering of the mevalonate and non‐mevalonate isopentenyl diphosphate‐forming pathways for the production of health‐promoting isoprenoids in tomato. Plant Biotechnol. J. 3, 17–27. [DOI] [PubMed] [Google Scholar]
- Enfissi, E.M.A. , Nogueira, M. , Bramley, P.M. and Fraser, P.D. (2017) The regulation of carotenoid formation in tomato fruit. Plant J. 89, 774–788. [DOI] [PubMed] [Google Scholar]
- Eriksson, E.M. , Bovy, A. , Manning, K. , Harrison, L. , Andrews, J. , Silva, J.De. , Tucker, G.A. et al (2004) Effect of the Colorless non‐ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136, 4184–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini, E. , Falcone, G. , Frusciante, S. , Giliberto, L. and Giuliano, G. (2013) Dissection of tomato lycopene biosynthesis through virus‐induced gene silencing. Plant Physiol. 163, 986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P.D. , Enfissi, E.M.A. and Bramley, P.M. (2009) Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch. Biochem. Biophys. 483, 196–204. [DOI] [PubMed] [Google Scholar]
- Fraser, P.D. , Enfissi, E.M.A. , Halket, J.M. , Truesdale, M.R. , Yu, D. , Gerrish, C. and Bramley, P.M. (2007) Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19, 3194–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P.D. , Romer, S. , Shipton, C.A. , Mills, P.B. , Kiano, J.W. , Misawa, N. , Drake, R.G. , et al. (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit‐specific manner. Proc. Natl Acad. Sci. USA 99, 1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci, P. , Hodgman, C. , Teyssier, E. and Seymour, G.B. (2016) DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front. Plant Sci. 7, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz, N. , Ronen, G. , Khalfa, Z. , Zamir, D. and Hirschberg, J. (2006) A chromoplast‐specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white‐flower locus. Plant Cell, 18, 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmecker, S. , Schaub, P. , Koschmieder, J. , Brausemann, A. , Drepper, F. , Rodriguez‐Franco, M. , Ghisla, S. et al (2015) Phytoene desaturase from Oryza sativa: oligomeric assembly, membrane association and preliminary 3D‐analysis B. T. Ayele, ed. PLoS ONE, 10, e0131717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, J. , Nguyen, C. , Ampofo, B. , Zhong, S. and Fei, Z. (2017) The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 68, 61–84. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992) A versatile binary vector system with a T‐DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Harris, W.M. and Spurr, A.R. (1969) Chromoplasts of tomato fruits. I. Ultrastructure of low‐pigment and high‐ beta mutants. Carotene analyses on JSTOR. Am. J. Bot. 56, 369–379. [Google Scholar]
- Haurogné, K. , Bach, J.‐M. and Lieubeau, B. (2007) Easy and rapid method of zygosity determination in transgenic mice by SYBR Green real‐time quantitative PCR with a simple data analysis. Transgenic Res. 16, 127–131. [DOI] [PubMed] [Google Scholar]
- Ip, B.C. , Hu, K.Q. , Liu, C. , Smith, D.E. , Obin, M.S. , Ausman, L.M. and Wang, X.D. (2013) Lycopene metabolite, apo‐10′‐lycopenoic acid, inhibits diethylnitrosamine‐initiated, high fat diet‐promoted hepatic inflammation and tumorigenesis in mice. Cancer Prev. Res. 6, 1304–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson, T. , Ronen, G. , Zamir, D. and Hirschberg, J. (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β‐carotene and xanthophylls in plants. Plant Cell, 14, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët, T. , Genty, B. , Josse, E.‐M. , Kuntz, M. , Cournac, L. and Peltier, G. (2002) Involvement of a plastid terminal oxidase in plastoquinone oxidation as evidenced by expression of the Arabidopsis thaliana enzyme in tobacco. J. Biol. Chem. 277, 31623–31630. [DOI] [PubMed] [Google Scholar]
- Josse, E.‐M. , Alcaraz, J.‐P. , Laboure, A.‐M. and Kuntz, M. (2003) In vitro characterization of a plastid terminal oxidase (PTOX). Eur. J. Biochem. 270, 3787–3794. [DOI] [PubMed] [Google Scholar]
- Kachanovsky, D.E. , Filler, S. , Isaacson, T. and Hirschberg, J. (2012) Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis‐carotenoids. Proc. Natl Acad. Sci. USA, 109, 19021–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lätari, K. , Wüst, F. , Hübner, M. , Schaub, P. , Beisel, K.G. , Matsubara, S. , Beyer, P. et al (2015) Tissue‐specific apocarotenoid glycosylation contributes to carotenoid homeostasis in arabidopsis leaves. Plant Physiol. 168, 1550–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, F. and Wang, X.‐D. (2008) Enzymatic metabolites of lycopene induce Nrf2‐mediated expression of phase II detoxifying/antioxidant enzymes in human bronchial epithelial cells. Int. J. Cancer, 123, 1262–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Roof, S. , Ye, Z. , Barry, C.S. , van Tuinen, A. , Vrebalov, J. , Bowler, C. et al (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl Acad. Sci. USA, 101, 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass, D. , Arango, J. , Wüst, F. , Beyer, P. and Welsch, R. (2009) Carotenoid crystal formation in arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE, 4, e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae, R. , Robinson, R.K. and Sadler, M.J. (1993) Encyclopaedia of Food Science, Food Technology, and Nutrition. Massachusetts: Academic Press. [Google Scholar]
- Martel, C. , Vrebalov, J. , Tafelmeyer, P. and Giovannoni, J.J. (2011) The tomato MADS‐box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING‐dependent manner. Plant Physiol. 157, 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuinn, R.P. , Giovannoni, J.J. and Pogson, B.J. (2015) More than meets the eye: from carotenoid biosynthesis, to new insights into apocarotenoid signaling. Curr. Opin. Plant Biol. 27, 172–179. [DOI] [PubMed] [Google Scholar]
- Melendez‐Martinez, A.J. , Stinco, C.M. , Liu, C. and Wang, X.D. (2013) A simple HPLC method for the comprehensive analysis of cis/trans (Z/E) geometrical isomers of carotenoids for nutritional studies. Food Chem. 138, 1341–1350. [DOI] [PubMed] [Google Scholar]
- Misawa, N. , Nakagawa, M. , Kobayashi, K. , Yamano, S. , Izawa, Y. , Nakamura, K. and Harashima, K. (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172, 6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar, N. , Li, L. , Lu, S. , Khin, N.C. and Pogson, B.J. (2015) Carotenoid metabolism in plants. Mol. Plant, 8, 68–82. [DOI] [PubMed] [Google Scholar]
- Nogueira, M. , Mora, L. , Enfissi, E.M.A. , Bramley, P.M. and Fraser, P.D. (2013) Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell, 25, 4560–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, S.R. , Barrette, T.R. and DellaPenna, D. (1995) Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell, 7, 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn, M. and Dollé, P. (2012) Retinoic acid signalling during development. Development, 139, 843–858. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Concepción, M. and Boronat, A. (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, S. , Fraser, P.D. , Kiano, J.W. , Shipton, C.A. , Misawa, N. , Schuch, W. and Bramley, P.M. (2000) Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18, 666–669. [DOI] [PubMed] [Google Scholar]
- Ronen, G. , Carmel‐Goren, L. , Zamir, D. and Hirschberg, J. (2000) An alternative pathway to beta ‐carotene formation in plant chromoplasts discovered by map‐based cloning of beta and old‐gold color mutations in tomato. Proc. Natl Acad. Sci. USA, 97, 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen, G. , Cohen, M. , Zamir, D. and Hirschberg, J. (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon‐cyclase is down‐regulated during ripening and is elevated in the mutantDelta. Plant J. 17, 341–351. [DOI] [PubMed] [Google Scholar]
- Sandmann, G. and Fraser, P.D. (1993) Differential inhibition of phytoene desaturases from diverse origins and analysis of resistant cyanobacterial mutants. Z. Naturforsch. C, 48, 307–311. [Google Scholar]
- Schaub, P. , Yu, Q. , Gemmecker, S. , Poussin‐Courmontagne, P. , Mailliot, J. , McEwen, A.G. , Ghisla, S. et al (2012) On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane‐peripheral and FAD‐dependent oxidase/isomerase. PLoS ONE, 7, e39550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiggert, R.M. , Kopec, R.E. , Villalobos‐Gutierrez, M.G. , Högel, J. , Quesada, S. , Esquivel, P. , Schwartz, S.J. , et al. (2014) Carotenoids are more bioavailable from papaya than from tomato and carrot in humans: a randomised crossover study. Br. J. Nutr. 111, 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiggert, R.M. , Mezger, D. , Schimpf, F. , Steingass, C.B. and Carle, R. (2012) Influence of chromoplast morphology on carotenoid bioaccessibility of carrot, mango, papaya, and tomato. Food Chem. 135, 2736–2742. [DOI] [PubMed] [Google Scholar]
- Shahbazi, M. , Gilbert, M. , Labouré, A.‐M. and Kuntz, M. (2007) Dual role of the plastid terminal oxidase in tomato. Plant Physiol. 145, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni, Y. , Linnewiel‐Hermoni, K. , Zango, G. , Khanin, M. , Salman, H. , Veprik, A. , Danilenko, M. et al (2012) The role of lycopene and its derivatives in the regulation of transcription systems: implications for cancer prevention. Am. J. Clin. Nutr. 96, 1173S–1178S. [DOI] [PubMed] [Google Scholar]
- Shumskaya, M. and Wurtzel, E.T. (2013) The carotenoid biosynthetic pathway: thinking in all dimensions. Plant Sci. 208, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck, J. , Kirk, D.D. and Walmsley, A.M. (2006) Tomato (Lycopersicum esculentum). Methods Mol. Biol. 343, 459–473. [DOI] [PubMed] [Google Scholar]
- Vrebalov, J. , Pan, I.L. , Arroyo, A.J.M. , McQuinn, R.P. , Chung, M.Y. , Poole, M. , Rose, J. et al (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell, 21, 3041–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov, J. , Ruezinsky, D. , Padmanabhan, V. , White, R. , Medrano, D. , Drake, R. , Schuch, W. et al (2002) A MADS‐box gene necessary for fruit ripening at the tomato ripening‐inhibitor (Rin) locus. Science, 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Ye, X. , Al‐Babili, S. , Klöti, A. , Zhang, J. , Lucca, P. , Beyer, P. and Potrykus, I. (2000) Engineering the provitamin A (beta‐carotene) biosynthetic pathway into (carotenoid‐free) rice endosperm. Science, 287, 303–305. [DOI] [PubMed] [Google Scholar]
- Yu, Q. , Ghisla, S. , Hirschberg, J. , Mann, V. and Beyer, P. (2011) Plant carotene cis‐trans isomerase CRTISO: a new member of the FAD(RED)‐dependent flavoproteins catalyzing non‐redox reactions. J. Biol. Chem. 286, 8666–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q. , Feilke, K. , Krieger‐Liszkay, A. and Beyer, P. (2014) Functional and molecular characterization of plastid terminal oxidase from rice (Oryza sativa). Biochim. Biophys. Acta, 1837, 1284–1292. [DOI] [PubMed] [Google Scholar]
- Zhong, S. , Fei, Z. , Chen, Y.‐R. , Zheng, Y. , Huang, H. , Vrebalov, J. , McQuinn, R.P. et al (2013) Single‐base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Welsch, R. , Yang, Y. , Alvarez, D. , Riediger, M. , Yuan, H. , Fish, T. et al (2015) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl Acad. Sci. USA, 112, 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Assessment of protein structure of tomato PDS; ZDS; and ZISO compared to CRTI from bacteria.
Figure S2 Representative chromatograph (285 nm—best wavelength to show all peaks) of carotenoids detected in the wild type ripe tomato.
Figure S3 Absence of phytoene (μg/g FW) accumulation in young leaves of AtPDS.OE.1A, AtPDS.OE.3, AtPDS.OE.4, and AtPDS.OE.6 (n = 5, Error bars ±SEM).
Figure S4 Fruit size in terms of mass (g) of AtPDS.OE.1A, AtPDS.OE.3, AtPDS.OE.4, and AtPDS.OE.6 ripe fruit compared to wild type ripe fruit (n > 10, Error bars ±SEM).
Figure S5 Chromatographs with carotenoids detected in the tangerine (t 3183); AtPDS3.2;t 3183; and hp1;t 3183 ripe tomatoes.
Figure S6 Tissue type negatively affects the extent of AtPDS over‐expression in tangerine mutant flower.
Figure S7 Representative chromatograph (454 nm) of stereoisomers of lycopene red, all‐trans‐lycopene standard (4 °C) and blue, stereomutated all‐trans‐lycopene (80 °C for 1 h).
Table S1 Primer sequences
Table S2 Identification of lycopene stereoisomers detected in the tangerine (t 3183) ripe fruit based on artificial stereomutation of All‐trans‐lycopene standard according to Melendez et al. (2013)


