Summary
Although hundreds of genetic male sterility (GMS) mutants have been identified in maize, few are commercially used due to a lack of effective methods to produce large quantities of pure male‐sterile seeds. Here, we develop a multicontrol sterility (MCS) system based on the maize male sterility 7 (ms7) mutant and its wild‐type Zea mays Male sterility 7 (ZmMs7) gene via a transgenic strategy, leading to the utilization of GMS in hybrid seed production. ZmMs7 is isolated by a map‐based cloning approach and encodes a PHD‐finger transcription factor orthologous to rice PTC1 and Arabidopsis MS1. The MCS transgenic maintainer lines are developed based on the ms7‐6007 mutant transformed with MCS constructs containing the (i) ZmMs7 gene to restore fertility, (ii) α‐amylase gene ZmAA and/or (iii) DNA adenine methylase gene Dam to devitalize transgenic pollen, (iv) red fluorescence protein gene DsRed2 or mCherry to mark transgenic seeds and (v) herbicide‐resistant gene Bar for transgenic seed selection. Self‐pollination of the MCS transgenic maintainer line produces transgenic red fluorescent seeds and nontransgenic normal colour seeds at a 1:1 ratio. Among them, all the fluorescent seeds are male fertile, but the seeds with a normal colour are male sterile. Cross‐pollination of the transgenic plants to male‐sterile plants propagates male‐sterile seeds with high purity. Moreover, the transgene transmission rate through pollen of transgenic plants harbouring two pollen‐disrupted genes is lower than that containing one pollen‐disrupted gene. The MCS system has great potential to enhance the efficiency of maize male‐sterile line propagation and commercial hybrid seed production.
Keywords: ZmMs7, genetic male sterility, hybrid seed production, transgenic maintainer line, nontransgenic progeny, maize
Introduction
Maize is one of the most important crops that have been successfully applied to achieve heterosis. Heterosis is a phenomenon in which heterozygous hybrid progeny are superior to both homozygous parents. To produce pure hybrid seeds, the female inbred parent line must be prevented from undergoing self‐pollination and should be directionally crossed to the male inbred parent because maize is a monoecious plant. During maize hybrid seed production, many methods can be used to prevent self‐pollination of the female parent line, such as manual or mechanical emasculation (detasseling), application of male‐specific gametocides and use of cytoplasmic or nuclear‐encoded male sterility (Perez‐Prat and van Lookeren Campagne, 2002).
Manual or mechanical detasseling, namely the physical removal of the male floral structure, remains the predominant method in commercial hybrid seed production in maize. However, this technique is not only time‐consuming, labour‐intensive and expensive but also detrimental to plant growth, thus reducing the yield of hybrid seeds because a portion of the plant photosynthetic body is destroyed during emasculation (Skibbe and Schnable, 2005; Wych, 1988). Although a number of chemicals have been investigated as potential gametocides for foliar sprays prior to flowering in commercial maize seed production (McRae, 1985), there has been little industrial use of chemical sterilants because of the risk of incomplete pollen sterility, a reduction in female fertility and its detrimental effect on the environment.
Male sterility can also be generated by either cytoplasmic or nuclear genes. Cytoplasmic male sterility (CMS) has been used in commercial hybrid maize production, but the CMS system suffers from several intrinsic problems, including the poor genetic diversity between the CMS lines and the restoration lines, increased disease susceptibility, a breakdown of sterility in certain environments or genetic backgrounds, and unreliable restoration (Williams, 1995). Nuclear‐encoded male sterility (NMS) or GMS results from mutations in the nuclear genome, which is a common spontaneous phenomenon in flowering plants (Kaul, 1988). To date, hundreds of genetic male‐sterile mutants have been identified and described in maize (Timofejeva et al., 2013). Most of these mutants are controlled by a recessive gene, which provides an excellent genetic means of emasculation for hybrid seed production in maize. However, it is not possible to obtain a pure increase in male‐sterile homozygous female inbred seeds through traditional self‐pollination because the plant is male sterile, and the largest percentage of male‐sterile seeds that can be produced is 50% (Williams, 1995). Fortunately, the rapid development of gene cloning, recombinant DNA and plant transformation techniques have provided new possibilities for creating genetically engineered male sterility.
Since the first transgenic male sterility system was described (Mariani et al., 1990), many strategies to produce male‐sterile plants have been reported,including chemical inducible male sterility systems (Feng et al., 2014; Singh et al., 2010; Venkatesh et al., 2014), transgenic maintainer systems (Chang et al., 2016; Fox et al., 2017; Perez‐Prat and van Lookeren Campagne, 2002; Williams, 1995; Wu et al., 2016) and other biotechnology‐based systems (Gils et al., 2008; Millwood et al., 2016; Mitsuda et al., 2006). Among them, the strategy of using transgenic maintainer lines to propagate nuclear male‐sterile plants is more desirable and reliable. The transgenic maintainer lines are obtained by transforming dominant or recessive male‐sterile plants with a construct comprising three modules: a male sterility complementary fertility restoration gene, a pollen‐lethality gene to disrupt the transgenic pollen and a seed colour marker gene for mechanical colour sorting of the seeds (Perez‐Prat and van Lookeren Campagne, 2002). Self‐pollination of the resulting maintainer lines propagates 50% of maintainer seeds and 50% of male‐sterile seeds, which can be sorted based on the seed colour marker. Cross‐pollination of the maintainer line to the male‐sterile line produces 100% male‐sterile seeds. To successfully apply the maintainer line, the basic prerequisite of this strategy is to identify and clone the male sterility gene, which is usually obtained from a male‐sterile mutant.
Despite the large collection of male‐sterile mutants in maize, only a handful of the mutants have been characterized cytologically, and even fewer genes have been cloned and studied in detail, such as ms8 (Wang et al., 2010, 2012b, 2013), ms9 (Albertsen et al., 2016), ms22/msca1 (Albertsen et al., 2009; Chaubal et al., 2003), ms23 (Nan et al., 2017), ms26 (Djukanovic et al., 2013), ms32 (Moon et al., 2013), Ms44 (Fox et al., 2017), ms45 (Cigan et al., 2001), apv1 (Somaratne et al., 2017), ipe1 (Chen et al., 2017), mac1 (Wang et al., 2012a) and ocl4 (Vernoud et al., 2009). The cloning and functional characterization of these male sterility genes have contributed significantly to our understanding of the molecular mechanism underlying anther development in maize and provided useful gene resources for genetic engineering of male‐sterile lines in hybrid seed production. However, compared with the related studies in rice and Arabidopsis, the molecular mechanism responsible for most maize male‐sterile mutants is largely unknown. Based on the phenotypic and cytological similarity of male‐sterile mutants among maize, rice and Arabidopsis, the cloned male sterility genes in rice and Arabidopsis can be used for reference in maize. For example, the Arabidopsis MALE STERILITY1 (AtMS1) gene encodes a PHD‐finger transcription factor and regulates pollen and tapetum development. AtMS1 is expressed specifically in the tapetum after meiosis and is involved in formation of the pollen exine and pollen cytosolic components as well as development of the tapetum (Ito and Shinozaki, 2002; Ito et al., 2007; Wilson et al., 2001; Yang et al., 2007). In rice, PERSISTENT TAPETAL CELL1 (OsPTC1), the ortholog of AtMS1, controls programmed tapetal cell death and functional pollen development. Additionally, the rice ptc1 mutant is phenotypically similar to the Arabidopsis ms1 mutant in terms of a lack of tapetal DNA fragmentation, delayed tapetal degeneration, abnormal pollen wall formation and aborted microspore development. The ptc1 mutant displays uniquely uncontrolled tapetal proliferation and subsequent commencement of necrosis‐like tapetal death (Li et al., 2011). However, to date, the ortholog of Arabidopsis MS1 and rice PTC1 in maize remains undescribed.
Based on the critical time points of male sterility conversion, the male‐sterile mutants can be classified as premeiotic, meiotic and postmeiotic. In previous reports, hundreds of maize male‐sterile mutants were identified and classified into the above three classes based on cytological characterization and allelism testing (Timofejeva et al., 2013). Twenty‐two maize male‐sterile mutants with premeiotic developmental defects were used to evaluate the detailed cytological characteristics. Four general types of premeiotic defects were classified: anther identity defects, abnormal anther structure, anther wall layer defects and premature cell layer degradation (Timofejeva et al., 2013). Among the male‐sterile genes mentioned above, ms8, ms22/msca1, ms23, ms32, mac1 and ocl4 were premeiotic mutants, while ms26, ms45, apv1 and ipe1 were postmeiotic mutants. Moreover, among the premeiotic mutants, ms8 was classified into the group of premature cell layer degradation, ms22/msca1 was classified into the group of anther identity defects, and ms23, ms32, mac1 and ocl4 were classified into the group of anther wall layer defects (Timofejeva et al., 2013). Furthermore, maize ms7 was a historic male‐sterile mutant and had been previously studied morphologically and cytologically (Albertsen and Phillips, 1981; Beadle, 1932; Morton et al., 1989). In 1981, the ms7 mutant was described as having a poorly developed, thin pollen wall and premature vacuolization and degradation of tapetal cells (Albertsen and Phillips, 1981). Morton et al. (1989) confirmed that the ms7 mutant had a relatively thin microspore wall, a poorly developed microspore aperture and several tapetal cell and orbicule morphological abnormalities. However, no visible defects were observed in ms7 anthers prior to tetrad formation (Morton et al., 1989). Therefore, the ms7 mutant should be classified as a postmeiotic developmental mutant with abnormal tapetal cell layer degeneration, in contrast to the above‐described mutants based on cytological observation. Nevertheless, the mutated ms7 gene and its role in anther development are largely unknown.
In this study, the maize wild‐type ZmMs7 gene was isolated by a map‐based cloning approach. ZmMs7 encodes a PHD‐finger transcription factor that is orthologous to rice PTC1 and Arabidopsis MS1. ZmMs7 was characterized with respect to spatio‐temporal expression patterns, cytological and molecular biological analyses and transgenic complementation. Based on the ZmMs7 gene, a MCS system was developed and preliminarily evaluated. This work will provide new insights into the molecular mechanism of male sterility in maize and may enhance the efficiency of maize male‐sterile line propagation and hybrid seed production.
Results
ms7 is a single recessive mutant that exhibits complete male sterility in maize
Two alleles of ms7 mutant, ms7‐6007 and ms7gl1, were obtained from the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu). Compared with the wild‐type male‐fertile sibling, ms7‐6007 displayed complete male sterility with no exserted anthers (Figure 1a and b) but normal vegetative growth and female fertility. The mutant anthers were small and whitish (Figure 1c), lacking pollen grains, and were resistant to I2‐KI staining (Figure 1d). Additionally, the mutant phenotype was stable in multiple environments including different years and locations. The ms7gl1 mutant also exhibited a similar male‐sterile phenotype to the ms7‐6007 mutant (Figure S1).
Figure 1.
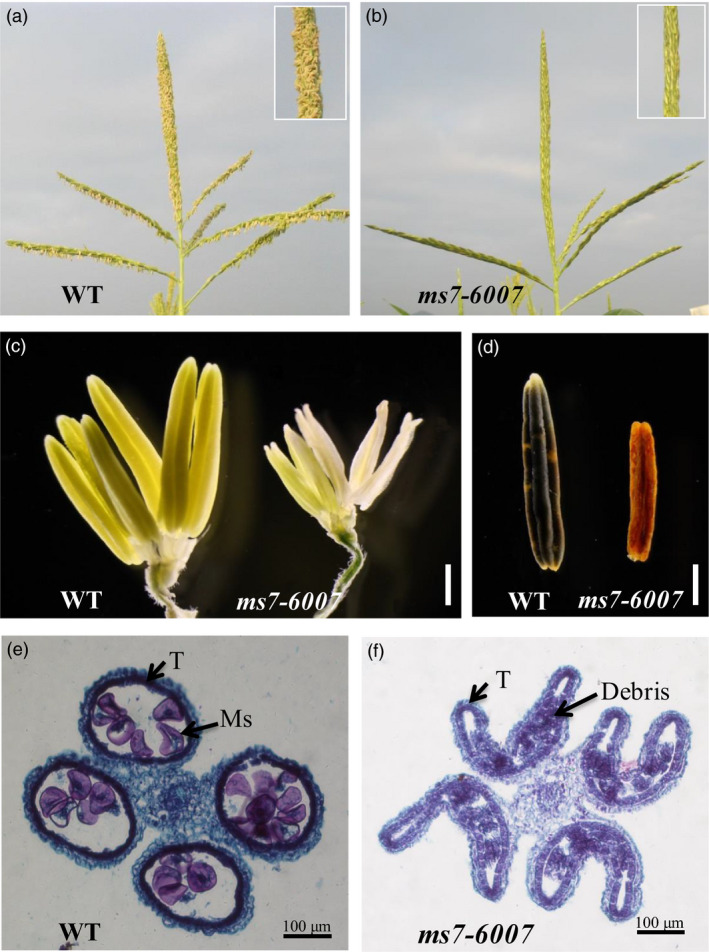
Phenotypic comparison of the wild type (WT) and ms7‐6007 mutant. (a and b) The tassel of WT (a) and ms7‐6007 (b). (c) The spikelet of WT (left) and ms7‐6007 (right) with the glume, lemma and palea removed. (d) The anther of WT (left) and ms7‐6007 (right) stained with I2‐KI. (e and f) Transverse sections of the entire anther of wild type (e) and ms7‐6007 (f) in the late microspore developmental stage. T, tapetum; Ms, microspore. Bars = 1 mm (c, d), 100 μm (E, F).
To investigate the cytological defects of the ms7 mutant in anther development, a microscopic comparison analysis between ms7‐6007 and wild type was conducted. During the late microspore developmental stage, the wild‐type tapetal cells became condensed and deeply stained, and they gradually degenerated via programmed cell death (PCD, Figure 1e). In contrast, the ms7‐6007 mutant tapetal cells were swollen and lightly stained, and the microspore was aborted, leaving debris in the shrinkage locule (Figure 1f) indicative of abnormal tapetal cell PCD.
When the two ms7 mutants were crossed with the normal maize inbred line Chang7‐2, all of the F1 progeny was male fertile and the F2 population displayed a 3:1 segregation of male fertile to sterile plants (Table S1), suggesting that both ms7‐6007 and ms7gl1 were single recessive mutations. Furthermore, when the ms7‐6007/ms7‐6007 homozygous plants were outcrossed using pollen from ms7gl1/+ heterozygous plants, the F1 progeny showed a 1:1 segregation ratio of male sterile to fertile plants, implying that the mutant gene of ms7‐6007 is allelic to that of ms7gl1.
Isolation of the ZmMs7 male fertility gene
The ms7 mutant gene was identified by the map‐based cloning approach. Using a F2 population that included 154 male‐sterile individuals, the ms7 locus was initially mapped to chromosome 7 between SSR markers umc2617 and bnlg1808 with a genetic distance of 4.8 cM (Figure 2a). Then, based on the maize genome sequence information, six CAPS markers in the interval were designed for fine mapping of the ms7 gene in an enlarged F2 population that included 611 male‐sterile individuals. Finally, the ms7 gene was defined to the interval with a 180‐kb length between EP299 and EP302 (Figure 2b). Six predicted genes are located in this region (Figure 2c), including GRMZM5G890224, which encodes a putative PHD‐finger protein.
Figure 2.
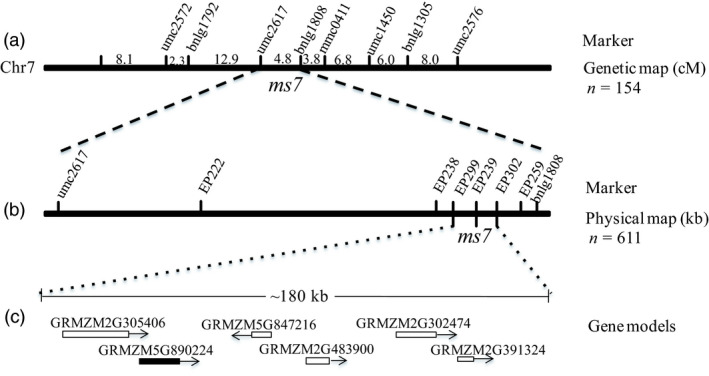
Map‐based cloning of the maize ms7 gene. (a) primary mapping of the ms7 gene on maize chromosome 7 between SSR markers umc2617 and bnlg1808. (b) Fine mapping of the ms7 gene to an interval of nearly 180 kb between CAPS markers EP299 and EP302. (c) The six putative gene models in the interval. Among them, GRMZM5G890224, similar to the rice PTC1 gene, is the candidate gene. n, The number of the male‐sterile plants used in the F2 mapping population.
The candidate gene (GRMZM5G890224) consists of three exons and two introns (Figure 3a) and encodes a 670‐amino acid protein. Sequencing of this gene in the ms7‐6007 mutant revealed a 3‐bp (CGA) insertion at the +22 nucleotide site in the first exon and a 7‐bp (GCTGCTG) insertion at the +814 nucleotide site in the second exon, respectively (Figure 3b). The latter insertion causes a frameshift mutation and alters the reading frame after amino acid residue 157, resulting in a lack of the leucine zipper region and PHD domain (Figure S2). Additionally, sequencing of the target gene in the ms7gl1 mutant showed that an 1136‐bp transposable element (DTA_ZM00023) is inserted at the +1179 nucleotide site in the third exon (Figure 3c), which also causes an in‐frame premature stop codon and leads to the lack of the leucine zipper region and PHD domain (Figure S2). These results suggested that GRMZM5G890224 is responsible for the male‐sterile phenotype in the ms7 mutant, and therefore, GRMZM5G890224 was tentatively designated ZmMs7.
Figure 3.
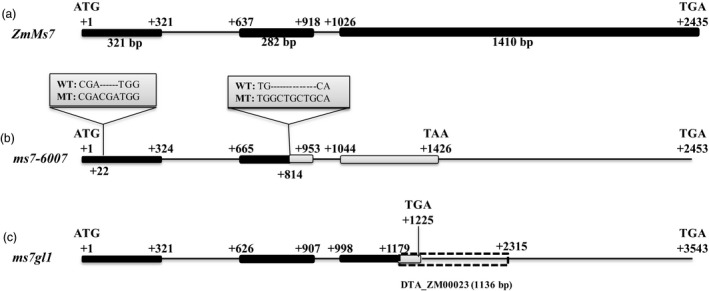
The gene structure of the maize ZmMs7 candidate gene ( GRMZM5G890224). A schematic representation of three exons and two introns of ZmMs7 in wild type (a) ms7‐6007 (b) and ms7gl1 (c). The +1 indicates the putative starting nucleotide of translation, and the stop codon (TGA) is +2435 in wild type. Black boxes indicate exons, and intervening lines indicate introns. Grey boxes indicate the mutant amino acid sequences. Numbers indicate the exon length (bp); the insertion sites in ms7‐6007 and ms7gl1 are shown, causing a frameshift mutation and premature stop codon, respectively.
To confirm the above prediction, the functional complementation experiment was performed. The maize Hi‐II hybrid line was transformed with Agrobacterium tumefaciens EHA105 containing a pZmMs7pro::ZmMs7 construct. The transgenic plants were then crossed with the ms7‐6007 mutant, and transgenic plants in the ms7‐6007 homozygous mutant background were gained by self‐pollination and marker‐assisted selection. The transgenic plants rescued the male‐sterile defect of ms7‐6007 and recovered the fertility phenotype (Figure 4), demonstrating that the male‐sterile phenotype of the ms7‐6007 mutant results from the ZmMs7 gene mutation.
Figure 4.

Functional complementation of the maize ms7‐6007 mutant. Phenotypes of tassels (a), spikelets (b) and pollen grains stained with I2‐KI (c) in the wild type, ms7‐6007 mutant and complemented transgenic line ( pMs7::Ms7/ms7‐6007). Bars = 1 mm (B), 100 μm (c).
ZmMs7 is an anther‐specific gene
To analyse the expression pattern of ZmMs7, semi‐quantitative RT–PCR and quantitative real‐time PCR (qRT–PCR) were performed. The obtained results revealed that ZmMs7 was specifically expressed in postmeiotic anthers from the quartet to late vacuolate microspore stages, with the highest expression levels in the early‐vacuolate microspore stage (Figure 5a and b). In contrast, ZmMs7 expression was not detected in anthers at other developmental stages or in the leaf, ear or root during any developmental stages (Figure 5a and b). Compared with the expression of ZmMs7 in Ms7/ms7‐6007 male‐fertile siblings, the ms7‐6007 male‐sterile mutant exhibited lower ZmMs7 expression at approximately one‐third the expression level (Figure 5c). Together, these results confirmed that ZmMs7 is an anther‐specific expression gene and may play a specific role during the development of anthers and microspores.
Figure 5.
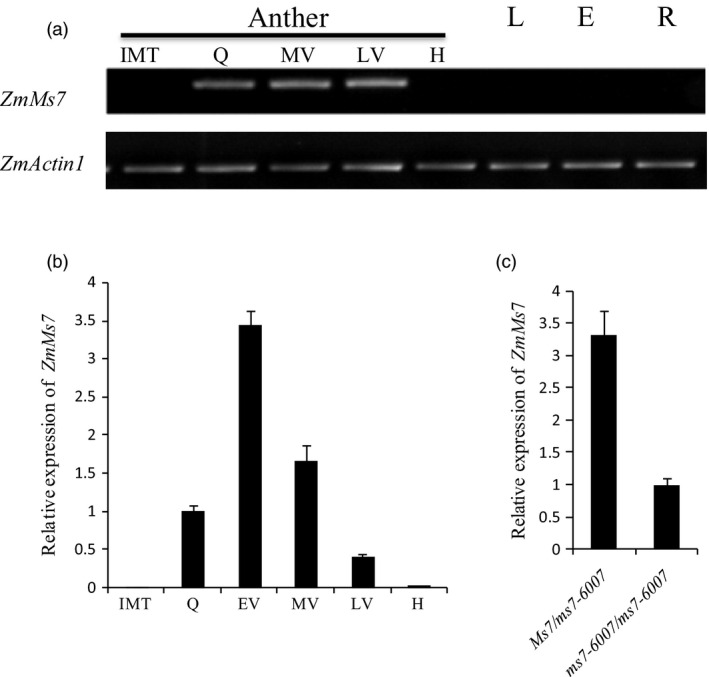
Expression pattern of maize ZmMs7. (a) Maximum expression was observed by RT–PCR in the anther during the meiosis quartet to late vacuolate microspore stages. (b) qRT–PCR analysis of ZmMs7 in wild type (WT) during different anther developmental stages. (c) qRT–PCR analysis of ZmMs7 in fertile (Ms7/ms7‐6007) and sterile (ms7‐6007/ms7‐6007) plants during the early‐vacuolate microspore stage. ZmMs7 expression data were normalized to ZmActin1. Different developmental stages of anther are shown: IMT, immature tassels; Q, quartets; EV, early‐vacuolate microspore; MV, middle vacuolate microspore; LV, late vacuolate microspore; H, heading. L, leaves; E, ears; R, roots.
ZmMs7 is the ortholog of rice PTC1 and Arabidopsis MS1
The full‐length genomic DNA and cDNA of ZmMs7 were cloned and sequenced from the B73 maize inbred line (GenBank accession numbers KY657532 and KY657533). The ZmMs7 protein was predicted to comprise 670 amino acids (73.5 kDa). To gain insight into the phylogenetic relationship between ZmMs7 and its close homologs, the ZmMs7 protein sequence was used in BLASTp queries in NCBI. A phylogenetic tree of ZmMs7 and its 9 homologs was constructed (Figure 6). The phylogenetic analysis indicated that maize ZmMs7 was located in the same clade with rice PTC1 (Li et al., 2011) and other monocot homologs, such as Sorghum bicolor, Hordeum vulgare, Oryza brachyantha, Dichanthelium oligosanthes, Setaria italica and Brachypodium distachyon (Figure 6a), whereas the Arabidopsis thaliana MS1 and Brassica napus homolog occupied a relatively distant dicot branch (Figure 6b).
Figure 6.
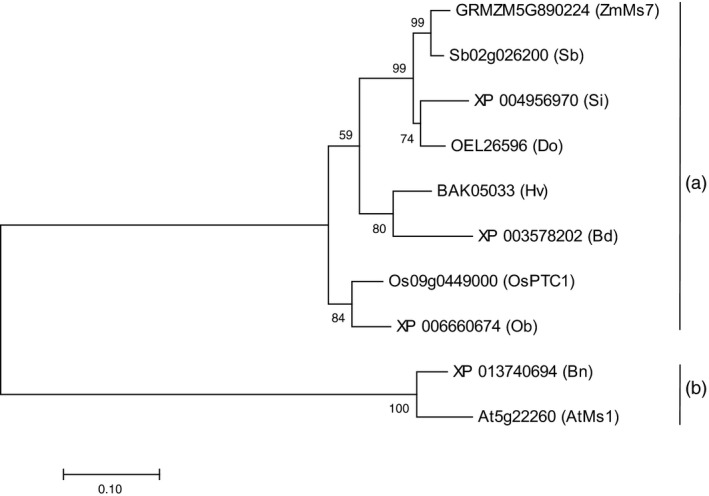
Phylogenetic analysis of ZmMs7 and its related proteins. The evolutionary analyses were conducted in MEGA7 using the maximum‐likelihood method based on the Poisson correction model. The tree with the greatest log likelihood is shown. The analysis involved 10 amino acid sequences from Zea mays (Zm), Sorghum bicolor (Sb), Oryza sativa (Os), Oryza brachyantha (Ob), Arabidopsis thaliana (At), Dichanthelium oligosanthes (Do), Setaria italica (Si), Brachypodium distachyon (Bd), Hordeum vulgare (Hv) and Brassica napus (Bn). Group A comprises eight proteins from monocots, and group B includes two proteins from dicots.
Based on the predicted amino acid sequence alignment, ZmMs7 shared 80.85% and 40.49% similarity with rice PTC1 and Arabidopsis MS1, respectively. Using the conserved domain BLAST in NCBI, the 624‐658‐amino acid interval of ZmMs7 was predicted as a plant homeodomain (PHD) domain (Figure 7) and was conserved among maize, rice and Arabidopsis. Furthermore, based on Arabidopsis MS1 (Wilson et al., 2001), a nuclear localization signal (RRRKR) was found at the N‐terminus, and the 260‐282‐amino acid interval of ZmMs7 was predicted to be a leucine zipper region (Figure 7), which is required for protein function (Ito et al., 2007). Interestingly, the region harbouring the PTC1 gene on rice chromosome 9 was syntenic to the maize chromosome 7 region containing ZmMs7 (Li et al., 2011). Considered together with the phylogenetic analysis (Figure 6), we concluded that ZmMs7 encodes a PHD‐finger transcription factor that is orthologous to rice PTC1 and Arabidopsis MS1.
Figure 7.
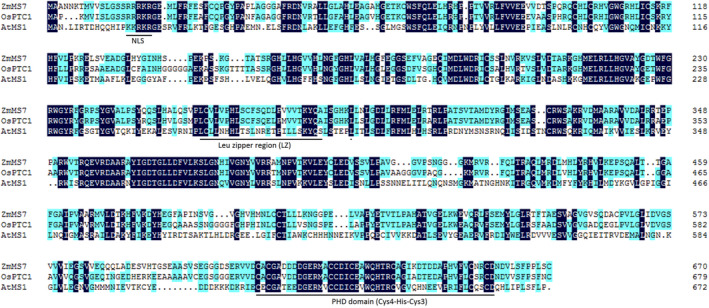
The amino acid sequence alignment of maize ZmMs7, rice PTC1 (OsPTC1) and Arabidopsis MS1 (AtMS1). The nuclear localization sequence (NLS), leucine zipper region (LZ) and putative conserved PHD domain (Cys4‐His‐Cys3) are underlined (refer to Li et al., 2011; Wilson et al., 2001).
Development of a multicontrol sterility system using ZmMs7
As the ms7‐6007 mutant exhibits complete male sterility and no pollen exsertion from its tassel (Figure 1; Beadle, 1932), it is desirable for use in the development of a male‐sterile line for hybrid seed production. Here, we developed a MCS system based on the ms7‐6007 mutant and ZmMs7 gene. The technical strategy is shown in Figure S3a.
First, three pMCS binary vectors (pMCS0701, 0702 and 0703) were constructed (Table 1 and Figure S3b). The T‐DNA region contained four or five functional modules: (i) ZmMs7 driven by its native promoter for restoration of male fertility, (ii) maize α‐amylase gene ZmAA (Wu et al., 2016) driven by the pollen‐specific PG47 promoter (Allen and Lonsdale, 1993) and/or (iii) the DNA adenine methylase gene Dam (Brooks et al., 1983; Unger et al., 2001) driven by the pollen‐specific Zm13 promoter (Hanson et al., 1989) to devitalize the transgenic pollen, (iv) red fluorescence protein gene DsRed2 or mCherry (Matz et al., 1999) driven by the aleurone‐specific LTP2 promoter (Kalla et al., 1994) to mark the transgenic seed and (v) the herbicide‐resistant gene Bar driven by the CaMV35S promoter for genetic transformation and transgenic seed selection.
Table 1.
Constructs for development of MCS maintainer lines using the ZmMs7 gene
| Construct | Promoter‐gene combination |
|---|---|
| pMCS0701 | p35S::Bar//pZmMs7::ZmMs7//pZm13::Dam//pPG47::Bt1:ZmAA//pLTP2::mCherry |
| pMCS0702 | p35S::Bar//pPG47::Bt1:ZmAA//pZmMs7::ZmMs7//pLTP2::DsRed2 |
| pMCS0703 | p35S::Bar//pPG47::Bt1:ZmAA//pZm13::Dam//pZmMs7::ZmMs7//pLTP2::DsRed2 |
ZmMs7, maize fertility restoration gene; ZmAA, α‐amylase gene; DsRed2 and mCherry, red fluorescence gene; 35S, cauliflower mosaic virus 35S promoter; Bar, herbicide resistance gene; Bt1, Brittle‐1 transit peptide; pZm13, Zm13 gene promoter; Dam, DNA adenine methylase gene; pLTP2, lipid transfer protein‐2 gene promoter; pZmMs7, ZmMs7 gene promoter; pPG47, polygalacturonase gene promoter.
Second, the pMCS constructs were introduced into the maize Hi‐II hybrid line via Agrobacterium tumefaciens‐mediated transformation to obtain ZmMs7 transgenic intermediate material. The T0 transgenic plants were pollinated using the pollen of heterozygous Ms7/ms7‐6007, and the T1 progenies were screened for events carrying both a single copy of the pMCS T‐DNA and the ms7‐6007 allele. Based on the phenotypic characterization, RT–PCR and Southern blotting analysis (Figure S4), an elite T1 transgenic event of pMCS0701 (M0701‐2) was selected as a representative and used for further study. The M0701‐2 plant exhibited normal vegetative and reproductive growth, and a nearly 1:1 (59:61) ratio of fluorescent seeds (with a hemizygous transgene) to nonfluorescent seeds (no transgene) on the ear (Figure 8). The seeds were sorted out manually based on red fluorescence and grown to obtain the next generation.
Figure 8.
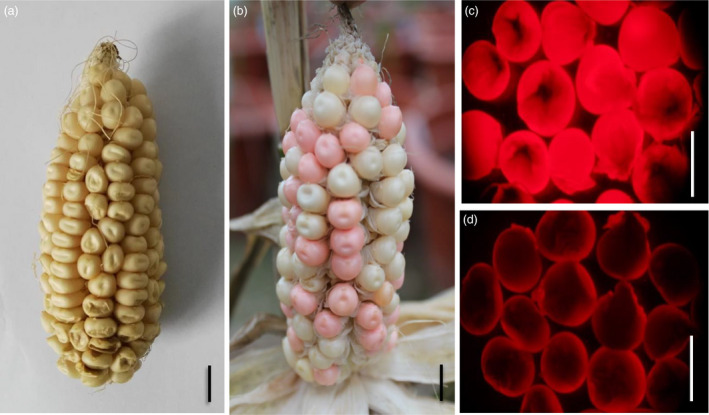
Phenotype of the ZmMs7 transgenic maintainer line (T1). (a and b), Ear phenotype of the maize Hi‐II hybrid line (a) and transgenic event (M0701‐2, b); (c and d), transgenic seeds (c) and nontransgenic seeds (d) from M0701‐2 under a red fluorescence (RF) filter. T1 is the first transformation generation. Bars = 1 cm.
Third, the transgenic event M0701‐2 was self‐pollinated and screened for the homozygous genotype of the ms7‐6007 allele, and then, a candidate male‐sterile maintainer line for ms7‐6007 was obtained. Because ZmMs7 is a sporophytic male fertility gene, a hemizygous ZmMs7 transgene in ms7‐6007 can fully restore male fertility. As ZmAA driven by a PG47 promoter is a gametophytic factor that disrupts starch accumulation only in the transgenic pollen (Allen and Lonsdale, 1993; Wu et al., 2016), and anther‐targeted expression of Dam in maize is used to inactivate a genetic region critical for pollen formation or function thereby causing abnormal development of tapetal cells and microspores (Cigan and Albertsen, 2001; Unger et al., 2001), all the transgenic pollen grains produced by the hemizygous transgenic plants were deactivated and infertile. The selected T2 plant was homozygous at the ms7‐6007 locus but hemizygous at the MCS T‐DNA locus. As pollen grains carrying the transgenic T‐DNA were all defective, the transgenic element was inherited only through the female gamete. All plants from the fluorescent seeds were male fertile, but all plants from the nonfluorescent seeds were male sterile. The fluorescence screening and fertility examination experiments were repeated for three generations, with a total of more than 500 000 seeds, all of which showed the same results as described above; namely, all fluorescent seed‐derived plants were male fertile, but all nonfluorescent seed‐derived plants were male sterile, indicating that both the transgenic pMCS0701 and ms7‐6007 mutant locus elements were inherited stably.
Confirmation of whether MCS progeny seeds inherit the transgene through pollen
To determine the transgene transmission risk through pollen, transgenic MCS maintainer plants from three constructs (differing by the number of pollen‐disrupted genes, the gene orientation and the seed colour marker gene; Table 1) were used to pollinate the maize inbred line Zheng58, which is the female parent line of Zhengdan 958, a famous maize hybrid with the largest planting area in the last decade in China. Ears were harvested from the female parent plants and examined under green excitation light for the presence of red fluorescent seeds resulting from the expression of DsRed2 or mCherry protein. The transgene transmission rate through pollen in seeds varied with different constructs and different transformant events (Table 2). The percentages of red fluorescent seeds in the four events of pMCS0701 were varied from 0.031% to 0.037% (averagely 0.033%), those in five events of pMCS0703 were varied from 0.032% to 0.043% (averagely 0.036%), while those in three events of pMCS0702 were varied from 0.236% to 0.301% (averagely 0.263%). In summary, the constructs of pMCS0701 and pMCS0703, containing two pollen‐disrupted modules (ZmAA and Dam), showed significantly lower rates of transgenic pollen transmission compared with the construct of pMCS0702 harbouring one pollen‐disrupted module (ZmAA). This result suggests that two pollen‐disrupted modules are more efficient than one pollen‐disrupted module in preventing transgene transmission through pollen in the genetic background of maize inbred line Zheng58. Subsequently, transformant M0701‐2 from the pMCS0701 construct (Table 1) was selected as the MCS maintainer line to produce the maize male‐sterile lines, which are very useful in maize breeding and hybrid seed production.
Table 2.
Transgene transmission rate through the pollen of different ZmMs7 transgenic maintainer lines
| Construct | Generation | Transformants | Number of red fluorescent seeds/total number of seeds produced by nontransgenic plants pollinated by transgenic plants | Per cent of red fluorescent seeds (%) |
|---|---|---|---|---|
| pMCS0701 | T2 | M0701‐2 | 58/157 552 | 0.037 |
| M0701‐9 | 26/83 871 | 0.031 | ||
| M0701‐25 | 14/41 176 | 0.034 | ||
| M0701‐37 | 11/34 874 | 0.0315 | ||
| pMCS0702 | T2 | M0702‐3 | 224/94 751 | 0.236 |
| M0702‐6 | 78/31 076 | 0.251 | ||
| M0702‐11 | 53/17 608 | 0.301 | ||
| pMCS0703 | T2 | M0703‐6 | 204/578 682 | 0.035 |
| M0703‐9 | 31/96 875 | 0.032 | ||
| M0703‐16 | 28/74 684 | 0.038 | ||
| M0703‐22 | 19/43 901 | 0.043 | ||
| M0703‐23 | 14/4 0967 | 0.034 |
T2 is the second‐generation transformant. There are two pollen‐disrupted genes (ZmAA and Dam) in the pMCS0701 and pMCS0703 constructs but only one pollen‐disrupted gene (ZmAA) in the pMCS0702 construct (refer to Table 1).
Application of the M0701‐2 maintainer line for breeding various male‐sterile lines in maize
To breed various male‐sterile lines by transferring the ms7‐6007 mutant locus and the MCS transgenic T‐DNA element into other germplasm, M0701‐2 (as the maternal parent) was crossed with five different maize elite inbred lines (including Zheng58) as the recurrent male parent, respectively. Progeny were analysed for the presence of the ms7‐6007 mutant locus and transgenic element by marker‐assisted selection, and seeds of two consecutive backcrossed and self‐pollinated generations (BC2F2) have been obtained to date. Both the ms7‐6007 mutant locus and the transgenic element were accurately associated with their respective phenotypes in the segregated progeny. The ears of three candidate MCS transgenic maintainer lines are shown in Figure S5. The ratio of red fluorescent seeds to nonfluorescent seeds in each ear was nearly 1:1, while all red fluorescent seeds were male fertile, and nonfluorescent seeds were male sterile. These results indicated that both the ms7‐6007 mutant locus and the transgenic element from the M0701‐2 event maintained their respective functions when transferred into different genetic backgrounds in maize, making it feasible to breed male‐sterile lines using traditional backcrossing and marker‐assisted selection methods.
Discussion
The ms7 mutant displays complete recessive male sterility, an abnormal microspore wall and tapetal cell development
ms7 is a complete recessive male‐sterile mutant with no exserted anthers (Figure 1b, c, d; Table S1) and has been characterized cytologically as having a poorly developed, thin microspore wall and aperture as well as abnormal tapetal cell development (Morton et al., 1989). The tapetal cells in the ms7 mutant did not become flattened tangentially to the anther wall layers as in normal tapetal cell development, and darkly staining material was observed within the ms7 mutant cells. Microsporogenesis was normal until the quartet stage when precocious chromosome condensation might occur (Albertsen and Phillips, 1981). Similarly, tapetal cells of the ms7 mutant appeared normal up to the quartet stage when they started to vacuolate and rapidly degenerate (Morton et al., 1989). Consistent with previous reports, our data showed that during the late microspore developmental stage, the tapetal cells of the ms7‐6007 mutant were swollen and lightly stained with aborted microspores, leaving debris in the shrinkage locule and indicating abnormal PCD of tapetal cells (Figure 1e, f). Furthermore, the cytological characteristics of the ms7 mutant were similar to the rice ptc1 and Arabidopsis ms1 mutants, which displayed delayed tapetum degeneration and defects in the development of the pollen exine (Ito and Shinozaki, 2002; Ito et al., 2007; Li et al., 2011; Yang et al., 2007). Together, these results suggested that ZmMs7 played important roles in microspore wall formation and tapetal cell PCD during late anther development. However, until now, the ZmMs7 gene has not been cloned or characterized.
ZmMs7 encodes a PHD‐finger protein similar to rice PTC1 and Arabidopsis MS1
Using a map‐based cloning approach, we found that ZmMs7 encodes a PHD‐finger protein with a N‐terminal nuclear location signal and a leucine zipper‐like region, which shares high similarity with rice PTC1 and Arabidopsis MS1 (Figure 7). All three mutants (maize ms7, rice ptc1 and Arabidopsis ms1) showed similar male‐sterile phenotypes, defects in pollen exine formation and tapetal cell development (Figure 1) (Ito and Shinozaki, 2002; Ito et al., 2007; Li et al., 2011; Morton et al., 1989; Yang et al., 2007). Moreover, similar to the spatio‐temporal expression patterns of rice PTC1 and Arabidopsis MS1 genes, ZmMs7 was also expressed specifically in maize anthers from the quartet to the late vacuolate microspore stages (Figure 5). Furthermore, the orthologs of ZmMs7 were found in nine flowering plants by a BLASTp search and phylogenetic analysis (Figure 6). Based on the cytological similarity of mutants, the same expression pattern of genes and the high identity of the proteins, we conclude that ZmMs7 gene is the ortholog of the rice PTC1 and Arabidopsis MS1 genes, and they are functionally conserved in flowering plants. Mutations in the three genes (maize ZmMs7, rice PTC1 and Arabidopsis MS1) led to a similar male‐sterile phenotype and cytological defect, demonstrating that this kind of PHD‐finger protein is required for plant male gametogenesis and has potential application value in crop hybrid seed production.
The technical strategy of the MCS system based on the ZmMs7 gene
To date, hundreds of recessive genetic male‐sterile mutants have been identified in maize, but their application in maize breeding and hybrid seed production has been limited because of the inability to propagate a pure male‐sterile line via self‐pollination. As more male‐sterile genes have been cloned and an efficient transgenic method becomes feasible in maize, several strategies for maintaining and propagating male‐sterile lines have been proposed based on the transgenic maintainer line (Perez‐Prat and van Lookeren Campagne, 2002; Williams, 1995; Wu et al., 2016). Among them, the Seed Production Technology (SPT) strategy devised by DuPont‐Pioneer has been used for commercial hybrid seed production (Wu et al., 2016). The core technology of SPT system is the transformation of the male‐sterile mutant ms45 with a SPT construct, which comprises three functional modules: (i) a male fertility gene, (ii) a pollen‐disrupted gene and (iii) a seed colour marker gene. Although many advantages of the SPT system have been suggested, such as no need detasseling, no limitations to its use in maize germplasm and an increased hybrid seed purity, and even enhanced hybrid seed production yields, the transgene transmission rate through pollen (based on red florescence seeds expressing the DsRed2 gene) has been observed to vary with different SPT constructs, transformant events and genetic backgrounds, with the range of transgene transmission rates from 0.002% to 0.518% (Wu et al., 2016).
To decrease the transgene transmission rate of the transgenic maintainer line through pollen, we developed a MCS system by transforming MCS constructs into the ms7‐6007 mutant (Figure S3). The MCS constructs contained (i) the cloned male fertility gene ZmMs7 to restore fertility, (ii) two pollen‐disrupted genes (ZmAA and Dam) to disrupt the transgenic pollens, (iii) the screenable fluorescent colour marker gene (DsRed2 or mCherry) to identify the transgenic seeds and facilitate purification of the transgenic MCS maintainer line and (iv) the herbicide‐resistant gene (Bar) to prevent sophistication of MCS maintainer line seeds (Figure S3). Because the MCS construct harbours two pollen‐disrupted modules, both of which can inhibit transgenic pollen formation or function, the transgene transmission rate through pollen can be greatly decreased. One is the ZmAA gene driven by the late pollen‐specific PG47 promoter (Allen and Lonsdale, 1993; Wu et al., 2016), which can interfere with normal starch accumulation in transgenic pollen and repress pollen development. The other is the Dam gene driven by the pollen‐specific Zm13 promoter (Brooks et al., 1983; Hamilton et al., 1992; Hanson et al., 1989; Unger et al., 2001), which catalyses the methylation of adenine residues in the DNA of pollen and affects the cell viability of transgenic pollen. Anther‐targeted expression of Dam gene results in abnormal tapetal cells and microspores and renders maize male sterile (Unger et al., 2001). Our data indicated that the transgene transmission rate through pollen in MCS transformants harbouring two pollen‐lethality genes (ZmAA and Dam) decreased significantly (1/8–1/7) compared with those harbouring one gene (ZmAA) in the genetic background of Zheng58 (Table 2), confirming that the MCS constructs containing two pollen‐lethality genes were more efficient and viable in the genetic background of Zheng58. Furthermore, there are several advantages of Bar in the MCS construct to propagate highly pure seeds of the MCS transgenic maintainer line and male‐sterile line. During the MCS transgenic maintainer line‐propagating phase, to assure the high purity (100%) of the MCS maintainer line seeds, appropriate herbicide spraying of seedlings of the MCS maintainer line in the field will efficiently eliminate any nontransgenic plants. During the male‐sterile line‐propagating phase, the male‐sterile line is cross‐pollinated with the transgenic maintainer line as pollen donor. Additionally, it is useful to spray herbicide specifically on the transgenic maintainer line to eliminate the mixed nontransgenic plants, which ensures a high purity of male parent pollen and male‐sterile line seeds. Compared with the SPT, the MCS constructs harbouring the five functional modules contain a Bar and a Dam gene, which can assure high purity of the male‐sterile parent line and hybrid seed production, and greatly decrease the transgene transmission rate as well as the transgene flowing risk. Therefore, this strategy was designed as a multicontrol sterility system.
The MCS system is superior to CMS and other biotechnology‐based systems in several aspects. First, compared to the CMS system, the MCS male‐sterile line is controlled by a single recessive nuclear gene, and any maize germplasm carrying the wild‐type ZmMs7 allele can complement the ms7‐6007 mutation. Moreover, the male sterility is genetically stable in various environments, greatly reducing the risk induced by weather changes. Second, compared with the SPT system, the transgenic MCS maintainer lines contain five functional modules, in particular, two pollen‐disrupted genes (ZmAA and Dam) driven by two pollen‐specific promoters, PG47 and Zm13, respectively, which significantly reduce the transgene transmission rate through pollen (Table 2). Third, the herbicide resistance gene Bar in the MCS system makes it easier to transfer and select transgenic constructs in maize germplasm with different genetic backgrounds by appropriate herbicide spraying of the seedlings to eliminate nontransgenic plants. Concurrently, herbicide resistance in the MCS system is beneficial for the propagation of high‐purity MCS transgenic maintainer line seeds and male‐sterile line seeds through herbicide spraying during specific stages of production. Finally, although the MCS system involves transgenic processes, only the MCS maintainer line carries the transgene. Thus, neither the obtained male‐sterile line seeds nor the hybrid seeds contain any transgenic elements.
Acknowledgements of the nontransgenic status of progeny produced by the SPT process have been supported by regulatory agencies in the USA (USDA‐APHIS, 2011), Australia (FSANZ, 2012) and Japan (MHLW, 2013). Compared with the SPT system, the transgene transmission rate through the MCS maintainer pollen may be reduced in a specific genetic background, and the herbicide resistance in MCS maintainer lines makes them safer and more efficient for the production of high‐purity male‐sterile parent lines and hybrid seeds. Therefore, the MCS system should also be considered for a nontransgenic status to facilitate the use of MCS male‐sterile lines in commercial maize hybrid seed production, especially in China. First, there are more than 200 000 hectares of maize hybrid seed production acreage every year in China, and commercial application of the MCS system will dramatically reduce the total labour costs. Second, hybrid seed production yields can be increased significantly due to the absence of a need for physical (manual and/or mechanical) detasseling, which often removes the tassel together with several leaves, resulting in reductions by as much as 40% of the potential seed yield (Wych, 1988). Third, based on the MCS male‐sterile line, the efficiency of hybrid maize breeding will be greatly accelerated by the increased level of hybrid combination per breeder through the use of the open‐pollination approach in different isolation fields. Finally, considering the huge planted acreage (approximately 34 million hectares) of maize in China, the total yield of commodity maize grain using MCS hybrid seeds will be significantly increased because of the high quality and purity of hybrid seeds produced from the MCS system. Therefore, commercial application of the MCS technology will greatly enhance the efficiency of maize hybrid breeding, increase hybrid seed yields and even raise maize grain commodity yields.
Potential applications of the MCS system in other major crops
Based on BLASTp and phylogenetic analyses, several function‐conserved orthologs of ZmMs7 have been found in other major crops, including sorghum, barley and oilseed rape (Figure 6), which have flowers that are not amenable to manual emasculation. Therefore, a mutation generated artificially in orthologous genes (Sb02 g026200, BAK05033 and XP_013740694, Figure 6) may also cause male sterility. Recently, MALE STERILITY1 (HvMS1), the ortholog of Arabidopsis MS1 and rice PTC1, has been isolated using RACE‐PCR and subsequent functional testing using RNAi lines in barley (Fernández Gómez and Wilson, 2014). Thus, a reverse genetic approach could be adopted using targeted mutagenesis technologies such as the programmable DNA endonuclease and CRISPR/Cas9 system (Cigan et al., 2017; Gomez et al., 2015; Svitashev et al., 2016), which will lead to site‐specific mutations in the corresponding orthologs of ZmMs7 and produce male‐sterile mutants in these crops. The MCS system could be transferred into other major crops using artificial ms mutants and the corresponding fertility restoration genes. In summary, the MCS system will greatly expand the potential to produce male‐sterile lines and hybrid seeds of important crops.
Experimental procedures
Plant materials and growth conditions
ms7‐6007 and ms7gl1 mutants were obtained from the Maize Genetics Cooperation Stock Center. The two F2 mapping populations were derived from crosses of ms7‐6007 × Chang7‐2 and ms7gl1 × Chang7‐2, respectively. All the plants were grown in the field in Beijing or Sanya, China. The T0 transgenic plants and their progeny were grown in a greenhouse in Beijing, China.
Phenotypic identification, histochemical analysis and microscopy
Tassels and spikelets were photographed using a Canon EOS 700D digital camera (Canon, Japan). To evaluate the pollen viability, anthers and pollen grains were stained with 1% I2‐KI and photographed under an Olympus SZ51 microscope (Olympus, Japan). For transverse section analysis, the spikelet was fixed in 3:1 ethanol: acetic acid. Fixed samples were dehydrated through an ethanol series and embedded in epoxy resin. Semi‐thin sections were obtained using an Ultracut E Ultramicrotome (Reichert), stained with 0.1% toluidine blue O and observed under an Olympus BX61 microscope (Olympus, Japan).
Map‐based cloning
Genomic DNA was extracted from maize leaves using the CTAB method with some modifications (Murray and Thompson, 1980). Nine SSR primers on maize chromosome 7L were chosen for ms7 primary mapping (Table S2). For fine mapping, several CAPS markers were designed with DNAMAN6.0 (LynnonBiosoft) (Table S3). All PCR primers for these SSR and CAPS markers were synthesized by Sangon Biotech (Shanghai, China) and were tested to identify polymorphic markers distinguishing fertile from sterile plants. By scoring the presence/absence of recombinants at diverse marker locations, the ms7 gene was narrowed to a 180‐kb interval on chromosome 7L.
Protein alignment and phylogenetic analysis
Ten homologs of ZmMs7 were obtained by BLASTp search at the NCBI website. The phylogenetic tree was generated in MEGA7.0 (Kumar et al., 2016) using the maximum‐likelihood method with the following parameters: Poisson model, complete deletion and 1000 bootstrap replications. The amino acid sequences of maize ZmMs7, rice PTC1 and Arabidopsis MS1 were aligned using DNAMAN6.0, and the conserved domains were analysed with a CD search in the NCBI website.
RNA extraction and expression analysis
Total maize RNA was isolated using TRIzol reagent (Invitrogen) from the following maize tissues: anthers during different stages, leaves, roots and immature ears. Total RNA (1 μg) was used to synthesize first‐strand cDNA using Superscript III RT (Invitrogen). RT–PCR analyses were conducted using 1 μL cDNA as template. qRT–PCR analyses were performed using SYBR Green PCR Master Mix (ABI). ZmActin1 was used as the internal control. The relative expression was calculated using the 2−▵▵Ct method. The SD was calculated with three biological replications. All primers used for RT–PCR and qRT–PCR are listed in Table S4.
Plasmid construction, transformation and transgene determination
For complementation, the ZmMs7 gene promoter (1.2 kb) was amplified from maize B73 using primer Ms7‐ProP, and the ZmMs7 coding sequence (2 kb) was amplified using primer Ms7‐CDSP from the cDNA of B73 anthers. The two fragments were fused together with HindIII/BamHI and then cloned into pBCXUN (Chen et al., 2009). The vector was named pZmMs7pro::ZmMs7.
The T‐DNA of the MCS constructs was inserted into the backbone of pCAMBIA3301 (www.cambia.org) and named pMCS0701, pMCS0702 and pMCS0703 respectively. The detailed procedure refers to Data S1.
All constructs were transformed into the maize Hi‐II hybrid line using the Agrobacterium‐mediated transformation method (Frame et al., 2002). The Bar gene was employed as a selectable marker, and the transformants were screened by PCR amplification using the primer Bar‐P. The transgenic T‐DNA region was then transferred into the ms7‐6007 mutant by the crossing and backcrossing method. To confirm the presence of the ms7‐6007 allele and transgenic T‐DNA sequence in the progeny, the transgenic plants were screened by PCR amplification using primers ms6007‐ID and Bar‐P. The genomic DNA of each transgenic plant was digested with HindIII and further analysed using a DIG High Prime DNA labelling and Detection Starter Kit II (Roche) with DIG‐labelled mCherry as the probe for Southern blotting analysis. Expression of the mCherry gene was determined by RT–PCR using mCherry‐P primer, and ZmActin1 was used as the native control. All the above‐described primers are listed in Table S4.
Seed colour sorting
A dual‐fluorescent protein (DFP) flashlight (www.nightsea.com/products/dfp) was used for seed colour sorting. With the matching barrier filter glasses, the red fluorescent seeds could be easily sorted from the nonred fluorescent seeds. The number of red fluorescent seeds in different transformants was counted manually.
Supporting information
Figure S1 Phenotypic comparison of the wild type (WT) and ms7gl1 mutant.
Figure S2 Predicted amino acid sequences and alignment of ZmMs7 in wild type and the ms‐6007 and ms7gl1 mutants.
Figure S3 A multi‐control sterility (MCS) system in maize via a transgenic approach.
Figure S4 Molecular analysis of ZmMs7 transgenic maintainer lines.
Figure S5 Phenotype of three ZmMs7 transgenic maize male‐sterile maintainer lines (BC2F1).
Table S1 The ratio of fertile to sterile plants in the F2 population of the ms7 mutant.
Table S2 The SSR marker information used for ZmMs7 primary mapping.
Table S3 The CAPS marker information used for ZmMs7 fine mapping.
Table S4 PCR primers used in this study.
Data S1 The detailed construction procedure used for the three MCS plasmids.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (06500060), the National Key R&D Program (2017YFD0102000, 2017YFD0101200), the International S&T Cooperation Program of China (2015DFA30640), the National Key Technology R&D Program (2014BAD01B02) and the Beijing Science and Technology Plan Program (Z161100000916013). The authors declare no conflict of interest.
ZmMs7 gene accession numbers: KY657532 and KY657533.
Contributor Information
Jiuran Zhao, Email: maizezhao@126.com.
Xiangyuan Wan, Email: wanxiangyuan@ustb.edu.cn.
References
- Albertsen, M.C. and Phillips, R.L. (1981) Developmental cytology of 13 genetic male sterile loci in maize. Can. J. Genet. Cytol. 23, 195–208. [Google Scholar]
- Albertsen, M.C. , Fox, T. , Trimnell, M. , Wu, Y. , Lowe, L. , Li, B. and Faller, M. (2009) Msca1 nucleotide sequences impacting plant male fertility and method of using same. US patent US20090038027A1.
- Albertsen, M. , Fox, T. , Leonard, A. , Li, B. , Loveland, B. and Trimnell, M. (2016) Cloning and use of the ms9 gene from maize. US patent US20160024520A1.
- Allen, R.L. and Lonsdale, D.M. (1993) Molecular characterization of one of the maize polygalacturonase gene family members which are expressed during late pollen development. Plant J. 3, 261–271. [DOI] [PubMed] [Google Scholar]
- Beadle, G.W. (1932) Genes in maize for pollen sterility. Genetics, 17, 413–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, J.E. , Blumenthal, R.M. and Gingeras, T.R. (1983) The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 11, 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Z. , Chen, Z. , Wang, N. , Xie, G. , Lu, J. , Yan, W. , Zhou, J. et al. (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. 113, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubal, R. , Anderson, J.R. , Trimnell, M.R. , Fox, T.W. , Albertsen, M.C. and Bedinger, P. (2003) The transformation of anthers in the msca1 mutant of maize. Planta, 216, 778–788. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Songkumarn, P. , Liu, J. and Wang, G.L. (2009) A versatile zero background T‐vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Zhang, H. , Sun, H. , Luo, H. , Zhao, L. , Dong, Z. , Yan, S. et al. (2017) IRREGULAR POLLEN EXINE1 Is a novel factor in anther cuticle and pollen exine formation. Plant Physiol. 173, 307–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan, A.M. and Albertsen, M.C. (2001) Reversible nuclear genetic system for male sterility in transgenic plants. US Patent US6281348B1
- Cigan, A.M. , Unger, E. , Xu, R.J. , Kendall, T. and Fox, T.W. (2001) Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex. Plant Reprod. 14, 135–142. [Google Scholar]
- Cigan, A.M. , Singh, M. , Benn, G. , Feigenbutz, L. , Kumar, M. , Cho, M.J. , Svitashev, S. et al. (2017) Targeted mutagenesis of a conserved anther‐expressed P450 gene confers male sterility in monocots. Plant Biotechnol. J. 15, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukanovic, V. , Smith, J. , Lowe, K. , Yang, M. , Gao, H. , Jones, S. , Nicholson, M.G. et al. (2013) Male‐sterile maize plants produced by targeted mutagenesis of the cytochrome P450‐like gene (MS26) using a re‐designed I‐CreI homing endonuclease. Plant J. 76, 888–899. [DOI] [PubMed] [Google Scholar]
- Feng, P.C. , Qi, Y. , Chiu, T. , Stoecker, M.A. , Schuster, C.L. , Johnson, S.C. , Fonseca, A.E. et al. (2014) Improving hybrid seed production in corn with glyphosate‐mediated male sterility. Pest Manag. Sci. 70, 212–218. [DOI] [PubMed] [Google Scholar]
- Fernández Gómez, J. and Wilson, Z.A. (2014) A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12, 765–777. [DOI] [PubMed] [Google Scholar]
- Fox, T. , DeBruin, J. , Haug Collet, K. , Trimnell, M. , Clapp, J. , Leonard, A. , Li, B. et al. (2017) A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 15, 942–952 10.1111/pbi.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, B.R. , Shou, H. , Chikwamba, R.K. , Zhang, Z. , Xiang, C. , Fonger, T.M. , Pegg, S.E. et al. (2002) Agrobacterium tumefaciens‐mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129, 13‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FSANZ (2012) New Plant Breeding Techniques. Report of a Workshop hosted by Food Standards Australia New Zealand (FSANZ). Available at: http://www.foodstandards.gov.au/publications/Documents/New%20Plant%20Breeding%20Techniques%20Workshop%20Report.pdf.
- Gils, M. , Marillonnet, S. , Werner, S. , Grutzner, R. , Giritch, A. , Engler, C. , Schachschneider, R. et al. (2008) A novel hybrid seed system for plants. Plant Biotechnol. J. 6, 226–235. [DOI] [PubMed] [Google Scholar]
- Gomez, J.F. , Talle, B. and Wilson, Z.A. (2015) Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 57, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, D.A. , Roy, M. , Rueda, J. , Sindhu, R.K. , Sanford, J. and Mascarenhas, J.P. (1992) Dissection of a pollen‐specific promoter from maize by transient transformation assays. Plant Mol. Biol. 18, 211–218. [DOI] [PubMed] [Google Scholar]
- Hanson, D.D. , Hamilton, D.A. , Travis, J.L. , Bashe, D.M. and Mascarenhas, J.P. (1989) Characterization of a pollen‐specific cDNA clone from Zea mays and its expression. Plant Cell, 1, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T. and Shinozaki, K. (2002) The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD‐finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Ito, T. , Nagata, N. , Yoshiba, Y. , Ohme‐Takagi, M. , Ma, H. and Shinozaki, K. (2007) Arabidopsis MALE STERILITY1 encodes a PHD‐type transcription factor and regulates pollen and tapetum development. Plant Cell, 19, 3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalla, R. , Shimamoto, K. , Potter, R. , Nielsen, P.S. , Linnestad, C. and Olsen, O.A. (1994) The promoter of the barley aleurone‐specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell‐specific expression in transgenic rice. Plant J. 6, 849–860. [DOI] [PubMed] [Google Scholar]
- Kaul, M.L.H. (1988) Male Sterility in Higher Plants: Monographs on Theoretical and Applied Genetics 10. Berlin: Springer Verlag. [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Yuan, Z. , Vizcay‐Barrena, G. , Yang, C. , Liang, W. , Zong, J. , Wilson, Z.A. et al. (2011) PERSISTENT TAPETAL CELL1 encodes a PHD‐finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani, C. , De Beuckeleer, M. , Truettner, J. , Leemans, J. and Goldberg, R.B. (1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature, 347, 737–741. [Google Scholar]
- Matz, M.V. , Fradkov, A.F. , Labas, Y.A. , Savitsky, A.P. , Zaraisky, A.G. , Markelov, M.L. and Lukyanov, S.A. (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973. [DOI] [PubMed] [Google Scholar]
- McRae, D.H. (1985) Advances in chemical hybridization. Plant Breed Rev. 3, 169–191. [Google Scholar]
- MHLW, J. (2013) The outcome of the discussion on F1 hybrid seeds produced with the DuPont's Seed Production Technology (SPT) using DP‐32138‐1. Japan Ministry of Health, Labor, and Welfare. Available at: http://www.mhlw.go.jp/stf/shingi/2r9852000002tccm-att/2r9852000002tck7.pdf.
- Millwood, R.J. , Moon, H.S. , Poovaiah, C.R. , Muthukumar, B. , Rice, J.H. , Abercrombie, J.M. , Abercrombie, L.L. et al. (2016) Engineered selective plant male sterility through pollen‐specific expression of the EcoRI restriction endonuclease. Plant Biotechnol. J. 14, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N. , Hiratsu, K. , Todaka, D. , Nakashima, K. , Yamaguchi‐Shinozaki, K. and Ohme‐Takagi, M. (2006) Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4, 325–332. [DOI] [PubMed] [Google Scholar]
- Moon, J. , Skibbe, D. , Timofejeva, L. , Wang, C.J. , Kelliher, T. , Kremling, K. , Walbot, V. et al. (2013) Regulation of cell divisions and differentiation by MALE STERILITY32 is required for anther development in maize. Plant J. 76, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, C.M. , Lawson, D.L. and Bedinger, P. (1989) Morphological study of the maize male sterile mutant ms7. Maydica, 34, 239–245. [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan, G.L. , Zhai, J. , Arikit, S. , Morrow, D. , Fernandes, J. , Mai, L. , Nguyen, N. et al. (2017) MS23, a master basic helix‐loop‐helix factor, regulates the specification and development of the tapetum in maize. Development, 144, 163–172. [DOI] [PubMed] [Google Scholar]
- Perez‐Prat, E. and van Lookeren Campagne, M.M. (2002) Hybrid seed production and the challenge of propagating male‐sterile plants. Trends Plant Sci. 7, 199–203. [DOI] [PubMed] [Google Scholar]
- Singh, S.P. , Pandey, T. , Srivastava, R. , Verma, P.C. , Singh, P.K. , Tuli, R. and Sawant, S.V. (2010) BECLIN1 from Arabidopsis thaliana under the generic control of regulated expression systems, a strategy for developing male sterile plants. Plant Biotechnol. J. 8, 1005–1022. [DOI] [PubMed] [Google Scholar]
- Skibbe, D.S. and Schnable, P.S. (2005) Male sterility in maize. Maydica, 50, 367–376. [Google Scholar]
- Somaratne, Y. , Tian, Y. , Zhang, H. , Wang, M. , Huo, Y. , Cao, F. , Zhao, L. et al. (2017) ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize. Plant J. 90, 96–110 10.1111/tpj.13476. [DOI] [PubMed] [Google Scholar]
- Svitashev, S. , Schwartz, C. , Lenderts, B. , Young, J.K. and Mark Cigan, A. (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofejeva, L. , Skibbe, D.S. , Lee, S. , Golubovskaya, I. , Wang, R. , Harper, L. , Walbot, V. et al. (2013) Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3: Genes ‐ Genomes – Genet. 3, 231–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger, E. , Betz, S. , Xu, R. and Cigan, A.M. (2001) Selection and orientation of adjacent genes influences DAM‐mediated male sterility in transformed maize. Transgenic Res. 10, 409–422. [DOI] [PubMed] [Google Scholar]
- USDA‐APHIS (2011) Pioneer Hi‐Bred International, Inc. Seed Production Technology (SPT) Process OECD Unique Identifier: DP‐32138‐1 Corn: Final Environmental Assessment. Riverdale, MD: United States Department of Agriculture Animal and Plant Health Inspection Service. Available at: http://www.aphis.usda.gov/brs/aphisdocs/08_33801p_fea.pdf [Google Scholar]
- Venkatesh, T.V. , Breeze, M.L. , Liu, K. , Harrigan, G.G. and Culler, A.H. (2014) Compositional analysis of grain and forage from MON 87427, an inducible male sterile and tissue selective glyphosate‐tolerant maize product for hybrid seed production. J. Agric. Food Chem. 62, 1964–1973. [DOI] [PubMed] [Google Scholar]
- Vernoud, V. , Laigle, G. , Rozier, F. , Meeley, R.B. , Perez, P. and Rogowsky, P.M. (2009) The HD‐ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 59, 883–894. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Oses‐Prieto, J.A. , Li, K.H. , Fernandes, J.F. , Burlingame, A.L. and Walbot, V. (2010) The male sterile 8 mutation of maize disrupts the temporal progression of the transcriptome and results in the mis‐regulation of metabolic functions. Plant J. 63, 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.J. , Nan, G.L. , Kelliher, T. , Timofejeva, L. , Vernoud, V. , Golubovskaya, I.N. , Harper, L. et al. (2012a) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development, 139, 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Adams, C.M. , Fernandes, J.F. , Egger, R.L. and Walbot, V. (2012b) A low molecular weight proteome comparison of fertile and male sterile 8 anthers of Zea mays. Plant Biotechnol. J. 10, 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.X. , Skibbe, D.S. and Walbot, V. (2013) Maize Male sterile 8 (Ms8), a putative β‐1,3‐galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod. 26, 329–338. [DOI] [PubMed] [Google Scholar]
- Williams, M.E. (1995) Genetic engineering for pollination control. Trends Biotech. 13, 344–349. [Google Scholar]
- Wilson, Z.A. , Morroll, S.M. , Dawson, J. , Swarup, R. and Tighe, P.J. (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD‐finger family of transcription factors. Plant J. 28, 27–39. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Fox, T.W. , Trimnell, M.R. , Wang, L. , Xu, R.J. , Cigan, A.M. , Huffman, G.A. et al. (2016) Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross‐pollinating crops. Plant Biotechnol. J. 14, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wych, R.D. (1988) Production of hybrid seed corn. In Corn and Corn Improvement ( Sprague, G.F. , ed), pp. 565–607. Madison, WI: American Society of Agronomy Inc, Crop Science Society of America, and Soil Science Society of America. [Google Scholar]
- Yang, C. , Vizcay‐Barrena, G. , Conner, K. and Wilson, Z.A. (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell, 19, 3530–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phenotypic comparison of the wild type (WT) and ms7gl1 mutant.
Figure S2 Predicted amino acid sequences and alignment of ZmMs7 in wild type and the ms‐6007 and ms7gl1 mutants.
Figure S3 A multi‐control sterility (MCS) system in maize via a transgenic approach.
Figure S4 Molecular analysis of ZmMs7 transgenic maintainer lines.
Figure S5 Phenotype of three ZmMs7 transgenic maize male‐sterile maintainer lines (BC2F1).
Table S1 The ratio of fertile to sterile plants in the F2 population of the ms7 mutant.
Table S2 The SSR marker information used for ZmMs7 primary mapping.
Table S3 The CAPS marker information used for ZmMs7 fine mapping.
Table S4 PCR primers used in this study.
Data S1 The detailed construction procedure used for the three MCS plasmids.


