Abstract
Chromosomal focal amplifications often cause an increase in gene copy number contributing to the pathogenesis of cancer. PRR14 overexpression is associated with recurrent locus amplification in lung cancer, and it correlates with a poor prognosis. We show that increased PRR14 expression promoted and reduced PRR14 expression impeded lung cancer cell proliferation. Interestingly, PRR14 cells were markedly enlarged in size and exhibited an elevated activity of PI3-kinase/Akt/m-TOR pathway, which was associated with a heightened sensitivity to the inhibitors of PI3K and mTOR. Biochemical analysis revealed that PRR14, as a proline-rich protein, binds to the Src homology 3 (SH3) domains of GRB2 resulting in PI3K activation. Significantly, 2 cancer patient-derived PRR14 mutants displayed considerably augmented GRB2-binding and enhanced ability of promoting cell proliferation. Together with the in vivo data demonstrating a strong tumor-promoting activity of PRR14 and the mutants, our work uncovered this proline-rich protein as a novel activator of the PI3K pathway that promoted tumorigenesis in lung cancer.
Keywords: PRR14, oncogene, PI3K/Akt/mTOR, lung cancer
Introduction
Lung cancer is a leading cause of cancer-related deaths claiming lives of over 25% of cancer patients (26% in woman and 28% in man)33. Basically, it is divided into two broad groups: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC, which accounts for about 85% of lung cancers, can be further subtyped to lung squamous cell carcinoma (LUSC), adenocarcinoma (LUAD), and large cell carcinoma. The majority of NSCLC patients are diagnosed at advanced stage, where chemotherapy may improve survival and palliation of symptoms, but has reached a plateau in efficacy with a median survival of 8–10 months12, raising a pressing need of identifying novel therapeutic targets.
A commonly activated pathway in NSCLC is the PI3K/AKT/mTOR pathway8. A broad range of cancer-related functions has been associated with this signaling pathway, including cell proliferation21, glucose transport and catabolism11, cell adhesion20, apoptosis19, RAS signaling5 and oncogene transformation6, 15, 28. Several subunits of PI3Ks23, 27 and their up or downstream effectors, including GRB29, 24, have been found to be amplified or activated in human tumors. Indeed, mTOR phosphorylation increased with the malignant progression and blocking mTOR was sufficient to block lung tumor progression36.
We report in this study the role of the Proline-Rich Protein 14 (PRR14), an understudied gene located in 16p11.2 region, which is recurrently increased in copy number and overexpressed in lung cancer. Using integrated approach consisting of genomics, in vitro and in vivo molecular studies we estimated PRR14 oncogenic features in lung cancer. Our observations suggest PRR14 is an oncogene that plays an important role in lung cancers and it is a first report linking PRR14 with carcinogenesis.
Results
PRR14 is amplified and overexpressed in lung cancer
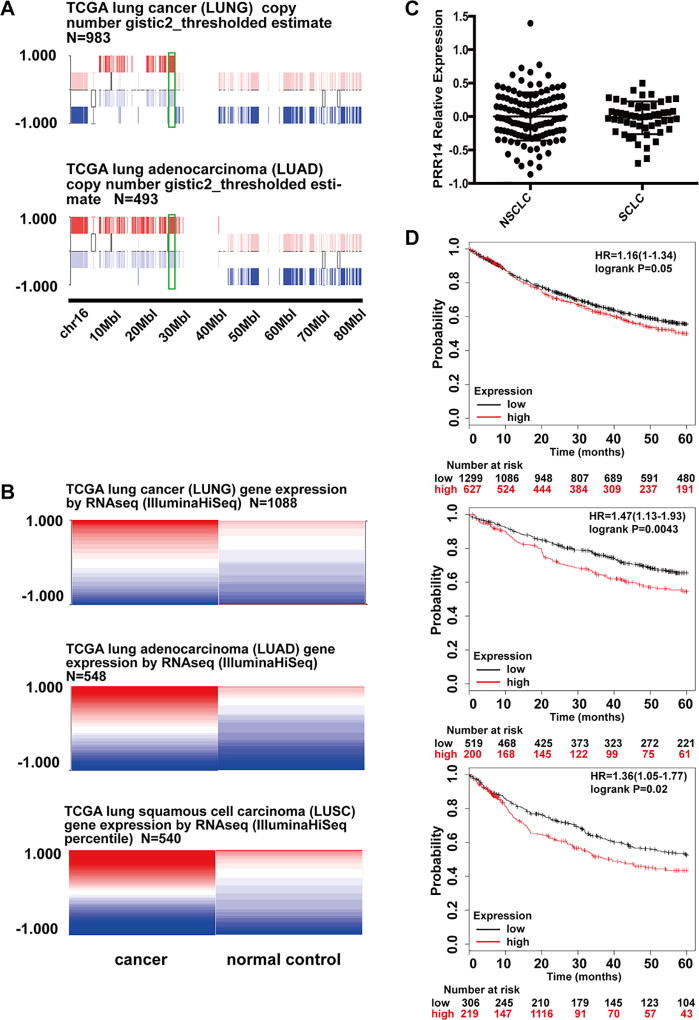
Studies using comparative genomic hybridization have revealed that the region 16p11.2, which contains PRR14, is recurrently increased in copy number in lung cancer16. Data mining from TCGA (The Cancer Genome Atlas) shows consistent copy number increase in lung cancer (Fig 1 A), which correlates with significantly enhanced PRR14 transcription in both adenocarcinoma and squamous cell carcinoma (Fig 1 B). Because of lack of SCLCs data, we analyzed the gene expression data from CCLE (Cancer Cell Line Encyclopedia) and compared the PRR14 expression level in different cell lines from either SCLCs or NSCLCs (Fig 1 C). There is no significant difference between cell lines from these 2 different types of lung cancer, suggesting that PRR14 expression is also increased in SCLCs. The high expression of PRR14 in lung cancer cases significantly associates with worse 5 year survival in all lung cancers (SCLC, LUSC and LUAD) (p=0.05) especially LUAD (p=0.0043) and LUSC (p=0.02) (Fig 1 D).
Fig 1. The amplification and overexpression of PRR14 in lung cancer.
(A) Gene copy number information was taken from UCSC Cancer Browser. (B) Gene expression level taken from UCSC Cancer Browser. The expression values are normalized by subtraction of the mean of PRR14 expression from each sample. The implemented in USCS Cancer Browser statistical analysis used student’s t test adjusting the false discovery rate using the Benjamini-Hochberg procedure identified p value as statistically different <0.05 with higher expression of PRR14 in cancer samples. (C) PRR14 expression comparison between cell lines of NSCLC AND SCLC. (D) Kaplan-Meier (KM) survival curves of lung cancer patients are stratified by their expression levels of PRR14. The analysis was done through online software Kaplan-Meier Plotter14.
Overexpression of PRR14 in cancer samples associates with increased gene expression
The PRR14 mRNA level was significantly increased in lung cancer samples compared to healthy controls and its expression levels varied across cancer samples. Therefore, we decided to explore what effect high PRR14 expression has within cancer samples deposited in TCGA database. Using Gitools 2.2.229 LUAD and LUSC datasets, we extracted the high-PRR14 (>0.5) and low-PRR14 expression samples (<−0.5) and performed differential expression analysis across deposited transcriptome (S1 A).
The comparative analysis of 42 high-PRR14 and 38 low-PRR14 expressing samples within TCGA-LUAD collection showed 714 differentially expressed (DE) transcripts (q<0.001) where 80% were overexpressed in high-PRR14 samples (Supplementary table 1). The analysis in TCGA-LUSC collection performed on 31 low-PRR14 and 32 high-PRR14 samples showed 480 transcripts where 71% was overexpressed in high-PRR14 samples (Supplementary table 2). In both LUAD and LUSC collections we observed increased expression of transcripts in high-PRR14 samples, and it was more profound in LUAD collection compared to LUSC dataset.
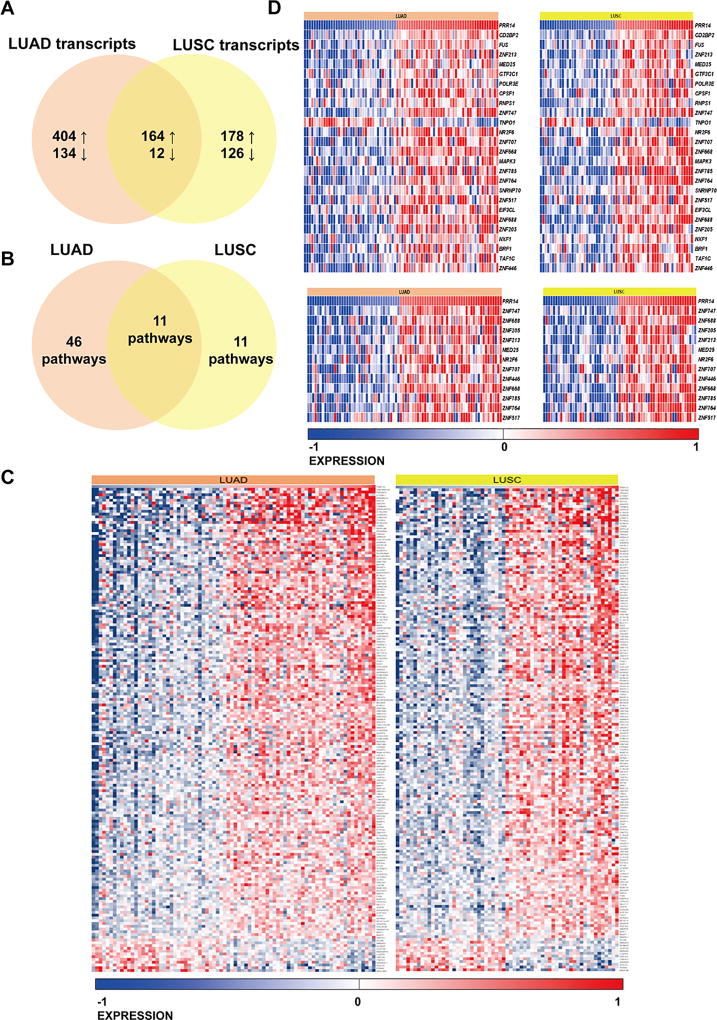
The high-PRR14 expression level associated with increased gene expression in both LUAD and LUSC types of cancer. We compared the DE transcripts in both collections discovering 176 common DE transcripts, out of which 164 were increased with high-PRR14 expression levels (Fig 2 A). Further, we performed over-representation analysis of common and unique DE transcripts in LUAD and LUSC collections with ConsensusPathDB17. Using 176 commonly DE genes in both cancers (Figure 2), 11 pathways were statistically significantly overrepresented (Fig 2 B, Supplementary Table 3). Two top hits pathways were Gene Expression (q< 0.0001) and Generic Transcription Pathway (q = 0.0024) (Figure 2 D). The overrepresentation analysis performed on unique DE transcripts separately in LUSC and LUAD collections showed 46 in LUAD and 11 in LUSC overrepresented pathways (Supplementary Table 3). Information derived from human TCGA data analysis suggests PRR14 as a potential lung cancer gene.
Fig 2.
Venn diagram presenting comparative analysis of differentially expressed genes between high- and low-PRR14 expressing LUAD and LUSC cases deposited in TCGA database. The cut-off of p<0.001 was considered significant to determinate differentially expressed transcripts (A). Venn diagram presenting pathways overrepresentation performed on DE transcripts unique for LUAD, common for LUAD and LUSC and unique for LUSC cases (B). (C) Heat map of common differentially expressed genes in LUAD and LUSC cases. (D) Heat maps presenting differentially expressed genes in two significantly enriched pathways Gene Expression and Generic Transcription Pathway (Reactome) in both LUAD and LUSC. The gene expression levels are normalized and log2 transformed reaching form −1 to +1 that corresponds to blue and red colors on heatmap.
PRR14 is positively associated with lung cancer cell proliferation
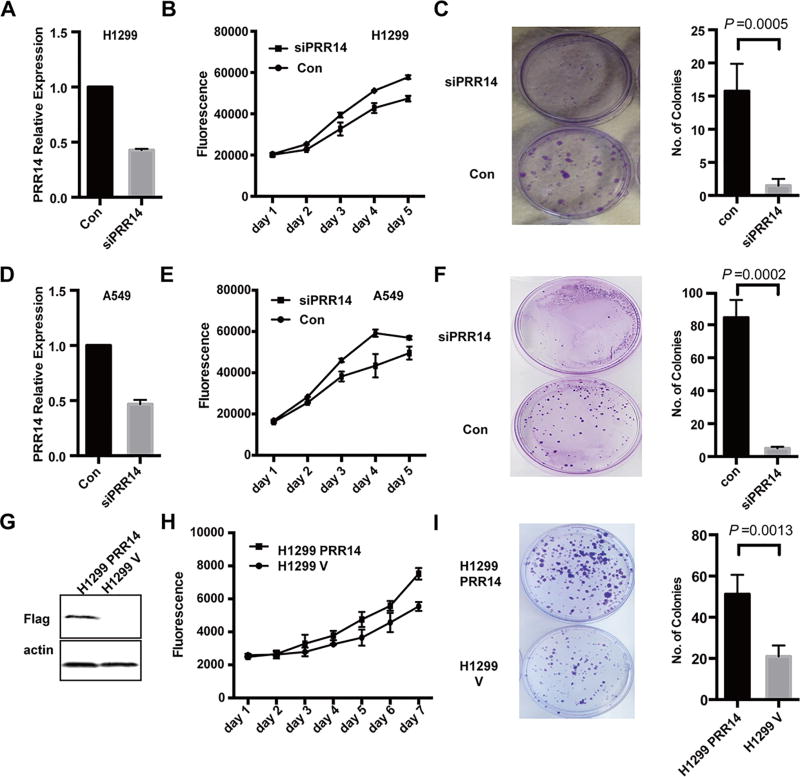
We next went on functionally characterizing PRR14. As PRR14 expression is increased in lung cancer, we asked whether PRR14 might affect lung cancer cell proliferation. To test this hypothesis, we employed 2 NSCLC cell lines, H1299 and A549, and performed knockdown experiments. A mixture of 3 different siRNAs targeting 3 sites of PRR14 was used to decrease its expression. qRT-PCR analysis showed that PRR14 mRNA level was reduced to <50% by the siRNAs (Fig 3 A, D). In both NSCLC cell lines decreased PRR14 expression resulted in slower proliferation rate (Fig 3 B, E) and significantly lower colony formation capacity (Fig 3 C, F).
Fig 3. PRR14 promotes lung cancer cells growth.
siRNA was transfected into lung cancer cell line: H1299 and A549 respectively. Compared with control sequence, PRR14 specific siRNA decreased its mRNA level to less than 50% (A, D). Growth curves following transfection were monitored by Alamar Blue Assay (B, E). Colony formation was performed at the same time (C, F). Colony numbers were calculated and statistically analyzed by two-tailed student's t-test. The numbers were presented as mean±S.D (n=3). The overexpression of PRR14 in H1299 cells was confirmed by immunostaining (G). Then growth curve (H) and colony formation assay (I) were performed. Colony numbers were calculated and statistically analyzed by two-tailed student's t-test. The numbers were presented as mean±S.D (n=3).
Consequently, we expected that the overexpression of PRR14 would increase the proliferation rate. We generated PRR14 overexpression stable cell line and its empty vector control cell line in H1299 through a retrovirus-mediated gene transfer. PRR14 overexpression was confirmed by immunoblot (Fig 3 G) and it correlated with higher proliferation rate (Fig 3 H) as well as significantly higher colony formation capacity (Fig 3 I). The ability of PRR14 to stimulate lung cancer cell proliferation was independently validated (S1 B–D). Altogether, these results indicate that PRR14 expression is critical for lung cancer cell proliferation in vitro
PRR14 activates Akt/mTOR signaling pathway
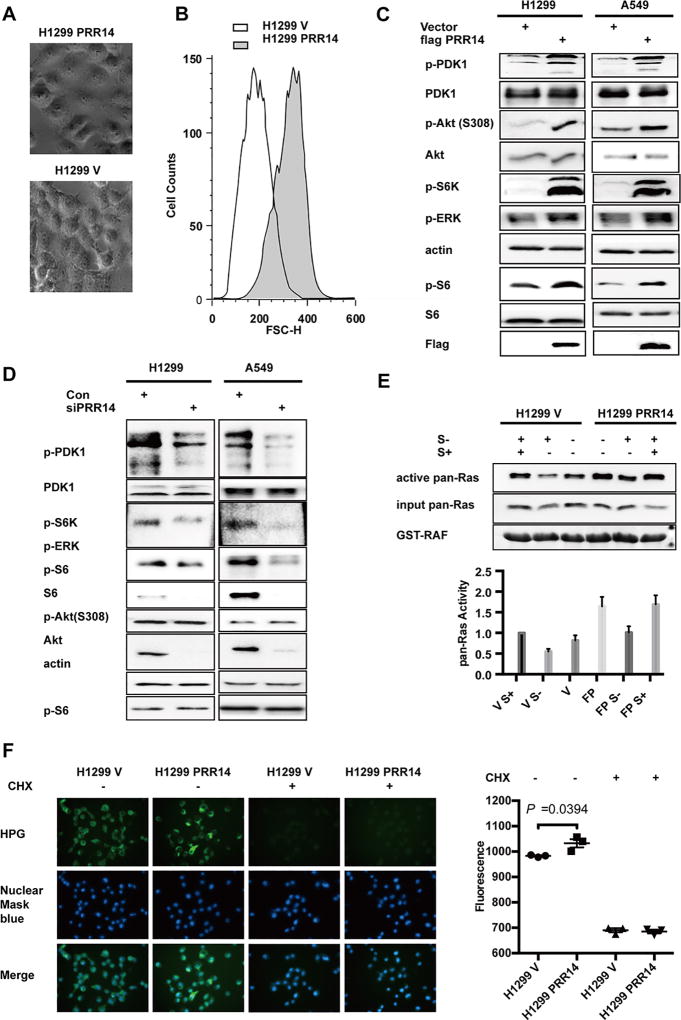
Interestingly, we noticed that the PRR14 overexpression cells are significantly larger in size than the control cells (Fig 4 A) which was confirmed with flow cytometry analysis (Fig 4 B). Since mTOR signal pathway is known to regulate mammalian cell size10, 38, the enlarged cell volume and global gene overexpression prompted us to probe a potential involvement of the mTOR pathway. The immunoblot result indicated that PRR14 overexpression was associated with an elevated phosphorylation in both H1299 and A549 cell lines of PDK1, AKT, S6K, ERK and S6, consistent with an increased activity of the mTOR pathway (Fig 4 C). To complement the result derived from PRR14-overexpressing cells, we depleted PRR14 expression using siRNA. In contrast to the elevated protein phosphorylation in PRR14 overexpressing cells, PRR14 underexpression resulted in marked decrease in phosphorylation of PDK1 and the downstream markers (Fig 4 D), which was consistent with decreased cell proliferation. Alternatively, the highly conserved RAS family of GTPases (HRAS, NRAS and KRAS) is known for its involvement of oncogenesis, the regulation of cell size and is one of the common upstream stimulators of the Akt/mTOR pathway4, 5, 38 To further pinpoint the exact effective target of PRR14 in PI3K/Akt/mTOR signaling pathway, we measured Ras activity in H1299 stable cell lines (Fig 4 E). As expected, serum starvation decreased Ras activity, which quickly recovered to original level upon serum replenishment. The PRR14-expressing cells displayed higher Ras activity under both serum-starved and stimulated conditions (Fig 4 E lanes 4–6), implicating a PRR14-mediated stimulation of Ras.
Fig 4. PRR14 increases PI3K/Akt/mTOR signaling pathway activity.
(A) Phase contrast images of H1299 PRR14 cell line and its control H1299 V; (B) FACS analysis of H1299 cell lines was performed using propidium Iodide with FACS Canto II. And representative distribution of cell size was shown. Several key components of the PI3K/Akt/mTOR signaling pathway were detected by immunostaining in both H1299 and A549 cell lines, when cells were transfected with either plasmid to increase PRR14 expression (C) or siRNA to reduce its expression (D). (E) Ras pull-down activation Assay was performed to compare total Ras activation between H1299 PRR14 and its control H1299 cell lines. The active Ras protein was normalized to total Ras protein and quantified. The numbers of relative abundance of active Ras protein are presented as the mean± S.D. (n = 3). (F) Newly synthesized proteins within 30 min were visualized and quantified in both H1299 PRR14 and its control cell lines in the presence or absence of CHX (50µg/ml) to compare the protein synthesis rate.
Further more, it was reported that PI3K/Akt/mTOR signaling pathway increased cell size is mediated through increased protein synthesis rate. Therefore, the protein synthesis rate was monitored and compared between PRR14 overexpressing and its control cell lines (Fig 4 F). As expected, protein translation inhibitor cycloheximide (CHX) dramatically decreased protein synthesis. Increased PRR14 expression significantly increased protein synthesis rate (P=0.0394), which is consistent with activated Gene Expression signal pathway in TCGA samples (Fig 2 D).
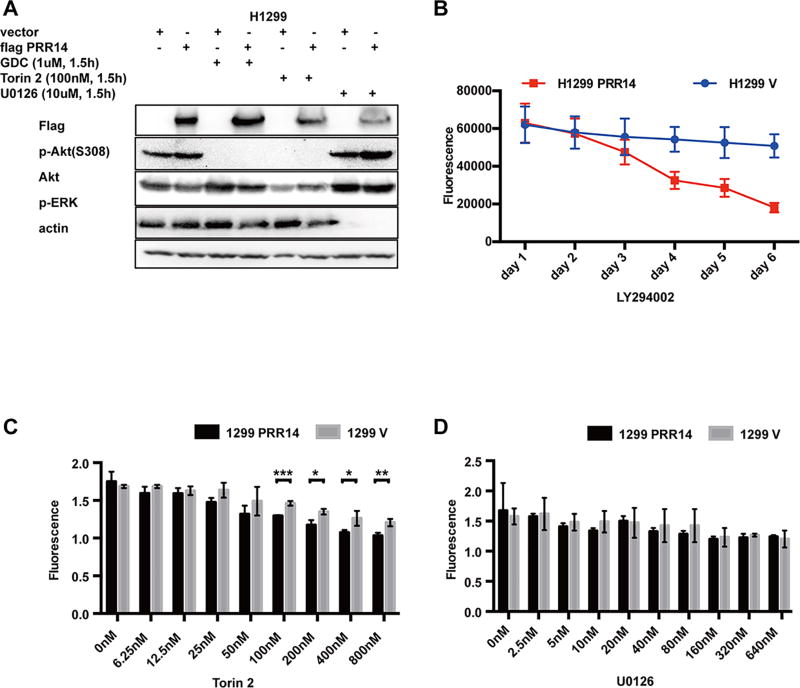
Additionally, PRR14-mediated increased activity of Akt/mTOR signaling pathway was completely blocked by either PI3K inhibitor GDC or mTOR inhibitor Torin 2; but, mitogen-activated protein kinases (MAPK) inhibitor U0126 showed no effect on the activation (Fig 5 A). The data together suggests that PRR14 targeted Ras or its upstream to stimulate the activity of PI3K/Akt/mTOR pathway.
Fig 5. Increased PRR14 makes cells more sensitive to PI3K/Akt/mTOR signaling pathway inhibitors.
(A) H1299 cells transfected with PRR14 or empty vector were treated with indicated inhibitors for 1.5h, and the PI3K/Akt/mTOR signaling pathway activity was detected through immunostaining p-AKT (S308). (B) Both H1299 PRR14 cell line and its control H1299 V were treated with 10uM mTOR inhibitor, LY294002, and analyzed for cell activity by Alamar Blue Assay at indicated time points. Alternatively, both cell lines were treated with mTOR inhibitor, Torin2 (C) or MAPK inhibitor, U0126 (D), at different doses for 48h. Then cell activity was measured by Alamar blue assay and statistically analyzed by two-tailed student's t-test Statistical analysis (*P<0.05, ** P<0.01, *** P<0.001).
We went on to test whether PI3K/Akt/m-TOR pathway was responsible for PRR14-induced cell proliferation. It has been shown that the PI3K inhibitor LY294002 alone rendered cells with highly activated Akt more sensitive to apoptosis and growth inhibition3. We assessed the contribution of PI3K/Akt/mTOR to PRR14-mediated cell proliferation by treating cells with 10uM LY294002. The PRR14 over-expressing cells were considerably more sensitive to LY294002-induced inhibition of cell proliferation (Fig 5 B). To complement with the treatment with LY294002, we treated cells with two additional inhibitors, Torin2 (Fig 5 C) and U0126 (Fig 5 D). Consistently, PRR14 showed a heightened sensitivity to PI3K/Akt/mTOR signal pathway inhibitors, LY294002 and Torin2. Of note, the cells did not show significantly different sensitivity to U0126 treatment (Fig 5 D), indicative of an effect specific to the PI3K/Akt/mTOR signaling pathway. Correspondingly, PRR14 overexpressing cell line with increased PI3K/Akt/mTOR signaling pathway activity showed more resistance to Bleomycin, a DNA damage inducing chemical (S2 A).
PRR14 stimulates PI3K/Akt/mTOR pathway via interacting with GRB2
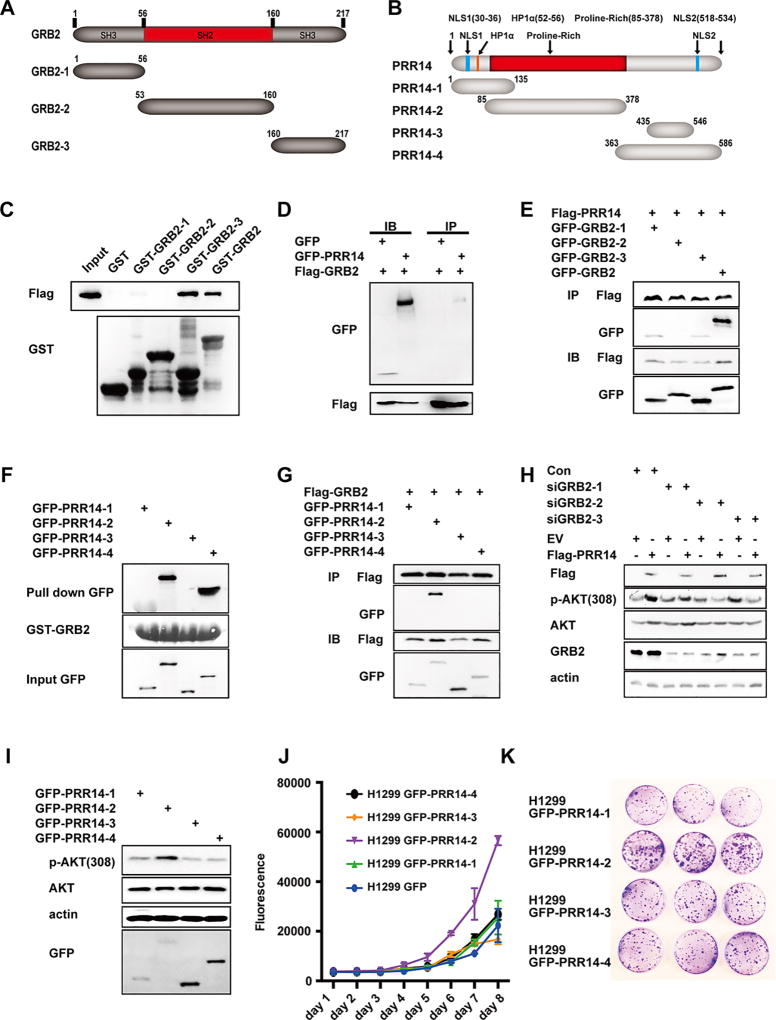
To explore how PRR14 activated Ras as well as PI3K, we focused on GRB2, a protein functioning upstream of both7, 22, 32. GRB2 is a docking protein containing an SH2 domain flanked by N- and C- terminal SH3 domains26. PRR14 on the other hand is a proline-rich protein. The interaction between SH3 domain and proline-rich domain is widely detected1, 18, 34, 40. We therefore tested if PRR14’s proline-rich sequence interacts with the SH3 domain of GRB2. We generated GST-fusion proteins of full-length, N-terminal SH3, middle part SH2, or C-terminal SH3 of GRB2 (Fig 6 A). The GST pull-down assay revealed a binding of PRR14 to full-length and both SH3 domains, albeit a significant weaker binding of the N-terminal SH3 domain (Fig 6 C). This data was confirmed by Co-IP (Fig 6 D and E). We also probed which domain of PRR14 binds to GRB2 by generating 4 fragments of PRR14 peptides as shown in Figure 4 B. The results from both GST pull-down (Fig 6 F) and co-IP experiments (Fig 6 G) revealed that PRR14-2 peptide, which contains the proline rich domain, binds specifically to GRB2. The binding of C-terminal of PRR14 (PRR14-4) to GRB2 was also detected in GST pull-down but not in Co-IP experiment, suggesting a non-specific binding in GST pull-down analysis37. Immunofluorescence staining showed the possibility of the 2 proteins interacting with each other spatially and temporally (S2 B).
Fig 6. PRR14 interacts with GRB2 to activate PI3K/Akt/mTOR signaling pathway.
Schematic diagram of the domain structures of GRB2 (A) and PRR14 (B) as well as the protein fragments used in this study. A direct interaction between PRR14 and full-length GRB2 or its fragments was shown by GST pull-down (C). Similarly, the direct interaction between GRB2 and PRR14 fragments was shown in (F). Co-immunoprecipitation of exogenous PRR14 and GRB2 as well as their fragments in HEK293 cells was shown in (D, E, G). (H) siRNA sequences targeting different sites of GRB2 were cotransfected with either PRR14 coding plasmid or its empty vector into H1299 cells and 48 hours later, cells were harvested for WB. (I) GFP-tagged PRR14 fragments 1–4 were transiently transfected into H1299 cells and cells were harvested for WB 24h later. (J) H1299 cell lines overexpressing GFP-tagged PRR14 fragments 1–4 were established and their proliferation rate and tumorigenesis were shown by growth curve (J) and colony formation assay (K).
We next asked whether the binding of PRR14 to GRB2 mediated its stimulation of Akt/mTOR activity. Indeed, in cells transfected with siRNA to decrease GRB2 expression, PRR14 mediated activation of Akt/mTOR signaling pathway was significantly compromised (Fig 6 H). The importance of this proline-rich domain-mediated, GRB2 binding was underscored by the observation that the proline-rich domain of PRR14 alone was sufficient to activate Akt/mTOR activity (Fig 6 I). The role for GRB2 in mediating the effect of PRR14 was further confirmed by examining cell proliferation. Remarkably, growth curve (Fig 6 J) and colony formation assay (Fig 6 K), showed that the proline-rich domain alone is adequate to increase cell proliferation and colony formation. The establishment of different cell lines was confirmed by immunofluorescence staining (S2 C). Altogether, the data supports a critical role of the proline-rich domain-mediated GRB2-binding in promoting PI3K/Akt/mTOR pathway activity and subsequent cell proliferation.
Characterization of PRR14 mutants derived from cancer patients
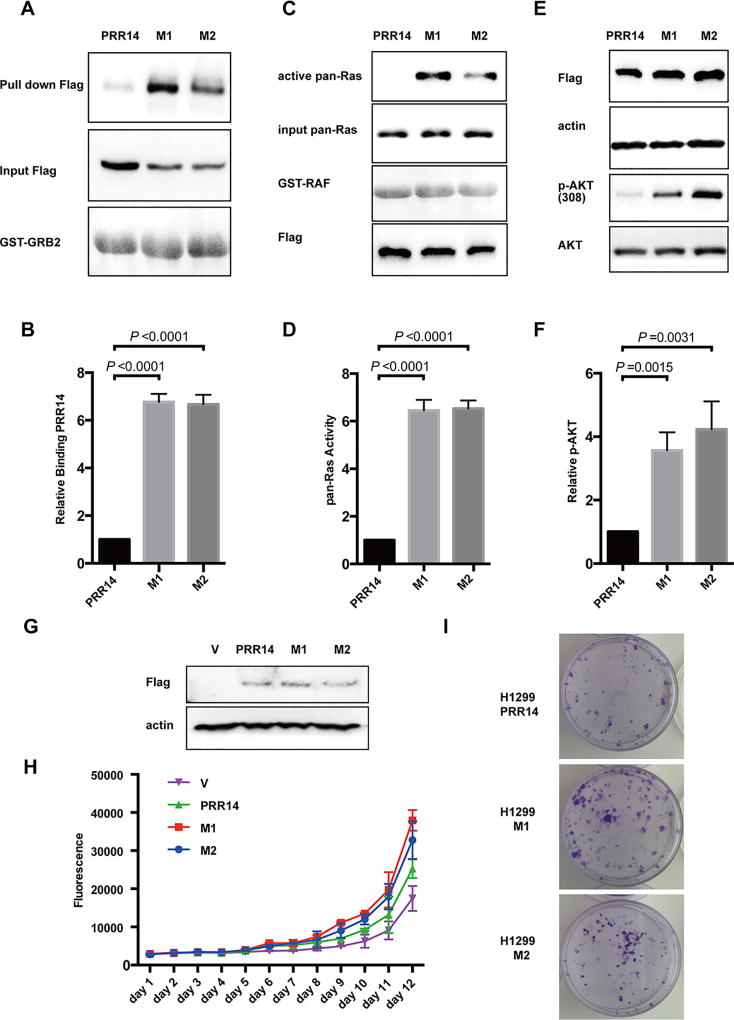
In addition to overexpression, TCGA database also revealed rare mutations of the PRR14 gene. We compared the mutants with wild-type PRR14 for their ability to bind to GRB2. Remarkably, 2 mutants (table 1) had a binding affinity considerably higher than wild-type PRR14 (Fig 7 A, B). As a consequence to this increased GRB2-binding, these 2 PRR14 mutants displayed a significantly greater potential of enhancing Ras (Fig 7 C, D) and AKT activity (Fig 7 E, F) than the wild-type counterpart. Growth curve and colony formation assays were performed to assess these 2 PRR14 mutants for their biological activity. At a comparable expression level (Fig 7 G), these 2 mutants induced a greater increase in cell proliferation (Fig 7 H) and colony formation (Fig 7 I) than wild-type PRR14, indicating an augmented ability of these 2 PRR14 mutants in promoting cancer formation.
Table 1.
TCGA-patients derived mutations in PRR14
| Name | Sample ID | CDS mutation | AA mutation |
domain |
|---|---|---|---|---|
| M1 | TCGA-EE-A3AB | c. 302c>g | p.S101C | PR |
| M2 | TCGA-91-6828 | c.1696g>a | p.E566K |
Fig 7. Mutants found in cancer patients increase interaction between PRR14 and GRB2.
(A) GST pull-down assay is employed to compare PRR14’s ability to interact with GRB2.The binding PRR14 protein was quantified and normalized to its total protein. The numbers of relative binding PRR14 protein are presented as the mean± S.D. (n = 3) and statistically analyzed by two-tailed student's t-test (B). (C) Ras pull-down activation Assay is used to compare PRR14’s ability to activate Ras in H1299 cells. The active Ras protein was quantified and normalized to total Ras protein. The numbers of relative abundance of active Ras protein are presented as the mean± S.D. (n = 3) and statistically analyzed by two-tailed student's t-test (D). (E) Mutants of PRR14 as well as their wild-type counterpart were transiently transfected into H1299 cells and cells were harvested for WB 24h later. The active Akt protein was quantified and normalized to its total protein, presented as the mean± S.D. (n = 3) and statistically analyzed by two-tailed student's t-test (F). (G) The overexpression of wild-type or mutant PRR14 is confirmed in H1299 PRR14, H1299 M1 and H1299 M2 cells lines by immunostaining. Growth curve (H) and colony formation assay (I) were also performed.
PRR14 promotes the xenograft tumor growth
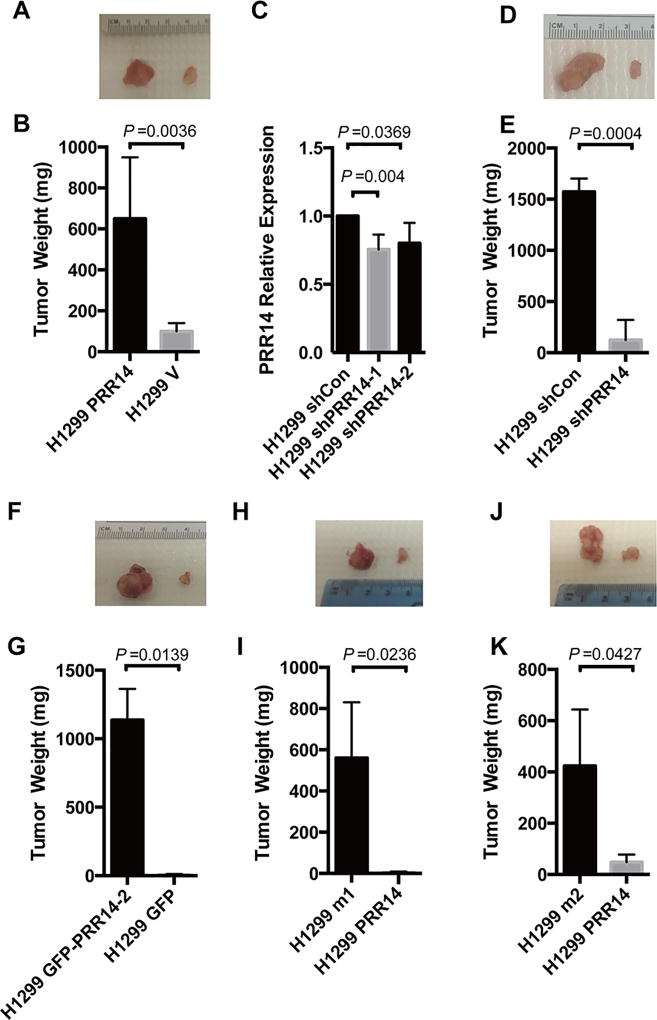
To confirm the effect of PRR14 on lung cancer cell tumorigenicity in vivo, we performed mouse xenograft experiments. H1299 cells stably expressing Flag-PRR14 or empty vector were subcutaneously implanted into either side of nude mice. Quantification of the tumor weights revealed a significantly larger tumor mass from PRR14- overexpressing cell line than the control cells (Fig 8 A, B). The tumor promoting activity of PRR14 was validated by a complementary knockdown experiment. The result showed that as little as 10% decrease of PRR14 expression by shRNA (Fig 8 C) reduced significantly tumor formation in mice (Fig 8 D, E). Consistent with the in vitro data, proline-rich domain of PRR14 itself is sufficient to promote tumor formation in mice (Fig 8 F, G).
Fig 8. PRR14 promotes lung cancer formation in vivo.
Representative image of tumors formed in either side of mice injected with H1299 PRR14 and its control H1299 V cells (A), H1299 shPRR14 and its control H1299 shCon cells (D), H1299 GFP-PRR14-2 and its control H1299 GFP cells (F), or either PRR14 mutants overexpressing cells (H1299 m1 or H1299 m2) and their wildtype overexpressing cells (H and J). Tumors were weight and presented as the mean± S.D (n=3). Two-tailed student's t test was employed to determine the significance of the difference between two groups (B, E, G, I, K). Totally 2 shRNAs targeting 2 different sites of PRR14 were employed to establish H1299 PRR14-deficient cell line and their efficiency were determined by RT-qPCR. PRR14 mRNA level were presented as the mean± S.D (n=3). Two-tailed student's t test was employed to determine the significance of the difference between two groups (C).
We also tested the tumorigenicity of the 2 PRR14 mutants that exhibited an enhanced ability of stimulating cell proliferation. H1299 M1 (Fig 8 H) or H1299 M2 (Fig 8 J) cells, together with wild-type PRR14 expressing H1299 cells, were subcutaneously injected into nude mice to induce tumor formation. Tumor xenografts were weighed and statistically analyzed (Fig 8 I and K). In agreement with the in vitro data, both PRR14 mutants showed greater tumorigenicity than wild-type PRR14.
Discussion
An elevated activity of the PI3K/AKT/mTOR pathway is a common driving force of lung tumorigenesis. Mutations, dysregulation and epigenetic alterations at various points in the cascade have been reported to contribute to the stimulation of this oncogenic pathway35. We demonstrate PRR14, which is upregulated in lung cancer, as a novel activator of the PI3K/AKT/mTOR pathway. Through in vitro and in vivo PRR14 gain-of-function and loss-of-function studies, we confirmed that PRR14 indeed acts as an oncogene in lung cancer.
PRR14 gene encodes a member of the proline-rich protein family, which contains one proline-rich region flanked by N- and C- terminal nuclear localization signal30. The proline-rich region is frequently involved in signaling events through binding with various domains, SH3 domain in particular1, 18, 34, 40. Here, we showed that PRR14 is able to directly bind with GRB2 and activate its downstream signal pathway (Fig 4 C–H). And the binding is mediated by the interaction between PRR14’s proline-rich region and both SH3 domains of GRB2. The importance of the proline-rich domain-mediated binding is highlighted by the findings that the PRR14 proline-rich peptide alone is sufficient to activate PI3K pathway and to promote cell proliferation in vitro and tumorigenesis in vivo. Of interest is that we also identified two PRR14 mutations from cancer patients, one in proline-rich region and the other at PRR14’s C-terminal. These two mutations enhance PRR14’s ability to interact with GRB2, and consequently further stimulate its activity. While it is unclear why the mutations at such distinct sites caused a similar increase in GRB2-binding, the data suggests that the conformation of PRR14 protein is very critical for its interaction with GRB2. Further structural studies will be necessary to address this possibility.
SH3 domains are noncatalytic peptide motifs found in many intracellular signaling proteins, including PI3K2, Ras GTPase (guanosine triphosphatase)-activating protein39, and Src-like tyrosine kinases31 and GRB213, 25. Together with SH2, it mediates protein-protein interaction and regulates signal transduction. GRB2 protein is composed of a SH2 domain flanked by N- and C- terminal SH3 domains. Co-immunoprecipitation experiment showed that both SH3 domains bind to PRR14 with a comparable affinity (Fig 4 E), whereas GST pull-down assay indicated that C-terminal SH3 domain has a much stronger binding affinity (Fig 4 C). The data suggests a potential involvement of post-translational modifications in regulating the interaction between the SH3 domains of GRB2 and PRR14. Nevertheless, our data indicates that through its proline-rich domain-mediated binding to GRB2, PRR14 stimulates the activity of PI3K/AKT/mTOR pathway promoting lung carcinogenesis.
Materials and Methods
Cell Culture, transfection and stable cell lines establishment
H1299, A549 and HEK293T cells were maintained using standard conditions. DNA transfection was carried out using Lipofectamine 2000 (Invitrogen). siRNA (SASI_Hs01_00196781, SASI_Hs01_00196782, SASI_Hs01_00196783, Sigma) transfection was mediated by Lipofectamine® RNAiMAX Transfection Reagent (Invitrogen).
Retroviral production and infection were done following the protocol from Clontech. Cells were allowed to recover for 24 hours prior to selection with hygromycin for 1 week. Cells transduced with the empty vector ware also established and used as control cell lines.
Lentivirus production and infection were done following the protocol from Addgene. After 24 hours recovery, cells were treated with puromycin for 1 week.
Plasmids
cDNA encoding full length human PRR14 and GRB2 was purchased from DF/HCC DNA Resource Core (Boston, MA) and cloned into pQCXIH (Clontech) and pEGFP-N1(Clontech) by Gateway Cloning system (Invitrogen). For deletion mutation, pEGFP-N1(Clontech) and pGEX-4T-2(Sigma) were used and the full-length plasmid was used as template for PCR.
Site-directed mutation was introduced by Q5® Site-Directed Mutagenesis Kit (NEB). All plasmids were sequenced to confirm the insertion. For Ras pull-down activation assay, plasmid Raf-1 GST RBD 1-149(#13338, addgene) was purchased.
RNA preparation, reverse transcription, and qPCR
Total RNA was extracted using the standard TRIzol method (Life Technologies) and used for the first-strand cDNA synthesis by iScript™ cDNA Synthesis Kit (Bio-Rad). Real-time PCR was performed with SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma) using the StepOnePlus™ Real-Time PCR System (Bio-Rad). All reactions were run in duplicate. After vortexing, 10 µl aliquots of the mixture were pipetted into each well of a 96-well thin-wall PCR plate (Bio-Rad). PCRs consisted of a denaturing cycle at 94°C for 2min, followed by 40 cycles of 15 sec at 94°C and 1min at 60°C. Relative mRNA amounts of target genes were calculated after normalization to an endogenous reference gene (18s) and relative to the negative control with the arithmetic formula 2−ΔΔCt.
Flow cytometry analysis and protein synthesis assay
Cells were trypsinized, harvested by centrifugation (500g,5min), washed in 4°C PBS once and gently resuspended in 1ml hypotonic fluorochrome solution (PI 50 µg/ml in 0.1% sodium citrate, 10mM NaCl, 0.3% NP40 plus 25mg/ml RNase) in polypropylene tubes, followed by incubation in 37°C for 30min. The tubes were placed at 4°C in darkness for flow cytometric analysis.
For protein synthesis assay, cells were treated with or without 50µg/ml CHX (#2112, Cell Signaling) for 30 min and the Click-iT HPG Alexa Fluor 488 protein synthesis assay kit (C10428, ThermoFisher) was employed for the visualization of newly synthesized proteins according to the manufacture’s instructions.
Immunohistochemistry, Western blotting, GST pull-down assay, immunoprecipitation and Ras pull-down activation assay
For western blot, cells were lysed in NP40 supplemented with phosphatase and protease inhibitors. Western analysis was performed using standard procedures. For immunohistochemistry, cells were fixed in paraformaldehyde, and blocked with 1% BSA/ 1% Triton X-100 / 1 × PBS. The GST pull-down and Co-IP was done in HEK293T cells. For Ras pull-down activation assay, either H1299 cell lines or transfected H1299 cells were used 48h after transfection, cells were harvested and lysed in RIPA buffer and incubated with purified GST beads (#20211,Life Technology) or flag beads (Anti-FLAG® M2 Magnetic Beads, Sigma), following the protocol.
Animal experiments
All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Harvard School of Public Health (Boston, MA). H1299 cells (5×105) suspended in 200ul sterile PBS were subcutaneously injected into both flanks of female nude mice (#007850, JAX). Palpable tumors were established in 15–30 days after injection.
Statistical analysis
The data are presented as mean±S.D. (standard deviation). Two-tailed student's t test was used to determine the significance of the difference between two groups. If not indicated otherwise, the criterion for significance was set at P<0.05 (Prism 6, GraphPad Software).
Differential expression analysis and cohorts
Gene copy number information was taken from USCS Cancer Genome Browser (https://genome-cancer.ucsc.edu/) for:
TCGA Lung cancer (LUNG) gistic2_tresholded (983 cases)
TCGA Lung adenocarcinoma (LUAD) gistic2_thresholded (493 cases)
The PRR14 expression level in cancer and control samples was generated with USCS Cancer Genome Browser (https://genome-cancer.ucsc.edu/) using:
TCGA Lung cancer (LUNG) gene expression (IlluminaHiSeq) (1088 of cases)
TCGA Lung adenocarcinoma (LUAD) gene expression (IlluminaHiSeq) (548 cases)
TCGA Lung squamous cell carcinoma (LUSC) gene expression (IlluminaHiSeq) (540 cases)
Using the expression data from blood derived normal, solid tissue normal, buccal cell normal, EVB immortalized normal, bone marrow normal cells.
The overall survival with follow up threshold of 5 year was performed using KMplotter. The expression level cut-off was auto-selected for 218714_at probe.14
The differential expression analysis within LUAD and LUSC samples was performed on TCGA data using Gitools (http://gitools.org/home)29. The workflow of an analysis is presented in Supplementary Figure 2
The molecular concept-based analysis of over-represented pathways was performed with ConsensusPathDB17 defining the significance level at 0.001 and minimum of 4 genes represented in the pathway.
Supplementary Material
Acknowledgments
We are grateful to current and former members of the Yuan lab for support, advice and helpful discussions. We thank Dr. Brendan Manning for reagents.
Funding
This work was supported in part by the Morningside Foundation, the Zhu Fund and grants from the National Cancer Institute at the National Institutes of Health (R01CA085679, RO1CA167814 and RO1CA125144).
Abbreviations List
- Co-IP
Co-Immunoprecipitation
- DE
Differentially Expressed
- GRB2
Growth Factor Receptor-Bound Protein 2
- LUAD
Lung Adenocarcinoma
- LUSC
Lung Squamous Cell Carcinoma
- MAPK
Mitogen-Activated Protein Kinases
- mTor
mammalian Target of rapamycin
- NSCLC
Non-Small Cell Lung Cancer
- PI3K
Phosphatidylinositol 3-Kinase
- PRR14
Proline-Rich Protein 14
- SCLC
Small Cell Lung Cancer
- SH2
Src Homology 2
- SH3
Src Homology 3
- TCGA
The Cancer Genome Atlas
Footnotes
Conflict of interest
All authors are aware of the content and declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Ball LJ, Kuhne R, Schneider-Mergener J, Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angewandte Chemie. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 2.Booker GW, Gout I, Downing AK, Driscoll PC, Boyd J, Waterfield MD, et al. Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase. Cell. 1993;73:813–822. doi: 10.1016/0092-8674(93)90259-s. [DOI] [PubMed] [Google Scholar]
- 3.Brognard J, Clark AS, Ni Y, Dennis PA. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer research. 2001;61:3986–3997. [PubMed] [Google Scholar]
- 4.Castellano E, Downward J. Role of RAS in the regulation of PI 3-kinase. Current topics in microbiology and immunology. 2010;346:143–169. doi: 10.1007/82_2010_56. [DOI] [PubMed] [Google Scholar]
- 5.Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes & cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, Tsichlis PN, et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 7.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, et al. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H, Shcherba M, Pendurti G, Liang Y, Piperdi B, Perez-Soler R. Targeting the PI3K/AKT/mTOR pathway: potential for lung cancer treatment. Lung cancer management. 2014;3:67–75. doi: 10.2217/lmt.13.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly RJ, Binder MD, Sutherland RL. Overexpression of the Grb2 gene in human breast cancer cell lines. Oncogene. 1994;9:2723–2727. [PubMed] [Google Scholar]
- 10.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes & development. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frevert EU, Kahn BB. Differential effects of constitutively active phosphatidylinositol 3-kinase on glucose transport, glycogen synthase activity, and DNA synthesis in 3T3-L1 adipocytes. Molecular and cellular biology. 1997;17:190–198. doi: 10.1128/mcb.17.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Group NM-AC. Chemotherapy in addition to supportive care improves survival in advanced non-small-cell lung cancer: a systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:4617–4625. doi: 10.1200/JCO.2008.17.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guruprasad L, Dhanaraj V, Timm D, Blundell TL, Gout I, Waterfield MD. The crystal structure of the N-terminal SH3 domain of Grb2. Journal of molecular biology. 1995;248:856–866. doi: 10.1006/jmbi.1995.0266. [DOI] [PubMed] [Google Scholar]
- 14.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez C, Jones DR, Rodriguez-Viciana P, Gonzalez-Garcia A, Leonardo E, Wennstrom S, et al. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. The EMBO journal. 1998;17:743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P, et al. Genomic aberrations in lung adenocarcinoma in never smokers. PloS one. 2010;5:e15145. doi: 10.1371/journal.pone.0015145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic acids research. 2013;41:D793–800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14:231–241. [PubMed] [Google Scholar]
- 19.Kennedy SG, Wagner AJ, Conzen SD, Jordan J, Bellacosa A, Tsichlis PN, et al. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes & development. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 20.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. The EMBO journal. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klippel A, Escobedo MA, Wachowicz MS, Apell G, Brown TW, Giedlin MA, et al. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Molecular and cellular biology. 1998;18:5699–5711. doi: 10.1128/mcb.18.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, et al. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 23.Lin X, Bohle AS, Dohrmann P, Leuschner I, Schulz A, Kremer B, et al. Overexpression of phosphatidylinositol 3-kinase in human lung cancer. Langenbeck's archives of surgery / Deutsche Gesellschaft fur Chirurgie. 2001;386:293–301. doi: 10.1007/s004230100203. [DOI] [PubMed] [Google Scholar]
- 24.Liu AX, Testa JR, Hamilton TC, Jove R, Nicosia SV, Cheng JQ. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer research. 1998;58:2973–2977. [PubMed] [Google Scholar]
- 25.Lysek DA, Wuthrich K. Prion protein interaction with the C-terminal SH3 domain of Grb2 studied using NMR and optical spectroscopy. Biochemistry. 2004;43:10393–10399. doi: 10.1021/bi0494828. [DOI] [PubMed] [Google Scholar]
- 26.Maignan S, Guilloteau JP, Fromage N, Arnoux B, Becquart J, Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- 27.Massion PP, Taflan PM, Shyr Y, Rahman SM, Yildiz P, Shakthour B, et al. Early involvement of the phosphatidylinositol 3-kinase/Akt pathway in lung cancer progression. American journal of respiratory and critical care medicine. 2004;170:1088–1094. doi: 10.1164/rccm.200404-487OC. [DOI] [PubMed] [Google Scholar]
- 28.Meili R, Cron P, Hemmings BA, Ballmer-Hofer K. Protein kinase B/Akt is activated by polyomavirus middle-T antigen via a phosphatidylinositol 3-kinase-dependent mechanism. Oncogene. 1998;16:903–907. doi: 10.1038/sj.onc.1201605. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PloS one. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, Katz RA. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell reports. 2013;5:292–301. doi: 10.1016/j.celrep.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roskoski R., Jr Src protein-tyrosine kinase structure and regulation. Biochemical and biophysical research communications. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 32.Shen X, Xi G, Radhakrishnan Y, Clemmons DR. PDK1 recruitment to the SHPS-1 signaling complex enhances insulin-like growth factor-i-stimulated AKT activation and vascular smooth muscle cell survival. The Journal of biological chemistry. 2010;285:29416–29424. doi: 10.1074/jbc.M110.155325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 34.Sparks AB, Rider JE, Hoffman NG, Fowlkes DM, Quillam LA, Kay BK. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spoerke JM, O'Brien C, Huw L, Koeppen H, Fridlyand J, Brachmann RK, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:6771–6783. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- 36.Wislez M, Spencer ML, Izzo JG, Juroske DM, Balhara K, Cody DD, et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer research. 2005;65:3226–3235. doi: 10.1158/0008-5472.CAN-04-4420. [DOI] [PubMed] [Google Scholar]
- 37.Wissmueller S, Font J, Liew CW, Cram E, Schroeder T, Turner J, et al. Protein-protein interactions: analysis of a false positive GST pulldown result. Proteins. 2011;79:2365–2371. doi: 10.1002/prot.23068. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Xu T. Molecular mechanism of size control in development and human diseases. Cell research. 2011;21:715–729. doi: 10.1038/cr.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang YS, Garbay C, Duchesne M, Cornille F, Jullian N, Fromage N, et al. Solution structure of GAP SH3 domain by 1H NMR and spatial arrangement of essential Ras signaling-involved sequence. The EMBO journal. 1994;13:1270–1279. doi: 10.1002/j.1460-2075.1994.tb06379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.