Key Points
NGS-based prognostic panels may identify individuals at risk for HHMs despite not being designed for this purpose.
Variant allele frequency >0.4 and gene of interest may be predictive of germ line origin.
Abstract
Next-generation sequencing (NGS)–based targeted gene capture panels are used to profile hematopoietic malignancies to guide prognostication and treatment decisions. Because these panels include genes associated with hereditary hematopoietic malignancies (HHMs), we hypothesized that these panels could identify pathogenic germ line variants in malignant cells, thereby identifying patients at risk for HHMs. In total, pathogenic or likely pathogenic variants in ANKRD26, CEBPA, DDX41, ETV6, GATA2, RUNX1, or TP53 were identified in 74 (21%) of 360 patients. Germ line tissue was available for 24 patients with 25 pathogenic or likely pathogenic variants with variant allele frequencies >0.4. Six (24%) of these 25 variants were of germ line origin. Three DDX41 variants, 2 GATA2 variants, and a TP53 variant previously implicated in Li-Fraumeni syndrome were of germ line origin. No likely pathogenic/pathogenic germ line variants possessed variant allele frequencies <0.4. This study demonstrates that NGS-based prognostic panels may identify individuals at risk for HHMs despite not being designed for this purpose. Furthermore, variants known to cause Li-Fraumeni syndrome as well as known pathogenic variants in genes such as DDX41 and GATA2 are especially likely to be of germ line origin. Thus, tumor-based panels may augment, but should not replace, comprehensive germ line–based testing and counseling.
Visual Abstract
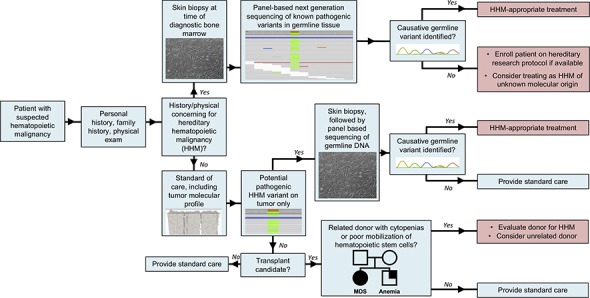
Introduction
Hereditary hematopoietic malignancies (HHMs) are caused by germ line mutations that increase an individual's risk for hematopoietic malignancies and are classically diagnosed based on personal or family history or when a related stem cell donor presents with cytopenia or mobilizes hematopoietic precursors poorly.1-3 HHMs may be misdiagnosed if physicians lack awareness of hereditary cancer syndromes or the significance of variants identified during molecular profiling of tumors. Therefore, it is important to identify additional means of diagnosing HHMs.
Tumor-based next-generation sequencing (NGS) panels identify acquired mutations that facilitate diagnosis, guide prognostication, and influence treatment decisions.4,5 These panels may identify variants in genes associated with HHMs,4,6 but tumor-only sequencing cannot differentiate between acquired and germ line variants. We hypothesized that we could identify some patients with HHMs using tumor-only NGS panels, and we determined the frequency at which this occurs.
Methods
We reviewed NGS panels from April 2014 through July 2017 for patients with hematopoietic malignancies on institutional review board–approved research protocols at our institution. We identified nonsynonymous variants in HHM-associated genes that were included on NGS panels at our institution: ANKRD26, CEBPA, DDX41, ETV6, GATA2, RUNX1, SRP72, TERT, and TP53. The NGS panels of our institution evolved in size over time (supplemental Table 1) and provided median sequencing depths of 785X (CEBPA, ETV6, GATA2, RUNX1, TP53) and 360X (ANKRD26, DDX41).5 We classified pathogenicity using American College of Medical Genetics and Genomics guidelines.7 We sequenced each variant in germ line tissue, including cultured skin fibroblasts, cultured mesenchymal stromal cells, or nonhematopoietic tissue biopsies from patients in complete remission.
Results
In total, 52 pathogenic or likely pathogenic (hereafter referred to as pathogenic) variants in HHM-associated genes were identified in tumors from 44 patients with germ line tissue available. Among these 52 variants, 6 (12%) were germ line (Figure 1). We consider a VAF threshold of 0.4 to be potentially indicative of a germ line variant, so we then examined variants on the basis of VAF. Among pathogenic variants with a VAF >0.4, 24% (6 of 25) were of germ line origin. No pathogenic germ line variants possessed VAFs <0.4 (Figure 1). All pathogenic germ line variants had Exome Aggregation Consortium frequencies <0.01% (Table 1).8 The diagnostic yield increased with the implementation of more comprehensive molecular panels (supplemental Table 1).
Figure 1.
Schematic summarizing the patient population, tissue availability, and general sequencing results for pathogenic/likely pathogenic variants only. VAF, variant allele frequency.
Table 1.
Patients for whom NGS-based tumor panels identified germ line variants in genes associated with HHMs
| Variant no. | Patient no. | Gene | Variant | VAF | Acquired/germ line | Diagnosis | Interpretation | Family history* | Tissue type | rsID | ExAC frequency |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 3 | ANKRD26 | p.Ser690Pro | 0.50 | Germ line | AML, prostate cancer | Likely benign | Prostate cancer, cervical cancer, ALL | Cultured skin fibroblasts | rs73596345 | 3.5 × 10−3 |
| 4 | 4 | ANKRD26 | p.Asp265Asn | 0.48 | Germ line | Breast cancer, t-AML | Likely benign | Brain, breast, gastric cancers | Cultured skin fibroblasts | rs61730101 | 5.0 × 10−3 |
| 10 | 9 | CEBPA | p.Pro192_His193insTyrPro | 0.41 | Germ line | AML | VUS | Lung cancer | Cultured skin fibroblasts | NA | NA |
| 10 | 9 | CEBPA | p.Pro192_His193insTyrPro | 0.41 | Germ line | AML | VUS | Lung cancer | Cultured skin fibroblasts | NA | NA |
| 12 | 11 | CEBPA | p.His195_Pro196dup | 0.33 | Germ line | Systemic mastocytosis, MDS | Likely benign | Hairy cell leukemia | Cultured skin fibroblasts | rs762459325 | 4.5 × 10−2 |
| 14 | 4 | CEBPA | p.His195_Pro196dup | 0.32 | Germ line | Breast cancer, t-AML | Likely benign | Brain, breast, gastric cancers | Cultured skin fibroblasts | rs762459325 | 4.5 × 10−2 |
| 15 | 13 | CEBPA | p.His195_Pro196dup | 0.31 | Germ line | Multiple myeloma, t-AML | Likely benign | Breast, colon cancers | Cultured skin fibroblasts | rs762459325 | 4.5 × 10−2 |
| 16 | 14 | CEBPA | p.His195_Pro196dup | 0.29 | Germ line | AML | Likely benign | MDS | Cultured skin fibroblasts | rs762459325 | 4.5 × 10−2 |
| 17 | 15 | CEBPA | p.His195_Pro196dup | 0.25 | Germ line | Breast cancer, t-AML | Likely benign | Esophageal cancer | Cultured skin fibroblasts | rs762459325 | 4.5 × 10−2 |
| 18 | 16 | DDX41 | p.Pro78GInfs*3 | 0.51 | Germ line | CML, AML | Pathogenic | NA | Cultured skin fibroblasts | NA | NA |
| 19 | 17 | DDX41 | p.Asp140Glyfs*2 | 0.43 | Germ line | t-MDS | Pathogenic‡ | NA | Cultured skin fibroblasts | rs762890562 | 8.0 × 10−5 |
| 20 | 18 | DDX41 | p.Asp140Glyfs*2 | 0.43 | Germ line | AML | Pathogenic‡ | MDS | Cultured skin fibroblasts | rs762890562 | 8.0 × 10−5 |
| 21 | 19 | ETV6 | p.Arg127Gln | 0.50 | Germ line | CML | VUS | NA | Cultured skin fibroblasts | rs140357643 | 5.3 × 10−4 |
| 28 | 25 | GATA2 | Exon 4 deletion | 0.50 | Germ line | MDS | Likely pathogenic | MDS | Cultured skin fibroblasts | NA | NA |
| 29 | 4 | GATA2 | 5′ UTR variant: c.-5C>G | 0.51 | Germ line | Breast cancer, t-AML | Likely benign | Brain, breast, gastric cancers | Cultured skin fibroblasts | rs374415484 | 8.7 × 10−4 |
| 30 | 26 | GATA2 | p.Pro161Ala | 0.51 | Germ line | Merkel cell carcinoma, basal cell carcinoma, t-AML | Likely benign | Pancreatic, uterine cancers | Cultured skin fibroblasts | rs34799090 | 9.6 × 10−3 |
| 31 | 27 | GATA2 | p.Pro161Ala | 0.50 | Germ line | AML | Likely benign | Bladder, colon, prostate cancers; NHL | Cultured skin fibroblasts | rs34799090 | 9.6 × 10−3 |
| 32 | 28 | GATA2 | p.Gly237Asp | 0.49 | Germ line | Hypereosinophilia | Likely benign | Prostate cancer | Cultured skin fibroblasts | rs191501191 | 2.3 × 10−3 |
| 33 | 29 | GATA2 | p.Pro161Ala | 0.49 | Germ line | AML | Likely benign | Renal cancer | Cultured skin fibroblasts | rs34799090 | 9.6 × 10−3 |
| 34 | 30 | GATA2 | p.Ala286Val | 0.48 | Germ line | CMML, AML | Likely pathogenic | AML, colorectal cancer, MDS | Cultured skin fibroblasts | NA | NA |
| 35 | 28 | GATA2 | p.Pro41Ala | 0.48 | Germ line | Hypereosinophilia | Likely benign | Prostate cancer | Cultured skin fibroblasts | rs143590990 | 8.1 × 10−4 |
| 42 | 9 | RUNX1 | p.Met52Lys | 0.53 | Germ line | AML | VUS | Lung cancer | Cultured skin fibroblasts | rs200431130 | 2.1 × 10−4 |
| 43 | 13 | RUNX1 | p.Gly217Arg | 0.53 | Germ line | Multiple myeloma, t-AML | VUS | Breast and colon cancers | Cultured skin fibroblasts | rs749004431 | 2.0 × 10−5 |
| 45 | 36 | RUNX1 | p.Leu56Ser | 0.51 | Germ line | AML | Likely benign | Basal cell carcinoma, AML | Cultured skin fibroblasts | rs111527738 | 1.6 × 10−2 |
| 46 | 37 | RUNX1 | p.Leu56Ser | 0.48 | Germ line | Endometrial cancer, AML | Likely benign | Breast and prostate cancers; CLL, NHL | Cultured skin fibroblasts | rs111527738 | 1.6 × 10−2 |
| 74 | 56 | TP53 | p.Arg290His | 0.48 | Germ line | AML | Likely pathogenic†‡ | Breast cancer, lymphoma | Cultured skin fibroblasts | rs55819519 | NA |
Variants may be pathogenic, likely pathogenic, a VUS, likely benign, or benign in nature.
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; CMML, chronic myelomonocytic leukemia; ExAC, Exome Aggregation Consortium; MDS, myelodysplastic syndrome; NA, not applicable; NHL, non-Hodgkin lymphoma; rsID, reference single-nucleotide polymorphism identification number; t, therapy related; VUS, variant of undetermined significance.
Family history for first-degree relatives. Accessions: NM_004364.8 (CEBPA), NM_016222.3 (DDX41), NM_001987.4 (ETV6), NM_032638.4 (GATA2), NM_001754.4 (RUNX1), and NM_000546.4 (TP53).
Known Li-Fraumeni variant.
Known pathogenic hereditary variant.
Five (83%) of 6 patients with pathogenic germ line variants had a family history of hematopoietic cancers but not solid tumors. All patients were enrolled on research protocols investigating the hereditability of hematopoietic diseases, a selection bias that potentially enriched our study for HHMs. It is unknown if the proportions of HHMs in this series are similar to those in broader patient populations. No pathogenic germ line variants were detected in ANKRD26, CEBPA, ETV6, RUNX1, SRP72, or TERT.
DDX41 had the highest diagnostic yield for pathogenic germ line variants (100%; n = 3; supplemental Figure 1C). Two individuals with germ line DDX41 variants acquired second-hit DDX41 mutations (p.Glu345Asp or p.Arg525His).9
Two pathogenic germ line GATA2 variants were identified (Table 1). The first was a heterozygous exon 4 deletion in a 23-year-old woman with MDS with monosomy 7. Her brother was diagnosed with MDS with monosomy 7 at age 23 years. A second GATA2 mutation (p.Ala286Val) was identified in a 41-year-old woman with chronic myelomonocytic leukemia whose sister was diagnosed with MDS at age 45 years.10
TP53 variants (n = 33; all VAFs) were the most common acquired variants but had the lowest germ line diagnostic yield; 1 (7%) of 15 variants with a VAF >0.4 was germ line. However, this yield doubled to 14% when TP53 variants previously found in patients with Li-Fraumeni syndrome and VAFs >0.4 were considered (n = 7; supplemental Figure 1).
Our experience with patient 17 (Table 1) demonstrates the utility of NGS-based tumor-only sequencing for diagnosing HHMs unexpectedly. At age 70 years, she was diagnosed with stage IIA gastric cancer, with normal blood counts, and received neoadjuvant cisplatin plus 5-fluorouracil, gastrectomy, and adjuvant 5- fluorouracil, oxaliplatin, and leucovorin. She developed a therapy-related myeloid neoplasm at age 72 years. She did not have a family history of hematopoietic malignancies. Sequencing of the therapy-related myeloid neoplasm demonstrated DDX41 p.Asp140Glyfs*2, a known pathogenic germ line frameshift that has not been observed as a somatic mutation.11,12 Sequencing of DNA from cultured skin fibroblasts confirmed the germ line origin of this variant. Individuals with pathogenic germ line DDX41 mutations develop hematopoietic malignancies at an age similar to the age at which individuals develop sporadic malignancies, so tumor-only sequencing may offer the initial evidence of an HHM for individuals with germ line DDX41 mutations.9,11-13
Discussion
The retrospective nature of this study demonstrates the limitations of performing tumor-only sequencing without matched normal controls. Tumor-only sequencing cannot identify hereditary variants without paired normal tissue.4,5,14 However, many patients in this study were unable to provide germ line samples secondary to death, travel constraints, or lack of follow-up. Therefore, an ideal diagnostic workflow would include counseling and culturing of skin fibroblasts at the time of initial bone marrow biopsy.1 This pipeline is feasible at any center with laboratories capable of culturing skin fibroblasts.
HHM detection via tumor-only sequencing creates potential patient counseling dilemmas. The American College of Medical Genetics and Genomics has deemed universal counseling before genomic tumor profiling impractical but emphasizes physicians must notify patients of incidental findings.15 Patients with suspected HHMs should be referred to a cancer risk specialist for comprehensive hereditary testing.1,10,16 Tumor-only sequencing identified pathogenic variants in HHM-associated genes in 74 (21%) of 360 patients in this study, a large number of patients for whom cancer risk referral would not be feasible at many institutions.
The diagnostic resources spent on these evaluations may be streamlined. First, prospective germ line tissue collection may expedite cancer risk evaluations. Second, known Li-Fraumeni variants and pathogenic variants in DDX41 and GATA2 were the genes in which germ line variants were identified most frequently in our study. Therefore, variants in these genes should be prioritized for germ line evaluations if the variants are detected via tumor-only sequencing.
No pathogenic germ line variants in this study possessed tumor VAFs <0.4. Therefore, variants with tumor VAFs <0.4 in tumor-only testing are less likely to be of germ line origin; however, low VAFs should not deter germ line testing. We caution against using VAF as an exclusion criterion for additional germ line testing, because structural aberrations, including uniparental disomy, loss of heterozygosity, or variants in primer hybridization sites, may skew VAFs. We perform a hereditary evaluation, including assays sensitive to structural aberrations, for all patients at high risk for HHMs regardless of tumor VAFs. This approach has been shown to be an important component of an HHM evaluation.10 Of note, NGS-based assays in this study detect insertions or deletions up to 52 base pairs in size, but they are typically unable to detect larger structural aberrations without specialized bioinformatic pipelines.5 Therefore, this study may have excluded patients affected by larger structural aberrations and may underestimate the proportion of patients affected by HHMs.
In conclusion, this study demonstrates that NGS-based tumor-only sequencing may provide another method by which we can identify patients at risk for HHMs. Overall, 25 pathogenic variants with VAFs >0.4 were detected using tumor-only sequencing, and 6 (24%) of these variants were of germ line origin. This yield is similar to that for adult patients with concerning personal or family histories who are referred for panel-based testing for hereditary MDS/acute leukemia predisposition.10 In addition to personal history, family history, and cytopenia or poor stem-cell mobilization in related donors, tumor profiling via NGS panels represents an emerging method by which patients with HHMs may be identified.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors appreciate the engagement of the patients and their families in their ongoing studies of inherited predisposition syndromes.
M.W.D. is funded by a Physician-Scientist Training Award from the Damon Runyon Cancer Research Foundation. This work was also funded by the Cancer Research Foundation.
Authorship
Contribution: M.W.D., G.R., Z.L., J.P.S., J.E.C., and L.A.G. designed the research; M.W.D., S.K., M.S., S.A.P., A.H.W., S.F., D.A.C., M.F.J., C.M.W., C.K.D., A.A.C.-L., M.W.L., and Z.L. performed the research; M.W.D., M.S., A.H.W., S.F., D.A.C., M.F.J., and Z.L. analyzed the data; M.W.D. and L.A.G. wrote the initial draft of the manuscript; and all authors edited and approved the final version.
Conflict-of-interest disclosure: J.E.C. and L.A.G. receive royalties from their coauthored article on inherited hematopoietic malignancies in UpToDate. A.A.C.-L. has received honoraria from Celgene as a speaker. The remaining authors declare no competing financial interests.
The current affiliation for G.R. is Department of Pathology, Keck School of Medicine, University of Southern California, Los Angeles, CA.
Correspondence: Lucy A. Godley, 5841 S. Maryland Ave, MC 2115, Chicago, IL 60637; e-mail: lgodley@medicine.bsd.uchicago.edu.
References
- 1.University of Chicago Hematopoietic Malignancies Cancer Risk Team. How I diagnose and manage individuals at risk for inherited myeloid malignancies. Blood. 2016;128(14):1800-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Churpek JE, Nickels E, Marquez R, et al. Identifying familial myelodysplastic/acute leukemia predisposition syndromes through hematopoietic stem cell transplantation donors with thrombocytopenia. Blood. 2012;120(26):5247-5249. [DOI] [PubMed] [Google Scholar]
- 3.Rojek K, Nickels E, Neistadt B, et al. Identifying inherited and acquired genetic factors involved in poor stem cell mobilization and donor-derived malignancy. Biol Blood Marrow Transplant. 2016;22(11):2100-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J, Abdel-Wahab O, Nahas MK, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127(24):3004-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadri S, Long BC, Mujacic I, et al. Clinical validation of a next-generation sequencing genomic oncology panel via cross-platform benchmarking against established Amplicon sequencing assays. J Mol Diagn. 2017;19(1):43-56. [DOI] [PubMed] [Google Scholar]
- 6.McKerrell T, Moreno T, Ponstingl H, et al. Development and validation of a comprehensive genomic diagnostic tool for myeloid malignancies. Blood. 2016;128(1):e1-e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germ line DDX41 mutation. Int J Hematol. 2017;106(2):163-174. [DOI] [PubMed] [Google Scholar]
- 10.Guidugli L, Johnson AK, Alkorta-Aranburu G, et al. Clinical utility of gene panel-based testing for hereditary myelodysplastic syndrome/acute leukemia predisposition syndromes. Leukemia. 2017;31(5):1226-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polprasert C, Schulze I, Sekeres MA, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27(5):658-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewinsohn M, Brown AL, Weinel LM, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127(8):1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tawana K, Fitzgibbon J. Inherited DDX41 mutations: 11 genes and counting. Blood. 2016;127(8):960-961. [DOI] [PubMed] [Google Scholar]
- 14.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green RC, Berg JS, Grody WW, et al. ; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yushak ML, Han G, Bouberhan S, et al. Patient preferences regarding incidental genomic findings discovered during tumor profiling. Cancer. 2016;122(10):1588-1597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



