Key Points
A subset of snoRNAs is expressed in a developmental- and lineage-specific manner during human hematopoiesis.
Neither host gene expression nor alternative splicing accounted for the observed differential expression of snoRNAs in a subset of AML.
Abstract
Small nucleolar RNAs (snoRNAs) are noncoding RNAs that contribute to ribosome biogenesis and RNA splicing by modifying ribosomal RNA and spliceosome RNAs, respectively. We optimized a next-generation sequencing approach and a custom analysis pipeline to identify and quantify expression of snoRNAs in acute myeloid leukemia (AML) and normal hematopoietic cell populations. We show that snoRNAs are expressed in a lineage- and development-specific fashion during hematopoiesis. The most striking examples involve snoRNAs located in 2 imprinted loci, which are highly expressed in hematopoietic progenitors and downregulated during myeloid differentiation. Although most snoRNAs are expressed at similar levels in AML cells compared with CD34+, a subset of snoRNAs showed consistent differential expression, with the great majority of these being decreased in the AML samples. Analysis of host gene expression, splicing patterns, and whole-genome sequence data for mutational events did not identify transcriptional patterns or genetic alterations that account for these expression differences. These data provide a comprehensive analysis of the snoRNA transcriptome in normal and leukemic cells and should be helpful in the design of studies to define the contribution of snoRNAs to normal and malignant hematopoiesis.
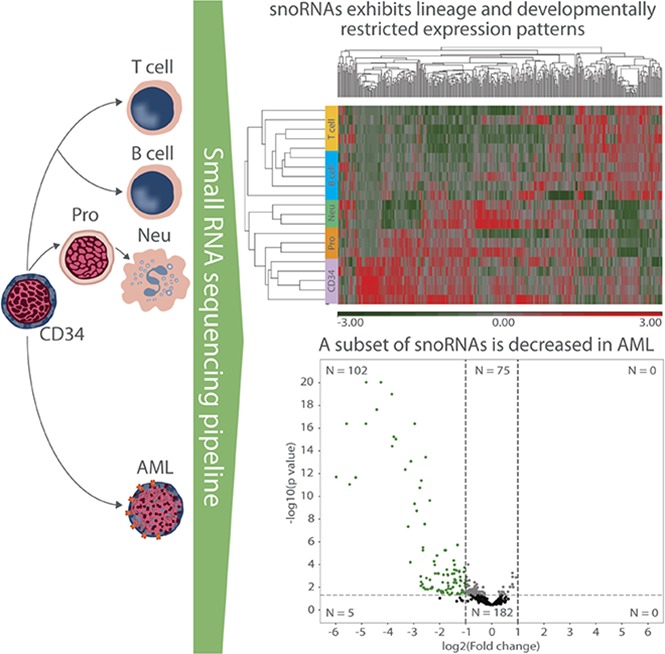
Visual Abstract
Introduction
There has been increasing interest in the contribution of the noncoding transcriptome to the regulation of normal and malignant hematopoiesis. Noncoding RNA (ncRNA) species are classified into 2 groups based on their sizes. Long noncoding RNAs (lncRNAs) are >200 nucleotides, and they are expressed in a lineage-specific fashion in hematopoiesis.1 Recent studies have implicated lncRNAs in hematopoietic lineage commitment and control of self-renewal.1 Small noncoding RNAs (sncRNAs) are <200 nucleotides and include a heterogeneous group of RNA species. Best characterized are microRNAs (miRNAs), which are 19-26 nucleotide RNAs that repress translation of target RNAs by targeting them to the RNA-induced silencing complex. MicroRNAs are also expressed in a lineage-specific fashion and have been shown to play key roles in the regulation of hematopoiesis.2-4 Other sncRNAs include small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), small interfering RNAs, and Piwi-interacting RNAs. With some exceptions, the expression of these other sncRNAs in hematopoietic cells and their contribution to the regulation of hematopoiesis are not well characterized.
snoRNAs are a subset of sncRNAs that are involved in the posttranscriptional modification of ribosomal RNAs (rRNAs) and snRNAs. These modifications are critical for a variety of cellular processes, including ribosomal biogenesis and splicing of RNAs. Classification of snoRNA species is based on the presence of highly conserved sequence elements that define 3 snoRNA families: H/ACA box (SNORAs), C/D box (SNORDs), or small Cajal body–specific RNAs (scaRNAs). H/ACA and CD box snoRNAs target specific ncRNA species with base pair complementarity for site-specific pseudouridylation5 or 2'-O-methylation,6 respectively. scaRNAs localize to RNA-containing Cajal bodies and are responsible for the methylation and pseudouridylation of spliceosomal RNAs U1, U2, U4, U5, and U12. There are also orphan snoRNAs, which lack known complementarity to rRNAs or snRNAs and therefore largely have unknown functions. Recent studies have suggested an expanded role for snoRNAs beyond ribosomal biogenesis and modifications to snRNA. For example, emerging data suggest that snoRNAs may contribute to alternative splicing,7 regulation of chromatin structure,8 metabolism,9 and neoplastic transformation.10
The contribution of snoRNAs to the regulation of normal and malignant hematopoiesis is largely unknown. Chu et al reported that overexpression of the H/ACA box snoRNA ACA11 in t(4;14)-associated multiple myeloma contributes to myeloma cell proliferation and resistance to chemotherapy.11 Several groups have reported marked increased expression of snoRNAs contained in the DLK-DIO3 locus in acute promyelocytic leukemia, although their contribution to leukemogenesis is unknown.12-14 The lack of a method to accurately and comprehensively assess snoRNA expression has limited research in this area. Array-based methods only interrogate a subset of snoRNAs and cannot distinguish between mature and precursor snoRNAs.15,16 To avoid sequencing very abundant rRNAs and transfer RNAs (tRNAs), most next-generation sequencing approaches to interrogate the transcriptome have focused on longer (>200 nucleotide) or very short (17-26 nucleotide) RNA species. Thus, there is a gap in current transcriptome sequencing that includes most snoRNAs. To address this gap, we developed a next-generation sequencing approach optimized to interrogate sncRNAs, including snoRNAs. We show that snoRNAs are expressed in a lineage- and development-specific fashion in human hematopoiesis with a subset of snoRNAs that show consistent differential expression in acute myeloid leukemia (AML). We further show that expression of snoRNAs does not correlate with expression or splicing of host genes, suggesting that other factors are determining cellular levels of mature snoRNAs.
Materials and methods
Fluorescence-activated cell sorting of hematopoietic populations
Bone marrow aspirate samples were obtained from normal healthy donors after obtaining informed consent (institutional review board approval Washington University Human Studies Committee #01-1014). Samples were processed via ammonium–chloride–potassium red cell lysis, washed once in phosphate-buffered saline, and then stained for flow cytometry using the following antibodies: CD34-phycoerythrin (PE) (PE-pool, Beckman Coulter, IM1459U), CD14-allophycocyanin (BD Biosciences, clone M5E2), CD15-fluorescein isothiocyanate (BD Biosciences, clone HI98), CD16-PE (BD Biosciences, clone 3G8), CD33-allophycocyanin (eBioscience, clone WM-53), CD3-V450 (eBioscience, clone OKT3), and CD19-PE (BD Biosciences, clone HIB19). Defined hematopoietic cell populations that were sorted included: promyelocytes (CD14−, CD15+, and CD16low/−),17 monocytes (CD14+), neutrophils (CD14−, CD15+, and CD16+),17 and CD34+ cells. Cells were sorted directly into lysis buffer, and RNA was isolated using the Quick-RNA Microprep Kit (Zymo Research).
Small RNA library construction and sequencing
The NEBNext Small RNA Library Prep Set for Illumina (New England BioLabs, Inc.) was used to prepare the libraries following the manufacturer’s specifications using 100 to 500 ng of total RNA as input.18 After adaptor ligation, reverse transcription, and polymerase chain reaction (PCR) amplification, the libraries were size selected on a Blue Pippin (Sage Science) to enrich for library molecules with inserts between ∼17 and 200 nucleotides. The resulting libraries were sequenced on a MiSeq instrument to generate 150 bp, single-end reads. All sequence data will be deposited in dbGaP.
Bioinformatic analysis
Sequencing data were trimmed to remove adapter sequences using cutadapt with the command “cutadapt -f fastq -a AGATCGGAAGAGCACACGTCT” and then mapped to the National Center for Biotechnology Information Build 37 human reference sequence using bwa mem19 with the custom parameters “bwa mem -M -k 15 -T 17” to obtain short alignments that result from small RNA species. These alignments were then used in the following analyses to characterize the spectrum of RNA species captured by the library approach, identify novel RNA species, and quantify the expression of annotated snoRNAs.
We first defined the distribution of RNA species captured in the library by annotating the sequencing reads from all samples with RNA biotypes from GENCODE version 19,20 mirBase version 21,21 and a previously described set of snoRNA annotations (snoRNAome22). Given that the library preparation method has a 3′ end bias, reads were assigned to a single RNA annotation in a strand-specific manner based on the proximity of the read start position to the 3′ end of overlapping annotations. Reads were called “unannotated” if the alignments were uncertain (ie, mapping quality of 0), or they did not map to any annotation in a strand-dependent manner.
Next, we identified potentially novel RNA species using a custom Practical Extraction and Report Language (PERL) script designed to detect and annotate aggregate read “clusters” using pooled sequence data from all normal hematopoietic cell and AML samples (N = 64). Briefly, mapped reads for all samples with a mapping quality >0 were merged into a single BAM file, and regions with a minimum strand-specific read depth of 50 were extracted. All reads spanning these regions were then merged to create strand-specific read clusters, which were trimmed such that the cluster edges were ≥20% of the maximum read depth (to separate closely spaced clusters that may have become merged by spurious “joining” reads), and subsequently filtered to retain those with an AT nucleotide content <80% to exclude low-complexity sequences. The total number of clusters that resulted from this procedure was 6231. Clusters were then annotated with read quality and mapping statistics (eg, mean mapping quality, number of unique read start positions, and mean number of mismatches with the reference sequence), the number of strand-specific read counts, and maximum depth across the cluster. Cluster coordinates were compared with the GENCODE, mirBase, and snoRNAome annotations, and “tagged” with the strand-specific transcript or gene annotation with the best reciprocal overlap. Potentially novel species from this set that demonstrated <50% reciprocal overlap with known annotations and total counts ≥500 (N = 340 clusters) were then manually analyzed with the programs snoGPS and snoSCAN, which identify H/ACA box and C/D box snoRNAs with reported rRNA targets,23,24 and with snoReport for identification of all snoRNAs, including orphans25 as well as a custom script. This produced a final list of 111 clusters that were identified as potential snoRNA species, which were manually reviewed using the Integrated Genome Viewer (version 2.3.40)26 and the sno/miRNA track of UCSC Genome Browser27 to exclude low-quality clusters or those that overlapped known snoRNAs.
Finally, expression levels for a comprehensive set of annotated sncRNA species were generated for each sample using annotations curated from snoRNAome22 and miRBase28 along with all GENCODE version 19 annotations with biotype “snoRNA.” These annotation databases were combined to produce a set of 4931 nonoverlapping annotations, with snoRNAome and mirBase entries superseding those from GENCODE version 19 with overlapping coordinates. Overlapping annotations from snoRNAome and mirBase were reviewed, and a single species was selected based on the correspondence between the sequencing reads at the locus and annotation; the other annotation was excluded. Expression values for these annotations were then obtained with the featureCounts program29 using parameters for strand-specific counting and including only reads with a mapping quality of ≥1. These counts were normalized to the total mapped reads × 106 for visualization and subsequent statistical analyses.
Quantitative reverse transcription PCR of selected snoRNAs
The extracted RNA was purified on a RNA Clean & Concentrator-5 column (Zymo Research, R1013) using the manufacturer's >17-nucleotide-long protocol and resuspended in 10.0 μL of nuclease-free water. The Qubit RNA HS Assay Kit (Life Technologies, Q32855) and the TapeStation system (Agilent) were used for quantification and quality assessment, respectively, according to the manufacturer's instructions. The RNA was reverse transcribed using iScript Reverse Transcriptase (BioRad, 1708841) at 42°C, according to the manufacturer's instructions. The complementary DNA (cDNA) was PCR amplified using forward and reverse primers containing sequences specific to the snoRNAs (supplemental Table 1). In a 20-μL reaction, 3.0 ng of cDNA template, 0.5 μM each of forward and reverse primers (IDT), 10 μL iTaq Universal SYBR Green 2× Supermix, (BioRad, 1725120), and nuclease-free water were cycled for 60 rounds at an annealing temperature of 60°C on a StepOnePlus Real-Time PCR System (Applied Biosystems). The 5S rRNA was used to normalize snoRNA expression.
Differential expression analysis
Differential expression and hierarchical clustering analyses were performed with the Partek Genomic Suite (Partek, Inc.) using log2 (read count per million mapped reads × 106 [RPM]) expression values for the curated sncRNA annotations as input30; only RNA species with mean normalized count values ≥5 were selected to produce reliable differential expression profiles. Data were first assessed for normality, and differential expression analysis was performed with the Partek Genomic Suite using 1-way analysis of variance (ANOVA) with estimation via the method of moments model.31 The differential expression of snoRNAs and miRNAs in the AML patients vs normal hematopoietic stem/progenitors was based on a fold change >2 and P < .05).
Splicing analysis
Intron junction counts for annotations in GENCODE version 19 were obtained from aligned BAM files using Tablemaker and Ballgown32 and normalized to the total number of junction reads observed × 106. The linear regression between normalized snoRNA expression (RPM) and the normalized expression for “host gene” junctions spanning each snoRNA were assembled in R.33 Correlations between all snoRNAs and junction expressions were similarly performed.
Somatic mutation of snoRNAs
The coordinates of the 344 651 introns in the genome (GRCh37) and that of 402 snoRNAs were intersected with the coordinates of 367 904 prevalidation indels from 49 Cancer Genome Atlas AML patient samples using BEDtools34 and R.
Statistical analysis
Statistical analysis and graphing were performed with Prism (GraphPad Software, Inc.) and R. Error bars represent the standard error of the mean (SEM). Significance was determined by 1-way ANOVA followed by Tukey multiple comparisons test. Significance is denoted as: *P < .05; ** < .01; ***P < .001; ****P < .0001; and ns, not significant.
Results
Small RNA-seq pipeline
We modified a previously described method for sequencing miRNAs to analyze more comprehensively the small RNA component of the transcriptome.18 A key aspect of this approach is the method used for cDNA library generation, which includes the addition of an oligonucleotide adaptor to the 3′-end of RNA molecules before reverse transcription. Importantly, this requires the presence of a free 3′ hydroxyl group on the RNA molecule. We then performed an expanded size selection to capture RNA species between 17 and 200 nucleotides, which includes miRNAs, snoRNAs, and other sncRNAs, but excludes most messenger RNA (mRNA) and lncRNA molecules. The sequence data obtained were analyzed using 2 complementary bioinformatic approaches to quantify both annotated and novel sno- and miRNAs (Figure 1A).
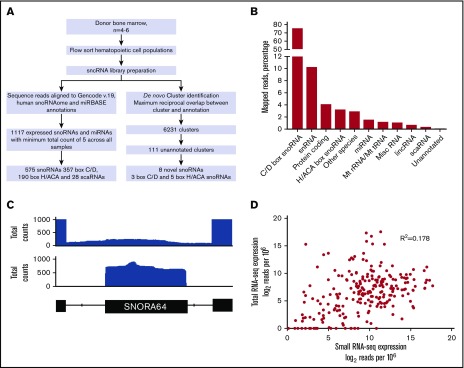
Figure 1.
Small RNA-Seq pipeline. (A) Schematic representation of the small RNA-seq pipeline. Sequence reads were aligned to the GENCODE version 19, miRBase version 21, and human snoRNAome, and reads corresponding to annotated sncRNAs were quantified (left pathway). Alternatively, aligned sequence reads were organized into genomic clusters; a total of 111 unannotated clusters were identified, of which 8 were classified as novel snoRNAs (right pathway). (B) Graph showing the distribution of annotation biotypes in GENCODE version 19 for mapped reads from all samples. The percentage of all mapped reads (mapping quality >0) is shown on the y-axis, and the annotated species is shown on the x-axis. Miscellaneous (Misc) snoRNAs include rRNAs, other species include unprocessed pseudogenes, immunoglobulin genes, T-cell receptor genes, sense introns, antisense transcripts, sense overlapping transcripts, retained introns, and processed transcripts. Protein-coding genes include nonsense mediated decay, and nonstop decay biotypes. (C) Integrated Genome Viewer browser view of SNORA64, which is embedded in an intron of the coding gene RPS2. The top panel shows the alignment of reads generated using total RNA-seq. The middle panel shows the alignment of reads from a small RNA-seq library produced from the same sample. The genomic boundaries of SNORA64 and exons 4 and 5 of host gene RPS2 are shown in the bottom panel. (D) Representative scatter plot showing log-transformed normalized read counts of annotated snoRNAs for a CD34 sample analyzed using total RNA-seq (y-axis) or our small RNA-seq pipeline (x-axis).
We used this approach to interrogate the small RNA transcriptome in human hematopoietic cell populations from normal hematopoietic stem/progenitors and from diagnostic AML samples. CD34 cells, promyelocytes, neutrophils, monocytes, T cells, and B cells were sorted by flow cytometry from the bone marrow of 6 healthy individuals. Data from primary AML samples were generated from bulk leukemic cells from 33 treatment-naive patients with AML (Table 1). Most of these cases (97%) had normal cytogenetics, and all were classified as intermediate-risk AML. An average of 3.2 × 106 reads was obtained across both normal and leukemic samples (supplemental Table 2). Mapping of sequencing reads from all samples to annotation features from GENCODE version 19 and snoRNA and miRNA annotations in the human snoRNAome and miRBase (see “Materials and methods”) demonstrated that snoRNAs were by far the most abundant small RNA species present in our data (Figure 1B). C/D box snoRNAs represented 74.95% of all reads; H/ACA box snoRNAs and scaRNAs represented another 3.19% and 0.34% of total mapped reads, respectively. Small nuclear RNAs, which are involved in RNA splicing, were the next most abundant class of sncRNA, representing 10.19% of reads. miRNAs represented a relatively small percentage of all sequenced reads (1.48% of all mapped reads). Reads mapping to unannotated regions of the genome accounted for 0.04% of all sequences.
Table 1.
Characteristics of the 33 AML patients
| Characteristic | Value |
|---|---|
| Age at study entry, mean ± standard deviation, y | 51.3 ± 15.0 |
| Race or ethnic group, n (%) | |
| White | 26 (78.8) |
| African American | 1 (3.03) |
| Other | 6 (18.18) |
| Male sex, n (%) | 14 (42.4) |
| Bone marrow blasts at diagnosis, mean ± standard deviation, % | 81.8 ± 13.4 |
| Normal cytogenetic profile, n (%) | 32 (97) |
| White blood cell count at diagnosis, ×109/L | |
| Mean ± standard deviation | 63.66 ± 70.75 |
| Median | 45.60 |
| Cytogenetic risk group, n (%) | |
| Intermediate | 33 (100.0) |
| AML FAB subtype, n (%) | |
| AML without maturation: M1 | 18 (54.5) |
| AML with maturation: M2 | 1 (3.03) |
| Acute myelomonocytic leukemia: M4 | 5 (15.1) |
| Acute monoblastic or monocytic leukemia: M5 | 9 (27.3) |
| Mutation, n (%) | |
| NPM1 | 27 (88.8) |
| FLT3 | 18 (54.5) |
| DNMT3A | 12 (36.4) |
| IDH1 or IDH2 | 10 (30.3) |
| NRAS or KRAS | 4 (12.1) |
| CEBPA | 3 (9.1) |
| TET2 | 2 (6.1) |
| WT1 | 2 (6.1) |
| PTPN11 | 2 (6.1) |
FAB, French-American-British.
We next compared the expression of snoRNAs using data from our modified library protocol with expression levels obtained by standard total RNA sequencing (RNA-seq) (Illumina Tru-seq) and of the same tissue sample. Relevant to this analysis, the majority of snoRNAs are embedded in the introns of host genes. We observed that standard transcriptome sequencing cannot reliably distinguish unspliced primary host gene RNA from correctly processed snoRNA. Typical results are shown for SNORA64, which is located in the intron of its host gene, RP32 (Figure 1C). Whereas sequence reads corresponding to mature SNOR64 were readily identified using our pipeline, only low-level reads that span the entire intron of RP32 were detected using total RNA-seq. Accordingly, the correlation of snoRNAs quantified using these 2 RNA-seq pipelines was poor (Figure 1D). These data demonstrate the superiority of our small sequencing pipeline to quantify mature, correctly processed snoRNA expression.
To provide orthogonal validation of the snoRNA expression data, we used commercially available reagents to perform quantitative reverse transcription PCR (RT-qPCR) on a set of 9 snoRNAs with a wide range of expression across 11 primary AML samples. Although some variability was observed, a significant correlation between snoRNA expression determined by small RNA Seq and RT-qPCR was observed (R2 = 0.5002; P < .0001; supplemental Figure 1).
To determine whether our sequencing approach identified any novel RNA species, we formed read clusters by merging overlapping reads and compared them with the RNA annotations as described above. The intersection of read cluster coordinates with our sncRNA annotation set demonstrated that a number of them did not overlap with known annotations, and could therefore represent novel RNA species. The genomic regions spanned by these clusters were then analyzed for features of snoRNAs, including the presence of conserved sequence motifs and secondary structure. Eight putative novel snoRNAs were identified, including 5 in the SNORA family and 3 in the SNORD family. One of the putative SNORDs lacked sequence complementarity to rRNAs or snRNAs and was therefore classified as an orphan snoRNA (supplemental Table 3). There was some degree of overlap (≤50%) with annotated species, but our analysis supports the characterization of these snoRNAs as putatively novel.
Developmental- and lineage-specific expression of snoRNAs in human hematopoiesis
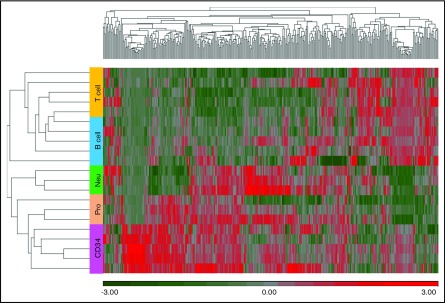
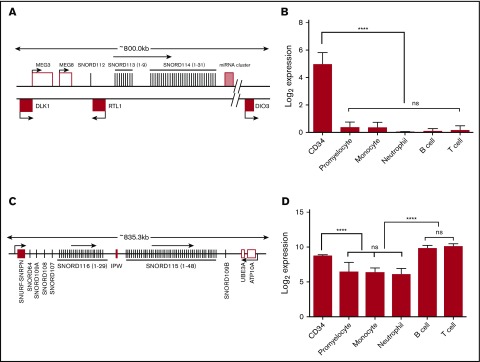
Because snoRNAs were the most abundant sncRNA detected, we focused our analysis on these RNA species. We first performed unsupervised hierarchical clustering of annotated snoRNAs with a normalized expression of ≥5 RPM (N = 378) to determine whether expression of snoRNAs is developmentally regulated during hematopoiesis. This demonstrated that snoRNAs exhibit lineage- and developmentally restricted expression patterns (Figure 2). The most striking examples were orphan snoRNAs contained in the imprinted DLK-DIO3 and SNURF/SNRPN loci. The DLK-DIO3 locus contained a large number of maternally expressed ncRNAs, including 41 snoRNAs, 11 lncRNAs, and 53 miRNAs (Figure 3A). Expression of snoRNAs in this locus was highest in CD34 cells and rapidly decreased with granulocytic differentiation, becoming nearly undetectable in mature neutrophils (Figure 3B). Expression of these snoRNAs was also markedly reduced in B cells and T cells. Expression of snoRNAs in the SNURF/SNRPN locus showed a similar, but distinct, pattern of snoRNA expression. This locus contained 82 paternally expressed snoRNAs that were expressed at a high level in CD34 cells and rapidly downregulated during granulocytic differentiation (Figure 3C). However, in contrast to the DLK-DIO3 locus snoRNAs, expression of these snoRNAs remained high in B and T cells (Figure 3D).
Figure 2.
Unsupervised hierarchical clustering of snoRNA expression. Small RNA-seq was performed on the indicated sorted hematopoietic cell populations obtained from 4-6 normal hematopoietic stem/progenitors. snoRNA expression (z-scored log2 RPM) is shown, with red indicating high expression, and green indicating low expression. Each column represents a unique snoRNA, and each row represents a sample.
Figure 3.
Expression of snoRNAs in the DLK-DIO3 and SNURF/SNRPN loci. (A) Organization of the DLK-DIO3 locus. Maternally expressed genes (open boxes) and paternally expressed genes (filled boxes) are shown. The miRNA cluster contains 54 miRNAs. (B) Normalized read counts for SNORD113-6 are shown; its expression is representative of other SNORDs in this locus. Significance was determined by 1-way ANOVA followed by Tukey multiple comparisons test. (C) Organization of the human SNURF-SNRPN locus (drawing is not to scale). Paternally expressed genes, including SNORDs, are shown as ▪, and maternally expressed genes are shown as □. (D) Normalized read counts for SNORD116-1, representative of SNORDs in the SNURF-SNRPN locus, are shown. ****P < .0001. ns, not significant.
Expression of a subset of snoRNAs is decreased in AML
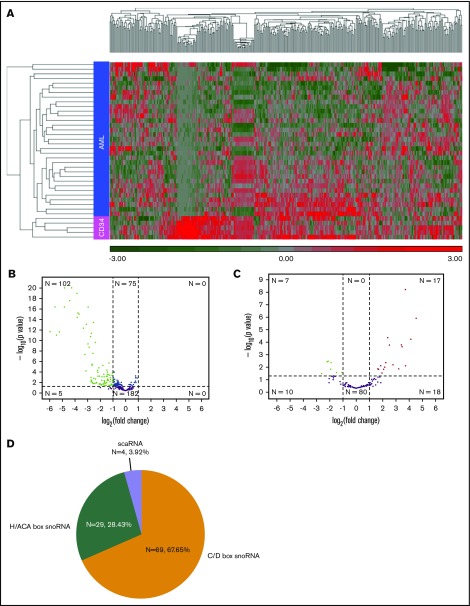
We next compared snoRNA expression in 33 de novo AML samples with normal CD34 cells. Analysis of expression across all annotated snoRNA species (N = 364) via unsupervised hierarchical clustering demonstrated that AMLs had distinct snoRNA expression patterns from normal CD34 cells (Figure 4A). We required a mean normalized expression of ≥5 counts across any AML and healthy donor samples to be considered for analysis. Differential expression analysis identified 102 snoRNAs that were differentially expressed (adjusted P ≤ .05; absolute log2-fold change > 1) (supplemental Table 4), all of which had decreased expression in the AML samples (Figure 4B). By comparison, 24 differentially expressed miRNAs were identified in a similar analysis using the same samples, which included 17 with increased expression in AML vs 7 that were decreased (Figure 4C). Although differentially expressed snoRNAs in AML spanned all RNA species, a disproportionate number of box C/D snoRNAs were observed (69 of 102, 67.65%; Figure 4D), with 37 located in the DLK-DIO3 or SNURF-SNRPN loci. There were 66 (64.71%) orphan snoRNAs with representation from all snoRNA classes. Of note, differential expression of snoRNAs that are known to play key roles in splicing as well as the modification of the peptidyl transferase center (PTC) and the intersubunit bridge (ISB) during ribosomal biogenesis was observed (supplemental Table 4). For example, expression of SNORA21 and -36C, which target crucial nucleotides in the PTC and ISB, respectively, were decreased 2.69- and 2.56-fold, respectively, in AML compared with CD34 cells, and expression of SCARNA15, which targets a key nucleotide in the U2 spliceosomal RNA, was decreased 2.81-fold.
Figure 4.
Differentially expressed snoRNAs. (A) Unsupervised hierarchical clustering of snoRNA expression in AML and CD34 from healthy adult donors based on z scores, with red indicating high expression, and green indicating low expression. Each column represents a specific snoRNA, and each row represents an individual sample. (B-C) Volcano plot illustrating significant differentially expressed snoRNAs (B) and miRNAs (C). The fold change difference in RNA expression between AML and normal CD34 samples is plotted on the x-axis, and P value significance is plotted on the y-axis. The horizontal line on the plot represents the α-level used for this analysis (0.05). Vertical lines represent the threshold for the log2 fold change (equivalent to a twofold difference). The y-axis reflects the −log10 (q value–corrected P value). Green and red colored dots represent snoRNAs showing significantly increased or decreased expression in AML, respectively. (D) Distribution of differentially expressed snoRNAs across snoRNA subtypes. H/ACA snoRNA distribution includes 4 AluACA snoRNAs.
Somatic mutation of snoRNAs is uncommon in AML
Whole-genome sequencing data were available for 14 of the 33 analyzed cases in this study. No somatic single nucleotide variants or small indels were detected in the snoRNA genes. In addition, for those snoRNAs located in a host gene, no recurrent indels in the introns harboring the snoRNA or mutations in splice donor sites for that intron were identified. We expanded this analysis to an additional 35 AML cases with whole-genome sequencing data available from the The Cancer Genome Atlas.35 Again, no somatic single nucleotide variants or small indels were detected in snoRNA genes, suggesting that genetic alterations in snoRNAs are uncommon in AML with normal cytogenetics and are not the cause of their differential expression in this disease.
There is minimal correlation between host gene and snoRNA expression
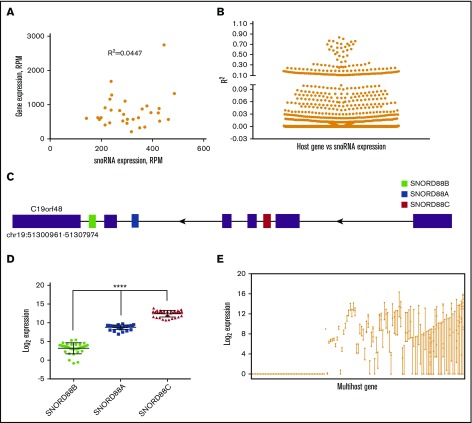
Because many snoRNAs are located in the introns of host genes,36 we next asked whether variation in snoRNA expression may be explained by differences in expression and/or processing of these host genes. We limited our analysis to the AML cases, where matching small RNA and total RNA-seq data were available. For most snoRNAs, there was minimal correlation between host gene and corresponding snoRNA expression, as illustrated by host gene RPL7A and its corresponding snoRNAs (Figure 5A). Across all 1379 snoRNAs contained in host genes, the average coefficient of determination (R2) was 0.037 ± 0.102 (Figure 5B). Multiple snoRNAs are often located within different introns of a single host gene, as shown for the C19orf48 gene (Figure 5C). If host gene expression is the primary determinant of snoRNA expression, then expression of each snoRNA located in a given multihost gene should be similar. However, we observed marked variability in the expression of snoRNAs contained within a single gene. For example, expression of the 3 snoRNAs hosted by C19orf48 varied by >32-fold (Figure 5D). Indeed, marked variability in the expression of snoRNAs contained within the same host gene was observed in the majority of cases (Figure 5E). These data show that host gene expression is not the primary determinant of snoRNA expression in AML.
Figure 5.
Correlation between host gene and snoRNAs expression. (A) Representative plot showing the correlation between the normalized expression of the host gene RPL7A and its encoded SNORD36B in AML. (B) Summary scatter plot showing the coefficient of determination (R2) of all host gene/snoRNAs pairs (N = 1379, mean ± SEM = 0.037 ± 0.102). (C) Organization of the human gene C19orf48 gene (drawing not to scale); exons are shown as ▪ and SNORD88B, −88A and −88C are shown in green-, blue-, or red-filled boxes, respectively. (D) Log-transformed normalized read counts in AML. Significance was determined by 1-way ANOVA followed by Tukey multiple comparisons test. (E) Plot showing the log-transformed median expression values for all expressed snoRNAs in a multihost gene (N = 130). Each line represents a distinct multihost gene.
Alternative splicing of host genes is not the primary determinant of snoRNA expression
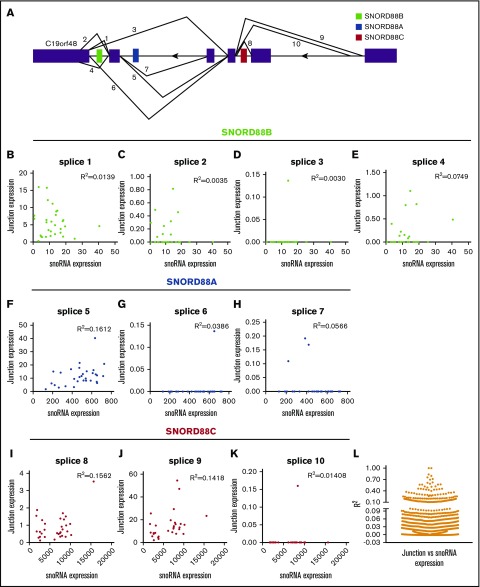
Mature snoRNAs are processed from excised introns after splicing of the host gene. Thus, we next asked whether alternative splicing of host genes is a major determinant of snoRNA expression. We assessed RNA splicing by measuring junction reads, as previously described.37 For example, the host gene C19orf48 has 10 predicted splice events that involve introns containing embedded snoRNAs (Figure 6A). Expression of junction reads corresponding to each splice event showed minimal correlation with expression of the relevant snoRNA (Figure 6B-K). This analysis was extended to look at snoRNA expression and encompassing junction expression across 858 snoRNAs spanning 1616 junctions (Figure 6L). For most cases, junction reads correlated minimally with snoRNA expression. Collectively, these data show that alternative splicing of host genes is unlikely to be the primary determinant of snoRNA expression.
Figure 6.
Correlation of host gene splicing to snoRNA expression. (A) Schematic of all splice junctions for C19orf48. Coding exons are shown as ▪ and embedded snoRNAs as color-filled boxes. Each numbered line represents a different splice event. Drawing not to scale. (B-K) RNA splicing was assessed by quantifying each of the junction reads (splice 1-10). Shown are scatter plots showing normalized junction reads (reads per million) vs snoRNA expression (reads per million). (L) Scatter plot showing R2 values (mean ± SEM = 0.041 ± 0.093) between snoRNA expression and host gene splicing (total of 1616 junction reads).
Discussion
The expression of snoRNAs has traditionally been determined by high-throughput techniques that rely on hybridization-based methods, such as microarray analysis or by standard RNA-seq technologies.12-14 Microarrays for snoRNAs are unable to effectively capture novel sequences or resolve the expression of snoRNAs in families with highly homologous members.38 Standard RNA-seq is generally limited to RNA species >200 nucleotides in length, and thus does not reliably detect most sncRNAs, including snoRNAs. In this study, we optimized both library preparation and bioinformatic analysis to address these challenges, which resulted in improved sensitivity for detecting novel sncRNAs, more accurate expression levels of annotated species, and efficient resolution of closely related snoRNA species, such as those in the DLK-DIO3 and SNURF-SNRPN loci. In addition, for those snoRNAs embedded in host genes, this approach can distinguish between host gene primary transcripts and mature, fully processed snoRNAs.
The best method to normalize small RNA-seq expression data is uncertain. For miRNAs, several studies have compared normalization methods, suggesting that the upper quartile, median, the DESeq normalization offered in the DESeq Bioconductor package, and the trimmed mean of M values offered in the edgeR Bioconductor package may be superior to the RPM normalization method.39-41 In the absence of a “gold standard” for snoRNA expression, a rigorous comparison of normalization strategies of our small RNA-seq snoRNA expression data was not possible. Thus, in this study, we normalized our small RNA-seq data with the widely used RPM method.
To our knowledge is the first study to comprehensively analyze snoRNA expression in human hematopoiesis. snoRNAs are the most highly expressed sncRNAs in all of the hematopoietic cell populations tested. Although snoRNAs have been considered to be housekeeping genes,42 we identified a subset of snoRNAs that exhibit marked differential expression in a lineage- and development-specific pattern. This is particularly true for orphan snoRNAs contained in the imprinted DLK-DIO3 and SNURF/SNRPN loci. The DLK-DIO3 locus contains 47 orphan CD box snoRNAs that are highly expressed in CD34 cells and downregulated during myeloid or lymphoid differentiation. This observation is consistent with previous reports showing high hematopoietic stem/progenitor expression of lncRNAs43 and miRNAs44 that are contained in the DLK-DIO3 locus. The SNURF/SNRPN locus contains 2 large orphan CD box snoRNA clusters, SNORD115 and SNORD116, that are expressed highly in CD34 cells and downregulated during myeloid differentiation. Loss of SNORD116 in the SNURF/SNRPN locus is thought to be key to the pathogenesis of Prader-Willi syndrome, a genetic disorder characterized by obesity and developmental delay.45,46 Of note, SNORD115 has been shown to promote alternative splicing of the serotonin receptor 2C.47 The contribution of the DLK-DIO3 and SNURF/SNRPN loci to hematopoietic stem/progenitor function is unknown, although it is interesting to note that expression of ncRNAs from the DLK-DIO3 locus correlates with pluripotency in both embryonic and inducible pluripotent stem cells.48
We observed no recurring mutations of snoRNA genes in our cohort of cytogenetically normal AML, suggesting that genetic alterations that specifically target snoRNAs in AML are uncommon. A previous study reported that snoRNAs are globally suppressed in AML relative to CD34 cells from normal hematopoietic stem/progenitors.14 Although we also observed a trend toward decreased expression in AML, this was limited to a small subset of snoRNAs (102 of 364, 28%). The reasons for this discrepancy are not certain, but the previous study primarily used a microarray approach to assess expression of a more limited set of snoRNAs. Of note, of the 102 snoRNAs with significantly reduced expression in AML, 37 are located in the DLK-DIO3 or SNURF-SNRPN loci. Because expression of these snoRNAs is suppressed during normal myeloid differentiation, it is possible that their decrease in AML reflects normal differentiation along the myeloid lineage. This is in sharp contrast to previous studies showing marked increased expression of DLK-DIO3 snoRNAs in acute promyelocytic leukemia.12-14 Of note, Valleron et al showed that enforced expression of SNORD114-1, which is contained in the DLK1-DIO3 locus, promotes cell growth in vitro, possibly by targeting the Rb pathway.14
We observed significant differential expression of snoRNAs that mediate pseudouridylation or 2′-O methylation of key sites in rRNA. Decreased expression of snoRNAs that target modifications of the PTC and ISB regions of the 60S ribosome was observed in AML patients vs normal hematopoietic stem/progenitors. The PTC is the catalytic site where peptide bonds are made during protein elongation and peptidyl-tRNAs are hydrolyzed during the termination of protein synthesis.49 The ISB forms multiple interactions between the ribosomal subunits, which maintain ribosome stability and modulate dynamics that are critical for translation, such as that between the tRNA and mRNA.50 Studies in yeast suggest that, although loss of pseudouridylation or 2′-O methylation at individual rRNA sites has only subtle effects on activity, loss at multiple sites is synergistic, resulting in reading frame changes, increased stop-codon read-through, and altered tRNA selection.51-53 We also identified several snoRNAs responsible for the pseudouridylation of snRNAs in regions critical for splicing. For example, scaRNA15, whose expression is reduced 2.81-fold in AML, targets the branch site recognition region of U2 snRNA. Studies in HeLa cells54 and yeast55 show that pseudouridylation at this site is required for the formation of early spliceosomal complexes and the catalytic phase of pre-mRNA splicing. Further study is needed to determine whether the observed decreases in snoRNA expression in AML are sufficient to induce biologically meaningful differences in translation or splicing.
The mechanisms regulating snoRNA expression are not well defined. Most snoRNAs and scaRNAs are embedded in the introns of host genes that produce proteins involved in nucleolar function, ribosome structure, or protein synthesis,56 providing a potential mechanism for the coordinated expression of snoRNAs and proteins targeting common pathways. Interestingly, we observed that in AML, snoRNA expression correlates minimally with host gene expression. Recent studies in yeast and human brain samples have reported a similar uncoupling of host gene and snoRNA expression.57-59 Indeed, we even observed striking variability in the expression of snoRNAs contained in the same host gene. Mature snoRNAs are produced from host genes by exonucleolytic processing of the debranched intron after splicing.60,61 A recent study suggested that alternative splicing of host genes contributes to the regulation of snoRNA expression and accounts, in part, for the variability in the expression of snoRNAs contained with the same host gene.62 However, in AML, snoRNA expression and alternative splicing correlate minimally. Thus, in AML, other mechanisms besides host gene expression or splicing are contributing to mature snoRNA expression. This may include alterations in snoRNA processing, secondary snoRNA structure stability, maturation, trans-acting protein accumulation factors, and intranuclear trafficking of the maturing snoRNPs to the nucleolus or Cajal body.63 Of note, many snoRNA host genes contain a characteristic terminal oligopyrimidine track in their 5′-untranslated region that has been shown to modulate the differential production of mRNA vs snoRNAs from that host gene.64,65 Given the critical role of snoRNAs in translation, the contribution of these various elements to the regulation of snoRNA expression warrants further study.
As outlined, array-based and qPCR-based approaches do not distinguish between mature snoRNAs and primary mRNA transcripts containing unprocessed snoRNAs. Without robust orthogonal validation technologies for generating gold standard expression values, optimal statistical procedures for expression normalization from count-based sequence data have not been established for snoRNAs. This contrasts with miRNA squencing data, for which qPCR provides robust orthogonal validation that has made it possible to evaluate and optimize expression normalization methods.40 In the absence of a consensus approach for snoRNA data, we used the total count method, which involves normalization of the read count for each snoRNA species for the total number of counts obtained for each experiment. Additional studies will be needed to determine the optimal normalization procedures for sequence data from this intermediate-sized RNA species.
In summary, we developed a small RNA-seq pipeline to quantify snoRNA and other sncRNA expression. We showed that a subset of snoRNAs are regulated in lineage- and development-specific expression. Although genetic alterations that specifically target snoRNA genes in AML appear to be uncommon, a subset of snoRNAs are differentially expressed. The contribution of these differentially expressed snoRNAs to the regulation of normal and malignant hematopoiesis represents an exciting new area of investigation.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported by National Cancer Institute (NCI), National Institutes of Health (NIH) grant PO1 CA101937 (D.C.L. and T.J.L.); Washington University School of Medicine Graduate School of Arts and Sciences/Chancellor's Graduate Fellowship Fund 94028C (W.A.W.); NIH, NCI grant K12CA167540 and Clinical and Translational Award UL1 TR000448 from the NIH, National Center for Advancing Translational Sciences (B.S.W.); and by NIH, NCI grant K08CA190815 and an American Society of Hematology Scholar Award (D.H.S.).
Authorship
Contribution: W.A.W. performed the experiments, analyzed the data, and wrote the manuscript; D.H.S. performed the bioinformatic analysis, analyzed the data, and edited the manuscript; M.T. and N.H. performed some experiments; B.S.W. performed the splicing analysis; T.J.L. provided some reagents, AML samples, and helped with data analysis; and D.C.L. designed and supervised the entire research project and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Washington University School of Medicine, 660 S Euclid Ave, Campus Box 8007, St. Louis, MO 63110; e-mail: danielclink@wustl.edu.
References
- 1.Luo M, Jeong M, Sun D, et al. . Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell. 2015;16(4):426-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raaijmakers MH, Mukherjee S, Guo S, et al. . Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464(7290):852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georgantas RW III, Hildreth R, Morisot S, et al. . CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104(8):2750-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu W, Dooley J, Chung SS, et al. . miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125(14):2206-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35(5):1452-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145-148. [DOI] [PubMed] [Google Scholar]
- 7.Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42(2):701-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubert T, Längst G. Changes in higher order structures of chromatin by RNP complexes. RNA Biol. 2013;10(2):175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel CI, Holley CL, Scruggs BS, et al. . Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14(1):33-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siprashvili Z, Webster DE, Johnston D, et al. . The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48(1):53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu L, Su MY, Maggi LB Jr, et al. . Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J Clin Invest. 2012;122(8):2793-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen Y, Hertzog K, Reish O, et al. . The increased expression of 14q32 small nucleolar RNA transcripts in promyelocytic leukemia cells is not dependent on PML-RARA fusion gene. Blood Cancer J. 2012;2(10):e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liuksiala T, Teittinen KJ, Granberg K, et al. . Overexpression of SNORD114-3 marks acute promyelocytic leukemia. Leukemia. 2014;28(1):233-236. [DOI] [PubMed] [Google Scholar]
- 14.Valleron W, Laprevotte E, Gautier EF, et al. . Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26(9):2052-2060. [DOI] [PubMed] [Google Scholar]
- 15.Teittinen KJ, Laiho A, Uusimäki A, Pursiheimo JP, Gyenesei A, Lohi O. Expression of small nucleolar RNAs in leukemic cells. Cell Oncol (Dordr). 2013;36(1):55-63. [DOI] [PubMed] [Google Scholar]
- 16.Ronchetti D, Todoerti K, Tuana G, et al. . The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012;2(11):e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elghetany MT, Patel J, Martinez J, Schwab H. CD87 as a marker for terminal granulocytic maturation: assessment of its expression during granulopoiesis. Cytometry B Clin Cytom. 2003;51(1):9-13. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Yuan T, Tschannen M, et al. . Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14(1):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ARXIV. 2013;arXiv:1303.3997.
- 20.Harrow J, Frankish A, Gonzalez JM, et al. . GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68-D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorjani H, Kehr S, Jedlinski DJ, et al. . An updated human snoRNAome. Nucleic Acids Res. 2016;44(11):5068-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA. 2006;12(1):15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283(5405):1168-1171. [DOI] [PubMed] [Google Scholar]
- 25.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24(2):158-164. [DOI] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. . Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent WJ, Sugnet CW, Furey TS, et al. . The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(suppl 1):D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. [DOI] [PubMed] [Google Scholar]
- 30.Partek Flow. Version 3.0. St. Louis, MO: Partek Inc.; 2014. [Google Scholar]
- 31.Eisenhart C. The assumptions underlying the analysis of variance. Biometrics. 1947;3(1):1-21. [PubMed] [Google Scholar]
- 32.Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33(3):243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 34.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley TJ, Miller C, Ding L, et al. ; Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94(2):83-88. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Head SR, Komori HK, LaMere SA, et al. . Library construction for next-generation sequencing: overviews and challenges. Biotechniques. 2014;56(2):61-64, 66, 68 passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillies MA, Rau A, Aubert J, et al. ; French StatOmique Consortium. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14(6):671-683. [DOI] [PubMed] [Google Scholar]
- 40.Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA-Seq data. RNA. 2012;18(6):1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garmire LX, Subramaniam S. The poor performance of TMM on microRNA-Seq. RNA. 2013;19(6):735-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galiveti CR, Rozhdestvensky TS, Brosius J, Lehrach H, Konthur Z. Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA. 2010;16(2):450-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez-Dominguez JR, Hu W, Gromatzky AA, Lodish HF. Long noncoding RNAs during normal and malignant hematopoiesis. Int J Hematol. 2014;99(5):531-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dostalova Merkerova M, Krejcik Z, Votavova H, Belickova M, Vasikova A, Cermak J. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. Eur J Hum Genet. 2011;19(3):313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Smith AJ, Purmann C, Walters RG, et al. . A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet. 2009;18(17):3257-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters J. Prader-Willi and snoRNAs. Nat Genet. 2008;40(6):688-689. [DOI] [PubMed] [Google Scholar]
- 47.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311(5758):230-232. [DOI] [PubMed] [Google Scholar]
- 48.Stadtfeld M, Apostolou E, Ferrari F, et al. . Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44(4):398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol Cell. 2007;26(3):311-321. [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Fredrick K. Intersubunit bridges of the bacterial ribosome. J Mol Biol. 2016;428(10 pt B):2146-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS One. 2007;2(1):e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11(2):425-435. [DOI] [PubMed] [Google Scholar]
- 53.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28(6):965-977. [DOI] [PubMed] [Google Scholar]
- 54.Dönmez G, Hartmuth K, Lührmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10(12):1925-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17(19):5783-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64(1):897-934. [DOI] [PubMed] [Google Scholar]
- 57.Zhang B, Han D, Korostelev Y, et al. . Changes in snoRNA and snRNA Abundance in the Human, Chimpanzee, Macaque, and Mouse Brain. Genome Biol Evol. 2016;8(3):840-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ge J, Crosby SD, Heinz ME, Bessler M, Mason PJ. SnoRNA microarray analysis reveals changes in H/ACA and C/D RNA levels caused by dyskerin ablation in mouse liver. Biochem J. 2010;429(1):33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He H, Cai L, Skogerbø G, et al. . Profiling Caenorhabditis elegans non-coding RNA expression with a combined microarray. Nucleic Acids Res. 2006;34(10):2976-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18(19):5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20(2):441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lykke-Andersen S, Chen Y, Ardal BR, et al. . Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014;28(22):2498-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiss T, Fayet E, Jády BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407-417. [DOI] [PubMed] [Google Scholar]
- 64.de Turris V, Di Leva G, Caldarola S, Loreni F, Amaldi F, Bozzoni I. TOP promoter elements control the relative ratio of intron-encoded snoRNA versus spliced mRNA biosynthesis. J Mol Biol. 2004;344(2):383-394. [DOI] [PubMed] [Google Scholar]
- 65.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.