Abstract
Background
Pseudomonas aeruginosa is an opportunistic pathogen problematic in causing nosocomial infections and is highly susceptible to development of resistance to multiple antibiotics. The gene encoding methionyl-tRNA synthetase (MetRS) from P. aeruginosa was cloned and the resulting protein characterized.
Methods
MetRS was kinetically evaluated and the KM for its three substrates, methionine, ATP and tRNAMet were determined to be 35, 515, and 29 μM, respectively. P. aeruginosa MetRS was used to screen two chemical compound libraries containing 1690 individual compounds.
Results
A natural product compound (BM01C11) was identified that inhibited the aminoacylation function. The compound inhibited P. aeruginosa MetRS with an IC50 of 70 μM. The minimum inhibitory concentration (MIC) of BM01C11 was determined against nine clinically relevant bacterial strains, including efflux pump mutants and hypersensitive strains of P. aeruginosa and E. coli. The MIC against the hypersensitive strain of P. aeruginosa was 16 μg/ml. However, the compound was not effective against the wild-type and efflux pump mutant strains, indicating that efflux may not be responsible for the lack of activity against the wild-type strains. When tested in human cell cultures, the cytotoxicity concentration (CC50) was observed to be 30 μg/ml. The compound did not compete with methionine or ATP for binding MetRS, indicating that the mechanism of action of the compound likely occurs outside the active site of aminoacylation.
Conclusion
An inhibitor of P. aeruginosa MetRS, BM01C11, was identified as a flavonoid compound named isopomiferin. Isopomiferin inhibited the enzymatic activity of MetRS and displayed broad spectrum antibacterial activity. These studies indicate that isopomiferin may be amenable to development as a therapeutic for bacterial infections.
Keywords: Protein synthesis, in vitro screening, tRNA aminoacylation, drug discovery, antibiotics, Pseudomonas.
INTRODUCTION
Antibiotic resistance is an issue of growing public concern. The rise of so-called ‘superbugs’ or bacteria that are resistant to multiple classes of antibiotics has marked the beginning of an era where current antibiotic treatments are no longer sufficient [1]. Pseudomonas aeruginosa, a Gram-negative pathogen, is a member of this group of bacteria and has a high intrinsic resistance to antibiotics due to numerous efflux systems and low permeability of its membrane. This bacterial pathogen is the leading cause of pulmonary infections in cystic fibrosis patients and a significant contributor to hospital acquired infections [2].
Methionyl-tRNA synthetase (MetRS), encoded by the metS gene, binds three natural substrates (ATP, methionine, and tRNA) and catalyzes the reaction resulting in the aminoacylation of tRNAMet. MetRS is a class I aminoacyl tRNA synthetase (aaRS) characterized by an active site structure formed by a Rossman fold containing two structural motifs (HIGH and KMSKS) [3]. MetRS is further grouped along with isoleucyl-, valyl-, leucyl-, cysteinyl- and arginyl-tRNA synthetases (IleRS, ValRS LeuRS, CysRS and ArgRS) into a subclass Ia based on sequence conservation. A unique property of MetRS is its ability to recognize and charge two tRNAMet substrates: the elongator tRNAMet and the initiator tRNAMet. MetRS thus plays a crucial role during the initiation phase and the elongation phase of protein biosynthesis.
Most bacterial isolates contain one of the two major forms of MetRS (MetRS1 and MetRS2) that are distinct at the amino acid sequence level and are encoded by two different genes, metS1 and metS2, respectively. MetRS1 is the form typically found in Gram-positive bacteria while MetRS2 appears to be the product of archaea and eukaryotic organisms as well as the form found in Gram-negative bacteria [4]. There are exceptions to this in that certain Gram-positive bacteria, in particular Streptococcus pneumoniae, contain genes encoding both forms of MetRS, which is likely the result of acquisition through horizontal gene transfer [5]. In addition, structural studies have divided MetRS into four additional families based on the number of Zn-binding knuckle motifs and the number of Zn2+ atoms bound to the motifs. MetRS2 contains two Zn-binding knuckles and either two Zn2+ (A family) or one Zn2+ atom bound (B family). MetRS1 orthologs contain only one Zn-binding knuckle motif and either one (C family) or no Zn2+ atoms bound (D family). Sequence analysis indicates that MetRS from P. aeruginosa is of the MetRS2 form and belongs to the B structural family based on Zn-binding knuckle motifs.
MetRS from P. aeruginosa was purified and the kinetic parameters (KM, Vmax and kcat) for interaction with its natural substrates were experimentally determined. It was then developed into a screening platform using scintillation proximity assay (SPA) technology [6] and used to screen 1690 natural and synthetic compounds for inhibitory activity. One compound was identified that inhibited the activity of the enzyme. This compound was further characterized for inhibition of enzymatic activity and bacterial growth, mode of action, mechanism of inhibition and toxicity issues.
METHODS AND MATERIALS
Materials
All chemicals were obtained from Fisher Scientific (Pittsburg, PA). Radioactive isotopes, SPA beads and 96-well screening plates were from PerkinElmer (Waltham, MA). E. coli tolC mutant, P. aeruginosa PAO200 (efflux pump mutant) and P. aeruginosa hypersensitive (ATCC® 35151™) strains were a kind gift from Urs Ochsner (Crestone Pharma, Boulder, CO). All other bacteria were from American Type Culture Collection (ATCC) (Manassas, VA). The synthetic compound library was from TimTec LLC (Newark, DE) and the natural product library was from MicroSourceDiscovery Systems, Inc. (Gaylordsville, CT). Compounds were supplied as 10 mM stocks dissolved in dimethyl sulfoxide (DMSO), stored at −20 °C and thawed immediately before analysis. The compounds have an average purity of 95%, and the minimum purity is at least 90%.
Cloning and Purification of P. aeruginosa MetRS
The gene encoding P. aeruginosa MetRS was obtained through PCR amplification (MJ Mini Thermo Cycler, Bio-Rad, Hercules, CA) using P. aeruginosa PAO1 (ATCC 47085) genomic DNA as a substrate. A forward primer (5′-ATATGCTAGCTCCGAACCACGCAAGATC-3′), designed to add an NheI restriction site to the 5′ end of the gene and the reverse primer (5′-CTCTAAGCTTTTACTTGACGCGCTGGC-3′) which was designed to add a HindIII restriction site to the 3′ end of the gene, were used in the PCR. The PCR product was inserted into a pET-28b(+) plasmid (Novagen) digested with NheI/HindIII. The recombinant plasmid was transformed into E. coli Rosetta 2(DE3) Singles Competent Cells (EMD Millipore, Danvers, MA).
The E. coli bacterial cultures were grown in Terrific Broth containing 25 μg/mL of kanamycin and 50 μg/mL of chloramphenicol at a temperature of 37 °C to an optical density (A600) of 0.6–0.8. The over-expression of P. aeruginosa MetRS in the cultures was induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a concentration of 0.25 mM. Growth of the culture was continued for 2 hours post-induction, and cells were harvested using centrifugation (10,000 g, 30 min, 4 °C). Fraction I lysates were prepared as previously described [7]. P. aeruginosa MetRS was purified to more than 98% homogeneity as previously described [6].
Gel Electrophoresis and Protein Analysis
Proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4–12% polyacrylamide precast gradient gels (Novex NuPAGE; Invitrogen, Grand Island, NY) with 3-(N-morpholino) propanesulfonic acid (MOPS) as the running buffer (Invitrogen). EZ-Run Rec Protein Ladder (Fisher Scientific) was used as a protein standard. Coomassie Protein Assay Reagent (Thermo Scientific, Waltham, MA) was used to determine protein concentrations with bovine serum albumin as a standard [8].
Timed tRNA Aminoacylation Assays
Aminoacylation was measured using filter binding assays (50 μL) containing 50 mM Tris-HCl (pH 7.5), 7.5 mM MgCl2, 2.5 mM ATP, 1 mM dithiothreitol (DTT), 75 μM [3H] methionine, and 0.1 μM P. aeruginosa MetRS as previously described [9]. Assays were stopped at time intervals between 1 and 5 min. The tRNA concentrations were varied in six different sets of assays and contained 10, 30, 50, 70, 90 or 110 μM total tRNA (0.5, 1.5, 2.5, 3, 4, 5 μM tRNAMet). From the timed assays, initial velocities were measured and the kinetic parameters (KM and kcat) were determined by fitting the data to the Michaelis-Menten steady-state model using XLfit (IDBS).
Phosphate Exchange Reactions
ATP:PPi exchange reactions (100 μl) were carried out at 37 ºC in 50.0 mM Tris-HCl (pH 7.5), 10 mM KF, 2 mM [32P]PPi, 7.5 mM MgCl2, 0.1 μM P. aeruginosa MetRS as previously described [10]. The reactions were stopped at 1, 2, 3, 4, and 5 min intervals. In the reactions in which the concentration of ATP was varied (25, 50, 100, 200, 300, and 400 μM), the methionine concentration was held constant at 2 mM; alternatively, when the concentration of the amino acid was varied (2.5, 5, 10, 20, 30 μM), the ATP concentration was at 2 mM. Initial velocities were determined for exchange of PPi and the kinetic parameters (KM, Vmax and Kcat) for the interactions of P. aeruginosa MetRS with ATP and methionine were determined and fit to the Michaelis-Menten steady-state model using XLfit (IDBS).
Chemical Compound Screening
In compound screening assays, the aminoacylation of tRNA was monitored using SPA technology as previously described [10]. The reactions were carried out in 96-well microtiter plates (Costar). Compounds were dissolved in 100% DMSO to a concentration of 3.3 mM, and the final concentration of compounds in the screening assay was 132 μM. The concentration of P. aeruginosa MetRS in the screening assays was set at 0.1 μM. To determine IC50 values, the compounds were serially diluted into the assay at concentration ranging from 200 μM down to 0.4 μM.
Microbiological Assays
Minimum inhibitory concentration (MIC) for the hit compound was obtained using broth microdilutions following Clinical and Laboratory Safety Standards Institute (CLSI) guideline M7-A7 [11]. MIC values were determined for E. coli (ATCC 25922), E. coli tolC mutant, Enterococcus faecalis (ATCC 29212), Haemophilus influenzae (ATCC 49766), P. aeruginosa (ATCC 47085), P. aeruginosa PAO200 (efflux pump mutant), P. aeruginosa hypersensitive strain (ATCC 35151), Staphylococcus aureus (ATCC 29213), and Streptococcus pneumonia (ATCC 49619). Quality control (QC) of MIC data and culture purity was maintained by MIC determination for antibiotics specific for each bacterial strain [10] as described in Table 1: Summary of Etest® Performance, Interpretive Criteria and Quality Control Ranges (Biomerieux).
Table 1.
P. aeruginosa MetRS amino acid conservation (average) compared with homologs from different phyla.
| MetRS | Similarity/Identity (%) |
|---|---|
| Gammaproteobacteria | 73/61 |
| Betaproteobacteria | 58/48 |
| Alphaproteobacteria | 31/21 |
| Delta-, Epsilonproteobacteria | 37/24 |
| Terrabacteria (not including Firmicutes) | 35/23 |
| Firmicutes | 40/25 |
| Spirochaetales | 33/22 |
| Chlamydiae | 36/25 |
| FCB | 43/28 |
| Human mitochondria | 28/18 |
| Human cytosolic | 29/18 |
Time-kill studies were performed to determine mode of inhibition of the hit compound using H. influenzae and S. aureus based on the MIC assay results, according to the CLSI document M26-A [12]. Growth media was Haemophilus Test Broth and Trypticase Soy Broth from Remel (Lenexa, KS).
Cytotoxicity Testing
To determine the effect of the hit compound on the growth of human cell cultures, in vitro cytotoxicity testing was carried out as described using human embryonic kidney 293 cells (HEK-293) [6]. The Trevigen TACS® MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Cell Proliferation Assay Kit (Gaithersburg, MD) was used to analyze the impact of the hit compound on human cell proliferation and/or viability. The control staurosporine was serially diluted in assays from 1 to 0.001 μg/ml and the hit compound was tested in assays ranging from 100 μg/ml to 1 μg/ml.
Binding Mode Assay
To determine the mechanism of action of the hit compound with respect to ATP and methionine, IC50 values were determined using SPA assays as described above. Final compound concentrations in each IC50 reaction ranged from 200 to 0.4 μM. Competition with ATP was determined at 25, 50, 100, 250, 500, 1000 μM ATP while the methionine concentration was 75 μM. Competition with the amino acid was determined at 25, 50, 100, 200, 300 μM methionine while the ATP concentration was 2.5 mM. Positive controls contained only DMSO (2 μl) without compound.
RESULTS AND DISCUSSION
Sequence Analysis
The crystal structures of bacterial MetRS from E. coli [13, 14] and Thermus thermophilus [15] and Brucella melitensis [16] in an apo-form and bound to its substrates have been solved. Also the MetRS structure from the archaea Pyrococcus abyssi is known [17]. The amino acid sequence of MetRS from P. aeruginosa exhibits a high degree of conservation when compared with corresponding enzymes of the γ-subdivision of the proteobacteria resulting in 73% sequence similarity and 61% sequence identity (Table 1). The level of conservation is reduced considerably when compared with MetRS from other phyla and when compared with either human mitochondrial MetRS (hmMetRS), or human cytosolic MetRS (hcMetRS), there is only 18% conservation of the amino acid residues.
MetRS binds methionine (Met) through an induced-fit mechanism [14] and the active site residues, Y15, W253 and H301 (E. coli numbering), that have been shown to directly interact with the amino acid substrate are contained within the Rossmann fold in the catalytic domain and are strictly conserved between E. coli, T. thermophilus and P. aeruginosa MetRS (Fig. 1). These residues, along with others, make up the methionine binding pocket of MetRS, in which the amino acid residues are conserved between E. coli and P. aeruginosa MetRS, but greater variation occurs in the T. thermophilus enzyme. MetRS must discriminate between the binding of methionine and homocysteine (a precursor of cysteine). Two amino acids, L13 and Y260, that are located in the methionine binding pocket form H-bonds with the δ-sulfur atom of methionine and, along with W253 and Y15, create the binding pocket surrounding the side-chain, resulting in specificity for methionine [18][14]. L13 and Y260 are conserved in MetRS from E. coli and P. aeruginosa.
Fig. 1.
Sequence alignment of P. aeruginosa MetRS and close analogs. The protein sequences were downloaded from the National Center for Biotechnology Information (NCBI). Abbreviations are: Ec, E. coli; Pa, P. aeruginosa; Tth, T. thermophiles. Accession numbers for MetRS protein sequences of E. coli, P. aeruginosa, and T. thermophiles are P00959, NP_252172 and P23395, respectively. Sequence alignments were performed using Vector NTI Advance (TM) 11.0 (Invitrogen). Identical amino acid residues are white on black and similar residues are black on grey. The positions of the HIGH and KMSKS motifs are indicated. Motifs 1–4 containing zinc binding knuckles in E. coli and P. aeruginosa are indicated above the motifs using solid brackets and zinc binding knuckles in T. thermophilus are indicated below the motifs using dashed brackets. Amino acid residues that directly interact with Met are indicated with a cross (✖), and additional residues that form the amino acid binding pocket are indicated with a bar (▬). Amino acids that directly interact with ATP (★) and tRNA (●) are indicated. Amino acid residues that function in proofreading cognate tRNA are indicated by open circles (○).
Binding of the ATP is stabilized by several residues. In E. coli MetRS, V326 and H23 appear to form H-bonds with the adenine base [14]. In both P. aeruginosa and T. thermophilus, the valine at position 326 is replaced by the similar leucine. The ribose sugar of ATP is stabilized by interactions with E27, G294, and D296. The amino acids at positions 294 and 296 are conserved in all three MetRS enzymes. However, the glutamic acid at position 27 is replaced with a threonine in T. thermophilus MetRS. The two histidine residues in the HLGH (HIGH) motif interact with the adenosine moiety of the ATP and H24 likely stabilizes the phosphate in the aminoacyladenylate [14]. These amino acids are also strictly conserved in the three MetRS proteins.
A number of amino acids have been shown to interact with and proofread the cognate tRNA [13]. In sequence alignments (Fig. 1) these amino acids are fairly well conserved between MetRS from E. coli, P. aeruginosa and T. thermophilus. At only one position, D456 (E. coli numbering), is there a lack of conservation. The aspartic acid in E. coli is replaced with an alanine and an asparagine in P. aeruginosa and T. thermophilus, respectively. This is interesting since D456 in E. coli is one of two amino acid residues behaving as “anti-determinants” contributing to rejection of non-methionine anticodon containing tRNAs [19].
MetRS is a known zinc metalloprotein, in which binding of Zn2+ is required for enzymatic activity [20][21]. Sequence alignment of the amino acids from various MetRS proteins identified four families in context of the number of zinc-binding ligands and the number of Zn2+ bound [13]. E. coli contains four zinc-binding motifs (1–4), but only motif 2 and motif 3 actively bind Zn2+ (Fig. 1). Alternatively, T. thermophilus MetRS contains only two zinc-binding motifs and binds one Zn2+ [15]. In the alignment in Fig. 1, MetRS from P. aeruginosa aligns more closely with that of E. coli, containing all four of the zinc-binding motifs, and with the lack of cysteine residues in motifs 1 and 3, like E. coli MetRS, likely only binds one Zn2+ in what is considered the distal knuckle.
Expression and Characterization of MetRS from P. aeruginosa
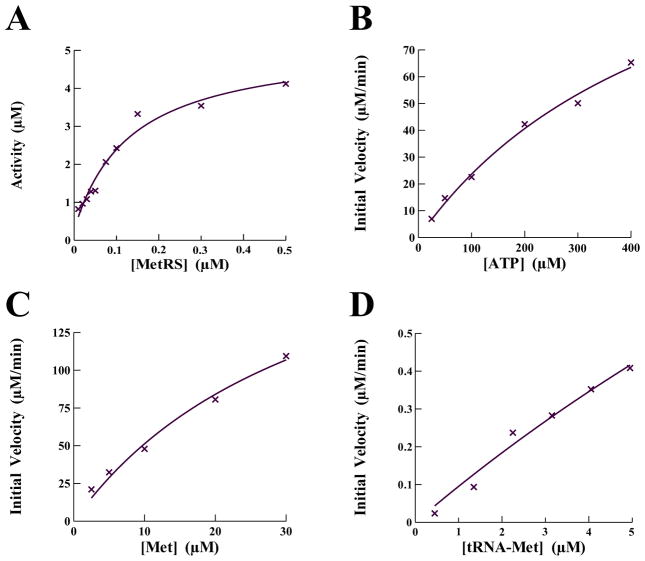
Methionyl-tRNA synthetase from P. aeruginosa was cloned, overexpressed in E. coli and purified to greater than 98% homogeneity as visualized by SDS-PAGE and shown to be active in the aminoacylation assay (Fig. 2A). The primary physiological function of an aminoacyl tRNA synthetase (aaRS) is the catalytic attachment of an amino acid to its cognate tRNA. This reaction occurs via two distinct enzymatic steps catalyzed without the amino acid substrate being released from the enzyme. In the first step the enzyme catalyzes the condensation of the amino acid and another substrate, ATP, forming an aminoacyl adenylate intermediate with the release of an inorganic pyrophosphate (PPi). This reaction is reversible in the absence of cognate tRNA and can be used to monitor the interaction of the enzyme with the amino acid and ATP. The kinetic parameters governing the interaction of P. aeruginosa MetRS with methionine and ATP were determined using the ATP:PPi exchange reaction as described under the “Methods and Material” section. To determine the kinetic parameters with respect to ATP, the amino acid concentration was held constant while ATP concentrations were varied between 25 and 400 μM. Alternatively, to determine the kinetic parameters with respect to the amino acid, the ATP concentration was held constant while the methionine concentrations were varied between 2.5 and 30 μM. The initial velocities were determined and fit to the Michaelis-Menten steady-state model using XLfit (IDBS) (Fig. 2B–C). The kinetic parameters, KM, kcat, and kcat/KM, for interaction of P. aeruginosa MetRS with ATP were determined to be 515 μM, 24.2 sec−1, and 0.05 s−1μM−1, respectively (Table 2). The same kinetic parameters for the interaction with methionine were 35 μM, 37.2 sec−1, and 1.1 s−1μM−1, also respectively. These values are similar to the kinetic interactions previously observed for E. coli MetRS with ATP (KM = 528 μM, kcat = 74 μM) and methionine (KM = 21 μM, kcat = 74 μM) [22].
Fig. 2.
Determination of activity and the kinetic parameters governing the catalytic enzymology of P. aeruginosa MetRS with the substrates tRNAMet, ATP, and methionine. (A) Titration of P. aeruginosa MetRS into the aminoacylation assay to determine activity and the optimal concentration in compound screening assays. Initial velocities for the interaction of P. aeruginosa MetRS with (B) ATP, (C) methionine and (D) tRNAMet were determined and the data were fit to a Michaelis-Menten steady-state model using XLfit (IDBS) to determine KM and Vmax.
Table 2.
The kinetic parameter governing the interaction of MetRS with its substrates.
| tRNA | Methionine | ATP | ||||||
|---|---|---|---|---|---|---|---|---|
| KM (μM) | kcat (s−1) | kcat/KM (s−1 μM) | KM (μM) | kcat (s−1) | kcat/KM (s−1 μM) | KM (μM) | kcat (s−1) | kcat/KM (s−1 μM) |
| 29 | 1.93 | 0.07 | 35 | 37.2 | 1.1 | 515 | 24.2 | 0.05 |
In the second step of the reaction the methionine is released from the aminoacyl adenylate and transferred to the acceptor end of the cognate tRNAMet. This step tends to be the rate limiting step and the turnover number for the entire aminoacylation reaction is well below that for formation of the aminoacyl adenylate. In these reactions the concentration of Met and ATP were saturating and the concentration of tRNAMet was varied between 0.5 and 5 μM. The initial velocities were determined and fit to the Michaelis-Menten steady-state model (Fig. 2D) and the KM, kcat, and kcat/KM values for tRNA were determined to be 29.0 μM, 1.90 sec−1, and 0.07 s−1μM−1 respectively.
Chemical Compound Screening
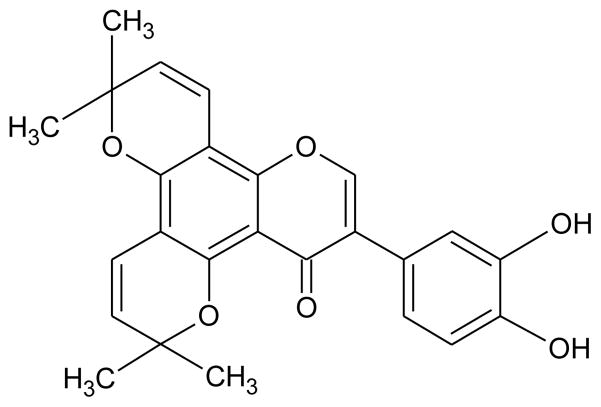
The chemical compound screening assays were based on the aminoacylation assay and modified for monitoring using SPA technology. Initially, the non-enzymatic components of the aminoacylation reaction were individually titrated into the assay to ensure optimal concentrations for maximum signal. Next, MetRS was titrated into the assay to determine the concentration yielding maximum sensitivity to enzymatic inhibition; this resulted in a screening concentration of 0.1 μM (Fig. 2A). Using this optimized assay, two chemical compound libraries were screened against the activity of P. aeruginosa MetRS. One library contained 800 compounds composed of natural products including simple and complex oxygen heterocycles, alkaloids, sesquiterpenes, diterpenes, pentacyclic triterpenes, and sterols from Microsource Discovery Systems (Gaylordsville, CT) [9]. The other library contained 890 low molecular weight synthetic compounds with scaffolds based on known anti-bacterial, anti-fungoid, and anti-microbial agents from TimTec LLC (Newark, DE) [23]. The chemical compounds (3.3 mM) were dissolved in DMSO and screening reactions (50 μl) contained 2 μl of compound resulting in a final compound concentration of 132 μM. This also resulted in 4% DMSO in each reaction. To determine the effect of DMSO on the activity of MetRS, it was titrated into the assay up to 10% final concentration. There was no decrease in enzymatic activity observed at 4% DMSO (data not shown). The screening reactions were very robust with an average signal to background ratio of 10:1. The Z′ and Z factors across all plates averaged 0.7 and 0.5, respectively. Screening assays were performed as single point assays and compounds observed to inhibit at least 50% of enzymatic activity were confirmed in additional assays performed in triplicate. The initial screen resulted in eleven confirmed hit compounds. These compounds were analyzed to determine IC50 values. Only two compounds, BM01C11 (70 μM) and BM01F04 (8.3 μM), both from the natural compound library, were observed to have an IC50 value below 200 μM. BM01F04 was identified as a compound named hieracin, also called Tricetin. The PubChem BioAssay database was searched for biological activity of hieracin and it was found to have inhibitory activity against a number of eukaryotic and viral targets as well as antibacterial activity [24]. This compound will not be analyzed further. BM01C11 was identified as a compound named isopomiferin (Fig. 3). The Pub-Chem BioAssay database was also searched for biological activity of isopomiferin and a close conformer, isopomiferin dimethyl ether. There was no inhibitory bioactivity found for this compound and it will be investigated further.
Fig. 3.
The chemical structure of the hit compound BM01C11 (isopomiferin).
Microbiological Assays
Isopomiferin was tested in broth microdilution assays to determine minimum inhibitory concentrations (MIC) against nine pathogenic bacteria (Table 3). The organisms used to determine the MICs were chosen using Clinical Laboratory Standards Institute guidelines [11] along with efflux and hypersensitive mutant strains of E. coli and P. aeruginosa. There was no inhibition of wild-type E. coli or P. aeruginosa observed at concentrations below 128 μg/ml. There was also no inhibitory activity observed with the efflux pump mutant of P. aeruginosa and only a slight decrease in the MIC with the efflux pump mutant of E. coli. However, isopomiferin inhibited the growth of cultures of the hypersensitive strain of P. aeruginosa with a moderately low MIC of 16 μg/ml, indicating that efflux may not be responsible for the lack of activity against the wild-type strains. The hypersensitive strain of P. aeruginosa is susceptible to numerous antibiotics because of an increase in outer membrane permeability partially due to a malfunction in porin protein F [25]. Therefore, the inability of isopomiferin to effect the growth of wild-type and efflux pump mutant strains of P. aeruginosa may be a permeability issue. The other Gram-negative bacteria, Haemophilus influenzae, does not contain a porin protein F and was susceptible to the compound. Isopomifer-in inhibited growth of cultures of the Gram-positive bacteria, Enterococcus faecalis and Staphylococcus aureus, but the compound did not inhibit the growth of Streptococcus pneumoniae at a concentration lower than 128 μg/ml. This may be due to the fact that S. pneumoniae contains two genes (metS1and metS2) encoding two distinctly different forms of MetRS [4] and both forms of the protein may not be compromised by the compound.
Table 3.
Minimum inhibitory concentration of the hit compound against clinically important bacterial.
| Bacteria | Isopomiferin |
|---|---|
| E. coli (ATCC 25922) | 128a |
| E. coli tolC (efflux mutant) | 64 |
| E. faecalis (ATCC 29212) | 32 |
| H. influenzae (ATCC 49766) | 32 |
| P. aeruginosa (ATCC 47085) | 128 |
| P. aeruginosa PAO200 (efflux mutant) | 128 |
| P. aeruginosa (hypersensitive) | 16 |
| S. aureus (ATCC 29213) | 32 |
| S. pneumoniae (ATCC 49619) | >128 |
The MIC (μg/ml) against each of the bacteria was determined in three separate sets of assays.
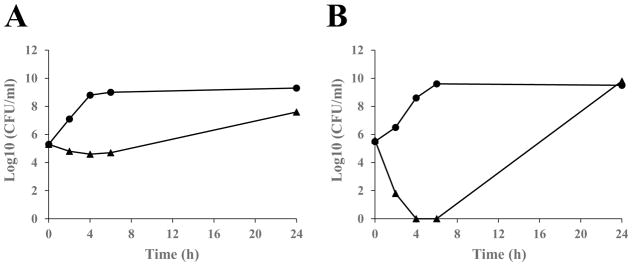
Next, to determine the mode of inhibition in bacterial cultures, time kill studies were performed with isopomiferin. Based on MIC results, isopomiferin was tested against cultures of H. influenzae and S. aureus. The cultures were analyzed between 0 and 24 hours and contained compounds at four times the MIC. H. influenzae grown in the presence of isopomiferin was observed to have constant growth in the presence of compounds, but a decrease of 2 to 5 log10 colony forming units (CFU) compared to the control during the twenty-four hour period (Fig. 4). This is indicative of bacteriostatic inhibition and occurs, in this case, because inhibition of aminoacylation by an amino-acyl tRNA synthetase produces the same results as amino acid starvation, and likely activates the stringent response, which results in static bacterial growth [26]. The effect of isopomiferin on cultures containing S. aureus were observed to be bactericidal and no CFUs were observed in the first six hours of growth. However, increased growth was observed at 24 hours, possibly because the compounds lost potency over extended time periods. S. aureus contains MetRS1 and H. influenzae contains MetRS2, and the different forms of the synthetase may contribute to the difference observed in the mode of inhibition. The reason for the bactericidal activity is not clear, but many aaRS proteins have secondary roles that could be affected by the compound leading to cell death. MetRS contains an editing function which prevents the misaminoacylation of tRNAMet with homocysteine (Hcy). Blocking this editing function could result in the incorporation of Hcy into nascent proteins, which in turn would inactivate the biological activity of these proteins, possibly leading to cell death [27]. Alternatively, inhibition of the editing function could result in the misaminoacylation of the initiator tRNAMet, thus blocking initiation of protein synthesis.
Fig. 4.
Time-kill kinetics of the hit compound isopomiferin against (A) H. influenzae and (B) S. aureus. Isopomiferin was added to bacterial cultures at 4×MIC. Samples were analyzed by plating and determination of colony forming units (CFU) at 0, 2, 4, 6, and 24 h. Filled circles (●) represent control cultures containing no compound but an equal amount of DMSO. Filled triangles (▲) represent cultures containing isopomiferin.
Cytotoxicity Assays
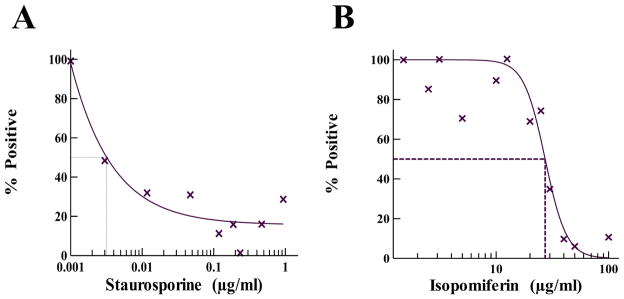
Mammalian cells contain a cytoplasmic as well as a mitochondrial form of MetRS, which have many structural similarities with bacterial MetRS. This presents the potential that a compound which is active in inhibition of bacterial MetRS may have cytotoxic effects on eukaryotic cells. It was therefore important to analyze the potential for toxicity by isopomiferin in eukaryotic cells. Predicting toxicity of chemicals using human cell systems instead of whole animal systems is a viable first step in drug discovery, therefore isopomiferin was tested for cytotoxicity in human embryonic kidney 293 (HEK-293) cell cultures using the Trevigen TACS® MTT Cell Proliferation Assay Kit (Fig. 5). Staurosporine, a potent inhibitor of human cell cultures [28], was used as a comparator in the studies. Staurosporine was observed to inhibit cell growth with a CC50 of 0.003 μg/ml. MTT assays were performed at isopomiferin concentrations between 1 and 100 μg/ml for 24 hours. Isopomiferin exhibited a CC50 of near 30 μg/ml, which when compared to the toxicity of many antibiotics appears problematic for therapeutic use. However, the CC50 of isopomiferin is 10,000-fold higher than that of staurosporine, and when compared with other antibiotics, erythromycin and tetracycline, the toxicity profile was similar [29].
Fig. 5.
Determination of the toxicity of isopomiferin in human cell cultures. MTT assays were performed for 24 hours under standard tissue culture conditions as described in ‘Materials and Methods’. The control (A) staurosporine was serially diluted in assays from 1 to 0.001 μg/ml to determine CC50 (0.003 μg/ml). Isopomiferin (B) was diluted in the assays from 100 to 1 μg/ml. The “% Positive” indicates the percent of activity observed relative to activity in assays where only DMSO was added to the assay in the absence of compound. The data points represents an average value of assays carried out in triplicate. The curve fits and CC50 values were determined using the Sigmoidal Dose-Response Model in XLfit 5.3 (IDBS).
Mechanism of Action
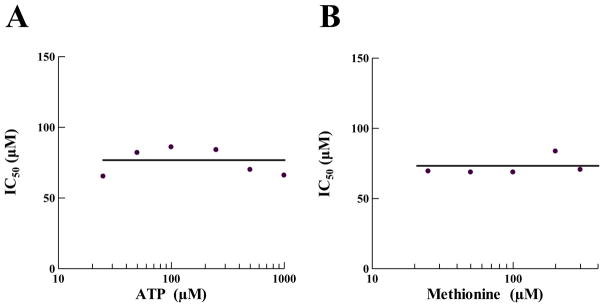
The binding sites of both ATP and methionine are located in the Rossman fold active site region of MetRS. Small molecules primarily used in drug screening have a molecular mass that is similar to both ATP and the amino acid. It is therefore prudent as an initial determination of the mechanism of action to ascertain if a hit compound interferes with binding of one or both of these substrates. An inhibitory compound that competes with ATP for binding may be a poor candidate for drug development, since ATPase enzymes are prevalent in both bacterial and eukaryotic cells. However, since methionine uniquely binds MetRS in an induced fit mechanism, competitive inhibition of this mechanism could be very specific. To understand the mechanism of action relative to substrate binding, isopomiferin was tested in competition assays with varying amounts of ATP or methionine (Fig. 6). The compound was not observed to compete with either ATP or methionine for binding, indicating that the binding region of isopomiferin is likely outside the active site for aminoacylation. In the assays, as the concentration of either ATP or methionine was increased the IC50 remained constant, indicating a non-competitive form of inhibition [30].
Fig. 6.
Determination of the mechanism of action of isopomiferin relative to (A) ATP and (B) methionine. IC50s were determined in the aminoacylation assay using the SPA format. To determine isopomiferin inhibition relative to ATP, the IC50s were determined at six different ATP concentrations ranging from 25 to 1000 μM. To determine inhibition relative to methionine, the IC50s were determined at five different methionine concentrations ranging from 25 to 300 μM. The data were fit to the Sigmoidal Dose-Response model using XLfit (IDBS).
CONCLUSION
The amino acid sequence of P. aeruginosa MetRS, when compared to homologs obtained from a variety of bacterial species was, as expected, more similar (greater than 60% sequence conservation) to that of other MetRS proteins from the gamma subdivision of the proteobacteria (Table 1). In particular, the critical amino acid residues in E. coli MetRS are strictly conserved in P. aeruginosa MetRS. However, when compared with human cytosolic or mitochondrial MetRS, there is little conservation of the primary sequence, indicating that a compound that inhibits the function of the bacterial MetRS may be ineffective against the human forms of MetRS. In the present work, P. aeruginosa MetRS was developed into a screening platform based on scintillation proximity assay technology and used to screen two chemical compound libraries (1690 compounds) for inhibitors of aminoacylation activity. The screening assays were robust and eleven compounds from the compound libraries were identified and confirmed as hit compounds. Two of the compounds, isopomiferin (BM01C11) and hieracin (BM01F04), both natural products, inhibited MetRS activity with IC50s of 70 μM and 8.3 μM, respectively. Hieracin was discarded due to promiscuous inhibitory activity observed in bioassay data (National Center for Biotechnology Information. PubChem Compound Database; CID=5281701, https://pubchem.ncbi.nlm.nih.gov/compound/5281701 (accessed Feb. 9, 2017). Isopomiferin displayed inhibition against bacterial growth from a panel of both Gram-negative and Gram-positive bacteria. Moderate activity was observed against S. aureus, E. faecalis, and H. influenzae. However, the wild-type and efflux pump mutant strains of E. coli and P. aeruginosa were not affected by the compounds, indicating that the mechanism of resistance may not be due to efflux. Cultures of the antibiotic hypersensitive strain of P. aeruginosa were inhibited by isopomiferin. This suggest that the lack of inhibitory activity against the wild-type and efflux pump mutant strains may be due to an inability to gain entry into the cells.
When isopomiferin was tested in human cell cultures, the CC50 was observed to be near 30 μg/ml. When compared with a small subset of antibiotics, this is not unusual. However, when compared with the majority of antibiotics in use, this is a relatively high toxicity level. The toxicity issue with isopomiferin will need to be addressed in future structure activity relationship (SAR) studies. Isopomiferin did not interfere with MetRS binding of methionine or with ATP, and would appear to inhibit the activity of MetRS by another mechanism. It is possible that the compound may interact with the Zn++ binding motif interfering with coordination of the metal due to the catechol moiety or it may bind at an unknown site thereby inducing a conformational change in the protein leading to inactivation of enzymatic function. In this era of multi-drug resistant bacteria, compounds that interact with molecular targets differently, possibly with a different mechanism of action, or with different binding sites, would be advantageous. This is evidenced by the low level of clinical resistance observed, even after many years of use, against mupirocin that occurs by mutations in the IleRS protein itself. Mupirocin blocks the binding site of the unique aminoacyl adenylate intermediary molecule [31]. Higher levels of resistance come from the acquisition of the mupA gene, normally from outside sources [32]. Also, the structure of isopomiferin offers several avenues in which chemical modifications can be made in attempts to increase potency against the growth of bacteria and to decrease toxicity observed in human cell lines.
Acknowledgments
The authors are grateful for the financial support provided by the National Institutes of Health (grant number: 2SC3GM098173-06). The contents of this article/publication/etc. are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. A portion of student support was from a Departmental Grant from the Robert A. Welch Foundation (Grant No. BG-0017).
LIST OF ABBREVIATIONS
- aaRS
Aminoacyl-tRNA synthetase
- AS
Ammonium sulfate
- ATP
Adenosine triphosphate
- DMSO
Dimethyl sulfoxide
- DTT
Dithiothreitol
- EDTA
Ethylene diamine tetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid
- IPTG
Isopropyl b-D-1-thiogalactopyranoside
- KF
Potassium floride
- KPi
Potassium phosphate
- Met
Methionine
- MgOAc
Magnesium acetate
- MOPS
3-(N-morpholino)propanesulfonic acid
- SAR
Structure-activity relationship
- SPA
Scintillation proximity assay
- TCA
Trichloroacetic acid
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
Send Orders for Print-Reprints and e-prints to reprints@benthamscience.ae
References
- 1.Breidenstein EB, Fuente-Nunez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19:419–26. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Giamarellou H, Antoniadou A. Antipseudomonal antibiotics. Med Clin North Am. 2001;85:19–42. doi: 10.1016/s0025-7125(05)70303-5. [DOI] [PubMed] [Google Scholar]
- 3.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–6. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 4.Gentry DR, Ingraham KA, Stanhope MJ, et al. Variable sensitivity to bacterial methionyl-tRNA synthetase inhibitors reveals subpopulations of Streptococcus pneumoniae with two distinct methionyl-tRNA synthetase genes. Antimicrob Agents Chemother. 2003;47:1784–9. doi: 10.1128/AAC.47.6.1784-1789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JR, Gentry D, Becker JA, Ingraham K, Holmes DJ, Stanhope MJ. Horizontal transfer of drug-resistant aminoacyl-transfer-RNA synthetases of anthrax and Gram-positive pathogens. EMBO Rep. 2003;4:692–8. doi: 10.1038/sj.embor.embor881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Guerrero E, Keniry M, Manrrique J, Bullard JM. Identification of chemical compounds that inhibit the function of glutamyl-tRNA synthetase from Pseudomonas aeruginosa. J Biomol Screen. 2015;20:1160–70. doi: 10.1177/1087057115591120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cull MG, McHenry CS. Purification of Escherichia coli DNA polymerase III holoenzyme. Methods Enzymol. 1995;262:22–35. doi: 10.1016/0076-6879(95)62005-2. [DOI] [PubMed] [Google Scholar]
- 8.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Keniry M, Palmer SO, Bullard JM. Discovery and analysis of natural product compounds inhibiting protein synthesis in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:4820–9. doi: 10.1128/AAC.00800-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Palmer SO, Munoz H, Bullard JM. High throughput screen identifies natural product inhibitor of phenylalanyl-tRNA synthetase from Pseudomonas aeruginosa and Streptococcus pneumoniae. Curr Drug Discov Technol. 2015;11:279–92. doi: 10.2174/1570163812666150120154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically: approved guidelines M7-A7. CLSI, Wayne PA. M7-A7. 2006. Clinical and Laboratory Standards Institute.
- 12.Clinical and Laboratory Standards Institute. Methods for determining bactericidal activity of antimicrobial agents: approved guideline M26-A. CLSI; Wayne, PA: 2002. p. M26-A. [Google Scholar]
- 13.Mechulam Y, Schmitt E, Maveyraud L, et al. Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J Mol Biol. 1999;294:1287–97. doi: 10.1006/jmbi.1999.3339. [DOI] [PubMed] [Google Scholar]
- 14.Crepin T, Schmitt E, Mechulam Y, et al. Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 2003;332:59–72. doi: 10.1016/s0022-2836(03)00917-3. [DOI] [PubMed] [Google Scholar]
- 15.Sugiura I, Nureki O, Ugaji-Yoshikawa Y, et al. The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Structure. 2000;8:197–208. doi: 10.1016/s0969-2126(00)00095-2. [DOI] [PubMed] [Google Scholar]
- 16.Ojo KK, Ranade RM, Zhang Z, et al. Brucella melitensis methionyl-tRNA-synthetase (MetRS), a potential drug target for Brucellosis. PLoS One. 2016;11:e0160350. doi: 10.1371/journal.pone.0160350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crepin T, Schmitt E, Blanquet S, Mechulam Y. Three-dimensional structure of methionyl-tRNA synthetase from Pyrococcus abyssi. Biochemistry. 2004;43:2635–44. doi: 10.1021/bi0356247. [DOI] [PubMed] [Google Scholar]
- 18.Serre L, Verdon G, Choinowski T, Hervouet N, Risler JL, Zelwer C. How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding. J Mol Biol. 2001;306:863–76. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt E, Meinnel T, Panvert M, Mechulam Y, Blanquet S. Two acidic residues of Escherichia coli methionyl-tRNA synthetase act as negative discriminants towards the binding of non-cognate tRNA anticodons. J Mol Biol. 1993;233:615–28. doi: 10.1006/jmbi.1993.1540. [DOI] [PubMed] [Google Scholar]
- 20.Nureki O, Kohno T, Sakamoto K, Miyazawa T, Yokoyama S. Chemical modification and mutagenesis studies on zinc binding of aminoacyl-tRNA synthetases. J Biol Chem. 1993;268:15368–73. [PubMed] [Google Scholar]
- 21.Fourmy D, Meinnel T, Mechulam Y, Blanquet S. Mapping of the zinc binding domain of Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 1993;231:1068–77. doi: 10.1006/jmbi.1993.1352. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh G, Pelka H, Schulman LH, Brunie S. Activation of methionine by Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1991;30:9569–75. doi: 10.1021/bi00104a002. [DOI] [PubMed] [Google Scholar]
- 23.Palmer SO, Hu Y, Keniry M, Bullard JM. Identification of chemical compounds that inhibit protein synthesis in Pseudomonas aeruginosa. J Biomol Screen. 2016 doi: 10.1177/1087057116679591. pii: 1087057116679591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pistelli L, Bertoli A, Noccioli C, et al. Antimicrobial activity of Inga fendleriana extracts and isolated flavonoids. Nat Prod Commun. 2009;4:1679–83. [PubMed] [Google Scholar]
- 25.Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cozzone AJ. Stringent control and protein synthesis in bacteria. Biochimie. 1980;62:647–64. doi: 10.1016/s0300-9084(80)80022-8. [DOI] [PubMed] [Google Scholar]
- 27.Deniziak MA, Barciszewski J. Methionyl-tRNA synthetase. Acta Biochim Pol. 2001;48:337–50. [PubMed] [Google Scholar]
- 28.Schnier JB, Gadbois DM, Nishi K, Bradbury EM. The kinase inhibitor staurosporine induces G1 arrest at two points: effect on retinoblastoma protein phosphorylation and cyclin-dependent kinase 2 in normal and transformed cells. Cancer Res. 1994;54:5959–63. [PubMed] [Google Scholar]
- 29.Inoue K, Kumakura S, Uchida M, Tsutsui T. Effects of eight antibacterial agents on cell survival and expression of epithelial-cell- or cell-adhesion-related genes in human gingival epithelial cells. J Periodontal Res. 2004;39:50–8. doi: 10.1111/j.1600-0765.2004.00704.x. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 31.Nakama T, Nureki O, Yokoyama S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J Biol Chem. 2001;276:47387–93. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 32.Patel JB, Gorwitz RJ, Jernigan JA. Mupirocin resistance. Clin Infect Dis. 2009;49:935–41. doi: 10.1086/605495. [DOI] [PubMed] [Google Scholar]