Graphical Abstract
Triacrylate polymers may enable novel regenerative dental therapies by both restoring dental tissue and providing a supportive niche for dental pulp stem cells (DPSCs) A polymer microarray of wide chemical diversity is used to rapidly identify triacrylate biomaterials that support DPSC adhesion, proliferation, and differentiation in vitro, and can directly restore the dentin-pulp interface in a model of severe dental pulp injury.
Keywords: dental materials, dental pulp stem cells, multi-functional acrylates, polymer microarray, differentiation, Synthetic bio-instructive material
Dental decay is the most prevalent oral disease worldwide, with an estimated 3 billion untreated cases of tooth decay, and is increasing despite improvements in prevention and treatments.[1] Dental decay is treated by surgically removing infected dentin and restoring tooth structure and function with a material. Exciting progress is being made in regenerative dentistry,[2] which may promote natural repair of dental tissues through dentin and pulp regeneration from stem cells. Previous work has shown the potential of endogenous dental pulp stem cells (DPSCs) for promoting dentin repair.[3] In addition, scaffolds for encapsulating stem cells[4] and delivering growth factors or drugs[5] are promising approaches for dentin and pulp regeneration. However, currently available dental scaffold materials do not adequately support regeneration.[6] Dental surgery is unique from other surgical sites, because teeth are directly exposed to the oral cavity, and therefore, must be filled with a permanent restoration rather than degradable materials. Existing synthetic polymer-based chemistries in dental materials cannot be used in direct contact with cells and vital tissue for regeneration, because they contain residual methacrylate monomers, such as bisphenol A glycidyl methacrylate (BisGMA), which negatively impact cells by disrupting regulatory cellular signaling.[7]
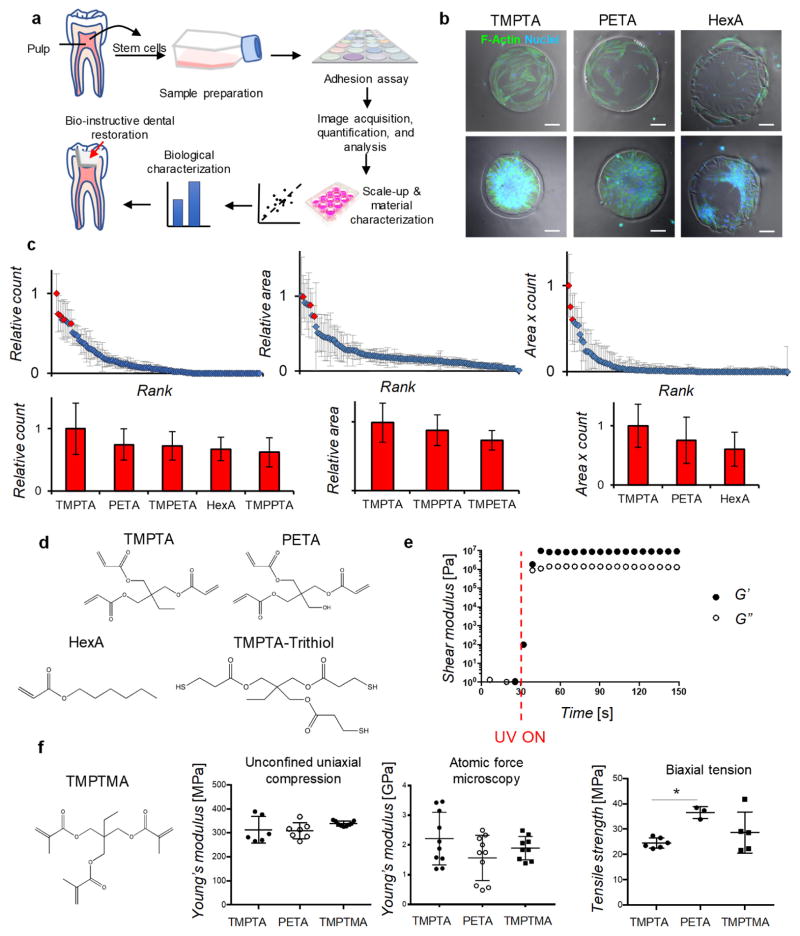
Regenerative therapies would benefit from a new class of bio-instructive restorative materials that support the underlying native function of dental pulp cells, such as DPSCs, to differentiate into dentin-producing cells, while also restoring the tooth to protect vital tissue from mechanical, microbial, thermal, and chemical injury.[8] Here we aim to identify synthetic polymeric materials that provide a supportive niche for DPSCs to differentiate to odontoblasts, which are the cells that regenerate dentin, as well as demonstrate potential clinical utility in a relevant animal model. Polymer microarrays have previously been used to identify novel materials for stem cell research,[9–11] and here we applied this approach to rapidly identify candidate chemistries for regenerative dental materials. The general strategy involved first determining the ability of DPSCs from human donor teeth to adhere in vitro on the various materials in the array, followed by scale-up of the lead materials, and a more complete characterization of materials properties and the biological response (Figure 1a). The adhesion assay allowed for rapid identification of top-performing polymers from a large and chemically diverse material library. The polymer microarray was prepared from commercially-available monomers of wide chemical diversity, including mono- and multi-functional acrylates, methacrylates, acrylamides, and methacrylamides with assorted side-chain chemistries totaling 119 unique homopolymers (Table S1) on a cell non-adherent poly(2-hydroxyethyl methacrylate) (pHEMA) background.[11] The monomers were selected from commercial vendors based on their low-viscosity for compatibility with the array-printing manufacturing process.[9] Adhesion was chosen for the screen, because cells must first be able to adhere to a substrate in order to be impacted functionally. DPSC adhesion and gene expression were examined in later experiments as metrics of the ability of the top-performing polymers to support dentinogenesis. Cells were seeded onto arrays in serum-free conditions to provide a more defined environment for examining the effects of polymer chemistries on cell adhesion. Serum contains a significant fraction of albumin and other carrier proteins, which can vary with each batch and may confound the specific material-cell interactions that we intend to study. Thus, serum-free media conditions allow for more specific examination of the intrinsic ability of polymers to enable cell adhesion. Cell adhesion was measured after 24 and 48 hours by fluorescence imaging of cells stained for cell nuclei (DAPI, blue) and F-actin (phalloidin, green) (Figure 1b). Quantification by an index of cell area multiplied by cell count yielded three top-performing polymers, which corresponded to hydrophobic acrylate monomer HexA (hexyl acrylate), and triacrylates TMPTA (trimethylolpropane triacrylate) and PETA (pentaerythritol triacrylate) (Figure 1c–d). Derivatives of TMPTA, TMPETA (trimethylolpropane ethoxylate triacrylate) and TMPPTA (trimethylolpropane propoxylate triacrylate), were also identified by count and area (Figure 1c). From the top-performing chemistries identified within the library, the triacrylate monomers, TMPTA and PETA, were chosen to investigate further, over the monofunctional acrylate HexA because triacrylates in general cure more rapidly, which is advantageous for in-situ bulk polymerization of a dental restorative material. Multifunctional acrylates polymerize by both chain-growth and network polymerization. Conversely, the monofunctional acrylate HexA polymerizes only via chain-growth into linear polymer chains of a physically crosslinked thermoplastic, rather than a more rigid covalently crosslinked polymer network.[12] Further, HexA performed lower than TMPTA in all cell adhesion metrics (Figure 1c).
Figure 1.
Rapid identification of triacrylate polymers as candidate bio-instructive dental restorative materials. a) Schematic of general strategy: human dental pulp stem cells (DPSCs) were prepared for screening a library of 119 monomers of diverse chemistries on a polymer microarray that enabled rapid identification of candidate chemistries for therapeutic use in dentistry. Candidate polymers were then scaled-up for further chemical and biological characterizations to assess their ability to support DPSCs, followed by application in a dental injury model (red arrow). b) Fluorescence imaging of cells on TMPTA, PETA, and HexA, stained for F-actin (green) and nuclei (blue), at high (80,000 cells per array, bottom-row) and low seeding density (8,000 cells per array, top-row), scale bar 100 μm. c) Image analysis was performed to quantify the mean cell count and area on each polymer, and the two metrics were also combined to provide a comparison of cell adhesion. Candidate polymers were identified by comparing the mean count, area, or index of each polymer to the overall mean. Red diamonds on the scatterplots and red histogram bars below correspond to candidate polymers in either cell count, area, or combined index, which are statistically significantly different from overall mean in each group, p<0.05, n=5 per marker. Blue diamonds on the scatterplots were not statistically significantly different from the overall mean in each group. Chemical abbreviations - TMPTA (trimethylolpropane triacrylate), PETA (pentaerythritol triacrylate), HexA (hexyl acrylate), TMPETA (trimethylolpropane ethoxylate triacrylate), and TMPPTA (trimethylolpropane propoxylate triacrylate). d) Chemical structures of best-performing monomers, TMPTA, PETA, and HexA, which were selected according to combined index, as well as structure of tri-thiol (trimethylolpropane tris(3-mercaptopropionate)) for thiol-ene polymerization to scale-up monomers into bulk polymer formulations. e) Storage (closed circles) and loss (open circles) moduli of thiol-ene polymerization of TMPTA during UV-irradiation (initiated at 30-seconds, red dashed line), measured by oscillatory shear rheology (1 Hz, 1% strain). f) Chemical structure of methyl-analog of TMPTA, trimethylol propane trimethacrylate (TMPTMA), and compressive moduli, measured by bulk compression and AFM, and tensile strength of bulk thiol-ene-crosslinked polymers TMPTA, PETA, and TMPTMA, * statistically significant difference, p<0.05.
Next, thiol-ene chemistry was employed to scale-up polymerization of the monomers TMPTA and PETA by adding a structurally-related tri-thiol compound and photoinitiator (Figure 1d). These rapidly cured by UV-irradiation into bulk polymers in approximately 10 seconds, with storage moduli of ~107 Pascals (Figure 1e). Rapid bulk crosslinking of TMPTA is consistent with previous findings that multifunctional acrylates polymerize faster than multifunctional methacrylates due to less steric hindrance and higher radical stability.[13] In order to directly confirm in later studies whether DPSC adhesion was specific to triacrylates, a structurally similar trimethacrylate, trimethylolpropane trimethacrylate (TMPTMA) (Figure 1f), was polymerized in a similar manner and tested in parallel. The bulk polymers exhibited similar compressive moduli, measured by unconfined uniaxial compression, of around 300 MPa (Figure 1f), which is consistent with the bulk compressive moduli of resins used in commercial dental materials.[14] Nanoscale atomic force microscopy (AFM) measurements showed the surface of the triacrylate and trimethacrylate polymers achieved an elastic modulus of ~2 GPa (Figure 1f and S1a), which is on the order of the range of dentin stiffness (~10 GPa) measured by nanoindentation.[15] Biaxial tension experiments showed PETA had a higher tensile strength, close to 40 MPa, as compared to a mean of between 20–30 MPa for TMPTMA and TMPTA (Figure 1f). Each of these analyses, which utilize distinct methodologies, are measuring different aspects of the mechanical properties, but together indicate these polymers have appropriate properties for dental restorative materials. After curing, the alkene and thiol groups were not detectable by Fourier-transform infrared spectroscopy (Figure S1b), and sulphur species from the tri-thiol were detected on the polymer surface using time-of-flight secondary-ion mass spectrometry (ToF-SIMS) (Figure S1c). Fast bulk curing, as well as high stiffness and strength, suggest these triacrylate polymeric materials are suitable for in-situ curing, which is ideal for clinical applications in dentistry.
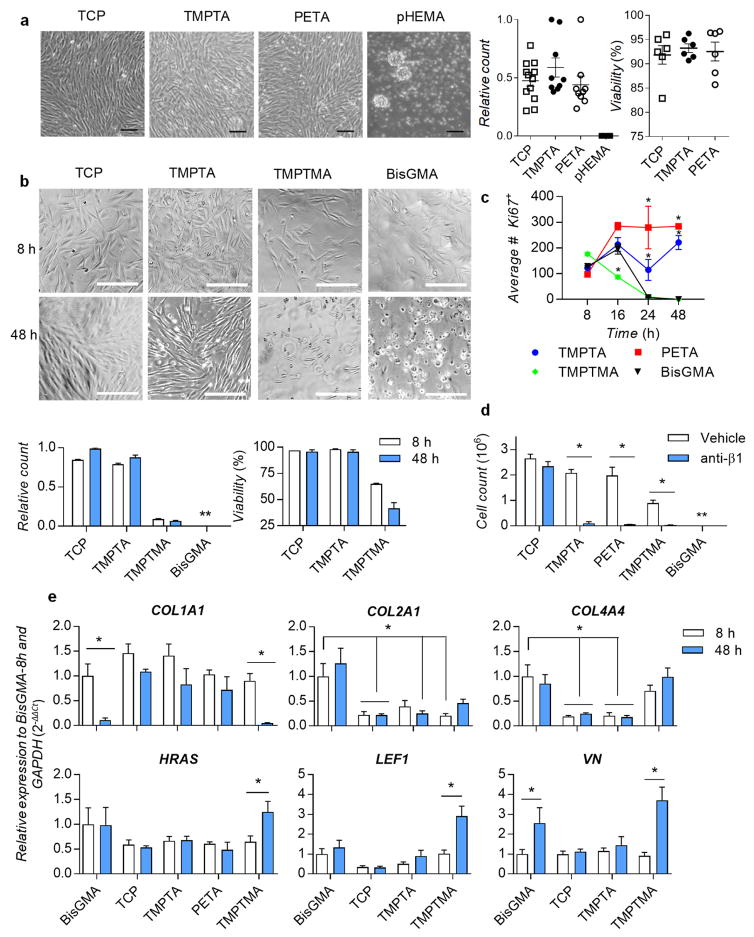
We confirmed thiol-ene polymerized bulk materials TMPTA and PETA adhered DPSCs in serum-free conditions over 48 hours in similar numbers and viability as tissue culture plastic (TCP); conversely, no cells adhered to pHEMA (Figure 2a), suggesting the thiol-ene chemistry maintained the adherent phenotype observed from the initial screen. Next, we determined DPSC adhesion and proliferation over 48 hours in serum-free media on the bulk polymers, compared to the commercial gold-standard methacrylate monomer used in dental fillings, BisGMA,[16] as well as a structurally-related trimethacrylate control, trimethylol propane trimethacrylate (TMPTMA). DPSC cell count and phase images over 48 hours showed DPSCs were adherent to TCP and TMPTA, but not to TMPTMA or BisGMA (Figure 2b). DPSCs initially adhered as viable single cells on all materials, but failed to spread, de-adhered and did not reach confluence on TMPTMA and BisGMA (Figure S2). These results suggested that cell adhesion was specific under these conditions to the selected triacrylate polymers, as TMPTA and its methyacrylate-analog TMPTMA shared the same branched trimethylolpropane core structure, were polymerized with the same thiol-ene chemistry, and exhibited similar physicochemical surface properties (Figure S1c). Further investigation is warranted to determine whether it is possible to rescue TMPTMA’s non-adherent phenotype, since it is structurally similar to TMPTA. DPSCs on TMPTA and TCP showed trends of increased cell number from 8-to-48 hours (Figure 2b). Significantly higher numbers of cells on TMPTA and PETA were positive for nuclear proliferation marker Ki67 at 24 and 48 hours, compared to BisGMA (Figure 2c and S3). Initially proliferating Ki67+ cells on TMPTMA and BisGMA (8 hours) lost adhesion over 48 hours (Figures 2c and S3). Subconfluent seeding of DPSCs on TMPTA and PETA in the presence of serum (10% fetal bovine serum, FBS) led to continued proliferation over 72 hours (Figure S4). Overall, these results are consistent with data from the initial screen, which showed all the 37 monofunctional or multi-functional methacrylates from the candidate chemistries were not one of the top polymers (Figure 1c and Table S1). Further, hexyl methacrylate, which is structurally-related to HexA, was not included in the top polymers. The integrin-β1 subunit is involved in cell adhesion, through its heterodimerization with integrin subunits such as α1, α2, α3, α5, and αV, and subsequent binding to extracellular ligands. We hypothesized that the integrin-β1 receptor subunit was required for DPSCs to adhere to the extracellular matrix that the cells express and deposit onto the triacrylates. A blocking anti-β1 antibody was used, in media containing 1% FBS to reduce non-specific adsorption of the blocking antibody, at a concentration of 5 μg/mL. Anti-β1 inhibited cell adhesion on TMPTA, PETA, TMPTMA and BisGMA, while it did not significantly affect cell adhesion on the TCP control (Figures 2d and S5). Together, these results demonstrate that DPSCs adhere and proliferate on TMPTA and PETA in an integrin-β1-dependent manner.
Figure 2.
DPSCs adhere to triacrylate polymers and proliferate in a β1-integrin-dependent manner. a) Images of thiol-ene-polymerized TMPTA and PETA with adhered DPSCs in serum-free medium at high confluence after 48 hours, with quantitation of relative cell numbers and viability, compared to tissue culture plastic control TCP and negative control polyHEMA. Scale bar 100 μm. b) Phase images (scale bar 200 μm), and relative DPSC cell count and viability in serum-free medium at 8- and 48-hours compared to negative control BisGMA and structurally-related trimethacrylate TMPTMA (** below counting limit). c) Quantification of proliferation marker Ki67 in adhered DPSCs in serum-free medium on TMPTA, PETA, TMPTMA, and BisGMA from 8 to 48 hours, * statistically significant difference from BisGMA control, p<0.05, n=3. d) Cell counts of DPSCs after 48-hour culture on TCP, TMPTA, PETA, TMPTMA, or BisGMA in media containing blocking anti-integrin-β1 receptor subunit antibody (anti-ITGβ1) or vehicle control, * statistically significant difference, p<0.05, n=6. e) Relative gene expression of DPSCs adhered in serum-free medium for 8 and 48 hours on TMPTA, PETA, TMPTMA, TCP, or BisGMA for collagen type I (COL1A1), collagen type II (COL2A1), collagen type IV (COL4A4), vitronectin (VN), HRAS, and LEF1, normalized to GAPDH and BisGMA-8h, * statistically significant difference, p<0.05, n≥4.
To investigate the impact of adhesion, a commercial gene screening array (Table S2) was used to measure the expression of a wide range of genes associated with β1-signaling (Figures S6–7). Gene expression of cells on TMPTA, PETA, TMPTMA, and TCP were compared to the BisGMA control at 8 and 48 hours to determine how triacrylates differentially regulate integrin-β1-signaling pathways. Significantly impacted genes were defined as those expressed by DPSCs on triacrylates at 8 hours higher than 2-fold (Figure S8a), and these were validated in three subsequent independent experiments at 8 and 48 hours by quantitative polymerase chain reaction (qPCR). The significantly impacted genes included matrix proteins collagen-1 (COL1A1), collagen-2 (COL2A1), collagen-4 (COL4A4), and vitronectin (VN), cell signaling genes HRAS, LEF1, and SRC, proliferation gene mitogen-associated protein kinase-3 (MAPK3), cytoskeletal marker G-actin-1 (ACTG1), and integrin receptor beta-1 subunit (ITGB1) (Figures 2e and S8b). Gene expression of COL1A1 was maintained on TCP, TMPTA, and PETA, but not on BisGMA and TMPTMA, and COL2A1, COL4A4, and VN increased on BisGMA and TMPTMA, but not on TCP and TMPTA (Figure 2e). Furthermore, DPSCs on BisGMA and TMPTMA upregulated expression of genes associated with cell survival, HRAS, LEF1 and SRC, compared to TCP and TMPTA (Figures 2e and S8b). LEF1 is a transcription factor involved in Wnt/beta-catenin signaling pathways, which can impair odontoblast differentiation.[17] It is important to note that these analyses measured gene expression in the very few numbers of cells that remained adhered on BisGMA and TMPTMA at 48 hours. We speculate these remaining cells upregulate COL2A1, COL4A4, and VN expression as a compensatory mechanism to remain adherent, even though they have a significant reduction in COL1A1 expression. ITGB1 expression slightly increased on TMPTA and PETA after 48 hours (Figure S8b). Together, these data suggest that collagen-1 expression and integrin-β1 receptor binding are correlated with DPSC adhesion to triacrylates TMPTA and PETA. This finding suggests these materials can functionally support DPSCs, because dental pulp stem cells express β1 integrin during differentiation.[18] Conversely, DPSCs fail to adhere well on BisGMA and the methacrylate-analog (TMPTMA) controls, and appear to compensate by upregulating other matrix genes and cell survival genes.
These data are consistent with previous literature that showed expression of the β1 heterodimer of integrin α5-β1 is required to promote cell survival in the absence of serum when adhered to fibronectin-coated surfaces.[19] Adhesion to the TMPTA and PETA polymers via integrin α5-β1 may help promote DPSCs to survive in serum-free conditions similar to TCP control, whereas DPSCs on BisGMA and TMPTMA materials may exhibit limited adhesion via integrin α5-β1. In addition, previous literature showed α1-β1 integrin expression is a negative regulator of collagen-I expression.[20] Initially when seeded without serum, presumably there is limited availability of collagen ligands and less activation of α1-β1 integrin. Collagen-1 expression is downregulated over 48 hours on BisGMA and TMPTMA, possibly due to the role of α1-β1 activation, whereas collagen-1 expression is maintained on TMPTA and PETA similar to TCP control. Further work investigating the role of integrins in regulating cell adhesion to these triacrylates is warranted in the future.
The triacrylate materials did not require any chemical modification or pre-adsorption of proteins after polymerization for direct signaling on cells, but rather the data are consistent with the hypothesis that the materials support cells to produce and adhere via their own adsorbed matrix proteins. Mammalian cells often can adhere to surfaces of synthetic materials even in the absence of serum proteins or precoating of the substrate with adhesive proteins – many cell types adhere very well to tissue culture plastic under these conditions. In these situations, it is believed that cells adhere via the proteins they actively synthesize and secrete or which are already present on the cell surface, which subsequently interact with the surface of the material.[21] It is possible that the higher reactivity and stability of triacrylate free-radicals[13] enhanced bulk polymerization compared to TMPTMA and BisGMA, which improved interaction with cells and extracellular matrix and other absorbed proteins. A goal of future work is to determine in detail how macromolecular, mechanical, and biochemical cues of these materials direct the cell behavior, including the specific role of adsorbed proteins.
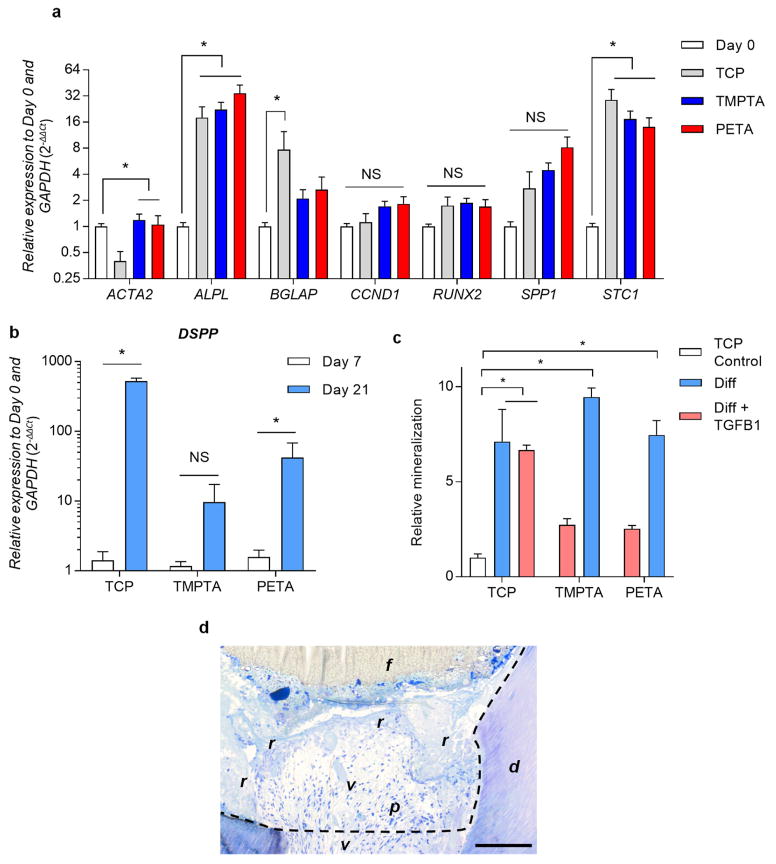
To further evaluate the compatibility of triacrylates with DPSCs, we investigated the relative gene expression of DPSCs in long-term serum-free culture on TMPTA and PETA compared to TCP. DPSCs expressed lower levels of osteocalcin (BGLAP), RUNX2, and TGFB1 on TMPTA after 21 days compared to TCP, indicating they differentiated less on TMPTA than TCP (Figure S9a). Trends were similar for PETA, but not statistically significant. We next confirmed that these effects were not due to residual monomer leaching into the media, as conditioned media from TMPTA and PETA added to cells on TCP did not cause significant differences in gene expression compared to TCP control (Figure S9a). Next, osteogenic conditions were used to determine whether triacrylates TMPTA and PETA are permissive for DPSC differentiation in an appropriate chemical environment, compared to TCP control. After 21 days, DPSCs on TMPTA and PETA expressed increased levels of the osteogenic differentiation markers[22] alkaline phosphatase (ALPL), BGLAP, osteopontin (SPP1), and staniocalcin (STC1), but the dentin-specific marker dentin sialophosphoprotein (DSPP)[23] was not detected (Figure 3a). With the addition of the growth factor TGFβ1, DPSCs were able to differentiate into odontoblasts, as confirmed by expression of DSPP on TMPTA and PETA (Figure 3b) at Day 21 compared to Day 7. The cells clustered into aggregates on TCP, TMPTA, and PETA in the presence of TGFβ1, indicative of a contractile phenotype, which was associated with a reduction in ALPL, COLA1A, and SPP1 (Figure S9b) that may have resulted from increased cell-cell contact and reduced cell-matrix interactions. TMPTA and PETA supported DPSC mineralization under osteogenic differentiation conditions (Figures 3c and S9c), which was quantified by alizarin red staining. Together, these differentiation data suggest that TMPTA and PETA-based materials are compatible with stem cell behavior, although material cues alone are not sufficient to induce differentiation. Rather, the polymeric materials provide a supportive niche that allows DPSCs to respond appropriately to biochemical cues. Thus, these triacrylate materials could comprise a new, first-generation regenerative dental filling that supports stem cells to respond to regenerative therapies. They can be rapidly cured into bulk materials with minimal residual monomer, have rigid mechanical properties similar to existing fillings and dentin, and provide a supportive niche for dental pulp stem cells. Triacrylates TMPTA and PETA are likely to be compatible with regenerative treatments, such as delivery of stem cells in degradable scaffolds or delivery of bioactive molecules that promote endogenous repair mechanisms.
Figure 3.
Triacrylate polymers supported DPSC osteogenic and odontogenic differentiation, and can be placed in direct contact with the dentin-pulp interface to mimic their use in regenerative dentistry. a) Relative gene expression of proliferation marker cyclin-D1 (CCND1) and differentiation markers smooth muscle actin (ACTA2), alkaline phosphatase (ALPL), osteocalcin (BGLAP), RUNX2, osteopontin (SPP1), and staniocalcin (STC1) by DPSCs before placement on polymer (D0), and on TCP, TMPTA, and PETA after culture for 21 days in osteogenic media, normalized to GAPDH and D0. Statistically significant differences (*) between D0 and the material conditions were determined by two-way ANOVA, p<0.05, n ≥ 3. b) DPSCs cultured in osteogenic media + TGFβ1 expressed elevated levels of dentin-specific marker dentin sialophosphoprotein (DSPP) after 21 days compared to 7 days, normalized to GAPDH and D0, * statistically significant difference, two-way ANOVA, p< 0.05. c) Relative mineralization of DPSCs after 14 days of culture in osteogenic media with (red) or without (blue) TGFβ1 on TCP, TMPTA, and PETA, normalized to cells on TCP in non-osteogenic conditions (TCP control, white), * statistically significant difference, p<0.05, n = 6. d) Hard tissue histological section of TMPTA in direct contact with molar pulp defect at the time of pulp injury, day 0. Scale bar 100 μm. Dotted line – dental preparation, f – composite filling, r – TMPTA resin layer, p – dental pulp, d – dentin, v – blood vessel.
Regenerative therapies in dentistry would be indicated for treating pulp exposure injuries in adult human teeth, because they cause irreversible damage that results in necrosis of the tissue and otherwise require root canal surgery[24] (ClinicalTrials.gov NCT00187837 and NCT00187850). Restoring the dentin-pulp complex, including DPSCs, odontoblasts, and dentin, with regenerative therapies will likely require multiple components, including a permanent restorative material that biochemically and mechanically interfaces with regenerating tissue and remaining dentin. The potential clinical utility of TMPTA in these types of traumatic irreversible dental pulp injuries was subsequently explored by examining if the triacrylate-trithiol monomer solution could be cured into bulk materials inside irreversibly-injured molars of 6-week male rats in direct contact with pulp tissue. Other animal models that cause reversible pulp injury and vital dentin regeneration do not accurately mimic clinical cases with non-vital injured teeth indicated for regenerative therapies. After proper hemostasis and isolation, the TMPTA-thiol bulk monomer solution was applied to the injury site and was cured inside the tooth with visible light irradiation. Conventional commercial dental materials, calcium hydroxide paste and a resin composite filling, were used as controls. All animals were ambulatory, ate and grew normally, and exhibited normal patterns of behavior, suggesting there were no systemic negative effects of the treatment. Animals were sacrificed immediately after injury and after 8-weeks for fixation, decalcification, resin-embedding, and hard tissue sectioning for histology. Hard tissue histological sections showed the TMPTA filling (r) cured in direct contact with pulp (p) and dentin (d) tissues at day 0, with a conventional dental composite filling (f) on top (Figure 3d). Further, the oral cavity showed no signs of pathology or infection, restored teeth were intact, TMPTA was retained in contact with the pulp tissue after 8-weeks, and the pulp tissue qualitatively appeared similar to that resulting from the use of the calcium hydroxide and resin composite control (Figure S10a). Similar to human clinical cases of pulp exposure restored with dental composites, the pulp tissue became calcified as a result of irreversible pulpal injury and necrosis, and thus suffered loss of potential for vital dentin regeneration. MicroCT imaging qualitatively showed no major differences in the mineralized-to-total volume ratio between conditions, and TMPTA alone performed similarly as TMPTA + resin composite restoration (Figure S10b). We propose the triacrylates TMPTA and PETA could be used clinically with stem cell therapies as therapeutic biomaterials in direct contact with pulp tissues to promote regeneration of the dentin-pulp complex, while also functioning as a dental filling material that mechanically supports tooth structure in the oral cavity (Figure S11). A key issue for the clinical use of the multifunctional acrylates is their compatibility in vivo. These types of materials are widely used as cross-linkers in commercial industrial applications such as UV-curable inks, coatings, and paints. There are reports of skin exposure to multifunctional acrylate monomers causing sensitization in humans. In mice, topical application only at high doses (above 1 mg/kg) was inflammatory and carcinogenic.[25] However, very small doses are used here (~ 0.1 μg/kg TMPTA), and the monomer is rapidly cured into a bulk polymer with minimal residual monomer. Together with the in vitro characterizations of their biological response, these data suggest the triacrylates TMPTA and PETA, unlike existing synthetic materials in dentistry, provide a stem cell-compatible restorative material that can be used in direct contact with pulpal tissues, and thus improve the state-of-the-art in clinical dentistry.
In summary, triacrylate-based materials have significant promise as novel, bio-instructive, restorative materials for regenerative dentistry. They can be cured rapidly into bulk polymers without detectable residual monomer, provide robust mechanical properties, support DPSC regenerative-related functions, and can be placed directly at the dentin-pulp interface in a severe model of dental pulp injuries. Additionally, human periodontal ligament fibroblasts adhered in vitro to TMPTA and PETA in a similar fashion as to TCP (Figure S12) suggesting these materials could be utilized more broadly beyond restorative dentistry. Furthermore, the triacrylate TMPTA provides antimicrobial properties, because TMPTA was previously discovered to resist biofilm formation of Staphylococcus aureus and uropathogenic Escherichia coli.[26] Thus, these triacrylate-based materials could be broadly useful as medical devices by instructing multiple biological functions.
Experimental Section
Cell culture
For adhesion experiments, human DPSCs were seeded at 75,000 cells/cm2 on either polymer-coated TCP or TCP alone in serum-free aMEM (Gibco) complete media (Supplementary Information). After 48-hours, bright-field microscopic images and cells were retrieved for counting. Live/dead staining assay was performed using Live/Dead Viability/Cytotoxicity kit (Life Technologies). Integrin inhibition experiments used an anti-β1 blocking antibody at 5 μg/mL (mab1959, Milipore) in 1% FBS. For differentiation experiments, DPSCs were cultured in serum-free media until they reached confluency, followed by supplementation with 10% fetal bovine serum, 285 μM L-ascorbic acid, 10 mM beta-glycerol phosphate, 0.01 μM dexamethasone, and +/− 10 ng/mL of recombinant human TGFβ1 (PeproTech). Fresh media was replaced every 2–3 days. Cells were lysed at 7- and 21-days for RNA isolation and subsequent quantitative polymerase chain reaction (qPCR) (Supplementary Information). The level of mineralization during differentiation was determined by an osteogenesis quantitation kit (EMD Millipore), which involved Alizarin Red staining, acetic acid extraction, and quantification based on absorbance at 405 nm.
High-throughput screening and scaling-up polymer chemistries
Polymeric microarrays were prepared as described.[11] Briefly, 119 commercially available monomers of 250–400 μm diameter were printed and photo-polymerized on polyHEMA coated slides in microarrays, with six replicates per array. DPSCs were seeded at high (80,000 cells per array) or low (8,000 cells per array) densities in serum-free media for 24- and 48-hours, respectively. For scale-up, candidate monomers from the screen were mixed with a thiol-ene crosslinking reagent, trimethylolpropane tris(3-mercaptopropionate), at a molar ratio of 1.4:1 (acrylate:thiol) and 1% w/v of photo-initiator 2,2-dimethoxy-2-phenylacetophenone. In a typical 12-well dish, mixed monomer solution (400 μL) was added to each well and irradiated with 365 nm UV-irriadiation for 5 minutes at an intensity of 20 mW/cm2. Polymer coatings were UV-sterilized for 30 minutes, then washed three times with Hanks’ balanced salt solution (HBSS).
Mechanical characterizations
Oscillatory rheology was performed using AR-G2 stress controlled rheometer (TA Instruments). Compressive and tensile tests were measured by an Instron 3342 single column apparatus. The elastic moduli of cylindrical bulk polymers with high aspect-ratio (10 mm width, 2 mm height) were measured by the initial slope of stress-strain during application of unconfined uniaxial compression to 15% strain with a deformation rate of 1 mm min−1. Bulk polymers were fabricated in dog-bone geometry for measuring ultimate tensile strength, with application of biaxial tension at strain rate of 1 mm min−1 until rupture.
Immunostaining and confocal imaging
Samples for immunocytochemical staining and subsequent confocal imaging were fixed in 4% paraformaldehyde for 10 minutes, washed three times with DPBS (+Ca/+Mg), permeabilized in 0.5% Triton X-100 for 10 minutes, washed in 0.1% Tween-20 in PBS, blocked in 5% goat serum and 1% BSA overnight at 4C, and stained at RT for 2-hours. Anti-Ki67 (350502, BioLegend), Alexa Fluor 488-phalloidin, anti-Ki67 Alexa Fluor-594 (350528, BioLegend), DyLight goat anti-mouse 594, and ProLong Gold with DAPI were used for staining. Fluorescent cell culture images were captured on an EVOS microscope, confocal images were captured using Zeiss LSM 710, polymer microarrays were imaged on an automated stage fluorescence microscope Zeiss Axio Observer Z1, and histological samples were imaged on a Zeiss Axio Imager Z2. Zen Black software was used for image acquisition and processing of confocal, microarray, and histologic images. Polymer microarray images were analyzed with 5 replicates per condition for manual cell counting on ImageJ and for cell area by MATLAB quantification. Staining for Ki67 was analyzed in at least three images per field with thresholding and counting on ImageJ.
Rodent model of dental pulp injury
All animal procedures were approved by Harvard IACUC and followed all state and federal regulations. The dental pulp injury model was adapted from a previous study.[3] Each male Sprague Dawley rat at 6-weeks old was properly anesthetized and cavity preparations to excavate pulpal tissue were made on the occlusal aspect of maxillary first and second molars with a high speed conventional handpiece and a #1/2 carbide round bur (0.8 mm diameter) with simultaneous irrigation of sterile saline solution. Adequate hemostasis was obtained by cotton-pellet pressure. Bulk monomer solution of TMPTA was placed at the base of the cavity and cured with visible-light irradiation. Subsequently, the tooth was restored with a composite resin (3M-ESPE) associated with a one-step adhesive system (Single-Bond, 3M-ESPE). Calcium hydroxide paste and resin composite without TMPTA, and resin composite alone without TMPTA were used as controls. Animals were sacrificed at 8-weeks and fixed via intracardiac perfusion of 4% paraformaldehyde (PFA) (Sigma Aldrich) in phosphate-buffered saline (PBS). Maxillary segments were dissected and subsequently fixed overnight in 4% PFA in PBS at 4°C. For histology, specimens were decalcified in 14% EDTA in PBS, pH 7.4 for 2 weeks with changes every 2–3 days. Resin embedding and hard tissue sectioning were performed by the Harvard Medical School Electron Microscopy Core.
Statistical analysis
Error bars represent standard error of mean. Statistical tests were performed on polymer microarray data using Kruskal-Wallis test with Dunn’s multiple comparisons test, and on scaled-up cell count and qPCR data using two-way ANOVA and Sidak post-hoc test, p<0.05. For qPCR, statistical tests were performed on the log-2 transform of the relative expression values.
Supplementary Material
Acknowledgments
Research reported in this manuscript was supported by the National Institute of Dental & Craniofacial Research of the National Institutes of Health under Award Numbers R01DE013033 (DM) and K08DE025292 (KV), a Marie Curie International Outgoing Fellowship from the European Commission under FP7 agreement number 629320 (AC), the Royal Society of Chemistry Emerging Technologies Prize (AC and KV), Anne Marcus Wedner Graduate Research and Henry M. Thornton/SCADA Fellowships (KV), Harvard-Amgen Scholars Program (AB), and Harvard PRISE Fellowship (JS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Part of this work was performed in part at the Harvard University Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), which is supported by the National Science Foundation under NSF ECCS award no. 1541959. At the Wyss Institute we thank Thomas Ferrante for assistance with imaging, as well as Amanda Graveline and Matthew Pezone for their assistance with animal surgery. We thank the Department of Cell Biology of Harvard Medical School in Boston, MA for the use of the Electron Microscopy Core, which provided histological processing services.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Kyle H. Vining, Wyss Institute for Biologically Inspired Engineering and Harvard John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
Jacob C. Scherba, Wyss Institute for Biologically Inspired Engineering and Harvard John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
Alaina M. Bever, Wyss Institute for Biologically Inspired Engineering and Harvard John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
Prof. Morgan R. Alexander, Advanced Materials and Healthcare Technologies Division, School of Pharmacy, University of Nottingham, Nottingham NG7 2RD, UK.
Dr. Adam D. Celiz, Wyss Institute for Biologically Inspired Engineering and Harvard John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA. Advanced Materials and Healthcare Technologies Division, School of Pharmacy, University of Nottingham, Nottingham NG7 2RD, UK
Prof. David J. Mooney, Wyss Institute for Biologically Inspired Engineering and Harvard John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
References
- 1.Kassebaum NJ, Smith AGC, Bernabé E, Fleming TD, Reynolds AE, Vos T, Murray CJL, Marcenes W. Journal of Dental Research. 2017;96:380. doi: 10.1177/0022034517693566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Syed-Picard FN, Ray HL, Kumta PN, Sfeir C. Journal of Dental Research. 2014;93:250. doi: 10.1177/0022034513517901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arany PR, Cho A, Hunt TD, Sidhu G, Shin K, Hahm E, Huang GX, Weaver J, Chen ACH, Padwa BL, Hamblin MR, Barcellos-Hoff MH, Kulkarni AB, Mooney DJ. Science Translational Medicine. 2014;6:238ra69. doi: 10.1126/scitranslmed.3008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang WB, Walboomers XF, van Kuppevelt TH, Daamen WF, Bian Z, Jansen JA. Biomaterials. 2006;27:5658. doi: 10.1016/j.biomaterials.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Lee JH, Lee DS, Choung HW, Shon WJ, Seo BM, Lee EH, Cho JY, Park JC. Biomaterials. 2011;32:9696. doi: 10.1016/j.biomaterials.2011.09.007. [DOI] [PubMed] [Google Scholar]; Neves VCM, Babb R, Chandrasekaran D, Sharpe PT. Scientific Reports. 2017;7:7. doi: 10.1038/srep39654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song JS, Takimoto K, Jeon M, Vadakekalam J, Ruparel NB, Diogenes A. Journal of Dental Research. 2017;96:640. doi: 10.1177/0022034517693606. [DOI] [PubMed] [Google Scholar]
- 7.Krifka S, Spagnuolo G, Schmalz G, Schweikl H. Biomaterials. 2013;34:4555. doi: 10.1016/j.biomaterials.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y, Song M, Kim E, Shon W, Chugal N, Bogen G, Lin L, Kim RH, Park NH, Kang MK. Journal of Dental Research. 2015;94:1544. doi: 10.1177/0022034515601658. [DOI] [PubMed] [Google Scholar]
- 9.Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, Van Vliet KJ, Davies MC, Alexander MR, Langer R, Jaenisch R, Anderson DG. Nature Materials. 2010;9:768. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celiz AD, Smith JGW, Langer R, Anderson DG, Winkler DA, Barrett DA, Davies MC, Young LE, Denning C, Alexander MR. Nature Materials. 2014;13:570. doi: 10.1038/nmat3972. [DOI] [PubMed] [Google Scholar]
- 11.Celiz AD, Smith JGW, Patel AK, Hook A, Epa V, Langer R, Anderson D, Winkler D, Barrett D, Young L, Davies M, Denning C, Alexander M. Advanced Materials. 2015;27:4006. doi: 10.1002/adma.201501351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratna D, Simon GP. Polymer. 2001;42:7739. [Google Scholar]
- 13.Nason C, Roper T, Hoyle C, Pojman JA. Macromolecules. 2005;38:5506. [Google Scholar]
- 14.Chun KJ, Lee JY. Journal of Dental Biomechanics. 2014;5:1. doi: 10.1177/1758736014555246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YR, Du W, Zhou XD, Yu HY. In J Oral Sci. 2014;6:61. doi: 10.1038/ijos.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cramer NB, Stansbury JW, Bowman CN. Journal of Dental Research. 2010;90:402. doi: 10.1177/0022034510381263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheller EL, Chang J, Wang CY. Journal of Dental Research. 2008;87:126. doi: 10.1177/154405910808700206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito K, Fukumoto E, Yamada A, Yuasa K, Yoshizaki K, Iwamoto T, Saito M, Nakamura T, Fukumotoet S. PLOS ONE. 2015;10:e0121667. doi: 10.1371/journal.pone.0121667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Vuori K, Reed JC, Ruoslahti E. Proceedings of the National Academy of Sciences. 1995;92:6161. doi: 10.1073/pnas.92.13.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner H, Broberg A, Pozzi A, Laato M, Heino J. Journal of Cell Science. 1999;112:263. doi: 10.1242/jcs.112.3.263. [DOI] [PubMed] [Google Scholar]
- 21.Curtis ASG, Forrester JV, Mclnnes C, Lawrie F. The Journal of Cell Biology. 1983;97:1500. doi: 10.1083/jcb.97.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tamada Y, Ikada Y. J Colloid Interface Sci. 1993;155:334. [Google Scholar]
- 22.Saito K, Nakatomi M, Ida-Yonemochi H, Ohshima H. Journal of Dental Research. 2016;95:1034. doi: 10.1177/0022034516645333. [DOI] [PubMed] [Google Scholar]
- 23.Guo S, Lim D, Dong Z, Saunders TL, Ma PX, Marcelo CL, Ritchie HH. Stem cells and development. 2014;23:2883. doi: 10.1089/scd.2014.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjørndal L, Fransson H, Bruun G, Markvart M, Kjældgaard M, Näsman P, Hedenbjörk-Lager A, Dige I, Thordrup M. Journal of Dental Research. 2017;96:747. doi: 10.1177/0022034517702620. [DOI] [PubMed] [Google Scholar]
- 25.Doi AM, Hailey JR, Hejtmancik M, Toft JD, Vallant M, Chhabra RS. Toxicol Pathol. 2005;33:631. doi: 10.1080/01926230500295615. [DOI] [PubMed] [Google Scholar]
- 26.Hook AL, Chang CY, Yang J, Atkinson S, Langer R, Anderson DG, Davies MC, Williams P, Alexander MR. Advanced Materials. 2013;25:2542. doi: 10.1002/adma.201204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.