Abstract
Purpose
NRAS mutations are now routinely included in RAS testing prior to EGFR (epidermal growth factor receptor) inhibitor therapy for metastatic colorectal cancer (mCRC). The clinical implications of NRAS mutation beyond lack of response to anti-EGFR therapy, however, are not known. We undertook this study to determine the clinical features and treatment outcomes of patients with NRAS mutant mCRC.
Experimental Design
We reviewed clinical characteristics, concurrent mutations, and outcomes for all mCRC cases with NRAS mutations undergoing standard genotyping at our institution from 2008–2015. Comparison groups consisted of RAS wild-type and KRAS mutant mCRC consecutive cases genotyped from 2008–2012.
Results
Three percent (87/2764) of mCRC patients had NRAS mutant tumors (45% exon 2, 55% exon 3), including three cases with concurrent NRAS and KRAS mutations. Left-sided primary site and African-American self-reported race were associated with NRAS mutation (p<0.01). Resection rate at 12 months was lower for NRAS mutant mCRC than for RAS wild-type or KRAS mutant mCRC. Median survival from time of first known metastasis was 33 months for NRAS mutant, 47 months for KRAS mutant, and 78 months for RAS wild-type cases (p<0.001). Multivariate analysis assigned a hazard ratio for overall survival of 2.0 for NRAS mutation and 1.5 for KRAS mutation (p<0.01).
Conclusions
NRAS defines a molecular subset with distinct clinical characteristics from KRAS mutant and wild-type mCRC. NRAS mutations are enriched in left-sided primary tumors and among African Americans. Mutations in NRAS are associated with poor survival and worse outcomes than either KRAS mutant or wild-type mCRC.
Keywords: NRAS, colorectal cancer, survival, race
Introduction
The RAS oncogenes (KRAS, NRAS, HRAS) encode a family of GTP-regulated switches that are recurrently mutated in human cancer. The four enzymes encoded by the three RAS genes are highly homologous to one another. Activating mutations render the RAS protein in the GTP-bound activated form by preventing hydrolysis of GTP and so preventing transition to the GDP-bound inactive state. RAS mutations occur at conserved hotspots, and oncogenic mutations have been shown to occur at codons 12, 13, 61, 117, and 146. Activation of the RAS proteins leads to pleiotropic effects in cells, including cellular proliferation, survival, and differentiation. The differences in signaling and downstream effectors that result from the different activated RAS isoforms are not currently clear. In colorectal cancer (CRC), RAS mutations predominantly occur in the KRAS gene; 45% of metastatic CRC (mCRC) harbor an activating KRAS mutation(1). NRAS mutations occur in 2–7% of mCRC cases(2–4).
NRAS mutations are now regularly identified in the clinical treatment of mCRC as part of routine RAS testing prior to EGFR (epidermal growth factor receptor) inhibitor therapy. The clinical implications of NRAS mutation beyond lack of response to anti-EGFR therapy(4) and whether these tumors behave similarly to KRAS mutant mCRC are not known. KRAS mutations have been associated with right-sided colon tumors, while NRAS mutations have been associated with left-sided primary tumors and female gender(3), suggesting a distinct biology for KRAS and NRAS mutant molecular subsets of mCRC.
Recent clinical series have examined all RAS mutant CRC or NRAS mutant CRC, but have been limited by a small number of NRAS mutant cases. KRAS and all RAS mutations have been associated with worse survival in mCRC(5, 6). KRAS and all RAS mutations have been associated with worse outcomes after hepatectomy with increased risk for recurrences in the lungs(7, 8). However, it is unknown if the NRAS mutant cases contributed to these poor outcomes, as they represented the minority of RAS mutant cases studied.
To better characterize the biology of NRAS mutant mCRC, we identified all NRAS mutant mCRC detected by genotyping of consecutive mCRC cases presenting at Memorial Sloan Kettering Cancer Center (MSKCC) between 2008 and 2015. We now report the analysis of the clinical characteristics, concurrent mutation spectrum, and outcomes in this large series of 87 NRAS mutant mCRC cases.
Materials and Methods
Patient population
Cases were derived from patients seen at MSKCC with mCRC who had their tumors submitted for standard genotyping for anti-EGFR treatment selection between 2008 and 2015. We performed a computerized search of electronic medical records to identify all cases that had an NRAS mutation in the sequencing report during this period. A comparison group of all RAS wild-type or KRAS mutant mCRC came from cases sequenced between 2008 and 2012(6). During this period 1095 unique patients with mCRC had their tumors genotyped, including 786 cases genotyped for extended RAS mutations. Cases with a KRAS exon 2 mutation or an extended KRAS mutation (exons 3 or 4) were analyzed together as a KRAS mutant mCRC comparison group (n=423) and cases with RAS wild-type status confirmed on extended testing formed a RAS wild-type mCRC comparison group (n=475). Supplementary Figure 1 diagrams the cases analyzed and their RAS status.
Sequencing
Genomic DNA was extracted from formalin fixed paraffin embedded (FFPE) tissue obtained from biopsies or resections. Sequencing was performed on a metastatic specimen in cases where tissue was available from metastasectomy or diagnostic biopsy (n=541; 55%), and on the primary tumor (n=444; 45%) in all other cases. For the NRAS mutant mCRC cases, genotyping was performed by a mass-spectrometry based assay (Sequenom) to detect hotspot mutations in a panel of 8 genes, MiSeq assay of 45 genes, or (starting in 2015) through an exon-capture next generation sequencing assay (MSK-IMPACT) of >300 cancer related genes. All known hotspots in NRAS were genotyped with these assays. Supplementary Figure 1 indicates the number of cases analyzed with each sequencing assay. The all RAS wild-type mCRC comparison group was genotyped with the Sequenom assay, and the KRAS mutant mCRC comparison group was genotyped with either Sanger sequencing for exon 2 mutations or the Sequenom assay.
Data Collection
All cases were by reviewed a medical oncologist (A.C., M.I.B., R.Y.) for patient characteristics, treatment history, and survival. Specific clinical characteristics collected included age, gender, race, primary tumor site, stage at diagnosis, sites of metastatic disease, metastasectomy, previous treatment, and survival. Stage was categorized by timing of metastasis as synchronous (stage IV at diagnosis) or metachronous (stage I–III at diagnosis). Tumors arising from the cecum to distal transverse colon were classified as right-sided, and tumors arising from the splenic flexure to rectosigmoid junction were classified as left-sided. New patient questionnaires included self-reported race with options of white, black, or asian, designated as Caucasian, African-American, and other, respectively, in our analysis. Metastatic sites were identified by review of medical records and/or imaging. Patients’ first and second line chemotherapy treatments were reviewed in detail including regimen, duration of therapy, and radiographic outcomes. All research was conducted under appropriate Institutional Review Board/Privacy Board protocols and waivers, and the study was conducted in accordance with recognized ethical guidelines.
Statistical Analysis
Associations between clinicopathologic characteristics and tumor mutation status were analyzed using the Fisher’s exact test for categorical variables and the Wilcoxon Rank-Sum test for continuous variables. Overall survival (OS) was examined from date of first known metastatic disease to date of death or last available follow up. Log-rank test was used to examine whether OS differed by mutation status. Cox proportional hazards model was used to evaluate the independent effect of mutation status on OS after adjusting for the following known confounders: age at diagnosis, gender, race, tumor location, synchronous tumor, and surgery. Surgery and treatment with hepatic arterial infusion were treated as a time-dependent covariate in the multivariate model.
Cumulative incidences of resection of gross metastatic disease were estimated using competing risks methods and compared between mutation status using Gray’s test. Recurrence free survival (RFS) was calculated from the date of complete liver resection to first recurrence or death, whichever occurred first. Recurrence was confirmed by routine CT scan. RFS and OS were estimated using the Kaplan-Meier methods.
All statistical analyses were performed using SAS version 9.3 (SAS Institute, INC., Cary, NC, USA) or R version 3.0.1 (R foundation for Statistical Computing, Vienna, Austria) using the cmprsk package. All p-values were two-sided. P-values of <0.05 were considered to indicate statistical significance.
All relevant Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria were followed in this study(9).
Results
RAS genotyping of 2764 sequential mCRC cases between 2008 and 2015 at our institution identified 87 (3.1%) mCRCs harboring activating NRAS mutations. Three cases had concurrent hotspot KRAS and NRAS mutations and were not included in the analysis of NRAS mutant mCRC. The clinical characteristics of the 84 NRAS mutant mCRC cases and of control groups of RAS wild-type mCRC and KRAS mutant mCRC, identified from cases genotyped between 2008 and 2012, are summarized in Table 1.
Table 1.
Patient characteristics.
| Total (n=982) | RAS WT (n=475) | KRAS MUT (n=423) | NRAS MUT (n=84) | p-valuea overall | p-valueb KRAS v NRAS | |
|---|---|---|---|---|---|---|
| Age at diagnosis | 0.99 | 0.92 | ||||
| Mean (years) | 58.3 | 58.2 | 58.4 | 58.3 | ||
| Gender | 0.23 | 0.28 | ||||
| Male | 527 (54%) | 265 (56%) | 214 (51%) | 48 (57%) | ||
| Female | 455 (46%) | 210 (44%) | 209 (49%) | 36 (43%) | ||
| Self-reported race* | <0.01 | 0.23 | ||||
| Caucasian | 839 (85%) | 419 (88%) | 356 (84%) | 64 (76%) | ||
| African-American | 68 (7%) | 19 (4%) | 37 (9%) | 12 (14%) | ||
| Other | 56 (6%) | 32 (7%) | 21 (5%) | 3 (4%) | ||
| Timing of metastasis | 0.98 | 0.90 | ||||
| Synchronous (stage IV at diagnosis) | 553 (56%) | 267 (56%) | 238 (56%) | 48 (57%) | ||
| Site of primary tumor** | <0.01 | <0.01 | ||||
| Right-colon | 280 (29%) | 100 (21%) | 161 (38%) | 19 (23%) | ||
| Left-colon | 149 (15%) | 71 (15%) | 46 (11%) | 32 (38%) | ||
| Rectosigmoid/rectum | 552 (56%) | 304 (64%) | 216 (51%) | 32 (38%) | ||
| MMR status | 0.96 | 1.00 | ||||
| Proficient (pMMR) | 547 (56%) | 259 (55%) | 260 (61%) | 48 (57%) | ||
| Deficient (dMMR) | 17 (2%) | 8 (2%) | 8 (2%) | 1 (1%) | ||
| Unknown/Missing | 417(42%) | 208 (44%) | 155 (37%) | 35 (42%) | ||
| First site of metastasis | 0.04 | 0.81 | ||||
| Liver | 580 (59%) | 300 (63%) | 233 (55%) | 47 (56%) | ||
| Lung | 86 (9%) | 31 (7%) | 47 (11%) | 8 (10%) | ||
| Liver + lung | 48 (5%) | 16 (3%) | 25 (6%) | 7 (8%) | ||
| Other | 268 (27%) | 128 (27%) | 118 (28%) | 22 (26%) | ||
Race was unknown for 19 patients (5 with RAS wild-type, 9 with KRAS mutant, and 5 with NRAS mutant tumors), and these cases were excluded in the p-value calculations.
Site of primary tumor was unknown for one patient with NRAS mutant colorectal cancer.
p-value comparing the 3 groups, Unknown/missing categories were excluded in the p-value calculations;
p-value comparing KRAS MUT and NRAS MUT patients.
WT – wild-type; MUT - mutant
Median age at diagnosis, gender distribution, and timing of metastasis (synchronous versus metachronous) did not significantly vary by mutation status. African-American self-reported race was enriched among the NRAS mutant mCRC cases (p<0.01), with a greater than two-fold increase in the frequency of African-American reported race in NRAS mutant mCRC compared to RAS wild-type cases (14% versus 4%). The race distribution of patients did not significantly vary between NRAS and KRAS mutant mCRC (p=0.23; African Americans 14% versus 9%). Among African Americans whose tumors were genotyped between 2008 and 2012 when all consecutive cases were reviewed, the distribution of tumor molecular subtypes was 32% RAS wild-type, 61% KRAS mutant, and 7% NRAS mutant. Seventeen patients were found to be mismatch repair protein deficient (dMRR) by immunohistochemistry. Of these five were germline mutations and 11 were somatic. One patient initiated germline testing but died shortly after and the results were not obtained.
The primary tumor site varied significantly by molecular status with the highest frequency of right-sided tumors among KRAS mutant cases (Table 1). Compared to all other cases and to the KRAS mutant mCRC, NRAS mutant cases had a significantly higher frequency of left-sided primary tumors.
The sites of metastasis involved at the time of diagnosis of metastasis also varied by molecular alteration (Table 1), but did not significantly differ between KRAS and NRAS mutant cases. Like KRAS mutant tumors, the NRAS mutant tumors had a lower frequency of liver limited metastases at diagnosis and a higher frequency of pulmonary metastases. At first diagnosis of metastatic disease, metastases were limited to the liver in 63% of RAS wild-type, 55% of KRAS mutant, and 56% of NRAS mutant cases (p=0.04). Metastases were limited to the lung in 7% of RAS wild-type, 11% of KRAS mutant, and 10% of NRAS mutant cases.
Mutation genotype and concurrent alterations
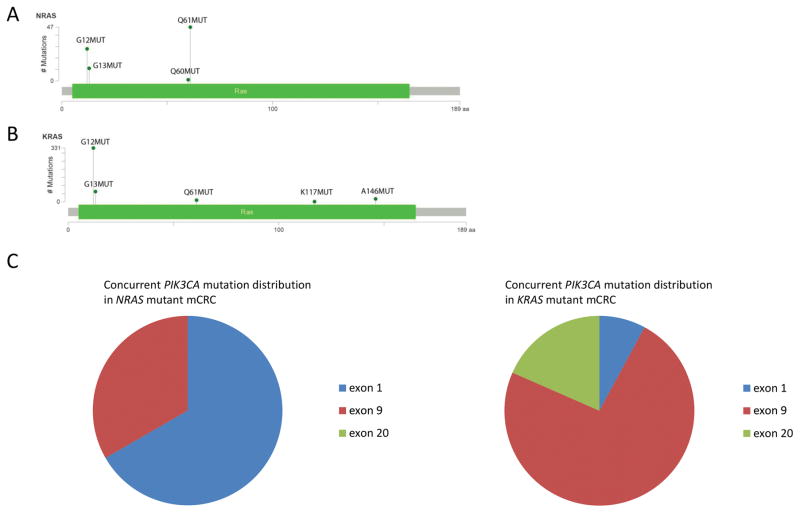
The distribution of mutations in the RAS genes and concurrent alterations in the phosphatidylinositol 3-kinase (PI3K) pathway varied significantly between the NRAS and KRAS mutant mCRC. Mutations in NRAS occurred in exon 2 in 45% of cases and in exon 3 in 55% of cases (Fig. 1A). In contrast, in the KRAS mutant mCRC cohort, 93% of mutations were in exon 2, 2% in exon 3, and 4% in exon 4 (Fig. 1B). Of the three cases with concurrent KRAS and NRAS mutations, NRAS mutations occurred in exon 2 (G12V, G13V) in 2 cases and in exon 3 (Q61K) in one case; concurrent KRAS mutations all occurred at codon 12. Overall the frequency of concurrent NRAS and KRAS mutations was low; 5% of NRAS exon 2 mutant cases and 2% of NRAS exon 3 mutant cases. Concurrent PIK3CA mutations were identified in four NRAS mutant cases (5%) (Supplementary Fig. S2), a significantly lower frequency than in KRAS mutant mCRC (18%) (p<0.01). Additionally, two NRAS mutant mCRC cases harbored concurrent AKT1 E17K activating mutations. The PI3K pathway alterations consisted of less common alterations, including three mutations in C2 (N345K twice, R357Q) and one mutation in the helical domain of PIK3CA (Fig. 1C).
Figure 1.
(A, B) Mutation map showing the location of mutations in (A) NRAS and (B) KRAS. The height of the lollipops in the plot corresponds to the number of cases with each RAS variant, as indicated on the y-axis. (C) Distribution of concurrent mutations in PIK3CA, which encodes the catalytic subunit of PI3K, in NRAS mutant colorectal cancer (left panel) and KRAS mutant colorectal cancer (right panel).
Outcomes by tumor mutational status
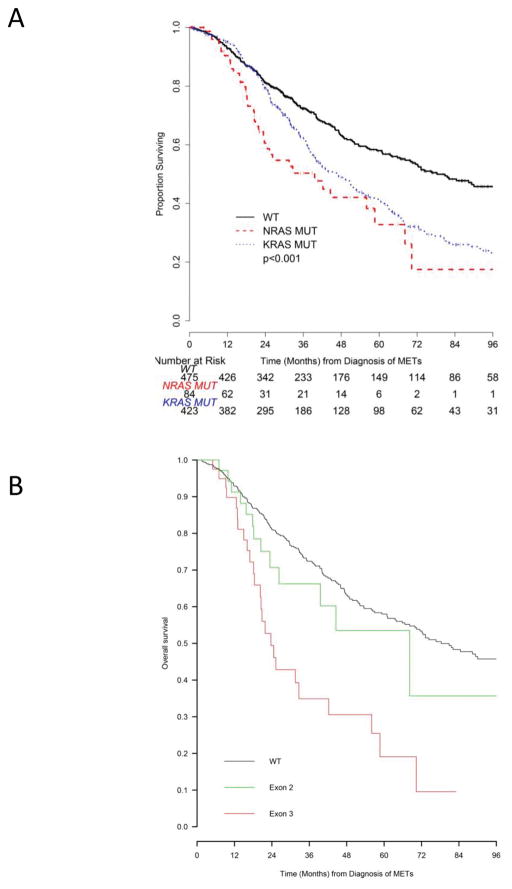
With a median follow up among survivors of 35.2 months (range 0.4–286), we observed a total of 483 deaths. OS varied by mutational status (Fig. 2A): median OS from diagnosis of first metastasis was 33 months (95% CI: 23–59) for NRAS mutant mCRC, 47 months (95% CI: 40–53) for KRAS mutant mCRC, and 78 months (95% CI: 67–98) for RAS wild-type mCRC (p<0.01). There was a trend towards worse OS between the NRAS mutant and KRAS mutant mCRC patients (logrank p=0.05). A Cox proportional model that adjusted for age at metastasis, gender, self-reported race, primary tumor site, synchronous disease, liver-limited metastases, and metastasectomy or hepatic arterial infusion treatment as time-dependent covariates, identified mutation status, the presence of extrahepatic disease, and liver resection as significant predictors of OS (Table 2). Compared to RAS wild-type mCRC, NRAS mutant and KRAS mutant mCRC had a hazard ratio (HR) of 2.0 (95% CI: 1.3–2.8, p<0.01) and 1.5 (95% CI: 1.2–1.8, p<0.01) for OS, respectively.
Figure 2.
Kaplan-Meier estimates of overall survival from diagnosis of metastatic disease (METs). (A) Overall survival by RAS mutation status: RAS wild-type (WT) cases are marked in black, KRAS mutant cases (KRAS MUT) are marked in blue, and NRAS mutant cases (NRAS MUT) are marked in red. (B) Overall survival by NRAS mutation genotype: NRAS WT cases are marked in black, NRAS exon 2 mutant cases (codon 12 or 13) in green, and NRAS exon 3 mutant cases (codon 60 or 61) in red.
Table 2.
Multivariate overall survival model.
| Characteristics | HR (95%CI) | p-value |
|---|---|---|
| Gene Group | <0.01 | |
| RAS WT | Reference | |
| NRAS MUT | 2.0 (1.3,2.8) | |
| KRAS MUT | 1.5 (1.2,1.8) | |
| Age at diagnosis* | 1.1 (0.9,1.2) | 0.12 |
| Gender | 0.06 | |
| Male | 1.2 (0.9,1.4) | |
| Female | Reference | |
| Race/ethnicity | 0.08 | |
| African-American | 1.5 (1.0, 2.1) | 0.03 |
| Others | 1.1 (0.7,1.6) | 0.70 |
| Caucasian | Reference | |
| Primary tumor site | 0.19 | |
| Right colon | 1.2 (0.9,1.5) | |
| Left colon | 1.1 (0.8,1.5) | |
| Rectosigmoid/rectum | Reference | |
| Synchronous metastatic disease | 0.30 | |
| Yes | 1.1 (0.9,1.3) | |
| No | Reference | |
| Extrahepatic disease | <0.01 | |
| Lung | 1.1 (0.7,1.6) | |
| Liver & Lung | 1.8 (1.2,2.7) | |
| Other location | 1.7 (1.4,2.2) | |
| Liver only | Reference | |
| HAI** | 0.8 (0.6,1.1) | 0.10 |
| Liver resection** | 0.5 (0.4,0.6) | <0.01 |
Hazard ratio reflects 10 years increase in age
HAI and liver resection were coded as time-dependent covariates in the multivariable model
WT – wild-type; MUT - mutant
We also evaluated OS by mutation genotype (Fig. 2B), comparing survival among three groups – NRAS exon 2 mutant mCRC (n=39), NRAS exon 3 mutant mCRC (n=48), and RAS wild-type mCRC (n=475). Among these three groups, patients whose mCRC harbored exon 3 NRAS mutant mCRC exhibited the shortest OS. OS did not vary significantly between NRAS exon 2 mutant mCRC patients and RAS wild-type mCRC patients (HR 1:33 [95% CI: 0.77–2.34], p=0.32). OS was significantly shorter for NRAS exon 3 mutant mCRC patients compared to RAS wild-type mCRC patients (HR 2.85 [95% CI: 1.87–4.36], p<0.01) and to NRAS exon 2 mutant mCRC patients (HR: 2.0 [95% CI: [95% CI: 1.04–4.0], p=0.039).
To address a potential referral bias to MSKCC for metastasectomy, we also evaluated OS among patients with mCRC who did not undergo complete resection of metastatic disease (n=397). In this group, median OS was 30 months (95% CI: 25.5–33.3) with a breakdown by mutational status of median OS 35 months (95% CI: 28–44) for RAS wild-type mCRC patients (n=169), 20 months (95% CI: 16–26) for NRAS mutant mCRC patients (n=52), and 28 months (95% CI 25–33) for KRAS mutant mCRC patients (n=176). Compared to RAS wild-type mCRC patients, NRAS mutation was associated with a HR of 1.7 (95% CI: 1.1–1.6, p=0.02) and KRAS mutation was associated with a HR of 1.5 (95% CI: 1.1–1.8, p<0,01).
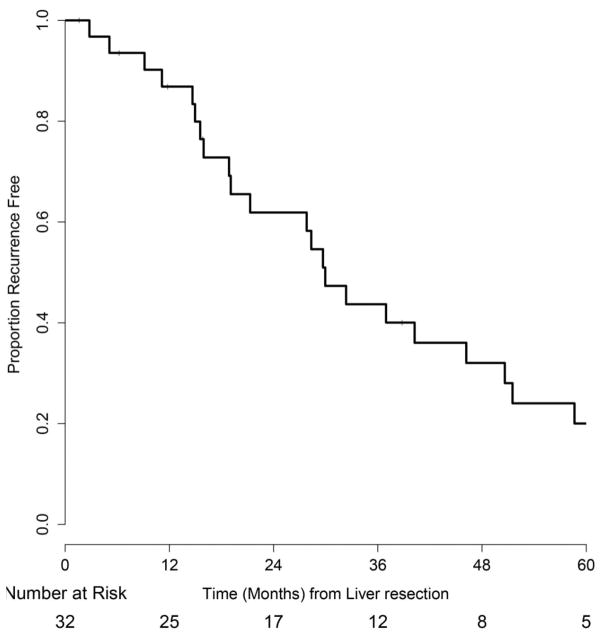
To understand the decreased survival among patients with NRAS mutant mCRC, several subset analyses were conducted. Recent data suggest that in RAS wild-type tumors, left-sided primary site is associated with significantly better prognosis than right-sided primary site(10, 11). In our study we saw a higher frequency of left-sided primary tumor sites in the NRAS mutant cases. However, among the NRAS mutant mCRC, there was no difference in OS for left-sided versus right-sided colon primary tumor site (p=0.31). We also evaluated survival among NRAS mutant mCRC patients based on outcomes after surgery or first line chemotherapy. Significantly fewer patients with NRAS mutant mCRC underwent resection of all gross metastatic disease by 12 months from time of diagnosis of metastasis compared to RAS wild-type or KRAS mutant mCRC cases (Supplementary Table 1). Thirty-four percent of NRAS mutant mCRC patients underwent metastasectomy by 12 months compared to 44% of KRAS mutant mCRC patients and 49% of RAS wild-type mCRC patients (p<0.01). Forty-seven patients with NRAS mutant mCRC had metastatic disease limited to the liver at the time of diagnosis of metastases. Thirty-two of these patients underwent hepatectomy. Recurrence-free survival at 24 months from resection of colorectal liver metastases was 62% (95% CI: 46–82%) (Fig. 3). Forty-four patients received first line 5-fluourouracil-based combination chemotherapy (with either the FOLFOX or FOLFIRI regimen) for unresectable metastatic disease; the other patients received other regimens (n=12), no chemotherapy for metastatic disease (n=10), preoperative chemotherapy before resection of metastasis (n=9), preoperative chemotherapy before radiation treatment for isolated metastasis (n=1), or were lost to follow-up (n=8). The median duration of this first line treatment was 4.7 months (range 0.5–17.9 months) in this population, a substantially shorter period than the 8-month duration for these first line regimens in clinical trials(12). Reasons for stopping first-line treatment were progression of disease (n=24), toxicity (n=7), palliative surgery for symptomatic metastasis (n=1), surgery for placement of hepatic arterial infusion pump (n=7), and treatment break (n=3). Two patients remain on first-line treatment at this time.
Figure 3.
Kaplan-Meier estimate of recurrence free survival from time of hepatectomy in patients with liver limited NRAS mutant metastatic colorectal cancer.
Response to targeted therapies
Sixteen patients (19%) received EGFR inhibitors at some point during their treatment course. Among them, three patients received EGFR inhibitors in the first line setting in combination with the FOLFOX or FOLFIRI regimens, two patients received EGFR inhibitors in combination with irinotecan on which they did not previously progress, and two patients received EGFR inhibitors together with a MEK inhibitor. The remaining nine patients received EGFR inhibitors alone or in combination with irinotecan-containing regimens after progression on irinotecan, allowing assessment of potential benefit of treatment; eight of these patients (89%) had progression of disease on first imaging and one patient had disease stability for about 8 months.
Discussion
In a large series of 84 patients with NRAS mutant mCRC, we have analyzed clinical characteristics, outcomes, and response to therapy. We find that NRAS mutant mCRC is an aggressive subset of CRC; OS for patients with NRAS mutant mCRC, particularly with exon 3 NRAS mutations, is worse than that for patients with RAS wild-type tumors or KRAS mutant mCRC. NRAS mutation defines a clinically distinct subgroup of mCRC, with increased left-sided colon primary tumors and more common in the African-American race. NRAS mutant mCRC may have a distinct molecular pathogenesis: concurrent alterations in the PI3K pathway were relatively uncommon and often involved less common pathway alterations (PIK3CA C2 domain mutations, AKT1 mutations).
As a retrospective study of patients undergoing molecular analyses and care at a single center, our study has some inherent limitations. It is possible that the prevalence of NRAS mutations in this population may not be reflective of the general population, but the incidence of NRAS mutations we found was in concordance with previously published data(2, 3). Several sequencing assays were used as technology improved during the period of the study. The sensitivity of the assay to detect hotspot mutations in RAS improved from about 5–10% to about 2% for our current multi-gene next generation sequencing panel MSK-IMPACT. Several cases were sequenced both by the sequenom assay and MSK-IMPACT, and we found no discordant cases. Genotyping for NRAS mutations began at our institution before the presence of these mutations was shown to cause resistance to EGFR inhibitors(4), and about a fifth of the patients with NRAS mutant tumors received an EGFR inhibitor at some point in their treatment. The management of these patients may be variable in other institutions which would affect the overall outcomes although first and second line therapies would be expected to be similar in clinical practice(13). The median OS in our cohort was better than would be expected based on published data(14, 15). This may be in part due to a referral bias for patients to undergo resection of colorectal liver metastases or other metastases and a focus on liver directed therapy, including hepatic arterial infusion, at our institution. To address this potential bias we analyzed OS in patients with unresectable metastatic disease and found median OS of 30 months, in line with recent OS estimates from large clinical trials(15). In this group, NRAS mutation remained a significant poor prognostic factor for OS.
Our study identifies a molecular marker which is enriched in African Americans and associated with a worse outcome. Several studies have reported on disparaging outcomes in patients with mCRC between Caucasians and African Americans(16). To date the reports have focused on socioeconomic standing including access to care and therefore a diagnosis in a more advanced stage of the disease(17, 18). Analyses of epigenetic and genetic differences in tumorigenesis in African-American patients describe discordant findings for the incidence of microsatellite instability with regards to possible differences in development of CRC in African-American patients. We saw an increase in the proportion of RAS mutant tumors, both KRAS and NRAS, among African-American patients compared to Caucasians, that was more pronounced for NRAS. Consistent with the analysis by Yoon et al of North American patients participating in a large adjuvant colon cancer trial (Alliance No147), we found that KRAS mutant tumors are the most common molecular subtype among African-American patients(19), and our data further suggest that about 68% of mCRCs in African Americans have a RAS mutation, possibly contributing to the poor outcomes seen among African Americans.
In other tumor types, differences in tumor molecular profile by patient race have been reported. Most notably, mutations in EGFR in non-small cell lung adenocarcinoma are more common among patients of East Asian ethnicity(20). Somatic NRAS mutations have not been previously associated with African-American race. Melanoma with NRAS mutation is rare among African Americans as NRAS mutations are associated with melanoma in sun-exposed skin. Review of the molecular profiles of melanoma cases seen at MSKCC suggests that 1% of NRAS mutant melanoma occur in African Americans. Similarly, NRAS mutant non-small cell lung cancer does not appear more common among African Americans; 5% of NRAS mutant non-small cell lung cancers genotyped at MSKCC occurred in African-American patients, a similar number to the relative proportion of African Americans in the patient population at MSKCC. The etiology for the increased proportion of RAS mutant mCRC among African Americans is unclear and future studies are needed to evaluate the biologic basis of this observation.
We found that NRAS mutation is associated with more aggressive disease and worse outcomes, a striking finding in the setting of a higher frequency of left-sided primaries with NRAS mutation. In our series, primary tumor site was not associated with outcome suggesting a complicated relationship between primary tumor site, tumor molecular profile, and outcomes. Several previous studies have looked at the prognostic effect of NRAS mutations. Wang et al reported that in a retrospective analysis of stage I–IV CRCs the presence of an NRAS mutation was associated with significantly shorter survival(21), and in the COIN trial of advanced CRC, irrespective of treatment, patients with KRAS or NRAS mutant tumors had shorter OS compared to those with wild-type tumors(22). A case series from Italy of mCRC patients that included 47 cases with NRAS mutant disease found a significant association between NRAS mutation and shorter OS compared to wild-type cases on univariate and multivariate analyses(23). A recent pooled analysis of five randomized trials in mCRC that included 39 patients with NRAS mutant tumors, however, did not find a significant effect of NRAS mutation status on survival(24). An analysis of NRAS mutation locus by Summers et al(25) suggests that prognostic effects of NRAS mutation in mCRC vary by mutation locus with no clear effect for exon 2 mutants, but shorter survival for codon 61 mutants. Consistent with this study, we find that exon 3 NRAS mutants are associated with a strong effect on survival (hazard ratio of 2.85 versus RAS mutant tumors and of 2.0 versus NRAS exon 2 mutant tumors) and find no significant effect for exon 2 NRAS mutants compared to RAS wild-type tumors. These data suggest exon 3 mutant NRAS tumors are an aggressive subset of mCRC and may be driving the poor outcomes we see among patients with NRAS mutant tumors. We found in our series, that despite the poor prognosis of NRAS mutant mCRC, a substantial portion of patients with liver-limited disease who were able to get to resection achieved long periods of disease control, in contrast to BRAF mutant mCRC(26). However, a lower proportion of NRAS mutant mCRC patients underwent resection of all metastases compared to patients with RAS wild-type or KRAS mutant mCRC. Additionally, patients with NRAS mutant mCRC often progressed quickly through first-line 5-fluorouracil-based combination chemotherapy, suggesting that new approaches are needed to better treat NRAS mutant mCRC.
In summary, mutations in NRAS occur in about 3% of mCRC cases and are being identified in routine clinical practice. The presence of an NRAS mutation serves as a marker of more aggressive mCRC and is enriched among African Americans. These tumors have a distinct biology, both in sites of development and co-mutation pattern. Further understanding of the biology of this aggressive subset of mCRC is needed to better target these tumors.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
NRAS mutations are being identified in routine clinical testing prior to treatment with epidermal growth factor receptor (EGFR) inhibitors with unknown clinical implications beyond lack of response to anti-EGFR therapy. In this study, we evaluated 87 consecutive cases of NRAS mutant metastatic colorectal cancer (mCRC) to determine the clinical features of NRAS mutant mCRC. We found that NRAS mutation, particularly in exon 3, is a strong, independent factor for poor overall survival in mCRC and NRAS mutant tumors are enriched among African Americans.
Acknowledgments
Supported by a Career Development Award from the Conquer Cancer Foundation of the American Society of Clinical Oncology (R. Yaeger) and the National Institutes of Health Memorial Sloan Kettering Cancer Center Core Grant (P30 CA 008748 to Craig Thompson).
Footnotes
Disclosures: There are no conflicts.
References
- 1.Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–11. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The lancet oncology. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 3.Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, et al. NRAS mutations are rare in colorectal cancer. Diagnostic molecular pathology: the American journal of surgical pathology, part B. 2010;19:157–63. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 5.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 6.Yaeger R, Cowell E, Chou JF, Gewirtz AN, Borsu L, Vakiani E, et al. RAS mutations affect pattern of metastatic spread and increase propensity for brain metastasis in colorectal cancer. Cancer. 2014 doi: 10.1002/cncr.29196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemeny NE, Chou JF, Capanu M, Gewirtz AN, Cercek A, Kingham TP, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer. 2014 doi: 10.1002/cncr.28954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vauthey JN, Zimmitti G, Kopetz SE, Shindoh J, Chen SS, Andreou A, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Annals of surgery. 2013;258:619–26. doi: 10.1097/SLA.0b013e3182a5025a. discussion 26–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–72. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 10.Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Van Cutsem E, Beier F, et al. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials. JAMA oncology. 2016 doi: 10.1001/jamaoncol.2016.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venook AP, Niedzwiecki D, Innocenti F, Fruth B, Greeene C, O’Neill BH, et al. Impact of pirmary tumor location on overall survival (OS) and progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance) Journal of Clinical Oncology. 2016;34 abstract 3504. [Google Scholar]
- 12.Tournigand C, Andre T, Achille E, Lledo G, Flesh M, Mery-Mignard D, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22:229–37. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 13.Benson AB, 3rd, Venook AP, Bekaii-Saab T, Chan E, Chen YJ, Cooper HS, et al. Colon cancer, version 3.2014. Journal of the National Comprehensive Cancer Network: JNCCN. 2014;12:1028–59. doi: 10.6004/jnccn.2014.0099. [DOI] [PubMed] [Google Scholar]
- 14.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. The lancet oncology. 2014;15:1065–75. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 15.Venook AP, Niedzwiecki D, Lenz H, Innocenti F, Mahoney MR, et al. CALGB/SWOG 80405: Phase III trial of irinotecan/5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild-type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC) J Clin Oncol. 2014;32(suppl) abstr LBA3. [Google Scholar]
- 16.Robbins AS, Siegel RL, Jemal A. Racial disparities in stage-specific colorectal cancer mortality rates from 1985 to 2008. J Clin Oncol. 2012;30:401–5. doi: 10.1200/JCO.2011.37.5527. [DOI] [PubMed] [Google Scholar]
- 17.Ashktorab H, Ahuja S, Kannan L, Llor X, Ellis NA, Xicola RM, et al. A meta-analysis of MSI frequency and race in colorectal cancer. Oncotarget. 2016;7:34546–57. doi: 10.18632/oncotarget.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams R, White P, Nieto J, Vieira D, Francois F, Hamilton F. Colorectal Cancer in African Americans: An Update. Clinical and translational gastroenterology. 2016;7:e185. doi: 10.1038/ctg.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon HH, Shi Q, Alberts SR, Goldberg RM, Thibodeau SN, Sargent DJ, et al. Racial Differences in BRAF/KRAS Mutation Rates and Survival in Stage III Colon Cancer Patients. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Velho S, Vakiani E, Peng S, Bass AJ, Chu GC, et al. Mutant N-RAS protects colorectal cancer cells from stress-induced apoptosis and contributes to cancer development and progression. Cancer discovery. 2013;3:294–307. doi: 10.1158/2159-8290.CD-12-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–14. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirripa M, Cremolini C, Loupakis F, Morvillo M, Bergamo F, Zoratto F, et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer. 2015;136:83–90. doi: 10.1002/ijc.28955. [DOI] [PubMed] [Google Scholar]
- 24.Modest DP, Ricard I, Heinemann V, Hegewisch-Becker S, Schmiegel W, Porschen R, et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol. 2016;27:1746–53. doi: 10.1093/annonc/mdw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summers M, Smith CG, Maughan TS, Kaplan R, Escott-Price V, Cheadle JP. BRAF and NRAS locus specific variants have different outcomes on survival to colorectal cancer. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1541. [DOI] [PubMed] [Google Scholar]
- 26.Yaeger R, Cercek A, Chou JF, Sylvester BE, Kemeny NE, Hechtman JF, et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer. 2014;120:2316–24. doi: 10.1002/cncr.28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.