Short abstract
Prenatal and postnatal myogenesis share many cellular and molecular aspects. Myogenic regulatory factors are basic Helix-Loop-Helix transcription factors that indispensably regulate both processes. These factors (Myf5, MyoD, Myogenin, and MRF4) function as an orchestrating cascade, with some overlapped actions. Prenatally, myogenic regulatory factors are restrictedly expressed in somite-derived myogenic progenitor cells and their derived myoblasts. Postnatally, myogenic regulatory factors are important in regulating the myogenesis process via satellite cells. Many positive and negative regulatory mechanisms exist either between myogenic regulatory factors themselves or between myogenic regulatory factors and other proteins. Upstream factors and signals are also involved in the control of myogenic regulatory factors expression within different prenatal and postnatal myogenic cells. Here, the authors have conducted a thorough and an up-to-date review of the myogenic regulatory factors since their discovery 30 years ago. This review discusses the myogenic regulatory factors structure, mechanism of action, and roles and regulations during prenatal and postnatal myogenesis.
Impact statement
Myogenic regulatory factors (MRFs) are key players in the process of myogenesis. Despite a considerable amount of literature regarding these factors, their exact mechanisms of actions are still incompletely understood with several overlapped functions. Herein, we revised what has hitherto been reported in the literature regarding MRF structures, molecular pathways that regulate their activities, and their roles during pre- and post-natal myogenesis. The work submitted in this review article is considered of great importance for researchers in the field of skeletal muscle formation and regeneration, as it provides a comprehensive summary of all the biological aspects of MRFs and advances a better understanding of the cellular and molecular mechanisms regulating myogenesis. Indeed, attaining a better understanding of MRFs could be utilized in developing novel therapeutic protocols for multiple myopathies.
Keywords: MRF4, Myf5, MyoD, myogenic determination, myogenin, myoblasts, satellite cells
Historical view
Since the seminal work of identifying the myogenic stem cells, satellite cells (SCs), in rats and frogs respectively by Alexander Mauro and Bernard Katz in 1961,1 many extensive studies have been conducted to further investigate the underlying mechanisms of skeletal muscle development and regeneration. It has become established that growth of post-natal skeletal muscle fibers is substantially dependent on SCs.2,3 However, the exact molecular mechanisms of myogenic program regulation are still incompletely understood.4
SCs are mononuclear cells located between the basal lamina and plasmalemma of the skeletal muscle fiber.3,5 These cells induce muscle growth either by fusing to preexisting myofibers (hypertrophy) or, less commonly, by fusing together to form new myofibers (hyperplasia).5–7
Surprisingly, studies on non-SCs were the first to reveal the molecular mechanisms of the myogenic program. At first, it was noticed that fraction of fibroblast cell line (C3H 10T1/2) was transformed into myogenic lineage cells after being treated with an anti-neoplastic nucleoside analogue, 5-Azacytidine.8 It was hence determined that demethylation action of 5-Azacytidine induces the whole myogenic machinery cascade.9 Thereafter, myogenic transformation was also achieved through transfection of a single gene locus.10 These preliminary steps opened the door for the identification of gene loci of four orchestrated transcription factors that regulate the process of myogenesis. These factors, which have some structural homologies and overlapped functional potentials, were appealingly designated as myogenic regulatory factors (MRFs).
Myogenic determination factor 1 (MyoD) was the first factor to be identified as a result of transfection of C3H 10T1/2 cells with cDNA containing MyoD gene loci.11,12 The other three factors were then independently identified based on the original concept of gene loci transfection. These were myogenic factor 5 (Myf5),13 myogenin14 and herculin (MRF4).15
Structure of MRFs
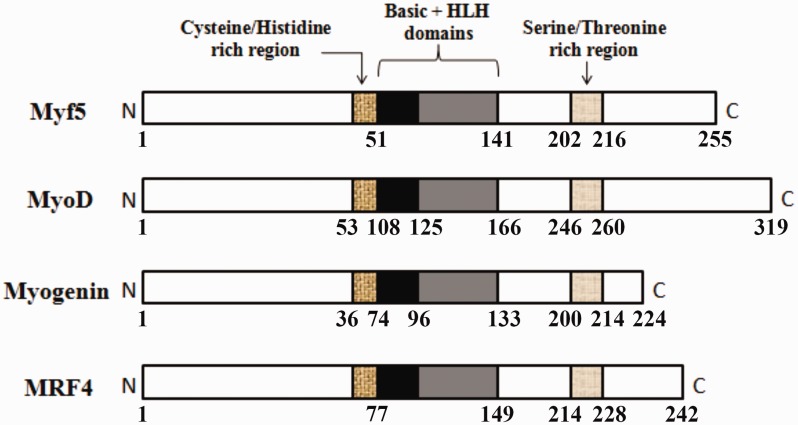
Despite variation in length and amino acid sequence, each MRF protein contains three structural domains that are almost homologous with that of other MRFs. The first one is a basic domain linked to a helix-loop-helix region, collectively called basic helix-loop-helix (bHLH) domain. The second one is a cysteine/histidine domain that lies on the N-terminal side adjacent to the bHLH. The third one is a serine/threonine-rich domain, which is located near the carboxyl terminal (Figure 1).16 It is, however, the bHLH domain that is considered the main contributor to myogenesis activation.17 The bHLH domain has been found in MRFs of many different species, such as humans, rodents, avian, and xenopus.16
Figure 1.
The primary structures for different MRFs. The three structural homologous domains of different MRFs are shown. These are; a basic domain linked to HLH region, a cysteine/histidine-rich domain that lies on the N-terminal side of the basic domain, and a serine/threonine-rich domain that is located near the C-terminal. The amino acid numbers are represented beneath each structure. (A color version of this figure is available in the online journal.)
In general, there are seven classes of HLH proteins. MRFs are considered class II proteins as their expression is restricted to a single tissue type (skeletal muscle).18 In addition, they have more tendency to heterodimerize, with proteins of other HLH classes, notably E2A proteins, before their binding to DNA.19 The E2A proteins (E12 and E47) belong to class I of HLH superfamily which is characterized by the nonspecific distribution in tissues.18 In addition, other proteins called inhibitor of DNA binding (ID) proteins can heterodimerize with either E2A or MRF proteins. These are HLH proteins belong to class V as they are characterized by their HLH regions which are uniquely not linked to basic domains.18
MRFs mechanism of action
The MRFs, via their bHLH domains, can bind specific sequence in DNA called E-box (CANNTG) that is ubiquitously found in promoter and enhancer regions of different downstream muscle and non-muscle specific genes.20 Before DNA binding, each MRF has to adopt either heterodimerization or homodimerization.19 Nonetheless, as they belong to class II of HLH proteins, MRFs have much more tendency to heterodimerize specifically with E2A proteins to proceed for myogenesis.18 However, ID proteins can act as potential competitors as they can heterodimerize with either E2A proteins or MRFs, reducing their E-box binding capacity and consequently downstream gene transcription.21
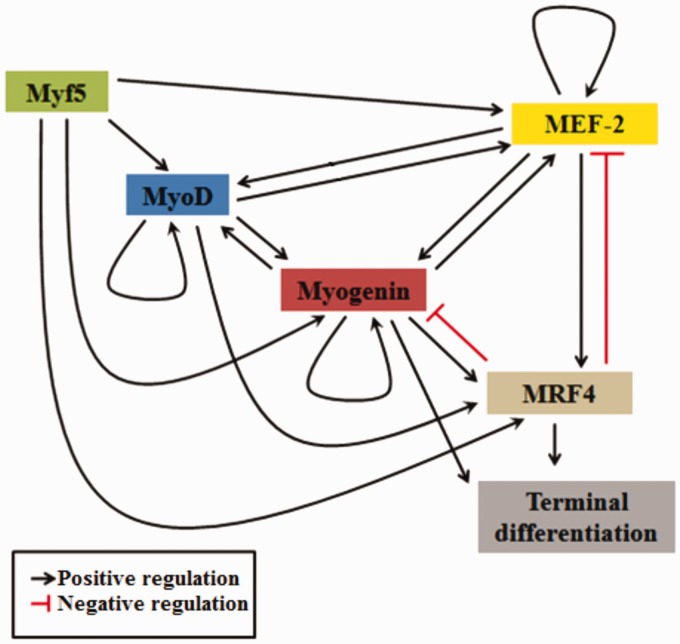
The collective action of MRFs appears highly concerted and cumulative where some factors are able to induce others, while others can further regulate their own level of expression (Figure 2).22 For example, MyoD and Myogenin have been long known to have both auto-regulatory and cross-regulatory mechanisms to modulate their respective level of expression (Figure 2).23 On the other hand, it has been shown that different portions of MRF4 promoter region can be trans-activated by all other MRFs, but MRF4 has no auto-activation role to enhance its level of expression (Figure 2).24 However, MRF4 is required to negatively regulate the level of Myogenin during terminal differentiation (Figure 2).25
Figure 2.
Signaling cascade of MRFs with their auto and cross-regulatory mechanisms. Myf5 activates all other MRFs (MyoD, Myogenin and MRF4) and MEF-2 proteins. MyoD has an auto-regulatory mechanism and cross-activation mechanism with Myogenin. Myogenin has an auto-regulatory mechanism and can induce terminal differentiation directly or/and indirectly via activation of MRF4. MRF4 directly induces terminal differentiation and can be activated by all other MRFs and MEF-2. MRF4 can inhibit both Myogenin and MEF-2 expressions. MEF-2 has an autoregulatory mechanism and can reciprocally activate both MyoD and Myogenin. (A color version of this figure is available in the online journal.)
In addition to the pivotal role of MRFs, myogenesis can be synergized by the action of another family of proteins called myocyte enhancer factor 2 (MEF-2) family. These proteins alone have almost no myogenesis potential. However, if present along with different MRFs, they can potentiate myogenesis.26 It was shown that MEF-2 proteins can positively regulate different MRFs, specifically MyoD, Myogenin, and MRF4 (Figure 2).27,28 Moreover, MEF-2 proteins level of expression can be positively regulated through both self-auto-activation and trans-activation by different MRFs.29 Unexpectedly, a recent study showed that MRF4 can target and reciprocally repress MEF-2, leading to decreased muscle growth (Figure 2).30
MRFs in prenatal myogenesis
Prenatal and postnatal myogenesis programs have many molecular features in common, wherein MRFs play indispensable roles.31 Indeed, MRFs are by far the master regulatory elements of the myogenic program as they participate in a complex network of very precisely arranged regulators with varying spatio-temporal expressions.31 Initially, groups of upstream regulators and signals have to direct undifferentiated cells to the myogenic program. Then, MRFs regulate these specified myogenic cells until their terminal differentiation.32
MRFs were thought to be simply divided into early factors (MyoD and Myf5) that are involved in the commitment and proliferation of the myogenic directed cells, and late factors (Myogenin and MRF4) which regulate the terminal differentiation of the committed cells.25 However, later accumulating evidences emphasized that MRFs are functionally overlapped. For example, MyoD can also be involved in the regulation of terminal differentiation, and MRF4 can, besides, has a role in the early commitment stage.33 Moreover, all MRFs except Myf5 can individually induce the transition of prenatal skeletal muscle precursors from proliferation to differentiation.34
Due to the incompletely defined head skeletal muscle origins and molecular regulation of their precursors, our review only discusses the role MRFs in skeletal muscles of the trunk and limbs.35 In addition, investigations on prenatal skeletal muscle development in vertebrates have been mostly conducted on avian, and murine embryos.36 Therefore, our upcoming discussion on prenatal myogenesis will be almost confined to these two species.
General overview of prenatal myogenesis
Myogenesis program indistinctly commences early during embryonic development in vertebrates. It virtually coincides with different phases of many other developmental programs, wherein numerous multi-program regulators and signals are shared. Generally, all body skeletal muscles originate from mesoderm which eventually divides into axial (notochord), paraxial, intermediate, and lateral plates.31 Specifically, limbs and trunk skeletal muscles originate restrictedly from paraxial mesoderm, whereas head and neck muscles are thought to be derived from the axial, paraxial, and lateral plates of mesoderm.37,38
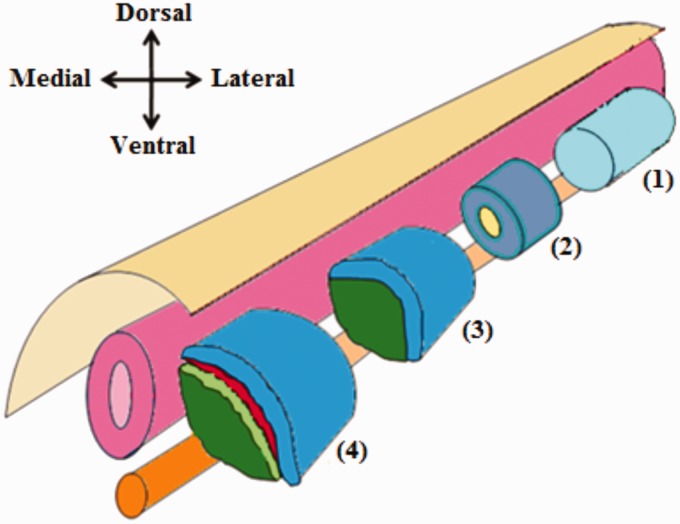
The paired box transcription factor 3 (PAX3) is diffusely detected as the first ever expressed myogenic transcription factor in paraxial mesoderm.39 PAX3 recruits and guides undifferentiated mesodermal cells toward the myogenic lineage, defining the seminal myogenic progenitor cells (MPCs).40 Paraxial mesoderm then gets segmented into epithelial blocks each with a mesenchymal core, collectively called somites (Figure 3).41 Myf5 henceforth appears in the somites as the first MRF to be expressed, mainly in their upper medial quadrants.42
Figure 3.
Differentiation of the paraxial mesoderm. Prior to somite formation, the paraxial mesoderm is composed of a single epithelial tube which flanks the neural tube and notochord (1). Later on, the paraxial mesoderm becomes segmented into somites. Each somite consists of epithelial cells with a mesenchymal core (2). Further differentiation of each somite results in the formation of two distinct layers; the dorsolateral dermomyotome, and the ventromedial sclerotome (3). After that additional two layers formed; the myotome which derived from the dermomyotome and the syndetome which derived from sclerotome (4). (A color version of this figure is available in the online journal.)
Differentiation within these somites results in two distinct portions. These two portions are the ventromedial portion, which comprises sclerotome, and dorsolateral portion, that comprises dermomyotome. Sclerotome undergoes mesenchymal transformation, lose Myf5 expression, and form another distinct layer called syndetome. Sclerotome forms the precursor cells of tendons, ribs, and vertebral column. Dermomyotome preserves its epithelial characteristics, maintain Myf5 expression, and form an underlying layer called myotome. Dermomyotome acts as a source of both precursor cells of dermis and MPCs of skeletal muscles (Figure 3).32
Undifferentiated mesodermal cells recruited toward the myogenic lineage are generally known as MPCs. These cells further acquire their myogenic identity by expressing different MRFs, and they are regarded henceforth as myoblasts.31 During embryonic period, embryonic myoblasts differentiate further and establish the basis for myogenesis by elongating and forming (Mono-nucleated) primary myofibers. After that, as fetal period commences, another class of myoblasts named fetal myoblasts, begin to either fuse with one another or with the already formed primary myofibers, which collectively results in multinucleated secondary myofibers formation.40
Detailed roles of MRFs during prenatal myogenesis
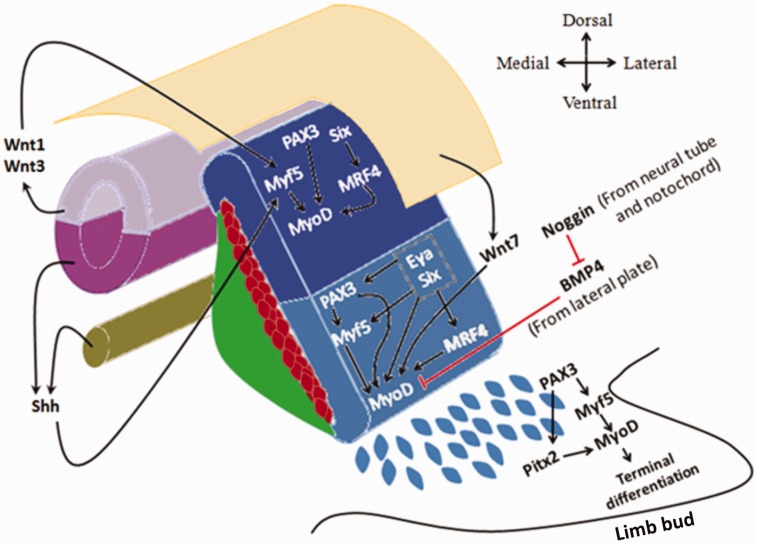
The PAX3+/MRFs- MPCs from the hypaxial lip of dermomyotome delaminate before further myogenic differentiation. These MPCs are designated for long migratory fates such as those committed toward forelimbs, hindlimbs, cervical, and occipital mesenchyme.43 These cells maintain their PAX3 expression and do not express MRFs while migrating.44 Once they reach their assigned destination, they proliferate and gradually upregulate their MRFs resulting in PAX3 down-regulation. Hence, these cells are now regarded as myoblasts (Figure 4).31 The transformation of migrating MPCs into myoblasts occurs through one of two pathways. The first canonical pathway is through PAX3 activation of Myf5, which in turn direct the expression of MyoD.45 The second pathway is through PAX3 activation of Pitx2 expression which directly activates MyoD expression (Figure 4).46
Figure 4.
Different epaxial and hypaxial myogenic factors in dermomyotome and the upstream signals. Wnt1, Wnt3, and Shh signals activate Myf5 in the epaxial dermomyotome which in turn activates MyoD expression. In addition, Six proteins and PAX3 can activate MyoD expression independent of Myf5. However, in the hypaxial dermomyotome, Myf5 expression is dependent on PAX3, Six, and Eya proteins. Never the less, all the aforementioned factors can directly activate MyoD expression, while BMP4 inhibits its expression in the hypaxial dermomyotome. In the limb bud, delaminated (PAX3+/MRFs−) cells do not express MyoD until they reach their destination. MyoD expression in these cells can be induced either canonically through PAX3 and Myf5, or non-canonically by PAX3 and Pitx2. (A color version of this figure is available in the online journal.)
Subsequently, Myf5 predominates in the epaxial part of dermomyotome, which then expresses MRF4 and MyoD. The Myf5 also appears in the hypaxial dermomyotome along with MyoD. Then, MyoD becomes the remarkably dominant MRF in the hypaxial dermomyotome as a result of Myf5 direct activation.45 However, MyoD expression in the hypaxial dermomyotome can occur even in the absence of Myf5. This indicates that Myf5 expression is dispensable for the expression of MyoD within hypaxial dermomyotome (Figure 4).47
Upstream signals from different sources in the embryo (i.e. neural tube, notochord and dorsal ectoderm) are responsible for the patterning of dermomyotome and its different MRFs spatial distribution. Signals of the Wnt family including Wnt1 and Wnt3 (originating from the dorsal neural tube) and Sonic hedgehog (originating from both ventral neural tube and notochord) directly induce Myf5 expression in the epaxial portion of the dermomyotome (Figure 4).48,49 Another signal of the Wnt family, Wnt7, from the dorsal ectoderm targets the hypaxial portion of the dermomyotome where it directly activates MyoD expression (Figure 4).50 Bone morphogenetic protein 4 (BMP4) expressed by the lateral mesoderm counteracts Wnt7 activity and inhibits premature MyoD expression. Yet, BMP4 is in turn negatively regulated by a protein called Noggin, which is Wnt 1 and Shh dependent, leaving Wnt7 activity unopposed.51 Other members of the Wnt signaling group such as Wnt4, Wnt5a, and Wnt6may also activate both Myf5 and MyoD expression (Figure 4).48
Other upstream regulators include PAX3 and PAX7 proteins. PAX3can be expressed throughout the dermomyotome, specifying all MPCs, while the expression of its paralogue, PAX7, is restricted to the central portion of the dermomyotome.31 However, Myf5 expression in the hypaxial dermomyotome appears to be PAX3 dependent, while it is not within the epaxial dermomyotome (Figure 4).45,52 In general, during the early embryonic period, both PAX3 and Myf5 lie upstream of MyoD in the myogenic cascade. Later on, as PAX7 upregulates, Myf5 and MyoD together lie downstream of PAX3 and PAX7.52 Nevertheless, PAX7 exerts much more important roles during postnatal myogenesis of growth and regeneration.53
Other upstream signaling proteins include the sine oculus family members Six1 and Six4, and eye absent family members Eya1 and Eya2.54 These signaling proteins can act directly on MRFs or indirectly through regulating PAX3 expression.55 In epaxial dermomyotome, PAX3 expression appears to be independent of Six or Eya proteins. However, in hypaxial dermomyotome, both Six and Eya are immensely required for PAX3 to be expressed.56 On the other hand, in epaxial dermomyotome, only MRF4 has been found to be under direct activation of Six proteins.54 Nevertheless, Myf5, MRF4, and MyoD can be directly activated by both Six and Eya proteins in the hypaxial dermomyotome (Figure 4).56
It has been revealed that the expressed Myf5 in epaxial dermomyotomal MPCs is required for the development of back muscles (epaxials). MyoD in the MPCs of the hypaxial dermomyotome is essential for development of limbs, tongue, and diaphragm muscles (hypaxials). However, intercostal and abdominal wall muscles have dual origins (epaxial–hypaxial).57 Furthermore, it has been disclosed that Myf5 and MyoD are not only MPCs fate determinants, but are also important requisites for their proliferation.58
Subsequently, groups of the MPCs, initially from the epaxial followed by hypaxial, caudal, and rostral lips of the dermomyotome, migrate underneath the dermomyotome forming the primary (early) myotome. Another waves of the dermomyotomal MPCs successively invade the primary myotome aiding its growth and differentiation leading to formation of the secondary (late) myotome.59 Myotome is regarded as the first differentiated skeletal muscles to occur in the body.60 The MPCs in the myotome will downregulate PAX3 expression and upregulate MRFs expression to become myoblasts. These cells eventually elongate, fuse with each other, and terminally differentiate forming myotubes and myofibers.61
The growing myotome remains in place serving as a source of myogenic cells for some axial muscles.62 The myogenic regulatory factor Myf5 has been found to be a substantial requisite for early myotome development, while it is not during further consolidation and differentiation of the early myotome.63 However, even in the absence of myotome, as a result of Myf5 and MRF4 inactivation, there will be no effects on the muscle development, and the myogenesis process can proceed normally.64
It has been revealed in mice, that loss of both MyoD and Myf5 during the embryonic period results in complete absence of myoblasts and consequently skeletal muscles postnatally. However, single mutation of either one has inconsiderable effects, raising the possibility of compensation of one another supposedly via their partial redundant action. Early MPCs lacking Myf5 can only be compensated by MyoD or to a lesser extent MRF4 to pursue their myogenic pathway, otherwise they adopt other non-myogenic fates.65 Unexpectedly, it has been shown that MRF4 can rescue myogenesis in the early stages even in complete absence of both Myf5 and MyoD.66 On the other hand, myogenin absence leads to loss of skeletal muscles, despite the presence of normal committed myoblasts and death shortly after birth.67 This indicates that there is no possible compensation for myogenin absence, and it has an indispensable role in terminal differentiation of myoblasts.68 In addition, previous studies reported the presence of a differential transcriptional activation among the four MRFs. For example, it has been shown that MyoD, Myf5, and Myogenin are capable of activating several muscle specific genes including Myosin light chain, muscle creatine kinase, and acetylcholine receptor α subunit chain, while MRF4 is inefficient in activating the transcription of the aforementioned genes.69,70
MRFs in postnatal myogenesis
As in prenatal skeletal muscle development, MRFs play a pivotal role in myogenesis during postnatal life.71 Postnatal growth, repair, and regeneration have nearly the same molecular mechanisms, wherein several growth factors activate quiescent SCs so they adopt myogenesis program that is precisely regulated by the MRFs orchestra.72 While prenatal development of skeletal muscles is achieved by the maturation of temporally different groups of myoblasts (embryonic and fetal), the postnatal growth and regeneration are indispensably dependent on SCs.40 Although, SCs are described to be heterogeneous with regard to their origin and functional status.73
During the early postnatal life, SCs are plentiful in skeletal muscle tissue where they account for nearly 30% of all nuclei identified underneath the basal lamina. This relative abundance of SCs gradually declines to reach 5% or less within adult skeletal muscles.74 SCs are small cells that are uniquely interposed between the myofiber plasma and basement membranes, while myonuclei are located within the myofiber just underneath the plasma membrane.7,75 SCs have distinct structural characteristics including their scant cytoplasm and relatively small nuclear size in comparison with that of myonuclei.75 In addition, these cells are characterized by their specific expression of PAX7 protein which is always upstream of MRFs. It is known that PAX7 has an anti-apoptotic role in SCs. In case of PAX7 absence, SCs are lost postnatally and skeletal muscles undergo atrophy and cannot regenerate even in the presence of PAX3. This indicates that PAX3 has no compensatory potential for PAX7 absence in SCs.76,77
General overview of postnatal myogenesis
Postnatal and adult skeletal muscle tissue, like any other living tissue, is in a dynamic state with continuous turnover due to daily minor traumas and small lesions.78 However, in the case of prominent or recurrent traumas, regenerative myogenesis is accompanied by fibrosis and adipogenesis. This occurs due to the activation of skeletal muscle resident cell population called fibro/adipogenic progenitors (FAPs) rather than SCs.79 However, signals coming from activated FAPs have been shown to be required during differentiation of SC-derived myogenic cells. Thus, a balanced action of both SC and FAP populations is important for normal myogenesis.80
In general, the process of postnatal myogenesis due to an injury of skeletal muscle can be divided into three main stages; degeneration, regeneration, and remodeling.81 During degeneration, necrosis accompanied by inflammatory process occurs in the injured tissue. This is followed by the overlapped stages of regeneration and remodeling mediated via the process of SC activation, proliferation, and differentiation which is orchestrated by the expression of different MRFs within SCs.82
Detailed roles of MRFs during postnatal myogenesis
MRFs during SC quiescence
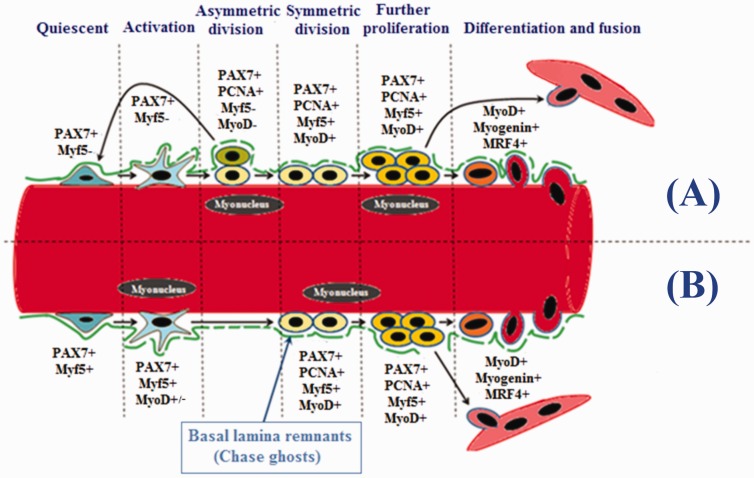
SCs are usually quiescent cells, which means that they are arrested in G0 phase.7 These quiescent SCs are almost negative for MRFs. This is achieved by their ubiquitous high expression of PAX7 and, to a lesser extent, PAX3.76 However, it has been, surprisingly, found that disparate levels of Myf5 protein are detectable within most of quiescent SCs (Figure 5).83 It was shown that about 90% of SCs have an active Myf5 transcriptional locus, which indicates that these quiescent SCs are already directed to the myogenic pathway.84 Significant levels of Myf5 translation are inhibited within quiescent SCs by miR31 which sequester myf5 mRNA in mRNP particles.85 The remaining (∼10%) of SCs are described as stem cells rather than myogenic progenitors, due to their lack of Myf5 expression (Figure 5).77
Figure 5.
Expression of myogenic regulatory factors during different stages of satellite cells. Satellite cells are divided into two groups according to their self-renewal ability; myogenic stem (∼10%, PAX7+/Myf5−) and myogenic precursor (∼90%, PAX7+/Myf5+) cells. (A) Myogenic stem cells can undergo asymmetric divisions where some daughter cells return back to quiescence. (B) Myogenic precursor cells undergo symmetrical divisions where all daughter cells are committed to the myogenic fate. Proliferating satellite cells can be identified by the expression of PCNA. Upregulation of MyoD along with the expression of Myogenin and MRF4 induces these cells into the differentiating stage. Terminally differentiated cells may either fuse into pre-existing myofibers, or into newly formed one. Chase ghost act as a scaffold for myogenic stem and precursor cells during activation, proliferation, and differentiation. (A color version of this figure is available in the online journal.)
Interestingly, it is the Myf5-ve SC pool which undergoes depletion in mdx mice leading to diminished SC self-renovation capacity. This occurs because Myf5-SCs tend to have a relatively higher self-renovation ability compared with Myf5+ ones.86 On the other hand, almost all SCs have been shown to be derived from prenatal MyoD+ precursors.87 However, neither the MyoD mRNA transcript nor MyoD protein are detectable within postnatal quiescent SCs.88 MyoD locus is maintained non-transcriptable within the peripheral SC nuclear heterochromatin by a dimethyltransferase protein called Suv4–20h1 to promote SC quiescence.89
MRFs during SC activation
Different stimuli can induce myogenic SC activation such as exercise,90 electrical stimulation,91 and some pharmaceutical preparations, notably anabolic androgenic steroids.2,3,5,6 In general, upon activation, most of SCs gradually downregulate their expression of PAX792 and, if present, PAX3.76 Meanwhile, they begin to upregulate their MRFs as the myogenic program ensues.93
The process of SC activation starts principally as the level of Myf5 protein gradually increases in most of the activated SCs leaving small number of them as Myf5-. This non inclusive Myf5 expression gives rise to the “PAX7+/Myf5+ SC majority” which can divide only symmetrically. However, the “PAX7+/Myf5- SC minority” can adopt either symmetrical or asymmetrical divisions (Figure 5). Progenies of “PAX7+/Myf5+ cells” are arranged in close proximity to the plasma membrane of myofibers to pursue their myogenic fate via adopting further proliferation and differentiation. On the other hand, PAX7+/Myf5- cells become adjacent to the basal lamina where they are capable to return to the quiescent state to maintain SC pool.77
In addition to Myf5 and PAX7 expression, SCs start to express MyoD protein in early stages of their activation.94 PAX7 is substantially required to bring about the activation of SCs by its direct binding to the enhancer and promoter regions of Myf5 and MyoD, respectively.95 Activated progeny of SCs that is committed for myogenesis is thought to be guided by basal lamina remnants (chase ghosts) to migrate and proliferate in the injured myofibers (Figure 5).96
The stability of both Myf5 and MyoD factors is regulated at the translational level by miRNAs and RNA binding proteins. During SC quiescence, miRNA-31 sequester the Myf5 mRNA in isolated granules, which will dissociate upon SC activation.85 Moreover, fragile X mental retardation protein, in cooperation with miRNA pathway, inhibits Myf5 translation by direct binding to Myf5 transcripts and impacting its deadenylation.97 The mRNA binding protein tristetraprolin (TTP) was found to promote the decay of MyoD mRNA through binding to MyoD mRNA 3′ region. However, upon SC activation, the p38 mitogen-activated protein kinases α and β were found to promote TTP inactivation and therefore stabilize MyoD mRNA.98
MRFs during SC proliferation
The most prominent myogenic factors expressed in proliferating SCs are Myf5 and MyoD.94 These two factors are also expressed in activated SCs. Yet, the best distinguishing marker for proliferating SCs is the expression of proliferating cell nuclear antigen (PCNA). After proliferation, the majority of cells start to downregulate their PAX7 and maintain MyoD to proceed for differentiation. The remaining cells maintain PAX7 expression and suppress MyoD to return back to quiescence (Figure 5).94
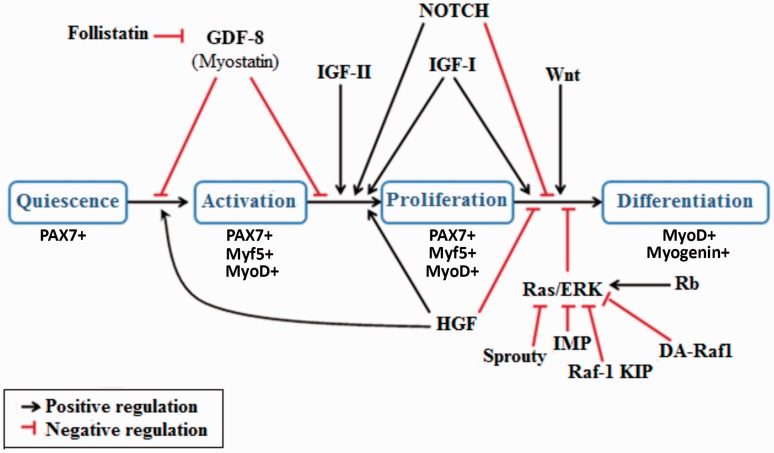
Activation of Ras-ERK pathway is important to maintain SCs in proliferative state by inducing the retinoblastoma protein expression which leads to the suppression of myogenin. Thus, to induce SCs differentiation through MRFs canonical expression, Ras-ERK pathway must be suppressed (Figure 6).99 Many proteins have been identified to block the Ras-ERK pathway including Sprouty,100 Impedes Mitogenic signal Propagation,101 Raf-1 Kinase Inhibitor Protein,102 and DA-Raf1.99 In addition, activated and intra-nuclear translocated Notch-1 protein can promote SC proliferation through keeping MyoD expression at low levels (Figure 6).103 It is important to note that upregulation of MyoD expression is required for SCs to exit the cell cycle and enter the differentiation state.104 Previous studies reported that MyoD expression induces differentiation by increasing the expression of cyclin-dependant kinase inhibitor protein p21.104
Figure 6.
Illustrating diagram for several growth factors and signaling molecules that regulate different stages of satellite cells. Transition from quiescence into activation and proliferation is stimulated by HGF and inhibited by myostatin (GDF-8). IGF-II induces proliferation. IGF-I can induce both proliferation and differentiation. HGF and NOTCH sustain proliferation and inhibit transition to differentiation. Transition to differentiation is also inhibited by Rb protein through activating Ras/ERK pathway. Transition to differentiation can be induced either directly by the Wnt family of proteins, or indirectly via inhibition of the Ras/ERK pathway by Sprouty, IMP, Raf-1 KIP, and DA-Raf1. (A color version of this figure is available in the online journal.)
Role of growth factors in SC activation and proliferation
Several growth factors are expected to play an important role in SC activation and proliferation. For example, fibroblast growth factor 2 (FGF-2) recruits more SCs to the cell cycle, providing more (PCNA+/MyoD+ SCs) to the myogenesis program (Figure 6). This occurs without affecting the transit time from proliferation (PCNA+/MyoD+ SCs) to differentiation (MyoD+/Myogenin+ SCs).105 Insulin-like growth factors (IGF-I and IGF-II) have been shown to induce proliferation (PCNA+/MyoD+ SCs). However, IGF-I appear to be much more potent in differentiation (MyoD+/Myogenin+ SCs).106 Hepatocyte growth factor (HGF) has been shown to induce SC activation and proliferation, while it inhibits heterodimerization process of MRF and E proteins. This results in inhibition of the progression toward differentiation (Figure 6).107 However, myostatin which is a member of the transforming growth factor β (TGF-β) superfamily, prevents activation of SCs and maintain their quiescence.108 Yet, follistatin antagonize the inhibitory action of many TGF-β family members, including myostatin which results in SC activation and progression into myogenesis (Figure 6).109
MRFs during SC differentiation
Transition of SCs from proliferation to differentiation is regulated principally at a level above the MRFs. This is achieved by progressive switch in the expression from Notch proteins in proliferating SCs to Wnt proteins in differentiated SCs. Wnt proteins promote stabilization of β-catenin which activates transcription of several muscle specific genes required for differentiation (Figure 6).110
At the level of MRFs, transition from proliferation to differentiation is marked by the progressive down-regulation of PAX7 expression with greater expression of MyoD. In addition, Myogenin expression upregulates and directs differentiating SCs to subsequently become terminally differentiated into myonuclei.7,75 It has been shown that the lack of MyoD delays proliferation to differentiation transition time.111
If SCs maintain PAX7 expression and suppress MyoD level, then the progression toward differentiation is impeded. However, once SCs express Myogenin, it can directly suppress PAX7 expression and differentiation ensues, which suggesting a reciprocal inhibitory mechanism between MRFs and PAX7.112 PAX7 is substantially required to bring about the activity of MRFs cascade.104 The levels of MRFs expression are adjusted by counter regulatory proteins like Id2 and Id3 which also require PAX7 to induce their expression. This provides adequate control of the myogenesis machinery that is achieved in the case of Id proteins via prevention of the unwanted early differentiation of proliferating SCs.113
MRFs are almost similarly expressed within both the prenatal MPCs-derived myoblasts and postnatal activated SCs. Interestingly, however, MRF4 is not expressed in SCs unless they become terminally differentiated. Moreover, the expression of MRF4 becomes more evident and detectable within myonuclei during the process of regeneration.114 On the other hand, Myf5 and MyoD expressing SCs contain a constellation of miRNAs, including miR206 and miR1, which are important to downregulate the expression of PAX proteins within proliferating and differentiating SCs.32 In addition, a recent study by Kim et al.115 demonstrated that miR1 and miR206 can induce MRFs expression, notably MyoD and Myogenin.
Summary
In conclusion, MRFs are considered key players in prenatal skeletal muscle formation and postnatal skeletal muscle growth and regeneration. Since their discovery 30 years ago, substantial research has been conducted to understand their exact signaling pathways and mechanisms of action. The vital role of MRFs in regulating myogenesis renders them as an excellent target for manipulation in potential stem cells therapeutic protocols for muscular degenerative disorders. For example, Tedesco et al.116 demonstrated that transduction of the MyoD gene by a lentiviral vector resulted in myogenic differentiation of pluripotent mesoangioblast-like stem cells derived from healthy individuals and from patients affected by limb-girdle muscular dystrophy type 2D (LGMD2D). Transplantation of these MyoD transducted cells into LGMD2D mice resulted in functional improvement of the dystrophic phenotype.116 Moreover, forced expression of MyoD in the human adipose-derived stem cells promoted them into the myogenic fate. These cells were able to fuse with Duchenne muscular dystrophy myoblasts and to improve dystrophin expression in vitro.117
Despite many research findings, several concepts pertaining to these factors are still incompletely understood. However, certain facts have been established regarding these factors and their role in myogenesis. All MRFs (Myf5, MyoD, Myogenin and MRF4) are members of class II bHLH superfamily of regulatory factors which are solely expressed in skeletal muscle tissue. In order to exert their role, MRFs can either heterodimerize with the class I bHLH E-proteins or, to a lesser degree, homodimerize with another MRF before binding to DNA. Prenatally, MRFs expression in the MPCs of the dermomyotome differs according to the cells location and eventual migratory route. Long fate migratory MPCs derived from the hypaxial lip of dermomyotome express PAX3 and do not express any MRF until they reach their final destination. These cells are considered the precursors of limbs, tongue, and diaphragm skeletal muscles. Other dermomyotomal cells delaminate and form a distinct layer called myotome starting from the epaxial (Myf5 dominant) and then hypaxial (MyoD dominant) regions. Myotome serves as the source of precursor cells of some prevertebral muscles. Many upstream factors and signals regulate Myf5 and MyoD expression in epaxial and hypaxial portions of the dermomyotome, including PAX3, PAX7, Shh, and Wnt family of proteins.
Postnatally, skeletal muscle growth and regeneration are mediated by activation, proliferation, and differentiation of SCs where they ultimately fuse to myofibers and their nuclei become new myonuclei. Many upstream growth factors and signals regulate the progression of SC from quiescence to activation, proliferation, and differentiation, including HGF, TGFβ, IGFs, Notch, and Ras-Erk pathway ligands. All quiescent SCs express PAX7; however, most of them (90%) also express Myf5 in addition to PAX7. The expression of Myf5 in these cells indicates that they are designated for symmetrical division giving rise to committed myogenic cells. Once activated, these SCs are characterized by their expression of MyoD. Activated SCs continue to express MyoD during subsequent stages even after their differentiation into myonuclei, which explains the high percentage (>90%) of MyoD+ Myonuclei. During proliferation, SCs can be further distinguished by their expression of PCNA. The terminally differentiated SCs can be identified by their expression of Myogenin, MRF4 as they cease PAX7 expression to become Myonuclei.
Authors’ contributions
HAA wrote the historical view and prenatal myogenesis sections, and sketched all figures. MZA wrote the postnatal myogenesis part of the review and critically revised the whole manuscript. RSS wrote the abstract and summary parts, and contributed to the editing process of the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was supported by the Deanship of Research at JUST via grant # 20160247.
References
- 1.Yablonka-Reuveni Z. The skeletal muscle satellite cell: still young and fascinating at 50. J Histochem Cytochem 2011; 59:1041–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allouh MZ, Rosser BW. Nandrolone decanoate increases satellite cell numbers in the chicken pectoralis muscle. Histol Histopathol 2010; 25:133–40 [DOI] [PubMed] [Google Scholar]
- 3.Allouh MZ, Jarrar AA, Asfour HA, Said RS, Shaqoura EI. Sustanon induces dose-independent hypertrophy and satellite cell proliferation in slow oxidative fibers of avian skeletal muscle. Histol Histopathol 2017; 19:11871. [DOI] [PubMed] [Google Scholar]
- 4.Waldemer-Streyer RJ, Chen J. Myocyte-derived Tnfsf14 is a survival factor necessary for myoblast differentiation and skeletal muscle regeneration. Cell Death Dis 2015; 6:e2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allouh MZ, Aldirawi MH. Influence of mesterolone on satellite cell distribution and fiber morphology within maturing chicken pectoralis muscle. Anat Rec 2012; 295:792–9 [DOI] [PubMed] [Google Scholar]
- 6.Allouh MZ, Aldirawi MH. Effects of sustanon on the distribution of satellite cells and the morphology of skeletal muscle fibers during maturation. Pak J Biol Sci 2012; 15:215–23 [DOI] [PubMed] [Google Scholar]
- 7.Allouh MZ, Yablonka-Reuveni Z, Rosser BW. Pax7 reveals a greater frequency and concentration of satellite cells at the ends of growing skeletal muscle fibers. J Histochem Cytochem 2008; 56:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constantinides PG, Jones PA, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature 1977; 267:364–6 [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell 1980; 20:85–93 [DOI] [PubMed] [Google Scholar]
- 10.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 1986; 47:649–56 [DOI] [PubMed] [Google Scholar]
- 11.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987; 51:987–1000 [DOI] [PubMed] [Google Scholar]
- 12.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A 1989; 86:5434–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. Embo J 1989; 8:701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 1989; 56:607–17 [DOI] [PubMed] [Google Scholar]
- 15.Rhodes SJ, Konieczny SF. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev 1989; 3:2050–6 [DOI] [PubMed] [Google Scholar]
- 16.Olson EN. MyoD family: a paradigm for development? Genes Dev 1990; 4:1454–61 [DOI] [PubMed] [Google Scholar]
- 17.Ozerniuk ID, Miuge NS. Evolutional principles of homology in regulatory genes of myogenesis. Izv Akad Nauk Ser Biol 2012; 4:383–90 [PubMed] [Google Scholar]
- 18.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 2000; 20:429–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucl Acids Res 2007; 35:7087–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 2005; 16:585–95 [DOI] [PubMed] [Google Scholar]
- 21.Ling F, Kang B, Sun XH. Id proteins: small molecules, mighty regulators. Curr Top Dev Biol 2014; 110:189–216 [DOI] [PubMed] [Google Scholar]
- 22.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005; 132:2685–95 [DOI] [PubMed] [Google Scholar]
- 23.Edmondson DG, Brennan TJ, Olson EN. Mitogenic repression of myogenin autoregulation. J Biol Chem 1991; 266:21343–6 [PubMed] [Google Scholar]
- 24.Sirri V, Leibovitch MP, Leibovitch SA. Muscle regulatory factor MRF4 activates differentiation in rhabdomyosarcoma RD cells through a positive-acting C-terminal protein domain. Oncogene 2003; 22:5658–66 [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev 1995; 9:1388–99 [DOI] [PubMed] [Google Scholar]
- 26.Jin W, Shang Y, Peng J, Jiang S. Histone H3 Methyltransferase Suv39h1 prevents myogenic terminal differentiation by repressing MEF2 activity in muscle cells. Int J Mol Sci 2016; 17:pii:E1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naidu PS, Ludolph DC, To RQ, Hinterberger TJ, Konieczny SF. Myogenin and MEF2 function synergistically to activate the MRF4 promoter during myogenesis. Mol Cell Biol 1995; 15:2707–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu W, de Folter S, Shen X, Zhang W, Tao S. Vertebrate paralogous MEF2 genes: origin, conservation, and evolution. PLoS One 2011; 6:e17334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran B, Yu G, Li S, Zhu B, Gulick T. Myocyte enhancer factor 2A is transcriptionally autoregulated. J Biol Chem 2008; 283:10318–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moretti I, Ciciliot S, Dyar KA, Abraham R, Murgia M, Agatea L, Akimoto T, Bicciato S, Forcato M, Pierre P, Uhlenhaut NH, Rigby PW, Carvajal JJ, Blaauw B, Calabria E, Schiaffino S. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nat Comms 2016; 7:12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo T. Molecular mechanisms of skeletal muscle development, regeneration, and osteogenic conversion. Bone 2015; 80:2–13 [DOI] [PubMed] [Google Scholar]
- 32.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell 2014; 28:225–38 [DOI] [PubMed] [Google Scholar]
- 33.Francetic T, Li Q. Skeletal myogenesis and Myf5 activation. Transcription 2011; 2:109–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev Biol 2000; 219:287–98 [DOI] [PubMed] [Google Scholar]
- 35.Nogueira JM, Hawrot K, Sharpe C, Noble A, Wood WM, Jorge EC, Goldhamer DJ, Kardon G, Dietrich S. The emergence of Pax7-expressing muscle stem cells during vertebrate head muscle development. Front Aging Neurosci 2015; 7:62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moncaut N, Rigby PW, Carvajal JJ. Dial M(RF) for myogenesis. FEBS J 2013; 280:3980–90 [DOI] [PubMed] [Google Scholar]
- 37.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S. The formation of skeletal muscle: from somite to limb. J Anat 2003; 202:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pu Q, Patel K, Huang R. The lateral plate mesoderm: a novel source of skeletal muscle. Results Probl Cell Differ 2015; 56:143–63 [DOI] [PubMed] [Google Scholar]
- 39.Magli A, Schnettler E, Rinaldi F, Bremer P, Perlingeiro RC. Functional dissection of Pax3 in paraxial mesoderm development and myogenesis. Stem Cells 2013; 31:59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev 2009; 23:997–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musumeci G, Castrogiovanni P, Coleman R, Szychlinska MA, Salvatorelli L, Parenti R, Magro G, Imbesi R. Somitogenesis: from somite to skeletal muscle. Acta Histochem 2015; 117:313–28 [DOI] [PubMed] [Google Scholar]
- 42.Mok GF, Mohammed RH, Sweetman D. Expression of myogenic regulatory factors in chicken embryos during somite and limb development. J Anat 2015; 227:352–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birchmeier C, Brohmann H. Genes that control the development of migrating muscle precursor cells. Curr Opin Cell Biol 2000; 12:725–30 [DOI] [PubMed] [Google Scholar]
- 44.Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 1999; 126:1621–9 [DOI] [PubMed] [Google Scholar]
- 45.Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev 2006; 20:2450–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L’honoré A, Ouimette JF, Lavertu-Jolin M, Drouin J. Pitx2 defines alternate pathways acting through MyoD during limb and somitic myogenesis. Development 2010; 137:3847–56 [DOI] [PubMed] [Google Scholar]
- 47.Brunelli S, Relaix F, Baesso S, Buckingham M, Cossu G. Beta catenin-independent activation of MyoD in presomitic mesoderm requires PKC and depends on Pax3 transcriptional activity. Dev Biol 2007; 304:604–14 [DOI] [PubMed] [Google Scholar]
- 48.Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development 1998; 125:4155–62 [DOI] [PubMed] [Google Scholar]
- 49.Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson CP., Jr., Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev 2002; 16:114–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cossu G, De Angelis L, Borello U, Berarducci B, Buffa V, Sonnino C, Coletta M, Vivarelli E, Bouche M, Lattanzi L, Tosoni D, Di Donna S, Berghella L, Salvatori G, Murphy P, Cusella-De Angelis MG, Molinaro M. Determination, diversification and multipotency of mammalian myogenic cells. Int J Dev Biol 2000; 44:699–706 [PubMed] [Google Scholar]
- 51.Hirsinger E, Duprez D, Jouve C, Malapert P, Cooke J, Pourquié O. Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development 1997; 124:4605–14 [DOI] [PubMed] [Google Scholar]
- 52.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005; 435:948–53 [DOI] [PubMed] [Google Scholar]
- 53.Buckingham M, Relaix F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin Cell Dev Biol 2015; 44:115–25 [DOI] [PubMed] [Google Scholar]
- 54.Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 2005; 132:2235–49 [DOI] [PubMed] [Google Scholar]
- 55.Relaix F, Demignon J, Laclef C, Pujol J, Santolini M, Niro C, Lagha M, Rocancourt D, Buckingham M, Maire P. Six homeoproteins directly activate Myod expression in the gene regulatory networks that control early myogenesis. PLoS Genet 2013; 9:e1003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu PX, Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol 2007; 302:602–16 [DOI] [PubMed] [Google Scholar]
- 57.Kablar B, Asakura A, Krastel K, Ying C, May LL, Goldhamer DJ, Rudnicki MA. MyoD and Myf-5 define the specification of musculature of distinct embryonic origin. Biochem Cell Biol 1998; 76:1079–91 [PubMed] [Google Scholar]
- 58.Montarras D, Lindon C, Pinset C, Domeyne P. Cultured myf5 null and myoD null muscle precursor cells display distinct growth defects. Biol Cell 2000; 92:565–72 [DOI] [PubMed] [Google Scholar]
- 59.Cheng L, Alvares LE, Ahmed MU, El-Hanfy AS, Dietrich S. The epaxial-hypaxial subdivision of the avian somite. Dev Biol 2004; 274:348–69 [DOI] [PubMed] [Google Scholar]
- 60.Balakrishnan-Renuka A, Morosan-Puopolo G, Yusuf F, Abduelmula A, Chen J, Zoidl G, Philippi S, Dai F, Brand-Saberi B. ATOH8, a regulator of skeletal myogenesis in the hypaxial myotome of the trunk. Histochem Cell Biol 2013; 141:289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yusuf F, Brand-Saberi B. The eventful somite: patterning, fate determination and cell division in the somite. Brain Struct Funct 2006; 211:21–30 [DOI] [PubMed] [Google Scholar]
- 62.Hollway GE, Currie PD. Myotome meanderings. Cellular morphogenesis and the making of muscle. EMBO Rep 2003; 4:855–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teboul L, Hadchouel J, Daubas P, Summerbell D, Buckingham M, Rigby PW. The early epaxial enhancer is essential for the initial expression of the skeletal muscle determination gene Myf5 but not for subsequent, multiple phases of somitic myogenesis. Development 2002; 129:4571–80 [DOI] [PubMed] [Google Scholar]
- 64.Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature 1996; 384:266–70 [DOI] [PubMed] [Google Scholar]
- 65.Gayraud-Morel B, Chrétien F, Flamant P, Gomès D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol 2007; 312:13–28 [DOI] [PubMed] [Google Scholar]
- 66.Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 2004; 431:466–71 [DOI] [PubMed] [Google Scholar]
- 67.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993; 364:501–6 [DOI] [PubMed] [Google Scholar]
- 68.Myer A, Olson EN, Klein WH. MyoD cannot compensate for the absence of myogenin during skeletal muscle differentiation in murine embryonic stem cells. Dev Biol 2001; 229:340–50 [DOI] [PubMed] [Google Scholar]
- 69.Chakraborty T, Brennan T, Olson E. Differential trans-activation of a muscle-specific enhancer by myogenic helix-loop-helix proteins is separable from DNA binding. J Biol Chem 1991; 266:2878–82 [PubMed] [Google Scholar]
- 70.Fujisawa-Sehara A, Nabeshima Y, Komiya T, Uetsuki T, Asakura A, Nabeshima Y. Differential trans-activation of muscle-specific regulatory elements including the mysosin light chain box by chicken MyoD, myogenin, and MRF4. J Biol Chem 1992; 267:10031–8 [PubMed] [Google Scholar]
- 71.Zhao P, Hoffman EP. Embryonic myogenesis pathways in muscle regeneration. Dev Dyn 2004; 229:380–92 [DOI] [PubMed] [Google Scholar]
- 72.Zanou N, Gailly P. Skeletal muscle hypertrophy and regeneration: interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell Mol Life Sci 2013; 70:4117–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cossu G, Tajbakhsh S. Oriented cell divisions and muscle satellite cell heterogeneity. Cell 2007; 129:859–61 [DOI] [PubMed] [Google Scholar]
- 74.Cheng W, Wang L, Yang B, Zhang R, Yao C, He L. Self-renewal and differentiation of muscle satellite cells are regulated by the Fas-associated death domain. J Biol Chem 2014; 289:5040–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Said RS, Mustafa AG, Asfour HA, Shaqoura EI. Myogenic satellite cells: biological milieu and possible clinical applications. Pak J Biol Sci 2017; 20:1–11 [DOI] [PubMed] [Google Scholar]
- 76.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol 2006; 172:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 2007; 129:999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmalbruch H, Lewis DM. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle Nerve 2000; 23:617–26 [DOI] [PubMed] [Google Scholar]
- 79.Lemos DR, Babaeijandaghi F, Low M, Chang CK, Lee ST, Fiore D. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat Med 2015; 21:786–94 [DOI] [PubMed] [Google Scholar]
- 80.Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol 2010; 12:153–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grefte S, Kuijpers-Jagtman AM, Torensma R, Von den Hoff JW. Skeletal muscle development and regeneration. Stem Cells Dev 2007; 16:857–68 [DOI] [PubMed] [Google Scholar]
- 82.Garg K, Corona BT, Walters TJ. Therapeutic strategies for preventing skeletal muscle fibrosis after injury. Front Pharmacol 2015; 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gayraud-Morel B, Chrétien F, Jory A, Sambasivan R, Negroni E, Flamant P. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci 2012; 125:1738–49 [DOI] [PubMed] [Google Scholar]
- 84.Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol 2000; 151:1221–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 2012; 11:118–26 [DOI] [PubMed] [Google Scholar]
- 86.Jiang C, Wen Y, Kuroda K, Hannon K, Rudnicki MA, Kuang S. Notch signaling deficiency underlies age-dependent depletion of satellite cells in muscular dystrophy. Dis Model Mech 2014; 7:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev Biol 2009; 332:131–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dhawan J, Rando TA. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol 2005; 15:666–73 [DOI] [PubMed] [Google Scholar]
- 89.Boonsanay V, Zhang T, Georgieva A, Kostin S, Qi H, Yuan X. Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-dependent facultative heterochromatin formation. Cell Stem Cell 2016; 18:229–42 [DOI] [PubMed] [Google Scholar]
- 90.Bazgir B, Fathi R, Rezazadeh Valojerdi M, Mozdziak P, Asgari A. Satellite cells contribution to exercise mediated muscle hypertrophy and repair. Cell J 2017; 18:473–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xing H, Zhou M, Assinck P, Liu N. Electrical stimulation influences satellite cell differentiation after sciatic nerve crush injury in rats. Muscle Nerve 2015; 51:400–11 [DOI] [PubMed] [Google Scholar]
- 92.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol 2004; 166:347–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol 2004; 275:375–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem 2006; 54:1177–91 [DOI] [PubMed] [Google Scholar]
- 95.Soleimani VD, Punch VG, Kawabe Y, Jones AE, Palidwor GA, Porter CJ, Cross JW, Carvajal JJ, Kockx CE, van IJcken WF, Perkins TJ, Rigby PW, Grosveld F, Rudnicki MA. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev Cell 2012; 22:1208–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mourikis P, Relaix F. Activated muscle satellite cells chase ghosts. Cell Stem Cell 2016; 18:160–2 [DOI] [PubMed] [Google Scholar]
- 97.Fujita R, Zismanov V, Jacob JM, Jamet S, Asiev K, Crist C. Fragile X mental retardation protein regulates skeletal muscle stem cell activity by regulating the stability of Myf5 mRNA. Skelet Muscle 2017; 7:18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hausburg MA, Doles JD, Clement SL, Cadwallader AB, Hall MN, Blackshear PJ, Lykke-Andersen J, Olwin BB. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife 2015; 4:e03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yokoyama T, Takano K, Yoshida A, Katada F, Sun P, Takenawa T, Andoh T, Endo T. DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras-ERK pathway, is required for myogenic differentiation. J Cell Biol 2007; 177:781–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 2006; 16:45–54 [DOI] [PubMed] [Google Scholar]
- 101.Ory S, Morrison DK. Signal transduction: implications for Ras-dependent ERK signaling. Curr Biol 2004; 14:R277–8 [DOI] [PubMed] [Google Scholar]
- 102.Odabaei G, Chatterjee D, Jazirehi AR, Goodglick L, Yeung K, Bonavida B. Raf-1 kinase inhibitor protein: structure, function, regulation of cell signaling, and pivotal role in apoptosis. Adv Cancer Res 2004; 91:169–200 [DOI] [PubMed] [Google Scholar]
- 103.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell 2002; 3:397–409 [DOI] [PubMed] [Google Scholar]
- 104.Allouh MZ. Satellite cell and myonuclear distribution within normal and hypertrophic models of skeletal muscle growth, and the expression of myogenic regulatory factors during growth. 2007. May 31. Ph.D. Thesis, Department of Anatomy and Cell Biology, University of Saskatchewan, Saskatoon. http://www.collectionscanada.gc.ca/obj/s4/f2/dsk3/SSU/TC-SSU-05312007090402.pdf
- 105.Kästner S, Elias MC, Rivera AJ, Yablonka-Reuveni Z. Gene expression patterns of the fibroblast growth factors and their receptors during myogenesis of rat satellite cells. J Histochem Cytochem 2000; 48:1079–96 [DOI] [PubMed] [Google Scholar]
- 106.Jiménez-Amilburu V, Salmerón C, Codina M, Navarro I, Capilla E, Gutiérrez J. Insulin-like growth factors effects on the expression of myogenic regulatory factors in gilthead sea bream muscle cells. Gen Comp Endocrinol 2013; 188:151–8 [DOI] [PubMed] [Google Scholar]
- 107.Gal-Levi R, Leshem Y, Aoki S, Nakamura T, Halevy O. Hepatocyte growth factor plays a dual role in regulating skeletal muscle satellite cell proliferation and differentiation. Biochim Biophys Acta 1998; 1402:39–51 [DOI] [PubMed] [Google Scholar]
- 108.Rathbone CR, Yamanouchi K, Chen XK, Nevoret-Bell CJ, Rhoads RP, Allen RE. Effects of transforming growth factor-beta (TGF-β1) on satellite cell activation and survival during oxidative stress. J Muscle Res Cell Motil 2011; 32:99–109 [DOI] [PubMed] [Google Scholar]
- 109.Gilson H, Schakman O, Kalista S, Lause P, Tsuchida K, Thissen JP. Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. Am J Physiol Endocrinol Metab 2009; 297:E157–64 [DOI] [PubMed] [Google Scholar]
- 110.von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol 2012; 22:602–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol 1999; 210:440–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol 2007; 177:769–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. MolBiol Cell 2009; 20:3170–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chang TH, Vincent SD, Buckingham ME, Zammit PS. The A17 enhancer directs expression of Myf5 to muscle satellite cells but Mrf4 to myonuclei. Dev Dyn 2007; 236:3419–26 [DOI] [PubMed] [Google Scholar]
- 115.Kim N, Yoo JJ, Atala A, Lee SJ. Combination of small RNAs for skeletal muscle regeneration. FASEB J 2016; 30:1198–206 [DOI] [PubMed] [Google Scholar]
- 116.Tedesco FS, Gerli MF, Perani L, Benedetti S, Ungaro F, Cassano M, Antonini S, Tagliafico E, Artusi V, Longa E, Tonlorenzi R, Ragazzi M, Calderazzi G, Hoshiya H, Cappellari O, Mora M, Schoser B, Schneiderat P, Oshimura M, Bottinelli R, Sampaolesi M, Torrente Y, Broccoli V, Cossu G. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci Transl Med 2012; 4:140ra89. [DOI] [PubMed] [Google Scholar]
- 117.Goudenege S, Pisani DF, Wdziekonski B, Di Santo JP, Bagnis C, Dani C, Dechesne CA. Enhancement of myogenic and muscle repair capacities of human adipose-derived stem cells with forced expression of MyoD. Mol Ther 2009; 17:1064–72 [DOI] [PMC free article] [PubMed] [Google Scholar]