Abstract
Duchenne and Becker muscular dystrophies (DMD/BMD) are neuromuscular disorders that primarily affect boys due to an X-linked mutation in the DMD gene, resulting in reduced to near absence of dystrophin or expression of truncated forms of dystrophin. Some newer therapeutic interventions aim to increase sarcolemmal dystrophin expression, and accurate dystrophin quantification is critical for demonstrating pharmacodynamic relationships in preclinical studies and clinical trials. Current challenges with measuring dystrophin include the variation in protein expression within individual muscle fibers and across whole muscle samples, the presence of pre-existing dystrophin-positive revertant fibers, and trace amounts of residual dystrophin. Immunofluorescence quantification of dystrophin can overcome many of these challenges, but manual quantification of protein expression may be complicated by variations in the collection of images, reproducible scoring of fluorescent intensity, and bias introduced by manual scoring of typically only a few high-power fields. This review highlights the pathology of DMD/BMD, discusses animal models of DMD/BMD, and describes dystrophin biomarker quantitation in DMD/BMD, with several image analysis approaches, including a new automated method, that evaluates protein expression of individual muscle fibers.
Keywords: Animal models, Computational tissue analysis, Duchenne muscular dystrophy, Dystrophin, Image analysis, Laminin, Utrophin
Introduction
Muscular dystrophy
Muscular dystrophies are a large, heterogeneous group of diseases that result in progressive weakness and degeneration of the skeletal muscles that control movement (Guiraud et al., 2015, Kornegay, 2017, Wicklund, 2013). Muscular dystrophy was first described in the early 1800s, and since then, many different forms have been identified, caused by an expanding list of genetic mutations (Kamdar and Garry, 2016, Tyler, 2003). More than 30 types of muscular dystrophy have been identified, and examples include Duchenne/Becker (DMD/BMD), myotonic, limb-girdle, facioscapulohumeral, oculopharyngeal, and Emery-Dreifuss (Berger and Currie, 2012, Plantie et al., 2015). The genetic mutations, major muscle groups affected by each mutation, clinical syndromes, and treatment options have been reviewed elsewhere (Bastian et al., 2015, Bonne and Quijano-Roy, 2013, Chau and Kalsotra, 2015, Fanin and Angelini, 2015, Gatica and Rosa, 2016, Luigetti et al., 2015, Madej-Pilarczyk and Kochanski, 2016, Meola and Cardani, 2015, Wang and Tawil, 2016, Wicklund, 2013). This review will focus on DMD and BMD.
Duchenne and Becker Muscular Dystrophies
DMD and BMD are X-linked disorders that have a combined estimated prevalence of 1 in 7250 males (Romitti et al., 2015). DMD affects approximately 1 in 3500–5000 live male births worldwide (Emery, 1991, Guiraud et al., 2015, Kamdar and Garry, 2016, Mendell et al., 2012, Nigro and Piluso, 2015, Robinson-Hamm and Gersbach, 2016, Romitti et al., 2015). Affected boys appear clinically normal at birth; however, signs and symptoms become apparent and a diagnosis is made by around 4–5 years of age (Guiraud et al., 2015, Mendell et al., 2012, Robinson-Hamm and Gersbach, 2016). Functional improvements due to natural growth are observed in boys younger than age 7, until the characteristic degeneration and loss of muscle tissue outpaces maturational development and physical growth (Mazzone et al., 2013, McDonald et al., 2010). As muscle deterioration overtakes muscle growth, patients with DMD have difficulty walking during childhood and become wheelchair dependent by their early teens(Bello et al., 2016, Flanigan, 2014, Guiraud et al., 2015, Kole and Krieg, 2015, Mazzone et al., 2011, Nigro et al., 1983). These patients continue to lose upper extremity function and require progressively more assisted ventilation during the teenage years (Guiraud et al., 2015). Cardiomyopathy is typically present by 14–15 years old (Viollet et al., 2012). Premature death typically occurs by 30 years of age, though recent improvements in management of cardiorespiratory function are improving life expectancy (Guiraud et al., 2015, Kohler et al., 2009, Nigro and Piluso, 2015, Zaharieva et al., 2013). BMD is milder and occurs less frequently than DMD, affecting 1:11,500–19,000 males (Kamdar and Garry, 2016). Typically, BMD manifests later in life with a mean onset of 12 years, loss of ambulation is delayed until the third decade of life, onset of cardiac involvement is variable, and the overall life expectancy is longer compared with DMD patients (Bushby et al., 1993, Finsterer and Stollberger, 2008, Flanigan, 2014, Nigro and Piluso, 2015). Though the disease is X-linked, 3–8% of female carriers can present with symptoms ranging from mild (symptomatic carriers) to rapidly-progressive DMD-like muscular dystrophy due to skewed inactivation of X chromosomes (Guiraud et al., 2015, Miyagoe-Suzuki et al., 2017).
DMD/BMD Gene Mutations
DMD/BMD results from an inherited or spontaneous mutation in the DMD gene (Kamdar and Garry, 2016). Approximately 1/3 of cases arise from spontaneous mutations (Nardes et al., 2012, Selsby et al., 2015). The correlation between mutation and phenotype is better explained by the reading frame theory of Monaco (Monaco et al., 1988). This theory argues that if a deletion leads to a shift in the open reading frame, then premature termination results in a truncated protein (or no protein), which is often associated with the DMD phenotype (Blake et al., 2002). By contrast, if a deletion does not result in a frameshift, then a partially functional, abnormally small dystrophin protein is often produced, resulting in the BMD phenotype (Blake et al., 2002). While the reading frame theory of Monaco explains >90% of cases, some cases contradict this model (Koenig et al., 1989). In this study, there were patients with milder phenotypes who had out-of-frame deletions of exons 3–7 and there were patients with deletions that did not disrupt the reading frame who exhibited more severe phenotypes; some of these latter patients had deletions large enough to likely result in a protein too small to be functional (Koenig et al., 1989). These contradictory cases suggest that there may be additional modifiers influencing the patient’s disease phenotype.
A variety of DMD mutations have been documented. Approximately two thirds are large deletions or duplications detectable by microarray analysis, while approximately one third are point mutations or small insertions/deletions detectable by gene sequencing; less than 5% of DMD mutations are in the non-coding regions of the gene (Gee et al., 2017, Koenig et al., 1989, Le Rumeur, 2015, Robinson-Hamm and Gersbach, 2016). Deletions can occur anywhere within the dystrophin gene, but are often concentrated between exons 44 and 55 (Aartsma-Rus et al., 2016). In-frame deletions in this region remove part of the rod domain (Blake et al., 2002). The DMD cases resulting from point mutations and very small insertions/deletions primarily introduce premature stop codons (Blake et al., 2002). These mutations seem to be evenly distributed throughout the gene (Blake et al., 2002).
Dystrophin and the dystrophin-associated protein complex (DAPC)
The dystrophin-associate protein complex (DAPC) is present within muscle fibers. Some components are subsarcolemmal, some are associated with the cell membrane and some are immediately outside the sarcolemma. The DAPC provides mechanical support during contraction/relaxation and likely functions in signal transduction (Heydemann et al., 2007). The DAPC is composed of dystrophin, cytoskeletal proteins, and proteins that form 3 separate complexes—the dystroglycan subcomplex, the sarcoglycan-sarcospan subcomplex and the cytoplasmic subcomplex (Ehmsen et al., 2002, Sancar et al., 2011). Proteins in these complexes include α- and β-dystroglycan, sarcoglycans, sarcospan, syntrophins, and dystrobrevin; abnormalities in these proteins may result in loss of membrane integrity, cell injury and myonecrosis (Kamdar and Garry, 2016, Quattrocelli et al., 2017). The functions of these proteins have been reviewed in (Allen et al., 2016). Mutations in some of the non-dystrophin DAPC-proteins or other glycosyltransferases that support O-mannosylation of α-dystroglycan cause non-DMD/BMD forms of muscular dystrophy (Guiraud et al., 2015, Heydemann et al., 2007).
Dystrophin is encoded by the DMD gene, which is the largest protein-coding gene in the human genome and is present on the X chromosome (Tuffery-Giraud et al., 2017). The gene is ~2.5 Mb, composed of 79 exons, and results in a 427 kDa protein predominantly expressed in cardiac, smooth, and skeletal muscle (Nardes et al., 2012, Sironi et al., 2002). Interestingly, splice variants of dystrophin are also expressed in the brain (cerebral cortical, hippocampal, and Purkinje cell neurons neurons as well as glial cells), retina, and Schwann cells (Bies et al., 1992, Sironi et al., 2002). Altered dystrophin expression in these neurons is thought to explain the cognitive impairment often observed in DMD and BMD (D’Angelo et al., 2011, Mazzone et al., 2011). In muscle, dystrophin is a protein of low abundance (0.002% of skeletal muscle protein) located on the intracellular side of the sarcolemma (Brown et al., 2012, Hoffman et al., 1987). Functionally, dystrophin is thought to act as a molecular shock absorber, as part of the DAPC, by transmitting forces generated by contraction to the extracellular matrix (Guiraud et al., 2015, Kamdar and Garry, 2016).
Utrophin, a dystrophin paralog
Utrophin is encoded by the UTRN gene on human chromosome 6 and mouse chromosome 10 (Helliwell et al., 1992, Nguyen et al., 1991, Schofield et al., 1993). It is a 395 kDa protein expressed in skeletal, smooth, and cardiac muscle (Helliwell et al., 1992, Pons et al., 1994, Schofield et al., 1993). Early in development, utrophin is present at the sarcolemma of skeletal muscle. It is progressively replaced by dystrophin, such that in adult muscle, utrophin is primarily expressed at myotendinous and neuromuscular junctions (Clerk et al., 1993, Janghra et al., 2016, Nguyen et al., 1991, Schofield et al., 1993, Tome et al., 1994). Utrophin is also expressed in vascular smooth muscle and nerves (Helliwell et al., 1992). In regenerating skeletal myofibers, utrophin is again expressed at the sarcolemma (Helliwell et al., 1992). Structurally, utrophin shares 80% sequence homology with dystrophin (Schofield et al., 1993, Tinsley et al., 1992). Despite functional differences where dystrophin, but not utrophin, plays a role in microtubule organization and nNOS localization to the sarcolemma, several studies have shown that utrophin improves function in dystrophic muscle (Anthony et al., 2014, Belanto et al., 2014, Janghra et al., 2016, Krag et al., 2004, Rafael et al., 1998, Squire et al., 2002, Tinsley et al., 1992).
Muscle pathology in DMD and BMD
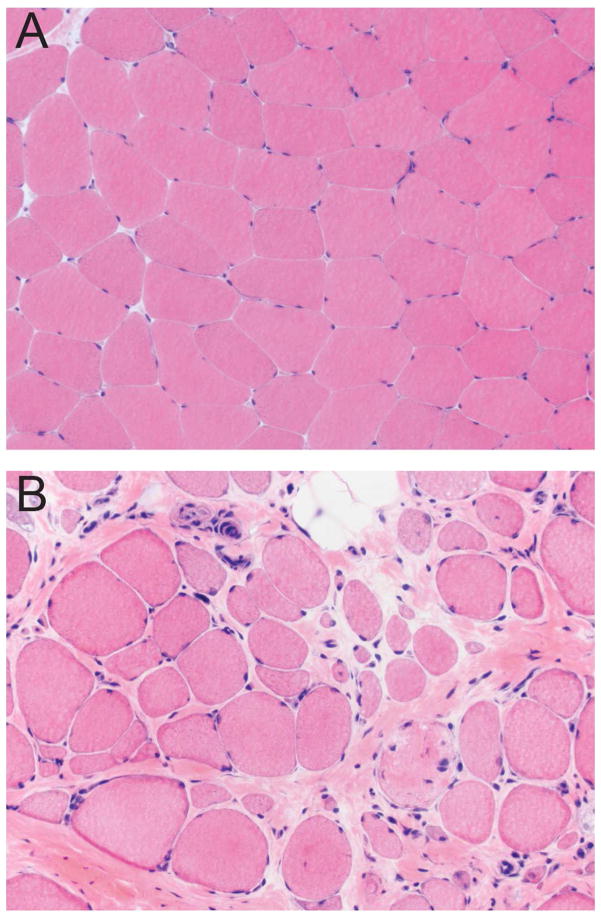
Non-diseased skeletal muscle is composed of fibers that in cross section have a relatively uniform size and polygonal shape; there is very little endomysial connective tissue separating adjacent muscle fibers (Figure 1A) (Blake et al., 2002). DMD muscle is characterized by variable degrees of atrophy, hypertrophy, myonecrosis, regeneration and endomysial fibrosis (Figure 1B) (Blake et al., 2002, Mah et al., 2016). The severity of histopathology ranges widely depending on the age of the patient and the muscle being evaluated. The absence of functional dystrophin prevents the connection between the intracellular cytoskeleton and the cell membrane, leading to repeated cycles of myonecrosis and regeneration associated with inflammation and repair (Allen et al., 2016, Blake et al., 2002, Falzarano et al., 2015, Mah et al., 2016, Zaharieva et al., 2013). Within individual muscle fibers, the necrosis is segmental and regeneration relies upon satellite cell proliferation to produce myoblasts that eventually fuse to bridge the necrotic segment (Blake et al., 2002). Macrophages are invariably present as they participate in phagocytosis of the necrotic muscle fibers (Blake et al., 2002, Madaro and Bouche, 2014). Lymphocyte infiltrates are variable, but only rarely are lymphocytes numerous enough to mimic myositis. Late in the disease process, myofibers are largely replaced by fibrofatty tissue (Blake et al., 2002, Collins and Morgan, 2003).
Figure 1. Histologic lesions of DMD muscle.
The section of normal muscle (A) demonstrates myofibers that are uniform in size, polygonal in shape, and evenly spaced with very little endomysial connective tissue. The section of DMD muscle (B) shows increased myofiber size variation (atrophy and hypertrophy), internal nuclei, myonecrosis, endomysial fibrosis, and fatty replacement. Hematoxylin and eosin.
In BMD, skeletal muscle pathology is similar to, though milder than that present in age-matched DMD patients (Kaido et al., 1991, Tachi et al., 1993). Interestingly, Kaido et al (1991) describe a distribution in BMD muscle pathology that segregates based on age at biopsy, not severity of disease presentation. In younger patients (≤15 years old), the pathology is predominantly active myonecrosis and regeneration, whereas, in older patients (>15 years old), chronic myopathic changes (increased internally-placed nuclei, split fibers, extremely hypertrophic muscle fibers, and endomysial fibrosis) were more likely to be observed (Ciciliot and Schiaffino, 2010, Kaido et al., 1991, Tachi et al., 1993). The entire spectrum of DMD and BMD histopathologic changes can also be observed in the various genotypes of congenital and limb-girdle muscular dystrophies (D’Angelo et al., 2011, Robinson-Hamm and Gersbach, 2016).
Mechanism of injury in DMD
The mechanism of injury in DMD/BMD is multifactorial and complex. Early studies suggested that membrane damage and the consequences of Ca2+ influx were key to the pathology (Allen et al., 2016, Blake et al., 2002, Deconinck and Dan, 2007, Nowak and Davies, 2004). This was supported by EM evidence of focal disruptions in the cell membrane and elevations in intracellular Ca2+ concentration (Allen et al., 2016). Later, studies in mdx mice, a mouse model that harbors a genetic mutation resulting in a DMD-like phenotype, demonstrated that muscle contraction is required for the initial muscle damage to occur (Allen et al., 2016, Shoji et al., 2015). In mdx mice with unlimited activity, histologic evidence of myonecrosis and regeneration first appears at 3–4 weeks of age and affects >50% of fibers by 8 weeks of age; whereas in muscle of limbs that were kept at a fixed length (short or long), evidence of damage (i.e. central nuclei) was not present (Allen et al., 2016, Mokhtarian et al., 1999). Similarly, in cultured mdx myotubes, resting intracellular Ca2+ levels were higher in spontaneously contracting myotubes versus those immobilized by tetrodotoxin (Allen et al., 2016, Hopf et al., 1996). Together, this suggests that dystrophin loss combined with calcium influx and muscle contraction results in muscle damage (Shoji et al., 2015).
Additional studies demonstrate a role for loss of nNOS and nitric oxide (NO) and increases in reactive oxygen (ROS) and nitrogen species (RNS) in the pathology of DMD and BMD, though the true importance of nNOS loss is not known (see (Allen et al., 2016) for detailed review). Neuronal nitric oxide synthase (nNOS), a signal transduction protein associated with the DAPC, produces NO, which may regulate local vascular perfusion, calcium mobilization, glucose metabolism and contractile function (Allen et al., 2016, Guiraud et al., 2015). In DMD patients, reduced nNOS activity results in impaired vasodilation and ischemia; however, some BMD patients lack the nNOS binding domain but remain mildly affected and ambulant, suggesting nNOS tethering at the sarcolemma is not an absolute requirement to have a mild phenotype (Anthony et al., 2011, Guiraud et al., 2015, Thomas, 2013, Torelli et al., 2004). Loss of nNOS from the sarcolemma is also associated with an increase in the ROSproducing NADPH oxidase (NOX2) (Allen et al., 2016). Increases in protein carbonyl groups and levels of isoprostanes, indicators of muscle oxidation and lipid peroxidation, respectively, have been demonstrated in muscle from mdx mice and DMD patients, suggesting a role for ROS in DMD pathology (Allen et al., 2016, Grosso et al., 2008, Hauser et al., 1995, Haycock et al., 1996, Messina et al., 2006). Finally, studies in mdx mice suggest that stretch activation of NOX2 rapidly produces ROS, triggering Ca2+ entry through stretch-activated channels, leading to increased mitochondrial Ca2+ and ROS production, ultimately resulting in muscle damage and weakness (Allen et al., 2016).
Experimental models of DMD
The number of experimental models for studying DMD is high, with more than 60 models available (McGreevy et al., 2015). Stem cell models and non-mammalian models also have a role in understanding disease pathogenesis and effects of treatments on disease pathophysiology. While each has its limitations, each of the following model systems has provided significant information in understanding DMD.
Stem cell models
Although embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) with dmd gene mutations have been used to study the skeletal and cardiac muscle pathology and drug efficacy, the focus thus far has been on modeling cardiomayopathy (Kalra et al., 2016, Smith et al., 2016). Kalra et al (2016) suggest that cardiomyocytes derived from iPSCs provide a superior alternative to the mdx mouse to study the cardiac complications of DMD. First, mdx mice develop mild disease late in their lifespan, and mdx mice with a second knockout (double knock out (dko)) that develop cardiac disease earlier have a second gene mutation not present in humans (Kalra et al., 2016). Second, mouse hearts differ physiologically from human hearts by having a faster heart rate (HR), a decrease in the force of contraction with increasing HR (versus an increase in force for humans), differences in ion channels that repolarize the plasma membrane, and differences in expression of α- vs β-myosin heavy chain (Kalra et al., 2016). Third, iPSCs can be genetically modified using the Cas9/CRISPR (clustered regularly interspaced short palindromic repeat) system to correct the mutation (Kalra et al., 2016). This provides an isogenic control in which the only difference between the two cell populations is the corrected gene sequence (Kalra et al., 2016).
Non-mammalian models
The dystrophin gene is highly conserved, and homologues have been identified in vertebrates and invertebrates, including Caenorhabditis elegans, Drosophila melanogaster, and Danio rerio (Collins and Morgan, 2003, McGreevy et al., 2015). While non-mammalian models have different musculature and pathology from their mammalian counterparts, their advantage is their physiologic simplicity and the ease of genetic manipulation (Collins and Morgan, 2003).
Zebrafish (Danio rerio) have rapid external development of translucent embryos that are easy to manipulate, which makes them attractive to researchers (Berger and Currie, 2012). They have abundant skeletal muscle and express orthologues of most DAPC proteins, that have similar membrane localization as in the human (Collins and Morgan, 2003). Dystrophin-deficient zebrafish have been identified (dmdta222a) and created by morpholino knockdown, and their histologic features include extensive muscle degeneration/necrosis, fibrosis, inflammation, activation of muscle stem cells and variation in fiber diameter (Berger et al., 2010, Berger and Currie, 2012). Similar to humans, zebrafish express dystrophin in embryos first at a junctional myotendinous location in the embryo and later at non-junctional sarcolemma (Berger et al., 2010). Failure of this transition is believed to be the major cause of the human pathology (Berger et al., 2010, Berger and Currie, 2012). Dystrophin-deficient zebrafish have been used to study exon-skipping therapies, and while they demonstrate that 20–30% of normal dystrophin levels are needed to rescue a severe phenotype in this context, they cannot be used to study issues related to systemic antisense oligonucleotide administration (Berger et al., 2011, Berger and Currie, 2012).
C. elegans are readily genetically manipulatable, and can be grown en masse, making them ideal for high-throughput genetic and pharmacologic studies (Collins and Morgan, 2003). They express the dystrophin homologue dys-1, and when in a mutated MyoD genetic background, dys-1 mutations lead to progressive impairment of locomotion (Collins and Morgan, 2003, Gaud et al., 2004). While affected nematodes have widespread degeneration of the body wall muscles, their muscular structure is dramatically different from that of mammals (Gaud et al., 2004). Their muscle fibers do not fuse and are incapable of regeneration (Gaud et al., 2004). Dystrophin-deficient C. elegans have been used to screen numerous compounds covering a wide spectrum of targets (Gaud et al., 2004). In these nematodes, prednisone reduces the number of degenerating fibers by 40%. Since they do not mount an inflammatory response to myonecrosis, this suggests that prednisone exerts a direct effect on muscle survival (Gaud et al., 2004). C. elegans has also been used to study Ca2+ signaling through their voltage-dependent Ca2+ channel (VDCC) (Zhan et al., 2014). In these studies, dystrophin was shown to be a load-bearing scaffold protein that influences membrane stiffness and transduces muscle tension to VDCCs (Zhan et al., 2014).
Drosophila melanogaster is useful because of its short generation time, large number of progeny, simple morphology, and segmental body plan (Kreipke et al., 2017, Plantie et al., 2015). The Drosophila DAPC also contains the major components present in the mammalian DAPC (Kreipke et al., 2017). Drosophila are useful for studying degenerative muscle diseases because their muscles lack satellite cells responsible for regeneration, allowing differentiation between phenotypes stemming directly from degeneration vs. those arising from failure of regeneration (Kreipke et al., 2017). Similar to C. elegans, Drosophila has also been used for small molecule therapeutics (Pantoja and Ruohola-Baker, 2013).
Rodent models
The mdx mouse is commonly used to study DMD and the dystrophic phenotype arises because of a point mutation (C to T transition) in exon 23, which results in a stop codon and truncated protein (De Luca, 2012, McGreevy et al., 2015, Selsby et al., 2015). While the mdx mouse is well-characterized (Table I), the disease phenotype in all commonly examined muscles except for the diaphragm is much milder than the human DMD phenotype (Selsby et al., 2015). These mice exhibit only a 25% decrease in lifespan (vs 75% decrease in humans) and display minimal clinical signs (McGreevy et al., 2015). Double knockout mice (dko) have been developed with varying effects on phenotype (McGreevy et al., 2015, Selsby et al., 2015). For example, dko mice lacking dystrophin and utrophin have a severe DMD-like phenotype with a significantly shortened lifespan (Deconinck et al., 1997). Second mutations have been introduced to reduce compensatory mechanisms (i.e. loss of utrophin), humanize mice (i.e. inactivation of cytidine monophosphate sialic acid hydrolase (Cmah)), reduce muscle regeneration (i.e. elimination of MyoD), and to mutate genes involved in cytoskeleton-ECM interactions (i.e. desmin and laminin); however, this introduces a second mutation not present in human DMD, complicating data interpretation (Deconinck et al., 1997, McGreevy et al., 2015, Selsby et al., 2015). Finally, mdx mice with second mutations to reduce inflammation have been developed (McGreevy et al., 2015). While mice are useful for studying pathogenesis, they are poor models of responses to gene therapy vectors. In mice, IM injection results in persistent adeno-associated virus (AAV) transduction (compared with nominal transduction in humans) (McGreevy et al., 2015).
Table I.
Comparison of clinical manifestations and histopathology of dystrophin-null mice, dogs and humans
| Feature | Humans | Mice | Dogs |
|---|---|---|---|
| Clinical manifestations | |||
| Birth body weight | Normal | Normal | Normal |
| Adult body weight | < Normal | ≥ Normal | < Normal |
| Clinical course | Severe, progressive | Mild, non-progressive | Severe, progressive |
| Lifespan | 25% of normal | 75% of normal | 25% of normal |
| Neonatal death | Rare | Rare | ~25% of affected dogs |
| Age at first clinical signs | 2–4 years old | ≥ 15 months | Birth to 3 months |
| Loss of ambulation | Common at early teenage | Rare | Uncommon |
| Muscle wasting | Progressive | Minimal until ≥ 15 months | Progressive |
| ECG abnormalities | Frequent | Frequent | Frequent |
| Cardiomyopathy | Evident at 16 years | ≥ 20 months; dilated (female) and hypertrophic (male) | Detectable at 6 months by echocardiography |
| Cognitive and CNS defects | 1/3 of affected individuals | Mild | No information available |
| Histopathology | |||
| At birth | Minimal | Minimal | Minimal |
| Acute necrosis | None | 2–6 weeks | None |
| Limb muscle fibrosis | Extensive and progressive | Minimal in adult | Extensive and progressive |
| Muscle regeneration | Poor | Robust | Poor |
Adapted from (McGreevy et al., 2015). Shading indicates characteristics that are similar to human disease.
Two different strains of dystrophin-deficient rats have been created and characterized (Larcher et al., 2014, Nakamura et al., 2014). These rats have deletions either between exons 3 and 16, or in exon 23 (Larcher et al., 2014, Nakamura et al., 2014). They have undetectable levels of dystrophin (by western blot) and <5% dystrophin positive fibers by IHC (Larcher et al., 2014, Nakamura et al., 2014). Rats with mutations in exon 23 display progressive myonecrosis and regeneration along with fibrofatty replacement (Larcher et al., 2014). Significant fibrosis arises within the heart, and they develop dilated cardiomyopathy as indicated by eccentric hypertrophy and diastolic dysfunction (Camacho et al., 2016). Because of the cardiac similarities of these rats with DMD patients, they are considered to be a better model than mdx mice for studying DMD cardiomyopathy (Camacho et al., 2016, Larcher et al., 2014).
Large animal models of DMD
DMD models have been established in the cat, dog, and pig (see Table II) (Klymiuk et al., 2013, Kornegay, 2017, McGreevy et al., 2015). Dystrophin gene disruption has been produced in a rhesus monkey, however a model has not been established (Chen et al., 2015). These models are useful for investigating responses to viral vectors. Canine models have similar nominal transduction rates as humans, and there is robust immune rejection following gene therapy as in DMD patients (Duan, 2015). These models are also useful for identifying issues from large-scale vector production and delivery (Duan, 2015). While some of these models more closely replicate the human disease than the non-mammalian and rodent models, they are costly and availability for research is limited. Additionally, the golden retriever muscular dystrophy dog (GRMD) and pig models have breeding issues. Finally, each of these large animal models is based on a single DMD mutation, which cannot model the heterogeneity of human mutations (McGreevy et al., 2015).
Table II.
Animal models of DMD
| Species | Comments |
|---|---|
| Non-mammalian | C. elegans, Drosophila, Zebrafish; rarely used in gene therapy studies |
| Mouse | Dystrophin-deficient mice; 17 models; most have point mutations, some have deletions or insertions |
| Mouse | Immune deficient mdx mice; 4 models; differing combinations of immune cell and cytokine deficiency; can be used to study cell or gene therapy without the compounding effects of the host immune response |
| Mouse | Phenotypic dko mice; 13 models; all result in severe dystrophic phenotype |
| Mouse | Dko mice with phenotype similar to mdx; 3 models |
| Mouse | Dko mice with reduced disease; 11 models |
| Mouse | Transgenic mdx mice; 13 models |
| Dog | 22 breeds; most are case reports with no colony established; 7 have established colonies |
| Rat | 2 models; exon deletion |
| Cat | 2 types; exon deletion; spontaneous mutation; rarely used in gene therapy studies |
| Pig | 1 BMD = missense mutation, 2 DMD = deletions of exon 52 |
Adapted from (McGreevy et al., 2015).
In the dog, dystrophin deficiency has been documented in 20 different dog breeds and colonies of affected animals have been established for 7 breeds (McGreevy et al., 2015). The GRMD is the most widely used dog model; these dogs have a point mutation in intron 6, one of the hotspots of mutation in DMD patients (McGreevy et al., 2015). This mutation results in exclusion of exon 7 and a frame shift that introduces a stop codon in exon 8 (Nakamura and Takeda, 2011, Selsby et al., 2015). The GRMD dogs display a similar disease course to that of DMD patients, with limb weakness and exercise intolerance at 2–3 months of age, muscle atrophy, dysphagia, abnormal weight gain, signs of heart disease by 6 months, and death at about 3 years (McGreevy et al., 2015, Nakamura and Takeda, 2011). Unfortunately, these dogs have a “honeymoon” period around 6–10 months, where disease progression stabilizes and the GRMD and human DMD phenotypes diverge (Kornegay, 2017, McGreevy et al., 2015). Though the genotype of these dogs remains consistent, phenotype can be variable and is likely due to genetic modifiers, including secreted phosphoprotein 1 (SPP1) and latent transforming growth factor β binding protein 4 (LTBP4) (Klymiuk et al., 2016, Kornegay, 2017). In the Cavalier King Charles spaniel (CKCS) with muscular dystrophy, the missense mutation was identified in exon 50, another hotspot for mutations in DMD patients, and these dogs are valuable for studying exon-skipping drugs (Nakamura and Takeda, 2011, Walmsley et al., 2010). Because of the genetic limitations of this breed, Professor Piercy at the Royal Veterinary College (United Kingdom) has engineered a model of DMD in the beagle with this same mutation, and the natural history study of these dogs is ongoing (Porter, 2015).
Dystrophin-deficiency with multifocal myonecrosis and regeneration has been identified in cats, and the large deletion mutations are located at the muscle and Purkinje promoters of the dystrophin gene (Carpenter et al., 1989, Gaschen and Burgunder, 2001, Klymiuk et al., 2016, Selsby et al., 2015). Affected cats develop hypertrophy of the tongue, neck and shoulder muscles, as well as megaesophagus, hepatosplenomegaly, and kidney failure (Nakamura and Takeda, 2011). In contrast to DMD patients, they develop hypertrophic cardiomyopathy (Gaschen et al., 1999, McGreevy et al., 2015). With the difference in skeletal and cardiac muscle phenotype as well as tongue and diaphragm hypertrophy leading to feeding difficulties and death, these cats are rarely used as a research animal (Nakamura and Takeda, 2011, Selsby et al., 2015).
Dystrophin-deficient pigs have been generated by deleting exon 52 (Klymiuk et al., 2013). These pigs demonstrate similar progressive multifocal myonecrosis and regeneration as in DMD patients (Klymiuk et al., 2013, Klymiuk et al., 2016). Interstitial fibrosis and mononuclear cellular infiltration is also present (Koenig et al., 1989, Le Rumeur, 2015). These pigs also demonstrate similar functional impairment, but in an accelerated manner. Similar to the CKCS and beagle models of DMD with deletions of exon 50–52, the pig model can be used to study exon-skipping drugs (Klymiuk et al., 2016). The pigs can also be used to study proteomic changes at the various disease stages (Frohlich et al., 2016). Affected pigs have a maximum life expectancy of 14 weeks and since the male pigs do not live to breeding age, these pigs cannot be propagated by breeding (Klymiuk et al., 2016). To overcome these breeding difficulties, female carriers have been generated and have been mated with wild type boars. Studies characterizing the offspring are in progress (Klymiuk et al., 2016).
Evaluating efficacy of therapeutic candidates
DMD pipeline
There currently is no cure for DMD/BMD and only two therapies are approved for use in these patients in the United States. Additional therapeutics are approved in Europe. One of the approved US therapies reduces inflammation (Emflaza™, the glucocorticoid deflazacort). Inflammation plays an important role in the pathogenesis of DMD. Early inflammation removes dead myofibers; however, without an increase in dystrophin, which is associated with a reduction in inflammatory marker expression, a chronic inflammatory state is established with overexpression of NF-kB, TNF-alpha and dysregulation of M1/M2 macrophages (De Luca, 2012, Madaro and Bouche, 2014). Glucocorticoids increase muscle strength, prolong independent ambulation, delay onset of cardiomyopathy, improve pulmonary function and reduce the incidence of scoliosis (Griggs et al., 2016). While clinical improvement is observed with these therapies, the improvement is temporary (disease progression is delayed), and treatment is associated with side effects, including weight gain, cataracts and bone fractures (Lim et al., 2017). Deflazacort is associated with less weight gain than prednisone (which is used off-label), and this helps reduce mechanical load on impaired muscles (Griggs et al., 2016). Because of the lack of long-term efficacy of glucocorticoids, potentially more curative therapies for DMD are in development. Strategies include cell-based strategies (transplantation of healthy myoblasts or stem cells into patients), gene-based strategies (gene therapy, exon-skipping drugs), utrophin modulators, and calcium modulators (Allen et al., 2016, Willcocks et al., 2016). Eteplirsen (Exondys 51™), the second US-approved drug, promotes dystrophin production by restoring the translational reading frame of the DMD gene by skipping translation of exon 51 (Lim et al., 2017). Ataluren (Translarna™), approved for use in the European Union, is used in patients with DMD caused by a nonsense mutation (nmDMD) (Bushby et al., 2014). Ataluren is believed to interact with ribosomes to read through premature stop codons, allowing production of the full-length protein (Welch et al., 2007).
End-points used for assessing clinical improvement
Methods for measuring efficacy can be broadly classified into two categories—clinically meaningful and surrogate. Clinically meaningful endpoints relate to quality of life, and in DMD patients, include the 6-minute walk test (6MWT), timed function tests (time to stand from a supine position, time to walk/run 10 m, time to climb or descend 4 standard-sized stairs) and quantitative strength using hand-held myometry (McDonald et al., 2013b). Surrogate endpoints are used to predict clinical benefit and include muscle enzymes, forced vital capacity and imaging (MRI-T2/H1-MRS, echocardiography), quantitative assessment of dystrophin in biopsies, and gene expression profiling. The 6MWT, MRI-T2/H1-MRS, and quantitative assessment of dystrophin in biopsies will be discussed below.
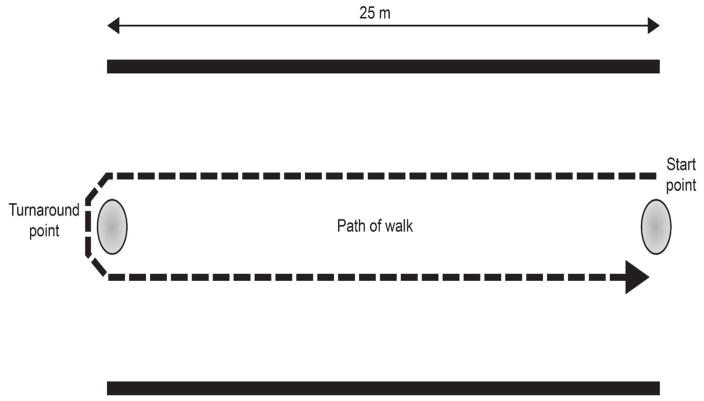
The 6MWT is an indicator of global muscle and cardiorespiratory function (Allen et al., 2016). In this test, boys walk down a flat, straight, well-lit, seldom-traveled corridor (see Figure 2) (McDonald et al., 2010, McDonald et al., 2013a). A tape line indicates the length of the course and is marked in 1m increments, and cones are used at the beginning and end of the course (McDonald et al., 2010, McDonald et al., 2013a). Distance traveled at each minute and location of falls are recorded (McDonald et al., 2010, McDonald et al., 2013a). Testing staff can provide verbal encouragement to maintain attention to the task, and “safety chasers” can help fallen boys stand (McDonald et al., 2013a). While the 6MWT is becoming the gold standard for assessing therapies, it does have practical difficulties (Allen et al., 2016). In normal boys, the distance traveled increases with age, but decreases as the disease becomes more debilitating (Allen et al., 2016). Thus, the test is insensitive to disease progression in younger boys. Also, distance increases in a drug trial are only unequivocally the result of the drug if the disease is in a stage of decline (Allen et al., 2016). This complicates data interpretation in younger boys, who might benefit most from the therapy under study.
Figure 2. Representative course for the 6-minute walk test.
This image shows the course for the 6-minute walk test (6MWT). A strip of tape 25 m long is placed in the center of a quiet, well-lit corridor at least 8 feet wide. Cones are positioned at the beginning and end of the course. Distance traveled at each minute and total are measured along with locations of falls.
Magnetic resonance imaging (MRI) and spectroscopy (MRS) are promising surrogate endpoints for DMD clinical trials (Willcocks et al., 2016). MRI-T2 images allow assessment of muscle damage, inflammation and fat infiltration, and 1H-MRS allows assessment of fat infiltration into muscle (i.e. fat fraction) (Willcocks et al., 2016). A study of 109 boys (ages 5.0–12.9 years old) demonstrated an increase in MRI-T2 signal and 1H-MRS fat fraction even in boys with stable or an improved 6-minute walk distance, suggesting that these MR biomarkers can detect subclinical disease progression (Willcocks et al., 2016).
While this technique requires trained personnel and careful calibration across test sites, it allows inclusion of younger test subjects (Willcocks et al., 2016). Additionally, these techniques allow for serial assessment of multiple muscles without needing multiple invasive muscle biopsies.
Dystrophin expression (protein and transcript) can be measured in excised muscle specimens with a variety of techniques, including western blot, mass spectrometry, ELISA, RT-qPCR and IHC/IF with scoring (Anthony et al., 2012, Anthony et al., 2014, Brown et al., 2012, Nguyen et al., 1990, Nicholson et al., 1990). Currently, quantifying dystrophin in muscle is the earliest indicator of efficacy; however, accurate assessment of dystrophin expression is challenging because of the low abundance and large size of the protein (Anthony et al., 2012, Merlini and Sabatelli, 2015, Taylor et al., 2012). Western blot, mass spectrometry, and ELISA excel at quantifying dystrophin in whole tissue samples, but lack fiber-by-fiber data and some techniques are challenged by limits of quantification (i.e. LLOQ of 3–5% for mass spectrometry is too high for DMD patients) (Anthony et al., 2014, Brown et al., 2012). While RT-qPCR does not measure dystrophin levels, this method can be used to measure dystrophin mRNA levels in patients treated with exon-skipping drugs (Anthony et al., 2012). IHC/IF excels at providing fiber-by-fiber data, exclusive assessment of muscle fibers (allowing exclusion of areas of necrosis, fibrofatty infiltrate), assessment of revertant fibers, measurement of expression heterogeneity across the tissue and within individual muscle fibers (completeness of membrane expression), and measurement of variation in staining intensity. Manual methods can provide robust data, but they must be carefully controlled or the quantification of dystrophin is likely to be affected by scoring biases (Aeffner et al., 2017, Arechavala-Gomeza et al., 2010b, Beekman et al., 2014). Despite these challenges, there is strong correlation of the number of positive fibers counted in IF-stained sections and protein quantified by western blot, suggesting that IF/IHC and western blot are comparable methods (Nicholson et al., 1993). Finally, care needs to be taken when selecting an antibody-based detection method, as inaccurate dystrophin levels will be reported if the antibody selected binds an epitope lost by a patient’s specific mutation.
Image analysis in non-clinical and clinical studies
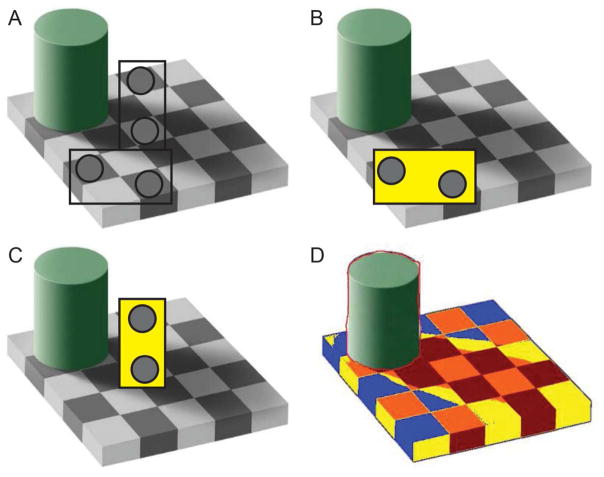
Image analysis methods quantify protein expression in IF or IHC images. Methods range from scoring small, high power fields (HPF) manually to scoring whole-slide images manually to scoring whole-slide images with image-analysis software. While HPFs are easier to score, this method is subject to bias, particularly in image selection. Scoring whole-slide images prevents the sampling bias, but manual evaluation can be time-consuming. Digital image analysis of whole sections is becoming the preferred method (Janghra et al., 2016). In addition to sampling bias, scoring biases can exist with manual scoring of either HPFs or whole-slide images. Unless scorers and scoring conditions are carefully controlled, scorers are prone to drift in threshold and estimating the percentage of membrane with positive staining, both resulting in an increase in number of positive fibers over time. The visual trap in Figure 3 demonstrates how context can impact perceived fiber intensity and why it is necessary to constantly reassess the performance of manual scorers. In this example, the shadow cast by the cylinder obscures the true intensity of the adjacent squares.
Figure 3. Adelson’s visual trap.
In this cognitive trap, two pairs of boxes are compared without (A) and with masks (B, C) to demonstrate how the brain is tricked into seeing boxes of equal intensity as different intensity (C). The image in D displays an image-analysis markup where boxes have been colored by intensity (i.e. all red boxes are the same intensity, etc.). Adapted from Adelson, EH, Lightness Perception and Lightness Illusions. In The New Cognitive Neurosciences, 2nd Edition. MIT Press (2001)
Several efforts have been made to objectively measure dystrophin intensity in DMD muscle biopsies. In the first two reports, sections were labeled with dystrophin only, circles were placed over select areas of the sarcolemma, and maximum membrane intensity within that circle was calculated (Arechavala- Gomeza et al., 2010a, Arechavala-Gomeza et al., 2010b). In the third report, sections were dual labeled with dystrophin and spectrin (a cytoskeletal protein lining the inner surface of the plasma membrane), and the average dystrophin intensity from all muscle fibers was calculated (Taylor et al., 2012). In the fourth report, sections were dual-labeled with dystrophin and spectrin, and the distribution of dystrophin intensity for each fiber was calculated (Beekman et al., 2014). Together, these methods present pros and cons of measuring dystrophin in IF-stained sections. Amongst the four reports, methods differed in measuring intensity in both positive and negative fibers, measuring mean vs. maximum intensity vs. distribution of intensity, and measuring intensity in a portion of the membrane vs. the entire membrane. While these early attempts provide reproducible data, they lack the sophistication of newer methods.
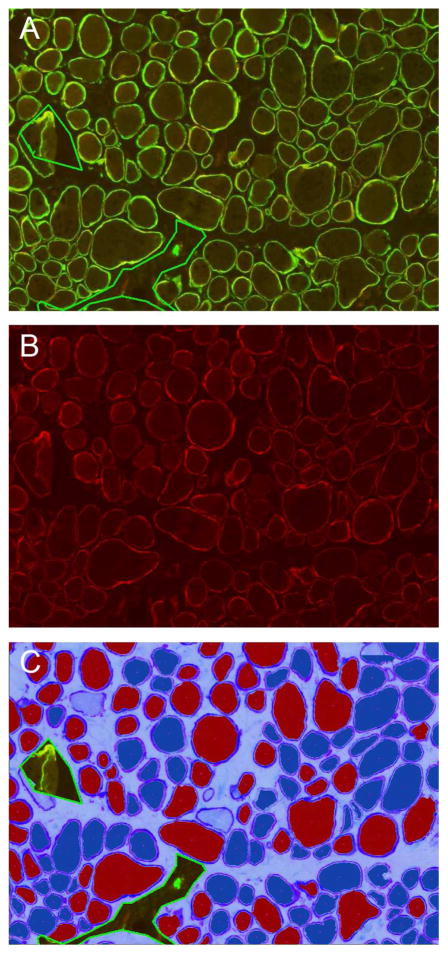
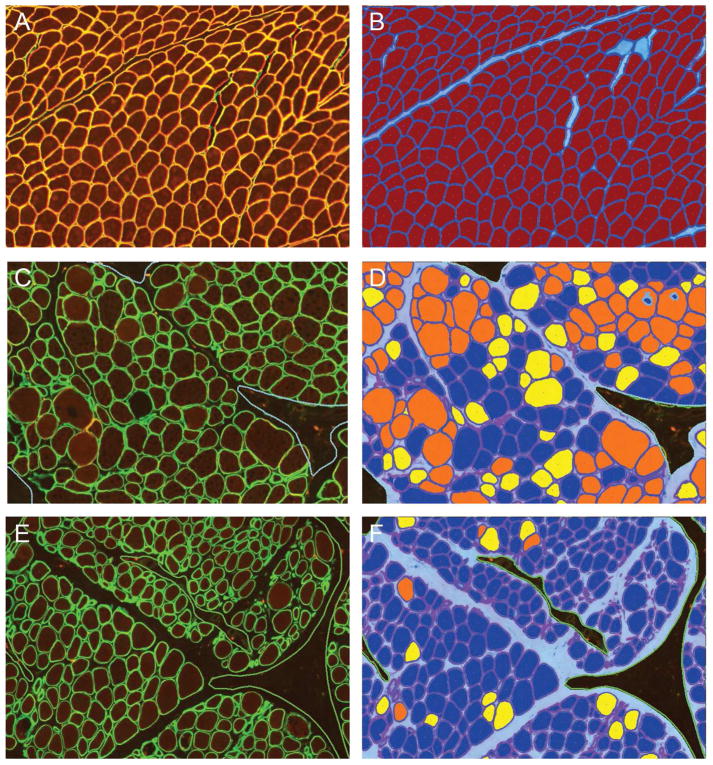
Considering the positives and negatives of the current solutions, our group has developed a software solution, MuscleMap™ that assesses dystrophin or utrophin intensity in individual fibers identified by a membrane-associated marker (i.e. laminin-α2 or spectrin) in a duplex immunostaining assay (manuscript submitted; Figure 4). Images are captured under identical conditions with a scanner qualified for reproducible image capture. MuscleMap identifies all muscle fibers using the membrane-associated marker, and then identifies and calculates the amount of dystrophin or utrophin. MuscleMap data outputs include membrane completeness (a measure of the circumference of the membrane with dystrophin or utrophin expression), biomarker intensity (which can be gated by thresholds into positive/negative or 0–3+ bins (Figure 5)), and a mathematic combination of these two values (the 4CC score). MuscleMap also calculates morphometric features (such as minimum fiber diameter), assesses levels of intra- and inter-fiber heterogeneity, and counts the number of revertant fibers. While first developed to analyze IF-stained images, MuscleMap can now be used to analyze IHC-stained images, as referenced in the future steps below and illustrated in Figure 6 (Tinsley et al., 2016). Overall, these image analysis techniques on images captured under controlled conditions help to overcome biases in sample selection as well as manual scoring. These standardized methods can be used to accurately and consistently measure biomarker changes to evaluate drug efficacy.
Figure 4. MuscleMap consistently scores protein expression in muscle fibers.
This panel shows the dual labeled assay in A (TRITC-dystrophin, FITC-spectrin), with just the TRITC-dystrophin image in B and the image analysis markup in C showing positive and negative fibers. A fiber is classified as negative (dark blue cytoplasm in C) or positive (red cytoplasm in C) based on achieving a minimum with respect to membrane intensity and % circumference at that threshold. The image in C shows three areas of artifact excluded from analysis (indicated by the green line) as well as several fibers not meeting the fiber definition (light blue center) and not included in the analysis.
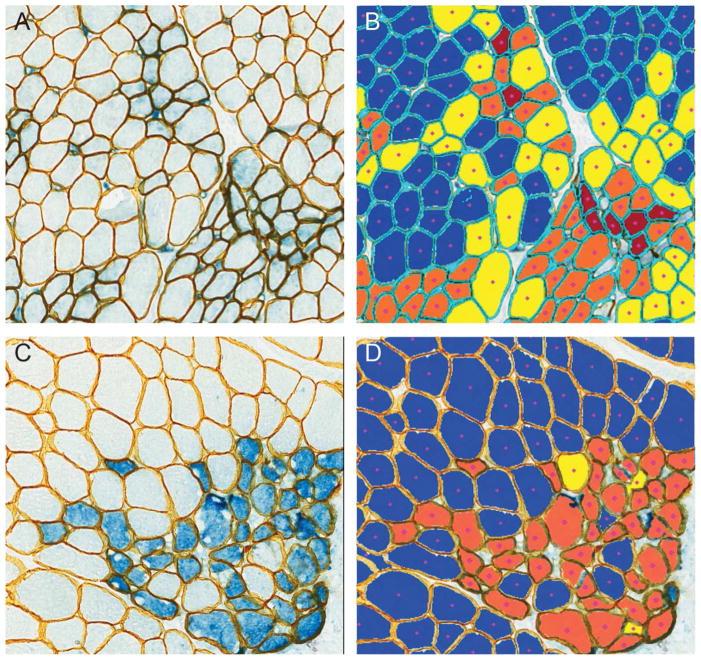
Figure 5. MuscleMap separates muscle fibers by expression intensity.
This panel shows the dual labeled images in A, C, and E and the respective image analysis markups in B, D and F. In B, D and F, muscle fibers have been classified by intensity, where dark blue = negative, yellow = low intensity, orange = medium intensity and red = high intensity. Similar to Figure 4, areas of artifact (dashed green line) and myofibers not meeting the fiber definition (light blue center) were not included in the analysis. Muscle in A, C and E are from a non-DMD/non-BMD control, BMD patient and DMD patient, respectively.
Figure 6. MuscleMap measures pathologist-impossible endpoints.
This panel shows DMD muscle samples stained with two duplex IHC assays: in A, utrophin is in blue and laminin-α2 is in yellow, and in C, developmental myosin heavy chain is in blue and laminin-α2 is in yellow. In images B and D, muscle fibers have been classified by intensity, where dark blue = negative, yellow = low intensity, orange = medium intensity and red = high intensity. Myofibers not meeting the fiber definition (uncolored) were not included in the analysis.
Future directions
While the number of potential therapies is increasing, <20% of Phase II trials are successful (Kornegay, 2017). These clinical trials often fail from either safety concerns or lack of efficacy, which results from the inability to accrue sufficient patients to achieve the necessary power (Kornegay, 2017). Because of the challenges in recruiting large numbers of patients, classical large clinical trial designs are not practical or even possible. This raises the question: are we using the best primary endpoint for these trials? Do we need to refine the methods of the 6MWT/timed functional tests or do we need to choose an alternative? In late 2016, the United States Congress passed the 21st Century Cures Act, which allows companies to seek accelerated approval using surrogate biomarkers that are likely to predict clinical benefit. One proposed alternative is using MR-based tools (i.e. MRI-T2 or 1H-MRS) to measure fat fraction. As part of their assessment of MRI/MRS techniques, Willcocks et al (2016) simulated patient numbers needed to detect stabilization of disease progression with 80% power. Their simulation suggests that only 13 patients/group would be required with this surrogate endpoint (Willcocks et al., 2016).
Similarly, tissue collection techniques and muscle based measurements of dystrophin may benefit from refinement. Currently, open biopsies are used to collect muscle specimens (Kinali et al., 2011). The advantages of open biopsy are direct visualization, a relatively large quantity of muscle obtained, and ease of freezing muscle in the correct orientation for IF/IHC studies. Needle biopsies might provide an alternative method of collection. The smaller incision size might make it more palatable for biopsies to be taken from multiple sites, allowing assessment of heterogeneity of disease. The downside to the small sample size obtained is that it may be insufficient for some of the in vitro techniques used, particularly IF/IHC. Needle biopsy sample size may obscure fiber orientation, which is particularly important for assessing dystrophin or other biomarker expression using IF/IHC-based assays.
Additionally, staining is darker at the edge of samples for some antibodies (“edge effect”), and this edge effect may hinder interpretation in small samples. Imprecise assay performance can result in under- or over-estimation of protein levels. Alternatively, transitioning IF assays into IHC assays that utilize more robust visualization components may be beneficial. While fluorescent detection allows for easier multiplexing signal separation, better target co-localization, and higher dynamic range, IF-stained slides have lower sensitivity and are susceptible to photobleaching. By contrast, IHC has greater sensitivity and a longer lasting signal, which may prove beneficial during clinical trials. Our lab has developed IHC methods for measuring utrophin/laminin-α2 and developmental myosin heavy chain (Figures 6A, 6B, respectively) and image analysis can reliably separate those signals to facilitate accurate quantification of multiplex assays (Tinsley et al., 2016).
Acknowledgments
The authors would like to thank the following people for their contributions to this work: Terese Nelson (Histology Laboratory at the University of Iowa), Alexander Moody (Flagship Biosciences Inc.), Sara K. Whitney (Flagship Biosciences Inc.), and Rebecca Kim (Flagship Biosciences Inc.).
Funding
This work was partially supported by NIH grant U54-NS053672, Iowa Wellstone Muscular Dystrophy Cooperative Research Center, and through cTA projects sponsored by Sarepta Therapeutics, Inc and Summit Therapeutics.
Abbreviations
- AAV
Adeno-associated virus
- BMD
Becker Muscular Dystrophy
- CV
Coefficient of variation
- DMD
Duchenne Muscular Dystrophy
- ELISA
Enzyme-Linked Immunosorbant Assay
- EM
Electron microscopy
- FITC
Fluorescein Isothiocyanate
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- IM
Intramuscular
- RT-qPCR
Quantitative Reverse Transcription Polymerase Chain Reaction
- TRITC
Tetramethylrhodamine
Footnotes
Declaration of Conflicting Interests
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Kristin Wilson, Crystal Faelan, Janet C. Patterson-Kane, Daniel G Rudmann, G. David Young, and Anthony J Milici are employed by Flagship Biosciences, Inc. Diane Frank and Jay Charleston are employed by Sarepta Therapeutics, Inc. Jon Tinsley is employed by Summit Therapeutics.
References
- Aartsma-Rus A, Ginjaar IB, Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. Journal of medical genetics. 2016;53:145–151. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeffner F, Wilson K, Martin NT, Black JC, Hendriks CLL, Bolon B, Rudmann DG, Gianani R, Koegler SR, Krueger J, Young GD. The Gold Standard Paradox in Digital Image Analysis: Manual Versus Automated Scoring as Ground Truth. Archives of pathology & laboratory medicine. 2017 doi: 10.5858/arpa.2016-0386-RA. [DOI] [PubMed] [Google Scholar]
- Allen DG, Whitehead NP, Froehner SC. Absence of Dystrophin Disrupts Skeletal Muscle Signaling: Roles of Ca2+, Reactive Oxygen Species, and Nitric Oxide in the Development of Muscular Dystrophy. Physiological reviews. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony K, Arechavala-Gomeza V, Taylor LE, Vulin A, Kaminoh Y, Torelli S, Feng L, Janghra N, Bonne G, Beuvin M, Barresi R, Henderson M, Laval S, Lourbakos A, Campion G, Straub V, Voit T, Sewry CA, Morgan JE, Flanigan KM, Muntoni F. Dystrophin quantification: Biological and translational research implications. Neurology. 2014;83:2062–2069. doi: 10.1212/WNL.0000000000001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony K, Cirak S, Torelli S, Tasca G, Feng L, Arechavala-Gomeza V, Armaroli A, Guglieri M, Straathof CS, Verschuuren JJ, Aartsma-Rus A, Helderman-van den Enden P, Bushby K, Straub V, Sewry C, Ferlini A, Ricci E, Morgan JE, Muntoni F. Dystrophin quantification and clinical correlations in Becker muscular dystrophy: implications for clinical trials. Brain : a journal of neurology. 2011;134:3547–3559. doi: 10.1093/brain/awr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony K, Feng L, Arechavala-Gomeza V, Guglieri M, Straub V, Bushby K, Cirak S, Morgan J, Muntoni F. Exon skipping quantification by quantitative reverse-transcription polymerase chain reaction in Duchenne muscular dystrophy patients treated with the antisense oligomer eteplirsen. Human gene therapy methods. 2012;23:336–345. doi: 10.1089/hgtb.2012.117. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Kinali M, Feng L, Brown SC, Sewry C, Morgan JE, Muntoni F. Immunohistological intensity measurements as a tool to assess sarcolemma-associated protein expression. Neuropathology and applied neurobiology. 2010a;36:265–274. doi: 10.1111/j.1365-2990.2009.01056.x. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Kinali M, Feng L, Guglieri M, Edge G, Main M, Hunt D, Lehovsky J, Straub V, Bushby K, Sewry CA, Morgan JE, Muntoni F. Revertant fibres and dystrophin traces in Duchenne muscular dystrophy: implication for clinical trials. Neuromuscular disorders : NMD. 2010b;20:295–301. doi: 10.1016/j.nmd.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Bastian A, Mageriu V, Micu G, Manole E. The Growing Family of Limb-Girdle Muscular Dystrophies: Old and Newly Identified Members. Romanian journal of internal medicine = Revue roumaine de medecine interne. 2015;53:13–24. doi: 10.1515/rjim-2015-0002. [DOI] [PubMed] [Google Scholar]
- Beekman C, Sipkens JA, Testerink J, Giannakopoulos S, Kreuger D, van Deutekom JC, Campion GV, de Kimpe SJ, Lourbakos A. A sensitive, reproducible and objective immunofluorescence analysis method of dystrophin in individual fibers in samples from patients with duchenne muscular dystrophy. PloS one. 2014;9:e107494. doi: 10.1371/journal.pone.0107494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanto JJ, Mader TL, Eckhoff MD, Strandjord DM, Banks GB, Gardner MK, Lowe DA, Ervasti JM. Microtubule binding distinguishes dystrophin from utrophin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5723–5728. doi: 10.1073/pnas.1323842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello L, Morgenroth LP, Gordish-Dressman H, Hoffman EP, McDonald CM, Cirak S. DMD genotypes and loss of ambulation in the CINRG Duchenne Natural History Study. Neurology. 2016;87:401–409. doi: 10.1212/WNL.0000000000002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Berger S, Hall TE, Lieschke GJ, Currie PD. Dystrophin-deficient zebrafish feature aspects of the Duchenne muscular dystrophy pathology. Neuromuscular disorders : NMD. 2010;20:826–832. doi: 10.1016/j.nmd.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Berger J, Berger S, Jacoby AS, Wilton SD, Currie PD. Evaluation of exon-skipping strategies for Duchenne muscular dystrophy utilizing dystrophin-deficient zebrafish. Journal of cellular and molecular medicine. 2011;15:2643–2651. doi: 10.1111/j.1582-4934.2011.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Currie PD. Zebrafish models flex their muscles to shed light on muscular dystrophies. Disease models & mechanisms. 2012;5:726–732. doi: 10.1242/dmm.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies RD, Phelps SF, Cortez MD, Roberts R, Caskey CT, Chamberlain JS. Human and murine dystrophin mRNA transcripts are differentially expressed during skeletal muscle, heart, and brain development. Nucleic acids research. 1992;20:1725–1731. doi: 10.1093/nar/20.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiological reviews. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- Bonne G, Quijano-Roy S. Emery-Dreifuss muscular dystrophy, laminopathies, and other nuclear envelopathies. Handbook of clinical neurology. 2013;113:1367–1376. doi: 10.1016/B978-0-444-59565-2.00007-1. [DOI] [PubMed] [Google Scholar]
- Brown KJ, Marathi R, Fiorillo AA, Ciccimaro EF, Sharma S, Rowlands DS, Rayavarapu S, Nagaraju K, Hoffman EP, Hathout Y. Accurate Quantitation of Dystrophin Protein in Human Skeletal Muscle Using Mass Spectrometry. Journal of bioanalysis & biomedicine. 2012;(Suppl 7) doi: 10.4172/1948-593X.S7-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushby K, Finkel R, Wong B, Barohn R, Campbell C, Comi GP, Connolly AM, Day JW, Flanigan KM, Goemans N, Jones KJ, Mercuri E, Quinlivan R, Renfroe JB, Russman B, Ryan MM, Tulinius M, Voit T, Moore SA, Lee Sweeney H, Abresch RT, Coleman KL, Eagle M, Florence J, Gappmaier E, Glanzman AM, Henricson E, Barth J, Elfring GL, Reha A, Spiegel RJ, O’Donnell MW, Peltz SW, McDonald CM. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle & nerve. 2014;50:477–487. doi: 10.1002/mus.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushby KM, Gardner-Medwin D, Nicholson LV, Johnson MA, Haggerty ID, Cleghorn NJ, Harris JB, Bhattacharya SS. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. II. Correlation of phenotype with genetic and protein abnormalities. Journal of neurology. 1993;240:105–112. doi: 10.1007/BF00858726. [DOI] [PubMed] [Google Scholar]
- Camacho P, Fan H, Liu Z, He JQ. Small mammalian animal models of heart disease. American journal of cardiovascular disease. 2016;6:70–80. [PMC free article] [PubMed] [Google Scholar]
- Carpenter JL, Hoffman EP, Romanul FC, Kunkel LM, Rosales RK, Ma NS, Dasbach JJ, Rae JF, Moore FM, McAfee MB, et al. Feline muscular dystrophy with dystrophin deficiency. The American journal of pathology. 1989;135:909–919. [PMC free article] [PubMed] [Google Scholar]
- Chau A, Kalsotra A. Developmental insights into the pathology of and therapeutic strategies for DM1: Back to the basics. Developmental dynamics : an official publication of the American Association of Anatomists. 2015;244:377–390. doi: 10.1002/dvdy.24240. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, Tu Z, Si C, Wang H, Xing R, Pu X, Yang SH, Li S, Ji W, Li XJ. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Human molecular genetics. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciliot S, Schiaffino S. Regeneration of mammalian skeletal muscle. Basic mechanisms and clinical implications. Current pharmaceutical design. 2010;16:906–914. doi: 10.2174/138161210790883453. [DOI] [PubMed] [Google Scholar]
- Clerk A, Morris GE, Dubowitz V, Davies KE, Sewry CA. Dystrophin-related protein, utrophin, in normal and dystrophic human fetal skeletal muscle. The Histochemical journal. 1993;25:554–561. [PubMed] [Google Scholar]
- Collins CA, Morgan JE. Duchenne’s muscular dystrophy: animal models used to investigate pathogenesis and develop therapeutic strategies. International journal of experimental pathology. 2003;84:165–172. doi: 10.1046/j.1365-2613.2003.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MG, Lorusso ML, Civati F, Comi GP, Magri F, Del Bo R, Guglieri M, Molteni M, Turconi AC, Bresolin N. Neurocognitive profiles in Duchenne muscular dystrophy and gene mutation site. Pediatric neurology. 2011;45:292–299. doi: 10.1016/j.pediatrneurol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A. Pre-clinical drug tests in the mdx mouse as a model of dystrophinopathies: an overview. Acta myologica : myopathies and cardiomyopathies : official journal of the Mediterranean Society of Myology. 2012;31:40–47. [PMC free article] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson JG, Tinsley JM, Davies KE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- Deconinck N, Dan B. Pathophysiology of duchenne muscular dystrophy: current hypotheses. Pediatric neurology. 2007;36:1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Duan D. Duchenne muscular dystrophy gene therapy in the canine model. Human gene therapy Clinical development. 2015;26:57–69. doi: 10.1089/humc.2015.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen J, Poon E, Davies K. The dystrophin-associated protein complex. Journal of cell science. 2002;115:2801–2803. doi: 10.1242/jcs.115.14.2801. [DOI] [PubMed] [Google Scholar]
- Emery AE. Population frequencies of neuromuscular diseases--II. Amyotrophic lateral sclerosis (motor neurone disease) Neuromuscular disorders : NMD. 1991;1:323–325. doi: 10.1016/0960-8966(91)90117-b. [DOI] [PubMed] [Google Scholar]
- Falzarano MS, Scotton C, Passarelli C, Ferlini A. Duchenne Muscular Dystrophy: From Diagnosis to Therapy. Molecules (Basel, Switzerland) 2015;20:18168–18184. doi: 10.3390/molecules201018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanin M, Angelini C. Protein and genetic diagnosis of limb girdle muscular dystrophy type 2A: The yield and the pitfalls. Muscle & nerve. 2015;52:163–173. doi: 10.1002/mus.24682. [DOI] [PubMed] [Google Scholar]
- Finsterer J, Stollberger C. Cardiac involvement in Becker muscular dystrophy. The Canadian journal of cardiology. 2008;24:786–792. doi: 10.1016/s0828-282x(08)70686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanigan KM. Duchenne and Becker muscular dystrophies. Neurologic clinics. 2014;32:671–688. viii. doi: 10.1016/j.ncl.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Frohlich T, Kemter E, Flenkenthaler F, Klymiuk N, Otte KA, Blutke A, Krause S, Walter MC, Wanke R, Wolf E, Arnold GJ. Progressive muscle proteome changes in a clinically relevant pig model of Duchenne muscular dystrophy. Scientific reports. 2016;6:33362. doi: 10.1038/srep33362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen F, Burgunder JM. Changes of skeletal muscle in young dystrophin-deficient cats: a morphological and morphometric study. Acta neuropathologica. 2001;101:591–600. doi: 10.1007/s004010000299. [DOI] [PubMed] [Google Scholar]
- Gaschen L, Lang J, Lin S, Ade-Damilano M, Busato A, Lombard CW, Gaschen FP. Cardiomyopathy in dystrophin-deficient hypertrophic feline muscular dystrophy. Journal of veterinary internal medicine. 1999;13:346–356. doi: 10.1892/0891-6640(1999)013<0346:ciddhf>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- Gatica LV, Rosa AL. A complex interplay of genetic and epigenetic events leads to abnormal expression of the DUX4 gene in facioscapulohumeral muscular dystrophy. Neuromuscular disorders : NMD. 2016;26:844–852. doi: 10.1016/j.nmd.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Gaud A, Simon JM, Witzel T, Carre-Pierrat M, Wermuth CG, Segalat L. Prednisone reduces muscle degeneration in dystrophin-deficient Caenorhabditis elegans. Neuromuscular disorders : NMD. 2004;14:365–370. doi: 10.1016/j.nmd.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Gee P, Xu H, Hotta A. Cellular Reprogramming, Genome Editing, and Alternative CRISPR Cas9 Technologies for Precise Gene Therapy of Duchenne Muscular Dystrophy. Stem cells international. 2017;2017:8765154. doi: 10.1155/2017/8765154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs RC, Miller JP, Greenberg CR, Fehlings DL, Pestronk A, Mendell JR, Moxley RT, 3rd, King W, Kissel JT, Cwik V, Vanasse M, Florence JM, Pandya S, Dubow JS, Meyer JM. Efficacy and safety of deflazacort vs prednisone and placebo for Duchenne muscular dystrophy. Neurology. 2016;87:2123–2131. doi: 10.1212/WNL.0000000000003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso S, Perrone S, Longini M, Bruno C, Minetti C, Gazzolo D, Balestri P, Buonocore G. Isoprostanes in dystrophinopathy: Evidence of increased oxidative stress. Brain & development. 2008;30:391–395. doi: 10.1016/j.braindev.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen GJ, Kunkel LM. The Pathogenesis and Therapy of Muscular Dystrophies. Annual review of genomics and human genetics. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- Hauser E, Hoger H, Bittner R, Widhalm K, Herkner K, Lubec G. Oxyradical damage and mitochondrial enzyme activities in the mdx mouse. Neuropediatrics. 1995;26:260–262. doi: 10.1055/s-2007-979768. [DOI] [PubMed] [Google Scholar]
- Haycock JW, MacNeil S, Jones P, Harris JB, Mantle D. Oxidative damage to muscle protein in Duchenne muscular dystrophy. Neuroreport. 1996;8:357–361. doi: 10.1097/00001756-199612200-00070. [DOI] [PubMed] [Google Scholar]
- Helliwell TR, Man NT, Morris GE, Davies KE. The dystrophin-related protein, utrophin, is expressed on the sarcolemma of regenerating human skeletal muscle fibres in dystrophies and inflammatory myopathies. Neuromuscular disorders : NMD. 1992;2:177–184. doi: 10.1016/0960-8966(92)90004-p. [DOI] [PubMed] [Google Scholar]
- Heydemann A, Doherty KR, McNally EM. Genetic modifiers of muscular dystrophy: implications for therapy. Biochimica et biophysica acta. 2007;1772:216–228. doi: 10.1016/j.bbadis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Hopf FW, Turner PR, Denetclaw WF, Jr, Reddy P, Steinhardt RA. A critical evaluation of resting intracellular free calcium regulation in dystrophic mdx muscle. The American journal of physiology. 1996;271:C1325–1339. doi: 10.1152/ajpcell.1996.271.4.C1325. [DOI] [PubMed] [Google Scholar]
- Janghra N, Morgan JE, Sewry CA, Wilson FX, Davies KE, Muntoni F, Tinsley J. Correlation of Utrophin Levels with the Dystrophin Protein Complex and Muscle Fibre Regeneration in Duchenne and Becker Muscular Dystrophy Muscle Biopsies. PloS one. 2016;11:e0150818. doi: 10.1371/journal.pone.0150818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaido M, Arahata K, Hoffman EP, Nonaka I, Sugita H. Muscle histology in Becker muscular dystrophy. Muscle & nerve. 1991;14:1067–1073. doi: 10.1002/mus.880141105. [DOI] [PubMed] [Google Scholar]
- Kalra S, Montanaro F, Denning C. Can Human Pluripotent Stem Cell-Derived Cardiomyocytes Advance Understanding of Muscular Dystrophies? Journal of neuromuscular diseases. 2016;3:309–332. doi: 10.3233/JND-150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar F, Garry DJ. Dystrophin-Deficient Cardiomyopathy. Journal of the American College of Cardiology. 2016;67:2533–2546. doi: 10.1016/j.jacc.2016.02.081. [DOI] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Cirak S, Glover A, Guglieri M, Feng L, Hollingsworth KG, Hunt D, Jungbluth H, Roper HP, Quinlivan RM, Gosalakkal JA, Jayawant S, Nadeau A, Hughes-Carre L, Manzur AY, Mercuri E, Morgan JE, Straub V, Bushby K, Sewry C, Rutherford M, Muntoni F. Muscle histology vs MRI in Duchenne muscular dystrophy. Neurology. 2011;76:346–353. doi: 10.1212/WNL.0b013e318208811f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymiuk N, Blutke A, Graf A, Krause S, Burkhardt K, Wuensch A, Krebs S, Kessler B, Zakhartchenko V, Kurome M, Kemter E, Nagashima H, Schoser B, Herbach N, Blum H, Wanke R, Aartsma-Rus A, Thirion C, Lochmuller H, Walter MC, Wolf E. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Human molecular genetics. 2013;22:4368–4382. doi: 10.1093/hmg/ddt287. [DOI] [PubMed] [Google Scholar]
- Klymiuk N, Seeliger F, Bohlooly YM, Blutke A, Rudmann DG, Wolf E. Tailored Pig Models for Preclinical Efficacy and Safety Testing of Targeted Therapies. Toxicologic pathology. 2016;44:346–357. doi: 10.1177/0192623315609688. [DOI] [PubMed] [Google Scholar]
- Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, Meng G, Muller CR, Lindlof M, Kaariainen H, de la Chapellet A, Kiuru A, Savontaus ML, Gilgenkrantz H, Recan D, Chelly J, Kaplan JC, Covone AE, Archidiacono N, Romeo G, Liechti-Gailati S, Schneider V, Braga S, Moser H, Darras BT, Murphy P, Francke U, Chen JD, Morgan G, Denton M, Greenberg CR, Wrogemann K, Blonden LA, van Paassen MB, van Ommen GJ, Kunkel LM. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. American journal of human genetics. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- Kohler M, Clarenbach CF, Bahler C, Brack T, Russi EW, Bloch KE. Disability and survival in Duchenne muscular dystrophy. Journal of neurology, neurosurgery, and psychiatry. 2009;80:320–325. doi: 10.1136/jnnp.2007.141721. [DOI] [PubMed] [Google Scholar]
- Kole R, Krieg AM. Exon skipping therapy for Duchenne muscular dystrophy. Advanced drug delivery reviews. 2015;87:104–107. doi: 10.1016/j.addr.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Kornegay JN. The golden retriever model of Duchenne muscular dystrophy. Skeletal muscle. 2017;7:9. doi: 10.1186/s13395-017-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krag TO, Bogdanovich S, Jensen CJ, Fischer MD, Hansen-Schwartz J, Javazon EH, Flake AW, Edvinsson L, Khurana TS. Heregulin ameliorates the dystrophic phenotype in mdx mice. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13856–13860. doi: 10.1073/pnas.0405972101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreipke RE, Kwon YV, Shcherbata HR, Ruohola-Baker H. Drosophila melanogaster as a Model of Muscle Degeneration Disorders. Current topics in developmental biology. 2017;121:83–109. doi: 10.1016/bs.ctdb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Larcher T, Lafoux A, Tesson L, Remy S, Thepenier V, Francois V, Le Guiner C, Goubin H, Dutilleul M, Guigand L, Toumaniantz G, De Cian A, Boix C, Renaud JB, Cherel Y, Giovannangeli C, Concordet JP, Anegon I, Huchet C. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PloS one. 2014;9:e110371. doi: 10.1371/journal.pone.0110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rumeur E. Dystrophin and the two related genetic diseases, Duchenne and Becker muscular dystrophies. Bosnian journal of basic medical sciences. 2015;15:14–20. doi: 10.17305/bjbms.2015.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KR, Maruyama R, Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug design, development and therapy. 2017;11:533–545. doi: 10.2147/DDDT.S97635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luigetti M, Lo Monaco M, Mirabella M, Primiano G, Lucchini M, Monforte M, Servidei S. Oculopharyngeal muscular dystrophy: Clinical and neurophysiological features. Clinical neurophysiology : official journal of the International Federation of Clinical Neurophysiology. 2015;126:2406–2408. doi: 10.1016/j.clinph.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Madaro L, Bouche M. From innate to adaptive immune response in muscular dystrophies and skeletal muscle regeneration: the role of lymphocytes. BioMed research international. 2014;2014:438675. doi: 10.1155/2014/438675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej-Pilarczyk A, Kochanski A. Emery-Dreifuss muscular dystrophy: the most recognizable laminopathy. Folia neuropathologica. 2016;54:1–8. doi: 10.5114/fn.2016.58910. [DOI] [PubMed] [Google Scholar]
- Mah JK, Korngut L, Fiest KM, Dykeman J, Day LJ, Pringsheim T, Jette N. A Systematic Review and Meta-analysis on the Epidemiology of the Muscular Dystrophies. The Canadian journal of neurological sciences. Le journal canadien des sciences neurologiques. 2016;43:163–177. doi: 10.1017/cjn.2015.311. [DOI] [PubMed] [Google Scholar]
- Mazzone E, Vasco G, Sormani MP, Torrente Y, Berardinelli A, Messina S, D’Amico A, Doglio L, Politano L, Cavallaro F, Frosini S, Bello L, Bonfiglio S, Zucchini E, De Sanctis R, Scutifero M, Bianco F, Rossi F, Motta MC, Sacco A, Donati MA, Mongini T, Pini A, Battini R, Pegoraro E, Pane M, Gasperini S, Previtali S, Napolitano S, Martinelli D, Bruno C, Vita G, Comi G, Bertini E, Mercuri E. Functional changes in Duchenne muscular dystrophy: a 12-month longitudinal cohort study. Neurology. 2011;77:250–256. doi: 10.1212/WNL.0b013e318225ab2e. [DOI] [PubMed] [Google Scholar]
- Mazzone ES, Pane M, Sormani MP, Scalise R, Berardinelli A, Messina S, Torrente Y, D’Amico A, Doglio L, Viggiano E, D’Ambrosio P, Cavallaro F, Frosini S, Bello L, Bonfiglio S, De Sanctis R, Rolle E, Bianco F, Magri F, Rossi F, Vasco G, Vita G, Motta MC, Donati MA, Sacchini M, Mongini T, Pini A, Battini R, Pegoraro E, Previtali S, Napolitano S, Bruno C, Politano L, Comi GP, Bertini E, Mercuri E. 24 month longitudinal data in ambulant boys with Duchenne muscular dystrophy. PloS one. 2013;8:e52512. doi: 10.1371/journal.pone.0052512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CM, Henricson EK, Abresch RT, Florence J, Eagle M, Gappmaier E, Glanzman AM, Spiegel R, Barth J, Elfring G, Reha A, Peltz SW. The 6-minute walk test and other clinical endpoints in duchenne muscular dystrophy: reliability, concurrent validity, and minimal clinically important differences from a multicenter study. Muscle & nerve. 2013a;48:357–368. doi: 10.1002/mus.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CM, Henricson EK, Abresch RT, Florence JM, Eagle M, Gappmaier E, Glanzman AM, Spiegel R, Barth J, Elfring G, Reha A, Peltz S. The 6-minute walk test and other endpoints in Duchenne muscular dystrophy: longitudinal natural history observations over 48 weeks from a multicenter study. Muscle & nerve. 2013b;48:343–356. doi: 10.1002/mus.23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CM, Henricson EK, Han JJ, Abresch RT, Nicorici A, Elfring GL, Atkinson L, Reha A, Hirawat S, Miller LL. The 6-minute walk test as a new outcome measure in Duchenne muscular dystrophy. Muscle & nerve. 2010;41:500–510. doi: 10.1002/mus.21544. [DOI] [PubMed] [Google Scholar]
- McGreevy JW, Hakim CH, McIntosh MA, Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Disease models & mechanisms. 2015;8:195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Shilling C, Leslie ND, Flanigan KM, al-Dahhak R, Gastier-Foster J, Kneile K, Dunn DM, Duval B, Aoyagi A, Hamil C, Mahmoud M, Roush K, Bird L, Rankin C, Lilly H, Street N, Chandrasekar R, Weiss RB. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Annals of neurology. 2012;71:304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- Meola G, Cardani R. Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochimica et biophysica acta. 2015;1852:594–606. doi: 10.1016/j.bbadis.2014.05.019. [DOI] [PubMed] [Google Scholar]
- Merlini L, Sabatelli P. Improving clinical trial design for Duchenne muscular dystrophy. BMC neurology. 2015;15:153. doi: 10.1186/s12883-015-0408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina S, Altavilla D, Aguennouz M, Seminara P, Minutoli L, Monici MC, Bitto A, Mazzeo A, Marini H, Squadrito F, Vita G. Lipid peroxidation inhibition blunts nuclear factor-kappaB activation, reduces skeletal muscle degeneration, and enhances muscle function in mdx mice. The American journal of pathology. 2006;168:918–926. doi: 10.2353/ajpath.2006.050673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagoe-Suzuki Y, Nishiyama T, Nakamura M, Narita A, Takemura F, Masuda S, Minami N, Murayama K, Komaki H, Goto YI, Takeda S. Induction of Pluripotent Stem Cells from a Manifesting Carrier of Duchenne Muscular Dystrophy and Characterization of Their X-Inactivation Status. Stem cells international. 2017;2017:7906843. doi: 10.1155/2017/7906843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhtarian A, Lefaucheur JP, Even PC, Sebille A. Hindlimb immobilization applied to 21- day-old mdx mice prevents the occurrence of muscle degeneration. Journal of applied physiology (Bethesda, Md: 1985) 1999;86:924–931. doi: 10.1152/jappl.1999.86.3.924. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Takeda S. Mammalian models of Duchenne Muscular Dystrophy: pathological characteristics and therapeutic applications. Journal of biomedicine & biotechnology. 2011;2011:184393. doi: 10.1155/2011/184393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Fujii W, Tsuboi M, Tanihata J, Teramoto N, Takeuchi S, Naito K, Yamanouchi K, Nishihara M. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Scientific reports. 2014;4:5635. doi: 10.1038/srep05635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardes F, Araujo AP, Ribeiro MG. Mental retardation in Duchenne muscular dystrophy. Jornal de pediatria. 2012;88:6–16. doi: 10.2223/JPED.2148. [DOI] [PubMed] [Google Scholar]
- Nguyen TM, Ellis JM, Ginjaar IB, van Paassen MM, van Ommen GJ, Moorman AF, Cartwright AJ, Morris GE. Monoclonal antibody evidence for structural similarities between the central rod regions of actinin and dystrophin. FEBS letters. 1990;272:109–112. doi: 10.1016/0014-5793(90)80460-z. [DOI] [PubMed] [Google Scholar]
- Nguyen TM, Ellis JM, Love DR, Davies KE, Gatter KC, Dickson G, Morris GE. Localization of the DMDL gene-encoded dystrophin-related protein using a panel of nineteen monoclonal antibodies: presence at neuromuscular junctions, in the sarcolemma of dystrophic skeletal muscle, in vascular and other smooth muscles, and in proliferating brain cell lines. The Journal of cell biology. 1991;115:1695–1700. doi: 10.1083/jcb.115.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson LV, Johnson MA, Bushby KM, Gardner-Medwin D, Curtis A, Ginjaar IB, den Dunnen JT, Welch JL, Butler TJ, Bakker E, et al. Integrated study of 100 patients with Xp21 linked muscular dystrophy using clinical, genetic, immunochemical, and histopathological data. Part 2. Correlations within individual patients. Journal of medical genetics. 1993;30:737–744. doi: 10.1136/jmg.30.9.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson LV, Johnson MA, Gardner-Medwin D, Bhattacharya S, Harris JB. Heterogeneity of dystrophin expression in patients with Duchenne and Becker muscular dystrophy. Acta neuropathologica. 1990;80:239–250. doi: 10.1007/BF00294640. [DOI] [PubMed] [Google Scholar]
- Nigro G, Comi LI, Limongelli FM, Giugliano MA, Politano L, Petretta V, Passamano L, Stefanelli S. Prospective study of X-linked progressive muscular dystrophy in Campania. Muscle & nerve. 1983;6:253–262. doi: 10.1002/mus.880060403. [DOI] [PubMed] [Google Scholar]
- Nigro V, Piluso G. Spectrum of muscular dystrophies associated with sarcolemmal-protein genetic defects. Biochimica et biophysica acta. 2015;1852:585–593. doi: 10.1016/j.bbadis.2014.07.023. [DOI] [PubMed] [Google Scholar]
- Nowak KJ, Davies KE. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO reports. 2004;5:872–876. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja M, Ruohola-Baker H. Drosophila as a starting point for developing therapeutics for the rare disease Duchenne Muscular Dystrophy. Rare diseases (Austin, Tex) 2013;1:e24995. doi: 10.4161/rdis.24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantie E, Migocka-Patrzalek M, Daczewska M, Jagla K. Model organisms in the fight against muscular dystrophy: lessons from drosophila and Zebrafish. Molecules (Basel, Switzerland) 2015;20:6237–6253. doi: 10.3390/molecules20046237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons F, Robert A, Fabbrizio E, Hugon G, Califano JC, Fehrentz JA, Martinez J, Mornet D. Utrophin localization in normal and dystrophin-deficient heart. Circulation. 1994;90:369–374. doi: 10.1161/01.cir.90.1.369. [DOI] [PubMed] [Google Scholar]
- Porter JD. A New Dog Model for Drug Development in Duchenne. 2015 Retrieved 7/18/2017 2017: http://community.parentprojectmd.org/profiles/blogs/a-new-dog-model-for-drug-development-induchenne.
- Quattrocelli M, Spencer MJ, McNally EM. Outside in: The matrix as a modifier of muscular dystrophy. Biochimica et biophysica acta. 2017;1864:572–579. doi: 10.1016/j.bbamcr.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nature genetics. 1998;19:79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- Robinson-Hamm JN, Gersbach CA. Gene therapies that restore dystrophin expression for the treatment of Duchenne muscular dystrophy. Human genetics. 2016;135:1029–1040. doi: 10.1007/s00439-016-1725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romitti PA, Zhu Y, Puzhankara S, James KA, Nabukera SK, Zamba GK, Ciafaloni E, Cunniff C, Druschel CM, Mathews KD, Matthews DJ, Meaney FJ, Andrews JG, Conway KM, Fox DJ, Street N, Adams MM, Bolen J. Prevalence of Duchenne and Becker muscular dystrophies in the United States. Pediatrics. 2015;135:513–521. doi: 10.1542/peds.2014-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar F, Touroutine D, Gao S, Oh HJ, Gendrel M, Bessereau JL, Kim H, Zhen M, Richmond JE. The dystrophin-associated protein complex maintains muscle excitability by regulating Ca(2+)-dependent K(+) (BK) channel localization. The Journal of biological chemistry. 2011;286:33501–33510. doi: 10.1074/jbc.M111.227678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield J, Houzelstein D, Davies K, Buckingham M, Edwards YH. Expression of the dystrophin-related protein (utrophin) gene during mouse embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 1993;198:254–264. doi: 10.1002/aja.1001980403. [DOI] [PubMed] [Google Scholar]
- Selsby JT, Ross JW, Nonneman D, Hollinger K. Porcine models of muscular dystrophy. ILAR journal. 2015;56:116–126. doi: 10.1093/ilar/ilv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji E, Sakurai H, Nishino T, Nakahata T, Heike T, Awaya T, Fujii N, Manabe Y, Matsuo M, Sehara-Fujisawa A. Early pathogenesis of Duchenne muscular dystrophy modelled in patient-derived human induced pluripotent stem cells. Scientific reports. 2015;5:12831. doi: 10.1038/srep12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Cagliani R, Pozzoli U, Bardoni A, Comi GP, Giorda R, Bresolin N. The dystrophin gene is alternatively spliced throughout its coding sequence. FEBS letters. 2002;517:163–166. doi: 10.1016/s0014-5793(02)02613-3. [DOI] [PubMed] [Google Scholar]
- Smith AS, Davis J, Lee G, Mack DL, Kim DH. Muscular dystrophy in a dish: engineered human skeletal muscle mimetics for disease modeling and drug discovery. Drug discovery today. 2016;21:1387–1398. doi: 10.1016/j.drudis.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire S, Raymackers JM, Vandebrouck C, Potter A, Tinsley J, Fisher R, Gillis JM, Davies KE. Prevention of pathology in mdx mice by expression of utrophin: analysis using an inducible transgenic expression system. Human molecular genetics. 2002;11:3333–3344. doi: 10.1093/hmg/11.26.3333. [DOI] [PubMed] [Google Scholar]
- Tachi N, Watanabe Y, Ohya K, Chiba S. Asymptomatic Becker muscular dystrophy: histological changes in biopsied muscles. Acta paediatrica Japonica : Overseas edition. 1993;35:409–411. doi: 10.1111/j.1442-200x.1993.tb03082.x. [DOI] [PubMed] [Google Scholar]
- Taylor LE, Kaminoh YJ, Rodesch CK, Flanigan KM. Quantification of dystrophin immunofluorescence in dystrophinopathy muscle specimens. Neuropathology and applied neurobiology. 2012;38:591–601. doi: 10.1111/j.1365-2990.2012.01250.x. [DOI] [PubMed] [Google Scholar]
- Thomas GD. Functional muscle ischemia in Duchenne and Becker muscular dystrophy. Frontiers in physiology. 2013;4:381. doi: 10.3389/fphys.2013.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley J, Pulliam C, Milici A, Aeffner F, Davies K, Moore S. Development of digital tissue image analysis solution for muscle biopsies in support of disease-modifying therapies for Duchenne muscular dystrophy. Neuromuscular Disorders. 2016;26:S155. [Google Scholar]
- Tinsley JM, Blake DJ, Roche A, Fairbrother U, Riss J, Byth BC, Knight AE, Kendrick-Jones J, Suthers GK, Love DR, et al. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- Tome FM, Matsumura K, Chevallay M, Campbell KP, Fardeau M. Expression of dystrophin-associated glycoproteins during human fetal muscle development: a preliminary immunocytochemical study. Neuromuscular disorders : NMD. 1994;4:343–348. doi: 10.1016/0960-8966(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Torelli S, Brown SC, Jimenez-Mallebrera C, Feng L, Muntoni F, Sewry CA. Absence of neuronal nitric oxide synthase (nNOS) as a pathological marker for the diagnosis of Becker muscular dystrophy with rod domain deletions. Neuropathology and applied neurobiology. 2004;30:540–545. doi: 10.1111/j.1365-2990.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Miro J, Koenig M, Claustres M. Normal and altered pre-mRNA processing in the DMD gene. Human genetics. 2017 doi: 10.1007/s00439-017-1820-9. [DOI] [PubMed] [Google Scholar]
- Tyler KL. Origins and early descriptions of “Duchenne muscular dystrophy”. Muscle & nerve. 2003;28:402–422. doi: 10.1002/mus.10435. [DOI] [PubMed] [Google Scholar]
- Viollet L, Thrush PT, Flanigan KM, Mendell JR, Allen HD. Effects of angiotensin-converting enzyme inhibitors and/or beta blockers on the cardiomyopathy in Duchenne muscular dystrophy. The American journal of cardiology. 2012;110:98–102. doi: 10.1016/j.amjcard.2012.02.064. [DOI] [PubMed] [Google Scholar]
- Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, Holder A, Stanley R, Chandler K, Marks SL, Muntoni F, Shelton GD, Piercy RJ. A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is amenable to exon 51 skipping. PloS one. 2010;5:e8647. doi: 10.1371/journal.pone.0008647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Tawil R. Facioscapulohumeral Dystrophy. Current neurology and neuroscience reports. 2016;16:66. doi: 10.1007/s11910-016-0667-0. [DOI] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA, Conn MM, Khan A, Almstead NG, Hedrick J, Mollin A, Risher N, Weetall M, Yeh S, Branstrom AA, Colacino JM, Babiak J, Ju WD, Hirawat S, Northcutt VJ, Miller LL, Spatrick P, He F, Kawana M, Feng H, Jacobson A, Peltz SW, Sweeney HL. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- Wicklund MP. The muscular dystrophies. Continuum (Minneapolis, Minn) 2013;19:1535–1570. doi: 10.1212/01.CON.0000440659.41675.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks RJ, Rooney WD, Triplett WT, Forbes SC, Lott DJ, Senesac CR, Daniels MJ, Wang DJ, Harrington AT, Tennekoon GI, Russman BS, Finanger EL, Byrne BJ, Finkel RS, Walter GA, Sweeney HL, Vandenborne K. Multicenter prospective longitudinal study of magnetic resonance biomarkers in a large duchenne muscular dystrophy cohort. Annals of neurology. 2016;79:535–547. doi: 10.1002/ana.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharieva IT, Calissano M, Scoto M, Preston M, Cirak S, Feng L, Collins J, Kole R, Guglieri M, Straub V, Bushby K, Ferlini A, Morgan JE, Muntoni F. Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular Dystrophy. PloS one. 2013;8:e80263. doi: 10.1371/journal.pone.0080263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H, Stanciauskas R, Stigloher C, Dizon KK, Jospin M, Bessereau JL, Pinaud F. In vivo single-molecule imaging identifies altered dynamics of calcium channels in dystrophin-mutant C. elegans. Nature communications. 2014;5:4974. doi: 10.1038/ncomms5974. [DOI] [PMC free article] [PubMed] [Google Scholar]