Abstract
Plasmodium vivax shows a strict host tropism for reticulocytes. We identify transferrin receptor 1 (TfR1) as the receptor for P. vivax reticulocyte-binding protein 2b (PvRBP2b). The structure of the N-terminal domain of PvRBP2b involved in red blood cell binding was determined, elucidating the molecular basis for TfR1 recognition. TfR1 was validated as the biological target of PvRBP2b engagement by TfR1 expression knockdown analysis. TfR1 mutant cells deficient in PvRBP2b binding were refractory to invasion of P. vivax, but not to invasion of P. falciparum. Using Brazilian and Thai clinical isolates, we show that PvRBP2b monoclonal antibodies that inhibit reticulocyte binding also block P. vivax entry into reticulocytes. These data show that TfR1-PvRBP2b invasion pathway is critical for the recognition of reticulocytes during P. vivax invasion.
Of the hundreds of Plasmodium species, only P. falciparum, P. vivax, P. ovale curtisi, P. ovale wallikeri, P. malariae and P. knowlesi are known to infect humans. Within the human host, malaria parasites invade liver and red blood cells for replication and transmission. Blood stage infection is the major cause of all clinical symptoms in malaria, and therefore the therapeutic prevention of parasite entry into red blood cells could alleviate malarial disease. Entry into red blood cells depends on the interactions between parasite invasion ligands and their cognate red blood cell receptors of which only a handful have been identified (1–7). These ligand-receptor interactions initiate a cascade of molecular events that progress from initial attachment, recognition, commitment and finally penetration of the parasite into red blood cells (8, 9).
P. vivax is the most widely distributed human malaria parasite. This parasite has a strict preference for invasion into reticulocytes, which are very young red blood cells that are formed in the bone marrow following enucleation and released into the circulation. The reticulocyte-specific receptor involved in P. vivax entry has not been identified (10). Most studies have focused on the interaction between the P. vivax Duffy binding protein (PvDBP) and the red blood cell Duffy antigen receptor for chemokines (DARC) as individuals from western and central Africa lacking DARC are resistant to P. vivax invasion (11). However, recent reports have highlighted the presence of P. vivax in apparently DARC negative individuals suggesting that P. vivax may enter reticulocytes by binding to other receptors (12–14). Furthermore, DARC is present on both normocytes and reticulocytes and therefore this ligand-receptor interaction cannot govern selective entry into reticulocytes (15). To identify other parasite proteins involved in reticulocyte recognition, we focused on the P. vivax reticulocyte-binding protein family (PvRBP). This protein family comprises 11 members of which several have been shown to bind reticulocytes; however, their cognate receptors have not been identified (16–19).
PvRBP2b binds transferrin receptor 1 to mediate recognition of reticulocytes
P. vivax preferentially invades reticulocytes expressing high levels of transferrin receptor 1 (TfR1 or CD71) (20). TfR1 is an essential housekeeping protein involved in cellular transport of iron into cells through binding of iron-loaded transferrin (Tf) (21). On circulating red blood cells, TfR1 is expressed only on reticulocytes and is progressively lost from their membranes as they mature into erythrocytes (22, 23). TfR1 is a type II transmembrane glycoprotein that forms a dimer and its ectodomain consists of three subdomains: a ‘protease-like domain’ resembling the structure of zinc metalloproteinases, an ‘apical domain’ and a ‘helical domain’ responsible for dimerization (24). TfR1 is also a cellular receptor for New World hemorrhagic fever arenaviruses including Machupo (MACV), Junin, Guanarito and Sabiá viruses (25, 26). Residues 208-212 of the TfR1 apical domain provide a critical recognition site for these viruses (25, 26).
PvRBP2b is expressed in late-stage P. vivax parasites and recombinant PvRBP2b (residues 161 to 1,454, PvRBP2b161-1454) binds preferentially to reticulocytes that express TfR1 (19, 27). We observed that binding by recombinant PvRBP2b was abolished when reticulocytes were treated with trypsin and chymotrypsin (fig. S1, A and B). We confirmed that the combination of these proteases cleaves TfR1 and CR1 from the surface of reticulocytes, with other known malaria receptors including glycophorin A, basigin and DARC being susceptible to different sets of protease treatment (fig. S1, A and B). The profile of PvRBP2b binding is strikingly similar to the TfR1 surface expression on reticulocytes (Fig. 1A, bottom panel) and we show that the level of PvRBP2b binding is directly correlated with the levels of TfR1 on the surface of reticulocytes (fig. S1, C and D).
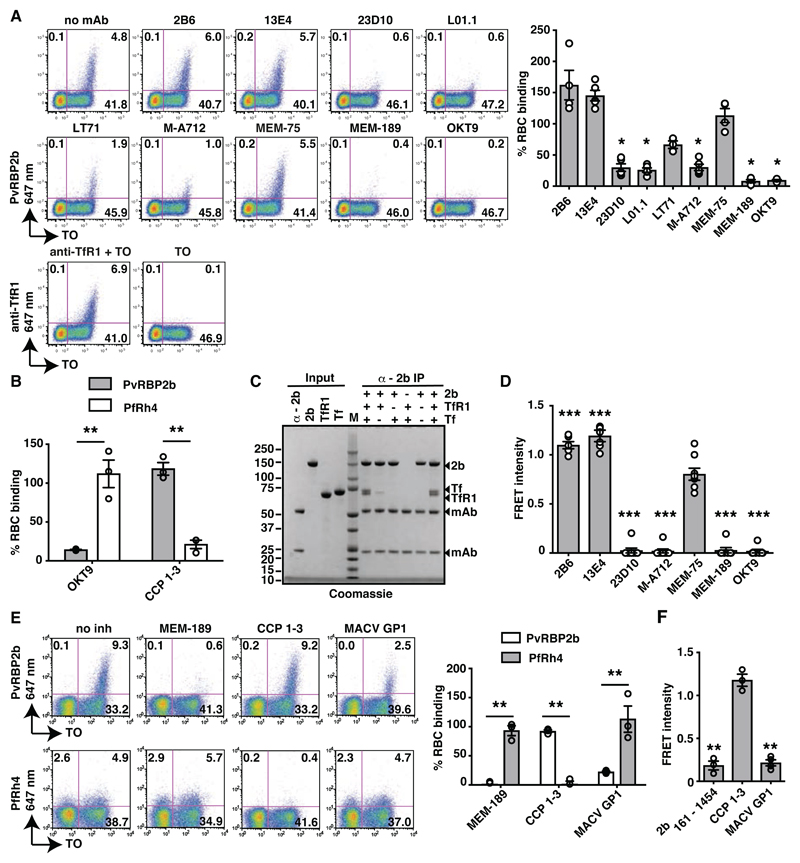
Fig. 1. PvRBP2b binds TfR1 on the reticulocyte surface.
(A). PvRBP2b161-1454 binding in the presence of anti-TfR1 mAbs analysed by flow cytometry. Left: Dot plots of PvRBP2b161-1454 binding (y-axis) to reticulocytes stained with thiazole orange (TO, x-axis). Right: normalized binding results where PvRBP2b161-1454 binding in the absence of mAbs was arbitrarily assigned to be 100%. (B) PvRBP2b161-1454 and PfRh428-766 binding were evaluated by flow cytometry with the addition of anti-TfR1 mAb OKT9 or CCP 1-3. PvRBP2b161-1454 and PfRh4 binding in buffer were arbitrarily assigned to be 100%. (C) Eluates of individual or mixtures of proteins immuno-precipitated with anti-PvRBP2b mAb analyzed by SDS-PAGE. + or – indicates protein present or absent. Molecular weight marker (M). (D) Anti-TfR1 mAbs inhibit PvRBP2b-TfR1 complex formation in the FRET-based assay. The FRET signal was relative to “no mAb” control. (E) Binding of PvRBP2b161-1454 and PfRh428-766 in the presence of anti-TfR1 mAb MEM-189, CCP 1-3 and MACV GP1. Left: dot plots showing PvRBP2b161-1454 (top) and PfRh428-766 binding (bottom). Right: normalized binding results where PvRBP2b161-1454 and PfRh428-766 binding in the presence of buffer was arbitrarily assigned to be 100%. (F) MACV GP1 inhibits PvRBP2b161-1454-TfR1 complex formation monitored by FRET assay. For (A), (B), (D), (E) and (F), Mean ± S.E.M., n ≥3, open dots represent biological replicates. Mann-Whitley test was used for (A) and (D) where MEM-75 was considered non-inhibitory and t-tests was used for (B), (E) and (F), * P ≤ 0.05, ** P ≤ 0.001.
To determine if PvRBP2b161-1454 binds to the population of reticulocytes that express TfR1 on their surfaces, we tested a panel of commercially available anti-TfR1 monoclonal antibodies (mAbs) for their ability to block recombinant PvRBP2b binding. Indeed, anti-TfR1 mAbs 23D10, L01.1, LT71, M-A712, MEM-189 and OKT9 inhibited PvRBP2b binding to reticulocytes by 78, 76, 33, 75, 92 and 90% respectively (Fig. 1A). M-A712 also prevents MACV pseudovirus entry (25, 28). Anti-TfR1 mAbs 2B6, 13E4 and MEM-75 did not inhibit PvRBP2b binding; while their epitopes have not been mapped, we propose that these three antibodies may bind to a site on TfR1 that is not involved in the PvRBP2b interaction (Fig. 1A). To determine if this inhibition was specific to PvRBP2b161-1454 binding, we analyzed the binding of P. falciparum reticulocyte binding protein-like homologue 4 (PfRh4) to its cognate receptor complement receptor 1 (CR1) (4). While addition of the first three complement control protein modules of CR1 (CCP 1–3) inhibited PfRh428-766 binding as expected (29), addition of anti-TfR1 mAb OKT9 did not significantly reduce PfRh4 binding (Fig. 1B). Since anti-TfR1 did not affect PfRh4 binding, these results show that TfR1 is a specific reticulocyte receptor for PvRBP2b.
To evaluate if PvRBP2b161-1454 interacts directly with TfR1, we performed immuno-precipitation experiments using purified recombinant TfR1, Tf, and PvRBP2b161-1454 proteins (Fig. 1C) (30). Using an anti-PvRBP2b mAb, we immuno-precipitated PvRBP2b in complex with TfR1 and Tf. PvRBP2b and TfR1 also formed a binary complex in the absence of Tf, demonstrating that PvRBP2b binds directly to TfR1 (Fig. 1C). The interaction between PvRBP2b and TfR1 is specific, as immuno-precipitation of PvRBP1a, PvRBP1b or PvRBP2a did not show evidence of complex formation with TfR1 (fig. S2A).
We developed a fluorescence resonance energy transfer (FRET)-based assay to monitor PvRBP2b-TfR1 complex formation where TfR1 labeled with DyLight-594 could be shown to interact with PvRBP2b161-1454 labeled with DyLight-488 (fig. S2B). The addition of 10-fold molar excess of unlabeled PvRBP2b161-1454 and TfR1 competed out the labeled proteins and reduced the signal of the PvRBP2b-TfR1 FRET pair. In contrast, proteins that were unable to bind TfR1, such as PfRh4, had no effect on the FRET signal. Using this assay, we observed that anti-TfR1 mAbs 23D10, M-A712, MEM-189 and OKT9 that inhibited PvRBP2b161-1454 reticulocyte binding also blocked PvRBP2b-TfR1 complex formation (Fig. 1D).
MACV GP1 and PvRBP2b bind to the apical domain of TfR1
The arenavirus envelope glycoprotein is the only protein on the virion surface and during maturation is processed into three subunits: the stable signal peptide (SSP), GP1 and GP2. The GP1 subunit interacts with cellular receptors and the structure of a MACV GP1-TfR1 complex shows that MACV GP1 binds to the apical domain of TfR1 (31, 32). To determine if PvRBP2b interacts with a similar surface on TfR1, we examined if soluble MACV GP1 competes with PvRBP2b161-1454 for binding to TfR1 on reticulocytes (Fig. 1E). Indeed, the addition of MACV GP1 reduced PvRBP2b161-1454 binding to reticulocytes, albeit at a lower level of inhibition compared to the addition of anti-TfR1 mAb MEM-189. This inhibition was specific as PfRh4 binding was unaffected by addition of MACV GP1 or MEM-189, but clearly reduced with the addition of CCP 1-3 (Fig. 1E). The addition of MACV GP1 inhibited PvRBP2b-TfR1 complex formation and reduced the FRET signal to a similar extent as unlabeled PvRBP2b161-1454 whereas addition of CCP 1-3 had negligible effect (Fig. 1F). These results indicate that MACV GP1 and PvRBP2b161-1454 bind to an overlapping site on TfR1.
Crystal structure of the N-terminal domain of PvRBP2b
PvRBP2b is a 326 kDa protein with a putative red blood cell binding domain and a C-terminal transmembrane region (Fig. 2). We determined the crystal structure of the N-terminal domain of PvRBP2b (residues 169 to 470, PvRBP2b169-470), refined to 1.71 Å resolution (Fig. 2A, fig. S3, A to D, and table S1). The surface of the domain is mostly positively charged (Fig. 2B). It is predominantly an α-helical protein, comprising ten α-helices and two very short antiparallel β-sheets, each comprising of two β-strands. The crystal structure of PvRBP2b169-470 has two disulfide bonds; one between Cys312 and Cys316 and the other between Cys240 and Cys284. This structure closely resembles the homologous domain of PvRBP2a and PfRh5, with a root mean square deviation (RMSD) of 1.7 Å and 3.7 Å over 268 and 225 aligned Cα atoms, respectively (Fig. 2C and fig. S4) (18, 33, 34). The theoretical X-ray solution scattering pattern calculated from the PvRBP2b169-470 crystal structure coordinates shows excellent agreement with the experimental small angle X-ray scattering (SAXS) data (χ = 0.35; fig. S5, A to F, and table S2), with concordance between the crystal and solution conformations apparent from the overlay of the crystal structure and the ab initio calculated molecular envelope (Fig. 2D, left). We also obtained SAXS data for a longer fragment of PvRBP2b including residues 169 to 652 (fig. S6 and table S2). The reconstructed molecular envelope has a rod-like shape with a C-terminal part forming a continuous extension of the N-terminal domain (Fig. 2D, right). SAXS data for a larger fragment of PvRBP2b encompassing residues 161 to 969 indicate that the molecule adopts an elongated, boomerang-like shape, similar to that previously reported for PvRBP2a (fig. S6, A to F, fig. S7, A to D, and table S2) (18).
Fig. 2. Crystal structure of the N-terminal domain of PvRBP2b and its functional requirement.
(A) Structure of the N-terminal domain of PvRBP2b from amino acid 169 to 470 shown in two orthogonal views. (B) Electrostatic surface potential on the PvRBP2b structure. (C) Superimposition of the PvRBP2b structure (green) with PvRBP2a (purple) and PfRh5 (orange). The PDB ID codes for PfRh5 and PvRBP2a are 4WAT and 4Z8N, respectively. (D) Crystal structure of the N-terminal domain superimposed with SAXS ab initio bead model of PvRBP2b169-470 (left) and PvRBP2b169-652 (right). (E) Sliding window analysis showing nucleotide diversity (π) values and Tajima’s D statistic in PvRBP2b. The grey box refers to a highly polymorphic region at amino acid positions 169 to 470 that appears to be under balancing selection. (F) Schematic representation of full-length PvRBP2b and recombinant protein fragments (left). Signal peptide (SP), transmembrane domain (TM) and N-terminal domain (yellow) are indicated. (G) PvRBP2b binding results by flow cytometry where PvRBP2b161-1454 binding was arbitrarily assigned to be 100%. (H) Unlabeled recombinant PvRBP2b fragments or PfRh4 were mixed at 10-fold molar excess relative to the labeled PvRBP2b161-1454-TfR1 FRET pair. The FRET intensity was relative to buffer control. For (G to H), Mean ± S.E.M, n = 4, open dots represent biological replicates. The Mann-Whitley test was used to calculate the P value using the binding of 2b474-1454 that was considered no binding, * P≤ 0.05, ** P ≤ 0.001.
We calculated nucleotide diversity (π) and Tajima’s D within PvRBP2b using data from the MalariaGEN P. vivax Genome Variation project (35). There was a peak in both metrics between amino acid positions 169 and 470, suggesting balancing selection within the N-terminal domain (Fig. 2E). Such signatures of balancing selection are often associated with genes or proteins expressed on the surface of merozoites, and are likely due to interaction with the immune system.
To determine the importance of the N-terminal domain for PvRBP2b function, we generated a series of purified recombinant PvRBP2b protein fragments (Fig. 2F, tables S3 and S4). All proteins were soluble and properly folded as indicated by high α-helical content in CD spectra, in agreement with the secondary structure predictions (fig. S8, A to D). We observed that all fragments with the N-terminal domain bound reticulocytes (PvRBP2b161-1454, PvRBP2b161-969, PvRBP2b169-813 and PvRBP2b169-652) whereas their corresponding fragments without the domain did not (PvRBP2b474-1454 and PvRBP2b474-969) (Fig. 2G). However, the isolated N-terminal domain PvRBP2b169-470 was unable to bind reticulocytes on its own (Fig. 2G), indicating that this fragment of PvRBP2b is necessary but not sufficient for reticulocyte binding. The shortest PvRBP2b fragment that showed binding to reticulocytes encompasses residues 169 to 652 (Fig. 2G). Our FRET-based assay showed that unlabeled recombinant fragments that bind reticulocytes inhibited PvRBP2b-TfR1 complex formation whereas recombinant fragments that did not bind reticulocytes had a negligible effect (Fig. 2H). Collectively our structural and functional analyses indicate that the N-terminal domain is necessary for binding, but requires the presence of the elongated C-terminal fragment to form a fully functional binding site.
PvRBP2b, TfR1 and Tf form a stable complex at nanomolar concentrations
Using surface plasmon resonance (SPR), we found that PvRBP2b161-1454 interacts with TfR1 alone or with the binary complex of TfR1-Tf (Fig. 3A, top and bottom panels, respectively). We also observed similar results for the PvRBP2b161-969 fragment with TfR1 and TfR1-Tf (Fig. 3B, top and bottom panels, respectively). These results indicate that Tf was not required for the PvRBP2b-TfR1 complex formation, since the addition of Tf resulted in similar binding responses than for TfR1 alone. We analyzed a PvRBP2b, TfR1 and Tf ternary complex using analytical size exclusion chromatography (SEC), and used SDS-PAGE analyses to confirm co-migration of complex components. The ternary complex was detected for PvRBP2b161-1454 and PvRBP2b161-969 (Fig. 3, C and D, top panels respectively, and table S5). In contrast, their corresponding fragments without the N-terminal domain (PvRBP2b474-1454 and PvRBP2b474-969) did not form any observable ternary complexes (Fig. 3, C and D, bottom panels). The interaction between PvRBP2b and TfR1-Tf binary complex is similar in the presence of either the iron-depleted or iron-loaded form of human transferrin (fig. S2C). Furthermore, the homologous member of the same protein family, PvRBP2a, did not form a ternary complex with TfR1-Tf (fig. S2D).
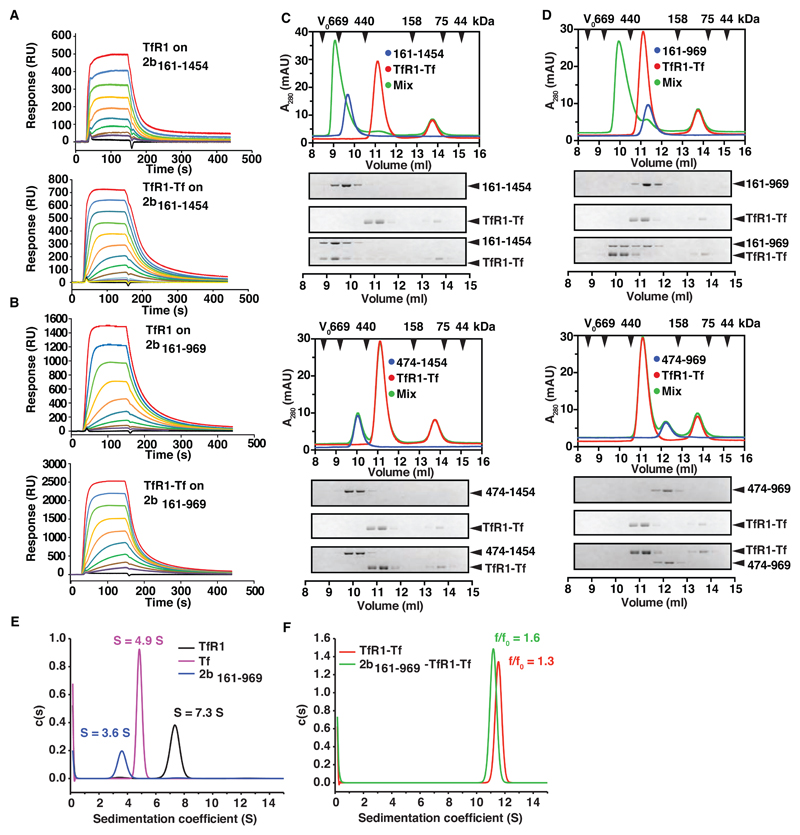
Fig. 3. PvRBP2b binds to TfR1-Tf to form a stable ternary complex.
PvRBP2b161-1454 (A) and PvRBP2b161-969 (B) were coupled covalently to a biosensor chip to probe binding of TfR1 (concentration range assayed: 2 µM to 7.5 nM, top panels) and TfR1-Tf complexes (concentration range of TfR1-Tf complexes assayed: 2 μM:4 μM to 1.8 nM:3.9 nM (A) and 2 μM:4 μM to 7.5 nM:15 nM (B), bottom panels). (C) and (D) Complex formation between PvRBP2b, TfR1 and Tf analyzed by analytical size exclusion chromatography (SEC). PvRBP2b-TfR1-Tf ternary complex can be observed for PvRBP2b161-1454 (C, top panel) and PvRBP2b161-969 (D, top panel). Two corresponding truncations of the N-terminal domain, PvRBP2b474-1454 (C, bottom panel) and PvRBP2b474-969 (D, bottom panel) do not interact with the TfR1-Tf binary complex. The exclusion volume (V0) of the columns and the elution volumes of selected marker proteins are indicated with black arrowheads. Lower part: Coomassie-Blue stained SDS-PAGE gels of the fractions obtained from SEC. (E-F) Continuous sedimentation coefficient distributions derived from fitting sedimentation velocity data to a c(s) sedimentation model. (E) c(s) distributions for TfR1 (black line), Tf (magenta line) and PvRBP2b161-969 (blue line). (F) c(s) distributions for the TfR1-Tf complex (red line), and PvRBP2b161-969-TfR1-Tf complex (green line).
Sedimentation velocity analyses of TfR1, Tf and PvRBP2b161-969 indicated that the isolated proteins are homogenous, with weight-average sedimentation coefficients of 7.3 S, 4.9 S and 3.6 S, respectively (Fig. 3E). These values are consistent with a stable dimer of TfR1 and monomeric forms of both Tf and PvRBP2b161-969. The empirically-fitted shape parameter value (frictional ratio) calculated for PvRBP2b161-969 was approximately 1.8, consistent with a highly elongated structure in solution. Mixtures of TfR1-Tf and PvRBP2b161-969-TfR1-Tf yielded single symmetrical peaks with weight-average sedimentation coefficients of 11.5 S and 11.2 S, respectively, with no peaks observed for the individual components in these samples (Fig. 3F and fig. S9). These results indicate that Tf and TfR1 form a stable binary complex in solution and that PvRBP2b161-969 binds to this binary complex. The frictional ratio (f/f0) for the ternary PvRBP2b161-969-TfR1-Tf was higher than for the binary TfR1-Tf complex, resulting in a reduction in the sedimentation coefficient on formation of the ternary complex and indicating that it has an elongated structure in solution.
Deletions in TfR1 generated via CRISPR/Cas9 abolishes PvRBP2b binding and P. vivax invasion
To investigate whether loss of TfR1 surface expression on red blood cells would affect PvRBP2b protein binding, we attempted to generate a knockout of the TFRC gene using CRISPR/Cas9 genome editing of the JK-1 erythroleukemia cell line. We obtained single cell clones that displayed reduced expression of TfR1 and validated the mutation in two independent clones (TfR1 mut1 and TfR1 mut2) (fig. S10, A to C). Both clones contained an identical -3 bp deletion that resulted in the loss of amino acid Gly217 in the TfR1 apical domain, but left the rest of the protein in-frame. TfR1 mut1 was homozygous for this deletion while TfR1 mut2 has a -3 bp as described above on one allele and a -11 bp deletion on the other allele, the latter leading to a premature stop codon. Deletion of TFRC in a mouse model is embryonic lethal and leads to severe disruption of erythropoiesis (36), suggesting that complete deletion of TFRC in erythroid-lineage cells, such as JK-1 may not be possible.
Differentiated polychromatic JK-1 cells (termed jkRBCs) express surface proteins (including TfR1) at levels comparable to differentiated CD34+ bone marrow-derived cultured red blood cells (cRBCs) (37). The jkRBCs, cRBCs and differentiated jkRBCs with a knockout within the basigin receptor (∆BSG) show normal levels of TfR1 whereas TfR1 mutant clones displayed an intermediate level of TfR1 surface staining with a panel of anti-TfR1 mAbs (Fig. 4A and fig. S11). Levels of glycophorin A (GypA) and basigin (BSG) on these TfR1 mutant clones were similar to all control cells, showing that only TfR1 surface expression is affected on these cells (Fig. 4A). To determine if deletion of Gly217 affects PvRBP2b binding, we generated a recombinant TfR1 protein lacking this amino acid (TfR1∆G217). Using SEC, we show that while TfR1∆G217 was still able to bind Tf, its binding to PvRBP2b was completely abolished (Fig. 4B). Gly217, which resides on the lateral surface of the TfR1 apical domain is close to the MACV GP1 interaction surface (fig. S10D) (31).
Fig. 4. Deletions in TFRC reduce TfR1 surface expression, abolish PvRBP2b binding and inhibit P. vivax invasion.
(A) Expression of TfR1, BSG and GYPA on the surface of jkRBCs, TfR1 mutants, ∆BSG null and cultured erythrocytes (cRBCs) as measured by flow cytometry. The right most panels show cytospin analysis of cells stained with May-Grünwald Giemsa staining technique. (B) TfR1∆G217 mutation in TfR1 abrogates PvRBP2b binding as observed using analytical SEC. (C) Quantitative surface proteomics demonstrate specific reduction in TfR1 protein levels in TfR1 mutants compared to wildtype jkRBCs. Levels of Tf, the binding partner for TfR1, are similarly reduced. Significance A was used to estimate p-values, and a minimum of 2 peptides were required for protein quantitation. (D) Binding of recombinant PvRBP2b fragments to jkRBCs, TfR1mutants, ∆BSG and cRBCs cells are shown in blue. Negative controls of unstained cells and isotype control stained cells are shown in the white and orange lines respectively. Compilation of results from PvRBP2b fragment binding to jkRBCs, TfR1 mutants, ∆BSG and cRBCs (right panel). Mean ± S.E.M, n = 3. (E) Comparison of invasion efficiency between jkRBCs and TfR1 mutant cell lines with either P. vivax or P. falciparum. The data shown are averages and the standard error of the mean from between four to five biological replicates shown as open dots. P value was calculated using a paired, two-tailed T test, **** P ≤ 0.0001 and ns is non-significant.
To confirm that the mutation in TFRC did not result in changes in expression of other red blood cells proteins, we compared the abundance of cell surface proteins between wildtype jkRBCs and the two TfR1 mutants using tandem mass tag-based quantitative surface proteomics (Fig. 4C). Out of 237 quantified surface proteins, only TfR1 and Tf were significantly modified, confirming the specificity of the TFRC mutations.
We next wanted to determine if PvRBP2b binding was affected in the TfR1 mutant clones. PvRBP2b161-1454 and PvRBP2b161-969 bound jkRBCs and cRBCs, whereas recombinant fragments PvRBP2b474-1454 and PvRBP2b474-969 that lacked the N-terminal domain did not (Fig. 4D). In contrast, we did not detect any PvRBP2b161-1454 and PvRBP2b161-969 binding to TfR1 mutant cells. This abolition of binding was specific to deletions in TFRC as PvRBP2b binding was unaffected on ∆BSG null cells or on cRBCs (Fig. 4D, right panel). We also compared the invasion efficiency between jkRBCs and TfR1 mutant cell lines with either Brazilian P. vivax isolates or P. falciparum 3D7 (Fig. S10E). A significant (> 10-fold) reduction in invasion efficiency was observed in the TfR1 mutant line compared to the jkRBCs line with P. vivax, while no significant difference was observed with P. falciparum (Fig. 4E). These results validate TfR1 as the cognate receptor for PvRBP2b and that TfR1 is an essential host factor for P. vivax invasion.
Anti-PvRBP2b antibodies block reticulocyte-binding and P. vivax invasion
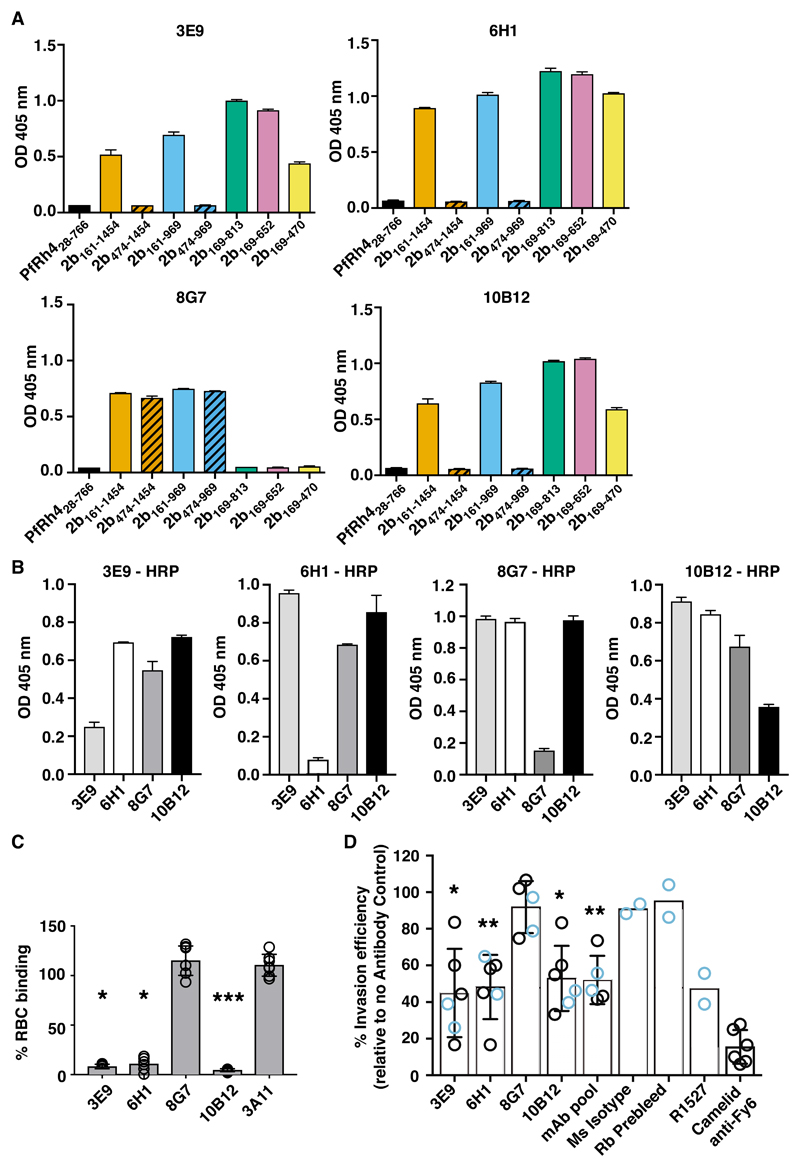
To examine if PvRBP2b antibodies could inhibit P. vivax invasion, we raised mouse monoclonal antibodies to PvRBP2b161-1454 and obtained four mAbs. 3E9, 6H1 and 10B12 bound epitopes within the N-terminal domain present in PvRBP2b169-470 with high affinities (Fig. 5A, fig. S12, A to C, and table S6), while mAb 8G7 recognized an epitope outside the N-terminal domain within amino acids 813 to 969 (Fig. 5A). Competition ELISA experiments using mAbs directly conjugated to horseradish peroxidase (HRP) show that each mAb only competed with itself for binding to PvRBP2b, showing that 3E9, 6H1, and 10B12 bind to distinct epitopes in the N-terminal domain (Fig. 5B). Neither polyclonal nor monoclonal anti-PvRBP2b antibodies recognize recombinant PfRh4 and five other recombinant PvRBPs indicating that these antibodies are specific to PvRBP2b (fig. S12B) (19). Using flow cytometry, we show that addition of anti-PvRBP2b mAbs 3E9, 6H1 and 10B12 abolished the PvRBP2b161-1454 binding to reticulocytes, whereas anti-PvRBP2b mAb 8G7 and anti-PvRBP2a mAb 3A11 had no effect (Fig. 5C).
Fig. 5. Anti-PvRBP2b mAbs inhibit reticulocyte binding and P. vivax invasion in Brazilian and Thai clinical isolates.
(A) ELISA plates were coated with equimolar concentrations of each recombinant fragment and detection with anti-PvRBP2b mAbs 3E9, 6H1, 8G7 and 10B12 are shown. (B) Competition ELISA using immobilized PvRBP2b incubated with un-conjugated anti-2b mAbs (x-axis) and detected with 3E9-HRP, 6H1-HRP, 8G7-HRP and 10B12-HRP as indicated. For (A) and (B) error bars represent range showing the variability of duplicate measures. (C) PvRBP2b161-1454 binding in the presence of anti-PvRBP2b mAbs 3E9, 6H1, 8G7 and 10B12 was analysed by flow cytometry. Normalized binding results where PvRBP2b161-1454 binding in the absence of mAbs was arbitrarily assigned to be 100%. The anti-PvRBP2a mAb 3A11 was used as a negative antibody control. Mean ± S.E.M, n = 5, open dots represent biological replicates. The Kruskal-Wallis test was used to calculate the P value using 8G7 binding as no inhibition, *P ≤ 0.05, ***P ≤ 0.0001. (D) Invasion of P. vivax in Brazilian (blue open dots) and Thai (black open dots) clinical isolates in the presence of anti-PvRBP2b 3E9, 6H1, 8G7 and 10B12, pooled mAbs (each mAb at one third of final concentration), mouse isotype control, purified rabbit prebleed IgG, purified total IgG of anti-PvRBP2b polyclonal antibody R1527 and camelid anti-Fy6 mAb. Antibodies were added in concentrations from 25 µg/ml to 125 µg/ml except for the camelid anti-Fy6 mAb which was used at 25 µg/ml. Mean ± SD, n from 2 to 6, open dots represent biological replicates. For experiments with n > 2, we used the Kolmogorov-Smirnov test to compare 8G7 to 3E9, 6H1 and 10B12, *P ≤ 0.05, **P ≤ 0.001.
We tested the ability of the anti-PvRBP2b antibodies to inhibit P. vivax invasion into human reticulocytes using a short-term P. vivax ex vivo assay with Brazilian and Thai clinical isolates (Fig. 5D, blue and black open dots respectively). As a control, we used a camelid anti-Fy6 antibody which is a single monovalent VHH domain (15 kDa) (38, 39), that targets a surface exposed epitope on DARC and blocks its interaction with PvDBP. The addition of the 25 µg/ml camelid anti-Fy6 antibody in ex vivo assays using Thai isolates resulted in 85% inhibition of P. vivax invasion (Fig. 5D). Using four Thai isolates, the addition of inhibitory anti-PvRBP mAbs 3E9, 6H1 and 10B12 at 25 µg/ml resulted in 49%, 45% and 42% inhibition of P. vivax invasion, respectively. To determine if inhibition could be improved by increasing the concentration of anti-PvRBP2b mAbs to match the molarity and valency of the single VHH domain, we used 125 µg/ml of inhibitory anti-PvRBP2b mAbs. Under these conditions, we tested the invasion efficiency of two Brazilian isolates. We observed that addition of inhibitory anti-PvRBP mAbs 3E9, 6H1 and 10B12 at 125 µg/ml resulted in 68%, 45% and 57% inhibition of P. vivax invasion in Brazilian isolates, respectively (Fig. 5D). To enable quantitative analyses of the ex vivo assays, we combined our initial results of Thai and Brazilian isolates at their respective mAb concentrations. We note that increased concentration of the inhibitory anti-PvRBP2b mAbs resulted in an equivalent or small increase in inhibition of P. vivax invasion (Thai at 25 µg/ml vs Brazilian at 125 µg/ml). Thus, our combined sample set underestimates the level of inhibition for antibody concentrations of 125 µg/ml. As additional controls, we included the non-inhibitory anti-PvRBP2b mAb 8G7 and an IgG1 mouse isotype control which displayed only 8% and 9% inhibition of P. vivax invasion, respectively (Fig. 5D). These results show that addition of anti-PvRBP2b inhibitory mAbs 3E9, 6H1 and 10B12 resulted in significant reduction of P. vivax invasion compared to the non-inhibitory anti-PvRBP2b mAb 8G7 (Fig. 5D).
We show that the inhibitory anti-PvRBP2b mAbs target a domain that appears to be under balancing selection (Fig. 2E and Fig. 5A), which may result in differences in inhibition between clinical isolates owing to the presence of polymorphic epitopes. To circumvent inter-isolate differences, we further tested the combination of all three inhibitory mAbs, 3E9, 6H1 and 10B12 pooled together (mAb pool) and polyclonal anti-PvRBP2b antibodies. The mAb pool resulted in significant 48% reduction in P. vivax invasion in both Thai and Brazilian isolates compared to anti-PvRBP2b mAb 8G7 (Fig. 5D). Addition of purified total IgG of polyclonal anti-PvRBP2b R1527 antibodies resulted in 53% reduction in P. vivax invasion whereas the rabbit prebleed IgG only showed only 5% inhibition (Fig. 5D). A previous study using rabbit anti-PvDBP antibodies shows that P. vivax invasion was reduced up to 64% (40), a level of inhibition comparable to what has been observed with our anti-PvRBP2b antibodies (Fig. 5D). These results show anti-PvRBP2b mAbs that block binding to reticulocytes also inhibit P. vivax invasion and highlight the important role of the PvRBP2b-TfR1 invasion pathway in P. vivax field isolates.
Our results reveal a stable interaction between PvRBP2b and TfR1, and that anti-PvRBP2b antibodies that block binding to reticulocytes also inhibit P. vivax invasion into human reticulocytes. P. vivax invasion is significantly inhibited in the presence of TfR1 mutant cells, showing that TfR1 is a critical host factor for entry into reticulocytes. We propose that the PvRBP2b-TfR1 interaction is important for the initial recognition of the target reticulocyte cells, which results in the commitment of P. vivax parasites for reticulocyte invasion and the subsequent engagement of PvDBP-DARC in tight junction formation leading to the successful completion of the invasion process. Identification of the molecular entities required for P. vivax invasion offer the possibility to target multiple invasion pathways for synergistic inhibition of P. vivax blood stage infection.
Supplementary Material
One Sentence Summary.
Invasion of immature red blood cells by the malaria parasite, Plasmodium vivax, is mediated by binding to the host’s transferrin receptor.
Acknowledgments
We thank Janet Newman from CSIRO Collaborative Crystallization Centre for assistance with setting up the crystallization screens, the Walter and Eliza Hall Institute’s Monoclonal Antibody Facility for production of antibodies, James Williamson for assistance with mass spectrometry and MX and SAXS beamline staff at the Australian Synchrotron for their assistance during data collection. We thank Prof. François Nosten, the staff and patients attending the Mae Sot Malaria Clinic, in Thailand and clinics associated with the Shoklo Malaria Research Unit (SMRU), Tak Province, Thailand. We thank also Professors Yves Colin and Olivier S. Bertrand (INSERM/University Paris 7) for the generous gift of the anti-DARC antibodies. WHT is a Howard Hughes Medical Institute-Wellcome Trust International Research Scholar. This work was supported in part by the Australian Research Council Future Fellowships to WHT and MDWG, a Speedy Innovation Grant to WHT and a National Health and Medical Research Council fellowship (1105754) to JMM. UK was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship. R.D.P. is funded by Wellcome Trust 090770. Genotypes were derived from sequence data generated at the Wellcome Trust Sanger Institute (Wellcome Trust 206194, 098051). MPW was supported by a Wellcome Trust Senior Clinical Research Fellowship (108070/Z/15/Z). This study received funding from Singapore National Medical Research Council (NMRC/CBRG/0047/2013) and the Agency for Science, Technology and Research (A*STAR, Singapore). SMRU is sponsored by The Wellcome Trust of Great Britain, as part of the Oxford Tropical Medicine Research Programme of Wellcome Trust-Mahidol University. The authors acknowledge the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. All data and code to understand and assess the conclusions of this research are available in the main text, supplementary materials and via the following repositories: The atomic coordinates and structure factors for PvRBP2b have been deposited in Protein Data Bank with accession number 5W53.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS
References
- 1.Maier AG, et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med. 2003;9:87–92. doi: 10.1038/nm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crosnier C, et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 4.Tham W-H, et al. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proc Natl Acad Sci U S A. 2010;107:17327–17332. doi: 10.1073/pnas.1008151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 6.Adams JH, et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 7.Horuk R, et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 8.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Weiss GE, et al. Revealing the sequence and resulting cellular morphology of receptor-ligand interactions during Plasmodium falciparum invasion of erythrocytes. PLoS Pathog. 2015;11:e1004670. doi: 10.1371/journal.ppat.1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller I, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–566. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- 11.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 12.Woldearegai TG, Kremsner PG, Kun JFJ, Mordmüller B. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans R Soc Trop Med Hyg. 2013;107:328–331. doi: 10.1093/trstmh/trt016. [DOI] [PubMed] [Google Scholar]
- 13.Mendes C, et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax--molecular evidences from the African West Coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5:e1192. doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ménard D, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A. 2010;107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malleret B, et al. Significant biochemical, biophysical and metabolic diversity in circulating human cord blood reticulocytes. PloS One. 2013;8:e76062. doi: 10.1371/journal.pone.0076062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlton JM, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 18.Gruszczyk J, et al. Structurally conserved erythrocyte-binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement. Proc Natl Acad Sci U S A. 2016;113:E191–E200. doi: 10.1073/pnas.1516512113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.França CT, et al. Plasmodium vivax reticulocyte binding proteins are key targets of naturally acquired immunity in young Papua New Guinean children. PLoS Negl Trop Dis. 2016;10:e0005014. doi: 10.1371/journal.pntd.0005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malleret B, et al. Plasmodium vivax: restricted tropism and rapid remodelling of CD71 positive reticulocytes. Blood. 2015;125:1314–24. doi: 10.1182/blood-2014-08-596015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 22.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 23.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence CM, et al. Crystal structure of the ectodomain of human transferrin receptor. Science. 1999;286:779–782. doi: 10.1126/science.286.5440.779. [DOI] [PubMed] [Google Scholar]
- 25.Radoshitzky SR, et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham J, et al. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog. 2009;5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bozdech Z, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helguera G, et al. An antibody recognizing the apical domain of human transferrin receptor 1 efficiently inhibits the entry of all New World hemorrhagic fever Arenaviruses. J Virol. 2012;86:4024–4028. doi: 10.1128/JVI.06397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tham W-H, et al. Plasmodium falciparum uses a key functional site in complement receptor type-1 for invasion of human erythrocytes. Blood. 2011;118:1923–1933. doi: 10.1182/blood-2011-03-341305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett MJ, Lebrón JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- 31.Abraham J, Corbett KD, Farzan M, Choe H, Harrison SC. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat Struct Mol Biol. 2010;17:438–444. doi: 10.1038/nsmb.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radoshitzky SR, et al. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc Natl Acad Sci U S A. 2008;105:2664–2669. doi: 10.1073/pnas.0709254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright KE, et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature. 2014;515:427–430. doi: 10.1038/nature13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, et al. Crystal structure of PfRh5, an essential P. falciparum ligand for invasion of human erythrocytes. eLife. 2014;3 doi: 10.7554/eLife.04187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson RD, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 2016;48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet. 1999;21:396–399. doi: 10.1038/7727. [DOI] [PubMed] [Google Scholar]
- 37.Bei AK, Brugnara C, Duraisingh MT. In vitro genetic analysis of an erythrocyte determinant of malaria infection. J Infect Dis. 2010;202:1722–1727. doi: 10.1086/657157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smolarek D, et al. A recombinant dromedary antibody fragment (VHH or nanobody) directed against human Duffy antigen receptor for chemokines. Cell Mol Life Sci CMLS. 2010;67:3371–3387. doi: 10.1007/s00018-010-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho JS, et al. Unambiguous determination of Plasmodium vivax reticulocyte invasion by flow cytometry. Int J Parasitol. 2016;46:31–39. doi: 10.1016/j.ijpara.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Grimberg BT, et al. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 2007;4:e337. doi: 10.1371/journal.pmed.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Ent F, Löwe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Hietanen J, et al. Gene models, expression repertoire, and immune response of Plasmodium vivax reticulocyte binding proteins. Infect Immun. 2016;84:677–685. doi: 10.1128/IAI.01117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebrón JA, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 44.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 45.James NG, Mason AB. Protocol to determine accurate absorption coefficients for iron-containing transferrins. Anal Biochem. 2008;378:202–207. doi: 10.1016/j.ab.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanjee U, et al. CRISPR/Cas9 knockouts reveal genetic interaction between strain-transcendent erythrocyte determinants of Plasmodium falciparum invasion. Proc Natl Acad Sci U S A. 2017;114:E9356–E9365. doi: 10.1073/pnas.1711310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 50.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egan ES, et al. Malaria. A forward genetic screen identifies erythrocyte CD55 as essential for Plasmodium falciparum invasion. Science. 2015;348:711–714. doi: 10.1126/science.aaa3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weekes MP, et al. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McAlister GC, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 55.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr Biol Crystallogr. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–71. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- 62.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 63.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 64.Liu L, Boldon L, Urquhart M, Wang X. Small and Wide Angle X-ray Scattering studies of biological macromolecules in solution. J Vis Exp JoVE. 2013 doi: 10.3791/4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svergun D, Barberato C, Koch MHJ. CRYSOL – a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 66.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chacón P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 69.DeLano WL. PyMOL molecular viewer: Updates and refinements. Abstr Pap Am Chem Soc. 2009;238 [Google Scholar]
- 70.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–41. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 71.Harding SE, Rowe AJ, Horton JC, editors. Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry; Cambridge [England]: c1992. [Google Scholar]
- 72.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 73.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 75.Abdul-Gader A, Miles AJ, Wallace BA. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinforma Oxf Engl. 2011;27:1630–1636. doi: 10.1093/bioinformatics/btr234. [DOI] [PubMed] [Google Scholar]
- 76.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinforma Oxf Engl. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russell B, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood. 2004;118:e74–81. doi: 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim C, et al. Expansion of host cellular niche can drive adaptation of a zoonotic malaria parasite to humans. Nat Commun. 2013;4:1638. doi: 10.1038/ncomms2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim C, et al. Improved light microscopy counting method for accurately counting Plasmodium parasitemia and reticulocytemia. Am J Hematol. 2016;91:852–855. doi: 10.1002/ajh.24383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arndt UW, Crowther RA, Mallett JF. A computer-linked cathode-ray tube microdensitometer for x-ray crystallography. J Sci Instrum. 1968;1:510–516. doi: 10.1088/0022-3735/1/5/303. [DOI] [PubMed] [Google Scholar]
- 81.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Crystallogr. 1997;30:203–205. [Google Scholar]
- 82.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weekes MP, et al. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res. 2012;11:1475–1484. doi: 10.1021/pr201135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.