ABSTRACT
Therapeutic strategies for esophageal cancer largely depend on histopathological assessment. To select appropriate treatments of individual patients, we examined the background molecular characteristics of tumor malignancy and sensitivity to multidisciplinary therapy. Seventy-eight surgically-resected esophageal squamous cell carcinoma (ESCC) cases during 2001–2013 were examined. PAX5, a novel gene methylation marker in ESCC, was evaluated in the specimens, as methylation of this gene was identified as an extremely tumor-specific event in squamous cell carcinogenesis of head and neck. PAX5 methylation status was evaluated by quantitative MSP (QMSP) assays. Mean QMSP value was 15.7 (0–136.3) in ESCCs and 0.3 (0–8.6) in adjacent normal tissues (P < 0.001). The 78 cases were divided into high QMSP value (high QMSP, n = 26) and low QMSP value (low QMSP, n = 52). High QMSP cases were significantly associated with downregulated PAX5 expression (P = 0.040), and showed significantly poor recurrence-free survival [Hazard Ratio (HR) = 2.84; P = 0.005; 95% Confidence Interval (CI): 1.39–5.81] and overall survival (HR = 3.23; P = 0.002; 95%CI: 1.52–7.01) in multivariable analyses with histopathological factors. PAX5-knockdown cells exhibited significantly increased cell proliferation and cisplatin resistance. PAX5 gene methylation can predict poor survival outcomes and cisplatin sensitivity in ESCCs and could be a useful diagnostic tool for cancer therapy selection.
KEYWORDS: CDDP, esophageal cancer, GLUT1, methylation, PAX5
Introduction
Esophageal cancer is one of the most difficult solid tumors to overcome. Nearly 90% of esophageal cancers in Asian countries are esophageal squamous cell carcinomas (ESCCs).1 Among Japanese patients diagnosed with ESCC from 2003–2005, the 5-year survival rates after the initial diagnosis were 32.3% in males and 41.3% in females (http://gdb.ganjoho.jp/graph_db/index). These rates are much lower than those for other digestive tract cancers (stomach: 64.2%/61.5%; colon: 70.3%/67.9%; rectum: 67.3%/67.8%) and almost equal to those for lung cancer (25.0%/41.0%) and hepatobiliary cancers (liver: 28.7%/26.2%; biliary tract/gall bladder: 22.5%/19.9%). Histopathologically, tumor budding phenomenon could be one of the key findings of poor prognostic ESCCs.2 To overcome this situation, multidisciplinary treatments are applied to ESCCs, which are generally chemoradiation therapy sensitive.
Diagnostic biomarkers of ESCCs have been widely examined and used clinically. Zhang et al.3 reviewed 5 serum markers (CEA, Cyfra21–1, p53, SCC, VEGF), and found that all 5 had favorable area under curve (AUC) values for receiver-operating characteristic (ROC) curves: CEA: 0.71; Cyfra21–1: 0.58; P53: 0.73; SCC: 0.69; VEGF: 0.81. However, these biomarkers did not generally have high sensitivity: CEA: 8–70%; Cyfra21–1: 36–63%; p53: 14–60%; SCC: 13–64%; VEGF: 64–85%. Biomarker combinations in panels are necessary for the majority of ESCC cases. Furthermore, gene or protein expressions must be evaluated according to various cutoff values depending on cohort characteristics or experimental assays. Prognostic markers of ESCC have also been extensively examined. Recently, Jia et al.4 reported TSP1 as an independent poor prognosis factor for ESCC based on high-throughput magnetic bead-based mass spectrometry. Although TSP1 clearly showed stage-dependent overexpression, cancer prognosis depends on not only histopathological findings but also the effectiveness of adjuvant therapy, including cisplatin-based chemotherapy. Focusing on biomarkers of chemoradiation sensitivity is another trend.
One of the authors previously reported that PAX5 gene methylation could be an excellent marker for head and neck squamous cell carcinoma (HNSCC) detection5 using methylated DNA-binding domain-based sequencing (MBD-seq) and fluorescence-based quantitative methylation-specific PCR (QMSP). He found that the marker had high sensitivity (80%) and high specificity (94%) (AUC: 0.86) in 76 tumors and 19 normal tissues. They further revealed that 79% of TP53-mutated samples exhibited PAX5 gene methylation. In another study, we used droplet digital PCR to show that PAX5 gene methylation can be used as a molecular marker for surgical margin analysis and as a prognostic marker of HNSCCs.6 PAX5, located at chromosome 19p13, was originally identified as a B-cell-specific transcription factor required for B-cell differentiation7 and neural development.8 PAX5 gene methylation has been reported in various neoplasms, including HNSCC,5 gastric cancer,9 hepatoma,10 breast cancer, and lung cancer.11 PAX5 is also mutated in human acute B-cell leukemia.12 To our knowledge, there are no previous reports on PAX5 in ESCC.
In this study, we applied this versatile marker to ESCCs, which have the same pathological characteristics as HNSCCs, to evaluate its potential as a marker for detection, prognosis, and cisplatin-based chemotherapy sensitivity.
Results
PAX5 gene methylation and expression in 78 clinical ESCC samples
QMSP assays were performed for tumors and adjacent normal tissues in the 78 ESCC samples (Fig. 1a). Median (interquartile range) PAX5 QMSP of tumors was 5.92 (1.28–23.27), while that of adjacent normal tissues was 0.08 (0.03–0.25). The difference between tumors and normal tissues was significant (P < 0.001, Mann–Whitney U test). Furthermore, 67 of 78 cases (85.9%) showed more methylation in tumors than in paired adjacent normal tissues.
Figure 1.
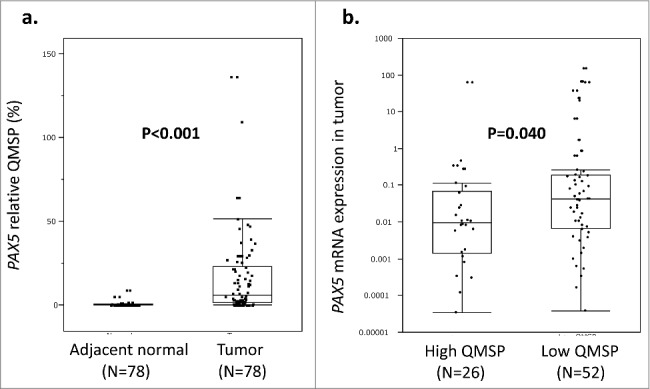
a: QMSP assay results of the 78 ESCCs. The tumor tissues showed significantly high relative QMSP values compared with the adjacent normal tissues (P < 0.001, Mann–Whitney U test). b: The 78 ESCC tumors were divided into PAX5 high QMSP (n = 26) and low QMSP (n = 52) groups by the optimal cutoff value (QMSP = 16.0). PAX5 mRNA expression in the high QMSP group was significantly lower than that in the low QMSP group (P = 0.040, Mann–Whitney U test). Gene hypermethylation appeared to be one of the major mechanisms for the downregulation of PAX5 mRNA expression.
To examine associations between PAX5 methylation and mRNA expression, all 78 cases were divided into PAX5 high QMSP group (n = 26) and PAX5 low QMSP group (n = 52) tumor groups by the optimal cutoff (QMSP = 17.4), which was calculated by ROC curve analysis of PAX5 QMSP values and high/low expression in the 78 ESCC tumors. High and low expression groups (n = 39 each) were divided by the median PAX5 expression value of tumors. qRT-PCR assays showed that PAX5 mRNA expression was significantly downregulated in the highly methylated tumor group (P = 0.040, Mann-Whitney U test; Fig. 1b). In the Spearman test, PAX5 mRNA expression showed marginal correlation with PAX5 QMSP (correlation coefficient -0.211, P = 0.064).
PAX5 QMSP and mRNA expression in esophageal cancer cell lines
PAX5 QMSP values and mRNA expression were also examined in 9 esophageal cancer cell lines (Supplementary Figure S1a). All cell lines except for WSSC had high QMSP values. Although PAX5 mRNA expression varied, most cell lines showed low expression (Supplementary Figure S1b) with significant upregulation after 5-Aza-dC treatment (Supplementary Figure S1c). Gene hypermethylation was confirmed to be modified after 5-Aza-dC treatment (Supplementary Figure S1d).
Influence of PAX5 inhibition on cell proliferation and cell cycle
To observe the effects of PAX5 knockdown, we transfected PAX5 siRNA into 2 esophageal cancer cell lines. We chose the NUEC1 and TE3 cell lines based on their relatively high PAX5 expression. PAX5 knockdown was confirmed by qRT-PCR (Fig. 2a). Cell proliferation ability and cell cycle activity were evaluated by WST-1 and BrdU assays. The results implied that PAX5 knockdown induced both cell proliferation and cell cycle promotion (Fig. 2b, c). However, invasion assays did not show a clear difference between control and PAX5 siRNA transfections (data not shown).
Figure 2.
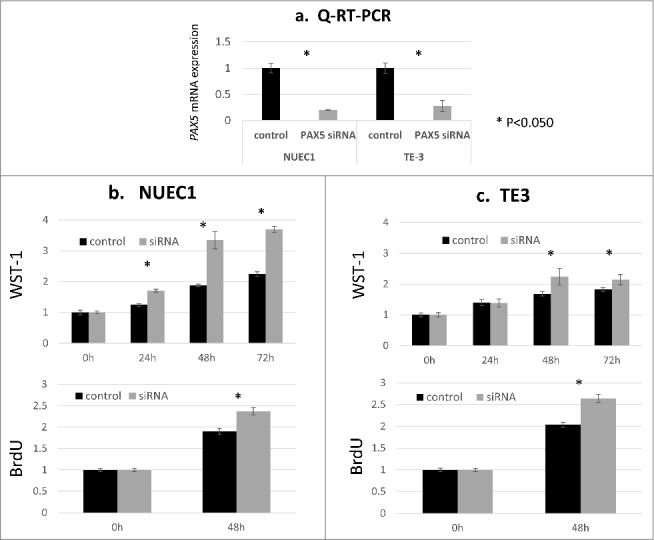
Functional analyses of the PAX5 gene using ESCC cell lines. PAX5 siRNA treatment was performed in NUEC1 and TE3 cells with relatively strong PAX5 mRNA expression. a: PAX5 mRNA expression was examined by qRT-PCR in negative control-transfected cells (control) and PAX5 siRNA-transfected cells (PAX5 siRNA). Significant downregulation of PAX5 was confirmed in both cell lines (P < 0.050, Student's t-test, 2-tailed). b, c: Results of WST-1 and BrdU assays. PAX5-silenced cells showed significantly increased cell proliferation ability in both NUEC1 and TE3 cell lines (P < 0.050, Student's t-test, 2-tailed).
Detection of PAX5 knockdown-related cancer pathways
To identify downstream targets in PAX5-related cancer pathways, we used the Human Cancer Pathway Finder array to compare well-known carcinogenetic gene expressions in control NUEC1 cells and PAX5-silenced NUEC1 cells. Using the optimal threshold (fold change >4.0 or <−4.0), CCL2, GSC, LPL, and SLC2A1 were significantly upregulated, while CCND2, IGFBP5, and TEK were significantly downregulated in PAX5-silenced cells (Fig. 3a). We then applied these findings to the 78 ESCC clinical samples. The results for SLC2A1 gene expression are shown in Fig. 3b as representative data. All cases were divided into PAX5 mRNA downregulated cases (n = 39) and upregulated cases (n = 39) in the tumor tissues when compared with matched normal tissues. Although not significant (P = 0.099, Mann–Whitney U test), downregulation of SLC2A1 expression (fold change <1.0) was mainly seen in PAX5-upregulated cases. Meanwhile, western blotting analysis revealed that SLC2A1 protein was upregulated in the PAX5-silenced TE3 cell line (Fig. 2a, Supplementary Figure S2a). Taken together, these data imply that PAX5 gene expression is inversely associated with SLC2A1 gene expression.
Figure 3.
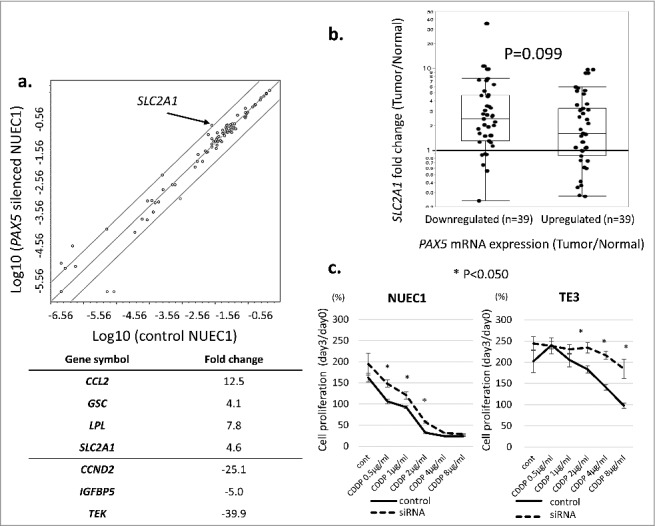
a: Results of the Human Cancer Pathway Finder array. PAX5-silenced NUEC1 cells (Y axis) were compared with negative control-transfected NUEC1 cells (X axis). We set the optimal threshold as fold change >4.0 or <−4.0. Significantly upregulated genes in PAX5-silenced cells were CCL2 (Fold change: 12.5), GSC (4.1), LPL (7.8), and SLC2A1 (4.6). Significantly downregulated genes in PAX5-silenced cells were CCND2 (Fold change: -25.1), IGFBP5 (-5.0), and TEK (-39.9). b: SLC2A1 gene expression in the 78 ESCC clinical samples. SLC2A1 overexpression in the tumor occurred in the majority of PAX5-downregulated tumor cases. c: WST-1 assays comparing PAX5 siRNA-transfected ESCC cells (NUEC1, TE3) and negative control-transfected cells under a wide range of cisplatin concentrations. In both cell lines, PAX5-silenced cells showed significantly higher cell proliferation than control cells.
RNA-seq and methylation array results from TCGA data
To confirm our findings for PAX5 and SLC2A1 more generally, we checked the Cancer Genome Browser website (https://genome-cancer.ucsc.edu/),13 which contains PAX5 methylation array data in 202 esophageal cancer cases (Illumina HumanMethylation450) (Supplementary Figure S2b) and PAX5 exon expression data in 198 esophageal cancer cases (IlluminaHiseq) (Supplementary Figure S2c). We sorted the data by histological cell type (adenocarcinoma or SCC) and sample type (cancer or adjacent normal). As expected, the majority of SCCs (lower part of each heat map) had PAX5 hypermethylation around the TSS site and low PAX5 expression. Moreover, SCCs in the same expression array cohort showed high SLC2A1 expression and low TEK expression (Supplementary Figure S2c). Meanwhile, esophageal adenocarcinoma (EAC) mainly showed PAX5 gene body methylation (red in upper middle area). This different methylation pattern appeared to affect PAX5 overexpression, SLC2A1 downregulation, and TEK overexpression in the 198 esophageal cancer cases.
Cisplatin resistance induced by PAX5 knockdown
SLC2A1, also known as glucose transporter 1 (GLUT1), is an established chemotherapy sensitivity-related gene. Overexpression of SLC2A1 was reported to cause adriamycin resistance14 and poor survival after preoperative chemoradiotherapy.15 Furthermore, inhibition of SLC2A1 was shown to overcome 5-fluorouracil resistance16 or cisplatin resistance.17 Thus, we examined whether PAX5 inhibition induced cisplatin resistance through SLC2A1 overexpression. For this, we performed WST1 assays comparing PAX5 siRNA-transfected ESCCs (NUEC1, TE3) and control cell lines under a wide range of cisplatin concentrations (Fig. 3c). In both cell lines, PAX5-silenced cells showed relatively higher cell proliferation, suggesting acquisition of cisplatin resistance.
PAX5 methylation and clinical outcomes
As PAX5-silenced ESCCs showed oncogenic behavior and cisplatin resistance in the in vitro experiments, we examined the associations between PAX5 expression or gene methylation and clinical outcomes of the 78 ESCC cases. The PAX5 low expression group showed poor RFS and OS (P = 0.082 and P = 0.018, respectively, log-rank test; Supplementary Figure S3). In particular, the PAX5 highly methylated group exhibited significantly poorer RFS and OS than the PAX5 low QMSP group (P = 0.004 and P < 0.001, respectively, log-rank test, Fig. 4). The Cox proportional hazards model was applied to outcome analyses to determine prognostic histological or molecular markers (Table 1). In multivariable analyses of RFS and OS, only positive lymphatic invasion and high PAX5 QMSP were significant poor prognostic factors. The HRs for high PAX5 QMSP were 2.54 (P = 0.006; 95%CI: 1.34–5.29) for RFS and 3.12 (P = 0.003; 95%CI: 1.48–6.70) for OS. Although lymph node metastasis could be one of the major prognostic factors, it is sometimes influenced by chemoradiotherapy or sampling error. Zhang et al. reported that the ratio of metastatic lymph node was superior to predict the survival than pN staging.18 Also, Chen et al. showed that lymphovascular invasion is the more important critical factor of patients’ survival.19 Even in our cohort, positive lymph node metastasis is the significant prognostic factor in univariate analysis, while that is marginally significant in multivariable analysis. We think our results are almost consistent with previous studies.
Figure 4.
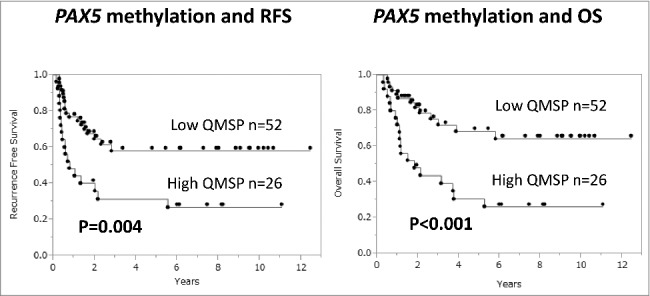
The PAX5 highly methylated group (n = 26) showed significantly poorer RFS and OS (P = 0.004 and P < 0.001, respectively, log-rank test) than the PAX5 low QMSP group (n = 52).
Table 1.
| Factors | Cases | RFS univariate | RFS multivariable | OS univariate | OS multivariable | |
|---|---|---|---|---|---|---|
| pT Stage | T3 / Tis-T2 | 51 / 27 | HR = 1.36 P = 0.392 95%CI:0.68–2.88 |
HR = 1.72 P = 0.169 95%CI:0.80–4.12 |
||
| pN Stage | N1 / N0 | 46 / 31 (1) |
HR = 2.39 P = 0.017 95%CI:1.16–5.39 |
HR = 1.99 P = 0.080 95%CI: 0.92–4.69 |
HR = 2.71 P = 0.013 95%CI:1.22–6.83 |
HR = 2.11 P = 0.089 95%CI:0.90–5.58 |
| Differentiation | Poor / Moderate-Well | 12 / 66 | HR = 1.75 P = 0.186 95%CI: 0.74–3.67 |
HR = 2.56 P = 0.036 95%CI:0.68–2.88 |
HR = 1.55 P = 0.349 95%CI:0.59–3.64 |
|
| Lymphatic invasion | Positive / Negative | 58 / 20 |
HR = 5.18 P < 0.001 95%CI: 1.86–21.55 |
HR = 3.56 P = 0.020 95%CI: 1.20–15.30 |
HR = 6.47 P < 0.001 95%CI:1.95–40.06 |
HR = 4.27 P = 0.019 95%CI:1.23–27.04 |
| Vessel invasion | Positive / Negative | 29 / 49 | HR = 1.89 P = 0.062 95%CI: 0.97–3.66 |
HR = 1.28 P = 0.500 95%CI:0.61–2.60 |
||
| Intraepithelial spread | Positive / Negative | 33 / 37 (8) | HR = 1.15 P = 0.690 95%CI: 0.57–2.33 |
HR = 1.12 P = 0.759 95%CI:0.53-2.43 |
||
| PAX5 QMSP | High / Low | 26 / 52 |
HR = 2.54 P = 0.006 95%CI:1.31–4.93 |
HR = 2.67 P = 0.006 95%CI: 1.34–5.29 |
HR = 3.16 P = 0.002 95%CI:1.55–6.59 |
HR = 3.12 P = 0.003 95%CI:1.48–6.70 |
(number): unknown
To evaluate cisplatin-resistant features of PAX5 methylation, we compared the PAX5 high QMSP and low QMSP groups in cisplatin-based adjuvant chemotherapy cases (n = 26) and no-chemotherapy cases (n = 52) (Fig. 5a). Notably, the PAX5 highly methylated group showed extremely poor RFS in cisplatin-treated cases (P < 0.001, log-rank test), while no significant difference was seen in no-chemotherapy cases (P = 0.091). We also compared the PAX5 high QMSP and low QMSP groups in postoperative radiotherapy cases (n = 15) and no-radiation cases (n = 63) regardless of chemotherapy combination (Fig. 5b). However, PAX5 methylation status did not appear to affect the prognostic outcome among the irradiated cases (P = 0.297). These clinical findings support the notion that PAX5 highly methylated tumors may have the potential of cisplatin resistance, although the case numbers in these subgroup analyses were very small.
Figure 5.
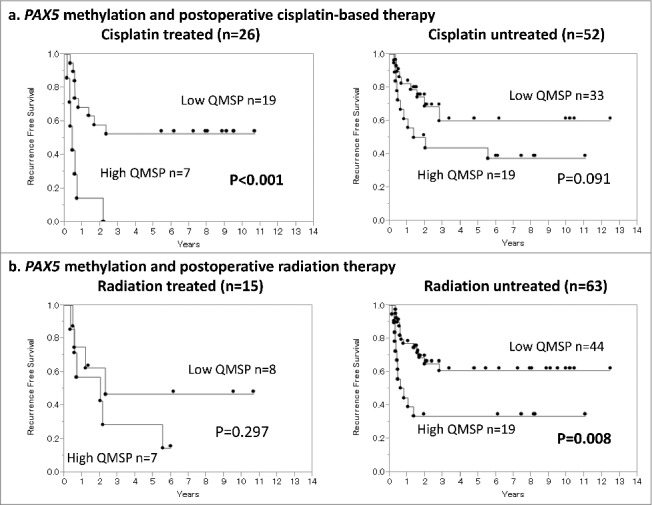
a: Influence of PAX5 methylation status on cisplatin-treated and untreated cases examined by Kaplan–Meier curves. Among the cisplatin-treated cases (n = 26), the PAX5 high QMSP group showed significantly poorer RFS (P < 0.001, log-rank test), while no significant difference was seen in the non-adjuvant chemotherapy cases (n = 52; P = 0.091, log-rank test). b: Influence of PAX5 methylation status on radiation-treated and untreated cases examined by Kaplan–Meier curves. The PAX5 methylation status did not affect the prognostic outcomes among the irradiated cases (P = 0.297, log-rank test).
Discussion
PAX5 methylation has been reported in several cancer types. The same MSP primer pairs were used for analyses of hepatoma,10 gastric cancer,20 and lung cancer,21 and are located at the promoter region of the PAX5 gene. A head and neck cancer study5 from our group defined a new quantitative MSP site around exon 2 containing the most tumor-specific methylated region. This site was also distinctively methylated in ESCCs (Supplementary Figure S2b, yellow rectangle). In addition, PAX5 highly methylated cases showed poorer overall survival than others in our esophageal SCCs. We also confirmed it using other head and neck SCC cohort from TCGA methylation data (Supplementary Figure S4). Interestingly, EACs showed a completely different methylation profile from ESCCs. Although EACs also exhibited high-level methylation, densely methylated sites were found in the gene body area and appeared to cause PAX5 overexpression (Supplementary Figure S2c). Cancer-specific PAX5 overexpression was reported in small-cell lung cancer (SCLC).22 In their cell line analysis, no PAX5 expression was clearly demonstrated in head and neck cancers (n = 12), esophageal cancers (n = 6), non-small cell lung cancers (n = 13), and pancreatic cancers (n = 6), while almost all SCLCs (n = 12) showed positive PAX5 expression. Thus, PAX5 expression patterns may depend on cancer pathological cell types, rather than cancer locations. PAX5 overexpression was also shown in oral squamous cell carcinoma23 and B-cell lymphomas.24 The latter paper described that PAX5 was upregulated through aberrant hypermutation or chromosomal translocation in lymphomas. The authors also thought that alternative mechanisms including gene body hypermethylation could exist.25 Liang et al. reported that the combination of histone deacetylase inhibitors and demethylating agent is necessary to induce PAX5 protein re-expression.26 It also implies that another inhibition mechanism exists. Anyway, although adenocarcinomas are thought to be less sensitive to chemotherapy,27 PAX5-overexpressing adenocarcinomas might be good candidates for cisplatin-based chemotherapy treatment.
The energy-related process of cancer cell growth is supported by increased glucose uptake through the plasma membrane and metabolism.28 SLC2A1, also known as GLUT1, is one of the major glucose transporters. It is frequently overexpressed in various tumors characterized by increased glycolysis rates to meet the requirements of rapidly growing cells29 and contributes to the Warburg effect, the most characterized metabolic change in tumor cells. Mutant p53 was reported to promote GLUT1 translocation from the cytoplasm to the cell surface. Meanwhile, GLUT1 knockdown abolished this stimulatory effect of mutant p53 and reduced glucose uptake.30 Therefore, GLUT1 expression is an essential factor to sustain cancer cell metabolism. Our Cancer Pathway Finder array revealed that PAX5 knockdown was inversely correlated with increasing GLUT1. Among the 78 clinical samples, PAX5 downregulation in tumors might be associated with GLUT1 upregulation (P = 0.099). Furthermore, increased GLUT1 was reported to be associated with mTOR activation, leading to increased glycolysis and reduced autophagy.31
Gene alterations of p53 have been widely examined in ESCCs. Mutant p53 proteins are strongly associated with tobacco smoking and alcohol consumption,32 bind to numerous downstream proteins to irregularly enhance or inhibit their functions and promote cell invasion and metastasis.33 Immunohistochemical staining showed that accumulation of mutant p53 proteins could be a poor prognostic marker in ESCCs.34,35 Zhang et al.36 performed a meta-analysis of p53 mutation status and chemotherapy sensitivity, and concluded that wild type p53 status was correlated with favorable chemotherapy response. Meanwhile, mutant p53 overexpression could be a key factor in ESCC carcinogenesis. In 2011, Liu and colleagues reported that PAX5 directly binds to the p53 promoter and activates p53 signaling in hepatocarcinogenesis.10 They demonstrated that PAX5 induction in a hepatoma cell line caused wild type p53 overexpression by binding to the p53 promoter. This may mean that epigenetic silencing of PAX5 decreases wild type p53 production, resulting in a mutant p53-dominant status that increases the oncogenic potential of tumor cells.
GLUT1 is also a known chemoresistance factor in cancer cells. Head and neck SCC cell lines with high GLUT1 levels showed significantly higher resistance to cisplatin.37 Meanwhile, GLUT1 inhibition chemosensitized cancer cells toward cisplatin.17 We did a confirmation experiment about this point in Supplementary Figure S3c. GLUT1 inhibition increased cis-diamminedichloroplatinum (CDDP) sensitivity, as we expected. Increased GLUT1 mRNA expression in response to PAX5 knockdown could be an explanation for our finding of PAX5 silencing as a biomarker of CDDP resistance (Fig. 3c, Supplementary Figure S2c). Although further mechanistic experiments are required, the association of PAX5 with GLUT1 may be mediated by the tumor suppressor p53, which is known to be regulated by PAX5 binding to its promoter site10 and to repress the expression of key regulators of metabolic genes including c-Myc and GLUT1.38 A possible network around PAX5-p53-SLC2A1 axis in esophageal carcinoma was drawn using cBioPortal,39,40 and is shown in Supplementary Figure S5.
A methylation marker can be a useful tool for cancer detection if the methylation is a very cancer-specific event.41 Although preoperative histological examination can distinguish SCC or adenocarcinoma, molecular diagnosis is necessary for evaluation of tumor malignancy, invasive nature, or chemotherapy sensitivity. The PAX5 QMSP assay might complement this information. Nowadays, molecular-based diagnosis has become less time-consuming. The rapid QMSP procedure requires <3 h from sample collection until data analysis,42 meaning that intraoperative judgments for additional therapy can be undertaken.
In conclusion, PAX5 methylation occurred frequently and was a very tumor-specific event in ESCCs. The PAX5 gene methylation was significantly associated with low expression in tumors. Since PAX5 silencing was correlated with increased cancer cell proliferation and cisplatin resistance, it might lead to poor RFS (HR = 2.67; P = 0.006; 95%CI: 1.34–5.29) and OS (HR = 3.12; P = 0.003; 95%CI: 1.48–6.70). PAX5 gene methylation could be useful as a strong prognostic factor for ESCCs.
Materials and methods
Sample collection
Surgical samples of ESCC were collected at Nagoya University Hospital (Nagoya, Japan) between December 2001 and October 2013. All resected specimens were histopathologically proven to be ESCC, and classified by the TNM classification 7th Edition (Union for International Cancer Control). The sample collection protocol was approved by the Review Board of Nagoya University, and written informed consent for surgical sample use was obtained from all patients. A total of 78 ESCC patients without any preoperative chemoradiotherapy were included in the study, and fresh frozen tissues were collected from their surgical samples. Cancers originated from Barrett's esophagus or esophagogastric junction was excluded. The background and clinicopathological features of the patients are summarized in Supplementary Table S1.
Cell culture
Seven esophageal cancer cell lines (TE1, TE2, TE3, NUEC1, NUEC2, T.Tn, WSSC) and one esophagus epithelial normal cell line (Het-1A) were propagated according to individual cell culture recommendations. TE1, TE2, and TE3 were donated by Tohoku University.43,44 NUEC1, NUEC2, and WSSC were established in our institute.45,46 T.Tn were obtained from the Japanese Collection of Research Bioresources. Het-1A was purchased from ATCC (CRL-2692). All cancer cell lines were cultured in RPMI-1640 medium supplemented with penicillin, streptomycin, and 10% fetal bovine serum at 37°C in a 5% CO2 incubator. Het-1A was cultured in BEBM medium kit without GA-1000 (CC-3170) following the ATCC protocol.
DNA extraction and bisulfite treatment
DNA was extracted from several 10-μm-thick dissected frozen tissues using a DNeasy Blood & Tissue Kit (Qiagen, 69504). Presence of >70% of cancer cells in all tumor samples was histologically confirmed in slides taken before and after sample harvesting. DNA samples (1 μg) extracted from the tissues were subjected to bisulfite treatment using an EpiTect Bisulfite Kit (Qiagen, 59104).
QMSP
The bisulfite-modified DNA was used as a template for fluorescence-based QMSP as described.5,6,47 The primer and probe sequences are shown in Supplementary Table S2. QMSP assays were performed in triplicate using StepOnePlus (Thermo Fisher Scientific). Thermal cycling was initiated with denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Each plate included patient DNA samples, serially diluted positive standards (Bisulfite Converted Universal Methylated Human DNA Standards; Zymo Research, D5015) for constructing the standard curve, and multiple water blanks as no-template controls. Mean values of triplicate samples were used for analyses. The methylation ratio (QMSP value) is defined as the ratio of the fluorescence emission intensity values for the target gene-specific PCR products to those of the ACTB (reference gene) and then multiplied by 100 for easier tabulation.
RNA extraction, cDNA conversion, and quantitative real-time PCR
Total RNA was isolated from frozen surgical samples or cell lines using an RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer's protocol. cDNA (cDNA) was synthesized with qScript cDNA SuperMix (Quanta BioSciences, 95048) with 1 μg of total RNA. PCR amplifications were performed in triplicate with SYBR Premix Ex Taq II (Takara Bio, RR820) as follows: 1 cycle of 95°C for 10 s, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Real-time detection of emission intensity was conducted with a StepOnePlus (Thermo Fisher Scientific). The primer pairs used for RT-PCR are also shown in Supplementary Table S2. ACTB was used as an internal control. PAX5 gene expression levels were calculated by the 2−ΔΔCT method.48 For esophageal cancer cell lines, we double-checked the expression using TaqMan expression assays (Hs00277134-m1 and Hs02758991-g1; Thermo Fisher Scientific).
Five-Aza-2′-deoxycytidine (5-Aza-dC) treatment of cell lines
At 24 h before treatment, cells were plated at low density (1–3 × 106 cells depending on growth characteristics of respective cell lines) in T75 75-cm3 Tissue Culture Flasks. Stock solutions of 100 mM 5-Aza-dC (Sigma, A-3656) and 3 mM Trichostatin A (Sigma, T1952) were prepared with Dimethyl Sulfoxide (Sigma, D2650). Immediately before addition to cell culture medium, appropriate aliquots of stock solutions were dissolved in phosphate-buffered saline (pH 7.5). Cells were treated with 5 µM 5-Aza-dC for 5 d, with daily changes of medium. Trichostatin A (300 nmol/L) was mixed into the medium for the final 24 h. The cells were harvested for RNA extraction.
PAX5 siRNA transfection and cell proliferation assay
Esophageal cancer cell lines with relatively high PAX5 expression (TE3 and NUEC1) were plated on a 96-well plate at 5 × 103 cells/well and incubated overnight at 37°C. The cells were then transfected with PAX5 small interfering RNA (siRNA) (Silencer Select PAX5), SLC2A1 siRNA (Silencer Select SLC2A1) or control siRNA (Silencer Select Negative Control No. 1 siRNA) and RNAiMAX transfection reagent (Thermo Fisher Scientific) (0 h). Cell proliferation was measured at different time points (24 h, 48 h, 72 h) using a Premix WST-1 Cell Proliferation Assay System (Takara Bio Inc., Shiga, Japan), according to the manufacturer's instructions. Briefly, 10 μl of Premix WST-1 was added to each well. After incubation in the dark for 4 h at 37°C, the absorbances were measured by spectrophotometric readings (440 nm). All assays were performed in triplicate, and each experiment was repeated at least twice. Cell proliferation values were calculated as percentages of the value at each time point relative to the value at 0 h (baseline).
BrdU assay
To assess DNA synthesis activity, BrdU cell proliferation enzyme-linked immunosorbent assays (Roche, Tokyo, Japan) were performed. Briefly, transfected cells in 96-well plates were labeled with BrdU for 2 h at each time point. BrdU incorporated into cellular DNA was quantified using the manufacturer's protocol.
Cancer pathway finder array
To find PAX5-related pathways, we used the RT2 Profiler PCR Array Cancer Pathway Finder (Qiagen) following the manufacturer's instructions. The NUEC1 and TE3 cell lines were selected as candidate cells with high PAX5 mRNA expression. PAX5 siRNA (Applied Biosystems) was transfected into both cell lines and resulted in significantly downregulated mRNA expression in both cell lines rather than negative control (Applied Biosystems) transfected cells. These cells were collected after 48 h. The results were analyzed using RT2 Profiler PCR Array Data Analysis v3.5 software. ACTB was used as a reference gene.
Western blotting
Cultured cells were washed and lysed with Pierce RIPA buffer (Thermo Fisher Scientific). Protein lysates were homogenized, and supernatants were collected after centrifugation. Protein concentrations were measured with a Pierce BCA Protein Assay Kit (Takara Bio, Otsu, Japan). NuPAGE LDS sample buffer (Thermo Fisher Scientific) was added to adjusted protein samples, and the samples were separated by 10% SDS polyacrylamide gel electrophoresis. The separated samples were electrotransferred onto polyvinylidene fluoride membranes using an iBlot Gel Transfer Device (Thermo Fisher Scientific) and blocked with 5% bovine serum albumin (Sigma-Aldrich). The membranes were immunoblotted overnight at 4°C with a mouse anti-GLUT1 antibody (Abcam, Tokyo, Japan) followed by Anti-mouse IgG, HRP-linked antibody (Cell Signaling Technology, Tokyo, Japan). For β-actin, a mouse monoclonal anti-β-actin antibody (Abcam) and the same mouse secondary antibody were used. Signals were detected by an LAS-4010 (GE Healthcare, Tokyo, Japan). Densitometry analysis was performed with ImageJ software.49
Statistical analysis
Continuous variables were analyzed by the Mann–Whitney U test as a non-parametric test and Student's t-test (2-tailed) as a parametric test. Categorical variables were analyzed by Fisher's exact test. Recurrence-free survival (RFS) was defined as the time from surgery to first documentation of disease recurrence. Overall survival (OS) was defined as the time from surgery to the date of death from any cause. Associations of gene methylation and other histopathological factors with RFS/OS were evaluated by the Cox proportional hazards model with hazard ratios (HRs) and 95% confidence intervals (95%CIs). Associations with P < 0.05 in univariate analyses were further evaluated in multivariate regression analyses. All tests were 2-sided and considered statistically significant and clinically promising for values of P < 0.05. Statistical analyses were performed using JMP 9 software (SAS Institute, Cary, NC, USA). Statistical significance was set at P < 0.05.
Supplementary Material
Funding Statement
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Scientific Research (C) Number 15K19850 and 15H06281
Conflict of interest
None.
References
- 1.Chen LQ, Hu CY, Ghadirian P, Duranceau A. Early detection of esophageal squamous cell carcinoma and its effects on therapy: an overview. Dis Esophagus. 1999;12:161-7. doi: 10.1046/j.1442-2050.1999.00039.x. PMID:10631905 [DOI] [PubMed] [Google Scholar]
- 2.Koike M, Kodera Y, Itoh Y, Nakayama G, Fujiwara M, Hamajima N, Nakao A. Multivariate analysis of the pathologic features of esophageal squamous cell cancer: tumor budding is a significant independent prognostic factor. Ann Surg Oncol. 2008;15:1977-82. doi: 10.1245/s10434-008-9901-6. PMID:18408975 [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Zhu Z, Liu Y, Jin X, Xu Z, Yu Q, Li K. Diagnostic value of multiple tumor markers for patients with esophageal carcinoma. PLoS One. 2015;10:e0116951. doi: 10.1371/journal.pone.0116951. PMID:25693076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia K, Li W, Wang F, Qu H, Qiao Y, Zhou L, Sun Y, Ma Q, Zhao X. Novel circulating peptide biomarkers for esophageal squamous cell carcinoma revealed by a magnetic bead-based MALDI-TOFMS assay. Oncotarget. 2016. Apr 26;7(17):23569-80. doi: 10.18632/oncotarget.8123. PMID:26993605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrero-Preston R, Michailidi C, Marchionni L, Pickering CR, Frederick MJ, Myers JN, Yegnasubramanian S, Hadar T, Noordhuis MG, Zizkova V, et al.. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9:1031-46. doi: 10.4161/epi.29025. PMID:24786473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi M, Guerrero-Preston R, Sidransky D, Koch WM. PAX5 methylation detection by droplet digital PCR for ultra-sensitive deep surgical margins analysis of head and neck squamous cell carcinoma. Cancer prevention research (Philadelphia, Pa). Cancer Prev Res (Phila). 2015 Nov;8(11):1017-26. doi: 10.1158/1940-6207.CAPR-15-0180. PMID:26304463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes ML, Pridans C, Nutt SL. The regulation of the B-cell gene expression programme by Pax5. Immunol Cell Biol. 2008;86:47-53. doi: 10.1038/sj.icb.7100134. PMID:17998914 [DOI] [PubMed] [Google Scholar]
- 8.Ohtsuka N, Badurek S, Busslinger M, Benes FM, Minichiello L, Rudolph U. GABAergic neurons regulate lateral ventricular development via transcription factor Pax5. Genesis (New York, NY: 2000). 2013;51:234-45. doi: 10.1002/dvg.22370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng J, Liang H, Zhang R, Dong Q, Hou Y, Yu J, Fan D, Hao X. Applicability of the methylated CpG sites of paired box 5 (PAX5) promoter for prediction the prognosis of gastric cancer. Oncotarget. 2014;5:7420-30. doi: 10.18632/oncotarget.1973. PMID:25277182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Li X, Chu ES, Go MY, Xu L, Zhao G, Li L, Dai N, Si J, Tao Q, et al.. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology (Baltimore, Md). 2011;53:843-53. doi: 10.1002/hep.24124. PMID:21319196 [DOI] [PubMed] [Google Scholar]
- 11.Palmisano WA, Crume KP, Grimes MJ, Winters SA, Toyota M, Esteller M, Joste N, Baylin SB, Belinsky SA. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res. 2003;63:4620-5. PMID:12907641 [PubMed] [Google Scholar]
- 12.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al.. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758-64. doi: 10.1038/nature05690. PMID:17344859 [DOI] [PubMed] [Google Scholar]
- 13.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, Zhu J. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. PMID:24084870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song K, Li M, Xu XJ, Xuan L, Huang GN, Song XL, Liu QF. HIF-1alpha and GLUT1 gene expression is associated with chemoresistance of acute myeloid leukemia. Asian Pac J Cancer prev: APJCP. 2014;15:1823-9. doi: 10.7314/APJCP.2014.15.4.1823. PMID:24641416 [DOI] [PubMed] [Google Scholar]
- 15.Saigusa S, Toiyama Y, Tanaka K, Okugawa Y, Fujikawa H, Matsushita K, Uchida K, Inoue Y, Kusunoki M. Prognostic significance of glucose transporter-1 (GLUT1) gene expression in rectal cancer after preoperative chemoradiotherapy. Surg Today. 2012;42:460-9. doi: 10.1007/s00595-011-0027-2. PMID:22072148 [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Fang Y, Wang XT, Liu J, Dan X, Sun LL. Overcoming 5-Fu resistance of colon cells through inhibition of Glut1 by the specific inhibitor WZB117. Asian Pac J Cancer prev: APJCP. 2014;15:7037-41. doi: 10.7314/APJCP.2014.15.17.7037. PMID:25227787 [DOI] [PubMed] [Google Scholar]
- 17.Wang YD, Li SJ, Liao JX. Inhibition of glucose transporter 1 (GLUT1) chemosensitized head and neck cancer cells to cisplatin. Technology in cancer research & treatment. 2013;12:525-35. doi: 10.7785/tcrt.2012.500343 [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Liang H, Gao Y, Shang X, Gong L, Ma Z, Sun K, Tang P, Yu Z. Metastatic lymph node ratio demonstrates better prognostic stratification than pN staging in patients with esophageal squamous cell carcinoma after esophagectomy. Scientific reports. 2016;6:38804. doi: 10.1038/srep38804. PMID:27941828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen WH, Huang YL, Chao YK, Yeh CJ, Chang HK, Tseng CK, Liu YH. Prognostic significance of lymphovascular invasion in patients with esophageal squamous cell carcinoma treated with neoadjuvant chemoradiotherapy. Ann Surg Oncol. 2015;22:338-43. doi: 10.1245/s10434-014-3881-5. PMID:25023545 [DOI] [PubMed] [Google Scholar]
- 20.Li X, Cheung KF, Ma X, Tian L, Zhao J, Go MY, Shen B, Cheng AS, Ying J, Tao Q, et al.. Epigenetic inactivation of paired box gene 5, a novel tumor suppressor gene, through direct upregulation of p53 is associated with prognosis in gastric cancer patients. Oncogene. 2012;31:3419-30. doi: 10.1038/onc.2011.511. PMID:22105368 [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Li S, Gan L, Li C, Qiu Z, Feng Y, Li J, Li L, Li C, Peng W, et al.. Paired box 5 is a frequently methylated lung cancer tumour suppressor gene interfering beta-catenin signalling and GADD45G expression. J Cell Mol Med. 2016 May;20(5):842-54. doi: 10.1111/jcmm.12768. PMID:26843424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanteti R, Nallasura V, Loganathan S, Tretiakova M, Kroll T, Krishnaswamy S, Faoro L, Cagle P, Husain AN, Vokes EE, et al.. PAX5 is expressed in small-cell lung cancer and positively regulates c-Met transcription. Laboratory investigation; a journal of technical methods and pathology. 2009;89:301-14. doi: 10.1038/labinvest.2008.168. PMID:19139719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norhany S, Kouzu Y, Uzawa K, Hayama M, Higo M, Koike H, Kasamatu A, Tanzawa H. Overexpression of PAX5 in oral carcinogenesis. Oncol Rep. 2006;16:1003-8. PMID:17016584 [PubMed] [Google Scholar]
- 24.Balasenthil S, Gururaj AE, Talukder AH, Bagheri-Yarmand R, Arrington T, Haas BJ, Braisted JC, Kim I, Lee NH, Kumar R. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res. 2007;67:7132-8. doi: 10.1158/0008-5472.CAN-07-0750. PMID:17671180 [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Han H, De Carvalho DD Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577-90. doi: 10.1016/j.ccr.2014.07.028. PMID:25263941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Y, Zeng J, Jelicks L, Ma S, Liu J, Mei J, Perez-Soler R, Zou Y. Pax5 Re-expression in H460 Cells Treated with the Combination of Demethylating Agent and Histone Deacetylase Inhibitor is Associated with the Enhancement of P53 Binding to Pax5 Promoter Region. Curr Cancer Drug Targets. 2017;17:169-76. doi: 10.2174/1568009616666160331124759. PMID:27029827 [DOI] [PubMed] [Google Scholar]
- 27.Sehdev V, Peng D, Soutto M, Washington MK, Revetta F, Ecsedy J, Zaika A, Rau TT, Schneider-Stock R, Belkhiri A, et al.. The aurora kinase A inhibitor MLN8237 enhances cisplatin-induced cell death in esophageal adenocarcinoma cells. Mol Cancer Ther. 2012;11:763-74. doi: 10.1158/1535-7163.MCT-11-0623. PMID:22302096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szablewski L. Expression of glucose transporters in cancers. Biochimica et biophysica acta. 2013;1835:164-9. PMID:23266512 [DOI] [PubMed] [Google Scholar]
- 29.Li SJ, Yang XN, Qian HY. Antitumor effects of WNT2B silencing in GLUT1 overexpressing cisplatin resistant head and neck squamous cell carcinoma. Am J Cancer Res. 2015;5:300-8. PMID:25628939 [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, Lin M, Yu H, Liu L, Levine AJ, et al.. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. PMID:24343302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya B, Mohd Omar MF, Soong R. The Warburg effect and drug resistance. Br J Pharmacol. 2016;173:970-9. doi: 10.1111/bph.13422. PMID:26750865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H, Yoshikawa M, Miyazaki T, Nakajima M, Fukai Y, Tajima K, Masuda N, Tsukada K, Fukuda T, Nakajima T, et al.. Expression of p53 protein related to smoking and alcoholic beverage drinking habits in patients with esophageal cancers. Cancer Lett. 2001;167:65-72. doi: 10.1016/S0304-3835(01)00461-X. PMID:11323100 [DOI] [PubMed] [Google Scholar]
- 33.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2-8. doi: 10.1038/ncb2641. PMID:23263379 [DOI] [PubMed] [Google Scholar]
- 34.Shang L, Liu HJ, Hao JJ, Jiang YY, Shi F, Zhang Y, Cai Y, Xu X, Jia XM, Zhan QM, et al.. A panel of overexpressed proteins for prognosis in esophageal squamous cell carcinoma. PLoS One. 2014;9:e111045. doi: 10.1371/journal.pone.0111045. PMID:25337715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bian YS, Osterheld MC, Bosman FT, Benhattar J, Fontolliet C. p53 gene mutation and protein accumulation during neoplastic progression in Barrett's esophagus. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2001;14:397-403. doi: 10.1038/modpathol.3880324. PMID:11353048 [DOI] [PubMed] [Google Scholar]
- 36.Zhang SS, Huang QY, Yang H, Xie X, Luo KJ, Wen J, Cai XL, Yang F, Hu Y, Fu JH. Correlation of p53 status with the response to chemotherapy-based treatment in esophageal cancer: a meta-analysis. Ann Surg Oncol. 2013;20:2419-27. doi: 10.1245/s10434-012-2859-4. PMID:23515910 [DOI] [PubMed] [Google Scholar]
- 37.Li S, Yang X, Wang P, Ran X. The effects of GLUT1 on the survival of head and neck squamous cell carcinoma. Cell Physiol Biochem. 2013;32:624-34. doi: 10.1159/000354466. PMID:24022001 [DOI] [PubMed] [Google Scholar]
- 38.Zawacka-Pankau J, Grinkevich VV, Hunten S, Nikulenkov F, Gluch A, Li H, Enge M, Kel A, Selivanova G. Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J Biol Chem. 2011;286:41600-15. doi: 10.1074/jbc.M111.240812. PMID:21862591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. PMID:23550210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4. doi: 10.1158/2159-8290.CD-12-0095. PMID:22588877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayashi M, Wu G, Roh JL, Chang X, Li X, Ahn J, Goldsmith M, Khan Z, Bishop J, Zhang Z, et al.. Correlation of gene methylation in surgical margin imprints with locoregional recurrence in head and neck squamous cell carcinoma. Cancer. 2015 Jun 15;121(12):1957-65. doi: 10.1002/cncr.29303. PubMed PMID:25773145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi M, Guerrero-Preston R, Okamura J, Michailidi C, Kahn Z, Li X, Ahn J, Goldsmith M, Koch W. Innovative Rapid Gene Methylation Analysis of Surgical Margin Tissues in Head and Neck Cancer. Ann Surg Oncol. 2014 Sep;21(9):3124-31. doi: 10.1245/s10434-014-3661-2. PubMed PMID:24671639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishihira T, Kasai M, Mori S, Watanabe T, Kuriya Y, Suda M, Kitamura M, Hirayama K, Akaishi T, Sasaki T. Characteristics of two cell lines (TE-1 and TE-2) derived from human squamous cell carcinoma of the esophagus. Gan. 1979;70:575-84. PMID:520749 [PubMed] [Google Scholar]
- 44.Kuriya Y, Kitamura M, Akaishi T, Hirayama K, Sekine Y, Nishihira T, Kasai M. A new cell line (TE-3) derived from human squamous cell carcinoma of the esophagus. The Tohoku J Exp Med. 1983;139:377-87. doi: 10.1620/tjem.139.377. PMID:6868074 [DOI] [PubMed] [Google Scholar]
- 45.Hibi K, Nakayama H, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. AIS overexpression in advanced esophageal cancer. Clin Cancer Res. 2001;7:469-72. PMID:11297235 [PubMed] [Google Scholar]
- 46.Tsunoo H, Komura S, Ohishi N, Yajima H, Akiyama S, Kasai Y, Ito K, Nakao A, Yagi K. Effect of transfection with human interferon-beta gene entrapped in cationic multilamellar liposomes in combination with 5-fluorouracil on the growth of human esophageal cancer cells in vitro. Anticancer Res. 2002;22:1537-43. PMID:12168834 [PubMed] [Google Scholar]
- 47.Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370-5. PMID:12684406 [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25:402-8. [DOI] [PubMed] [Google Scholar]
- 49.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671-5. doi: 10.1038/nmeth.2089. PMID:22930834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


