ABSTRACT
Lung carcinoma tops the categories of cancer related motility, and has been treated as the main threat to human health. The functions and related mechanism of FBXW7 controlled lung cancer stem cells' signatures is barely unknown, and the miR-367 regulations of FBXW7 via Wnt signaling have not been explored. Cancer stem cells of either ALDH1+ or CD133+ phenotype were found to be referred to advanced stages in patients with NSCLC (non-small cell lung carcinoma). To study the roles of miR-367, we found greater miR-367 level or FBXW7 level was reserved in NSCLC than that of paired adjacent normal tissues, and their upregulations were positively correlated with Wnt signaling activation. On the contrary, increased miR-367 was correlated with Let-7 repression. MiR-367 was related to stronger sphere forming ability in stem cells of NSCLC. We then explored the functions of the endogenous miR-367 in stem-like cells isolated from NSCLC cell lines. In HEK-293 cells, we identified FBXW7 as the direct downstream gene of miR-367, which consequently released the LIN-28 dependent inhibition of suppressive Let-7. Through informatics analysis, miR-367 was predicated to function through Wnt signaling, and decreased Let-7 played the pivotal role to maintain TCF-4/Wnt pathway activity. The reintroduction of FBXW7 abolished the oncogenic stimulation of miR-367 on TCF-4 activity, with Wnt signaling factors depression. In conclusion, our findings demonstrated the oncogenic roles of miR-367 exerting on the self-renewal ability of cancer stem-like cells through degrading the suppressive FBXW7, eventually helping to maintain Wnt signaling activation through a LIN28B/Let-7 dependent manner.
KEYWORDS: Cancer stem cells, FBXW7, Let-7, MiR-367, Wnt signaling
1. Introduction
Although the origin and plasticity remain controversial, cancer stem-like cells (cancer stem cells, CSCs) have been increasingly identified in many malignancies, and were accused for the roots of tumor generation, drug resistant hibernation, and long-term recurrence.1-3 Persistent activation of one or more pluripotency factors, EMT promoters, and stem cells' distinctive pathways have been identified and involved in development and tissue homeostasis,4 among which, the Wnt/β-catenin signaling is activated in multiple malignancies, especially in subgroup of CSCs.5-9 Targeted elimination of cancer roots of CSCs via Wnt signaling regulations have been put into practice in certain fields.10-13 Clinical trials design was tentatively put into effort with rational combinations of agents to inhibit Wnt, Notch and Hedgehog pathways, which could be of particular importance.2,14
Despite the substantial progress in the treatment of Non-small cell lung carcinoma (NSCLC), the prognoses of patients with NSCLC remain poor, let along those in advanced stages.15-17 The hibernated stem cells should be eliminated for curing cancer, and the strategies of regulating non-coding RNAs have been tentatively applied in multiple cancer. However, the precise controlling of certain non-coding RNA and its downstream signaling could not be predicated accurately, due to the signatures of not complete complementary paired of 3'UTR +binding.3 The non-coding genes of lnc-RNAs, miRNAs, piwi-RNAs, and nearly discovered ceRNAs have been extensively explored, especially for their signatures in in cancer biology and progression,18,19 yet how they are integrated to influence cell pluripotency and identity remains poorly understood.20 For now, little is known for their integration and interaction involving in CSCs' network with post-transcriptional gene, the regulatory machinery of which was critical for controlling cell identity and guiding cell fates.20,21 MiR-367 is one of the most abundant miRNAs in human embryonic stem cells (hESCs), being mainly involved in maintaining pluripotency of stem cells,22 however, how miR-367 was integrated with the CSCs' pluripotency network remains unclear.
Being frequently mutated in many human malignancies, the gene coding E3-Ligase Enzyme F-box and WD repeat domain-containing 7 (FBXW7) participates in ubiquitination and degradation of targeted oncoproteins,23 and was included in the affections of stem cells' renewal and EMT.24-27 David gene analysis indicated the downstream genes of miR-367 were enriched in Wnt signaling, and the possibility of the binding site between miR-367 and FBXW7 was strong and highly confidential. In this study, we were plunged into the interactions of miR-367 and FBXW7 in stem cells of NSCLC, which will help to uncover the miR-367 and FBXW7 dominated signaling pathways in controlling the CSCs' fates.
2. Material and method
2.1. Clinical specimens and Cell culturing
Sixty-three NSCLC cancer samples and paired normal tumor-adjacent samples were obtained from the First Affiliated Hospital of Xi'an Jiaotong University between the year 2012 and 2016. None of the patients had been treated with neoadjuvant radio-/chemotherapy. In order to verify the preliminary diagnosis, the histologic type of each specimen was identified by two experienced pathologists based on the 7th edition of the AJCC cancer staging manual. Tissue samples were frozen immediately in liquid nitrogen and stored at −80°C for later use. Written informed consent was obtained from all patients in according with the Declaration of Helsinki before sample collection. This study was approved and supervised by the Ethics committee of the First Affiliated Hospital of Xi'an Jiaotong University.
The three NSCLC cell lines (A549, H460 and H1299) and HEK-293T were purchased from American Type Culture Collection. Cell lines of A549, H460 and HEK-293T were maintained in DMEM (Gibco), and H1299 was grown in RPMI 1640 Medium (Hyclone). 10% FBS (Gibco), 1% penicillin and 1% streptomycin (Hyclone) were be supplemented into these medium. All Cells were incubated at 37°C a humidified incubator with 5% CO2. Moreover, the demographic and clinic-pathological features of the patients are listed in Table 1.
Table 1.
Clinicopathological characteristics
| Characteristics | Cases (63) |
|---|---|
| Age (years) | |
| <60 | 23 |
| ≥60 | 40 |
| Gender | |
| Male | 51 |
| Female | 12 |
| Tumor size(cm) | |
| ≤ 3 | 21 |
| >3 | 42 |
| TNM tumor stage | |
| I+II | 44 |
| III+IV | 19 |
| Histology | |
| Adenocarcinoma | 30 |
| Squamous cell carcinoma | 25 |
| Other | 8 |
| Degree of differentiation | |
| Well | 18 |
| moderately | 27 |
| Poorly | 18 |
| Lymph node metastasis | |
| Yes | 43 |
| No | 20 |
| Smoking history | |
| Yes | 33 |
| No | 30 |
2.2. Oligonucleotides, plasmids and transfection
miR-367-3p mimic (miR-367-3p), miR-367-3p inhibitor (anti-miR-367-3p), Let-7c mimic (Let-7c) and the negative control were obtained from RiboBio. The siRNAs targeting human FBXW7 and nonspecific control (NC) were obtained from Genechem. To overexpress FBXW7, the cDNA of FBXW7 was amplified and subcloned into pcDNA3.1 vector. The vector was validated by sequencing. PcDNA-FBXW7 was used for FBXW7 overexpression. The 3′-UTR sequence of FBXW7 predicted to target miR-367 or the mutated sequence within the seed sequence was synthesized and inserted into the pGL3 control vector to construct recombinant vectors (wt FOXA1-3′UTR or mt FOXA1-3′UTR). All Oligonucleotides and /or plasmids were transfected into NSCLC cells using Lipofectamine 3000 (Invitrogen).
2.3. Quantitative real-time PCR (qRT-PCR)
Purified RNA and miRNA from NSCLC tissues or cell lines was extracted using the miRNeasy kit (Qiagen) following the manufacturer's protocol; Total mRNA was reverse-transcribed into cDNA by using a RT-PCR kit (AT301 TransGen Biotech) according to the manufacturer's instructions. With the use of a CFX96 Real-Time PCR Detection System (Bio-Rid), real-time quantitative PCR (RT-qPCR) was performed as described in the method of SYBR Premix ExTaqTM II Kit (Takara). mRNA and miRNA levels were normalized to GAPDH and U6, respectively. Expression analysis was calculated using using the 2-ddCt method. The primers specific for miRNA and mRNA were obtained from RiboBio.
2.4. Immunohistochemistry/immunofluorescence and western blot
Immunohistochemistry analysis was performed using antibodies against Fbxw7 (1:250 dilution; ab105752, Abcam), ALDH1A1 (1:200 dilution; #54135, Cell Signaling Technology), CD133 (1:500 dilution; #64326, Cell Signaling Technology), TCF-4 (1:100 dilution; ab217668, Abcam). The evaluation was performed independently by three coworkers in a blind fashion as to the origins of samples. For immunofluorescence assay, formalinfixed and paraffin-embedded tissue sections fixed in 4% paraformaldehyde, and then permeabilized in 0.2% Triton X-100 for 30min at room temperature (RT). After that, tissue sections was blocked 1% BSA in PBS for 1h, and incubated with primary antibodies for 1h. Finally, FITC-labeled anti-rabbit-IgG antibody, or FITC-labeled anti-mouse-IgG antibody was used as the second antibody, and stained with DAPI (Sigma) for 6 min. For western blot analysis, the protein from cell extracts were separated by 10% SDS-PAGE electrophoresis and transferred onto a PVDF membrane. Membranes were incubated with antibodies against the following primary antibodies: Fbxw7 (1:1000 dilution; ab105752, Abcam), Lin28 (1:1000 dilution; ab109751, Abcam), TCF-4 (1:1000 dilution; ab217668, Abcam), Wnt1 (1:1000 dilution; ab15251, Abcam), Vinculin (1:1500 dilution; #18799, Cell Signaling Technology) and then detected using ECL Blotting Detection Reagents (Merck Millipore).
2.5. Sphere-formation assays
The pretreated cells were resuspended in DMEM/F12 Medium and supplemented with 20 ng/mL EGF (BD Biosciences), bFGF and 4 μg/mL insulin (Sigma),and then were plated at 500–1000cells/mL in 6-well ultra-low attachment dishes (Corning Incorporated) for 12 days. To analyze tumor sphere growth, the sphere number and size of two generation spheres were counted and measured by using phase contrast microscope (Nikon) at the 7th to 10th day.
2.6. Luciferase reporter assays and TOPFlash/FOPFlash reporter assay
For analysis of luciferase activity, HEK293 cells (2 × 105 per well) were seeded into 24-well plates, and then cultured in OptimMEM reduced serum media (GIBCO) for 24h before transfection. The cells were co-transfected with 300 ng luciferase reporter vectors (FBXW7-3′UTR-wt or FBXW7-3′UTR-mt), 30 nM miR-367-3p mimics using Lipofectamine 3000 (Invitrogen). Renilla luciferase plasmid (100 ng/well, Promega) was used as an internal control and co-transfected with the described vectors. The promoter reporter plasmid pGL3-Lin28B-WT or pGL3-Lin28B -Mut was cotransfected with miR-367-3p mimic into HEK293T cells. After 48 h, Luciferase activities (Firefly and Renilla) were determined using a luciferase assay kit (Promega). For the TOPFlash/FOPFlash Reporter assays, HEK-293T cells were seeded in 96 well-plates and co-transfected with 200 ngTOPFlash or FOPFlash plasmid, 25 nM miR-367mimics or negative control, 20 ng pRL-TK Renilla luciferase vector (Promega) as normalization vector, and 300 ng pcDNA3.0-FBXW7 or 25 nM siRNA-FBXW7 using Lipofectamine 3000 (Invitrogen). 48 h after transfection, cells were harvested and lysed for standard luciferase assay. Luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity. Each luciferase assay was performed in triplicate.
2.7. Statistical analysis
All numerical data were presented as mean ± standard deviation (SD) of three independent experiments, and analyzed by Graph Pad Prism 5 (Graph Pad) and SPSS13.0 software. Comparison significant differences between samples using Student's t test, or ANOVA, as appropriate. Pearson correlation coefficient was used to examine the correlation between miRNAs and mRNAs. P < 0.05 was considered to be statistically significant.
3. Results
3.1. Ratios of cancer stem cells in NSCLC correlates with prognosis
Stem cells group harboring in tissues of patients with NSCLC were detected by using markers of either ALDH1 or CD133, as were reported and applied before.28-35 Higher ratios of both ALDH1+ cells (Fig. 1A) and CD133+ (Fig. 1B) cells were correlated with advanced clinical phases of NSCLC, with significant differences, which were detected and defining by immunofluorescence (Fig. 1A-B, above) and immunochemistry (Fig. 1A-B, below) staining. Representative images referred to immunofluorescence staining and immunochemistry staining of samples classified at different stages were shown in Fig. 1C.
Figure 1.
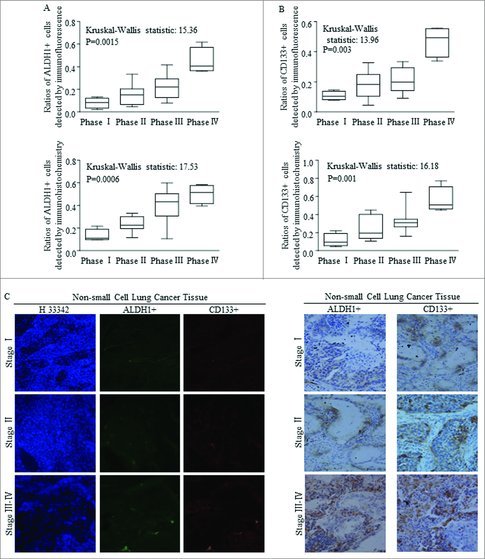
The expression of ALDH1 and CD133 in NSCLC tissues are positively correlated with the clinical features. (A-B) Statistical analysis of ratios of ALDH1+ and CD133+ in NSCLC specimens at different clinical stage using immunofluorescence (IF) and immunohistochemistry (IHC). (C) Representative images of IHC and IF assay of 33 primary NSCLC specimens, including stage I–IV lesions(200 ×). Each bar represents the mean ± SD of three independent experiments. Statistical significance level: p < 0.05.
3.2. The clinical association of FBXW7 with clinical parameters of NSCLC
To explore the significance of FBXW7 in NSCLS, the cBioPortal database was used for clinical analysis. Data of sample from TCGA NSCLC with RNA-seq v2 data was acquired. Kaplan-Meier plots were applied to generate results based on data from http://www.cbioportal.org/36,37 and http://kmplot.com/analysis/.38,39 The genomic alternation is nearly 8% among 119 patients (Fig. 2A), and Kaplan-Meier plots comparing the disease free survival in 119 cases with or without FBXW7 alternations. Data recruited from cBioPortal showed that the FBXW7 mutation and deletion were in accordance with shorter disease free surviva (Fig. 2B). Higher level of FBXW7 was correlated with improved overall survival time, and was also referred to longer survival time before first progression occurred (Fig. 2C-D). Kaplan-Meier plots results of clinic-pathologic parameter of NSCLC showed no significant differences in cases with or without Let-7c alternations, although improved survival time was detected (Fig. 2E-F).
Figure 2.

Clinical roles of FBXW7 in patients with NSCLC. (A) The copy number variation of FBXW7 in NSCLC patients, and the genomic alternation is nearly 8% among 119 patients (http://www.cbioportal.org/). (B) The alternation of both mutation and deletion of FBXW7 was referred to shorter disease free survival (http://www.cbioportal.org/). (C-D) Higher level of FBXW7 was correlated with improved overall survival time in about 2000 cases, and means longer survival time before first progression occurred in about 1500 cases (http://kmplot.com/analysis/index.php?p = service&cancer = lung). (E-F) Kaplan-Meier plots results of clinic-pathologic parameter of NSCLC showed no significant differences in cases with or without Let-7c alternations.
3.3. Inhibition of FBXW7 in NSCLC was accompanied with TCF-4 increasing
FBXW7 was decreased in tissues of NSCLC, comparing to adjacent lung tissues (Fig. 3A, above), while TCF-4 acted the opposite way, and was strengthened significantly in cancer (Fig. 3A, below). Decreased FBXW7 expression level accompanied with later clinical phases (Fig. 3B), but TCF-4 was activated continuously as cancer progression occurred (Fig. 3C). The different expression levels and the inverse correlation between FBXW7 and TCF-4 were identified and the results was significant (Fig. 3D).
Figure 3.
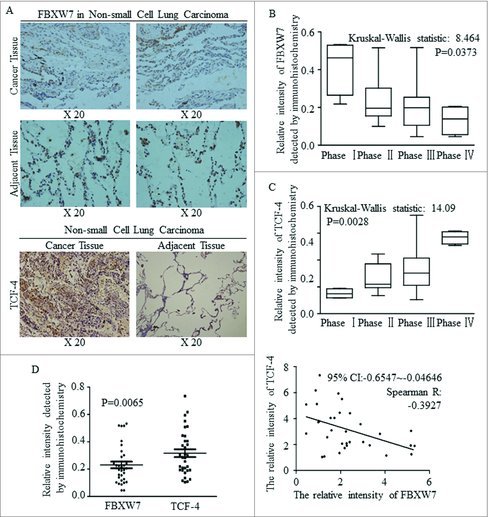
A negative correlation between the expression of FBXW7 and TCF-4 in tissue level. (A) IHC staining for Fbxw7 and TCF4 in tumor-adjacent tissues and NSCLC tissues, original magnification, 200 ×. (B-C) Statistical analysis of Fbxw7 (B) and TCF-4 (C) staining in NSCLC specimens at the different clinical stages (Kruskal-Wallis statistic: 8.464 and14.09, P: 0.0373 and 0.0028, respectively). (D) Using IHC staining, the relative expression and linear correlation of FBXW7 and TCF-4 were analyzed in NSCLC tissues respectively (P < 0.0065, Spearman R: -0.3927).
3.4. Higher miR-367 expression is associated with Wnt signaling activation
The extracted RNAs from NSCLC tissues and paired adjacent tissues of 33 patients were examined by qRT-PCR, and results revealed that miR-367-3p was much higher in tumor tissues compared to matched adjacent non-tumor tissues (Fig. 4A). Comparing analysis indicated that higher miR-367 level was positively correlated with TCF-4 (Fig. 4B-C) expression level and Wnt1 (Fig. 4D-E) expression level. The inverse correlation was detected between miR-367 and Wnt signaling repressor of Let-7c in cancer tissues (Fig. 4F-G). Level of let-7c was much higher in paired adjacent normal tissues, but no significant inverse correlation with miR-367-3p was detected (Fig. 4H-I).
Figure 4.

High expression of miR-367 in cancer tissues are closely related to Wnt signaling and let-7c. (A) qRT-PCR analysis of miR-367 and let-7c expression in NSCLC tissues and adjacent non-tumor lung tissues. (P = 0.0015, Student's t-test). (B-C) The relative expression and linear correlation of miR-367 and TCF4 in NSCLC tissues respectively (Spearman R: 0.7286, p < 0.0001, Spearman rank correlation test). (D-E) Analysis of relationship between miR-367 and Wnt1 in NSCLC tissues using the same statistical method (Spearman R: 0.7286, p < 0.0001,). (F-I) Linear regression and qRT-PCR analysis of miR-367 and let-7c expression in tumor tissue (F-G) and adjacent tissue (H-I) respectively (*p < 0.05, both of assay). These experiments were repeated three times, statistical significance level: *p < 0.05.
3.5. MiR-367 stimulated self-renewal of CSCs in NSCLC cells via blocking FBXW7 and Let-7c
Through using spheres forming assay, of which the spheres number was used as the criterial for assessing the self-renewal ability of cancer stem cells, we found in NSCLC cell lines of A549, H1299 and H460, overexpressed miR-367 increased the number of CSCs (p < 0.01). On the contrary, regulations of anti-miR-367 (p < 0.001), overexpressing FBXW7 (Fig. 5A-C), and overexpressing Let-7c (p < 0.001) all decreased the spheres number of the 1st generation significantly, and no significant differences were found among these three groups (Fig. 5A-C, p = 0.7091), as compared to the controlled group. Further, miR-367-3p overexpression stimulated the self-renewal ability effectively when seeding at the 2nd generation (Fig. 5D-F), and anti-miR-367 treatment, FBXW7 enforcement, and Let-7c overexpression all decreased the spheres number of the 2nd generation (Fig. 5D-F).
Figure 5.
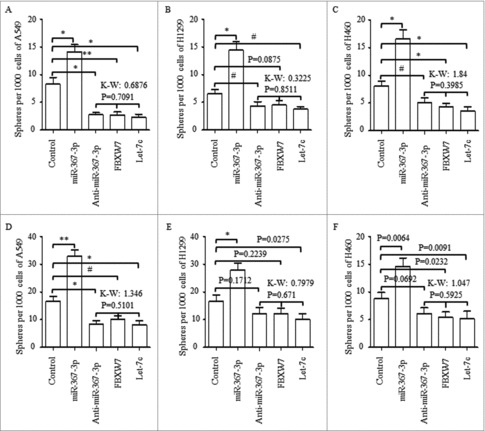
Effect of miR-367, FBXW7 and let-7c on tumor spheroid formation in NSCLC lines. (A-C) Number of spheres per well was quantified on the first generation from A549 (A), H1299 (B), H460 (C) cell lines. (D) Representative images of tumor spheroid form three NSCLC cells with different treatment. (E-G) Number of spheres per well was quantified on the second generation from A549 (E), H1299 (F), H460 (G) cell lines. (H) Representative images of tumor spheroid form three NSCLC cells with different treatment. *P < 0.05 and ** P < 0.01 were based on Student's t-test. ** p < 0.001,* p < 0.01, # p < 0.05.
To identify whether FBXW7 or Let-7 inhibition was necessary for miR-367 induction of Wnt activation, we tested the sphere forming efficiency in lung cancer cells with certain regulations. Reintroducing FBXW7, Let-7c or sh-TCF-4 all abolished the oncogenic roles of miR-367 in self-renewing (Fig. 6A-C). TCF-4 inhibition exerted similar effect as the combination of sh-TCF-4 and miR-367 did (Fig. 6A-C). Spheres from the 1st and 2nd generations were shown in Fig. 6D and Fig. 6E.
Figure 6.
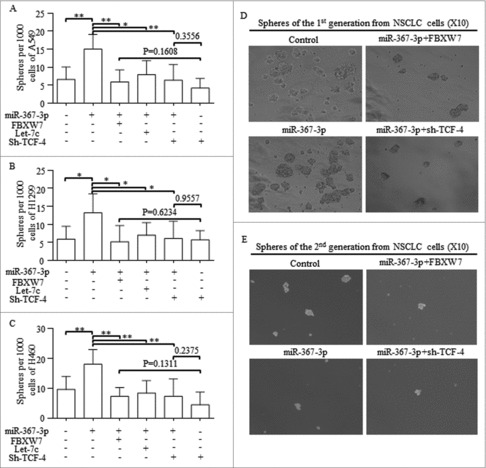
MiR-367 stimulated self-renewal of CSCs via blocking FBXW7 and releasing Let-7c repressed Wnt pathway, A-C. Reintroducing FBXW7, Let-7c or sh-TCF-4 into miR-367 overexpressed spheres cells all abolished its oncogenic roles in self-renewing. TCF-4 inhibition exerted similar effect as the combination of sh-TCF-4 and miR-367 did. D-E. Representative images of spheres from the 1st and 2nd generations were shown. ** p < 0.001,* p < 0.01, # p < 0.05.
3.6. MiR-367 directly degraded FBXW7 and suppressed Let-7 maturation
Let-7 was predicated to be downstream affecter of FBXW7, and we tried to prove that the miR-367 may function through its direct downstream gene of FBXW7 to achieve Let-7 degradation. FBXW7 was predicated to be one potential targeted gene of miR-367 through informatics analysis, and the possible binding site between the 3′UTR of miR-367 and FBXW7 was shown (Fig. 7A). Luciferase results confirmed the direct degradation of FBXW7 mRNA when introducing miR-367, and the mutant binding site of the 3′ UTR abolished this regulation (Fig. 7B). Informatics analysis also predicated the downstream genes of miR-367 may be involved in Wnt signaling pathways (data not shown), and considering that we previously proved the suppression of Let-7c exerted on Wnt activation, we hypothesized that Let-7 may act in miR-367 resulted Wnt dysregulation. LIN28 dominated and functioned through controlling Let-7 family,40,41 and we therefore hypothesized that miR-367 may affect Let-7 via LIN28B. We then proved that FBXW7 suppressed the promoter activity of LIN28B, which also substantially restrain the oncogenic role of miR-367 (Fig. 7C). Overexpressed miR-367 decreased Let-7c level effectively, which also was rescued by FBXW7 enforcement (Fig. 7D).
Figure 7.
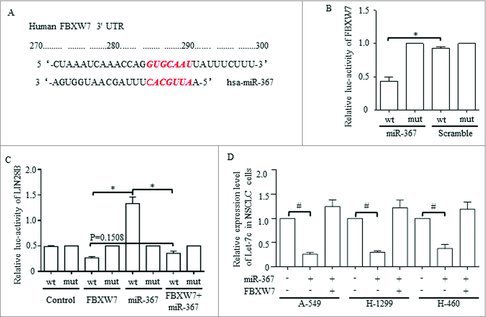
MiR-367 directly targets FBXW7 and represses the level of let-7c through activating Lin28B, (A) The predicted binding sites for miR-367-3p in the 3′-UTR of FBXW7. (B) Luciferase activity was performed in 293 T cells co-transfected with miR-367 mimics or negative control and pGL3 vector containing the wild-type or mutant FBXW7 3′-UTR (p = 0.002, Student's t-test). (C) Dual luciferase activity assays were performed using the promoter reporter plasmid (pGL3-Lin28B-WT or pGL3-Lin28B –Mut) and miR-367 mimics (p = 0.008, Student's t-test). (D) miR-367 significantly repressed the expression of Let-7c in NSCLC cells, p < 0.05 was based on Student's t-test. ** p < 0.001,* p < 0.01, # p < 0.05.
3.7. MiR-367 regulations of TCF-4/Wnt pathway through collateral alternations of FBXW7 and Let-7
Let-7c was proved as one of the non-coding connector of miR-367, decreasing due to LIN28 increasing. We used TOP/FOP plasmid as previously reported (Fig. 8A), and miR-367 activated Wnt activity effectively (Fig. 8B). The inhibition of FBXW7 was required for functions of miR-367 (Fig. 8B). No significant difference was detected between groups of miR-367 overexpression and FBXW7 inhibition (Fig. 8B). To test the influence of miR-367 on Wnt signaling, the key functional actors of Wnt were detected, and the potential mechanism was drafted and illustrated in Fig. 8C. Western was used to illustrate the Let-7 dominated pathway, and in lung cancer stem cells isolated from three cell lines, miR-367 inhibited FBXW7 and consequently released LIN28 level (Fig. 8D). Factors of Wnt signaling of TCF-4 and Wnt1 increased when miR-367 overexpressed and LIN28 increased (Fig. 8E). Reintroducing Let-7c into lung cancer stem cells attenuated the induction of Wnt signaling by miR-367 (Fig. 8E). The restoration of either FBXW7 or Let-7 exerted effective inhibition on Wnt activity (Fig. 8D-E).
Figure 8.
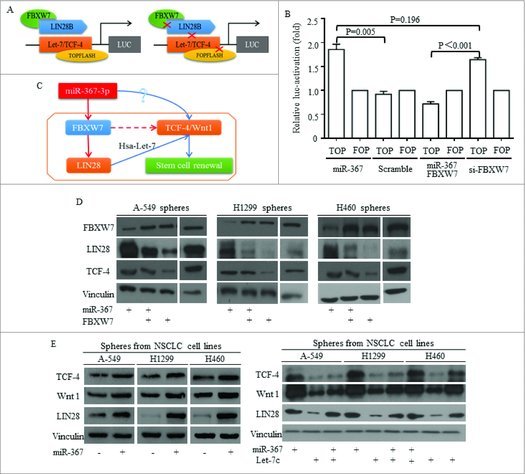
MiR-367 induction of Wnt signaling activation required FBXW7/Let-7c repression. TOP/FOP plasmid was used to detect the FBXW7 controlling of Wnt signaling activity (A), and miR-367 activated Wnt activity effectively (B) No significant difference was detected between groups of miR-367 overexpression and FBXW7 inhibition (B). (C) The hypothesized signaling pathway was drafted and illustrated. (D) Western was used to illustrate the Let-7 dominated pathway, and in lung cancer sphere cells, miR-367 inhibited FBXW7 and consequently released LIN28 and TCF-4 levels. Overexpressing FBXW7 abolished miR-367 stimulation of LIN28 and TCF-4. (E) Factors of Wnt signaling of TCF-4 and Wnt1 increased when miR-367 overexpressed and LIN28 increased, while, reintroducing Let-7c into stem cells attenuated the induction of Wnt signaling by miR-367.
4. Discussion
MiRNAs controlled physiological and pathophysiological processes of development, differentiation and cancer initiation uncovered their central roles in malignant phenotype, dominated in malignancy changes.42 MiR-367 was previously involved in the maintenance of pluripotency of stem cells, however was rarely introduced into cancer stem-like cells' renewal regulations. Moreover, the connections between FBXW7 and the non-coding miR-367 had been explored very little in cancer stem-like cells, the base of cancer counter attract. Non-coding Let-7 has been involved in various regulative mechanisms related malignancy inhibition, and had a crucial part in Wnt signaling interacted stem cells' signatures directly or bypass.1,43-45 HMGA2 activated Wnt signaling was explored in certain kinds of cancer, however the detailed mechanisms were rarely studied.43,46,47 HMGA2 integrated LIN28 level alternations master the post-transcriptional regulation of cell fate, coordinating the embryonic development,20,48 in addition to its role as blockade of let-7 microRNA biogenesis and direct modulation of mRNA translation,49 which we mentioned in activation of Wnt pathway.44
In Non-small cell lung cancer (NSCLCs), more cancer stem-like cells of CD133+ or ALDH1A1+ phenotype were testified to be correlated to advanced cancer progression, suggesting the strategy targeting at eliminating the stem cells group will be prospect in curing cancer. Public data of clinical pathological results confirmed the correlation between FBXW7 expression level and improved survival. We further found that FBXW7 decreasing was related to malignancy and later stages in patients of NSCLCs, as the same of the key Wnt signaling factor TCF-4. What's more, the correlation between miR-367 and TCF-4/Wnt signaling activity was reconfirmed in clinical cancer tissues of NSCLCs by immunohistochemically study, and the inverse relationship between let-7c and miR-367 was testified due to our findings of Let-7c blockade Wnt activity. In bench study, stem-like cells' renewal of NSCLCs was stimulated by miR-367, but could be blocked by FBXW7 and Let-7c, and the difference among groups of anti-miR-367, FBXW7 and Let-7c enforcement were not significant. Through informatic analysis, FBXW7 was predicated to be hunted by miR-367, and functions through interaction of Wnt signaling factors. MiR-367 suppressed FBXW7 expression level failed to sustain Let-7c induction of Wnt blockade, due to loss control of HMGA2, which was proved to be ubiquitinated by FBXW7. In summary, we concluded a FBXW7-centric signaling that miR-367 function through, and the downstream Wnt activation was responsible for miR-367 oncogenic role of self-renewal, in a LIN28B dependent manner. All of the implicated members in this complicated signaling should be given attention to when considering targeted therapy of Wnt signaling, and therefore the precise regulation could never be achieved until every factor in this chain stand steady.
Funding Statement
This experiment was mainly supported by National Science Foundation for Young Scientists of China, grant No. 81602597 (Referred to Xin Sun). This work was also supported in part by National Natural Science Foundation of China, grant No. 81272418 (Referred to Hong Ren), and Natural Science Foundation of Shaanxi Province, grant No. 2016JM8007 (Referred to Jing Zhang).
Conflict of interest
All co-authors implicated in this research approved this article to be published. The authors declare that they have no conflict of interest.
Acknowledgments
The authors acknowledge assistants in the Center for Translational Medicine of The First Affiliated Hospital of Xi'an Jiaotong University, for their technical assistance. The team appreciated prof Peijun Liu's help in experiment conduction and technique guidance. This experiment was mainly supported by National Science Foundation for Young Scientists of China, grant No. 81602597 (Referred to Xin Sun). This work was also supported in part by National Natural Science Foundation of China, grant No. 81272418 (Referred to Hong Ren), and Natural Science Foundation of Shaanxi Province, grant No. 2016JM8007 (Referred to Jing Zhang).
References
- [1].Sun X, Liu J, Xu C, Tang SC, Ren H. The insights of Let‐7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med. 2016;20:1779-88. doi: 10.1111/jcmm.12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445-64. doi: 10.1038/nrclinonc.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sun X, Jiao X, Pestell TG, Fan C, Qin S, Mirabelli E, Ren H, Pestell RG. MicroRNAs and cancer stem cells: the sword and the shield. Oncogene. 2014;33:4967-77. doi: 10.1038/onc.2013.492 [DOI] [PubMed] [Google Scholar]
- [4].Ni T, Li X-Y, Lu N, An T, Liu Z-P, Fu R, Lv W-C, Zhang Y-W, Xu X-J, Grant Rowe R, et al.. Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol. 2016;18:1221-32. doi: 10.1038/ncb3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254-64. doi: 10.1016/j.ceb.2013.01.004 [DOI] [PubMed] [Google Scholar]
- [6].Su J, Wu S, Wu H, Li L, Guo T. CD44 is functionally crucial for driving lung cancer stem cells metastasis through Wnt/β-catenin-FoxM1-Twist signaling. Mol Carcinog. 2016;55:1962-73. doi: 10.1002/mc.22443 [DOI] [PubMed] [Google Scholar]
- [7].Stewart DJ. Wnt signaling pathway in non–small cell lung cancer. J Nat Cancer Inst. 2014;106:1-11. doi: 10.1093/jnci/djt356 [DOI] [PubMed] [Google Scholar]
- [8].Basu S, Haase G, Ben-Ze'ev A. Wnt signaling in cancer stem cells and colon cancer metastasis. F1000Research. 2016;5:F1000 Faculty Rev-699. doi: 10.12688/f1000research.7579.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang J, Zhang B, Meng J, Xiao G, Li X, Li G, Qin S, Du N, Zhang J, Zhang J, et al.. Analysis of risk factors for post-operative complications and prognostic predictors of disease recurrence following definitive treatment of patients with esophageal cancer from two medical centers in Northwest China. Experimental and Therapeutic Medicine. 2017;14:2584-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-Catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Blagodatski A, Poteryaev D, Katanaev VL. Targeting the Wnt pathways for therapies. Mol Cell Ther. 2014;2:28. doi: 10.1186/2052-8426-2-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mao J, Fan S, Ma W, Fan P, Wang B, Zhang J, Wang H, Tang B, Zhang Q, Yu X, et al.. Roles of Wnt/[beta]-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis. 2014;5:e1039. doi: 10.1038/cddis.2013.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang X, Lou Y, Wang H, Zheng X, Dong Q, Sun J, Han B. Wnt signaling regulates the stemness of lung cancer stem cells and its inhibitors exert anticancer effect on lung cancer SPC-A1 cells. Medical Oncology. 2015;32:95. doi: 10.1007/s12032-014-0462-1 [DOI] [PubMed] [Google Scholar]
- [14].Pang Y, Liu J, Li X, Zhang Y, Zhang B, Zhang J, Du N, Xu C, Liang R, Ren H, et al.. Nano Let-7b sensitization of eliminating esophageal cancer stem-like cells is dependent on blockade of Wnt activation of symmetric division. Int J Oncol. 2017;51:1077–88. doi: 10.3892/ijo.2017.4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li L, Cole J, Margolin DA. Cancer stem cell and stromal microenvironment. Ochsner J. 2013;13:109-18. [PMC free article] [PubMed] [Google Scholar]
- [16].Sette G, Salvati V, Mottolese M, Visca P, Gallo E, Fecchi K, Pilozzi E, Duranti E, Policicchio E, Tartaglia M, et al.. Tyr1068-phosphorylated epidermal growth factor receptor (EGFR) predicts cancer stem cell targeting by erlotinib in preclinical models of wild-type EGFR lung cancer. Cell Death Dis. 2015;6:e1850. doi: 10.1038/cddis.2015.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zakaria N, Yusoff NM, Zakaria Z, Lim MN, Baharuddin PJN, Fakiruddin KS, Yahaya B. Human non-small cell lung cancer expresses putative cancer stem cell markers and exhibits the transcriptomic profile of multipotent cells. BMC Cancer. 2015;15:84. doi: 10.1186/s12885-015-1086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Koch L. Functional genomics: Screening for lncRNA function. Nat Rev Genet. 2017;18:70. doi: 10.1038/nrg.2016.168 [DOI] [PubMed] [Google Scholar]
- [19].Schmitt Adam M, Chang Howard Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452-63. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tsanov K, Pearson D, Wu Z, Han A, Triboulet R, Seligson M, Powers J, Osborne J, Kane S, Gygi S, et al.. LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency. Nat Cell Biol. 2017;19:60-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15:271-80. doi: 10.1016/j.stem.2014.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kaid C, Silva PB, Cortez BA, Rodini CO, Semedo-Kuriki P, Okamoto OK. miR-367 promotes proliferation and stem-like traits in medulloblastoma cells. Cancer Sci. 2015;106:1188-95. doi: 10.1111/cas.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yumimoto K, Akiyoshi S, Ueo H, Sagara Y, Onoyama I, Ohno S, Mori M, Mimori K, Nakayama KI. F-box protein FBXW7 inhibits cancer metastasis in a non-cell-autonomous manner. J Clin Invest. 2015;125:621-35. doi: 10.1172/JCI78782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Takeishi S, Nakayama KI. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br J Cancer. 2014;111:1054-9. doi: 10.1038/bjc.2014.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, Paquis P, Preynat-Seauve O, Krause KH, Chneiweiss H, et al.. The miR 302–367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232-44. doi: 10.1038/cdd.2011.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang SL, Yang M, Herrlinger S, Liang C, Lai F, Chen JF. MiR-302/367 regulate neural progenitor proliferation, differentiation timing, and survival in neurulation. Dev Biol. 2015;408:140-50. doi: 10.1016/j.ydbio.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu Z, Xu Y, Zhao J, Liu Q, Feng W, Fan J, Wang P. miR-367 promotes epithelial-to-mesenchymal transition and invasion of pancreatic ductal adenocarcinoma cells by targeting the Smad7-TGF-beta signalling pathway. Br J Cancer. 2015;112:1367-75. doi: 10.1038/bjc.2015.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nosrati A, Naghshvar F, Khanari S. Cancer stem cell markers CD44, CD133 in primary gastric adenocarcinoma. Int J Mol Cell Med. 2014;3:279-86. [PMC free article] [PubMed] [Google Scholar]
- [29].Huang M, Zhu H, Feng J, Ni S, Huang J. High CD133 expression in the nucleus and cytoplasm predicts poor prognosis in non-small cell lung cancer. Dis Markers. 2015;2015:986095. doi: 10.1155/2015/986095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu QF, Zhang ZF, Hou GJ, Yang GY, He Y. Polymorphisms of the stem cell marker gene CD133 and the risk of lung cancer in Chinese population. Lung. 2016;194:393-400. doi: 10.1007/s00408-016-9876-1 [DOI] [PubMed] [Google Scholar]
- [31].Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S, et al.. Highly tumorigenic lung cancer CD133(+) cells display stem-like features and are spared by cisplatin treatment. Proc Nat Acad Sci U S A. 2009;106:16281-6. doi: 10.1073/pnas.0905653106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang P, Suo Z, Wang M, Høifødt HK, Fodstad Ø, Gaudernack G, Kvalheim G. In vitro and in vivo properties of CD133 expressing cells from human lung cancer cell lines. Exp Hemat Oncol. 2013;2:16-. doi: 10.1186/2162-3619-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sarvi S, Mackinnon AC, Avlonitis N, Bradley M, Rintoul RC, Rassl DM, Wang W, Forbes SJ, Gregory CD, Sethi T. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res. 2014;74:1554-65. doi: 10.1158/0008-5472.CAN-13-1541 [DOI] [PubMed] [Google Scholar]
- [34].Roudi R, Korourian A, Shariftabrizi A, Madjd Z. Differential expression of cancer stem cell markers ALDH1 and CD133 in various lung cancer subtypes. Cancer Invest. 2015;33:294-302. doi: 10.3109/07357907.2015.1034869 [DOI] [PubMed] [Google Scholar]
- [35].Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018-32. doi: 10.18632/oncotarget.6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al.. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439-46. doi: 10.1007/s10549-016-4013-7 [DOI] [PubMed] [Google Scholar]
- [39].Győrffy B, Bottai G, Lehmann-Che J, Kéri G, Őrfi L, Iwamoto T, Desmedt C, Bianchini G, Turner NC, de Thè H, et al.. TP53 mutation-correlated genes predict the risk of tumor relapse and identify MPS1 as a potential therapeutic kinase in TP53-mutated breast cancers. Mol Oncol. 2014;8:508-19. doi: 10.1016/j.molonc.2013.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cimadamore F, Amador-Arjona A, Chen C, Huang C-T, Terskikh AV. SOX2–LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Nat Acad Sci. 2013;110:E3017-E26. doi: 10.1073/pnas.1220176110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends in Cell Biology. 2012;22:474-82. doi: 10.1016/j.tcb.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dror S, Sander L, Schwartz H, Sheinboim D, Barzilai A, Dishon Y, Apcher S, Golan T, Greenberger S, Barshack I, et al.. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol. 2016;18:1006-17. doi: 10.1038/ncb3399 [DOI] [PubMed] [Google Scholar]
- [43].Copley MR, Babovic S, Benz C, Knapp DJHF, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, et al.. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916-25. doi: 10.1038/ncb2783 [DOI] [PubMed] [Google Scholar]
- [44].Sun X, Xu C, Tang SC, Wang J, Wang H, Wang P, Du N, Qin S, Li G, Xu S, et al.. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83-9. doi: 10.1038/cgt.2016.3 [DOI] [PubMed] [Google Scholar]
- [45].Madison BB, Jeganathan AN, Mizuno R, Winslow MM, Castells A, Cuatrecasas M, Rustgi AK. Let-7 represses carcinogenesis and a stem cell phenotype in the intestine via regulation of Hmga2. PLoS Genet. 2015;11:e1005408. doi: 10.1371/journal.pgen.1005408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zha L, Zhang J, Tang W, Zhang N, He M, Guo Y, Wang Z. HMGA2 elicits EMT by activating the Wnt/β-catenin pathway in gastric cancer. Dig Dis Sci. 2013;58:724-33. doi: 10.1007/s10620-012-2399-6 [DOI] [PubMed] [Google Scholar]
- [47].Wend P, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS, et al.. WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med. 2013;5:264-79. doi: 10.1002/emmm.201201320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shyh-Chang N, Daley George Q. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell.2013;12:395-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sun X, Qin S, Fan C, Xu C, Du N, Ren H. Let-7: a regulator of the ERalpha signaling pathway in human breast tumors and breast cancer stem cells. Oncol Rep. 2013;29:2079-87. doi: 10.3892/or.2013.2330 [DOI] [PubMed] [Google Scholar]


