ABSTRACT
Accumulating evidence demonstrates that a series of differentially expressed lncRNAs is important in tumorigenesis. However, the function of many of the lncRNAs in lung cancer remains elusive. In the present study, we used microarray analysis to identify lncRNAs that are dysregulated in non-small-cell lung cancer (NSCLC) as compared with normal tissues. Among the dysregulated lncRNAs, we identified TFPI2AS1, an antisense transcript of the tumor suppressor TFPI2 (tissue factor pathway inhibitor 2). TFPI2AS1 was shown to be markedly upregulated in NSCLC patient tumors as compared to paired non-tumor samples. TFPI2AS1 knockdown increased NSCLC cell proliferation and migration, which was associated with enhanced G1/S transition and downregulation of cyclin D1 and cyclin-dependent kinases 2 (CDK2), while TFPI2AS1 overexpression had the opposite effect. Knockdown and overexpression experiments also suggested that TFPI2AS1 regulates NSCLC cell migration and AKT activation. Moreover, TFPI2AS1 is a positive regulator of TFPI2. Our findings bring new insights for understanding the role of TFPI2AS1 in mediating the proliferation and migration of NSCLC cells by regulating TFPI2 expression.
KEYWORDS: lncRNA, lung cancer, migration, proliferation, TFPI2AS1
Introduction
Lung cancer, one of the most frequent malignancies, has become the leading cause of cancer-associated mortality, with approximately 1.8 million new cases annually throughout the world.1 Among all lung cancer cases, non-small-cell lung cancer (NSCLC) constitutes approximately 85%.2 Despite considerable advances in curative protocols, the prognosis for lung cancer remains unfavorable, with a patient 5-year survival rate of about 15%.3 Therefore, it is urgent to identify the underlying mechanisms of lung cancer development and exploit new diagnostic biomarkers and therapeutic strategies for treating lung cancer.
Long non-coding RNAs (lncRNAs) are evolutionarily conserved transcripts of greater than 200 nucleotides in length with no capacity to code proteins. They are involved in a wide range of biologic functions such as cell cycle progression, cell growth, cell proliferation and apoptosis.4 Therefore, dysregulated lncRNAs play pivotal roles in a variety of human diseases, and especially cancer, for which they are associated with development, invasion, and metastasis of tumors.5 For example, upregulated lncRNA HOTAIR contributes to the metastasis of breast cancer cells by transcriptionally modulating important developmental genes6; the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is highly expressed in multiple types of cancer, including lung adenocarcinoma and hepatocellular carcinoma, and can promote proliferation and metastasis of cancer cells, suggesting that MALAT1 exerts oncogenic effects7; GAS5 (growth arrest-specific transcript 5) functions as a tumor suppressor and is decreased in breast cancer.8,9 Aberrant lncRNA expression also plays a regulatory role in lung cancer. It has been reported that 2,420 lncRNAs are expressed differentially between lung adenocarcinoma and normal lung tissue, suggesting that lncRNAs may represent novel molecular biomarkers for diagnosing lung adenocarcinoma.10 At present, however, only a few lncRNAs have been identified in lung cancer, including MALAT1, ANRIL, H19, and HOTAIR 11, GAS6-AS1 (GAS6 antisense RNA1)12, and AK126698 13, lncRNA CAR intergenic 10 (CAR10)14, lncRNA ZNRD1-AS115, and linc0067316. Therefore, identification of additional lncRNAs with roles in lung cancer needs further exploration.
In the present study, by analyzing lncRNA microarray data from human lung cancer tissues, we characterized a series of cancer-related lncRNAs. Among the differentially expressed lncRNAs, we identified TFPI2 antisense RNA 1 (TFPI2AS1; AC002076.1), which is an antisense transcript of tissue factor pathway inhibitor 2 (TFPI2). TFPI2 is a Kunitz-type serine protease inhibitor with inhibitory activity against tumor growth and metastasis.17 Analysis of lncRNA microarray data verified that TFPI2AS1 was overexpressed in human lung cancer tissues compared with matched non-tumor tissues. To explore TFPI2AS1 function in regulating the metastatic properties of NSCLC cells, the effects of TFPI2AS1 siRNA-mediated silencing and overexpression were assessed. Furthermore, we explored the relationship between TFPI2AS1 and TFPI2 expression. Our observations may provide novel direction for understanding the underlying mechanisms in lung cancer.
Results
TFPI2AS1 is upregulated in NSCLC tissues
To investigate the potential biological functions of lncRNAs in lung cancer, we examined the lncRNA expression profile in human lung cancer tissues by microarray analysis. Several lncRNAs were identified that were differentially expressed between lung cancer tissues and non-tumorous tissue specimens (data not published). One previously uncharacterized lncRNA (AC002076.1) was markedly upregulated in three sets of tumor tissues compared with matched non-tumor tissues (Figure 1A). Bioinformatics analysis showed that AC002076.1 is an antisense partner of tissue factor pathway inhibitor 2 (TFPI2), a putative tumor suppressor gene.18,19 Therefore, we named AC002076.1 as TFPI2 antisense RNA 1 (TFPI2AS1; AC002076.1). Subsequently, we also measured the background expression of TFPI2AS1 in HBE cells, 293T cells, HeLa cells as well as lung cancer cell lines including A549, H1299 and H441. We found that there existed background expression of TFPI2AS1 in all three NSCLC cell lines; HBE cells, 293T as well as HeLa cells (Figure 1B). Additionally, we selected A549 cell line, the relative expression level of TFPI2AS1 in which is higher than the two other NSCLC cell lines including H1299 and H441 cells, for further study to determine whether TFPI2AS1 expression may correlate with malignant behavior in lung cancer.
Figure 1.
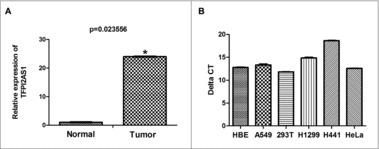
Relative expression levels TFPI2AS1 in lung cancer tissues and cell lines. (A) TFPI2AS1 expression was assessed by microarray analysis of lung cancer tumor specimens as compared to normal adjacent lung tissue (n = 3). (B) The expression of TFPI2AS1 was measured by qRT-PCR in A549, H1299, H441 lung cancer cell lines and HBE cells, 293T cells and HeLa cells. *P< 0.05 vs normal group. Data are expressed as the means ± SD of triplicate experiments.
TFPI2AS1 knockdown promotes growth of A549 cells
To further explore the role of TFPI2AS1 in lung cancer, we transiently transfected TFPI2AS1 siRNA into A549 cells to reduce the expression of TFPI2AS1. The knockdown efficiency was verified by qRT-PCR (Figure 2A). Subsequently, we investigated the effect of TFPI2AS1 knockdown on the proliferation of A549 cells by MTT assay. As shown in Figure 2B, the TFPI2AS1 siRNA significantly augmented the proliferation of A549 cells. We further studied the effect of downregulating TFPI2AS1 on the capacity of colony formation in A549 cells. The number of colonies was increased in TFPI2AS1 siRNA-transfected A549 cells (Figure 2C). We also measured cell proliferation using Brdu incorporation assays. The results showed that TFPI2AS1 deficiency markedly increased DNA synthesis in A549 cells. Collectively, these results suggest that knockdown of TFPI2AS1 can promote cell proliferation of lung cancer.
Figure 2.
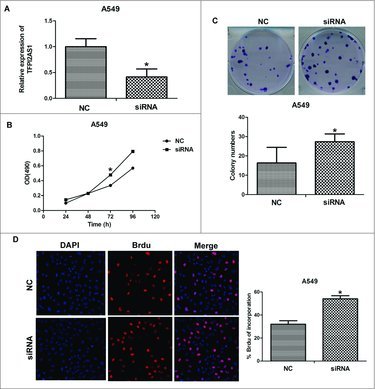
TFPI2AS1 knockdown promotes cell proliferation in A549 cells. (A) A549 cells were transfected with control siRNA or TFPI2AS1 siRNA for 48 h and then cells were harvested for examining efficiency of knockdown. (B) A549 cells transfected with control siRNA or TFPI2AS1 were trypsinized and plated into 96-well plates for MTT assay. (C) Colony formation of A549 cells 14 days after transfection with the indicated oligonucleotides. (D) After transfection with control siRNA or TFPI2AS1 siRNA for 48 h, A549 cell proliferation was measured by a Brdu incorporation assay. The proliferating cells that recently underwent DNA synthesis were labeled with Brdu (red), and total nuclei were visualized by DAPI (blue) staining. % Brdu presented as percent of Brdu incorporated cells within the siRNA-treated A549 cell population. *P < 0.05 vs NC. Data are expressed as the mean ± SD of the experiments performed in triplicate.
TFPI2AS1 overexpression suppresses cell growth in A549 cells
To confirm the function of TFPI2AS1 in mediating cell proliferation, we established TFPI2AS1 overexpression A549 cells. Full length TFPI2AS1 was recombined with eukaryotic expression plasmid pcDNA3.1 (+) and transfected into A549 cells with Lipofectamine 2000 (Figure 3A). MTT assay indicated that the proliferation ability of A549 cells was markedly reduced after transfection of TFPI2AS1 expression vector for 72 h (Figure 3B). Furthermore, TFPI2AS1 overexpressing cells had decreased number of colonies in the colony formation assay (Figure 3C). Consistently, upregulation of TFPI2AS1 in A549 cells inhibited DNA synthesis (Figure 3D). Taken together, these data confirm that TFPI2AS1 suppresses cell growth of A549 cells.
Figure 3.
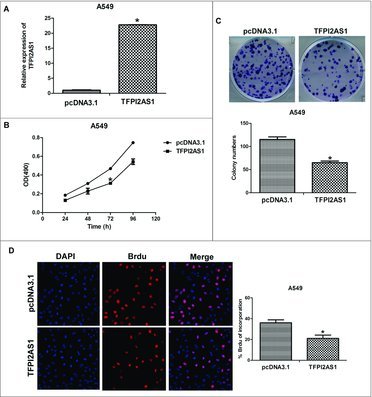
TFPI2AS1 overexpression inhibits cell proliferation in A549 cells. (A) The expression of TFPI2AS1 in A549 cells at 24 h-96 h post-transfection with pcDNA3.1/ TFPI2AS1-full was measured by qRT-PCR. (B) A549 cells transfected with pcDNA3.1/ TFPI2AS1-full were trypsinized and plated into 96-well plates for MTT assay. (C) Colony formation of A549 cells 14 days after transfection with pcDNA3.1/TFPI2AS1-full. (D) After transfection with pcDNA3.1/TFPI2AS1-full for 48 h, A549 cell proliferation was measured by a Brdu incorporation assay. The proliferating cells that recently underwent DNA synthesis were labeled with Brdu (red), and total nuclei were visualized by DAPI (blue) staining. % Brdu presented as percent of Brdu incorporated cells within the transfected A549 cell population. *P < 0.05 vs control group. Data are expressed as the mean ± SD. of the experiments performed in triplicate.
TFPI2AS1 suppresses cell cycle progression of A549 cells
To explore the mechanism by which TFPI2AS1 modulates cell growth, we evaluated the cell cycle progression of TFPI2AS1 siRNA-transfected A549 cells by flow cytometry. TFPI2AS1 knockdown obviously decreased the population of cells in G1 phase and had a tendency to increase the population of cells in S phase (Figure 4A). We further evaluated the effects of TFPI2AS1 knockdown on CyclinD1 and CDK2, each of which is involved in the regulation of G1 phase progression.20,21 Western blotting analyses (Figure 4B) demonstrated that the expression levels of each of these proteins were significantly decreased by TFPI2AS1 downregulation in A549 cells.
Figure 4.
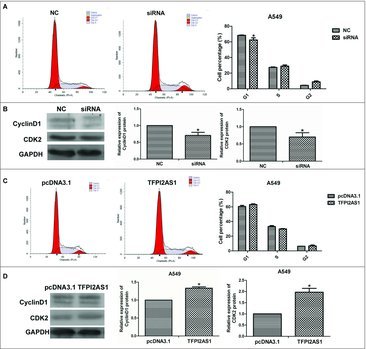
TFPI2AS1 knockdown or overexpression modulates the G1/S transition in A549 cells. A549 cells were transfected with siRNA or pcDNA3.1/TFPI2AS1-full for 48 h. The population of cells at the G1, S and G2/M phase was detected by flow cytometry (A and C). The levels of CyclinD1 and CDK2 were detected by Western blotting assay (B and D). GAPDH protein was used as endogenous normalize. *P < 0.05 vs control group. Data are expressed as the mean ± SD of the experiments performed in triplicate.
To further support these findings, we studied the effect of TFPI2AS1 upregulation on cell cycle progression in A549 cells. TFPI2AS1 overexpressing cells had a tendency to accumulate cells in the G1 phase with a reduced population of cells in S phase (Figure 4C). In addition, the expression levels of CyclinD1 and CDK2 were increased following TFPI2AS1 overexpression in A549 cells (Figure 4D). These data suggested that upregulating TFPI2AS1 arrests cell cycle progression by arresting G1/S transition.
TFPI2AS1 suppresses cell migration of A549 cells
One of the features of metastasis, the main cause of death among patients diagnosed with cancer, is the conversion to a malignant cell phenotype.22 To evaluate the effect of silencing TFPI2AS1 on cell metastasis, we carried out transwell assays in A549 cells after transfection of TFPI2AS1-specific siRNA. The number of migrated was clearly augmented for TFPI2AS1 knockdown cells (Figure 5A), indicating that knockdown of TFPI2AS1 promotes the migration capacity of NSCLC cells.
Figure 5.
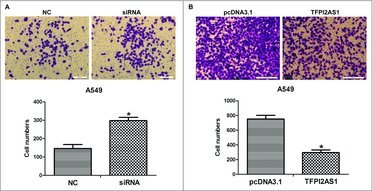
TFPI2AS1 knockdown or overexpression modulates cell migration in A549 cells. (A) A549 cells were transfected with control siRNA or TFPI2AS1 siRNA for 48 h. Representative images of the migration transwell assay; the number of migrated cells was randomly measured by photograph at 200 × magnification in five different views each chamber. (B) A549 cells were transfected with pcDNA3.1/TFPI2AS1-full for 48 h migrated cells was randomly measured by photograph at 200 × magnification in five different views each chamber. *P < 0.05 vs control group. Data are defined as mean ± SD of independent experiments.
We also explored the effect of TFPI2AS1 overexpression on cell metastasis of A549 cells. The result showed that the number of migrated A549 cells with TFPI2AS1 overexpression was significantly decreased (Figure 5B), indicating that the overexpression of TFPI2AS1 inhibits the migration capacity of A549 cells.
TFPI2AS1 modestly inhibits AKT signaling in A549 cells
To explore the influence of TFPI2AS1 on the AKT signaling pathway, which has been implicated in the proliferation and migration of cancer cells, we measured the expression levels of phosphorylated AKT and total AKT by Western blotting. As shown in Figure 6A, the ratio of phosphorylated AKT/total AKT in A549 cells was increased after TFPI2AS1 downregulation. Furthermore, the ratio of phosphorylated AKT/total AKT in A549 cells was modestly decreased after TFPI2AS1 overexpression (Figure 6B). These observations suggested that TFPI2AS1 mediates a slight inhibition of AKT activation, which could contribute to its ability to suppress proliferation and migration of A549 cancer cells.
Figure 6.
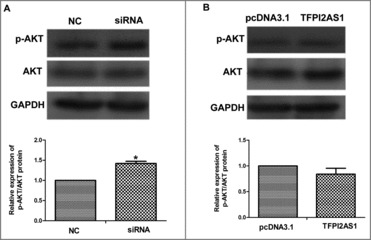
TFPI2AS1 mildly attenuates activation of the AKT signaling pathway in A549 cells. After transfected with control siRNAs or TFPI2AS1 siRNA (A) or with pcDNA3.1/TFPI2AS1-full (B) for 48 h, the protein level of p-AKT and AKT in A549 cells was examined by Western blotting analysis. GAPDH protein was used as endogenous normalize. *P < 0.05 vs control group. Data are expressed as the mean ± SD of the experiments performed in triplicate.
TFPI2AS1 expression is positively corrected with TFPI2 expression
Antisense oligonucleotides often serve to regulate their complementary sense mRNAs. Therefore, we examined the possibility that TFPI2 may be a downstream effector that mediates the function of TFPI2AS1 on the cell proliferation and migration of NSCLC cells in a feedback loop. To this end, we determined the impact of TFPI2AS1 knockdown or overexpression on the levels of TFPI2 mRNA and protein expression. Our results showed that downregulation of TFPI2AS1 obviously decreased the levels of TFPI2 mRNA and protein in A549 cells, and overexpression of TFPI2AS1 obviously increased the levels of TFPI2 mRNA and protein in A549 cells (Figure 7). These results indicate that TFPI2 expression is positively correlated with TFPI2AS1 expression.
Figure 7.
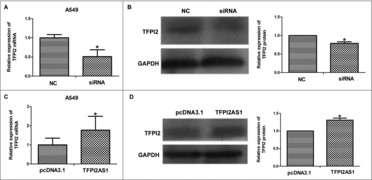
TFPI2AS1 is positively associated with the expression level of TFPI2 in A549 cells. A549 cells were transfected with siRNA or pcDNA3.1/TFPI2AS1-full. TFPI2 mRNA level was measured by qRT-PCR assay (A and C). TFPI2 protein level was determined by Western blotting assay (B and D). GAPDH was regarded as endogenous normalize. *P < 0.05 vs control group. Data are expressed as the mean ± SD of the experiments performed in triplicate.
Discussion
In the present study, we found that a novel lncRNA, TFPI2AS1, is upregulated in NSCLC tissues. Downregulation of TFPI2AS1 by siRNA transfection promoted the growth of A549 cells by facilitating the G1/S transition, while overexpression of TFPI2AS1 by pcDNA3.1-TFPI2AS1 had the opposite effect. We also observed effects of modulating TFPI2AS1 expression on migration and AKT activation. Silencing of TFPI2AS1 caused a decrease in the expression of the tumor suppressor TFPI2, whereas overexpression of TFPI2AS1 increased TFPI2 expression. Collectively, these findings indicate that the effects of TFPI2AS1 on cell proliferation and migration of A549 cells may be achieved through targeting of TFPI2 expression.
Lung cancer is the primary cause of cancer-associated mortality around the world, accounting for more than 25% of the total deaths caused by cancer23. It is a persistent challenge to develop effective therapeutics for treating lung cancer, which is characterized by shared features of other cancers, including unlimited proliferation, metastasis and invasion, and resistance to apoptosis24. Therefore, it is of great importance to find novel molecular biomarkers for early diagnosis, prognosis and potentially, treatment of lung cancer.
lncRNAs, which are non-coding transcripts with a length of greater than 200 nt and ranging up to 100 kilobases25, are well documented for their involvement in a variety of cellular processes, including proliferation26, apoptosis27 cell cycle regulation28, survival29, and migration.6,30 Mounting studies have demonstrated that aberrant lncRNAs have critical roles in many complicated human diseases including cancer. 31-33 Furthermore, dysregulated lncRNAs in cancer often function as oncogenes or tumor suppressor genes. 34,35 However, the mechanisms of biological activity and the related signaling pathways for most lncRNAs are not known. In the current study, we analyzed the expression profiles of lncRNAs in lung cancer tissues to find the potential biological function of lncRNAs in the pathogenesis of lung cancer. The resulting group of differentially expressed lncRNAs included TFPI2AS1. Bioinformatics analysis further showed that TFPI2AS1 is an antisense lncRNA of TFPI2 gene, suggesting that TFPI2 gene may be a target gene of TFPI2AS1. TFPI2 is a Kunitz-type serine protease inhibitor that has been reported to have inhibitory activity against tumor growth and metastasis.19,36 The results of microarray analysis showed its upregulation in NSCLC tumor compared to normal lung tissues. These findings suggested that TFPI2AS1 may have a potential role in the development of lung cancer. Thus, we focused on the role and the possible mechanism of TFPI2AS1 in NSCLC.
To understand the biological action of TFPI2AS1 in NSCLC cell lines, we established siRNA-mediated TFPI2AS1 knockdown and TFPI2AS1 overexpression induced by pcDNA3.1/TFPI2AS1 expression vector. Our results show that downregulation of TFPI2AS1 by siRNA transfection promoted cell proliferation and migration in A549 cells, while upregulation had the opposite effects. Loss of control of the cell cycle is a common mechanism of tumorigenesis, and dysregulation of cell cycle progression plays a key role in the development of cancer.37 We further explored the effect of TFPI2AS1 on the cell cycle in A549 cells and determined that TFPI2AS1 downregulation promotes cell cycle progression, and TFPI2AS1 upregulation arrests cell cycle progression at G1.It has been reported that CyclinD1 and CDK2 play regulatory roles in the G1 phase of cell cycle.20,21 Thus, we further investigated the impact of TFPI2AS1 on the expression of CyclinD1 and CDK2 protein. Our results demonstrate that TFPI2AS1 knockdown induces downregulation of both CyclinD1 and CDK2 protein, whereas TFPI2AS1 overexpression exerts the opposite effects. These data suggest that the TFPI2AS1-arrested G1/S transition may contribute to the inhibitory effects of TFPI2AS1 against lung cancer cell growth.
To unravel the possible pathway involved in the mediatory role of TFPI2AS1 on A549 cell proliferation and metastasis, we measured the expression levels of phosphorylated AKT and total AKT by Western blotting. The PI3K-AKT pathway is known to play a significant role in cancer progression, cell growth, proliferation, invasion and metastasis of tumor cells.38,39 Our results show that TFPI2AS1 knockdown dramatically enhances the level of phosphorylated AKT, while TFPI2AS1 upregulation slightly decreases the level of phosphorylated AKT. Taken together, these findings suggest that the role of TFPI2AS1 on cell proliferation and metastasis of A549 cells may be mediated, at least in part, through the AKT signaling pathway.
Tissue factor pathway inhibitor-2 (TFPI-2) is secreted within the extracellular matrix and is downregulated in most aggressive tumors including non-small cell lung cancer (NSCLC).40,41 To unravel whether the possible mechanism underlying the impact of TFPI2AS1 on A549 cell proliferation and metastasis involves modulation of TFPI2, we assessed the effects of TFPI2AS knockdown and overexpression on the TFPI2 expression. Knockdown of TFPI2AS1 decreased levels of TFPI2 mRNA and protein, while TFPI2AS1 overexpression had the opposite effects. These observations indicate that TFPI2AS1 positively regulates TFPI2; this positive regulation of antisense lncRNA on its target gene was previously reported by Zhang CL et al..42 and Liu W et al.43 Taken together, TFPI2AS1 inhibits the proliferation and migration of NSCLC cells by regulating TFPI2 expression. However, the precise mechanism of TFPI2AS1 in modulating TFPI2 is unclear.
Collectively, in this study, a series of in vitro assays demonstrated that TFPI2AS1 acted as tumor suppressor genes in NSCLC cells. Additionally, these findings are surprising in that its upregulation in lung cancer tissues in our primal findings, which usually suggest the oncogene specificity of TFPI2AS1 in the development of lung cancer. We speculate that the increases in TFPI2AS1 in NSCLC tissues are analogous to the role of insulin on blood sugar. When the level of blood sugar is elevated, insulin release from beta cells in the Islets of Langerhans in the pancreas is increased, resulting in a decrease in the levels of blood sugar.44 Thus, our results might be explained by a paradigm in which increased numbers of lung cancer cells induce an increase in TFPI2AS1 to modulate the growth of lung cancer cells, whereas the function of oncogenes is far stronger than the impact of TFPI2AS1 on lung cancer cells. There is another explanation that the expression of TFPI2AS1 might be associated with the stage of tumors and thus TFPI2AS1 upregulation in lung cancer tissues cannot simply suggest its oncogene specificity. This is a limitation of our study that did not the study the relation of TFPI2AS1 expression levels with stage of lung tumors. Another limitation is that the role of TFPI2AS1 in lung cancer should be verified in animal models.
In conclusion, we identified TFPI2AS1 as an upregulated lncRNA in NSCLC tissues and determined that downregulation of TFPI2AS1 increases cell proliferation and metastasis of A549 cells, whereas upregulation of TFPI2AS1 inhibits cell proliferation and metastasis of A549 cells. We also provided evidence demonstrating the probable association between lncRNA TFPI2AS1 and TFPI2. According to these findings, we propose that TFPI2AS1 and TFPI2 may provide a novel direction for future research of NSCLC. More elaborated studies are needed for determining the mechanism of TFPI2AS1-modulated TFPI2 gene transcription as well as the potential function of their interaction in the development of lung cancer.
Materials and methods
Patients and specimens
Lung cancer samples and matched adjacent normal lung tissues samples were obtained from patients at the Affiliated Hospital of Guangdong Medical University, China. None of the patients were subjected to preoperative chemotherapy and/or radiotherapy and all patients were diagnosed with NSCLC according to histopathology. Of the samples, three were used for lncRNAs microarray analysis. All obtained samples were immediately snap-frozen in liquid nitrogen after resection. This work was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical College, and all patients provided written informed consent prior to sample collection.
Reagents
Oligonucleotides, including negative control siRNA and TFPI2AS1 siRNA were synthesized by GenePharma (Shanghai, China). 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) was obtained from Sigma (St Louis, USA). Primary antibodies against p-AKT, AKT and Brdu (5-bromo-2-deoxyuridine) were purchased from Cell Signaling Technology, Inc (Beverly, MA, USA). anti-cyclin D1 antibody and anti-cyclin-dependent kinases 2 (CDK2) antibody were purchased from Invitrogen (California, USA). Anti-TFPI2 antibody was obtained from Sangon Biotech (Shanghai, China). Anti- glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was purchased from Abgent (San Diego, USA). Annexin V-FITC/ propidium iodide (PI) apoptosis test kit was obtained from MultiSciences Biotech (Hangzhou, China).
Cell culture and transfection
Human lung cancer-derived cell line A549 was obtained from the Shanghai Cell Institute Country Cell Bank (Shanghai, China). Cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (Gibco/BRL, MD) and grown at 37 °C in a humidified atmosphere containing 5% CO2. Negative control siRNA, TFPI2AS1 siRNA, pcDNA3.1 vector, TFPI2AS1 overexpression vector were transfected to cells using Lipofectamine 2000 Reagent (Invitrogen, California, USA) according to the manufacturer's standard protocol.
Microarray analysis
Three pairs of matched samples were used for microarray analysis. RNA was purified from total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was transcribed into fluorescent cRNA along the entire length of the transcripts without 3’ bias utilizing a random priming method. The labeled cRNAs were hybridized onto the Human lncRNA Array v2.0 (8 × 60 K, Arraystar)
RNA extraction and quantitative real-time PCR
Total RNA was extracted from cultured cells according to the manufacturer's protocol using Trizol reagent (Invitrogen, California, USA). 2 μg of total RNA were subsequently reverse transcribed using a PrimeScriptTM RT reagent kit with gDNA Eraser (Takara, Otsu, Shiga, Japan) in a total volume of 20 μL according to the manufacturer's protocol. Real-time quantitative polymerase chain reaction (qRT-PCR) was then conducted using SYBR Premix Ex TaqTM II PCR Kit (Takara, Otsu, Shiga, Japan) following the manufacturer's instrument. Results were normalized to the content of GAPDH. All experiments were performed in triplicate. The gene expression fold-change values are represented using the2−ΔΔCt method. Primers used in qRT-PCR were as follows: TFPI2AS1 5’-CCAGCAATCGGAAGTGAGC-3’ (forward) and 5’-TTCAAGAGGTGGATTCCGGC-3’ (reverse); TFPI2 5’-TCCATCATTTTGCTACAGTCCA-3’ (forward) and 5’-ATTGTCATTCCCTCCACAGC-3’(reverse). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 5’-GACCTGACCTGCCGTCTA-3’(forward) and 5’-AGGAGTGGGTGTCGCTGT-3’(reverse).
Cell proliferation assay
Cell proliferation was measured using MTT assay. In brief, 5000 cells per well were plated into 96-well plates. MTT stock solution (0.5 mg/ml in PBS) was added to each well, and the cells were cultured for an additional 4 h. Dimethyl sulfoxide (DMSO) was added to dissolve the resulting crystals before reading the absorbance at a wavelength of 490 nm in a plate reader. Three independent experiments were performed.
Brdu immunofluorescence assay
Cells were plated on laser confocal dishes. Brdu Sigma (St Louis, USA) stock solution (10 mg/ml) dissolved in PBS was added at 1:1000 to each dish. After 1 h incubation, the cells were washed with PBS and subsequently fixed with 4% paraformaldehyde. 20 min later, the samples were denatured with HCl at 37°C for 15 min and incubated with blocking solution for 1 h. The cells were then incubated with a primary mouse antibody against Brdu (1:200) at 4°C overnight, followed by treatment with Cy5-conjugated secondary antibody for 2 h. After staining with 4',6-diamidino-2-phenylindole (DAPI) and washing with PBS, the dishes were incubated with fluorescence quenching agent and visualized using confocal microscopy. Eight microscopic fields were randomly selected for calculating Brdu.
Cell cycle analysis
Cells were plated onto a 6-well plate at a density of 5 × 105 cells/well and grown for 24 h. The cells were then starved with serum-free culture medium for 24 h to synchronize them at the G1/S boundary, followed by transfection. 48 h later, the cells were collected, washed twice with ice‑cold PBS, and fixed with 70% ice-cold ethanol at 4°C overnight. After rehydration in PBS for 15 min, the cells were stained for 30 min in the dark with PI solution and then analyzed by flow cytometry (BD Biosciences, USA).
Colony formation assay
500 cells were plated in 6-well plates in triplicate. After 14-days of incubation, the cells were washed with PBS, fixed with 4% paraformaldehyde and subsequently stained using 0.1% crystal violet Sigma (St Louis, USA). The colony numbers were counted, and colonies consisting of more than 50 cells were defined as effective.
Transwell assay
Cell migration was evaluated using 8-μm pore size polycarbonate filter transwell inserts on the upper chamber with non-coated membranes. After transfection for 24 h, the cells were digested with trypsin and then 5 × 104 transfected cells were diluted with serum-free culture medium and plated onto the upper chamber in 200 μL RPMI 1640 without fetal bovine serum. 600-μL of 10% fetal bovine serum medium was added to the bottom chamber. After incubating for 18 h, the non-migrated cells in the upper chamber were removed with a cotton swab and the migrated cells in the lower chamber were fixed with 4% paraformaldehyde. All cells were stained using 0.1% crystal violet Sigma (St Louis, USA), and the cell numbers were counted in five random views of each chamber under a phase contrast microscope. The migrated cells were determined by photographing at 200 × magnification with an Olympus Microscope. Each experiment was repeated three times.
Western blotting analysis
Treated cells were collected and lysed in Radio-Immunoprecipitation Assay (RIPA) buffer containing 1 mM Phenylmethanesulfonyl fluoride (PMSF). After centrifugation at 12000 rpm for 15 min, supernatants were collected. Then the total protein concentration of each sample was determined by Bicinchoninic acid (BCA) protein assay according to the manufacturer's protocols. Equal amounts of cell extracts were dissolved in SDS-PAGE loading buffer and boiled at 100°C for 5 min. The samples were separated on 12% SDS-PAGE gels and then transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking with 5% nonfat milk, the membranes were incubated with rabbit anti-p-AKT, rabbit anti-AKT, rabbit anti-TFPI2, rabbit anti-cyclin D1 and rabbit anti- CDK2 or mouse anti-GAPDH antibodies with gentle agitation at 4°C overnight. After washing with Tris-buffered Saline with Tween 20 (TBST) three times, the membranes were incubated with secondary antibodies (1:5000) for 60 min. The signals of the target proteins were visualized by enhanced chemiluminescence (Pierce), with GAPDH as a control.
Statistical analysis
Statistical analyses were performed using SPSS 18.0. Data are presented as the mean ± SD. The Student's t-test was used for analyzing statistical differences between groups. Differences were considered significant at P < 0.05.
Funding Statement
This work was supported by the Natural Science Foundation of China (grant No: NSFC81570062, 81600049 and NSFC81172615); Guangdong Natural Science Foundation (grant No:2016A030313681); Guangdong medical University scientific research fund (grant No: M2016022).
Disclosure of interest
The authors report no conflict of interest.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2014;64:9-29. doi: 10.3322/caac.21208. PMID:24399786 [DOI] [PubMed] [Google Scholar]
- [2].Seve P, Reiman T, Dumontet C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67:136-43. doi: 10.1016/j.lungcan.2009.09.007. PMID:19828208 [DOI] [PubMed] [Google Scholar]
- [3].Raungrut P, Wongkotsila A, Lirdprapamongkol K, Svasti J, Geater SL, Phukaoloun M, Suwiwat S, Thongsuksai P. Prognostic significance of 14-3-3gamma overexpression in advanced non-small cell lung cancer. Asian Pacific journal of cancer prevention : APJCP. 2014;15:3513-8. doi: 10.7314/APJCP.2014.15.8.3513. PMID:24870749 [DOI] [PubMed] [Google Scholar]
- [4].Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA biology. 2012;9:703-19. doi: 10.4161/rna.20481. PMID:22664915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kazemzadeh M, Safaralizadeh R, Orang AV. LncRNAs: emerging players in gene regulation and disease pathogenesis. J Genet. 2015;94:771-84. doi: 10.1007/s12041-015-0561-6. PMID:26690535 [DOI] [PubMed] [Google Scholar]
- [6].Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. . Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. doi: 10.1038/nature08975. PMID:20393566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-61. doi: 10.1038/nm.3981. PMID:26540387 [DOI] [PubMed] [Google Scholar]
- [8].Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195-208. doi: 10.1038/onc.2008.373. PMID:18836484 [DOI] [PubMed] [Google Scholar]
- [9].Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. PMID:20124551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PloS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. PMID:25089627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. PMID:21489289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Han L, Kong R, Yin DD, Zhang EB, Xu TP, De W Shu YQ. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol. 2013;30:694. doi: 10.1007/s12032-013-0694-5. PMID:23979857 [DOI] [PubMed] [Google Scholar]
- [13].Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PloS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. PMID:23741487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wei MM, Zhou YC, Wen ZS, Zhou B, Huang YC, Wang GZ, Zhao XC, Pan HL, Qu LW, Zhang J, et al. . Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget. 2016;7:59556-71. doi: 10.18632/oncotarget.10006. PMID:27322209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li D, Song L, Wen Z, Li X, Jie J, Wang Y, Peng L. Strong evidence for LncRNA ZNRD1-AS1, and its functional Cis- eQTL locus contributing more to the susceptibility of lung cancer. Oncotarget. 2016;7:35813-7. doi: 10.18632/oncotarget.8411. PMID:27166266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X, Wu G, Lv T, Song Y. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558-75. doi: 10.18632/oncotarget.8338. PMID:27027352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lavergne M, Jourdan ML, Blechet C, Guyetant S, Pape AL, Heuze-Vourc'h N, Courty Y, Lerondel S, Sobilo J, Iochmann S, et al. . Beneficial role of overexpression of TFPI-2 on tumour progression in human small cell lung cancer. FEBS Open Bio. 2013;3:291-301. doi: 10.1016/j.fob.2013.06.004. PMID:23905012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang S, Xiao X, Zhou X, Huang T, Du C, Yu N, Mo Y, Lin L, Zhang J, Ma N, et al. . TFPI-2 is a putative tumor suppressor gene frequently inactivated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer. 2010;10:617. doi: 10.1186/1471-2407-10-617. PMID:21062455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sierko E, Wojtukiewicz MZ, Kisiel W. The role of tissue factor pathway inhibitor-2 in cancer biology. Semin Throm Hemost. 2007;33:653-9. doi: 10.1055/s-2007-991532. PMID:18000791 [DOI] [PubMed] [Google Scholar]
- [20].Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812-21. doi: 10.1101/gad.7.5.812. PMID:8491378 [DOI] [PubMed] [Google Scholar]
- [21].Tsai LH, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593-602. PMID:8502482 [PubMed] [Google Scholar]
- [22].Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. . Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580-4. doi: 10.1126/science.1228522. PMID:23372014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca A Cancer Journal for Clinicians. 2016;66:10-29. [DOI] [PubMed] [Google Scholar]
- [24].Fu X, Li H, Liu C, Hu B, Li T, Wang Y. Long noncoding RNA AK126698 inhibits proliferation and migration of non-small cell lung cancer cells by targeting Frizzled-8 and suppressing Wnt/beta-catenin signaling pathway. OncoTargets Ther. 2016;9:3815-27. doi: 10.2147/OTT.S100633. PMID:27445486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Gen. 2009;10:155-9. doi: 10.1038/nrg2521. PMID:19188922 [DOI] [PubMed] [Google Scholar]
- [26].Zhao X, Liu M, Zhang J, Zhang R, Zhang Q, Tan X. Long noncoding RNA CPS1-IT1 suppresses cell proliferation and metastasis in human lung cancer. Oncol Res. 2017;25:373-380. doi: 10.3727/096504016X14741486659473. PMID:27662619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- [28].Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18:111-23. doi: 10.1261/rna.029454.111. PMID:22128341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. . Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621-9. doi: 10.1038/ng.848. PMID:21642992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jimenez B, Tiana M, Del Peso L. lnc RNAs, hypoxia and metastasis. Oncoscience 2015;2:795-6. PMID:26682249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-61. doi: 10.1038/nm.3981. PMID:26540387 [DOI] [PubMed] [Google Scholar]
- [32].Niland CN, Merry CR, Khalil AM. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Front Genet. 2012;3:25. doi: 10.3389/fgene.2012.00025. PMID:22375145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:155-65. doi: 10.1038/modpathol.2012.160. PMID:22996375 [DOI] [PubMed] [Google Scholar]
- [35].Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407. doi: 10.1158/2159-8290.CD-11-0209. PMID:22096659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chand HS, Du X, Ma D, Inzunza HD, Kamei S, Foster D, Brodie S, Kisiel W. The effect of human tissue factor pathway inhibitor-2 on the growth and metastasis of fibrosarcoma tumors in athymic mice. Blood. 2004;103:1069-77. doi: 10.1182/blood-2003-06-1930. PMID:14525759 [DOI] [PubMed] [Google Scholar]
- [37].Visconti R, Della Monica R, Grieco D. Cell cycle checkpoint in cancer: a therapeutically targetable double-edged sword. J Exp Clin Cancer Res. 2016;35:153. doi: 10.1186/s13046-016-0433-9. PMID:27670139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Toker A, Yoeli-Lerner M. Akt signaling and cancer: surviving but not moving on. Cancer Res. 2006;66:3963-6. doi: 10.1158/0008-5472.CAN-06-0743. PMID:16618711 [DOI] [PubMed] [Google Scholar]
- [39].Faes S, Dormond O. PI3K and AKT: Unfaithful Partners in Cancer. Int J Mol Sci. 2015;16:21138-52. doi: 10.3390/ijms160921138. PMID:26404259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rollin J, Iochmann S, Blechet C, Hube F, Regina S, Guyetant S, Lemarie E, Reverdiau P, Gruel Y. Expression and methylation status of tissue factor pathway inhibitor-2 gene in non-small-cell lung cancer. Br J Cancer. 2005;92:775-83. doi: 10.1038/sj.bjc.6602298. PMID:15685245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wu D, Xiong L, Wu S, Jiang M, Lian G, Wang M. TFPI-2 methylation predicts poor prognosis in non-small cell lung cancer. Lung Cancer. 2012;76:106-11. doi: 10.1016/j.lungcan.2011.09.005. PMID:21983100 [DOI] [PubMed] [Google Scholar]
- [42].Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66-75. doi: 10.1016/j.canlet.2017.03.018. PMID:28323030 [DOI] [PubMed] [Google Scholar]
- [43].Liu W, Ma J, Cheng Y, Zhang H, Luo W, Zhang H. HMMR antisense RNA 1, a novel long noncoding RNA, regulates the progression of basal-like breast cancer cells. Breast Cancer (Dove Med Press). 2016;8:223-9. PMID:27920576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Koeslag JH, Saunders PT, Terblanche E. A reappraisal of the blood glucose homeostat which comprehensively explains the type 2 diabetes mellitus-syndrome X complex. J Physiol. 2003;549:333-46. doi: 10.1113/jphysiol.2002.037895. PMID:12717005 [DOI] [PMC free article] [PubMed] [Google Scholar]


