ABSTRACT
Macroautophagy/autophagy is a starvation and stress-induced catabolic process critical for cellular homeostasis and adaptation. Several Atg proteins are involved in the formation of the autophagosome and subsequent degradation of cytoplasmic components, a process termed autophagy flux. Additionally, the expression of several Atg proteins, in particular Atg8, is modulated transcriptionally, yet the regulatory mechanisms involved remain poorly understood. Here we demonstrate that the AGC kinase Ypk1, target of the rapamycin-insensitive TORC2 signaling pathway, controls ATG8 expression by repressing the heterodimeric Zinc-finger transcription factors Msn2 and Msn4. We find that Msn2 and Msn4 promote ATG8 expression downstream of the histone deacetylase complex (HDAC) subunit Ume6, a previously identified negative regulator of ATG8 expression. Moreover, we demonstrate that TORC2-Ypk1 signaling is functionally linked to distinct mitochondrial respiratory complexes. Surprisingly, we find that autophagy flux during amino acid starvation is also dependent upon Msn2-Msn4 activity, revealing a broad role for these transcription factors in the autophagy response.
KEYWORDS: autophagy, electron transport chain (ETC) complexes, gene expression, mitochondrial respiration, signal transduction, TOR signaling, Ypk1
Introduction
Autophagy is a catabolic cellular process that functions as a quality control mechanism that promotes homeostasis and adaptation. It does so through the formation of a double-membrane compartment, the phagophore, that captures cytoplasmic cargo, including damaged proteins and organelles, ultimately maturing into an autophagosome, which fuses with the cellular lytic compartment (vacuole or lysosome) for degradation and recycling.1-4 In several different cell types, autophagy often occurs selectively and at a basal rate, but can increase markedly upon nutrient starvation or other forms of metabolic stress, an event conventionally referred to as macroautophagy.1,5,6 Autophagy is involved in a myriad of cellular processes and its dysregulation can lead to metabolic and degenerative diseases including, but not limited to, cancer, osteoarthritis, Alzheimer and Parkinson diseases, as well as several cardiomyopathies.7-9
Many studies over the years have focused on defining the molecular mechanisms and machinery involved in autophagosomal biogenesis and subsequent degradation of cargo, a process commonly referred to as autophagy flux.2,10-13 Indeed, pioneering studies in the yeast S. cerevisiae led to the discovery of more than 40 autophagy genes (ATG genes) essential for this process, many of which are conserved in higher eukaryotes.11,12,14-17 Recently, it has also become clear that the transcription of a select number of autophagy-related genes is coupled to autophagy induction and that regulated expression of these genes is important for the proper control of autophagy.1,18-20 By comparison to autophagy flux, the molecular mechanism(s) regulating the transcriptional induction of these ATG genes remain(s) relatively poorly uncharacterized.
Of these transcriptionally regulated ATG genes, ATG8 is unique in that it is one of the few genes encoding an Atg protein that is physically incorporated into the phagophore membrane.10,13,21 Accordingly, Atg8 is suggested to regulate both autophagosome size as well as the binding of selective cargo receptor proteins.4,10,22-24 Our current understanding of ATG8 transcriptional regulation is defined primarily by recent studies showing that it is negatively regulated by Ume6, a subunit of the Rpd3 histone deacetylase complex (HDAC) (Fig. 1A).25,26 Significantly, the mechanism by which ATG8 expression is positively controlled downstream of Ume6, including the transcription factor(s) subject to Ume6 regulation, remains unknown.
Figure 1.
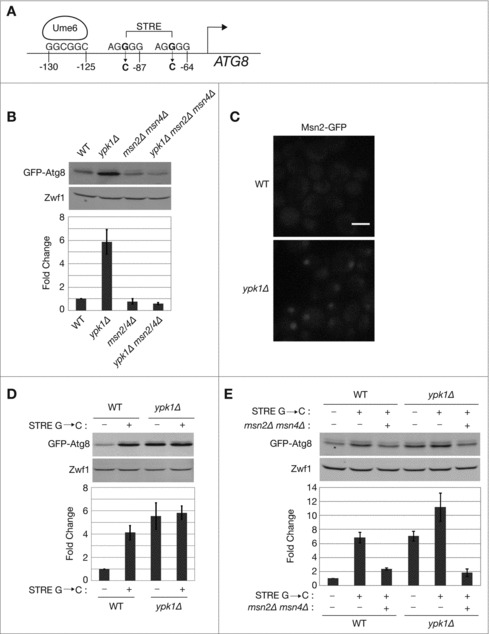
TORC2-Ypk1 signaling regulates ATG8 expression via the heterodimeric zinc-finger transcription factor Msn2-Msn4. (A) Schematic representation of the ATG8 promoter region, indicating the identified recognition site for the HDAC subunit Ume625 as well as potential stress response elements (STRE) as determined by YEASTRACT.30 Also indicated are positions of G to C mutations introduced into the STRE sites. (B) Indicated strains carrying pRS416 prATG8-GFP-ATG8 were grown to log phase in SCD medium without uracil. GFP-Atg8 protein was visualized via western blot analysis as described in Materials and Methods. GFP-Atg8 protein expression was normalized to the Zwf1 loading control and quantification represents Atg8 protein levels relative to WT, presented as the mean ± standard deviation (SD) of at least 3 independent experiments. (C) Indicated strains expressing endogenously tagged MSN2-GFP were visualized by fluorescence microscopy as described in Materials and Methods. Scale bar: 5 μm. ((D)and E) Indicated strains carrying pRS416 prATG8-GFP-ATG8 or pRS416 prATG8-GFP-ATG8 possessing STRE mutations depicted in (A) were grown to log phase in SCD medium without uracil. GFP-Atg8 protein levels were detected by western blot and analyzed as described in (B).
Our understanding of the regulation of autophagy has also expanded in recent years with the discovery that different environmental stresses initiate autophagy through distinct signaling pathways.5,20,27 For example, we have demonstrated that the TORC2 signaling pathway, via its target kinase Ypk1, functions as a positive regulator of autophagy flux during amino acid-limited growth conditions.27,28 In this context, TORC2-Ypk1 signaling promotes autophagy flux by negatively regulating the calcium-regulated phosphatase calcineurin and promoting the general amino acid control (GAAC) response. More recently, we have demonstrated that aberrant mitochondrial respiration, as a result of decreased TORC2-Ypk1 signaling, activates calcineurin and inhibits autophagy.27-29
In addition to its role as a positive regulator of autophagy flux, TORC2-Ypk1 also negatively regulates ATG8 expression.29 In particular, we have demonstrated that in both nutrient-rich and starvation conditions, loss of Ypk1 activity (in ypk1Δ cells) results in increased levels of Atg8 protein. This increase is not due to a block in autophagy flux as the expression of a GFP reporter gene fused to the ATG8 promoter is also elevated in Ypk1-deficient cells.27 Moreover, this regulation is independent from calcineurin, because increased ATG8 expression is still observed in ypk1Δ cells following deletion of CNB1, encoding the calcineurin B regulatory subunit.27,28 Similarly, no change in ATG8 expression is observed in ypk1Δ cells following deletion of CRZ1, encoding a stress-responsive transcription factor that functions downstream of calcineurin. Together, these findings point to a unique regulation for ATG8 induction downstream of TORC2-Ypk1 signaling that is independent from calcineurin.27
In this study, we demonstrate that the stress-responsive transcription factor complex composed of proteins Msn2 and Msn4 is a positive regulator of ATG8 expression. We describe functional interactions between TORC2-Ypk1 signaling and mitochondrial respiration that contribute to ATG8 transcriptional regulation by Msn2-Msn4. Finally, we demonstrate that Msn2 and Msn4 are also involved in regulating mitochondrial respiration and autophagy flux in collaboration with TORC2-Ypk1 signaling following amino acid starvation, highlighting broad roles for TORC2 signaling and Msn2-Msn4 within this regulatory pathway.
Results
TORC2-Ypk1 signaling regulates ATG8 expression via the heterodimeric zinc-finger transcription factor Msn2-Msn4
Inhibition of TORC2 or Ypk1 results in increased basal expression of ATG8 that is distinct from defects observed in autophagy flux during amino acid starvation.27 Inspection of the promoter region of the ATG8 gene revealed the presence of 2 stress response elements (STREs) (Fig. 1A), predicted to be targets of the Msn2-Msn4 heterodimeric zinc-finger transcription factor complex.30,31 Significantly, several genes that are negatively regulated by TORC2-Ypk1 signaling have identifiable STRE motifs within their promoters.32 Therefore, we tested whether ATG8 induction in ypk1Δ cells requires Msn2-Msn4 activity by deleting MSN2 and MSN4 within these cells. We observed that elevated GFP-Atg8 expression in ypk1Δ cells was indeed absent in ypk1Δ msn2Δ msn4Δ cells (Fig. 1B). We confirmed that Msn2-Msn4 activity was induced in ypk1Δ cells by examining the intracellular location of GFP-tagged Msn2, which adopted a punctate nuclear localization pattern specifically in ypk1Δ but not in WT cells (Fig. 1C).
We next tested whether Msn2-Msn4 utilize the 2 STRE elements within the ATG8 promoter, by introducing a single G to C base change within each element (Fig. 1A), based upon prior studies demonstrating this single mutation is sufficient to disrupt recognition by the Msn2-Msn4 complex.31 Surprisingly, we observed that a reporter construct harboring these base changes maintained elevated expression of ATG8 in ypk1Δ cells (Fig. 1D, compare lanes 3 and 4). Moreover, these mutations resulted in increased ATG8 expression even when introduced into WT cells alone (Fig. 1D, compare lanes 1 and 2). Importantly, however, increased expression of ATG8 in either WT or ypk1Δ cells required the presence of Msn2-Msn4 (Fig. 1E). From these results we conclude that Msn2-Msn4 is unlikely to bind directly or solely to the STRE elements and control expression of ATG8, at least compared with mechanisms described for other Msn2-Msn4 target genes. Nevertheless, these findings confirm that ATG8 expression is regulated by TORC2-Ypk1 signaling in an Msn2-Msn4-dependent manner.
Msn2-Msn4 link the HDAC subunit Ume6 to ATG8 transcription
Recently, Klionsky and colleagues demonstrated that inhibition of the HDAC subunit Ume6 results in a dramatic increase in ATG8 induction in nutrient-rich conditions.25,26 They demonstrated further that Ume6 is inactivated following nutrient deprivation, in part via phosphorylation by the nutrient-responsive kinase Rim15, which results in increased expression of ATG8.25 However, the transcription factor(s) involved in promoting ATG8 expression under these conditions was not identified.
Given our present results, we tested whether functional interactions exist between Ume6 and Msn2-Msn4, with respect to ATG8 expression. In agreement with prior findings, we observed that deletion of UME6 resulted in significant (∼20-fold) levels of ATG8 induction (Fig. 2A). Remarkably, this expression was completely dependent upon Msn2-Msn4, as ATG8 expression returned to basal levels in ume6Δ msn2Δ msn4Δ cells (Fig. 2A). Moreover, the additive effects of deleting both YPK1 and UME6, with respect to increased ATG8 expression, were also reversed completely when combined with deletions of MSN2 and MSN4 (Fig. 2A). These findings demonstrate an important role for Msn2-Msn4 in the transcriptional control of ATG8 in response to both TORC2-Ypk1 signaling as well as HDAC-regulated expression. Based upon our above analyses of the STRE site mutations, one possibility is that these base changes within the ATG8 promoter alleviate Ume6-dependent repression of ATG8 via an Msn2-Msn4-dependent mechanism.
Figure 2.
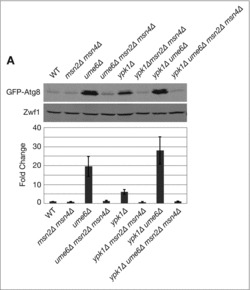
Msn2-Msn4 link the HDAC subunit Ume6 to ATG8 transcription. Indicated strains expressing pRS416 prATG8-GFP-ATG8 were grown to log phase in SCD medium without uracil and western blot analysis was performed as described in Materials and Methods. GFP-Atg8 protein expression was normalized to the Zwf1 loading control and quantification represents Atg8 protein levels relative to WT, presented as the mean ± SD of at least 3 independent experiments.
Mitochondrial respiration links TORC2-Ypk1 and Msn2-Msn4 to ATG8 expression
We have demonstrated recently that TORC2-Ypk1 signaling regulates autophagy flux in part by controlling mitochondrial respiration. In particular, Ypk1 represses the accumulation of mitochondria-derived reactive oxygen species (ROS), which play a key role in activating calcineurin.29 Accordingly, we explored the relationship between TORC2-Ypk1 signaling and mitochondrial respiration with respect to ATG8 induction. We systematically tested the role of individual electron transport chain (ETC) complexes in regulating ATG8 expression in Ypk1-deficient cells by deleting specific nuclear-encoded genes important for the assembly of ETC complex III (cbs1Δ), complex IV (mss51Δ), or complex V (atp10Δ). We observed that deletion of MSS51 or ATP10 significantly reduced ATG8 expression in ypk1Δ cells but had no significant effect on their own (Fig. 3A). By contrast, we found that deletion of CBS1 alone resulted in increased ATG8 expression that was exacerbated in ypk1Δ cbs1Δ cells, demonstrating a unique behavior for complex III in ATG8 expression (Fig. 3A).
Figure 3.
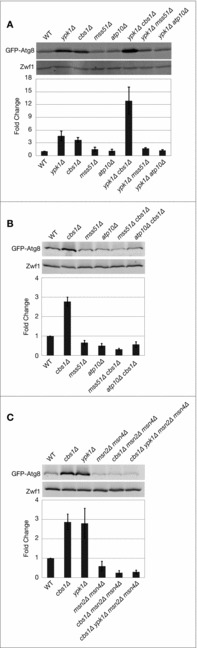
Specific mitochondrial respiratory complexes link TORC2-Ypk1 and Msn2-Msn4 to ATG8 expression. (A-C) Indicated strains expressing pRS416 prATG8-GFP-ATG8 were grown to log phase in SCD medium without uracil. Western blot analysis was performed as described in Materials and Methods. GFP-Atg8 protein expression was normalized to Zwf1 loading control and quantification represents Atg8 protein levels relative to WT, presented as the mean ± SD of at least 3 independent experiments.
To explore these differences further, we performed epistasis analyses by examining ATG8 expression in strains deleted for both CBS1 as well as one of the other genes encoding ETC components, MSS51 or ATP10. We observed that increased expression of ATG8 was abolished in both double mutants, demonstrating that induction of ATG8 in the absence of complex III activity requires functional complexes IV and V (Fig. 3B). Importantly, we observed that ATG8 induction in cbs1Δ cells also required Msn2-Msn4 activity, confirming that perturbation of mitochondrial respiration affects ATG8 via a common regulatory pathway involving Msn2-Msn4 (Fig. 3C). Thus, the status of mitochondrial respiration serves as a branch-point that mediates the impact of TORC2 and Ypk1 on both arms of the autophagy response: transcriptional induction of ATG8 as well as autophagy flux.
Msn2-Msn4 function upstream of mitochondria to regulate ROS and autophagy flux
As ATG8 expression is responsive to both Msn2-Msn4 activity as well as mitochondrial respiration, we next examined the relationship between Msn2-Msn4 and mitochondria in the context of the regulatory scheme we have described recently, wherein autophagy flux is controlled by TORC2-Ypk1 signaling, mitochondrial oxidative stress, and calcineurin.27-29 We have demonstrated previously that ROS accumulation in ypk1Δ cells occurs strictly through mitochondrial-derived sources.29 Accordingly, we tested whether Msn2-Msn4 regulates accumulation of mitochondrial ROS, using the ROS-reactive fluorescent dye H2DCF-DA (DCF).33,34 Here we observed that ROS was absent in ypk1Δ msn2Δ msn4Δ cells (Fig. 4A). These results indicate that in addition to being activated by aberrant mitochondrial respiration, Msn2 and Msn4 also contribute to the accumulation of ROS when TORC2-Ypk1 signaling is impaired.
Figure 4.
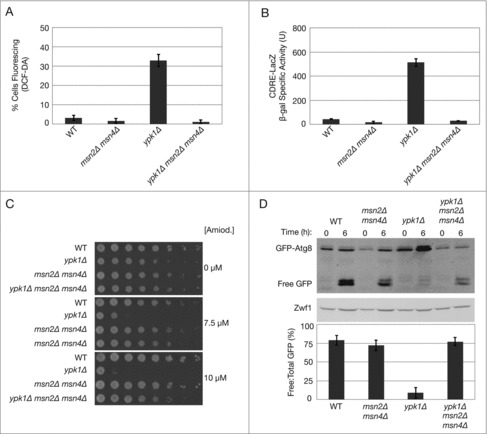
Msn2-Msn4 function upstream of mitochondria to regulate ROS and autophagy flux. (A) Indicated strains were grown to log phase and treated with 10 μM DCF for 30 min at 30°C before imaging by fluorescence microscopy, as described in Materials and Methods. Quantification represents the percentage of DCF-positive fluorescing cells, where a minimum of 250 cells for each strain was counted. (B) Indicated strains carrying plasmid pAMS363 that expresses 2xCDRE:LacZ were grown to log phase in SCD medium without uracil, and β-galactosidase activity was measured as described in Materials and Methods. β-galactosidase activity is represented as units of ONPG (nmol) converted/min/mg of protein. Data are presented as means ± SD of 3 independent experiments. (C) Indicated strains were grown to log phase and serial dilutions were plated onto agar plates containing YPD and the indicated concentrations of amiodarone. Cells were grown for ∼2–3 d at 30°C. (D) Indicated strains carrying pRS416 prATG8-GFP-ATG8 were grown to log phase in SCD medium without uracil medium and transferred to amino acid starvation medium (0.05% yeast extract, 2% dextrose) for 6 h. GFP and GFP-Atg8 protein levels were visualized by western blot analysis as described in Materials and Methods. Quantification of autophagy flux at 6 h of starvation is represented as a percentage of the ratio of free GFP to total GFP (GFP + GFP-Atg8) signal. Data are presented as means ± SD of 3 independent experiments.
We have shown previously that accumulation of mitochondria-derived ROS is essential for activation of calcineurin.29 Thus, based on our above findings we predicted that increased calcineurin activity in ypk1Δ cells might also require Msn2 and Msn4. To test this, we examined calcineurin activity using a calcineurin-dependent response element (CDRE)-driven lacZ reporter in both ypk1Δ as well as ypk1Δ msn2Δ msn4Δ cells, as described previously.35 Indeed, we observed that calcineurin activity was elevated in ypk1Δ cells, as reported previously, but that it returned to basal levels in the triple mutant (Fig. 4B).
As an independent approach for testing the role of Msn2-Msn4 in regulating both ROS and calcineurin, we used the anti-athrymic and antifungal drug amiodarone, which we have shown inhibits growth of ypk1Δ cells in a manner that correlates with activation of calcineurin in the presence of mitochondrial ROS.29,36 Indeed, we used this drug to characterize the importance of the calcium channel regulator Mid1 as a key intermediate for calcineurin activation in the presence of ROS-derived signals, as increased Mid1 activity results in amiodarone hypersensitivity.29,36 Because loss of Msn2-Msn4 activity prevented both accumulation of ROS as well as induction of calcineurin in ypk1Δ cells, we predicted that ypk1Δ msn2Δ msn4Δ cells may possess decreased Mid1 activity, which would be manifest as amiodarone-resistant growth. Indeed, we observed that while ypk1Δ cells were hypersensitive to sublethal concentrations of the drug, the triple mutant displayed significant resistance under these conditions (Fig. 4C). We conclude therefore that Msn2-Msn4 promotes Mid1 and calcineurin activity by facilitating dysfunctional mitochondrial respiration and ROS production in ypk1Δ cells.
Because inhibition of Msn2-Msn4 activity in ypk1Δ cells reduced calcineurin activity, we next tested whether they function in autophagy flux following amino acid starvation. Accordingly, we used western blotting to monitor processing of GFP-Atg8 during autophagy, where free GFP accumulates within the vacuole and autophagy flux is determined as the percentage of free GFP relative to total GFP signal.37 Indeed, we observed that while autophagy flux was severely compromised in ypk1Δ cells, it was restored in ypk1Δ msn2Δ msn4Δ cells to levels comparable to WT (Fig. 4D). Importantly, we observed that overall levels of GFP-Atg8, and hence levels of free GFP, were significantly reduced in the triple mutant, consistent with ATG8 induction also being impaired in the absence of Msn2-Msn4 activity. From these results we conclude that Msn2-Msn4 activity plays a dual role in both the induction of ATG8 expression as well as in autophagy flux following amino acid starvation in ypk1Δ cells.
Discussion
Our key findings described here are summarized in a model presented in Fig. 5. First, we have extended our understanding of the transcriptional regulation of the key autophagy gene ATG8. In particular, we have demonstrated that the stress-inducible transcription factor heterodimer Msn2-Msn4 functions as a positive regulator of ATG8 expression. Importantly, we find that this transcription factor complex interacts functionally with the HDAC subunit Ume6, where increased expression of ATG8 observed in ume6Δ cells requires Msn2-Msn4. In addition, we find that Msn2-Msn4 activity is also linked to TORC2-Ypk1 signaling, where increased ATG8 expression in ypk1Δ cells occurs in an Msn2-Msn4-dependent manner. By promoting autophagosome turnover while in turn limiting autophagy induction, TORC2-Ypk1 signaling plays opposing roles in autophagy that may provide a balanced response to cellular stress. Interestingly, both Msn2-Msn4 as well as Ume6 are controlled by direct phosphorylation by the nutrient-responsive kinase Rim15, suggesting TORC2-Ypk1 signaling may interface with Rim15 regulation (Fig. 4).25,26,38 While Ume6 has been shown to interact directly with the ATG8 promoter, our findings suggest that Msn2-Msn4 may regulate ATG8 indirectly, as mutations in 2 predicted STRE sites within the ATG8 promoter did not abolish Msn2-Msn4-dependent expression of ATG8. The possibility remains, however, that Msn2-Msn4 may interact with Ume6 or bind alternatively to the ATG8 promoter by a previously uncharacterized mechanism.
Figure 5.
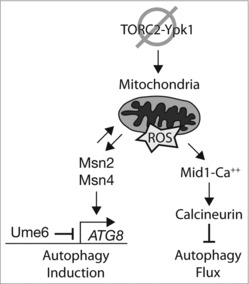
Model for regulation of ATG8 expression and autophagy flux by TORC2-Ypk1, mitochondrial respiration, and Msn2-Msn4 during amino acid starvation. Model depicts the consequences of decreased TORC2-Ypk1 signaling and impaired mitochondrial function with respect to Msn2-Msn4 activity during both autophagy induction as well as autophagy flux. See text for details.
Our findings also demonstrate that, in addition to TORC2-Ypk1 signaling, activation of ATG8 expression by Msn2-Msn4 is responsive to the respiratory state of mitochondria. We recently determined that activation of calcineurin and inhibition of the GAAC response and autophagy flux in Ypk1-deficient cells requires functional mitochondrial respiratory complexes and is associated with accumulation of mitochondrial-derived ROS.29 Similarly, here we demonstrate that Msn2-Msn4-dependent induction of ATG8 in Ypk1-deficient cells is also linked to mitochondrial respiration, in that increased ATG8 expression is abolished when genes encoding components of ETC complexes IV or V are deleted from ypk1Δ cells. Interestingly, we observe that deletion of CBS1, a component of ETC complex III, results in increased ATG8 expression, either alone or in combination with deletion of YPK1. However, this expression is both dependent upon functional ETC complex IV and complex V as well as Msn2-Msn4, indicating these complexes converge on a common mechanism to regulate ATG8 expression.
This complexity in the behavior of different ETC components suggests that each ETC complex is capable of possessing distinct signaling properties that affect autophagy induction. This notion is consistent with recent reports in human cells where diverse neurodegenerative diseases are linked to mutations in different ETC complex subunits and/or their specific assembly factors.39 Relevant to this conclusion, a study in MELAS patient cell lines demonstrates that autophagy degrades ETC complex I, linking defects in specific ETC complexes to the induction of autophagy in this neuronal disease.40 Thus, further investigation into the roles of each ETC complex in mediating autophagy may uncover novel aspects of neuronal pathogenesis and lead to unique avenues for treatment of autophagy-related diseases.
In addition to a positive role in ATG8 induction, we have also found that Msn2-Msn4 activity inhibits autophagy flux during amino acid starvation in Ypk1-deficient cells (Fig. 5). Our analyses indicate that this regulation is additionally connected to mitochondrial respiration, where accumulation of ROS in ypk1Δ cells requires both the activity of mitochondrial ETC complexes as well as Msn2-Msn4. Thus, in the absence of Msn2-Msn4, ROS fail to accumulate in ypk1Δ cells and calcineurin activity is not stimulated by a Mid1-dependent mechanism. Under these conditions, the GAAC response senses amino acid starvation and autophagy is induced. These findings highlight the complexity that exists between Msn2-Msn4 and mitochondria, where reciprocal regulation governs their behavior to control ATG8 expression as well as autophagy flux.
This complexity is reminiscent of the relationship between protein kinase A (PKA) activity and mitochondrial respiration during autophagy, where PKA both regulates the accumulation of ROS as well as being controlled by the respiratory state of mitochondria, according to the precise nutritional state of the cell.41,42 Consistently, a prior study revealed that in addition to the TORC1 downstream substrate Sch9, PKA negatively regulates autophagy flux via a pathway that requires Msn2 and Msn4.18 Given that PKA is an established negative regulator of Msn2-Msn4,18,43,44 it will be essential to understand the precise relationship between PKA and TORC2-Ypk1 signaling in regulating Msn2-Msn4 activity under different autophagy-inducing conditions.
Materials and methods
Yeast strains, media, and plasmids
Strains of yeast used in this study are derivatives of W303α (leu2-3,112; ura3-1; his3-11,15;trp1-1; ade2-1; can1-100) and are listed in Table S1. PCR-based targeted homologous recombination was used to replace complete open reading frames with kanMX6, HIS3MX6, NAT or TRP1 cassettes where indicated.45 Strains expressing endogenously tagged Msn2-GFP were constructed using a GFP:HIS3MX6 PCR product generated from the yeast GFP collection and targeted homologous recombination.46 Yeast transformations were performed using lithium acetate as previously reported.47 All strains were grown to log phase (OD600 = approximately 1) in synthetic complete dextrose (SCD) yeast medium (0.8% yeast nitrogen base without amino acids [Becton Dickinson, 291940], 2% [wt:vol] dextrose [Fisher Scientific, D16-3], pH 5.5) supplemented with amino acids as described previously.48 Where indicated, yeast strains contained the described previously plasmid prATG8-GFP-ATG8.37 Mutations in the predicted STRE sites within the promoter of ATG8 were introduced by site-directed mutagenesis of prATG8-GFP-ATG8 using “QuikChange” (Agilent technologies, 200523), according to the manufacturer's instructions.
Whole-cell extraction, antibodies, and western blot analysis
Whole cell protein extracts were prepared from yeast using the NaOH cell lysis method, loaded onto SDS-PAGE gels and transferred to nitrocellulose membrane for western blot analysis.49 Membranes were probed with anti-GFP N86/8 mouse monoclonal antibody (1 μg/mL; UC Davis/NeuroMab Facility, 73–131:RRID AB_10671444) and rabbit anti-Zwf1/G6PDH (1:1,000; Sigma-Aldrich, HPA000247), and visualized using the appropriate secondary antibodies conjugated to IR Dye (1:5,000; LI-COR Biosciences, 926–32210 and 926–68071). All western blot images were quantified using Fiji software.50 Averages are presented with means ± standard deviation (SD) of at least 3 independent experiments.
Fluorescence microscopy
Fluorescence microscopy was performed using a Nikon E600 fluorescence microscope and an Orca ER charge-coupled device camera (Hamamatsu) controlled by Micro Manager 1.2 ImageJ software. Strains were grown to log phase (OD600 = approximately 1) in SCD medium supplemented with amino acids as described above. Image capture and processing were done using Fiji and Photoshop (Adobe). For Msn2-GFP microscopy, precautions were taken to prevent Msn2 from becoming activated by aberrant exposure to blue light, specifically, by using proper UV filters and fast exposure times and proper controls, as outlined previously.51 Visualization of cellular ROS via fluorescence microscopy was performed using log phase growing cells in SCD complete medium that had been treated with 10 μM 2′,7′-dichlorofluorescin diacetate (DCFDA; Cayman Chemical, 20656) for 30 min at 30°C.
β-Galactosidase activity assay
Exponentially growing cells were incubated at 30°C in SCD medium supplemented with amino acids as required. β-Galactosidase activity was measured at 30°C using the substrate O-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma-Aldrich, 73600) as described previously.52 In brief, 10 OD600 units of a yeast cell pellet was acquired per sample and washed with 1 ml of 4°C sterile water. Cell pellets were resuspended in 250 µl of 4°C breaking buffer (100 mM Tris-HCl, pH 8.0, [Trizma base; Sigma-Aldrich, T1503], 1 mM dithiothreitol [Fisher Scientific, BP 172–25], 20% glycerol [Sigma-Aldrich, G5516]) on ice, and 12.5 µl of 100 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, P7620; diluted in ethanol) was added. Cells were broken and homogenized using glass beads and a mini–bead beater (Biospec Products, Bartlesville, OK, USA) for 1 min in the cold. After homogenization, an additional 250 µl of breaking buffer was added, and extract was removed from glass beads and clarified by centrifugation at 14,000 × g for 15 min at 4°C. Supernatant (cell lysate) was removed from the pellet for subsequent β-galactosidase measurement. To measure β-galactosidase activity, the following were added per sample in a 96-well plate: 14.2 µl of clarified cell lysate, 128.57 µl of z-buffer with fresh β-mercaptoethanol (60 mM Na2HPO4–7H2O, 40 mM NaH2PO4-H2O, 10 mM KCl, 1 mM MgSO4–7H2O, 50 mM β-mercaptoethanol [Sigma-Aldrich, M-3148], pH 7), and 28.57 µl ONPG stock solution (4 mg/ml ONPG in z-buffer). Before adding ONPG, samples were incubated at 30°C to ensure that the reaction occurred at this temperature. The reaction was incubated at 30°C, and time was recorded upon addition of ONPG. When the reaction turned a faint yellow color, the OD of samples was measured at 420 nm using a 96-well plate reader (SpectraMax M5; Molecular Devices). The protein concentration of cell lysate was quantified using a BSA assay with Bradford reagent (Sigma-Aldrich, B6916). β-Galactosidase activity is given in units of nanomoles of ONPG converted per min per mg of protein. Data are presented as means ± SD of 3 independent experiments.
Amiodarone growth assay
Amiodarone (Cayman Chemical, 15213) was prepared as a 50 mM stock in DMSO and then diluted into sterile water as appropriate and overlaid onto solid agar plates containing YPD medium. Exponentially growing yeast cells were diluted and plated onto these agar plates and allowed to grow at 30°C for 2–3 d.
Statistical analyes
To calculate autophagy flux, the ratio of free versus total GFP was determined for 3 independent biologic samples and Microsoft Excel was used to calculate and plot the mean and standard deviation. For CDRE-LacZ reporter assays, the mean of 3 independent biologic samples was similarly calculated and plotted using Excel.
Supplementary Material
Funding Statement
This work was supported by National Institutes of Health Grant GM086387 (To T. P.) and T-32 Training Grant in Molecular and Cellular Biology (to A. V.).
Abbreviations
- ATG
autophagy related
- CDRE
calcineurin-dependent response element
- ETC
electron transport chain
- GFP
green fluorescent protein
- HDAC
histone deacetylase complex
- PKA
protein kinase A
- ROS
reactive oxygen species
- STRE
stress response element
- TORC2
target of rapamycin complex 2
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs. Martin Graef and Jodi Nunnari for helpful discussions and for providing strains and plasmids used in this study. We also thank Samya Faiq, Elizabeth Iliuta, and Allison Gabbert for technical assistance.
References
- [1].He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67 93. doi: 10.1146/annurev-genet-102808-114910. PMID:19653858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102 9. doi: 10.1038/ncb1007-1102. PMID:17909521 [DOI] [PubMed] [Google Scholar]
- [3].Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65 76. PMID:12865942 [PMC free article] [PubMed] [Google Scholar]
- [4].Abeliovich H, Dunn WA Jr., Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025 34. doi: 10.1083/jcb.151.5.1025. PMID:11086004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ecker N, Mor A, Journo D, Abeliovich H. Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy. 2010;6:879 90. doi: 10.4161/auto.6.7.12753. PMID:20647741 [DOI] [PubMed] [Google Scholar]
- [6].Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582 6. doi: 10.1074/jbc.M506736200. PMID:16027116 [DOI] [PubMed] [Google Scholar]
- [7].Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651 62. doi: 10.1056/NEJMra1205406. PMID:23406030 [DOI] [PubMed] [Google Scholar]
- [8].Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27 42. doi: 10.1016/j.cell.2007.12.018. PMID:18191218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang EY, Biala AK, Gordon JW, Kirshenbaum LA. Autophagy in the heart: too much of a good thing? J Cardiovasc Pharmacol. 2012;60:110 7. doi: 10.1097/FJC.0b013e31824cc427. PMID:22343372 [DOI] [PubMed] [Google Scholar]
- [10].Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290 8. doi: 10.1091/mbc.E07-12-1292. PMID:18508918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Suzuki K, Noda T, Ohsumi Y. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast. 2004;21:1057 65. doi: 10.1002/yea.1152. PMID:15449304 [DOI] [PubMed] [Google Scholar]
- [12].Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes to cells: devoted to molecular & cellular mechanisms. 2007;12:209 18. doi: 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- [13].Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918 31. doi: 10.1091/mbc.E13-07-0381. PMID:23904270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Current opinion in cell biology. 2010;22:124 31. doi: 10.1016/j.ceb.2009.11.014. PMID:20034776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS letters. 1993;333:169 74. doi: 10.1016/0014-5793(93)80398-E. PMID:8224160 [DOI] [PubMed] [Google Scholar]
- [16].Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301 11. doi: 10.1083/jcb.119.2.301. PMID:1400575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971 81. doi: 10.1093/emboj/20.21.5971. PMID:11689437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180 9. doi: 10.1091/mbc.E07-05-0485. PMID:17699586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049 54. doi: 10.1073/pnas.0903316106. PMID:19805182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 2011;30:2101 14. doi: 10.1038/emboj.2011.104. PMID:21468027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al.. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488 92. doi: 10.1038/35044114. PMID:11100732 [DOI] [PubMed] [Google Scholar]
- [22].Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87 97. doi: 10.1016/j.devcel.2009.06.013. PMID:19619494 [DOI] [PubMed] [Google Scholar]
- [23].Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365 76. doi: 10.1016/j.devcel.2007.12.011. PMID:18331717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165 78. doi: 10.1016/j.cell.2007.05.021. PMID:17632063 [DOI] [PubMed] [Google Scholar]
- [25].Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al.. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012;109:11206 10. doi: 10.1073/pnas.1200313109. PMID:22733735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Backues SK, Lynch-Day MA, Klionsky DJ. The Ume6-Sin3-Rpd3 complex regulates ATG8 transcription to control autophagosome size. Autophagy. 2012;8:1835 6. doi: 10.4161/auto.21845. PMID:22960621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vlahakis A, Graef M, Nunnari J, Powers T. TOR complex 2-Ypk1 signaling is an essential positive regulator of the general amino acid control response and autophagy. Proc Natl Acad Sci U S A. 2014;111:10586 91. doi: 10.1073/pnas.1406305111. PMID:25002487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vlahakis A, Powers T. A role for TOR complex 2 signaling in promoting autophagy. Autophagy. 2014;10:2085 6. doi: 10.4161/auto.36262. PMID:25426890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vlahakis A, Lopez Muniozguren N, Powers T. Calcium channel regulator Mid1 links TORC2-mediated changes in mitochondrial respiration to autophagy. J Cell Biol. 2016;215:779 88. PMID:27899413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Teixeira MC, Monteiro PT, Guerreiro JF, Goncalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, et al.. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D161 6. doi: 10.1093/nar/gkt1015. PMID:24170807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996;15:2227 35. PMID:8641288 [PMC free article] [PubMed] [Google Scholar]
- [32].Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci U S A. 2012;109:1536 41. doi: 10.1073/pnas.1117563109. PMID:22307609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee ME, Singh K, Snider J, Shenoy A, Paumi CM, Stagljar I, Park HO. The Rho1 GTPase acts together with a vacuolar glutathione S-conjugate transporter to protect yeast cells from oxidative stress. Genetics. 2011;188:859 70. doi: 10.1534/genetics.111.130724. PMID:21625004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].James J, Fiji N, Roy D, Andrew Mg D, Shihabudeen MS, Chattopadhyay D, Thirumurugan K. A rapid method to assess reactive oxygen species in yeast using H2DCF-DA. Analytical Methods. 2015;7:8572 5. doi: 10.1039/C5AY02278A [DOI] [Google Scholar]
- [35].Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432 44. doi: 10.1101/gad.11.24.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Courchesne WE, Ozturk S. Amiodarone induces a caffeine-inhibited, MID1-depedent rise in free cytoplasmic calcium in Saccharomyces cerevisiae. Mol Microbiol. 2003;47:223 34. doi: 10.1046/j.1365-2958.2003.03291.x. PMID:12492866 [DOI] [PubMed] [Google Scholar]
- [37].Abeliovich H, Zhang C, Dunn WA Jr., Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477 90. doi: 10.1091/mbc.E02-07-0413. PMID:12589048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee P, Kim MS, Paik SM, Choi SH, Cho BR, Hahn JS. Rim15-dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae. FEBS letters. 2013;587:3648 55. doi: 10.1016/j.febslet.2013.10.004. PMID:24140345 [DOI] [PubMed] [Google Scholar]
- [39].Koopman WJ, Distelmaier F, Smeitink JA, Willems PH. OXPHOS mutations and neurodegeneration. EMBO J. 2013;32:9 29. doi: 10.1038/emboj.2012.300. PMID:23149385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hamalainen RH, Manninen T, Koivumaki H, Kislin M, Otonkoski T, Suomalainen A. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc Natl Acad Sci U S A. 2013;110:E3622 30. doi: 10.1073/pnas.1311660110. PMID:24003133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Niles BJ, Joslin AC, Fresques T, Powers T. TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Rep. 2014;6:541 52. doi: 10.1016/j.celrep.2013.12.040. PMID:24462291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Niles BJ, Powers T. TOR complex 2-Ypk1 signaling regulates actin polarization via reactive oxygen species. Mol Biol Cell. 2014;25:3962 72. doi: 10.1091/mbc.E14-06-1122. PMID:25253719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586 97. doi: 10.1101/gad.12.4.586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556 64. doi: 10.1093/emboj/17.13.3556. PMID:9649426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Longtine MS, McKenzie A, Demarini DJ 3rd, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953 61. doi: 10.1002/(SICI)1097-0061(199807)14:10. PMID:9717241<. 953::AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- [46].Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686 91. doi: 10.1038/nature02026. PMID:14562095 [DOI] [PubMed] [Google Scholar]
- [47].Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87 96. doi: 10.1016/S0076-6879(02)50957-5. PMID:12073338 [DOI] [PubMed] [Google Scholar]
- [48].Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3 21. doi: 10.1016/0076-6879(91)94004-V. PMID:2005794 [DOI] [PubMed] [Google Scholar]
- [49].Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103 15. doi: 10.1091/mbc.11.6.2103. PMID:10848632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al.. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676 82. doi: 10.1038/nmeth.2019. PMID:22743772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bodvard K, Wrangborg D, Tapani S, Logg K, Sliwa P, Blomberg A, Kvarnstrom M, Kall M. Continuous light exposure causes cumulative stress that affects the localization oscillation dynamics of the transcription factor Msn2p. Biochim Biophys Acta. 2011;1813:358 66. doi: 10.1016/j.bbamcr.2010.12.004. PMID:21167216 [DOI] [PubMed] [Google Scholar]
- [52].Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167 80. doi: 10.1016/0076-6879(83)01012-5. PMID:6310320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


