ABSTRACT
DICER1 plays a central role in the biogenesis of microRNAs and it is important for normal development. Altered microRNA expression and DICER1 dysregulation have been described in several types of tumors, including thyroid carcinomas. Recently, our group identified a new somatic mutation (c.5438A>G; E1813G) within DICER1 gene of an unknown function. Herein, we show that DICER1 is overexpressed, at mRNA level, in a significant-relative number of papillary (70%) and anaplastic (42%) thyroid carcinoma samples, whereas is drastically downregulated in all the analyzed human thyroid carcinoma cell lines (TPC-1, BCPAP, FRO and 8505c) in comparison with normal thyroid tissue samples. Conversely, DICER1 is downregulated, at protein level, in PTC in comparison with normal thyroid tissues. Our data also reveals that DICER1 overexpression positively regulates thyroid cell proliferation, whereas its silencing impairs thyroid cell differentiation. The expression of DICER1 gene mutation (c.5438A>G; E1813G) negatively affects the microRNA machinery and cell proliferation as well as upregulates DICER1 protein levels of thyroid cells but has no impact on thyroid differentiation. In conclusion, DICER1 protein is downregulated in papillary thyroid carcinomas and affects thyroid proliferation and differentiation, while DICER1 gene mutation (c.5438A>G; E1813G) compromises the DICER1 wild-type-mediated microRNA processing and cell proliferation.
KEYWORDS: Dicer1, microRNA, papillary thyroid carcinoma, thyroid cells
Introduction
Mature microRNAs (miRNAs or miRs) are small regulatory RNAs of 21–24 nucleotides (nt) in length that exert a crucial role on a variety of biological processes such as differentiation, proliferation, apoptosis, survival, growth, senescence and migration in vertebrates.1 They lead to decreased protein levels via repression of translation and/ or mRNA decay via deadenylation when miRNA pairs with target mRNA.2 A central role in the biogenesis of miRNAs is played by DICER1 that recognizes and cleaves the miRNAs precursors (50–70 nt) into mature miRNAs.2
Therefore, DICER1 gene is fundamental for normal development. Indeed, conditional DICER1 knockout models unraveled its importance for normal cerebellar3 and female reproductive system4 development as well as thyroid organogenesis and function.5
Moreover, recent studies have already demonstrated the dysregulation of DICER1 gene expression and/or mutations in human cancer. In fact, the downregulation of DICER1 expression has been associated to lung,6 breast7 and ovarian8 cancer progression and worse patient prognosis. Conversely, its overexpression has been described in prostate,9 colorectal10 and thyroid cancer.11 Somatic mutations in the metal-binding sites within the RNase IIIb catalytic domain (c.5438A>G, c.5429A>T and c.5429A>G) have been also described in human carcinomas. In particular, the mutation c.5438A>G (E1813G) has been reported in several human neoplasias, including non-epithelial ovarian,12 childhood cystic nephroma13 and thyroid cancer14 as well as Wilms tumors:15 it is predicted to impair the RNase IIIb function, critical for miRNA interaction and cleavage. Interestingly, this mutation has been also identified by our group in papillary thyroid carcinoma (PTC) samples16 and then further confirmed by Yoo et al. (2016)11 and associated with DICER1 overexpression. Noteworthy, the germline DICER1 mutations, concerning the coding sequence, have also been identified.17 They result in truncated protein nearby RNase III domain (i.e. c.3579_3580delCA), with an increased risk of multinodular thyroid hyperplasia and differentiated thyroid carcinoma for the patients carrying these mutations.14
In this study, we aimed at evaluating the role of DICER1 on thyroid proliferation and differentiation using rat normal and human carcinoma thyroid cell lines. Our data reveals that DICER1 overexpression positively regulates thyroid cell proliferation, whereas its silencing impairs thyroid cell differentiation. Finally, the expression of DICER1 gene mutation c.5438A>G (E1813G) in thyroid cells negatively affects miRNA processing and also thyroid cell proliferation.
Material and methods
Human thyroid samples
The human thyroid biopsies – 7 normal thyroid tissues (NT), 31 papillary thyroid carcinomas (PTC) and 14 anaplastic thyroid carcinomas (ATC) – were provided by the service of Pathological Anatomy of the Centre Hospitalier Lyon Sud, Pierre Bénite, France. Informed written consent was obtained from the patients.
Cell culture and transfection
PCCl 3 rat thyroid cells, derived from 18-month-old Fisher rats, were grown in Coon's modified Ham's F-12 medium (Euroclone), supplemented with 5% calf-serum and a six-hormone mixture (1 mU/ml TSH, 10 µg/ml insulin, 5 µg/ml transferrin, 10 nM hydrocortisone, 10 ng/ml somatostatin, and 10 ng/ml glycyl-L-histidyl-L-lysine acetate).18 Kras-transformed PCCl 3 (kiki) were cultured in Ham's F12 medium (Euroclone), supplemented with 10% calf serum.18 The human papillary thyroid carcinoma cell lines TPC-1 (RET/PTC) and BCPAP (BRAFV600E) were grown in DMEM medium (Life Technologies), supplemented with 10% fetal bovine serum.
For the inhibition of DICER1 expression in PCCl 3 and PCCl 3 kiki, cells were transfected with a short interfering RNA (siRNA) specific for DICER1 (NM_001195573-1/2, Ribox life science) and Nonsilencing Control siRNA (IBONI control N3, Ribox life science) using Lipofectamine RNAi MAX (Life Technologies), according to the manufacturer's recommendations. The siRNAs were used at a final concentration of 50 nM.
For overexpression of DICER1, transfections procedures were performed using Fugene HD reagent (Promega) for TPC-1 and BCPAP cells and, Lipofectamine 2000 (Life Technologies) for PCCl 3, following manufacturer's instructions.
Plasmids
The plasmid pFRT/TO/FLAG/HA-DEST DICER19 (pDICER1wt; #19881; Addgene) encodes human DICER1 protein (5772 bp; NM_177438) fused to the epitope of FLAG/HA in the N-terminal region. The vector containing the c.5438A>G (E1813G) mutation was constructed by excising the 788 bp fragment, flanking the mutation site, using the restriction enzymes XmaI (#R0180S; New England BioLabs) and PspXI (#R0656L; New England BioLabs) and, further, inserting the synthetized fragment containing the c.5438A>G (E1813G) mutation (Integrated DNA Technologies) into the linearized pDICER1wt, generating a plasmid encoding human mutated DICER1 (pDICER1mut). The plasmid was sequenced (Eurofins Genomics) and DICER1 expression was validated by q-RT-PCR and western blot analysis.
Cell proliferation
Cells were counted 48 hours post transfection using trypan blue. In parallel, as an index of cell viability, we used the commercially available MTT assay (Sigma-Aldrich). MTT reagent was diluted at final concentration of 0.5 mg/mL in cell medium and then, solubilized in DMSO. Measures were performed at 570 nm using ELx800 microplate Reader (BIO-TEK).
Flow cytometry
Cell cycle profile was evaluated using propidium iodide (2 µg/mL) on FACScan flow cytometer (Becton Dickinson) and analyzed on CELL-FIT software (Becton Dickinson).
q-RT-PCR
Total RNA was extracted from thyroid cell lines using the Trizol reagent (Life Technologies) according to the manufacturer's instruction. 1 µg of total RNA of each sample was used to obtained single strand cDNA with the QuantiTect Reverse Transcription Kit (Qiagen) using an optimized blend of oligo-dT and random primers according to the manufacturer's instruction.
Quantitative Real-Time PCR (q-RT-PCR) was performed with the CFX96 thermocycler (Bio-Rad) in 96-well plates using a final volume of 20 µl. For each of the PCR reaction, it was used 10 µl of 2X Sybr Green (Bio-Rad), 200 nM of each primer, and 20 ng of the cDNA previously generated. The oligonucleotides for q-RT-PCR, comprising exon-exon junctions, were purchased from Integrated DNA Technologies and designed with Primer-BLAST software,20 are listed in Table 1. Relative gene expression was determined using comparative C(T) method.21 RP18S and Rpl4 were used as housekeeping gene for human and rat samples, respectively.
Table 1.
Sequences of the primers used for q-RT-PCR.
| Target | Sequences |
|---|---|
| hs/rno_Dicer1 | F: CACATGCCTCCTACCACTACAAT |
| R: TGCTTGGTTATGAGGTAGTCCA | |
| hs_RP18S | F: TGCGAGTACTCAACACCAA |
| R: TTGGTGAGGTCAATGTCTGC | |
| rno_Rpl4 | F: GATGAATTGTACGGCACTTGG |
| R: TCTTTGGATCTCTGGGCTTTTTC | |
| rno-Ttf1 | F: CTACTGCAACGGCAACCTG |
| R: CCCATGCCATCATATATTCAT | |
| rno_Pax8 | F: GCCATGGCTGTGTAAGCAAGA |
| R: GCTTGGAGCCCCCTATCACT | |
| rno_Tg | F: CATGGAATCTAATGCCAAGAACTG |
| R: TCCCTGTGAGCTTTTGGAATG | |
| rno_Tpo | F: CAAAGGCTGGAACCCTAATTTCT |
| R: AACTTGAATGAGGTGCCTTGTCA | |
| rno_Nis | F: TCCACAGGAATCATCTGCACC |
| R: CCACGGCCTTCATACCACC |
To assess miRNA expression, 1 µg of total RNA of each sample was reverse transcribed with the miScript reverse transcription Kit (Qiagen), according to the manufacturer's instruction. For q-RT-PCR analysis, it was used miScript System Kits (Qiagen) and the following specific primers for mature miR (miScript Primer Sets; Qiagen): miR-21-5p (5′UAGCUUAUCAGACUGAUGUUGA); miR-33-5p (5′GUGCAUUGUAGUUGCAUUGCA); miR-125b-1-5p (5′UCCCUGAGACCCUAACUUGUGA); miR-296-5p ('AGGGCCCCCCCUCAAUCCUGU); miR-362 (5′AAUCCUUGGAACCUAGGUGUGAAU). RNU6 (MS00033740) was used for normalization.
Western blot
Cells were homogenized in RIPA buffer lysis (20 mM Tris-HCl pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Nonidet P40, and a mix of protease inhibitors) and then centrifuged at 13000 rpm at 4°C for 10 min. Cell lysate proteins (50–100 µg) were then subjected to SDS/PAGE electrophoresis, transferred onto Immobilon-P Transfer membranes (Millipore), membranes were blocked with 5% non-fat milk proteins and probed with the indicated antibodies at the appropriate dilutions: DICER1 (1:1000; sc-136981), Cyclin D1 (1:1000; sc-718), Cyclin E (1:1000; sc-248), Cyclin B1 (1:1000; sc-254), Vinculin (1:1000; sc-7649) and γ-tubulin (1:1000; sc-8035), all from Santa Cruz Biotechnology. Thyroglobulin (1:5000), TTF-1 (1:500) and PAX8 (1:5000) are rabbit polyclonal antibodies and described elsewhere.5 Membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (1:3000) for 60 min at room temperature and the signals were detected by western blotting detection system (ECL) (Thermo Scientific).
Statistical analysis
All results were expressed as mean ± SD. Data were analyzed by non-parametric Mann Whitney's test (when comparing two groups) or by the non-parametric Kruskal-Wallis test followed by Dunn's multiple comparison test (when comparing three or more groups). Statistical analyses were performed using the software Graphpad Prism (Version 5, Graphpad Software Inc.) and the difference was considered significant when p < 0.05.
Results
DICER1 expression in human thyroid carcinoma cell lines and tissues
First, we analyzed the expression of DICER1 in human thyroid carcinoma tissues and cell lines. The results shown in Fig. 1A demonstrate that DICER1 mRNA levels are increased in 22 out of 31 PTC and 6 out of 14 anaplastic thyroid carcinoma (ATC) samples when compared to normal thyroid tissue (NT) (p < 0.05) (fold-change ≥ 2), exhibiting a heterogeneous expression profile among thyroid carcinomas. Interestingly, DICER1 is drastically downregulated in all the analyzed thyroid carcinoma cell lines (TPC-1, BCPAP, FRO and 8505c; p < 0.001). Conversely, DICER1 protein levels were reduced in PTC in comparison with NT (0.31 ± 0.2) (Fig. 1B, C).
Figure 1.
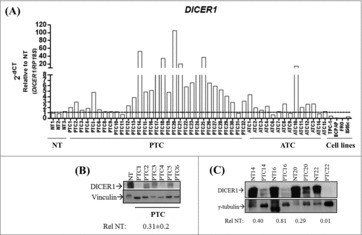
Expression of DICER1 in human thyroid carcinoma tissues and cell lines. (A) Expression of DICER1 was evaluated by q-RT-PCR in normal thyroid tissues (NT, n = 3), papillary thyroid carcinomas (PTC, n = 31), anaplastic thyroid carcinomas (ATC, n = 14) and human thyroid carcinoma cell lines (TPC-1, BCPAP, FRO, 8505c). RP18S was used as housekeeping gene. Expression levels were relative to NT. (PTC vs. NT, p = 0.049; ATC vs. NT, p = 0.048; cell lines vs. NT, p = 0.002). The DICER1 protein levels were assessed by western blot in (B) NT (1) and PTC (6) samples and, (C) four normal/ PTC paired samples. The results of the densitometric analysis were normalized by vinculin and γ-tubulin levels and relative to NT ± SD.
Silencing of Dicer1 affects proliferation and differentiation of normal thyroid cell line PCCl 3
Then, we investigated whether Dicer1 has a role in thyroid cell proliferation and differentiation using the normal rat thyroid cell line PCCl 3. This cell line is not tumorigenic and keeps in vitro all the markers of thyroid differentiation, i.e. thyroglobulin synthesis and secretion, ability to trap iodide and the dependency on thyrotropin for the growth.18 We knocked down (KD) Dicer1 expression by transfecting the PCCl 3 cells with Dicer1 siRNAs. First, we validated the silencing of Dicer1 mRNA (Fig. 2A) and protein levels (Fig. 2B) by q-RT-PCR and western blot analysis, respectively. Accordingly, decreased expression levels of miR-21-5p and miR-125-5p (Fig. 2C), previously shown to be modulated by Dicer1,22 in comparison with the control transfected PCCl 3 cells were observed. The Dicer1 downregulation results in a decreased number of cells (76 vs. 24, p<0.01) (Fig. 2D) and a reduction of cell viability (25%) with respect to the control cells (Fig. 2E). Consistently, the analysis of cyclin expression reveals decreased levels of cyclin E and cyclin B1 in Dicer1-silenced PCCl 3 cells in comparison with the control cells (Fig. 2F). Unexpectedly, an increased cyclin D1 expression was found in Dicer1-silenced PCCl 3 cells. However, a recent study showed that the cyclin D1 expression in breast cancer cells does not influence cell proliferation in absence of Dicer1 expression.23
Figure 2.
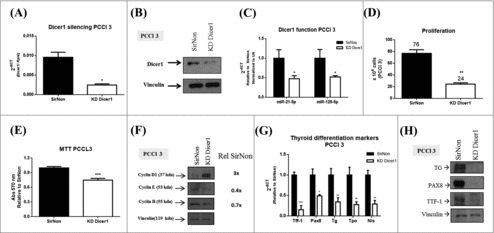
Silencing of Dicer1 in PCCl 3 cells. Cells were transfected with nonsilencing control siRNA (SirNon) or specific Dicer1 siRNA (KD Dicer1) and (A) Dicer1 mRNA and (B) protein levels were assessed by q-RT-PCR and western blot, respectively. (C) Expression of miR-21-5p and miR-125-5p to evaluate Dicer1 function. (D) Number of PCCl 3 cells was counted using trypan blue 48 hours post transfection with SirNon or specific Dicer1 siRNA. (E) In parallel, cell viability was assessed using MTT assay in the same conditions. (F) Expression of cyclin D1, E and B by western blot 48 hours after the transfection of PCCl 3 cells with SirNon or specific Dicer1 siRNA. Densitometry intensity quantification was calculated as ratio target: vinculin and relative to SirNon. (G) mRNA levels of the thyroid differentiation markers Ttf-1, Pax8, Tg, Tpo and Nis 48 hours after the transfection of PCCl 3 cells with SirNon or specific Dicer1 siRNA were evaluated by q-RT-PCR. (H) Protein levels of thyroglobulin (TG), PAX8 and TTF-1 were assessed by western blot in the same conditions. Data were relative to SirNon. Vinculin was used as loading control for western blot. Rpl4 and pseudogene U6 were used as housekeeping for q-RT-PCR of mRNA and miR levels, respectively. p < 0.05, p < 0.01; p < 0.001.
Since Dicer1 has been previously reported to have a critical role in thyroid organogenesis,5 we next investigated the expression of thyroid differentiation markers in the Dicer1-silenced PCCl 3 cells. As shown in Fig. 2G, the Ttf-1, Pax8, Tg, Tpo and Nis mRNA levels were drastically downregulated by silencing of Dicer1 expression and, further confirmed by western blot analysis for Ttf-1, Pax8 and Tg protein levels (Fig. 2H). These results suggest that Dicer1 and, consequently miRNA maturation, plays a critical role in the regulation of thyroid cell proliferation and differentiation.
Silencing of Dicer1 inhibits also the growth of Kras-transformed PCCl 3 cells (PCCl 3 kiki cells)
Subsequently, we silenced Dicer1 expression in the PCCl 3 cells transformed by the Kirsten murine sarcoma virus carrying the Kras oncogene. The q-RT-PCR (Fig. 3A) and western blot (Fig. 3B) analysis confirmed the silencing of Dicer1 expression at mRNA and protein level, respectively. In order to assess Dicer1 function, we analyzed the expression of miR-21-5p and miR-125-5p. As shown in Fig. 3C, the inhibition of Dicer1 expression results in a reduction of these miRNA levels. As shown above for the normal PCCl 3 cells, the number of cells diminished in comparison to SirNon (44 vs. 129, p < 0.05) (Fig. 3D) as well as attenuated the number of viable cells (20%) (Fig. 3E). Accordingly, both of the expression of cyclin D1 and E was downregulated in Dicer1-silenced cells (Fig. 3F). These data evidence that suppression of Dicer1 expression may also affect transformed thyroid cell proliferation.
Figure 3.
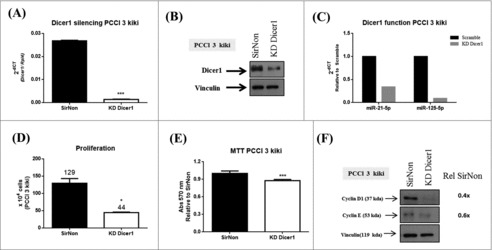
Dicer1 impacts the proliferation of KRAS-transformed PCCl 3 cells (PCCl 3 kiki). Cells were transfected with SirNon or Dicer1 specific siRNAs and (A) Dicer1 mRNA and (B) protein levels were assessed by q-RT-PCR and western blot, respectively. (C) To assess Dicer1 function, miR-21-5p and miR-125-5p expression were evaluated by q-RT-PCR in the same conditions. (D) Number of PCCl 3 kiki cells was counted using trypan blue 48 hours post transfection with SirNon or specific Dicer1 siRNAs. (E) Cell viability was assessed using MTT assay in the same conditions. (F) Expression of cyclin D1 and E by western blot 48 hours after the transfection of PCCl 3 kiki cells with SirNon or specific Dicer1 siRNA. Densitometry intensity quantification was calculated as ratio target: vinculin and relative to SirNon. Vinculin was used a loading control for western blot. Rpl4 was used as housekeeping gene. *p < 0.05; ***p < 0.001.
The mutation c.5438A>G (E1813G) within DICER1 gene affects DICER1 activity
In order to investigate the role of the DICER1 mutation c.5438A>G (E1813G), that was previously identified in PTC samples,11,14,16 we transfected two human thyroid papillary carcinomas cell lines, BCPAP and TPC-1, with the expressing vectors, containing the wild-type (pDICER1wt) and the mutated DICER1 (pDICER1mut) cDNAs. Firstly, we validated the overexpression of both expressing vectors by q-RT-PCR (Fig. 4A, D) and western blot (Fig. 4B, E) in comparison with the empty vector (EV). Since the expression of miR-21-5p and miR-125-5p was not regulated by pDICER1wt or pDICER1mut with respect to EV in BCPAP cells (data not shown), we searched for miRNA involved in thyroid differentiation and cancer: miR-33-5p and mir-296 target several thyroid differentiation markers such as NIS, thyroglobulin and TSH receptor.22 MiR-362 is downregulated in variant papillary thyroid carcinomas.24 Interestingly, the overexpression of pDICER1wt resulted in an increased expression of the miR-33-5p and miR-362 in BCPAP (Fig. 4C) and miR-21-5p and miR-296-5p in TPC-1 cells (Fig. 4F) and, the expression of the mutated DICER1 (pDICER1mut) reversed these phenomena, suggesting that this mutation might affect miRNA machinery.
Figure 4.
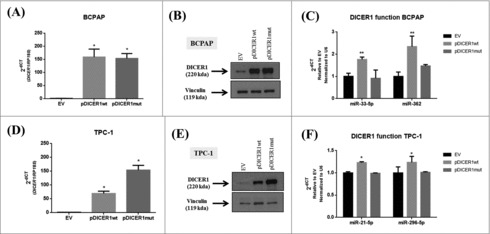
Effect of wild-type and mutated c.5438A>G (E1813G) DICER1 function in BCPAP and TPC-1 cells. Cells were transfected with empty vector (EV) and constructs expressing DICER1 wild-type (pDICER1wt) or mutated c.5438A>G (E1813G) (pDICER1mut) and 48 hours after transfection: (A) DICER1 mRNA and (B) protein levels were assessed in BCPAP cells by q-RT-PCR and western blot, respectively. (C) Expression of miR-33-5p and miR-362 to evaluate DICER1 function 48 hours post transfection in BCPAP cells. (D) mRNA and (E) protein levels of DICER1 in TPC-1 cells 48 hours after transfection with EV, pDICER1wt or pDICER1mut. (F) Expression of miR-21-5p and miR-296-5p to evaluate DICER1 function 48 hours post transfection in TPC-1 cells. Vinculin was used as loading control for western blot. RP18S and pseudogene U6 were used as housekeeping for q-RT-PCR of gene and miR expression, respectively. *p < 0.05, **p < 0.01.
Then, we wondered whether the growth rate of thyroid cells was affected by pDICER1wt and pDICER1mut. As shown in Fig. 5, when BCPAP and TPC-1 cells were transfected with pDICER1wt construct, but not with the pDICER1mut, they displayed a higher growth rate (80%, BCPAP, p < 0.001; 30%, TPC-1, p < 0.01) in comparison with the empty vector transfected cells (Fig. 5A, D). Moreover, pDICER1wt construct increased cell viability (13%, BCPAP, p < 0.001; 42%, TPC-1, p < 0.05) with respect to pDICER1mut (Fig. 5B, E). In agreement with that, an increment of the cyclins D1 and E levels was observed when cells were transfected with pDICER1wt construct, in comparison with pDICER1mut or EV (Fig. 5C, F). Cell cycle analysis unraveled that DICER1-ovexpressing vectors induced in pDICER1wt-transfected TPC-1 cells, but not pDICER1mut, a decreased cell number in G1 (40% vs. 45%, p < 0.05) and an increased one in S phase (45% vs. 40%, p < 0.05) of the cell cycle in comparison to EV (Fig. 5G).
Figure 5.
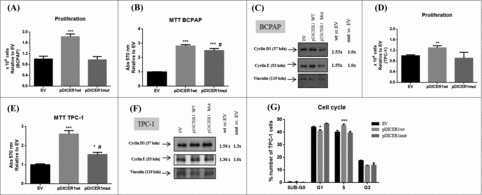
Effect of wild-type and mutated c.5438A>G (E1813G) DICER1 on BCPAP and TPC-1 cell proliferation. Cells were transfected with empty vector (EV) and DICER1-overexpression vectors, containing DICER1 wild-type (pDICER1wt) or mutated c.5438A>G (E1813G) DICER1 (pDICER1mut) cDNA and, 48 hours post transfection: (A) the number of BCPAP cells were counted using trypan blue and the results were relative to EV; (B) cell viability was evaluated using the MTT assay in BCPAP cells; (C) expression of cyclins D1 and E by western blot. Densitometry analyses were performed in comparison with EV in BCPAP cells. The same assays and conditions were performed for TPC-1 cells. (D) the number of TPC-1 cells were counted using trypan blue and the results were relative to EV; (E) cell viability was evaluated using the MTT assay in TPC-1 cells; (F) expression of cyclins D1 and E. Densitometry analyses were performed in comparison with EV; (G) cell cycle by flow cytometry using propidium iodide and data were represented as the percentage of cells in each phase of the cell cycle. Vinculin was used as loading control for western blot. *p < 0.05; **p < 0.01; ***p < 0.001 compared to EV. #p < 0.05, compared to pDICER1wt.
These data suggest that DICER1 wild-type overexpression induces human thyroid carcinoma cell line proliferation and enhances miRNA processing, while the mutated DICER1 form negatively impacts on these parameters.
Dicer1 overexpression increases the proliferation rate of the PCCl 3 cells, but does not affect thyroid differentiation
To investigate the role of the mutated DICER1 in normal thyroid cells, we transfected the PCCl 3 cells with pDICER1wt or pDICER1mut. We confirmed the overexpression of both constructs by q-RT-PCR (Fig. S1A) and western blot (Fig. S1B) analysis, forty-eight hours post transfection. Unexpectedly, the expression of miR-21-5p and miR-125-5p were downregulated by pDICER1wt with respect to EV and a weak or no effect was observed after the transfection with pDICER1mut (Fig. S1C). Then, we observed that pDICER1wt slightly increased the cell growth rate in comparison with EV (63 vs. 45, p < 0.001) and pDICER1mut reversed this effect (Fig. S1D). Accordingly, pDICER1wt weakly increased the number of viable cells in comparison with EV (27%, p < 0.001) and pDICER1mut reversed it (Fig. S1E). Above all, the transfection of DICER1-overexpressing vectors had no effect on the expression of the thyroid differentiation markers (Ttf-1, Pax8, Tg, Tpo and Nis) (Fig. S1F), suggesting that basal Dicer1 levels are important to maintain thyroid differentiation.
Discussion
DICER1 is a key player in miRNA processing and its role on development and cancer has been widely explored. Herein, we assess DICER1 expression in thyroid carcinoma samples and then, we focused on DICER1 impact on proliferation and differentiation of normal and cancer thyroid cell lines as well as the function of the mutation c.5438A>G (E1813G) on these parameters.
DICER1 dysregulation has been reported in thyroid cancer11,14,16 and its upregulation was associated with aggressive behavior (i.e, extrathyroidal extension and distant metastasis) of PTC.25 Our results demonstrate that DICER1 mRNA levels are upregulated in almost 70% of PTC and 42% of ATC analyzed. The limited sample number might account for the differences found at DICER1 mRNA levels of thyroid carcinoma samples between our results and those present in the TGCA database26 as well as the different technical approaches to assess DICER1 expression might account for the magnitude of its mRNA levels. Conversely, DICER1 protein levels were reduced in PTC samples. The downregulation of DICER1, at least at protein level, is in accordance with TGCA database.26 The discrepancy between mRNA and protein levels of DICER1 has been already reported by several studies,25,27 indicating that post-transcriptional mechanisms regulate DICER1 protein levels in neoplastic tissues.
Interestingly, a drastic DICER1 downregulation was found in all thyroid carcinoma cell lines. This result may be associated to the critical role of miRNA and, consequently, DICER1 on thyroid differentiation.5 Besides, thyroid carcinoma cell lines exhibit a dedifferentiated phenotype when compared to their original in vivo thyroid tumors28 or may be simply due to the very high number of passages of these cell lines in culture.
The effect of DICER1 on cell proliferation is still controversial. Our data point out that DICER1 stimulates thyrocyte proliferation. These results are in accordance with knockout DICER1 mice models, in which the loss of DICER1 led to a smaller sized thyroid gland.5,29 Paradoxically, in the same model the authors observed an increased BrdU positive cells,29 a proliferation marker,30 which could be partially explained by the severe hypothyroidism and increased TSH plasma levels induced by DICER1 loss. Moreover, it has been reported that the monoallelic but not biallelic loss of DICER1 promotes tumorigenesis in vivo, which means that the effect of DICER1 on cell proliferation depends on its levels.31,32 Thus, while the partial loss is advantageous to tumors, the massive decrease of DICER1 expression could compromise thyroid cell viability and proliferation. Accordingly, our results are in agreement with those reported by Frezzetti et al.,5 in which the thyroid glands of Dicer1 knockout mice were smaller than control and heterozygous mice. Interestingly, miR-21-5p and miR-125-5p, previously shown to target thyroid differentiation markers and to be modulated by DICER1,22 are downregulated in DICER1-silenced PCCl 3 cells, however, they are also downregulated in DICER1-overexpressing PCCl 3 cells, suggesting that the tight regulation of DICER1 levels are important to maintain miRNA processing machinery.
Noteworthy, DICER1-silencing PCCl 3 cells exhibit a dedifferentiated phenotype, whereas the differentiation markers do not show any change in their expression when DICER1 wild-type or mutated is overexpressed, indicating that basal expression of DICER1 is important to the maintenance of thyroid differentiation. In fact, DICER1 expression is crucial to thyroid homeostasis and its loss has been reported to disturb thyroid organogenesis and differentiation and might prompt cells to acquire cancer-like features.5,29
The RNase IIIb domain of DICER1 is a mutational hotspot in non-epithelial gonadal33 and endometrial tumors.34 The mutation E1813G of DICER1, located within RNase IIIb domain, has been consistently identified in thyroid carcinoma (5- 8% of the cases) by several studies.11,14,16 Our data point out that the mutation E1813G of DICER1 negatively affects the proliferation and miRNA processing machinery of thyroid cells. Indeed, this mutation could partially reduce DICER1 activity since it is predicted to impair the RNase IIIb function, failing to cleave miRNAs from the 5′-arm of pre-miRNA hairpins,2 which is in agreement with the inability of mutated DICER1 form to induce the miRNA expression in comparison with the DICER1 wild-type. Moreover, we could not exclude the contribution of other mutations within DICER1 gene for cell transformation. In fact, the presence of DICER1 germline mutations and additional somatic mutations such as E1813G were associated to well-differentiated thyroid carcinoma development in DICER1 syndrome patients.14
In summary, DICER1 protein is downregulated in papillary thyroid carcinomas and its basal expression is fundamental to thyroid differentiation. Moreover, the mutation E1813G within DICER1 gene affects DICER1 wild-type-mediated proliferation and activity.
Supplementary Material
1380127_Figure_S1.pptx
Funding Statement
This study has been supported by grants from: PNR-CNR Aging Program 2012–2014, CNR Flagship Projects (Epigenomics-EPIGEN), Associazione Italiana per la Ricerca sul Cancro (AIRC IG 11477). AF and RCCP have a scholarship from CAPES (Brazil).
Disclosure of interest
The authors report no conflict of interest.
References
- [1].Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803(11):1231-43. doi: 10.1016/j.bbamcr.2010.06.013. PMID:20619301 [DOI] [PubMed] [Google Scholar]
- [2].Kurzynska-Kokorniak A, Koralewska N, Pokornowska M, Urbanowicz A, Tworak A, Mickiewicz A, Figlerowicz M. The many faces of Dicer: the complexity of the mechanisms regulating Dicer gene expression and enzyme activities. Nucleic Acids Res. 2015;43(9):4365-80. doi: 10.1093/nar/gkv328. PMID:25883138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zindy F, Lee Y, Kawauchi D, Ayrault O, Merzoug LB, Li Y, McKinnon PJ, Roussel MF. Dicer is required for normal cerebellar development and to restrain medulloblastoma formation. PLoS One. 2015;10(6):e0129642. doi: 10.1371/journal.pone.0129642. PMID:26091048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149(12):6207-12. doi: 10.1210/en.2008-0294. PMID:18703631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Frezzetti D, Reale C, Calì G, Nitsch L, Fagman H, Nilsson O, Scarfò M, De Vita G, Di Lauro R. The microRNA-processing enzyme Dicer is essential for thyroid function. PLoS One. 2011;6(11):e27648. doi: 10.1371/journal.pone.0027648. PMID:22132122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al.. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111-5. doi: 10.1111/j.1349-7006.2005.00015.x. PMID:15723655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khoshnaw SM, Rakha EA, Abdel-Fatah TM, Nolan CC, Hodi Z, Macmillan DR, Ellis IO, Green AR. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135(2):403-13. doi: 10.1007/s10549-012-2169-3. PMID:22821364 [DOI] [PubMed] [Google Scholar]
- [8].Faggad A, Budczies J, Tchernitsa O, Darb-Esfahani S, Sehouli J, Müller BM, Wirtz R, Chekerov R, Weichert W, Sinn B, et al.. Prognostic significance of Dicer expression in ovarian cancer-link to global microRNA changes and oestrogen receptor expression. J Pathol. 2010;220(3):382-91. PMID:19960504 [DOI] [PubMed] [Google Scholar]
- [9].Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al.. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162-70. doi: 10.1158/0008-5472.CAN-08-0144. PMID:18676839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47(9):1414-9. doi: 10.1016/j.ejca.2011.01.006. PMID:21345667 [DOI] [PubMed] [Google Scholar]
- [11].Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, Song YS, Cho SW, Won JK, Shin JY, et al.. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet. 2016;12(8):e1006239. doi: 10.1371/journal.pgen.1006239. PMID:27494611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, Fejes AP, Chow C, Tone A, Kalloger SE, et al.. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366(3):234-42. doi: 10.1056/NEJMoa1102903. PMID:22187960 [DOI] [PubMed] [Google Scholar]
- [13].Doros LA, Rossi CT, Yang J, Field A, Williams GM, Messinger Y, Cajaiba MM, Perlman EJ, A Schultz K, Cathro HP, et al.. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod Pathol. 2014;27(9):1267-80. doi: 10.1038/modpathol.2013.242. PMID:24481001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].de Kock L, Sabbaghian N, Soglio DB, Guillerman RP, Park BK, Chami R, Deal CL, Priest JR, Foulkes WD. Exploring the association Between DICER1 mutations and differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2014;99(6):E1072-7. doi: 10.1210/jc.2013-4206. PMID:24617712 [DOI] [PubMed] [Google Scholar]
- [15].Wu MK, Sabbaghian N, Xu B, Addidou-Kalucki S, Bernard C, Zou D, Reeve AE, Eccles MR, Cole C, Choong CS, et al.. Biallelic DICER1 mutations occur in Wilms tumours. J Pathol. 2013;230(2):154-64. doi: 10.1002/path.4196. PMID:23620094 [DOI] [PubMed] [Google Scholar]
- [16].Costa V, Esposito R, Ziviello C, Sepe R, Bim LV, Cacciola NA, Decaussin-Petrucci M, Pallante P, Fusco A, Ciccodicola A. New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget. 2015;6(13):11242-51. doi: 10.18632/oncotarget.3593. PMID:25803323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14(10):662-72. doi: 10.1038/nrc3802. PMID:25176334 [DOI] [PubMed] [Google Scholar]
- [18].Fusco A, Berlingieri MT, Di Fiore PP, Portella G, Grieco M, Vecchio G. One- and two-step transformations of rat thyroid epithelial cells by retroviral oncogenes. Mol Cell Biol. 1987;7(9):3365-70. doi: 10.1128/MCB.7.9.3365. PMID:3670314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14(12):2580-96. doi: 10.1261/rna.1351608. PMID:18978028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. PMID:22708584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101-8. doi: 10.1038/nprot.2008.73. PMID:18546601 [DOI] [PubMed] [Google Scholar]
- [22].Fuziwara CS, Kimura ET. MicroRNAs in thyroid development, function and tumorigenesis. Mol Cell Endocrinol. 2017;456:44-50. doi: 10.1016/j.mce.2016.12.017. [DOI] [PubMed] [Google Scholar]
- [23].Yu Z, Wang L, Wang C, Ju X, Wang M, Chen K, Loro E, Li Z, Zhang Y, Wu K, et al.. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun. 2013;4:2812. doi: 10.1038/ncomms3812. PMID:24287487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P, Miccoli P, Basolo F. miRNA expression profiling of ‘noninvasive follicular thyroid neoplasms with papillary-like nuclear features’ compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Modern Pathology. 2016;30(1):39-51. doi: 10.1038/modpathol.2016.157. [DOI] [PubMed] [Google Scholar]
- [25].Erler P, Keutgen XM, Crowley MJ, Zetoune T, Kundel A, Kleiman D, Beninato T, Scognamiglio T, Elemento O, Zarnegar R, et al.. Dicer expression and microRNA dysregulation associate with aggressive features in thyroid cancer. Surgery. 2014;156(6):1342-50. doi: 10.1016/j.surg.2014.08.007. PMID:25456905 [DOI] [PubMed] [Google Scholar]
- [26].Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676-90. doi: 10.1016/j.cell.2014.09.050. PMID:25417114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jakymiw A, Patel RS, Deming N, Bhattacharyya I, Shah P, Lamont RJ, Stewart CM, Cohen DM, Chan EK. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49(6):549-59. doi: 10.1002/gcc.20765. PMID:20232482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saiselet M, Floor S, Tarabichi M, Dom G, Hébrant A, van Staveren WC, Maenhaut C. Thyroid cancer cell lines: an overview. Front Endocrinol (Lausanne). 2012;3:133. PMID:23162534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rodriguez W, Jin L, Janssens V, Pierreux C, Hick AC, Urizar E, Costagliola S. Deletion of the RNaseIII enzyme dicer in thyroid follicular cells causes hypothyroidism with signs of neoplastic alterations. PLoS One. 2012;7(1):e29929. doi: 10.1371/journal.pone.0029929. PMID:22242190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Muskhelishvili L, Latendresse JR, Kodell RL, Henderson EB. Evaluation of cell proliferation in rat tissues with BrdU, PCNA, Ki-67(MIB-5) immunohistochemistry and in situ hybridization for histone mRNA. J Histochem Cytochem. 2003;51(12):1681-8. doi: 10.1177/002215540305101212. PMID:14623936 [DOI] [PubMed] [Google Scholar]
- [31].Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23(23):2700-4. doi: 10.1101/gad.1848209. PMID:19903759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, Marine JC. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17(4):633-41. doi: 10.1038/cdd.2009.202. PMID:20019750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Witkowski L, Mattina J, Schönberger S, Murray MJ, Choong CS, Huntsman DG, Reis-Filho JS, McCluggage WG, Nicholson JC, Coleman N, et al.. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br J Cancer. 2013;109(10):2744-50. doi: 10.1038/bjc.2013.637. PMID:24136150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen J, Wang Y, McMonechy MK, Anglesio MS, Yang W, Senz J, Maines-Bandiera S, Rosner J, Trigo-Gonzalez G, Grace Cheng SW, et al.. Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis. J Pathol. 2015;237(2):215-25. doi: 10.1002/path.4569. PMID:26033159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1380127_Figure_S1.pptx


