ABSTRACT
Autophagy, a major catabolic process in eukaryotes, was initially related to cell tolerance to nutrient depletion. In plants autophagy has also been widely related to tolerance to biotic and abiotic stresses (through the induction or repression of programmed cell death, PCD) as well as to promotion of developmentally regulated PCD, starch degradation or caloric restriction important for life span. Much less is known regarding its role in plant cell differentiation. Here we show that macroautophagy, the autophagy pathway driven by engulfment of cytoplasmic components by autophagosomes and its subsequent degradation in vacuoles, is highly active during germ cell differentiation in the early diverging land plant Physcomitrella patens. Our data provide evidence that suppression of ATG5-mediated autophagy results in reduced density of the egg cell-mediated mucilage that surrounds the mature egg, pointing toward a potential role of autophagy in extracellular mucilage formation. In addition, we found that ATG5- and ATG7-mediated autophagy is essential for the differentiation and cytoplasmic reduction of the flagellated motile sperm and hence for sperm fertility. The similarities between the need of macroautophagy for sperm differentiation in moss and mouse are striking, strongly pointing toward an ancestral function of autophagy not only as a protector against nutrient stress, but also in gamete differentiation.
KEYWORDS: autophagy, cell differentiation, development, egg, fertilization, gamete, Physcomitrella patens, secretion, spermatogenesis
Introduction
Autophagy is an important process driving the self-consumption of cytoplasmic content in lytic compartments. It counteracts nutrient starvation by recycling cellular components and enables adaptation to various biotic and abiotic stresses. Macroautophagy, the best-studied form of autophagy, hereafter referred to as autophagy, is driven by a set of core autophagy-related genes (ATG) conserved in all eukaryotes.1–3 ATG proteins orchestrate the biogenesis of autophagosomes, spherical double-membrane structures engulfing portions of cytoplasmic material, which are delivered to lytic compartments for degradation.4
In the model plant Arabidopsis thaliana, autophagy is activated by environmental stresses or during certain stages of development. Lines mutated in ATG genes are hypersensitive to abiotic and biotic stresses and show accelerated senescence even under normal growth conditions.5–11 Although the majority of Arabidopsis atg mutants appear to develop normally, some defects related to cell expansion, life span and PCD have been identified. For instance, it has been suggested that ATG5-mediated autophagy contributes to tracheary element formation, a process that is PCD-dependent.12 Autophagy is also required for PCD in embryo-suspensor cells in the Norway spruce.13 Furthermore, mutations in the Arabidopsis ATG6 gene result in retarded growth, pollen germination failure and male sterility.14–16 However, as mutations in any other Arabidopsis ATG genes do not affect male fertility, it is unclear if ATG6 acts in an autophagy-dependent manner during pollen germination or in processes such as vesicle trafficking or vacuolar sorting. Finally, autophagic degradation of metabolic components in the anther tapetum layer, which supplies pollen with nutrients, is dependent on the ATG7 gene in rice (Oryza sativa/Os).17 Consequently, mutation in OsATG7 results in pollen germination failure.
Interestingly, autophagy is required during reproductive cell development and maintenance in some animals. Impaired autophagic flux in the medaka fish results in reduced cytoplasmic clearance during sperm specification, causing reduced fertility success.18 In mice, Atg7 mediates cytoplasmic reduction and flagella cytoskeleton organization during spermatogenesis, and germ-cell specific atg7 knockout (KO) also results in premature ovarian failure in female mice.19–21
Since cytoplasmic reduction is also important during sperm maturation in early diverging land plants,22,23 we hypothesized that the role of autophagy in germ cell differentiation may have been conserved across kingdoms. The early land plant model bryophyte Physcomitrella patens (P. patens) initiates its life cycle with the germination of a gametophytic haploid spore, which gives rise to filamentous protonema. Protonemal side branches occasionally develop into gametophores (leafy shoots), which produce apical male and female reproductive organs. The male reproductive organ, the antheridium, is composed of a sterile single cell layer, the jacket, surrounding a mass of spermatogenous cells that give rise to spermatids that differentiate into sperm (Fig. 1A). The female archegonium is composed of a neck with an inner canal that ends up in a mucilage-filled cavity where the egg resides. In the presence of water, the mature archegonium opens at its tip and flagellated sperm released by the antheridium swim through the canal to fertilize the egg. The resulting diploid zygote eventually develops into a sporophyte filled with haploid spores.23 It has been recently reported that protonema of P. patens atg5 KO lines show premature senescence and hypersensitivity to nutrient starvation, suggesting that moss ATG proteins have similar functions as those in Arabidopsis and yeast.24
Figure 1.
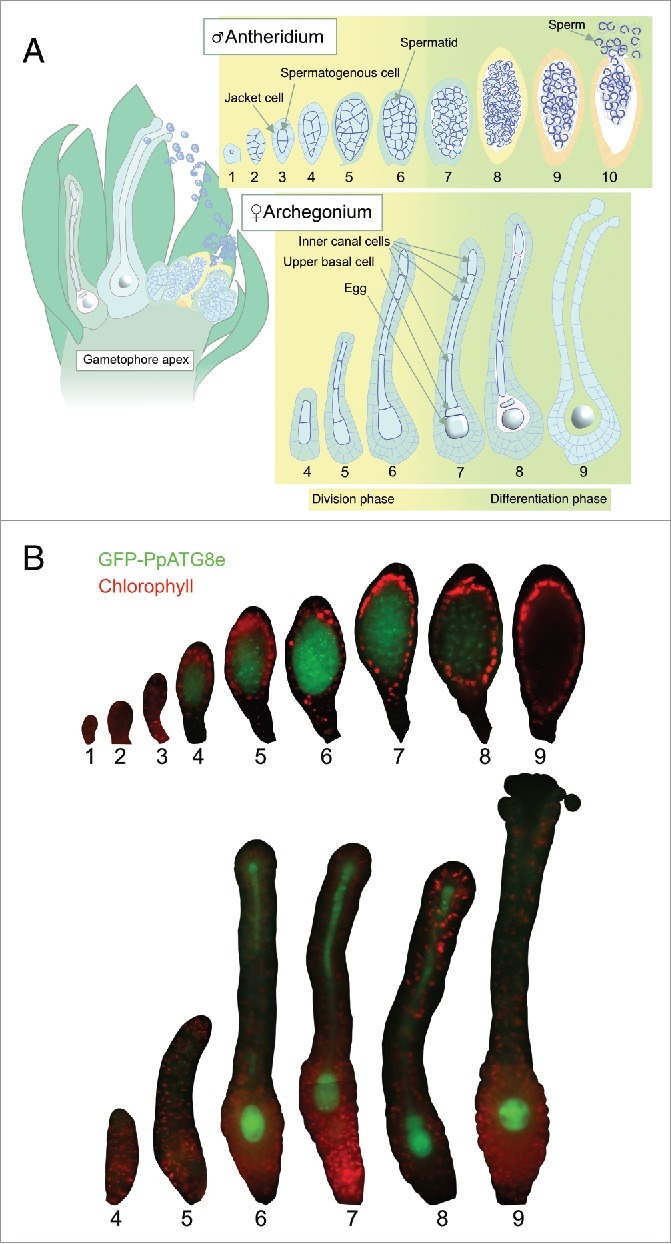
Autophagy is an active process during germ cell differentiation in moss. (A) Schematic representation of exposed reproductive organs in a gametophore apex at the moment of fertilization (left) and different stages of antheridium and archegonium development as described previously.23 (B) Representative epifluorescence pictures of WT reproductive organs expressing GFP-PpATG8e from the PpATG8e promoter in a cell- and stage-specific manner. GFP signals are shown in green, while red is chlorophyll autofluorescence. The red signal marks primarily the jacket cells in antheridia and the neck or body cells in archegonia. Due to large organ size, some archegonia images have been mounted from 2 complementary pictures.
We thus investigated the role of autophagy in P. patens germ cell development by using P. patens ATG8 (PpATG8) translational reporter lines and by comparing the progression of sperm, egg, and egg cavity development in wild type (WT) and lines lacking P. patens ATG5 (PpATG5) or ATG7 (PpATG7). This revealed that autophagy is highly active in germ cells and that autophagosome formation is needed for fertility, spermatid cytoplasmic clearance and egg cavity mucilage formation.
Results
Autophagy is induced during gamete maturation in moss
Autophagy-inducing conditions trigger transcription of ATG8-family genes and the formation of autophagosomes containing Atg8-family proteins in eukaryotes.25–27 Based on this characteristic, transgenic lines expressing Atg8-family proteins fused to fluorescent peptides have been broadly used as a tool to assess the dynamics of autophagosome formation in different kingdoms, including plants.28 To generate a moss-specific autophagy marker we selected PpATG8e, out of the 6 P. patens ATG8 orthologs29 based on its high expression during reproductive development (Fig. S1A). PpATG8epro::GFP-PpATG8e reporter lines were generated in the ecotype Gransden, showing a continuous production of reproductive organs probably due to low fertilization success, and in the highly fertile ecotype Reute,30 allowing sporophytic development to be analyzed. Correct integration was verified by PCR (Fig. S1B). Three Reute lines with signal strengths ranging from weak to strong and one Gransden line with intermediate signal strength, were chosen for further analysis.
Epifluorescence microscopy revealed consistent cell- and stage-specific expression of all 4 reporter lines in reproductive organs. In this and the following sections, we use a previously described stage specification (see also Fig. 1A).23 GFP signals in antheridia were restricted to the spermatogenic cell mass (Fig. 1B). PpATG8e expression synchronously comes on at stage 4 (when spermatogenous cells divide in all directions), peaks at stage 6 (when the shift from cell divisions to cell differentiation occurs), and is gradually reduced to below detection level at around transition from stage 8 to 9 (when spermatids complete their isolation and reshaping to give slender and crescent-shaped cells, see Fig. 1A for details). In archegonia, GFP signals are detected in the central cell file from stage 6, just before the division of the egg cell precursor, and onwards (Fig. 1B). Hence, canal cells and the egg cell precursor, as well as the progenies of the latter, the egg and the upper basal cell, all show GFP-PpATG8e signals throughout their development.
Ppatg5 lines show early gametophore senescence and disturbed autophagic flux
To analyze the importance of autophagy in moss reproductive development, we produced several lines mutated in the 2 single copy genes PpATG5 and PpATG7 needed for ATG8 lipidation and hence autophagosome formation (Fig. S2A and Table S1). Two Gransden (Ppatg5#1, Ppatg5#2) and 3 Reute (Ppatg5#3, Ppatg5#4, Ppatg5#5) atg5 KO lines were generated, as well as 3 Reute (Ppatg7#1, Ppatg7#2, and Ppatg7#3) atg7 KO lines. All Ppatg5 and Ppatg7 lines exhibit premature senescence of protonemata (not shown), as previously shown for independent Ppatg5 KO lines,24 as well as of gametophores (Fig. 2A and S2B).
Figure 2.
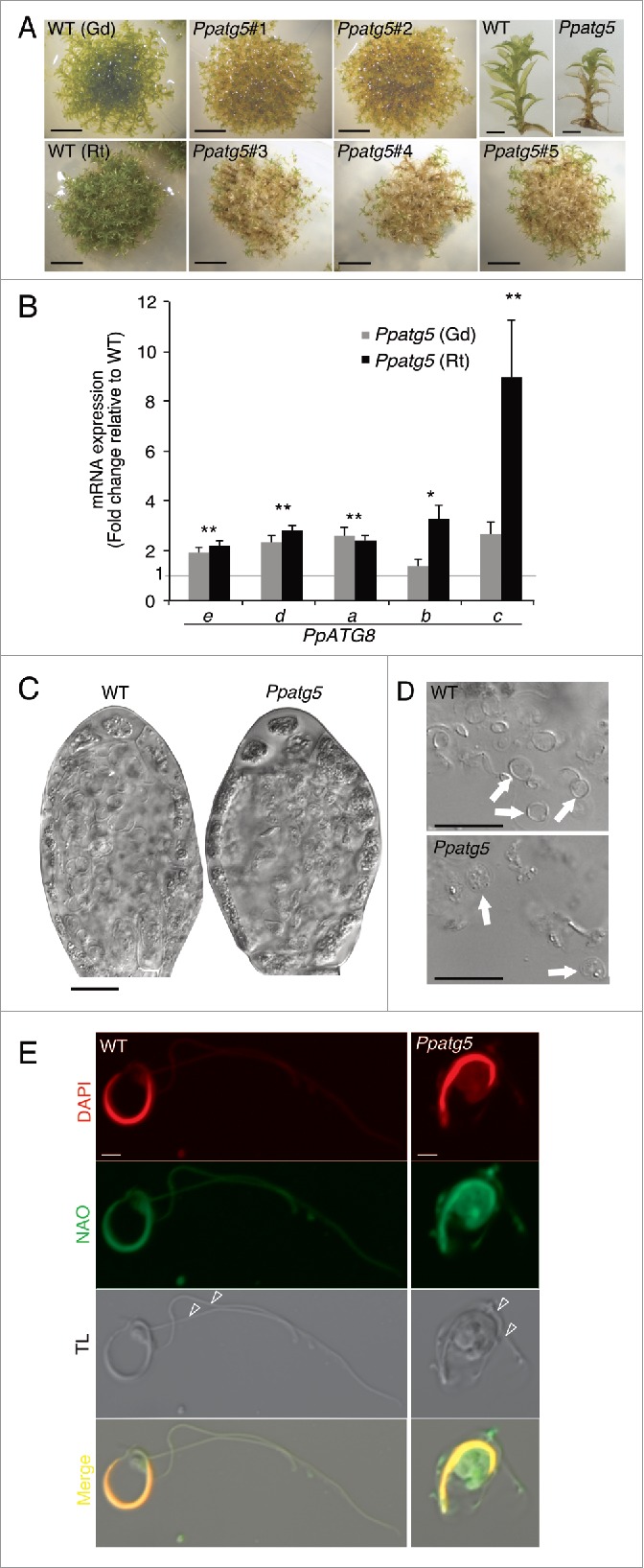
Ppatg5 lines show clear signs of blocked autophagy and morphologically defective sperm. (A) While 60-d-old gametophore producing WT colonies remain green and healthy, corresponding colonies of Ppatg5 KO lines have turned yellowish. Scale bar: 1 cm. Ppatg5 gametophore shoots show premature senescence in the basal leaves and stem-sections. Scale bar: 2 mm. (B) QRT-PCR analysis of the transcript levels of the different PpATG8 genes in the Ppatg5 lines compared with WT levels. Data are represented as mean of 3 biological replicates. **, statistically significant Student t test P< 0.05 for both ecotypes; * statistically significant Student t test P< 0.05 only for one ecotype. (C) Overview of representative antheridia at late stage of maturation, just before sperm release. (D) Mature sperm released from Ppatg5 antheridia appear malformed. (E) Mature sperm cells stained with DAPI and NAO. TL: Transmitted light. (C and D) are differential interference contrast micrographs, scale bar: 20 µm, (E) is confocal image, scale bar: 2 µm.
ATG8 overexpression is a hallmark of many Arabidopsis autophagy-deficient mutants,31,32 possibly reflecting a futile feedback cycle whereby cells try to deal with the starvation caused by autophagy deficiency by inducing autophagy machinery components. Therefore, we compared transcript levels of the PpATG8 genes in the Ppatg5 mutant and WT. This confirmed that all 5 PpATG8 genes tested show a significant upregulation in the Ppatg5 lines (Fig. 2B).
Sperm morphology is impaired in Ppatg5 and Ppatg7 lines
Previous studies of other moss species demonstrate that the mature motile sperm are thin, elongated and coiled with a small cytoplasmic belly and 2 flagella.33 While sperm of P. patens WT display this characteristic appearance, sperm of all Ppatg5 and Ppatg7 mutant lines are irregular, bulky and appear to have a larger cytoplasmic volume (Fig. 2C and D; Figs. S2C and D). As 10N-nonyl acridine orange (NAO) stain the whole cytoplasm of mature P. patens sperm, instead of marking the inner mitochondrial membrane as it does in other species,34 we took advantage of this to compare the cytoplasmic volume of mature WT, Ppatg5 and Ppatg7 sperm. Confocal images clearly show that the cytoplasm of WT sperm is extremely reduced at maturation, whereas Ppatg5 and Ppatg7 sperm cells in comparison retain a large cytoplasmic volume (Fig. 2E and Fig. S2E). DAPI staining of mature antheridia further revealed that instead of the highly regular crescent shape of WT sperm nuclei, Ppatg5 and Ppatg7 sperm nuclei show some irregularities, specifically toward the ends (Fig. 3I, J and Fig. S2D).
Figure 3.
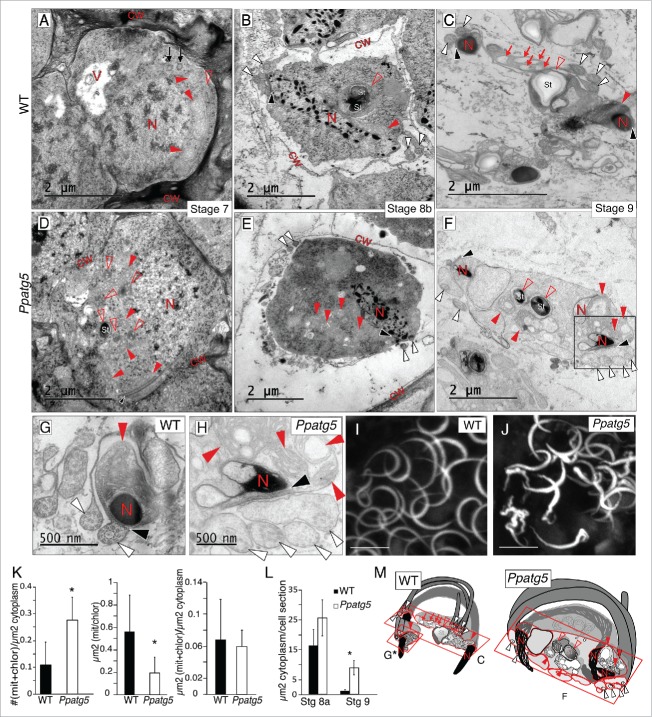
Reduction of cytoplasm, organelle morphology, nuclei condensation and flagella structure are affected in Ppatg5 sperm. (A to H) TEM micrographs of representative WT and Ppatg5 spermatids at successive developmental stages and at similar plane sections. (G) Details of nuclei, flagella and anterior mitochondrion in WT and (H) Higher magnification of the boxed area in (F). (I and J) Maximum projection images from confocal microscopy of DAPI-stained sperm nuclei of WT and Ppatg5 respectively. Scale bar: 5 µm. (K) Quantification of the number of mitochondria and chloroplasts sections per µm2 of cytoplasm, error bar represents the SD for n = 10, statistically significant Student t test, * P< 0.001. Quantification of the area of independent mitochondria or chloroplast sections (µm2), error bar represents the SD for n = 21, statistically significant Student t test, * P< 0,001. Quantification of total µm2 of mitochondria + chloroplasts per µm2 of cytoplasm, error bar represents the SD for n = 10, Student t test showed no significant differences. All quantifications were made in spermatids of stage 8a antheridia. (L) Quantification of the cytoplasmic area per spermatid section at stages 8a and 9 of WT and Ppatg5. The area difference is most striking at stage 9. Error bar represents the SD for n = 10. Statistically significant Student t test, * P< 0.001. Measurements in (K and L) were performed with ImageJ. (M) Schematic representation of a mature sperm in WT and Ppatg5. The red squares show the plane of sections in micrographs (C), (F and G). V, vacuole; N, nucleus; cw, cell wall; St, starch granule; filled red arrowhead, mitochondrion; empty red arrowhead, plastid; white arrowheads, cross-section of flagella; red arrow, empty vesicle; black arrowhead, spline.
Ppatg5 sperm is affected in cytoplasmic reduction, nuclear condensation and flagella formation
To study the detailed effects of impaired autophagy on sperm maturation, we compared the ultrastructure of developing WT and Ppatg5 spermatogenous cells using transmission electron microscopy (TEM). We analyzed antheridia from stage 5 to 9 and found that the key spermatogenic hallmarks described for other moss species35–38 are also shared by the P. patens WT. These include: i) reduction in organelle numbers to one plastid and 2 mitochondria, one anterior and one posterior (Compare Figs. S3A to S3B, filled and empty arrowheads), ii) nuclear condensation, first seen at early stage 8, allowing us to differentiate between stages 8a and 8b (Fig. S3B and Fig. 3B, red N letter), iii) de novo formation of a locomotory machinery composed of a band of parallel microtubule filaments (spline), the anterior part of which overlays a multilayered structure in which the 2 flagella are anchored via their basal bodies (Fig. 3A to C, black arrows, black and white arrowheads). As the spline elongates pressed to the plasma membrane (PM), it adopts a coiled shape dictated by the spherical spermatid (Fig. 3M, see WT).39 Other hallmarks include a drastic and continuous reduction of cytoplasm volume, which eventually fits the PM tightly around the spline and its aligned organelles, thereby sculpting the mature sperm (Fig. 3A to C).
Although we could not detect any major disturbance in spermatids of stage 5 Ppatg5 antheridia (Figs. S3A and S3C), the deviations are striking from stage 7, when sperm differentiation has initiated, to stage 9, when mature sperm are formed. At WT stage 7 the plastid number is already reduced to one, and the number of mitochondria is reduced, but has not yet reached the final number of 2 (Fig. 3A, filled and empty red arrowheads). TEM of Ppatg5 stage 7 spermatids show multiple plastid and mitochondria sections per cell, and the number of these sections per cytoplasmic area is higher compared with WT even at maturity (Fig. 3D to F, filled and empty red arrowheads, and 3K), pointing toward a defect in organelle dynamics. The area of these individual organelle sections is smaller in the mutant, but not the total organelle area per cell (Fig. 3K). This suggests that the organelle number and size could be affected in the KO background. Another likely alternative is that the number is not affected and that several organelle sections together represent one pleiomorphic organelle, a phenomenon described before in autophagy mutants.40 In addition, even if nuclear condensation in both WT and Ppatg5 spermatids initiates at stage 8a (Figs. S3B and S3D) and continues progressively (Fig. 3A to F, red letter N), the final nuclear morphology of Ppatg5 sperm is more irregular compared with the WT (Fig. 3G and H, letter N). The flagella are also affected and show disturbed microtubule arrangement and impaired fitting of the PM (Fig. 3G and H, white arrowheads). Finally, the reduction in cytoplasm is not as efficient in Ppatg5 (Fig. 3L), which can be the cause of the less slender sperm and the impaired fitting of the PM around the flagella, potentially affecting flagella function (Fig. 3G and H, white arrowheads). All these abnormalities contribute to the formation of an aberrant-shaped sperm schematically represented in Fig. 3M.
Ppatg5 shows altered electron density of the extracellular mucilage produced by the egg and canal cells
We next used TEM to compare WT and Ppatg5 archegonia and detected clear differences in the extracellular mucilage surrounding the egg and canal cells. Stage 8 WT egg and canal cells contain a large number of small vesicles (∼90 nm) with highly electron-dense material often localized close to the PM, or fusing to it (Fig. 4A and B, empty arrowheads). Furthermore, the mucilage, observed as an electron-dense fibrillar network, fills the egg cavity and the canal (Fig. 4A and B, red C letter), suggesting that both egg and canal cells are responsible for mucilage production. In contrast, the mucilage in Ppatg5 is significantly less electron-dense (Fig. 4A, red C letter and 4D) compared with WT, and the Ppatg5 egg and canal cell PM-associated vesicles are much fewer and often appear less electron dense (Fig. 4A, empty and filled arrowheads, and 4C). This suggests that PpATG5 is important for extracellular mucilage formation. In addition, nonfused double-membrane structures were identified in the Ppatg5 egg cells, but not in WT (Fig. 4A, filled arrowheads). As a control for the electron-density measurements we compared the electron density of the WT and Ppatg5 egg cell cytoplasm, which appeared very similar (Fig. S4).
Figure 4.
Density of the canal and egg cell produced matrix is altered in Ppatg5. (A and B) TEM micrographs of the edge of the egg (A) and canal cell (B) of stage 8 archegonia in WT compared with Ppatg5. Empty arrowheads mark vesicles with electron-dense material, filled arrowheads mark nonfused autophagosomes. Red inlets in drawings indicate the areas shown in the TEM micrographs in the egg. E, egg; C, cavity; cc, canal cell; cw, cell wall. (C) Number of electron-dense vesicles per µm2 of cytoplasm in egg sections of Ppatg5 and WT stage 8 archegonia. (D) Histogram of mucilage electron-density in egg cavity of Ppatg5 and WT stage 8 archegonia. Data presented are the mean of 2 independent measurements. Measurements were performed with ImageJ.
Abnormal Ppatg5 sperm is infertile whereas Ppatg5 eggs are able to develop into a functional sporophyte upon fertilization
To assess whether ATG5 affects fertility we self- and cross-fertilized lines in the highly fertile Reute background. While self-fertilization was successful in 99% of the WT gametophores, not a single sporophyte developed in the 100 gametophores analyzed in each of the 3 KO lines (Fig. 5A). However, all 3 Ppatg5 lines were capable of initiating sporophyte formation at a low frequency after successful cross-fertilization with WT sperm, exemplified by Ppatg5#3 in Fig. 5A. This result clearly demonstrates that PpATG5 is crucial for sperm fertility, whereas its role in female fertility is not essential. The development of the heterozygous Ppatg5 and PpATG5 sporophytes was delayed at least 2 wk compared with WT, and many arrested before maturation (Fig. 5B). This arrest may be due to the involvement of PpATG5 in sporophyte development, or to secondary effects caused by premature gametophore senescence.
Figure 5.
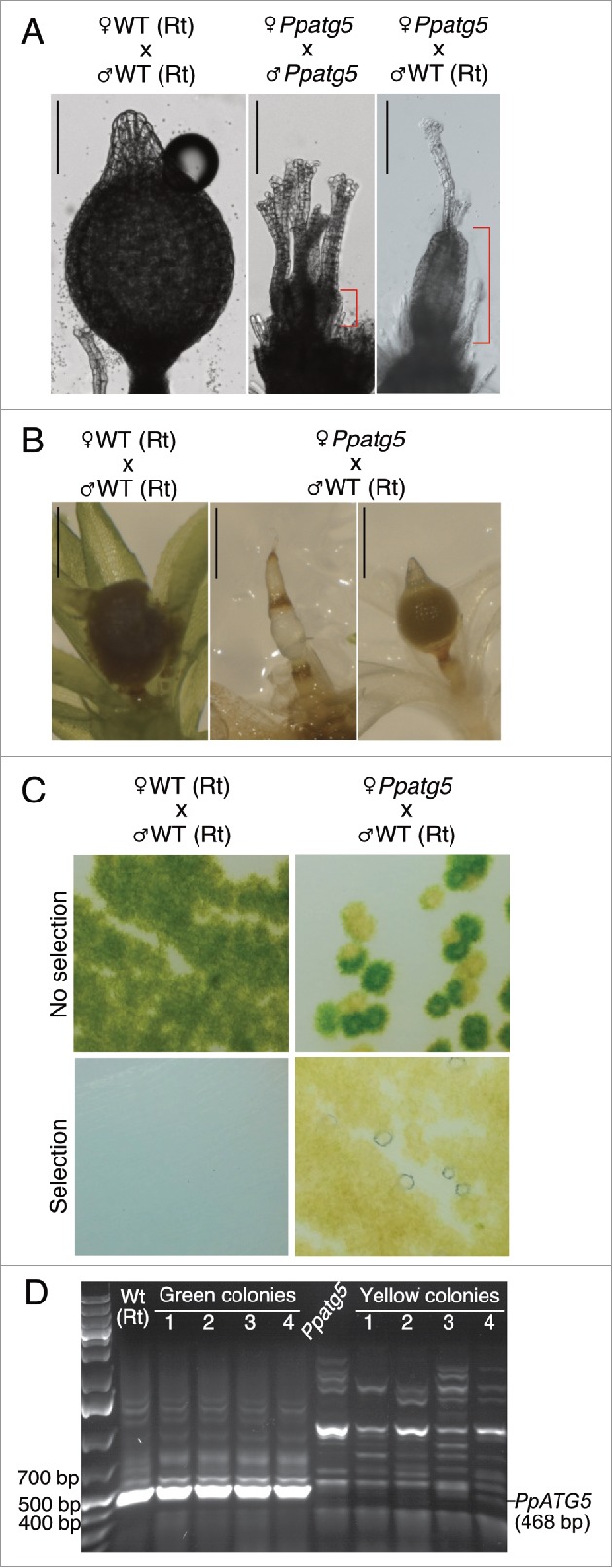
Sperm of Ppatg5 lines are infertile whereas the Ppatg5 egg upon fertilization, can develop into a functional sporophyte that produces viable spores. (A) differential interference contrast micrographs of representative gametophore apices 40 d after induction of reproductive development. Self-fertilization of Ppatg5 lines always fails. The red mark highlights the difference in size between a nonfertilized archegonium and a young sporophyte from a successful fertilization. Sporophyte maturation after successful female Ppatg5 x male WT crosses progresses much slower than after self-fertilization of WT. Scale bar: 200 µm (B) Sporophyte development comparison when WT is fully mature and has already burst. Scale bar: 1 mm. (C) Germinated spores in the presence or absence of selection marker (geneticin), which selects for genotypes carrying the Ppatg5 KO construct. (D) RT-PCR confirmation of colony genotype using primers p5 and p6. For principal positions of annealing sites and sequence of primers, see Figure S2A and Table S1.
To test whether ATG5 is essential for spore viability, we germinated haploid spores from the heterozygous sporophytes. Two colony types were produced; green, resembling WT and yellow, resembling the Ppatg5 early senescent genotype (Fig. 5C). Upon transfer to geneticin-supplemented medium, to which only the Ppatg5 genotype is resistant, only yellow colonies continued to grow and RT-PCR confirmed their Ppatg5 genotype (Fig. 5D). Thus, PpATG5 is not essential for spore viability or germination.
The occurrence and nature of autophagy-related structures depend on PpATG5
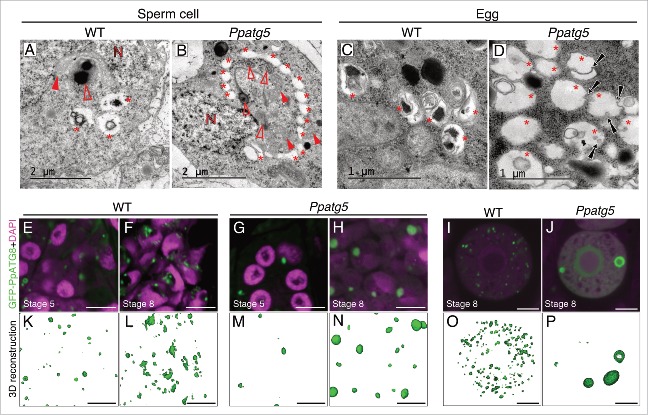
We next mapped the occurrence of presumptive autophagy-related structures during gametogenesis in WT and Ppatg5. TEM of stage 8 WT spermatids and egg cells revealed vesicles (diameter of around 0.6 µm in both spermatids and egg cells) delimited by a single membrane and with degrading material inside (Fig. 6A and C, asterisks). These vesicles are found in close proximity to each other and it cannot be ruled out that they are part of the same superstructures. In contrast to WT the vesicles detected in Ppatg5 egg and spermatids appear mostly empty (Fig. 6B and D, asterisks). In the egg cell they appear to engulf each other or small parts of the cytoplasm (Fig. 6D), whereas in spermatids vesicles gather in a collar-like structure around a portion of cytoplasm usually filled with organelles (Fig. 6B, filled and empty arrowheads). These spermatid vesicles progressively grow and fuse to sequester a large cytoplasmic portion (Figs. S5A and S5B).
Figure 6.
Autophagy-related structures and Atg8 family-associated structures can be observed in the Ppatg5 lines. (A to D) TEM micrographs of WT and Ppatg5 stage 8 spermatids and egg cells revealing details of autophagic related structures. N, nucleus; filled red arrowhead, mitochondrion; empty red arrowhead, plastid; asterisks, autophagy-related structures; black arrows, microautophagy events. (E to J) Single confocal plane images of WT and Ppatg5 spermatids (E to H) or single egg cells at stage 8 (I and J) expressing GFP-PpATG8e. (E and G) are spermatogenous cells at stage 5, (F and H) are spermatids at stage 8. Nuclei are stained with DAPI. Scale bar: 5 µm. (K to P) 3D reconstructions of confocal images from z-stacks including the images shown in (E to J). Reconstructions were performed with the Zen black software from Carl-Zeiss. Scale bar: 5 µm
We next used confocal imaging to analyze the subcellular localization of GFP-PpATG8e in PpATG8epro::GFP-PpATG8e lines in WT and Ppatg5 backgrounds. In WT, GFP-labeled dots (around 0.6 µm) and in some cases ring-like structures, were seen in both spermatids and egg (Fig. 6E, K, F, L, I and O). In stage 8 spermatids, when most of the cytoplasmic reduction occurs, GFP-labeled structures were more abundant and, apart from dots, irregular and larger structures (up to 2 µm long; Fig. 6F and L) could be observed. We hypothesize that these irregular structures are the vesicles observed by TEM in late spermatids and that they are actually clusters or branches of the same superstructure (Fig. 6A).
Compared to WT, GFP-PpATG8e-labeled structures in Ppatg5 are fewer (often only 1 or 2 per cell) but significantly larger and they grow as development progresses (Fig. 6G, M, H, N, J, P; Figs. S5C and S5D). These structures appear globular and in no case could the irregular structures seen in the WT spermatids be observed. Compared to WT, fewer and larger RFP-ATG8 structures were also detected in several Dictyostelium atg KO mutants and these were colocalized with ubiquitin, suggesting that they represent protein aggregates.41,42 In the Arabidopsis atg4a atg4b double-mutant, globular GFP-ATG8 accumulations are soluble and were also suggested to be protein aggregates.27 The most plausible interpretation is thus, that the GFP-PpATG8e fusion protein in Ppatg5 forms aggregates increasing in size over time. Thus, the reporter data support the autophagy deficiency of the Ppatg5 mutant.
Discussion
Isoforms of the highly conserved Atg8 family of proteins have been characterized, and their cellular dynamics studied through the generation of translational reporter lines in different plant species.13,27,29,43,44 Here we showed that the P. patens ATG8e and ATG8d isoforms are highly expressed in moss reproductive organs, and that they accumulate in developing germ cells, suggesting that autophagy is induced during sperm and egg development.
Indeed, reduced autophagy through KO of ATG5 or ATG7 severely affects the extreme shape modifications normally occurring in the P. patens male germ line during their differentiation, leading to male infertility. The incomplete reduction of cytoplasmic content in the atg5 mutant impairs the acquisition of the slender shape and proper flagella development resulting in restrained sperm movement. Here we provide evidence for a direct link between autophagy and cell reshaping during plant cell differentiation and our results provide a proof of concept for the requirement of autophagy in plant cell differentiation independent of PCD, just as described before for animal cells.45,46 Although autophagy has previously been reported to be upregulated in root cells entering the differentiation phase, the effects of blocked autophagy was limited to changes in vacuolar volume.47,48
The maturing egg germ cell does not undergo dramatic shape remodelling, although it is a site of active autophagy. Previous reports from other moss species show that the formation of the extracellular mucilage correlates with 2 observations: i) the disappearance of egg plastid starch, which may be converted into mucilage during egg maturation and ii) the presence of vesicles discharging their content to the cavity.49,50 The finding that mucilage accumulation is reduced in atg5 mutant egg cells suggests that PpATG5-mediated processes could be involved in mucilage formation. As autophagy is known to contribute to leaf starch degradation51 it is not unlikely that egg plastid starch degradation is an ATG5-dependent process that provides the building blocks important for the mucilage production in the egg and canal cells. Hypothetically, PpATG5-mediated processes could also be involved in mucilage secretion. In animals and yeast it has been suggested that a subset of autophagosomes escape degradation and instead fuse with multivesicular bodies to form amphisomes which subsequently fuse with the PM to secrete their content.52,53 It has recently been shown that mice lacking Atg5 in the intestinal epithelium suffer from diminished mucus secretion.54 In addition, endothelial secretion in mice is related to autophagosomes and dependent on functional Atg5 and Atg7.55 Thus, our data could be a starting point to evolutionary studies of a putatively conserved role of ATG proteins in secretion.
Based on our results and recent publications on the specific importance of autophagy in cytoplasmic reduction and morphogenesis in animal sperm and in the differentiation of female reproductive organs,18–21 we affirm that autophagy is a common player in plant and animal germ cell development. The similarity between observed sperm defects in a germ cell-specific atg7 KO mouse21 and in our moss atg5 and atg7 KO mutants is striking. First, both mouse and moss mutant sperm suffer from impaired autophagy and cytoplasmic clearance. In addition, some mutant sperm flagella of both species display a loss of proper microtubule structure, which was suggested to result from reduced autophagosomal-mediated degradation of a repressor of flagella formation in mice.21 In both moss and mouse mutant-sperm, nuclear packaging is also defective, which also may be attributed to reduced clearance of factors that repress this process. All these defects result in the loss of sperm motility and fertility and suggest that autophagy could have an ancestral function not only as a protector against nutrient stress, but also in gamete differentiation. This is intriguing, as it has been suggested that sexes and differently sized gametes (anisogamy) have evolved independently in the 2 kingdoms.56
The most prevalent autophagy-related structure detected in WT spermatids and egg cells resemble small vesicles delimited by a single-membrane with degrading material inside rather than canonical autophagosomes. These vesicles may represent one of 2 types of lytic way stations described previously: i) “autophagic vacuoles,” formed through the progressive maturation of autophagosomes into lytic compartments57 or ii) “autolysosome-like vesicles,” which are formed by the fusion of autophagosomes with small pre-existing lytic vesicles.46,58–62 We hypothesize that the empty vesicles identified in the atg5 germ cells are pre-existing lytic vesicles that, without the presence of autophagosomes, largely remain without cytoplasmic material. This suggests that the vesicles identified in WT germ cells should be regarded as autolysosome-like vesicles.
Despite the fact that we show that autophagy is needed for proper cytoplasmic reduction during spermatogenesis, it should also be emphasized that autophagy-deficient Ppatg5 spermatids undergo a limited reduction of cytoplasmic material. This suggests that there are additional mechanisms to reduce the cytoplasm that are independent of PpATG5 and the canonical macroautophagy pathway. While future work may reveal the details of such mechanisms, we suggest that 2 ultrastructural observations made in the Ppatg5 mutant could serve as a starting point for such studies. First, microautophagy mediated by lytic vesicles could be a contributing factor since it appears active in mutant egg cells and is believed to be independent of ATG5 activity in both animals and plants.63,64 Second, lytic vesicles in mutant spermatids might cluster and fuse to trap cytoplasmic portions destined for degradation and/or secretion. This is in analogy with an alternative autophagic pathway described previously in which small tubules containing hydrolases form in the cytoplasm in a collar-like structure that eventually merges to sequester the part of the cytoplasm that remains in the middle and that, in a final step, is degraded.65 The fact that these 2 phenomena were observed only in the Ppatg5 mutant is intriguing. It could indicate that they are slow processes detectable only in cells with an increased half-life of the lytic vesicles involved due to reduced macroautophagic flux. Alternatively, they may represent backup mechanisms induced only when canonical macroautophagy becomes a limiting factor.
Our data clearly indicate that the moss reproductive organs constitute a very suitable system for future in-depth studies of developmentally controlled autophagy in plants. In addition, as neither autophagosomes nor ATG proteins have previously been implicated in unconventional secretion in plants, our work indicates this is an area that should be given more attention. Our data also add further fuel to the interesting debate on the evolution of gametes and anisogamy in the animal and plant kingdoms.
Materials and Methods
Plant material and growth conditions
The P. patens subspecies patens WT strains Gransden and Reute were used in this study. Gransden 2014 is the background of lines GFP-PpATG8e-4, Ppatg5–1 and Ppatg5–2, while Reute 201430 is the background of lines GFP-PpATG8e-1, GFP-PpATG8e-2 and GFP-PpATG8e-3, Ppatg5–3, Ppatg5–4, Ppatg5–5 and all the Ppatg7 lines. Protonemal tissue was subcultivated on cellophane-covered plates containing routine basal medium (BCD)66 supplemented with 5 mM ammonium tartrate (Sigma Aldrich, A2956) and 0.8% agar (Sigma Aldrich, A1296) and grown at 25°C under constant white light from fluorescent tubes (FL40SS W/37; Toshiba) at 30 µmol m−2 s−1 in a Sanyo MLR-350 light chamber with side irradiation. For induction of reproductive organs, small pieces of protonemal tissue were shaped into 3 mm balls and inoculated on solid BCD medium in 25-mm-deep petri dishes (Phoenix Biomedical, 002). After growth under the above conditions for 5 wk, plates with gametophores already formed were transferred to gametangia-inducing conditions, 15°C and 8 h of light (30 µmol m−2 s−1) per day.
Gametophores were harvested between 19 and 25 d after induction of reproductive organs formation. For light microscopy, DAPI staining and TEM, leaves of the gametophores were detached under a dissecting microscope (Leica MZ16, Leica Biosystems, Heidelberg, Germany) to expose the reproductive organs.
Generation of PpATG8epro::GFP-PpATG8e reporter lines
First, the PpATG8e coding sequence was PCR amplified using the primers PpATG849_F_AscI and PpATG849_R_NotI (Table S1) and P. patens cDNA as the template. The resulting 394-base pair (bp) fragment was cloned by sticky end ligation in frame with the GFP coding sequence in the vector pTM13 after both the insert and vector had been cut with AscI and NotI (Thermo Fisher Scientific, FD1894 and FD0596). The pTM13 vector contains homology stretches for targeted integration into the neutral Pp108’ locus67 that flank the aforementioned GFP coding sequence and a hygromycin-resistance cassette driven by a 35S promoter. After this, the PpATG8e promoter was PCR-amplified using the primers PpATG849pro_F_SbfI and PpATG849pro_R_ BamHI (Table S1) and gDNA as the template. The resulting 1546 bp fragment was cloned by sticky-end ligation upstream of the GFP coding sequence in the previous product after both insert and vector had been cut with SbfI and BamHI (Thermo Fisher Scientific, FD1194 and FD0055). The resulting plasmid pCSK1 was linearized with SfiI (Thermo fisher, FD1824) before transformation68 into Reute WT, Gransden WT, Ppatg5–1 (Gransden background) and Ppatg5–3 (Reute background). Transformants were selected on 50 µg/ml hygromycin (Duchefa, H0192). List of primers used for confirmation of correct integration in stable lines can be found in Table S1 and location of the primers in the construct is shown in Figure S1B.
PpATG5 and PpATG7 gene disruption
PCR amplification of the 5’ and 3’flanking regions (1253 bp and 1260 bp respectively) of the PpATG5 gene (Table S2) using USER compatible primers pairs p21/p22 and p23/p24, respectively, (Table S1) rendered 2 PCR products, which were combined in a single, 4 fragment USER cloning step into pMBLU cut with the 2 restriction enzymes PacI and AsiSI (Thermo fisher Scientific, FD2204 and FD2094) and nicked with Nt.BbvCI.69 PCR amplification of 5′ and 3′ flanking regions (941 bp and 932 bp, respectively) of the PpATG7 gene was performed using primers p17/p18 and p19/p20 shown in Table S1. Fragments were digested with BamHI/XhoI and SpeI/NsiI respectively (Thermo Fisher Scientific, FD0055, FD0694, FD1254 and FD0734), and cloned by sequential sticky end ligation into the pMT164 plasmid.66 In both KO constructs, flanking regions were cloned upstream and downstream of the nptII gene driven by a 35S promoter (for geneticin resistance). Flanking regions ensure specific targeting of the PpATG5 and PpATG7 genes and their exchange for the nptII gene in the genome by homologous recombination. Overviews of PpATG5, PpATG7 and the resulting knockout constructs are depicted in Figure S2A. Constructs were linearized with SacI and BamHI/NsiI respectively before transformation into Reute WT and Gransden WT. Transformation was made as described previously.68 Stable Ppatg5 KO transformants were obtained in both the Gransden and Reute background and Ppatg7 KO in the Reute background. Transformants were selected on MM plates supplemented with geneticin (Thermo Fisher Scientific, 11811023) at a concentration of 50 µg/ml. List of primers used for confirmation of correct integration in stable lines can be found in Table S1 and annealing sites of the primers in the construct are shown in Figure S2A.
Quantitative real-time PCR
Based on sequence similarity to their Arabidopsis counterparts, 5 ATG8 genes were identified in the P. patens genome and selected for expression studies. To distinguish between different isoforms, primers annealing to less conserved regions were designed, such as parts of 5´UTR or 3´UTR depending on the gene. Primer3 software (primer3.ut.ee) was used for primer design (Table S1). Sizes of the PCR products varied between 85 bp and 154 bp. For analysis of PpATG8 expression in reproductive organs, 3 types of samples were harvested in the Gransden WT line: 80 tips of reproductive shoots carrying both archegonia and antheridia, ∼1000 isolated archegonia and 45 antheridia bundles (∼8 antheridia per bundle). Each type of sample was harvested in a microcentrifuge tube and flash frozen in liquid nitrogen. Tissue was grounded with a pestle in liquid nitrogen. RNA was isolated from the whole resulting ground tissue using the picoPure RNA kit (Thermo Fisher Scientific, KIT0204) according to the manufacturer's protocol and RNA concentration was measured with a nanodrop instrument (Thermo Fisher Scientific, Gothenburg, Sweden). For the analysis of gene expression in the Ppatg5 lines compared with WT lines (both the Gransden and Reute background), 3 independent protonema colonies were harvested separately in microcentrifuge tubes, as independent biological replicates. Tissue was flash frozen in liquid nitrogen and grounded with a pestle. RNA was extracted with the RNeasy plant mini kit (Qiagen, 74903) according to the manufacturer's protocol and RNA concentration was measured with a nanodrop (Thermo Fisher Scientific). On column DNAse treatment was performed in all samples (Qiagen, 79254). 250 ng of RNA was used for cDNA synthesis using the Superscript III reverse transcript kit (Thermo Fisher Scientific, 18080051) following manufacturer's instructions and 1/80th of each reverse-transcription reaction was used for quantitative real-time PCR (QRT-PCR) analysis. Maxima SYBR green fluorescein QRT-PCR master mix (Thermo Fisher Scientific, K0241) was used for QRT-PCR. Reactions were run on an iQ5 Real-Time PCR system (Bio-Rad, Sundbyberg, Sweden) with the following protocol: 95°C, 10 min, 40 cycles of 95°C, 15 sec and 60°, 1 min, 95°C, 1 min. iCycler iQ Optical System Software Version 3.1 and ∆∆CT was used to quantify fold expression of Ppatg8 in KO lines relative to WT. Three genes, previously shown to be stably expressed in P. patens,70 were tested as reference genes: actin, adenine phosphoribosyl transferase (Ade-PRT) and serine threonine protein phosphatase 2a regulatory subunit (ST-P2a). Analysis applying NormFinder71 showed Ade-PRT and actin as the pair with the most stable expression (smallest stability value, SV) in our samples, so normalization was done against them (actin SV: 0,017, Ade-PRT SV: 0,016, ST-P2a: 0,023; combined SV Actin-Ade-PRT: 0,009). For analysis of PpATG8 gene expression in reproductive organs, ∆Ct was performed using actin as a reference.
Light microscopy
A Leica DMI4000B microscope (Leica Biosystems, Heidelberg, Germany) with differential interference contrast (DIC, Nomarski) optics was used for reproductive organ observations. Photographs were taken with a Leica DFC360FX camera (Leica Biosystems, Heidelberg, Germany) and the LAS AF (Leica Microsystems) software.
Sample staining and confocal microscopy
Samples were fixed in 4% paraformaldehyde (Sigma Aldrich, 158127) and 50 mM sodium cacodylate (Sigma Aldrich, C0250) under vacuum for 2 × 15 min on ice followed by overnight incubation at 4°C. Three washes with phosphate-buffered saline were performed. For visualization of sperm cytoplasmic content, samples were mounted in 10 µl phosphate-buffered saline (1.37 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4, pH 7.4) supplemented with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma Aldrich, D9542) and 100 µM nonyl-acridine orange (NAO; Themo Fisher Scientific, A1372) on a glass slide and scanned after 30 min. For stage specification of GFP-ATG8 expressing cells, fixed samples were stained with DAPI overnight and washed 3 times in phosphate buffer, then they were mounted in a drop of Vectashield mounting medium for fluorescence detection (Vector Laboratories, H1000). Samples were scanned making z-stack scanning using a Zeiss 780 confocal microscope (Carl-Zeiss AB, Sweden), with excitation laser of 488 nm and 405 nm and emission filters of 499 to 561 nm and 410 to 492 nm, for NAO or GFP and DAPI, respectively. Maximum projection and 3D reconstruction was made with ZEN Black software from Carl-Zeiss.
Transmission electron microscopy
For TEM, 8 different WT shoots and 4 shoots per KO line in the Gransden background (8 in total) were analyzed. Samples were fixed with 2.5% glutaraldehyde (Merck, 104239) and 50 mM sodium cacodylate buffer (pH 7.4) under vacuum for 2 × 15 min at 4°C, then left in fixative for at least 24 h. Fixed samples were washed with buffer for 10 min and then post-fixed in buffered 1% OsO4 (Sigma Aldrich, 419494) for 60 min. After washing them in buffer, samples were dehydrated through an ethanol series (Solveco, 10018; 70%, 95% each step 30 min; 100% for 60 min and 100% for 30 min) and incubated in propylene oxide (TAAB, P021) for 10 min and in a solution of Spurr epoxy resin (Sigma Aldrich, EM0300) and propylene oxide (1:1) in a desiccator for 1 h; the Spurr epoxy resin was prepared with the following recipe; 4.1 g 3,4-Epoxycyclohexylmethyl-3’,4’-epoxycyclohexane carboxylate (ERL4221), 1.2 g diglycil ether of polypropylene glycol (D.E.R. 736), 5.9 g nonenyl succinic anhydride (NSA) and 0.1 g dimethylaminoethanol (DMAE). Samples were incubated in pure Spurr epoxy resin overnight and polymerization was performed at 60°C for 48 h. Ultrathin sections (50 to 70 nm) were obtained and were stained with uranyl acetate (Sigma Aldrich Aldrich, 73943) and lead citrate (Acros Organics, 1830) for 20 min each. Sections were examined with a Technai G2 (FEI, Trondheim, Norway) transmission electron microscope.
Fertility studies
For self-fertilization experiments, gametophore colonies were fully covered with water for 2 min on d 20 after transition to reproductive organ induction conditions. After that time, excess of water was removed with a pipette. For cross-fertilization experiments, half a colony of WT (Reute) was located together with half a colony of each of the different Reute KO lines (Ppatg5–3, Ppatg5–4 and Ppatg5–5) in a petri dish (one combination per petri dish) and covered with water on d 20 after reproductive organ induction. To enhance cross-fertilization, the water was not removed from the plate until the next day. The presence of sporophytes in the gametophores was analyzed 40 d after induction of reproductive organs.
For spore analysis, sporophytes were harvested at the stage of full maturation. We considered that this stage was reached when the structure had brown color and totally round shape. Mature sporophytes were then excised, incubated for 4 min in 70% ethanol and rinsed 3 times with sterile water at room temperature to surface sterilize. Sporophytes were kept at 4ºC for at least 2 wk and then spores were released by crushing the sporophyte capsule with a pipette tip. The spores were planted on plates with BCD medium supplemented with 5 mM ammonium tartrate, covered by cellophane and eventually transferred to 50 µg/ml geneticin supplemented plates once germination happened.
Statistical analysis
Microsoft Excel was used to calculate means and standard deviations for all data presented. Microsoft Excel was also used to perform a Student t test of data presented in Fig. 2B, Fig. 3K, L and S5D. The software R was used to make a Dunn´s test based on Kruskal-Wallis with a Bonferroni adjustment for data presented in Fig. S5C.
Supplementary Material
Abbreviations
- Ade-PRT
adenine phosphoribosyl transferase
- ATG
autophagy related
- BCD
routine basal medium for moss
- bp
base pair
- DAPI
4′,6-diamidino-2-phenylindole
- Gd
Gransden
- GFP
green fluorescent protein
- KO
knockout
- P. patens
Physcomitrella patens, Physcomytrella patens
- PCD
programmed cell death
- PCR
polymerase chain reaction
- PM
plasma membrane
- QRT-PCR
quantitative real-time PCR
- NAO
10N-nonyl-acridine orange
- Rt
Reute
- ST-P2a
serine threonine protein phosphatase 2a regulatory subunit
- SV
stability value
- TEM
transmission electron microscopy
- WT
wild type
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank Professors Daniel Hofius and Peter Bozhkov and 3 anonymous reviewers for scientific advice and critical reading of the manuscript. We thank Professor Stefan Rensing for kindly providing the Reute ecotype. We also thank BioVis at Uppsala University and SciLifeLab for access to their TEM facility and for technical support.
Funding
This work was supported by grants to ES from the Knut and Alice Wallenberg Foundation (# 2012.0087) and the Swedish Research Council (# 621–2014–4941), to MT from the Carl Trygger Foundation (12:493, 14:481), the Nilsson-Ehle Endowments and the Olle Engkvist Foundation and to JM from the Danish Research Council on Nature and the Universe (#1323–00267) and the Indo-Danish Research Program (2 research-exchange grants).
References
- [1].Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. PMID:8224160 [DOI] [PubMed] [Google Scholar]
- [2].Thompson AR, Vierstra RD. Autophagic recycling : lessons from yeast help define the process in plants. Curr Opin Plant Biol.2005;8:165–73. doi: 10.1016/j.pbi.2005.01.013. PMID:15752997 [DOI] [PubMed] [Google Scholar]
- [3].Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. PMID:17204848 [DOI] [PubMed] [Google Scholar]
- [4].Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016;428:1681–99. doi: 10.1016/j.jmb.2016.02.021. PMID:26908221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–93. doi: 10.1104/pp.011024. PMID:12114572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–14. doi: 10.1074/jbc.M204630200. PMID:12070171 [DOI] [PubMed] [Google Scholar]
- [7].Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in arabidopsis. Plant Physiol. 2007;143:291–9. doi: 10.1104/pp.106.092106. PMID:17098847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–7. doi: 10.4161/auto.5056. PMID:17932459 [DOI] [PubMed] [Google Scholar]
- [9].Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NHT, Mattsson O, Jørgensen LB, Jones JDG, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in arabidopsis. Cell. 2009;137:773–83. doi: 10.1016/j.cell.2009.02.036. PMID:19450522 [DOI] [PubMed] [Google Scholar]
- [10].Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux-Daubresse C. Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 2013;199:683–94. doi: 10.1111/nph.12307. PMID:23647084 [DOI] [PubMed] [Google Scholar]
- [11].Zhou J, Wang J, Cheng Y, Chi Y, Fan B, Yu J, Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. 2013;9:e1003196. doi: 10.1371/journal.pgen.1003196. PMID:2334177920659276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kwon S Il, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010;64:151–64. doi: 10.1111/j.1365-313X.2010.04315.x. PMID:20659276 [DOI] [PubMed] [Google Scholar]
- [13].Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, et al.. Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol. 2013;203:917–27. doi: 10.1083/jcb.201307082. PMID:24344187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis Homolog of Yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143:1132–9. doi: 10.1104/pp.106.093864. PMID:17259285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qin G, Ma Z, Zhang L, Xing S, Hou X, Deng J, Liu J, Chen Z, Qu L-J, Gu H. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007;17:249–63. doi: 10.1038/cr.2007.7. PMID:17339883 [DOI] [PubMed] [Google Scholar]
- [16].Harrison-Lowe NJ, Olsen LJ. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy. 2008;4:339–48. doi: 10.4161/auto.5629. PMID:18227644 [DOI] [PubMed] [Google Scholar]
- [17].Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, et al.. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014;10:878–88. doi: 10.4161/auto.28279. PMID:24674921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Herpin A, Englberger E, Zehner M, Wacker R, Gessler M, Schartl AM. Defective autophagy through epg5 mutation results in failure to reduce germ plasm and mitochondria. FASEB J. 2015;29:4145–61. doi: 10.1096/fj.14-265462. PMID:26183773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, Yang L, Tang H, Zhang X, Duan E, et al.. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:1–18. doi: 10.1038/cr.2014.70. PMID:24384820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Song Z-H, Yu H-Y, Wang P, Mao G-K, Liu W-X, Li M-N, Wang H-N, Shang Y-L, Liu C, Xu Z-L, et al.. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis. 2015;6:e1589. doi: 10.1038/cddis.2014.559. PMID:25590799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shang Y, Wang H, Jia P, Zhao H, Liu C, Liu W, Song Z, Xu Z, Yang L, Wang Y, et al.. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy. 2016;12:1–18. doi: 10.1080/15548627.2016.1192750. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Miller CCJ, Duckett JG. Cytoplasmic deletion processes during spermatogenesis in mosses. Gamete Res. 1986;13:253–70. doi: 10.1002/mrd.1120130308 [DOI] [Google Scholar]
- [23].Landberg K, Pederson ER a, Viaene T, Bozorg B, Friml J, Jönsson H, Thelander M, Sundberg E. The MOSS Physcomitrella patens reproductive organ development is highly organized, affected by the 2 SHI/STY genes and by the level of active auxin in the SHI/STY expression domain. Plant Physiol. 2013;162:1406–19. doi: 10.1104/pp.113.214023. PMID:23669745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mukae K, Inoue Y, Moriyasu Y. ATG5-knockout mutants of Physcomitrella provide a platform for analyzing the involvement of autophagy in senescence processes in plant cells. Plant Signal Behav. 2015;10:e1086859. doi: 10.1080/15592324.2015.1086859. PMID:26368055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is Traced with Apg8 / Aut7p in yeast. Jcb. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. PMID:10525546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. Erratum: LC3, a mammalian homolog of yeast Apg8p, is localized in autophagosome membranes after processing (EMBO Journal (2000) 19 (5720-5728)). EMBO J. 2003;22:4577. doi: 10.1093/emboj/cdg454. PMID:11060023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–83. doi: 10.1105/tpc.104.025395.et. PMID:15494556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Klionsky DJ, Abdelmohsen K, Abe A, Abedin J, Abeliovich H, Arozena AA, Adachi H, Adams M, Adams PD, Adeli K, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xia T, Xiao D, Liu D, Chai W, Gong Q, Wang NN. Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS One. 2012;7:e37217. doi: 10.1371/journal.pone.0037217. PMID:22629371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hiss M, Meyberg R, Westermann J, Haas FB, Schneider L, Schallenberg-Rüdinger M, Ullrich KK, Rensing SA. Sexual reproduction, sporophyte development and molecular variation in the model moss Physcomitrella patens: introducing the ecotype Reute. Plant J. 2017;606:20–110. doi: 10.1104/pp.105.060673. PMID:28161906 [DOI] [PubMed] [Google Scholar]
- [31].Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–110. doi: 10.1104/pp.105.060673. PMID:16040659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Minina EA, Sanchez-Vera V, Moschou PN, Suarez MF, Sundberg E, Weih M, Bozhkov PV. Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging Cell. 2013;12:327–9. doi: 10.1111/acel.12048. PMID:23331488 [DOI] [PubMed] [Google Scholar]
- [33].Renzaglia KS, Garbary DJ. Motile gametes of land plants: diversity, development, and evolution. CRC Crit Rev Plant Sci. 2001;20:107–213. doi: 10.1080/20013591099209 [DOI] [Google Scholar]
- [34].Petit JM, Maftah A, Ratinaud MH, Julien R. 1ON-Nonyl acridine orange interacts with cardiolipin and allows the quantification. Eur J Biochem. 1992;209:267–73. doi: 10.1016/0006-291X(89)91700-2. PMID:1396703 [DOI] [PubMed] [Google Scholar]
- [35].Bernhard DL, Renzaglia KS. Spermiogenesis in the Moss Aulacomnium palustre. Bryologist. 1995;98:52–70. http://www.jstor.org/stable/3243642. doi: 10.2307/3243642 [DOI] [Google Scholar]
- [36].Paolillo DJ, Kreitner GL, Reighard JA. Spermatogenesis in Polytrichum juniperinum – I. The origin of the apical body and the elongation of the nucleus. Planta. 1968;78:248–61. doi: 10.1007/BF00386424. PMID:24522733 [DOI] [PubMed] [Google Scholar]
- [37].Paolillo DJ, Kreitner GL, Reighard JA. Spermatogenesis in Polytrichum juniperinum – II. The mature sperm. Planta. 1968;78:248–61. http://www.jstor.org/stable/23367021. doi: 10.1007/BF00386425. PMID:24522733 [DOI] [PubMed] [Google Scholar]
- [38].Paolillo DJ Jr., Cukierski M. Wall developments and coordinated cytoplasmic changes in spermatogenous cells of Polytrichum (Musci). Bryologist. 1976;79:466–79. http://www.jstor.org/stable/3241940. doi: 10.2307/3241940 [DOI] [Google Scholar]
- [39].Carothers ZB, Duckett JG. The bryophite spermatozoid: a source of new phylogenetic information. Bull Torrey Bot Club. 1980;107:281–97. http://www.jstor.org/stable/2484152. doi: 10.2307/2484152 [DOI] [Google Scholar]
- [40].Spitzer C, Li F, Buono R, Roschzttardtz H, Chung T, Zhang M, Osteryoung KW, Vierstra RD, Otegui MS. The Endosomal Protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in arabidopsis. Plant Cell. 2015;27:391–402. http://www.ncbi.nlm.nih.gov/pubmed/25649438. doi: 10.1105/tpc.114.135939. PMID:25649438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Calvo-Garrido J, Escalante R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy. 2010;6:100–9. doi: 10.4161/auto.6.1.10697. PMID:20009561 [DOI] [PubMed] [Google Scholar]
- [42].Matthias J, Meßling S, Eichinger L. The two Dictyostelium autophagy eight proteins, ATG8a and ATG8b, associate with the autophagosome in succession. Eur J Cell Biol. 2016;95:15–25. doi: 10.1016/j.ejcb.2015.10.007. PMID:26697781 [DOI] [PubMed] [Google Scholar]
- [43].Su W, Ma H, Liu C, Wu J, Yang J. Identification and characterization of two rice autophagy associated genes, OsAtg8 and OsAtg4. Mol Biol Rep. 2006;33:273–8. doi: 10.1007/s11033-006-9011-0. PMID:17082902 [DOI] [PubMed] [Google Scholar]
- [44].Kuzuoglu-Ozturk D, Yalcinkaya OC, Akpinar BA, Mitou G, Korkmaz G, Gozuacik D, Budak H. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta. 2012;236:1081–92. doi: 10.1007/s00425-012-1657-3. PMID:22569921 [DOI] [PubMed] [Google Scholar]
- [45].Kadandale P, Stender JD, Glass CK, Kiger AA. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc Natl Acad Sci U S A. 2010;107:10502–7. doi: 10.1073/pnas.0914168107. PMID:20498061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. PMID:18454133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47:1641–52. doi: 10.1093/pcp/pcl031. PMID:17085765 [DOI] [PubMed] [Google Scholar]
- [48].Yano K, Suzuki T, Moriyasu Y. Constitutive autophagy in plant root cells. Autophagy. 2007;3:360–2. doi: 10.4161/auto.4158%0A. PMID:17426438 [DOI] [PubMed] [Google Scholar]
- [49].Lal M, Bell PR. Aspects of the Differentiation of the egg of the moss physcomitrium coorgense broth. Ann Bot. 1977;41:127–31. http://www.jstor.org/stable/42770131. doi: 10.1093/oxfordjournals.aob.a085260 [DOI] [Google Scholar]
- [50].Lal M, Kraur G, Chauhan E. Ultrastructural studies on archegonial development in the moss Physcomitrium cyathicarpum. New Phytol. 1982;92:441–52. http://www.jstor.org/stable/2432254. doi: 10.1111/j.1469-8137.1982.tb03402.x [DOI] [Google Scholar]
- [51].Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al.. Autophagy contributes to leaf starch degradation. Plant Cell. 2013;25:1383–99. doi: 10.1105/tpc.112.108993. PMID:23564204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bruns C, Mccaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol. 2011;195:979–92. doi: 10.1083/jcb.201106098. PMID:22144692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Manjithaya R, Subramani S. Autophagy: A broad role in unconventional protein secretion? Trends Cell Biol. 2011;21:67–73. doi: 10.1016/j.tcb.2010.09.009. PMID:20961762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan J-L, Saitoh T, Akira S, Seglen PO, et al.. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130–44. doi: 10.1038/emboj.2013.233. PMID:24185898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Torisu T, Torisu K, Lee IH, Liu J, Malide D, Combs CA, Wu XS, Rovira II, Fergusson MM, Weigert R, et al.. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med. 2013;19:1281–7. doi: 10.1038/nm.3288. PMID:24056772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Umen JG. Evolution of sexes from an ancestral mating-type specification pathway. PLoS Biol. 2014;12:e1001904. doi: 10.1371/journal.pbio.1001904. PMID:25003332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell. 2006; 98:53–67. doi: 10.1042/BC20040516. PMID:16354162 [DOI] [PubMed] [Google Scholar]
- [58].Toyooka K, Moriyasu Y, Goto Y, Takeuchi M, Fukuda H, Matsuoka K. Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy. 2006;2:96–106. doi: 10.4161/auto.2.2.2366. PMID:16874101 [DOI] [PubMed] [Google Scholar]
- [59].Kwon S Il, Cho HJ, Kim SR, Park OK. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in arabidopsis. Plant Physiol. 2013;161:1722–36. doi: 10.1104/pp.112.208108. PMID:23404918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Oh-ye Y, Inoue Y, Moriyasu Y. Detecting autophagy in Arabidopsis roots by membrane-permeable cysteine protease inhibitor E-64d and endocytosis tracer FM4-64. Plant Signal Behav. 2011;6:1946–9. doi: 10.4161/psb.6.12.18297. PMID:22105025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–77. doi: 10.1016/j.cell.2005.03.007. PMID:15907470 [DOI] [PubMed] [Google Scholar]
- [62].Bassham DC. Plant autophagy-more than a starvation response. Curr Opin Plant Biol. 2007;10:587–93. doi: 10.1016/j.pbi.2007.06.006. PMID:17702643 [DOI] [PubMed] [Google Scholar]
- [63].Mukherjee A, Patel B, Koga H, Cuervo AM, Jenny A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016;12:1–16. doi: 10.1080/15548627.2016.1208887. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chanoca A, Kovinich N, Burkel B, Stecha S, Bohorquez-Restrepo A, Ueda T, Eliceiri KW, Grotewold E, Otegui MS. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell. 2015;27:2545–59. doi: 10.1105/tpc.15.00589. PMID:26342015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Van Doorn WG, Papini A. Ultrastructure of autophagy in plant cells: A review. Autophagy. 2013;9:1922–36. doi: 10.4161/auto.26275. PMID:24145319 [DOI] [PubMed] [Google Scholar]
- [66].Thelander M, Nilsson A, Olsson T, Johansson M, PA G, DG S, JP Z, Ronne H. The moss genes PpSKI1 and PpSKI2 encode nuclear SnRK1 interacting proteins with homologues in vascular plants. Plant Mol Biol. 2007;64:559–73. doi: 10.1007/s11103-007-9176-5. PMID:17533513 [DOI] [PubMed] [Google Scholar]
- [67].Schaefer DG, Zryd J-P. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–206. doi: 10.1046/j.1365-313X.1997.11061195.x. PMID:9225463 [DOI] [PubMed] [Google Scholar]
- [68].Schaefer D, Zryd JP, Knight CD, Cove DJ. Stable transformation of the moss Physcomitrella patens. Mol Gen Genet. 1991;226:418–24. doi: 10.1007/BF00260654. PMID:2038304 [DOI] [PubMed] [Google Scholar]
- [69].Bressendorff S, Azevedo R, Kenchappa CS, Ponce de Leon I, Olsen J V, Rasmussen MW, Erbs G, Newman M-A, Petersen M, Mundy J. An innate immunity pathway in the moss Physcomitrella patens. Plant Cell. 2016;28:1328–42:tpc.00774.2015. doi: 10.1105/tpc.15.00774. PMID:27268428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Le Bail A, Scholz S, Kost B. Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in physcomitrella patens gametophytes. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0070998. PMID: 2395106315289330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR Data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245. LP-5250. http://cancerres.aacrjournals.org/content/64/15/5245.abstract. doi: 10.1158/0008-5472.CAN-04-0496. PMID:15289330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.