Abstract
Chromatin is organized and compacted in the nucleus through the association of histones and other proteins, which together control genomic activity. Two broad types of chromatin can be distinguished: euchromatin, which is generally transcriptionally active, and heterochromatin, which is repressed. Here we examine the current state of our understanding of repressed chromatin in Caenorhabditis elegans, focusing on roles of histone modifications associated with repression, such as methylation of histone H3 lysine 9 (H3K9me2/3) or the Polycomb Repressive Complex 2 (MES-2/3/6)-deposited modification H3K27me3, and on proteins that recognize these modifications. Proteins involved in chromatin repression are important for development, and have demonstrated roles in nuclear organization, repetitive element silencing, genome integrity, and the regulation of euchromatin. Additionally, chromatin factors participate in repression with small RNA pathways. Recent findings shed light on heterochromatin function and regulation in C. elegans, and should inform our understanding of repressed chromatin in other animals.
Keywords: C. elegans, chromatin, heterochromatin, histone methylation, H3K9me, H3K27me, WormBook
EUKARYOTIC DNA is organized and compacted in the nucleus through its association with histones and nonhistone proteins, forming a complex called chromatin. The N-terminal tails of all histones, as well as the C-terminal tails of histones H2A and H2B, are subject to post-translational modifications that selectively impact many aspects of nuclear function. The most common histone modifications include methylation, acetylation, ubiquitination, sumoylation, and phosphorylation.
Two broad classes of chromatin, euchromatin and heterochromatin, can be distinguished based on protein composition, characteristic post-translational modifications on histones, and transcriptional activity. Euchromatin is either potentially or actively transcribed, and is enriched for RNA polymerase, histone tail acetylation, and trimethylation on histone H3 lysines 4 and 36 (H3K4me3 and H3K36me3). In contrast, heterochromatin tends to be transcriptionally repressed and is associated with histone methylations such as H3K9me3 or the Polycomb-deposited modification H3K27me3. The proteins that recognize these modified histones and the association of heterochromatin with the nuclear envelope help hold chromatin in a compact conformation, and promote the spread of the repressed chromatin state (Eskeland et al. 2007; Towbin et al. 2010; Elgin and Reuter 2013; Simon and Kingston 2013; Wiles and Selker 2016). Despite its heritable nature, heterochromatin is nonetheless dynamically regulated. Moreover, its tendency to aggregate and create repressive subnuclear compartments means that it can also indirectly influence the organization of euchromatin and gene expression (Francastel et al. 2000; Sexton et al. 2007).
Although there are many types of repressed chromatin, reflecting a range of modifications and ligands, heterochromatin was traditionally split into two classes that largely reflect the properties of the underlying DNA. On one hand, constitutive heterochromatin covered regions of the genome that are repeat rich and gene poor, and that are kept in a silent state throughout cell division and cell differentiation by H3K9me2/3 and its ligand, Heterochromatin Protein 1 (HP1) (Eissenberg and Elgin 2014; Saksouk et al. 2015; Wang et al. 2016). Facultative heterochromatin, on the other hand, encompasses genes that are potentially active, such as those with spatial, temporal, or other types of context-specific expression. Its hallmark is H3K27me3, which is deposited by Polycomb Repressive Complex 2 (PRC2), and which defines a pathway that maintains transcriptional repression (Wiles and Selker 2016).
Recent findings suggest that these classical distinctions are inadequate to describe the complexity of heterochromatin types. For instance, chromatin immunoprecipitation (ChIP) analyses in Caenorhabditis elegans have shown that much of the H3K9me3-marked chromatin coincides with H3K27me3 (Liu et al. 2011). In mammals, H3K9me3 and H3K27me3 are negatively correlated when scored for coincidence on the same histone tail, yet some genomic regions carry both modifications, as detected by ChIP. Additionally, cooperation between H3K9 and H3K27 methylation in heterochromatin formation has been reported (Hawkins et al. 2010; Boros et al. 2014; Schwammle et al. 2016). Finally, H3K23me2 has been reported to coincide independently with K9 and K27 methylation on histone H3 tails in C. elegans (Vandamme et al. 2015; Sidoli et al. 2016), much like the trimethylation of H4K20 in mammals, which accumulates on both facultative and constitutive heterochromatin during cellular senescence (Nelson et al. 2016). Here, we focus on histone H3K9 and H3K27 methylations, as they are the best-understood heterochromatin marks, and are involved in genetically distinct but highly conserved pathways of transcriptional repression.
Histone Methyltransferases and Recognition of Modifications
The enzymes responsible for histone lysine methylation are called histone methyltransferases (HMTs). HMTs typically contain a conserved catalytic domain called SET, which stems from Su(var)3-9, Enhancer of zeste, and Trithorax, the first HMTs known to carry this domain (Tschiersch et al. 1994). The SET domain contains a S-adenosylmethionine (SAM)-binding site and a catalytic center (Yeates 2002). The C. elegans genome encodes 38 SET domain-containing, putative HMTs (Andersen and Horvitz 2007). A loss-of-function mutant has been isolated for 30 of these, of which five are essential for viability (Andersen and Horvitz 2007; Ni et al. 2012). The preferred substrates of most HMTs have not yet been identified in C. elegans, apart from: SET-25 and MET-2/SETDB1, which target H3K9 (Bessler et al. 2010; Towbin et al. 2012); MES-2/EZH2, which modifies H3K27 (Bender et al. 2004); SET-1 and SET-4, which modify H4K20 (Vielle et al. 2012); and MET-1 and MES-4, which are responsible for H3K36 methylation (Bender et al. 2006; Furuhashi and Kelly 2010; Rechtsteiner et al. 2010). One cannot exclude the possibility that these HMTs (summarized in Table 1 and Table 2) modify lysines in other proteins as well.
Table 1. Chromatin proteins discussed in this review.
| Protein | Ortholog | Domains | Description |
|---|---|---|---|
| Histone methyltransferases | |||
| MET-2 | SETDB1 | SET, MBD, Pre-SET, Post-SET | H3K9me1/2 HMT |
| SET-25 | G9a/SUV39 SET) | SET Post-SET | H3K9me1/2/3 HMT |
| SET-32 | SET | Putative H3K9me3 HMT | |
| SET-1 | PR-Set7/SETD8 | SET | H4K20me1 HMT |
| SET-4 | SET4-20 | SET | H4K20me2/3 HMT |
| MES-4 | NSD1-3 | SET, Post-SET, PHD | H3K36me2/3 HMT |
| MET-1 | SET-2 | SET | H3K36me3 HMT |
| PRC2-like complex | |||
| MES-2 | EZH2 | SET, CXC | PRC2 complex/H3K27 HMT |
| MES-3 | component of PRC2 complex | ||
| MES-6 | ESC/EED | WD40 | Component of PRC2 complex |
| Histone demethylases | |||
| SPR-5 | KDM1A, LSD1 | SWRIM, amino oxidase | H3K4 demethylase |
| JMJD-1.2 | PHF8 | PHD, JmjC | H3K9me2/H3K27me2 and H3K23 demethylase |
| JMJD-3.1 | KDM6B, JMJD3 | JmjC | H3K27me2/3 demethylase |
| UTX-1 | KDM6A, UTX | JmjC, TPR repeat | H3K27me2/3 demethylase |
| Heterochromatin-associated proteins | |||
| HPL-1 | HP1 | Chromo, chromoshadow | Binds HIS-24/H1K14me1 in vitro |
| HPL-2 | HP1 | Chromo, chromoshadow | Binds H3K9me1/2/3 in vitro |
| CEC-3 | Chromo | Binds H3K9me1/2/3 in vitro | |
| CEC-4 | Chromo | Binds H3K9me1/2/3 in vitro and in vivo | |
| LIN-61 | MBT | Binds H3K9me1/2/3 in vitro | |
| LIN-13 | C2H2 and RING/FYVE/PHD-type zinc fingers | In complex with HPL-2 and LIN-61 | |
| LET-418 | Mi-2, CHD3 | PHD, chromo, Helicase_C, SNF2_N, CHDCT2 | Nucleosome remodelling component of NuRD and Mec complexes |
| Nuclear lamina proteins | |||
| LMN-1 | Lamin A and B | Coiled-coil, Ig, and CAAX box | Nuclear intermediate filament protein |
| LEM-2 | MAN1 | LEM, Man1-Src1p-C-term | Lamin-binding INM protein |
| EMR-1 | Emerin | LEM, Man1-Src1p-C-term | Lamin-binding INM protein |
| SUN-1 | SUN1,2,3,5 | SUN | INM-spanning protein that binds KASH domain |
| UNC-84 | SUN1,2,3,5 | SUN | INM-spanning protein that binds KASH domain |
| BAF-1 | BANF1, BAF | BAF | dsDNA and lamin and LEM domain ligand |
| Small RNA pathway proteins | |||
| PRG-1 | Piwi | PAZ, Piwi | piRNA pathway argonaute |
| NRDE-1 | Novel nuclear RNAi factor | ||
| NRDE-2 | NRDE2 | NRDE-2 | Nuclear RNAi factor, interacts with NRDE-3 |
| NRDE-3 | PAZ, Piwi | Somatic nuclear RNAi argonaute | |
| NRDE-4 | Novel nuclear RNAi factor | ||
| HRDE-1 | PAZ, Piwi | Germ line nuclear RNAi argonaute | |
| MORC-1 | MORC1, MORC2 | HATPase_c | Nuclear RNAi pathway effector |
The C. elegans genome contains 38 SET domain proteins, 6 amino oxidase-type putative histone demethylases, 14 jmjC domain proteins, 67 putative histone mark readers (bearing either a chromodomain, Tudor, MBT, PHD, or WD-40 domain), 27 argonaute domain proteins, and an as yet undetermined number of nuclear lamina-associated proteins [for a more complete survey of nuclear envelope proteins see Dobrzynska et al. (2016)]. See text for references and types of data supporting these definitions. In the case of HMTs and histone mark readers, only a few are supported by mass spectrometric data, point mutations within the active domain, and/or an exhaustive analysis of potential ligands. Data based on genetic phenotypes and colocalization should be considered suggestive but not conclusive. SET, Su(var)3-9, Enhancer of zeste, and Trithorax; HMT, histone methytransferases; PHD, plant homeodomain; TPR, tetratricopeptide repeat; MBT, malignant brain tumor; RING; FYVE, Fab 1, YOTB, Vac 1 and EEA1; LEM, Lamin and Emerin; INM, inner nuclear membrane; SUN, Sad1/UNC-84-homology; KASH, Klarsicht/ANC-1/Syne-1 homology; BAF, barrier-to-autointegration factor; PAZ, Piwi Argonaut and Zwille; RNAi, RNA interference; piRNA, piwi-interacting RNA.
Table 2. Loss-of-function phenotypes of genes discussed in this review.
| Loss-of-function phenotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Superficially wild-type | Sterile | Lethal | Increased germ line apoptosis | L1 arresta | synMuv | Increased mutation rate | Somatic expression of germ line genes | Heterochromatic array desilencing | Other phenotypes | References |
| Histone methyltransferases | |||||||||||
| met-2 | x | mrt | x | x | x | Yes | Satellite repeat transcription | Poulin et al. (2005), Andersen and Horvitz (2007), Bessler et al. (2010), Koester-Eiserfunke and Fischle (2011), Towbin et al. (2012), Wu et al. (2012), Zeller et al. (2016), J. Padeken, P. Zeller, and S. M. Gasser, (unpublished results) | |||
| set-25 | x | x | Yes | Retrotransposon expression | Pothof et al. (2003), Andersen and Horvitz (2007), Towbin et al. (2012), Zeller et al. (2016), McMurchy et al. (2017), J. Padeken, P. Zeller, and S. M. Gasser, (unpublished results) | ||||||
| set-32 | x | mrt | Nuclear RNAi and piRNA pathway effector | Andersen and Horvitz (2007), Ashe et al. (2012), Kalinava et al. (2017), Spracklin et al. (2017) | |||||||
| set-1 | ste | Dosage compensation defect | Vielle et al. (2012), Wells et al. (2012), Kramer et al. (2016) | ||||||||
| set-4 | x | Vielle et al. (2012), Wells et al. (2012), Kramer et al. (2016) | |||||||||
| mes-4 | mes | Yes | synMuv suppressor | Capowski et al. (1991), Cui et al. (2006), Towbin et al. (2012) | |||||||
| met-1 | x | x | No | Andersen and Horvitz (2007), Towbin et al. (2012), Wu et al. (2012) | |||||||
| met-2 set-25 | x | mrt | x | x | Yes | Loss of heterochromatin anchoring; repeat element expression | Towbin et al. (2012), Zeller et al. (2016), McMurchy et al. (2017) | ||||
| PRC2-like complex | |||||||||||
| mes-2 | mes | Yes | Capowski et al. (1991), Towbin et al. (2012) | ||||||||
| mes-3 | mes | Yes | Capowski et al. (1991), Towbin et al. (2012) | ||||||||
| mes-6 | mes | Yes | Capowski et al. (1991), Towbin et al. (2012) | ||||||||
| Histone demethylases | |||||||||||
| spr-5 | x | mrt | Katz et al. (2009), Greer et al. (2016) | ||||||||
| jmjd-1.2 | x | Locomotion; mitochondrial stress-induced longevity defect; DNA repair defect | Lee et al. (2015), Merkwirth et al. (2016), Riveiro et al. (2017) | ||||||||
| jmjd-3.1 | Abnormal gonad development; transdifferentiation defect | Agger et al. (2007), Zuryn et al. (2014) | |||||||||
| utx-1 | steb | x | Wang et al. (2010), Vandamme et al. (2012) | ||||||||
| Heterochromatin-associated proteins | |||||||||||
| hpl-1 | x | Schott et al. (2006) | |||||||||
| hpl-2 | stec | x | x | x | x | Yes | Capowski et al. (1991), Couteau et al. (2002), Wang et al. (2005), Schott et al. (2006), Petrella et al. (2011), Towbin et al. (2012), Wu et al. (2012), McMurchy et al. (2017) | ||||
| cec-3 | x | Suppresses spr-5 Transgenerational sterility; unc-4 ectopic expression | Zheng et al. (2013), Greer et al. (2014) | ||||||||
| cec-4 | x | No | Loss of perinuclear chromatin anchoring | Gonzalez-Sandoval et al. (2015) | |||||||
| lin-61 | x | x | x | x | x | x | Yes | Compromised homology-driven repair | Pothof et al. (2003), Poulin et al. (2005), Harrison et al. (2007), Koester-Eiserfunke and Fischle (2011), Petrella et al. (2011), Towbin et al. (2012), Wu et al. (2012), Johnson et al. (2013), McMurchy et al. (2017) | ||
| lin-13 | ste | x | x | x | x | Yes | Ferguson and Horvitz (1985), Melendez and Greenwald (2000), Wang et al. (2005), Petrella et al. (2011), Wu et al. (2012), McMurchy et al. (2017) | ||||
| let-418 | ste | x | x | x | No | Solari and Ahringer (2000), von Zelewsky et al. (2000), Unhavaithaya et al. (2002), Passannante et al. (2010), Wu et al. (2012) | |||||
| Small RNA pathway proteins | |||||||||||
| prg-1 | x | mrt | x | Batista et al. (2008), Das et al. (2008), Simon et al. (2014), McMurchy et al. (2017) | |||||||
| nrde-1 | x | mrt | Yes | Burkhart et al. (2011), Buckley et al. (2012), J. Padeken, P. Zeller, and S. M. Gasser, (unpublished results) | |||||||
| nrde-2 | x | mrt | Guang et al. (2010), Buckley et al. (2012) | ||||||||
| nrde-3 | x | mrt | Yes | Guang et al. (2008), J. Padeken, P. Zeller, and S. M. Gasser, (unpublished results) | |||||||
| nrde-4 | x | mrt | Burkhart et al. (2011), Buckley et al. (2012), J. Padeken, P. Zeller and S. M. Gasser, (unpublished results) | ||||||||
| hrde-1 | x | mrt | Ashe et al. (2012), Buckley et al. (2012), Luteijn et al. (2012), Shirayama et al. (2012) | ||||||||
| morc-1 | x | mrt | Spracklin et al. (2017), Weiser et al. (2017) | ||||||||
Common phenotypes observed in chromatin-modulating mutants are listed. Phenotypes listed are not comprehensive and were not assessed in all mutants. synMuv, synthetic multivulval; mrt, mortal germ line (transgenerationally sterile); RNAi, RNA interference; piRNA, piwi-interacting RNA; ste, zygotic sterile; mes, maternal-effect sterile.
Some mutants undergo L1 arrest only at high temperature (26°).
Homozygotes are nearly sterile, and the few embryos produced die.
Null mutant is sterile at 25°.
Co-RNAi or double mutant of lem-2 and emr-1 is lethal.
Histone modifications can directly alter nucleosome–nucleosome or nucleosome–DNA interactions by changing the charge of the highly basic histone tail or disrupting contact sites between DNA and the nucleosomal core particle. Alternatively, specific histone modifications can create binding sites for proteins that specifically recognize a given modified amino acid. These “readers” of post-translational histone modifications can in turn alter the chromatin compaction state, or recruit additional transcriptional regulators or chromatin-modifying enzymes. A growing list of structural motifs have been shown to recognize modified histones, the most common being Bromo, Chromo, Tudor, malignant brain tumour (MBT), plant homeodomain (PHD)PHD, WD40 repeat (∼40 amino acid terminating in Trp-Asp) 14-3-3, and BRCT (BRCA1 C Terminus) domains (Taverna et al. 2007). C. elegans has 67 proteins containing such domains, which are predicted to be readers of histone modifications (Towbin et al. 2012; Gonzalez-Sandoval et al. 2015). Histone H3 tail methylations alone are known to be recognized by Chromo, MBT, PWWP (Pro-TrpTrp-Pro motif) or Tudor domains, as well as by specialized WD40 repeat structures (Margueron et al. 2009; Xu et al. 2010; Khorasanizadeh 2011).
The C. elegans Genome and the Distribution of Heterochromatin
The 100-Mbp genome of C. elegans is separated into five autosomes and an X chromosome (C. elegans Sequencing Consortium 1998). Hermaphrodites are diploid for all six chromosomes, whereas males have five pairs of autosomes and only a single X chromosome. The regulation and characteristics of the autosomal chromosomes differ from the X chromosome, in part due to dosage compensation in the soma, which represses gene expression approximately twofold on the two hermaphrodite X chromosomes to match expression on the single male X. Here, we do not cover dosage compensation, but refer the reader to reviews of this subject (Meyer 2010; Strome et al. 2014).
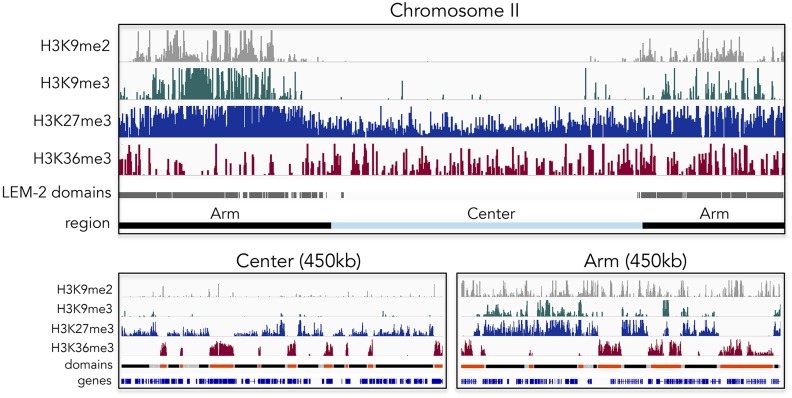
Like most nematodes, C. elegans chromosomes are holocentric (Albertson and Thomson 1982; Maddox et al. 2004). That is, centromere activity is distributed along chromosomes at sites of incorporation of a variant histone H3 CENP-A instead of being focused in a single, heritable centromeric site (Gassmann et al. 2012). The distal arms and central regions of the five autosomes have different characteristics. Most meiotic recombination occurs in the distal arm regions, where genes are on average longer and have larger introns. Genes in central regions are generally more highly expressed and show higher evolutionary conservation (C. elegans Sequencing Consortium 1998). These differences are reflected in the distributions of chromatin modifications. For example, average levels of modifications associated with gene activity are higher in central regions (Liu et al. 2011). Di- and trimethylation of H3K9, hallmarks of constitutive heterochromatin, are both predominantly found on distal chromosome arms, as is H3K9me1, although they have distinct distributions, reflecting distinct modes of recruitment of the relevant HMTs (Gu and Fire 2010; Liu et al. 2011) (J. Padeken, P. Zeller, and S. M. Gasser, unpublished results; Figure 1). Nonetheless, the distal arms are not devoid of transcribed genes, and those found interspersed among heterochromatic domains carry the same histone modifications as actively transcribed genes in central chromosomal domains (Liu et al. 2011).
Figure 1.
Histone modifications associated with heterochromatin and genome domains. Top: pattern of histone modifications associated with heterochromatin (H3K9me2, H3K9me3, and H3K27me3), H3K36me3, and LEM-2 domains (indicating nuclear lamina association) in embryos across C. elegans chromosome II. The majority of H3K9me marks and LEM-2 domains are found on the chromosome arms. H3K27me3 levels are higher on arm regions compared to the center. H3K36me3 shows a more uniform pattern, with a slight enrichment in the central region. Bottom left: 450-kb segment of a central region showing low H3K9 methylation, and anticorrelated domains marked by H3K27me3 and H3K36me3. H3K36me3 is found in active chromatin domains (orange) and H3K27me3 in regulated chromatin domains (black), which are separated by border regions (gray). Bottom right: 450-kb segment of an arm region. The distributions of H3K9me3 and H3K27me3 are largely similar to each other, but differ from H3K9me2. Patterns of H3K36me3 and H3K27me3 and chromatin domains are similar to those in central regions.
In contrast to constitutive heterochromatin, facultative heterochromatin carries H3K27 trimethylation, the most abundant histone methylation mark as measured by mass spectrometry in C. elegans. H3K27me3 is found on 67% of histones in embryos (Vandamme et al. 2015), and maps both to distal arms and central chromosome regions (Liu et al. 2011; Figure 1). Profiling early embryos and L3 larvae, levels were found to be higher on chromosome arms and higher on the X chromosome than on autosomes. On distal chromosome arms, H3K27me3 often colocalizes by ChIP sequencing with H3K9me3, as it does on large integrated arrays of transgenes, whereas in central regions of autosomes H3K27me3 is found without H3K9me3 (Meister et al. 2010; Liu et al. 2011) (Figure 1). Consistent with a role in transcriptional repression, the genome-wide distribution of H3K27me3 is anticorrelated with RNA levels and the presence of RNA polymerase (Liu et al. 2011).
While C. elegans has the repressive histone methyl marks and the ligands that are present in other organisms, it lacks 5-methyl cytosine on DNA and the 5meC-binding proteins that repress transcription in vertebrates. A very low level of adenine N(6)-methylation (6mA) has been reported on DNA in C. elegans (Greer et al. 2015), yet given that 6mA is the most abundant RNA modification, and no dedicated adenine methyltransferase for DNA has been identified, it is unclear whether 6mA-DNA has any physiological significance.
The Spatial Organization of Repetitive DNA
Roughly 20% of the C. elegans genome is repetitive DNA (Hillier et al. 2008; Hubley et al. 2016), including tandem repeats and sequences derived from DNA or RNA transposons. These repetitive elements are enriched in the distal arm regions, and are generally marked by nucleosomes bearing H3K9me2 and/or H3K9me3 (C. elegans Sequencing Consortium 1998; Gerstein et al. 2010; Zeller et al. 2016; McMurchy et al. 2017). Among repetitive sequences, those derived from DNA transposons are particularly abundant (covering 12.6% of the genome). Some DNA transposons can be activated, yet only a small minority of elements encode full-length transposases (Bessereau 2006; Hubley et al. 2016). LTR-containing and LTR-free sequences derived from RNA transposons (retrotransposons) cover ∼1% of the genome, but there are few full-length elements, and none appear to be active under wild-type conditions (Bessereau 2006; Hubley et al. 2016).
In organisms with localized centromeres, pericentric domains contain large arrays of tandem satellite repeats, which can occupy up to 1 Mbp in vertebrates (Plohl et al. 2008). Difficulties in sequencing and mapping these regions unambiguously have meant that the exact extent and composition of repetitive domains are unclear for most vertebrate genomes, yet pericentric satellite sequences are estimated to account for 5–10% of the human genome (Schueler and Sullivan 2006).
The C. elegans genome contains smaller arrays of tandem simple repeats (microsatellites of 1–6 bp), as well as minisatellite repeats (10–200 bp) that are distributed rather than clustered in large head-to-tail arrays (Subirana et al. 2015; Hubley et al. 2016). This is reminiscent of the dispersed nature of C. elegans centromeres. The shortness of the repeat clusters has allowed for high-quality sequencing of nearly all C. elegans repetitive DNA (C. elegans Sequencing Consortium 1998; Hillier et al. 2005). Additionally, 71% of the repeats are uniquely mappable with 50-bp single-end reads. Interestingly, several microsatellite families have been shown to be enriched in CENP-A, suggesting that they may be associated with centromere function (Subirana et al. 2015), although centromere function in worms is independent of H3K9 methylation (Towbin et al. 2012; Ho et al. 2014; Garrigues et al. 2015; Zeller et al. 2016).
The Nuclear Lamina and Chromatin Association
The mapping of chromatin regions associated with proteins of the nuclear lamina has shown that the central and distal arm regions of C. elegans chromosomes have distinct distributions with respect to the nuclear envelope (Ikegami et al. 2010; Towbin et al. 2012; Gonzalez-Aguilera et al. 2014). C. elegans expresses a single lamin protein (LMN-1), that forms a stable meshwork underlying the nucleoplasmic side of the nuclear envelope together with a number of well-characterized lamin-associated proteins (Dobrzynska et al. 2016). These lamin-associated factors include the LEM domain proteins LEM-2 (MAN1) and EMR-1 (Emerin), the SUN domain proteins UNC-84 and SUN-1, the accessory protein BAF, and four KASH domain proteins that form a bridge to the cytoskeleton (Bank and Gruenbaum 2011, see Table 1 for abbreviations). While this is undoubtedly a nonexhaustive list, this set of proteins and functions is conserved across animal species.
The genome-wide mapping profiles of LEM-2, LMN-1, and EMR-1 all show strong enrichment on distal chromosome arms, indicating that these regions are associated with the nuclear envelope (Ikegami et al. 2010; Towbin et al. 2012; Gonzalez-Aguilera et al. 2014; Gonzalez-Sandoval et al. 2015). A detailed analysis of LEM-2 binding showed that these domains are not continuous along the chromosome arms, but are interspersed with gaps that bear expressed genes (Ikegami et al. 2010), which are thought to extend inwards from the peripheral lamin- or LEM-2-associated domains. The LEM-2-associated domains are enriched for H3K9-methylated histones, and their perinuclear positioning is largely dependent on this mark, which is specifically recognized by a nuclear envelope-associated chromodomain protein, CEC-4 (Towbin et al. 2012; Gonzalez-Sandoval et al. 2015). The LEM-2-bound regions are not always transcriptionally silent, and those with gene expression are enriched for the HP1 homolog HPL-2 (Garrigues et al. 2015).
Phenotypes of Chromatin Repressor Mutants
In C. elegans, the development of the vulva has proven to be a powerful system to identify regulators of cell fate, as it is dispensable for survival and defects are easily observable (Horvitz and Sternberg 1991). Many genes with presumed roles in chromatin repression, including those encoding proteins that generate or bind methylated H3K9, were originally identified based on their synthetic multivulval (synMuv) phenotype (Fay and Yochem 2007). Single mutants of synMuv genes have a normal vulva, but double mutants between synMuv genes of different genetic classes develop extra vulvae, arguing for a partial functional redundancy between synMuv gene classes. The synMuv genes were found to encode proteins with a wide range of chromatin-modifying activities, including enzymes that deposit or remove methylation and acetylation on histone tails, ligands for the methylated residues, and nucleosome remodelers. Interestingly, ectopic vulval development in synMuv mutants was shown to be due to a failure to repress expression of a single gene, lin-3/EGF, in the epidermis (Cui et al. 2006). Of note, most synMuv genetic interactions occur between genes that encode different biochemical activities (e.g., between an acetyltransferase and a deacetylase). This functional redundancy reflects the inherent complexity found in chromatin regulatory mechanisms, and argues for redundancy in repression mechanisms. Although synMuv genes were identified based on their roles in the repression of vulval development, most are widely expressed, and the single mutants often show pleiotropic developmental defects and genetic interactions in nonvulval processes. Common phenotypes among them are impaired fertility, altered regulation of repetitive transgenes, L1 arrest at high temperature, and ectopic expression of germ line genes in somatic tissues (Table 2, references therein). We refer readers to chapters on developmental roles of chromatin factors and the synMuv genes for more detailed information (Cui and Han 2007; Fay and Yochem 2007).
Histone H3K9 Methyltransferases
Over the years, a number of C. elegans genes have been postulated to encode HMTs that target H3K9me [e.g., MET-2, SET-9, SET-26, SET-25, and SET-32; (Bessler et al. 2010; Towbin et al. 2012; Greer et al. 2014; Ni et al. 2016; Kalinava et al. 2017)]. However, it has been shown recently that the vast majority of H3K9me1, me2, and me3 depend on two key enzymes: MET-2, which is able to deposit me1 and me2 on H3K9, and SET-25, the major, if not only, HMT that deposits H3K9me3 in somatic cells of larvae and embryos (Towbin et al. 2012). In a double knockout for these two SET domain genes, embryos and L1 stage larvae lacked all detectable H3K9me when analyzed by mass spectrometry (Towbin et al. 2012; Garrigues et al. 2015). Moreover, immunofluorescence (IF) of embryos, L2 larvae, and dissected gonads of the double mutant showed no H3K9me signal (Towbin et al. 2012; Ho et al. 2014; Garrigues et al. 2015; Zeller et al. 2016). Thus, if other H3K9-modifying HMTs exist in worms, either they are expressed under very select conditions or in very few cells, or else their activity requires SET-25 or MET-2. Based on the lower limit of detection by the mass spectroscopy method used, we estimate that < 5% of embryonic histone H3K9 methylation is retained in a set-25met-2 double mutant.
SET-32, a germ line-specific protein whose SET domain has weak homology with EHMT1/G9a of humans, may provide a low level of H3K9me3 methylation in the germ line. Two recent studies found that set-32 mutants are transgenerationally sterile and had reduced H3K9me3 on nuclear RNA interference (RNAi) targets in young adults (Kalinava et al. 2017; Spracklin et al. 2017). However, at most genomic locations, H3K9me3 levels were as low in met-2set-25 double as in met-2set-25; set-32 triple mutants (Kalinava et al. 2017), confirming previous studies reporting loss of detectable H3K9 methylation in met-2set-25 mutants, both in gonads and somatic cells (Towbin et al. 2012; Garrigues et al. 2015; Zeller et al. 2016). MES-2, the EZH2-like H3K27 methyltransferase, was also initially suggested to modify H3K9 in the germ line (Bessler et al. 2010). However, the alterations detected were likely due to antibody cross-reactivity between methylated H3K27 and H3K9. Using validated H3K9me3 antibodies, mes-2 mutant germ lines have normal levels of H3K9me3 staining, while met-2set-25 double-mutant germ lines have none (Ho et al. 2014; Zeller et al. 2016).
MET-2, the C. elegans homolog of mammalian SETDB1/ESET, was first described as a potential transcriptional repressor based on its synMuv phenotype (Poulin et al. 2005; Andersen and Horvitz 2007). SET-25 is less conserved, yet its catalytic SET domain shares 28.8% identity and 44.6% similarity in protein sequence with mammalian EHMT1/G9a, as well as 27.9% identity and 45.7% similarity with Suv39h1/2, although SET-25 lacks both the chromodomain found in Suv39h and the Ankyrin repeats present in G9a. Because the relative abundance of other common H3 tail methylation marks did not change in the set-25met-2 mutant (Towbin et al. 2012), it is likely that these enzymes are specific for histone H3K9. However, one cannot exclude the possibility that the HMTs modify nonhistone targets as well.
Using a fluorescent heterochromatin reporter that allows the quantification of transcriptional silencing, nuclear position, and nucleosome modifications, it was shown that MET-2 works together with SET-25 to silence constitutively expressed promoters, and to tether silent transgene arrays and endogenous repeat sequences at the nuclear envelope (Towbin et al. 2012). SET-25 was shown to be essential for all H3K9me3 in embryos and L1 larvae, and to maintain ∼20% of wild-type levels of H3K9me1 and me2 in met-2 mutants. MET-2, on the other hand, is the main H3K9 mono- and dimethyltransferase, and it can compensate for SET-25 to maintain wild-type levels of H3K9me1 and me2 in set-25 embryos and L1 larvae (Towbin et al. 2012). Dissecting their individual contributions to chromatin localization using null alleles, it was shown that MET-2 is sufficient to confer anchoring of integrated transgene arrays and endogenous heterochromatin at the nuclear envelope, while SET-25 activity in a met-2 mutant was sufficient to anchor the integrated transgene arrays only.
There is evidence that both MET-2 and SET-25 associate with their own enzymatic products in the nucleus. Importantly, whereas a highly overexpressed MET-2-GFP fusion protein is primarily cytoplasmic (Towbin et al., 2012), and a MET-2-mCherry fusion expressed as a single-copy gene from the met-2 promoter gives a spotty nuclear signal (M. Guidi and S. M. Gasser, unpublished results). Moreover, the nuclear MET-2 ChIP pattern in adults is very similar to that of its product H3K9me2 (McMurchy et al. 2017). SET-25 is exclusively nuclear, and it binds to H3K9me3 in a SET domain-independent manner, marking heterochromatin- and H3K9me3-enriched foci (Towbin et al. 2012). In other words, once SET-25 trimethylates H3K9, it either recognizes its product or else binds another reader that recognizes this mark, such that it remains associated with the chromatin that it modified. The association of SET-25 with silent chromatin means that it can act to extend methylation to nearby histone H3 tails, ensuring the spread or potentially self-maintenance of heterochromatic domains.
H3K23me2 was recently shown to be strongly associated with H3K27- and H3K9-methylated heterochromatin in C. elegans (Vandamme et al. 2015; Sidoli et al. 2016). Methylated forms of H3K23 are also found in mouse, where the levels of modification seem to correlate with H3K27 methylation in a Suz12 (PRC2) mutant (Schwammle et al. 2016). Hints regarding its function are suggested by the affinity of the mammalian HP1β chromodomain for H3K23me1/2/3 in vitro (Liu et al. 2010) and the apparent affinity of C. elegans HPL-1 for H3K23me1/2 in binding assays in vitro (Vandamme et al. 2015). This association suggests that H3K23 methylation may function in transcriptional repression, although functional analysis requires identification of the HMT and demethylase that are responsible for the deposition and removal, respectively, of this mark. Interestingly, the H3K9me2 and H3K27me2 histone demethylase JMJD-1.2 appears to act on H3K23me2, as jmjd-1.2 mutants show increased H3K23me2 by IF and recombinant JMJD-1.2 can demethylate H3K23me2 in vitro (Lin et al. 2010). Intriguingly, this interaction appears to be conserved in mouse (Liu et al. 2010).
H3K9me Readers
Given that lysine methylation does not mask the positive charge of the side chain, but instead deposits a bulky adduct, this modification is thought to work primarily through the recruitment of specific ligands or readers. The prototype H3K9me reader is Drosophila HP1a, which has been shown to bind H3K9me through its chromodomain (Clark and Elgin 1992; Nielsen et al. 2002; Fischle et al. 2003). HP1a is essential for centromeric satellite heterochromatin compaction and silencing (Lachner et al. 2001; Nakayama et al. 2001). Additionally, HP1 proteins contain a C-terminal chromo-shadow domain that contributes to dimerization, and a hinge region that binds RNA and promotes HP1 association with chromatin (Muchardt et al. 2002; Meehan et al. 2003; Maison and Almouzni 2004; Keller et al. 2012; Eissenberg and Elgin 2014). So far, five C. elegans proteins (Table 1) have been shown to recognize methylated H3K9, although only two, CEC-4 and LIN-61, were tested against a wide range of methylated substrates in vitro. Four of these, HPL-1, HPL-2, CEC-3/EAP-1, and CEC-4, contain chromodomains, whereas LIN-61 has four MBT motifs (Koester-Eiserfunke and Fischle 2011; Greer et al. 2014; Garrigues et al. 2015; Gonzalez-Sandoval et al. 2015).
HPL-1 and HPL-2 are the C. elegans orthologs of HP1, as they bear both a chromodomain and a chromoshadow domain, separated by a less well-conserved hinge region (Couteau et al. 2002). Mutants lacking hpl-1 are phenotypically wild-type, whereas hpl-2 mutants have pleiotropic defects including loss of repression of a heterochromatic reporter, slow growth, abnormal germ line development and sterility, somatic expression of germ line genes, and a synMuv phenotype (Couteau et al. 2002; Coustham et al. 2006; Schott et al. 2006, 2009; Simonet et al. 2007; Meister et al. 2011; Petrella et al. 2011; Towbin et al. 2012; McMurchy et al. 2017). While the two orthologs clearly have different functions, HPL-1 is partially redundant with HPL-2, as evidenced by enhanced sterility and growth defects of double mutants (Schott et al. 2006). HPL-1 and HPL-2 were also shown to promote fertility and vulval development synergistically with HIS-24, an H1 linker histone, which, when methylated on K14, has been shown by in vitro assays to be a target of HPL-1 recognition (Studencka et al. 2012).
HPL-1 and HPL-2 GFP fusion proteins are both widely expressed, yet the two fusions localize to different nuclear foci (Couteau et al. 2002; Schott et al. 2006). LIN-13, a multi-Zn-finger protein, was shown to be essential for the localization of HPL-2::GFP into foci and to physically interact with HPL-2, but had no effect on HPL-1::GFP (Coustham et al. 2006). HPL-1::GFP binds integrated transgene arrays, while HPL-2::GFP does not (Towbin et al. 2012). Additional studies of the HPL-2::GFP fusion showed that its foci became more peripheral upon disruption of euchromatic regulators such as the TIP60/NuA4 remodeling complex (Grant et al. 2010). Because it is unclear if the HPL-1::GFP and HPL-2::GFP fusions are fully functional, these localization studies should be repeated with HPL-1- and HPL-2-specific antibodies. Nonetheless, it is clear that these two HP1 proteins do not have identical binding sites or functions.
In vitro, HPL-2 can bind all three methylated forms of H3K9 as well as H3K27me3, but in vivo mapping by ChIP-chip in embryos showed that it binds primarily on the distal arms of autosomes in a pattern that correlates well with that of H3K9me1 and me2, but not with H3K9me3 or H3K27me3 (Garrigues et al. 2015). The HPL-2 signal is reduced, but not lost, in the met-2set-25 double mutant, indicating that HPL-2 can associate with chromatin independently of H3K9 methylation (Garrigues et al. 2015), for example through binding another protein, methylation mark, or RNA. Genetic analyses also indicate that hpl-2 has roles independent of H3K9 methylation, because hpl-2 mutants have stronger sterility phenotypes at high temperature than met-2set-25 mutants (Garrigues et al. 2015). Intriguingly, HPL-2 is found at many expressed genes, even in the distal arms of autosomes, and 94% of HPL-2-bound genes on chromosome arms also show binding by the nuclear envelope protein LEM-2 (Garrigues et al. 2015). This suggests that neither association with HPL-2 nor association with the nuclear envelope is sufficient to repress transcription.
CEC-3/EAP-1 is a chromodomain-containing protein that can bind to all methylated forms of H3K9 in vitro. Its association with germ line chromatin in vivo depends on MET-2, suggesting that it binds H3K9me1 or me2 (Greer et al. 2014). Loss of CEC-3 suppresses the transgenerational sterility observed in worms lacking the H3K4 demethylase SPR-5. Strains lacking SPR-5 accumulate H3K4me2 and lose H3K9me3 over multiple generations (Katz et al. 2009; Greer et al. 2014). Interestingly, met-2 mutants also display transgenerational sterility, and the loss of MET-2 enhances spr-5 sterility, while the loss of SET-25 has no effect (Katz et al. 2009). Given that spr-5 mutants also accumulate high levels of H3K9me2 at several tested targets (Katz et al. 2009), it may be that an imbalance between H3K9me2 and H3K4me2 leads to transgenerational sterility in spr-5 mutants. Interaction between these two methylation events is also suggested by the finding that loss of the H3K4 methyl transferase SET-2 rescues hpl-2 somatic defects (Simonet et al. 2007).
LIN-61 is an MBT domain-containing protein that recognizes H3K9me2 and me3 (Harrison et al. 2007; Koester-Eiserfunke and Fischle 2011). Like hpl-2, lin-61 is a synMuv gene, and mutants lacking LIN-61 have a reduced brood size (Harrison et al. 2007; Koester-Eiserfunke and Fischle 2011). LIN-61 has also been shown to form a complex with HPL-2 and LIN-13 (Wu et al. 2012). Both hpl-2 and lin-61 are hypersensitive to ionizing radiation (Johnson et al. 2013; McMurchy et al. 2017), contribute to the repression of transgene arrays (Towbin et al. 2012), and play roles in cell type-specific control of gene expression (Studencka et al. 2012; Zheng et al. 2013). Specifically, mutations in CEC-3, HPL-2, or LIN-61 alleviate the tissue-specific repression of unc-4, a transcription factor expressed in a subset of ventral cord (VC) neurons. Loss of the H3K9 methyl-transferase MET-2, HPL-2, LIN-61, or CEC-3 leads to the ectopic expression of unc-4 in all VC neurons, while loss of the JMJD-2 histone demethylase reduces this unscheduled expression (Zheng et al. 2013). Such results underscore the partial redundancy found among H3K9me ligands (Schott et al. 2006; McMurchy et al. 2017), and suggests that H3K9 methylation is involved in the proper repression of some cell type-specific promoters (Zeller et al. 2016).
CEC-4- and H3K9me-Mediated Anchoring at the Nuclear Periphery
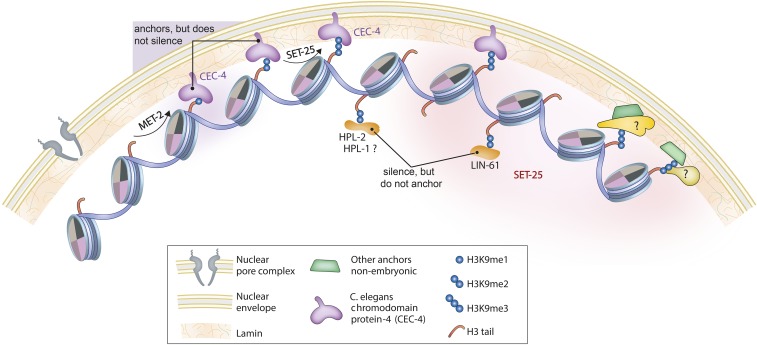
CEC-4 is a worm-specific chromodomain protein necessary for perinuclear anchoring of heterochromatin (Gonzalez-Sandoval et al. 2015). It has a canonical HP1β-like chromodomain that recognizes histone H3 tails methylated on K9, but it lacks the chromoshadow motif (Gonzalez-Sandoval et al. 2015). Its C-terminal domain lacks all significant homology to other characterized chromatin-binding proteins, yet it is required for CEC-4 localization to the inner face of the nuclear envelope. Its perinuclear localization is recapitulated when CEC-4 is expressed as a fusion protein both in yeast and in worms, where it ensures the peripheral tethering of H3K9-methylated chromatin throughout early development (Gonzalez-Sandoval et al. 2015). The chromodomain of CEC-4 binds mono-, di-, and trimethylated H3K9 with affinities similar to mammalian HP1β, and with high specificity: out of 188 methylated histone peptides tested, only histone H3 tail peptides bearing K9 methylation, and, more weakly, H3K37me2, were bound. Because the latter modification has never been detected in C. elegans, the H3K9me is probably the most important ligand. Indeed, ablation of two conserved aromatic residues in the CEC-4 chromodomain was sufficient to compromise the tethering of heterochromatin at the nuclear envelope in C. elegans embryos (Gonzalez-Sandoval et al. 2015). This CEC-4-H3K9me interaction is the first chromodomain–ligand interaction shown to be necessary for chromatin positioning in the interphase nucleus in any species (Figure 2).
Figure 2.
Histone H3K9 methylation triggers peripheral localization of chromatin independently of HPL-1/-2 or LIN-61 binding. In C. elegans early embryos, CEC-4 recognizes and binds H3K9 me1, me2, or me3 to mediate the anchoring of appropriately modified nucleosomes to the nuclear periphery, without necessarily repressing transcription (Gonzalez-Sandoval et al. 2015). The H3K9me ligands HPL-2 and LIN-61 mediate transcriptional repression by binding H3K9 methylation, but do not anchor chromatin. HPL-1 is associated with repressed chromatin, but its role in repression remains unclear. SET-25 colocalizes with heterochromatic transgene arrays bearing H3K9me3, and its activity, along with HPL-2 and LIN-61, leads to repression. MET-2 and/or SET-25 deposit H3K9me1 and me2, while only SET-25 deposits H3K9me3 in somatic cells (Towbin et al. 2012). Alternative anchors may be present in differentiated cells, although their identity is unknown.
Although dependent on CEC-4, perinuclear heterochromatin tethering in embryos is independent of HPL-1, HPL-2, and LIN-61 and of H3K27me3 (Figure 2). Indeed, the tethering of large transgene arrays is independent of their transcriptional state, although it requires H3K9 methylation (Gonzalez-Sandoval et al. 2015). Consistently, the loss of CEC-4 had very little effect on gene expression under unchallenged growth conditions. Nonetheless, CEC-4 was needed for a full response to the ectopic expression of a cell fate regulator in pregastrulation embryos (Gonzalez-Sandoval et al. 2015). Following challenge by the induction of the myogenic transcription factor HLH-1 before gastrulation, wild-type embryos all arrest as differentiated masses of muscle cells. In the cec-4 mutant, on the other hand, ∼25% of the embryos fail to arrest with a muscle-like phenotype, even though the induction of HLH-1 leads to muscle-specific protein expression. In other words, in the absence of CEC-4, other tissue lineages were not efficiently repressed, arguing that perinuclear anchorage helps restrict gene expression to that of the HLH-1-induced lineage (Gonzalez-Sandoval et al. 2015).
In another study, it was shown that CEC-4, H3K9 methyltransferases, and LEM-2 contribute to the condensed perinuclear status of the dosage-compensated X chromosomes in terminally differentiated postmitotic cells of adult animals (Snyder et al. 2016). This result suggests that nuclear organization, and specifically anchoring of chromosomal regions to the nuclear lamina, may affect dosage compensation. Nonetheless, during normal development, cec-4 embryos develop into fertile adults.
H3K9 Demethylases
Histone methyl marks are removed by demethylases that generally fall into two structural classes (Kooistra and Helin 2012). One class contains amine oxidases, of which LSD1 (a homolog of the C. elegans H3K4 demethylase SPR-5) is the founding member. These have been shown to be able to demethylate mono- and dimethylated lysine residues, but they are unable to act on trimethylated lysine. The second class contains the Jumonji C (JmjC) domain, which can act on all three methylation states.
C. elegans contains 13 JmjC domain-containing proteins (Klose et al. 2006), of which two (JMJD-2/JMJD2a and JMJD-1.2/CeKDM7a) are involved in H3K9 demethylation (Whetstine et al. 2006; Kleine-Kohlbrecher et al. 2010; Lin et al. 2010). IF of meiotic chromosomes showed that depletion of jmjd-2 by RNAi led to increased H3K36me3 on the X chromosome and increased H3K9me3 on the autosomes (Whetstine et al. 2006). Reduction of JMJD-2 also led to replication stress, as indicated by a replication checkpoint-dependent increase in germ cell apoptosis, slow DNA replication fork progression (incorporation of Cy3-dUTP), and an accumulation of RAD-51 foci in the germ line, indicative of DNA breaks (Whetstine et al. 2006; Black et al. 2010). Interestingly, these germ line phenotypes could be rescued by deletion of the H3K9me reader HPL-2, suggesting that mistargeting of HPL-2 might be responsible for the phenotypes (Black et al. 2010). Loss of JMJD-2 would presumably lead to an accumulation of H3K9me, which may facilitate ectopic HPL-2 binding. Although H3K9me2 or me3 have been shown to correlate with late replication in other organisms (Schwaiger et al. 2010; Lubelsky et al. 2014), it is not yet clear if H3K9 methylation directly influences replication timing in C. elegans. Alternatively, perturbations in H3K9me levels may lead to replication stress that arrests replication (Zeller et al. 2016).
JMJD-1.2/CeKDM7a is a bispecific H3K9me2/H3K27me2 demethylase (Kleine-Kohlbrecher et al. 2010; Lin et al. 2010). Recombinant JMJD-1.2 has been shown to demethylate H3K9me2 and H3K27me2 in vitro, and jmjd-1.2 mutants have increased levels of H3K9me2 and H3K27me2, by western blot analysis (Kleine-Kohlbrecher et al. 2010; Lin et al. 2010). ChIP of JMJD-1.2 has shown binding at H3K4me3-marked promoters, which are depleted for H3K9me2 and H3K27me2, and deletion of jmjd-1.2 leads to decreased expression of a subset of tested targets (Lin et al. 2010). This may be due to the local acquisition of heterochromatic marks, although this has not yet been demonstrated. JMJD-1.2 appears to have a role in the nervous system, as a JMJD-1.2 transgene is predominantly expressed in neurons and jmjd-1.2 mutants display movement defects (Kleine-Kohlbrecher et al. 2010).
Phenotypes Caused by the Loss of H3K9 Methyltransferases or H3K9me Readers
C. elegans provides an opportunity to characterize the effects of a complete loss of H3K9 methylation during development of a multicellular organism, given that met-2set-25 mutant embryos, larvae, and germ lines lack detectable H3K9 methylation, and are viable and fertile at 15 or 20° (Towbin et al. 2012; Ho et al. 2014; Garrigues et al. 2015; Zeller et al. 2016). Two recent studies investigated the consequences of an absence of H3K9 methylation or of interacting heterochromatin factors including H3K9me readers (Zeller et al. 2016; McMurchy et al. 2017). In adults, the heterochromatin factors studied were HPL-2 and LIN-61, the multi-zinc finger protein LIN-13 (Melendez and Greenwald 2000; Coustham et al. 2006; Harrison et al. 2007; Koester-Eiserfunke and Fischle 2011; Wu et al. 2012; Garrigues et al. 2015), and LET-418, an Mi-2 homolog that is part of the repressive NuRD (Nucleosome remodeling and histone deacetylase) and MEC (Mi-2 and MEP-1) complexes (von Zelewsky et al. 2000; Unhavaithaya et al. 2002; Passannante et al. 2010).
Unlike the situation in other metazoans, chromosome segregation is normal in the absence of H3K9 methylation (Zeller et al. 2016), possibly due to the holocentric nature of worm chromosomes. However, the loss of the MET-2 and SET-25 HMTs shares some phenotypes with the loss of the studied heterochromatin factors, such as the transcription of repetitive elements, an accumulation of DNA damage, reduced fertility, and increased germ cell apoptosis (Zeller et al. 2016; McMurchy et al. 2017). Not surprisingly, loss of the histone modification itself often showed more pronounced phenotypes than loss of a single reader.
Similar to other organisms, in C. elegans H3K9me2 and me3 are enriched on repetitive elements, with the majority of repetitive elements being marked by one or both modifications. Importantly, a detailed analysis of the two marks shows that they have different distributions: H3K9me2 is more significantly associated with a subset of DNA transposons and satellite repeats, while H3K9me3 was more prominent on retrotransposons and a second subset of DNA transposons (Zeller et al. 2016; McMurchy et al. 2017). With respect to genes, H3K9me3 is enriched on pseudogenes and silent cell type-specific genes, whereas H3K9me2 marks genes independently of their transcriptional activity (Ho et al. 2014; Garrigues et al. 2015; Zeller et al. 2016). H3K9me2, but not H3K9me3, is also associated with telomeres (McMurchy et al. 2017), although it is not required for their interaction with the nuclear envelope (Ferreira et al. 2013).
The genomic distributions of HPL-2, LIN-13, LIN-61, MET-2, and LET-418 are strikingly similar and highly correlated with H3K9me2, but not H3K9me3 (Garrigues et al. 2015; McMurchy et al. 2017). This begs the question of whether there are readers with a selective affinity for H3K9me3. While no direct binding studies have been reported to date, SET-25 colocalizes with repetitive arrays bearing H3K9me3, but not those with H3K9me2, suggesting that it might directly or indirectly associate with the me3 mark (Towbin et al. 2012). For HPL-1, its definitive binding specificity and endogenous genomic distribution remain to be determined. Preliminary evidence indicates that HPL-1 can recognize all three methylated states of H3K9 in vitro (W. Fischle, personal communication), and it bound methylated H3K23 in a peptide pull-down assay (Vandamme et al. 2015).
As mentioned above, a major function of H3K9me and heterochromatin proteins is transcriptional silencing of genes and repetitive elements. Previous work showed that deletion of the methyltransferases MET-2 and SET-25, or of HPL-2 or LIN-61, derepressed heterochromatic reporters (Towbin et al. 2012) and endogenous genes (Gonzalez-Sandoval et al. 2015). Mutants lacking H3K9me or any of the four studied heterochromatin proteins (HPL-2, LIN-61, LIN-13, or LET-418) derepressed genes and repetitive elements of all classes and families, including satellite repeats, simple repeats, and RNA and DNA transposons (Zeller et al. 2016; McMurchy et al. 2017). The majority of derepressed sequences are enriched for the relevant histone marks or chromatin factors in wild-type animals, suggesting that the effect is direct.
The observed derepression of repetitive elements is temperature-dependent, with many more repeat families being transcribed at 25° than at 20° (Zeller et al. 2016). This temperature dependence is particularly pronounced for tandem repeats. Temperature effects were also reported for the silencing of repetitive elements by the nuclear RNAi pathway (Ni et al. 2016), and for phenotypes of null alleles of other heterochromatin mutants and small RNA pathway factors (e.g., Batista et al. 2008; Wang and Reinke 2008; Schott et al. 2009). Whether these reflect temperature-dependent hyperactivity of RNA polymerase II at 25°, a heat-stress response, or a sensitivity of other factors to heat is unknown. Interestingly, a recent report suggests that wild-type SET-25 activity may be temperature sensitive, and that its effect in silencing repetitive sequences can be transgenerationally inherited (Klosin et al. 2017). However, loss of set-25 does not derepress endogenous micro- or mini-satellite repeats (J. Padeken and S. M. Gasser, unpublished results).
The detection of reproducible but low levels of repeat element transcription in met-2set-25 mutant embryos argues for a broad misregulation of RNA polymerase II-mediated transcription caused by the loss of H3K9 methylation (J. Padeken, P. Zeller, and S. M. G., unpublished results). For simple repeats, transcriptional start sites are not yet mapped, but it is possible that cryptic transcription factor binding sites become exposed by the loss of repressive heterochromatin. Importantly, the loss of individual HMTs, i.e., single met-2 and set-25 mutants, shows that different classes of repeats become expressed in the two mutants, consistent with their differential effects on H3K9me marks (J. Padeken, P. Zeller, and S. M. G., unpublished results).
The genes that carry H3K9 methylation in wild-type larvae and adults are generally pseudogenes or silent tissue-specific genes, and these too are derepressed in met-2set-25 double mutants (Zeller et al. 2016; McMurchy et al. 2017). The expressed repetitive elements are highly enriched for full-length DNA transposases and LTR-containing elements derived from RNA transposons, which carry functional Pol II promoter sequences (McMurchy et al. 2017). Nonetheless, few elements marked by H3K9 methylation become expressed in its absence, suggesting that other mechanisms prevent repetitive element expression. For instance, loss of the nuclear RNAi component nrde-2 leads to derepression of many elements not affected by the loss of H3K9 methylation (McMurchy et al. 2017). Nuclear RNA degradation mechanisms may be involved in repeat repression, as exosomes have been shown to play a role in heterochromatic silencing in other organisms (Shin et al. 2013; Sugiyama et al. 2016; Tucker et al. 2016). There may also be functional redundancy in repetitive element silencing among the heterochromatin factors studied, given that the mutants show redundancy in the promotion of fertility (McMurchy et al. 2017). In addition to transcriptional control, H3K9 methylation may prevent the movement of nonautonomous transposons or inhibit homologous recombination between repeats.
A striking phenotype associated with the loss of heterochromatin over repeat elements is the loss of genomic integrity. Increased germ line apoptosis in met-2set-25 mutants was shown to be cep-1/p53-dependent and therefore linked to the DNA damage response (Zeller et al. 2016; McMurchy et al. 2017). Activation of the DNA damage response pathway contributes to the sterility of heterochromatin factor mutants, as sterility is partially suppressed by mutation of cep-1/p53, although this is not the case for met-2set-25 (Zeller et al. 2016; McMurchy et al. 2017). Interestingly, resilencing of the MIRAGE1 DNA transposon by RNAi partially restored fertility in hpl-2, let-418, and lin-13 mutants, potentially by suppressing expression of its transposase activity (McMurchy et al. 2017).
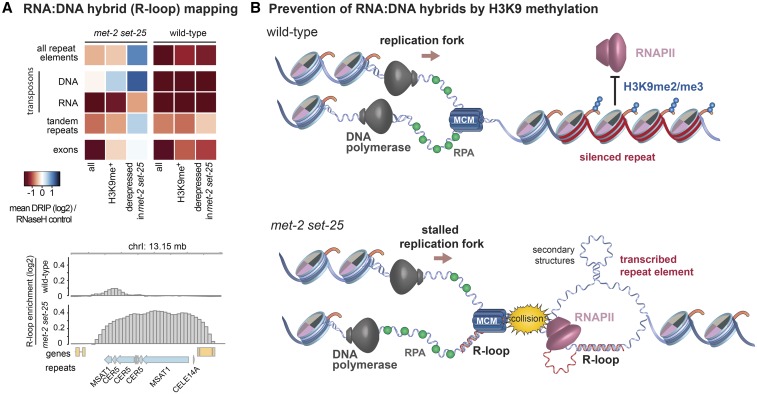
The expression of repetitive sequences in met-2set-25 mutant worms led to an increase in the formation of RNA:DNA hybrids or R-loops, a pathological annealing of RNA with DNA that generates a ssDNA loop where the RNA is bound (Zeller et al. 2016) (Figure 3). R-loops are the most common cause of replication fork-associated DNA damage (Aguilera and Garcia-Muse 2012). The RNA:DNA hybrids map specifically to the tandem repeats and DNA transposons that are derepressed by the loss of H3K9 methylation and, like transcription, the levels of RNA:DNA hybrids are enhanced at 25° vs. 20°, and are barely detectable at 15°, a temperature at which fertility defects are also suppressed (Zeller et al. 2016). The appearance of R-loops at repeat elements correlates with frequent small insertions and deletions at these sites in both somatic cells and the germ line, as detected by whole-genome sequencing (Zeller et al. 2016). Enhanced levels of Rad51 foci, indicative of spontaneous DNA damage, were observed in the germ lines of the met-2set-25 double mutant, as in heterochromatic regions in Drosophila lacking the H3K9 HMT Su(var)3-9 (Peng and Karpen 2009). Importantly, met-2set-25 mutant embryos and larvae are not hypersensitive to ionizing radiation, but are sensitive to replication stress induced by hydroxyurea (Zeller et al. 2016).
Figure 3.
Larvae lacking MET-2 and SET-25 show an accumulation of RNA:DNA hybrids or R-loops at transcribed repeat elements. (A) Genome-wide distribution of R-loops determined by RNA:DNA immunoprecipitation (DRIP) with antibody S9.6, followed by deep sequencing of recovered DNA [see Zeller et al. (2016) for details]. Heat map of an S9.6 DRIP sequencing (DRIP-seq) experiment showing mean log2 enrichment over the corresponding RNaseH-treated controls. Loci are segregated based on the type of repeat element (vertical legend) and whether or not the sequences carry H3K9 methylation in wild-type cells (by chromatin immunoprecipitation analysis, labeled H3K9me+), or whether they were derepressed in set-25 met-2 vs. wild-type embryos by RNA sequencing (horizontal legend). The lower panel is a DRIP-seq example showing the R-loop signal over a repeat element cluster. The immunoprecipitation signal is normalized to the input and the RNaseH control values were subtracted. (B) Transcribed repeat elements in H3K9me-deficient strains can exacerbate replication stress, provoking insertions and deletions in repetitive parts of the genome. This model suggests that the loss of H3K9me leads to R-loops, which allows the formation of secondary DNA structures that engender fork arrest, slippage, and breakage as forks deal with replication stress (Aguilera and Garcia-Muse 2012). These may perturb genome integrity, especially at heterochromatic repeats. Modified from Zeller et al. (2016). Chrl, Chromosome 1; RNAPII, RNA polymerase II.
Some H3K9me readers play additional roles in DNA damage repair. Namely, LIN-61 mutants are sensitive to ionizing radiation and LIN-61 is needed for double-strand break repair by homologous recombination, but not by nonhomologous end-joining or single-strand annealing (Johnson et al. 2013). This leads to enhanced chromosome fragmentation and lower germ cell survival rates. Similarly, ionizing radiation in hpl-2 mutants leads to increased checkpoint activation and oocyte chromosome fragmentation (McMurchy et al. 2017). Finally, RNAi-mediated reduction of the SPO-11 endonuclease partially rescues the fertility defect of K9me readers (McMurchy et al. 2017). The difference in hypersensitivities between H3K9me-deficient worms and hpl-2 or lin-61 mutants, suggests that the roles of HPL-2 and LIN-61 in double-strand break repair may be independent of H3K9 methylation. Indeed, HPL-2 binds chromatin both in the presence and absence of H3K9 methylation (Garrigues et al. 2015), and in mammals the recruitment of HP1 proteins to sites of damage is H3K9me-independent (Luijsterburg et al. 2009; Baldeyron et al. 2011).
PRC2/H3K27me3 and Interactions with MES-4/H3K36me3
The conserved PRC2 complex catalyzes methylation of lysine 27 of histone H3 and functions in the maintenance of transcriptional repression in metazoans (Steffen and Ringrose 2014; Piunti and Shilatifard 2016). The C. elegans PRC2-like complex is composed of MES-2, MES-3, and MES-6 (Bender et al. 2004). The SET domain protein MES-2, an ortholog of EZH2, provides H3K27 methylation activity. MES-6 is an ortholog of ESC/EED, and MES-3 is a novel protein.
The genes encoding PRC2 components were initially identified through genetic screens for genes required for germ cell development (Capowski et al. 1991). mes-2, mes-3, and mes-6 mutants are maternal-effect sterile; homozygous mutants derived from heterozygous mothers are viable and fertile, but their progeny are sterile because germ cells die. The activities and functions of these genes are primarily germ line-specific. Somatic development of mes-2, mes-3, and mes-6 mutants is overtly normal, although weak somatic defects in the expression of Hox genes has been observed (Capowski et al. 1991; Ross and Zarkower 2003). At least one somatically active H3K27 HMT exists because somatic H3K27 methylation is present in mes-2 mutants, but it has not yet been identified (Bender et al. 2004).
In addition to components of PRC2, the mes screens also identified mes-4, which encodes a germ line-expressed H3K36 HMT with functions in both the germ line and in early embryos as a maternal product (Capowski et al. 1991; Bender et al. 2006). MES-4 is a nuclear receptor binding SET domain (NSD)-type HMT, that also bears PHD and post-SET domains, and methylates H3K36me in a transcription-independent manner. (Bender et al. 2006; Furuhashi and Kelly 2010; Rechtsteiner et al. 2010). In addition to MES-4, C. elegans also contains MET-1, an ortholog of the transcription-coupled H3K36 HMT SET-1 (Andersen and Horvitz 2007; Furuhashi and Kelly 2010; Rechtsteiner et al. 2010). The loss of MES-4 also results in phenotypes in somatic cells of L1 larvae derived from homozygous adults (D. Cabianca and S.M.G., unpublished results). Mass spectrometry of histones isolated from L1 larvae shows that the loss of MET-1 eliminates 95% of the H3K36me3, while the full complement of me1/me2 is retained. RNAi depletion of MES-4 in met-1 mutants reduces H3K36me1/me2 and the residual H3K36me3 in an additive manner (D. Cabianca and S. M. Gasser, unpublished results).
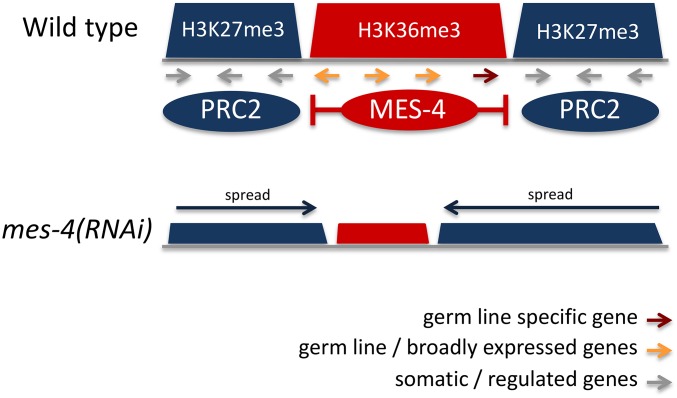
Studies of PRC2 and mes-4 mutants revealed an intimate relationship between H3K27 and H3K36 methylation. Genome-wide profiling in embryos and L3 larvae showed that H3K36me3 and H3K27me3 occupy mutually exclusive domains on autosomes, consistent with the finding that methylation of H3K36 inhibits EZH2 activity (Schmitges et al. 2011; Yuan et al. 2011; Gaydos et al. 2012). Additionally, the X chromosome has higher levels of H3K27me3 and lower levels of H3K36me3 than autosomes (Liu et al. 2011; Gaydos et al. 2012). Intriguingly, despite their marking different genomic regions, gene expression profiling in the germ line showed that loss of either MES-4 or PRC2 had similar consequences: reduced expression of germ line genes and increased expression of somatic and X chromosome genes (Gaydos et al. 2012). This was explained by showing that these two marking systems functionally antagonize each other. Loss of MES-4 causes spreading of H3K27me3 to germ line genes, and a concomitant reduction of H3K27me3 on somatic and X chromosome genes (Gaydos et al. 2012). Therefore, H3K27me3 marking by PRC2 and H3K36me3 by MES-4 cooperate to ensure correct gene expression in the germ line.
The chromatin modifications generated by MES-4 and PRC2 are transgenerationally inherited. MES-4 marks genes expressed in the germ line with H3K36me2/3, and then maternally contributed MES-4 maintains H3K36me2/3 marking of these genes in the early embryo independently of transcription, providing an epigenetic memory of germ line transcription to progeny (Furuhashi et al. 2010; Rechtsteiner et al. 2010). Similarly, H3K27 methylation generated by PRC2 in the germ line is inherited by progeny (Gaydos et al. 2014). Together, H3K27 methylation and maternal PRC2 components provide a memory of transcriptional repression (Gaydos et al. 2014).
A recent study used patterns of chromatin states in early embryos and L3 larvae to investigate domain properties of the C. elegans genome, defining two types of chromatin domain: active and regulated (Evans et al. 2016). It was found that active domains are associated with H3K36me3 and regulated domains with H3K27me3, which overlap the mutually exclusive patterns of these modifications noted by Gaydos et al. (2012). The domains separate genes of different types: active domains contain genes expressed in the germ line and broadly expressed across development and cell types, whereas regulated domains predominantly contain genes under spatial, temporal, or conditional control, lacking germ line expression. Regulated expression is consistent with the repressive role of H3K27me3 in facultative heterochromatin in other organisms. The locations of H3K36me3- and H3K27me3-marked domains in undifferentiated early embryonic cells were similar to those in differentiated L3 larvae, indicating that domain positions are a core property of the genome. The mechanism of domain definition is not yet understood, but appears to involve interactions between PRC2 and MES-4 (Gaydos et al. 2012; Evans et al. 2016) (Figure 4). Additionally, the finding that border regions between active and regulated domains contain long intergenic regions enriched for transcription factor binding suggests that transcriptional activity may play a role in defining the boundaries of active and regulated domains (Evans et al. 2016). A future challenge will be to investigate possible tissue-specific domain regulation by analyzing histone mark distribution in individual tissues rather than whole animals.
Figure 4.
Functional antagonism between MES-4 and PRC2. The genome is organized into domains of genes expressed only in the germ line, or both in the germ line and broadly across cell types, and domains of genes with somatic cell and regulated expression. MES-4 marks genes transcribed in the germ line with H3K36me2/3. This includes germ line-specific genes (red) and broadly expressed genes (orange). A PRC2-like complex composed of MES-2, MES-3, and MES-6 marks somatic genes (gray) with H3K27me3. MES-4 inhibits PRC2: in mes-4(RNAi) embryos (derived from RNA interference-treated mothers), H3K27me3 marking spreads into regions previously occupied by H3K36me3.
PRC2 in Reprogramming and Maintenance of Cell Fate
PRC2 is important for the maintenance of a repressed state, and this may be due in part to inhibition of developmental plasticity. Early evidence for this in C. elegans came from a study showing that loss of mes-2 in early (undifferentiated) embryos causes prolonged sensitivity to ectopic expression of developmental regulators that can cause cell fate transformations (Yuzyuk et al. 2009). This study also showed that loss of mes-2 causes a change in chromosome conformation suggestive of a loss of compaction. Studies in germ cells similarly found that loss of PRC2 components makes germ cells susceptible to conversion to a somatic fate when challenged by expression of cell type-inducing transcription factors (Patel et al. 2012). Sensitivity to somatic conversion was also observed in response to increased Notch activity (Seelk et al. 2016). Notch appears to act by antagonizing PRC2 repression of numerous genes, including the H3K27me3 demethylase UTX-1. These studies support a role for PRC2 in preventing inappropriate responses to regulatory inputs, perhaps by increasing the barrier to response. Consistent with this role, the H3K27me2/3 demethylase JMJD-3.1 is necessary for a natural C. elegans transdifferentiation event, where a hindgut cell is transformed into a motor neuron (Zuryn et al. 2014).
Chromatin Regulators and Small RNA Pathways
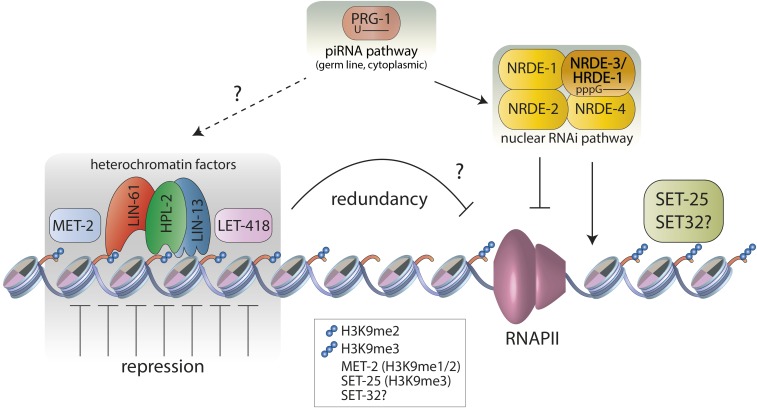
In addition to classical RNAi that carries out post-transcriptional silencing in the cytoplasm, C. elegans also has a nuclear RNAi pathway (called the Nrde pathway) that directs transcriptional silencing, whereby small RNAs provide sequence specificity that directs heterochromatin assembly at target loci and downregulation of RNA polymerase activity (Guang et al. 2008, 2010; Burkhart et al. 2011; Buckley et al. 2012; Gu et al. 2012; Mao et al. 2015). Although the precise mechanism of repression is still being worked out, a series of experiments showed that argonaute proteins bound to small interfering (siRNAs) (NRDE-3 in the soma or HRDE-1 in the germ line) recruit NRDE-1, NRDE-2, and NRDE-4 to target loci, leading to an accumulation of H3K9me3 in a MET-2- and SET-25-dependent manner, and the stalling of RNA polymerase II (Guang et al. 2008, 2010; Burkhart et al. 2011; Buckley et al. 2012; Gu et al. 2012; Mao et al. 2015) (Figure 5). Recently, SET-32 was also shown to affect HRDE-1-dependent H3K9 methylation (Kalinava et al. 2017; Spracklin et al. 2017), and MORC-1 has been implicated as a downstream effector needed to maintain H3K9me3 at HRDE-1 targets (Spracklin et al. 2017; Weiser et al. 2017). Additionally, it was found that MES-2-dependent H3K27me3 is also induced at Nrde targets (Mao et al. 2015), although the relationship between PRC2 and Nrde silencing is not yet known. The results support a model whereby the small RNA-targeted Nrde pathway represses transcription by triggering the generation of repressed chromatin. Interestingly, mutants affecting nuclear RNAi processes often show progressive sterility over generations (Table 2).
Figure 5.
Heterochromatin proteins and small RNA pathways repress transcription of repetitive elements and genes. Repressed chromatin is marked by methylation of H3K9me2 and/or H3K9me3. The nuclear RNAi (Nrde) pathway uses small RNAs bound by argonaute proteins HRDE-1 (germ line) or NRDE-3 (soma) to target NRDE proteins to chromatin, leading to inhibition of transcription elongation and H3K9me3 marking, dependent on SET-25. Some H3K9me3 induced by exogenous application of RNAi is dependent on SET-32. Loss of NRDE function leads to derepression of repetitive elements (enriched for those derived from retrotransposons) and genes. The piRNA pathway, initiated in the germ line cytoplasm, targets genes and repetitive elements for repression by transcriptional and post-transcriptional mechanisms. The piRNA pathway engages the Nrde pathway for transcriptional silencing through the generation of small RNAs that bind HRDE-1. A set of heterochromatin factors are found together at many genomic locations and are expressed in the germ line and soma. They repress common elements by a mechanism that is not yet understood. Repetitive elements that require the heterochromatin proteins for repression (enriched for DNA transposons) largely differ from those requiring the Nrde pathway. This apparent difference appears to be at least partially due to functional redundancy between the Nrde pathway and heterochromatin factors, the nature of which is not yet known. piRNA, piwi-interacting RNA; RNAi, RNA interference; RNAPII, RNA polymerase II.
The Nrde pathway is also engaged by the piwi-interacting RNA (piRNA) pathway, a small RNA pathway active in the germ line, which is important for the repression of transposable elements as well as endogenous genes (Ashe et al. 2012; Bagijn et al. 2012; Lee et al. 2012) (Figure 5). piRNAs are nuclear-encoded 21-nucleotide RNAs (starting with a U) that are bound by the argonaute PRG-1 in the cytoplasm (Ruby et al. 2006; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008). These direct the generation of secondary siRNAs that become bound by the germ line Nrde argonaute, HRDE-1, to engage the Nrde pathway in transcriptional silencing (Ashe et al. 2012; Buckley et al. 2012; Luteijn et al. 2012). Silencing of a piRNA pathway reporter was shown to require the H3K9 HMTs SET-25, MET-2, and SET-32, as well as the heterochromatin proteins HPL-2, LIN-61, and LET-418 (Ashe et al. 2012; McMurchy et al. 2017). In adult worms, both prg-1 mutants and mutants lacking these heterochromatin proteins derepress a common spectrum of repetitive elements, suggesting that these may work together in the germ line (McMurchy et al. 2017) (Figure 5).
Interestingly, loss of the Nrde pathway leads to the derepression of a different set of repetitive elements than loss of the piRNA pathway or of the above subset of heterochromatin factors (Kalinava et al. 2017; McMurchy et al. 2017). Retrotransposons are more affected in nrde-2 mutants, whereas a bias for DNA transposons was observed for prg-1 and heterochromatin mutants. Moreover, redundancy appears to underlie at least part of this apparent separation of function, because nrde-2 genetically interacts with let-418, lin-13, and hpl-2. Additionally, nrde-2; let-418 double mutants derepress many more repetitive elements than either single mutant (McMurchy et al. 2017) (Figure 5). Consistent with redundancy, we note that although the Nrde pathway induces H3K9 methylation, endogenous Nrde targets are still repressed in the absence of H3K9 methylation, indicating that H3K9 methylation is not necessary for repression by the Nrde pathway (Kalinava et al. 2017; McMurchy et al. 2017). The observed complexity and redundancy of silencing mechanisms underscores the importance of repeat element repression for genome stability.
Perspectives
Heterochromatin fulfills many important functions in the genome of complex organisms. In this respect, C. elegans is no exception, as indicated by the diverse physiological defects of mutants lacking the enzymes that deposit heterochromatic marks or the proteins that recognize them. Repressive chromatin is essential for nuclear organization, genome domain structure, repression of repetitive elements, genome stability, and regulation of protein-coding genes. Cross talk and redundancy between the proteins and pathways involved has been observed, but these interactions are poorly understood. Furthermore, the specificity of most HMTs and histone mark readers remains to be determined, as does the nature and importance of the interactions observed between heterochromatin and small RNA interference mechanisms. Above all, the role of heterochromatin in controlling or shaping developmental programs is still an open question, and the genetics and rapid development of C. elegans are particularly useful for its investigation. As demonstrated in the past, C. elegans is an excellent organism for gene expression studies, given its genetic flexibility, rapid developmental time, and well-defined cell differentiation pathways.
Acknowledgments
We thank Jan Padeken, Peter Zeller, Anna Mattout, Jennifer Harr, and Tessa Gaarenstroom for critical reading of the article. S.M.G. thanks the Swiss National Science Foundation and the Novartis Research Foundation for continued support. J.A. was supported by a Wellcome Trust Senior Research Fellowship (101863).
Footnotes
Communicating editor: S. Strome
Literature Cited
- Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., et al. , 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. [DOI] [PubMed] [Google Scholar]
- Aguilera A., Garcia-Muse T., 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46: 115–124. [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N., 1982. The kinetochores of Caenorhabditis elegans. Chromosoma 86: 409–428. [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Horvitz H. R., 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. [DOI] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E. M., Mitchell J., Bagijn M. P., et al. , 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn M. P., Goldstein L. D., Sapetschnig A., Weick E. M., Bouasker S., et al. , 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeyron C., Soria G., Roche D., Cook A. J., Almouzni G., 2011. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 193: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank E. M., Gruenbaum Y., 2011. Caenorhabditis elegans as a model system for studying the nuclear lamina and laminopathic diseases. Nucleus 2: 350–357. [DOI] [PubMed] [Google Scholar]
- Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N., et al. , 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L. B., Cao R., Zhang Y., Strome S., 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14: 1639–1643. [DOI] [PubMed] [Google Scholar]
- Bender L. B., Suh J., Carroll C. R., Fong Y., Fingerman I. M., et al. , 2006. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133: 3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessereau J.-L., 2006. Transposons in C. elegans (January 18, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.70.1, http://www.wormbook.org. [Google Scholar]
- Bessler J. B., Andersen E. C., Villeneuve A. M., 2010. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J. C., Allen A., Van Rechem C., Forbes E., Longworth M., et al. , 2010. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell 40: 736–748. [DOI] [PubMed] [Google Scholar]
- Boros J., Arnoult N., Stroobant V., Collet J. F., Decottignies A., 2014. Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1alpha at chromatin. Mol. Cell. Biol. 34: 3662–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., et al. , 2012. A nuclear argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart K. B., Guang S., Buckley B. A., Wong L., Bochner A. F., et al. , 2011. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 7: e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998 Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Capowski E. E., Martin P., Garvin C., Strome S., 1991. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics 129: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. F., Elgin S. C., 1992. Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res. 20: 6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V., Bedet C., Monier K., Schott S., Karali M., et al. , 2006. The C. elegans HP1 homologue HPL-2 and the LIN-13 zinc finger protein form a complex implicated in vulval development. Dev. Biol. 297: 308–322. [DOI] [PubMed] [Google Scholar]
- Couteau F., Guerry F., Muller F., Palladino F., 2002. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Han M., 2007. Roles of chromatin factors in C. elegans development (May 3, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.139.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Chen J., Myers T. R., Hwang B. J., Sternberg P. W., et al. , 2006. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10: 667–672. [DOI] [PubMed] [Google Scholar]
- Das P. P., Bagijn M. P., Goldstein L. D., Woolford J. R., Lehrbach N. J., et al. , 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynska A., Askjaer P., Gruenbaum Y., 2016. Lamin-binding proteins in Caenorhabditis elegans. Methods Enzymol. 569: 455–483. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Elgin S. C., 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 30: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C., Reuter G., 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R., Eberharter A., Imhof A., 2007. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 27: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K. J., Huang N., Stempor P., Chesney M. A., Down T. A., et al. , 2016. Stable Caenorhabditis elegans chromatin domains separate broadly expressed and developmentally regulated genes. Proc. Natl. Acad. Sci. USA 113: E7020–E7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., Yochem J., 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 306: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira H. C., Towbin B. D., Jegou T., Gasser S. M., 2013. The shelterin protein POT-1 anchors Caenorhabditis elegans telomeres through SUN-1 at the nuclear periphery. J. Cell Biol. 203: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., et al. , 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17: 1870–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C., Schubeler D., Martin D. I., Groudine M., 2000. Nuclear compartmentalization and gene activity. Nat. Rev. Mol. Cell Biol. 1: 137–143. [DOI] [PubMed] [Google Scholar]
- Furuhashi H., Kelly W. G., 2010. The epigenetics of germ-line immortality: lessons from an elegant model system. Dev. Growth Differ. 52: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi H., Takasaki T., Rechtsteiner A., Li T., Kimura H., et al. , 2010. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenet. Chromatin 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues J. M., Sidoli S., Garcia B. A., Strome S., 2015. Defining heterochromatin in C. elegans through genome-wide analysis of the heterochromatin protein 1 homolog HPL-2. Genome Res. 25: 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Rechtsteiner A., Yuen K. W., Muroyama A., Egelhofer T., et al. , 2012. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484: 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., Strome S., 2012. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Wang W., Strome S., 2014. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345: 1515–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C., Ikegami K., Ayuso C., de Luis A., Iniguez M., et al. , 2014. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol. 15: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Towbin B. D., Kalck V., Cabianca D. S., Gaidatzis D., et al. , 2015. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163: 1333–1347. [DOI] [PubMed] [Google Scholar]
- Grant J., Verrill C., Coustham V., Arneodo A., Palladino F., et al. , 2010. Perinuclear distribution of heterochromatin in developing C. elegans embryos. Chromosome Res. 18: 873–885. [DOI] [PubMed] [Google Scholar]
- Greer E. L., Beese-Sims S. E., Brookes E., Spadafora R., Zhu Y., et al. , 2014. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep. 7: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Blanco M. A., Gu L., Sendinc E., Liu J., et al. , 2015. DNA methylation on N6-adenine in C. elegans. Cell 161: 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Becker B., Latza C., Antebi A., Shi Y., 2016. Mutation of C. elegans demethylase spr-5 extends transgenerational longevity. Cell Res. 26: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. G., Fire A., 2010. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma 119: 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. G., Pak J., Guang S., Maniar J. M., Kennedy S., et al. , 2012. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet. 44: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Pavelec D. M., Burkhart K. B., Harding S., et al. , 2008. An argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Burkhart K. B., Burton N., Pavelec D. M., et al. , 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Lu X., Horvitz H. R., 2007. LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics 176: 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R. D., Hon G. C., Lee L. K., Ngo Q., Lister R., et al. , 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Coulson A., Murray J. I., Bao Z., Sulston J. E., et al. , 2005. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 15: 1651–1660. [DOI] [PubMed] [Google Scholar]
- Hillier L. W., Marth G. T., Quinlan A. R., Dooling D., Fewell G., et al. , 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5: 183–188. [DOI] [PubMed] [Google Scholar]
- Ho J. W., Jung Y. L., Liu T., Alver B. H., Lee S., et al. , 2014. Comparative analysis of metazoan chromatin organization. Nature 512: 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Sternberg P. W., 1991. Multiple intercellular signalling systems control the development of the Caenorhabditis elegans vulva. Nature 351: 535–541. [DOI] [PubMed] [Google Scholar]
- Hubley R., Finn R. D., Clements J., Eddy S. R., Jones T. A., et al. , 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44: D81–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Egelhofer T. A., Strome S., Lieb J. D., 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. M., Lemmens B. B., Tijsterman M., 2013. A role for the malignant brain tumour (MBT) domain protein LIN-61 in DNA double-strand break repair by homologous recombination. PLoS Genet. 9: e1003339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinava N., Ni J. Z., Peterman K., Chen E., Gu S. G., 2017. Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenet. Chromatin 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D. J., Edwards T. M., Reinke V., Kelly W. G., 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C., Adaixo R., Stunnenberg R., Woolcock K. J., Hiller S., et al. , 2012. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol. Cell 47: 215–227. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S., 2011. Recognition of methylated histones: new twists and variations. Curr. Opin. Struct. Biol. 21: 744–749. [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., et al. , 2010. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Kallin E. M., Zhang Y., 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7: 715–727. [DOI] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B., 2017. Transgenerational transmission of environmental information in C. elegans. Science 356: 320–323. [DOI] [PubMed] [Google Scholar]
- Koester-Eiserfunke N., Fischle W., 2011. H3K9me2/3 binding of the MBT domain protein LIN-61 is essential for Caenorhabditis elegans vulva development. PLoS Genet. 7: e1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra S. M., Helin K., 2012. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13: 297–311. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Lee H. C., Gu W., Shirayama M., Youngman E., Conte D., Jr, et al. , 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Hong S., Lee M. H., Koo H. S., 2015. A PHF8 homolog in C. elegans promotes DNA repair via homologous recombination. PLoS One 10: e0123865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Wang Y., Wang Y., Tian F., Pu P., et al. , 2010. Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res. 20: 899–907. [DOI] [PubMed] [Google Scholar]
- Liu H., Galka M., Iberg A., Wang Z., Li L., et al. , 2010. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets. J. Proteome Res. 9: 5827–5836. [DOI] [PubMed] [Google Scholar]
- Liu T., Rechtsteiner A., Egelhofer T. A., Vielle A., Latorre I., et al. , 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y., Prinz J. A., DeNapoli L., Li Y., Belsky J. A., et al. , 2014. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 24: 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg M. S., Dinant C., Lans H., Stap J., Wiernasz E., et al. , 2009. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 185: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M. J., van Bergeijk P., Kaaij L. J., Almeida M. V., Roovers E. F., et al. , 2012. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31: 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P. S., Oegema K., Desai A., Cheeseman I. M., 2004. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 12: 641–653. [DOI] [PubMed] [Google Scholar]
- Maison C., Almouzni G., 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5: 296–304. [DOI] [PubMed] [Google Scholar]
- Mao H., Zhu C., Zong D., Weng C., Yang X., et al. , 2015. The Nrde pathway mediates small-RNA-directed histone H3 lysine 27 trimethylation in Caenorhabditis elegans. Curr. Biol. 25: 2398–2403. [DOI] [PubMed] [Google Scholar]
- Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., et al. , 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchy A. N., Stempor P., Gaarenstroom T., Wysolmerski B., Dong Y., et al. , 2017. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. Elife 6: e21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Kao C. F., Pennings S., 2003. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 22: 3164–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Towbin B. D., Pike B. L., Ponti A., Gasser S. M., 2010. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 24: 766–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Schott S., Bedet C., Xiao Y., Rohner S., et al. , 2011. Caenorhabditis elegans heterochromatin protein 1 (HPL-2) links developmental plasticity, longevity and lipid metabolism. Genome Biol. 12: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A., Greenwald I., 2000. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics 155: 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C., Jovaisaite V., Durieux J., Matilainen O., Jordan S. D., et al. , 2016. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J., 2010. Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 20: 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Guilleme M., Seeler J. S., Trouche D., Dejean A., et al. , 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3: 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I., 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Nelson D. M., Jaber-Hijazi F., Cole J. J., Robertson N. A., Pawlikowski J. S., et al. , 2016. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 17: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Z., Kalinava N., Chen E., Huang A., Trinh T., et al. , 2016. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Ebata A., Alipanahiramandi E., Lee S. S., 2012. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 11: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., et al. , 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107. [DOI] [PubMed] [Google Scholar]
- Passannante M., Marti C. O., Pfefferli C., Moroni P. S., Kaeser-Pebernard S., et al. , 2010. Different Mi-2 complexes for various developmental functions in Caenorhabditis elegans. PLoS One 5: e13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T., Tursun B., Rahe D. P., Hobert O., 2012. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. C., Karpen G. H., 2009. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5: e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V., et al. , 2011. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138: 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A., Shilatifard A., 2016. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352: aad9780. [DOI] [PubMed] [Google Scholar]
- Plohl M., Luchetti A., Mestrovic N., Mantovani B., 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409: 72–82. [DOI] [PubMed] [Google Scholar]
- Poulin G., Dong Y., Fraser A. G., Hopper N. A., Ahringer J., 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24: 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtsteiner A., Ercan S., Takasaki T., Phippen T. M., Egelhofer T. A., et al. , 2010. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 6: e1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveiro A. R., Mariani L., Malmberg E., Amendola P. G., Peltonen J., et al. , 2017. JMJD-1.2/PHF8 controls axon guidance by regulating Hedgehog-like signaling. Development 144: 856–865. [DOI] [PubMed] [Google Scholar]
- Ross J. M., Zarkower D., 2003. Polycomb group regulation of Hox gene expression in C. elegans. Dev. Cell 4: 891–901. [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Jan C., Player C., Axtell M. J., Lee W., et al. , 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207. [DOI] [PubMed] [Google Scholar]
- Saksouk N., Simboeck E., Dejardin J., 2015. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges F. W., Prusty A. B., Faty M., Stutzer A., Lingaraju G. M., et al. , 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42: 330–341. [DOI] [PubMed] [Google Scholar]
- Schott S., Coustham V., Simonet T., Bedet C., Palladino F., 2006. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev. Biol. 298: 176–187. [DOI] [PubMed] [Google Scholar]
- Schott S., Ramos F., Coustham V., Palladino F., 2009. HPL-2/HP1 prevents inappropriate vulval induction in Caenorhabditis elegans by acting in both HYP7 and vulval precursor cells. Genetics 181: 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler M. G., Sullivan B. A., 2006. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 7: 301–313. [DOI] [PubMed] [Google Scholar]
- Schwaiger M., Kohler H., Oakeley E. J., Stadler M. B., Schubeler D., 2010. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 20: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwammle V., Sidoli S., Ruminowicz C., Wu X., Lee C. F., et al. , 2016. Systems level analysis of histone H3 post-translational modifications (PTMs) reveals features of PTM crosstalk in chromatin regulation. Mol. Cell. Proteomics 15: 2715–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelk S., Adrian-Kalchhauser I., Hargitai B., Hajduskova M., Gutnik S., et al. , 2016. Increasing Notch signaling antagonizes PRC2-mediated silencing to promote reprograming of germ cells into neurons. Elife 5: e15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Schober H., Fraser P., Gasser S. M., 2007. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 14: 1049–1055. [DOI] [PubMed] [Google Scholar]
- Shin J. H., Wang H. L., Lee J., Dinwiddie B. L., Belostotsky D. A., et al. , 2013. The role of the Arabidopsis Exosome in siRNA-independent silencing of heterochromatic loci. PLoS Genet. 9: e1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H. C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S., Vandamme J., Salcini A. E., Jensen O. N., 2016. Dynamic changes of histone H3 marks during Caenorhabditis elegans lifecycle revealed by middle-down proteomics. Proteomics 16: 459–464. [DOI] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E., 2013. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Sarkies P., Ikegami K., Doebley A. L., Goldstein L. D., et al. , 2014. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T., Dulermo R., Schott S., Palladino F., 2007. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev. Biol. 312: 367–383. [DOI] [PubMed] [Google Scholar]
- Snyder M. J., Lau A. C., Brouhard E. A., Davis M. B., Jiang J., et al. , 2016. Anchoring of heterochromatin to the nuclear lamina reinforces dosage compensation-mediated gene repression. PLoS Genet. 12: e1006341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G., Fields B., Wan G., Becker D., Wallig A., et al. , 2017. The RNAi inheritance machinery of Caenorhabditis elegans. Genetics 206: 1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G., Fields B., Wan G., Becker D., Wallig A., et al. , 2017. The RNAi inheritance machinery of C. elegans. Genetics 206: 1403–1416. DOI: 10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P. A., Ringrose L., 2014. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 15: 340–356. [DOI] [PubMed] [Google Scholar]
- Strome S., Kelly W. G., Ercan S., Lieb J. D., 2014. Regulation of the X chromosomes in Caenorhabditis elegans. Cold Spring Harb. Perspect. Biol. 6: a018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studencka M., Wesolowski R., Opitz L., Salinas-Riester G., Wisniewski J. R., et al. , 2012. Transcriptional repression of Hox genes by C. elegans HP1/HPL and H1/HIS-24. PLoS Genet. 8: e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subirana J. A., Alba M. M., Messeguer X., 2015. High evolutionary turnover of satellite families in Caenorhabditis. BMC Evol. Biol. 15: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Thillainadesan G., Chalamcharla V. R., Meng Z., Balachandran V., et al. , 2016. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell 61: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J., 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14: 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B. D., Meister P., Pike B. L., Gasser S. M., 2010. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb. Symp. Quant. Biol. 75: 555–565. [DOI] [PubMed] [Google Scholar]
- Towbin B. D., Gonzalez-Aguilera C., Sack R., Gaidatzis D., Kalck V., et al. , 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. [DOI] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G., et al. , 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J. F., Ohle C., Schermann G., Bendrin K., Zhang W., et al. , 2016. A novel epigenetic silencing pathway involving the highly conserved 5′-3′ exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe. PLoS Genet. 12: e1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., Shin T. H., Miliaras N., Lee J., Oyama T., et al. , 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002. [DOI] [PubMed] [Google Scholar]
- Vandamme J., Lettier G., Sidoli S., Di Schiavi E., Norregaard Jensen O., et al. , 2012. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet. 8: e1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme J., Sidoli S., Mariani L., Friis C., Christensen J., et al. , 2015. H3K23me2 is a new heterochromatic mark in Caenorhabditis elegans. Nucleic Acids Res. 43: 9694–9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A., Lang J., Dong Y., Ercan S., Kotwaliwale C., et al. , 2012. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet. 8: e1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky T., Palladino F., Brunschwig K., Tobler H., Hajnal A., et al. , 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127: 5277–5284. [DOI] [PubMed] [Google Scholar]
- Wang G., Reinke V., 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18: 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jia S. T., Jia S., 2016. New insights into the regulation of heterochromatin. Trends Genet. 32: 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D., Jr., Kim J. K., Gabel H. W., et al. , 2005. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436: 593–597. [DOI] [PubMed] [Google Scholar]
- Wang J. K., Tsai M. C., Poulin G., Adler A. S., Chen S., et al. , 2010. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 24: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser N. E., Yang D. X., Feng S., Kalinava N., Brown K. C., et al. , 2017. MORC-1 integrates nuclear RNAi and transgenerational chromatin architecture to promote germline immortality. Dev. Cell 41: 408–423.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., et al. , 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481. [DOI] [PubMed] [Google Scholar]
- Wiles E. T., Selker E. U., 2016. H3K27 methylation: a promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43: 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Shi Z., Cui M., Han M., Ruvkun G., 2012. Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 8: e1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Bian C., Yang W., Galka M., Ouyang H., et al. , 2010. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc. Natl. Acad. Sci. USA 107: 19266–19271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates T. O., 2002. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell 111: 5–7. [DOI] [PubMed] [Google Scholar]
- Yuan W., Xu M., Huang C., Liu N., Chen S., et al. , 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286: 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T., Fakhouri T. H., Kiefer J., Mango S. E., 2009. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev. Cell 16: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M., et al. , 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Zheng C., Karimzadegan S., Chiang V., Chalfie M., 2013. Histone methylation restrains the expression of subtype-specific genes during terminal neuronal differentiation in Caenorhabditis elegans. PLoS Genet. 9: e1004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Ahier A., Portoso M., White E. R., Morin M. C., et al. , 2014. Transdifferentiation. Sequential histone-modifying activities determine the robustness of transdifferentiation. Science 345: 826–829. [DOI] [PubMed] [Google Scholar]