Development of a eukaryotic karyotype relies on identification of individual chromosomes in the species, which has been accomplished only in a limited...
Keywords: chromosome identification, karyotype, oligo-FISH, chromosome painting, translocation
Abstract
Developing the karyotype of a eukaryotic species relies on identification of individual chromosomes, which has been a major challenge for most nonmodel plant and animal species. We developed a novel chromosome identification system by selecting and labeling oligonucleotides (oligos) located in specific regions on every chromosome. We selected a set of 54,672 oligos (45 nt) based on single copy DNA sequences in the potato genome. These oligos generated 26 distinct FISH signals that can be used as a “bar code” or “banding pattern” to uniquely label each of the 12 chromosomes from both diploid and polyploid (4× and 6×) potato species. Remarkably, the same bar code can be used to identify the 12 homeologous chromosomes among distantly related Solanum species, including tomato and eggplant. Accurate karyotypes based on individually identified chromosomes were established in six Solanum species that have diverged for >15 MY. These six species have maintained a similar karyotype; however, modifications to the FISH signal bar code led to the discovery of two reciprocal chromosomal translocations in Solanum etuberosum and S. caripense. We also validated these translocations by oligo-based chromosome painting. We demonstrate that the oligo-based FISH techniques are powerful new tools for chromosome identification and karyotyping research, especially for nonmodel plant species.
THE karyotype of a eukaryotic species represents the number, size, and shape of all chromosomes in the nucleus. Karyotype has long been used as the most general description of the basic genetic makeup of individual eukaryotic species. In most lineages, closely related species share a similar karyotype. For example, gorilla (Gorilla gorilla) diverged from the human/chimpanzee (Pan troglodytes) lineages >10 MYA and human and chimpanzee have been separated by 7–8 MY (Langergraber et al. 2012). These three species, however, have maintained a similar karyotype, except that human chromosome 2 was fused from two different chromosomes, resulting in the reduction of chromosome number from 2n = 48 in chimpanzee and gorilla to 2n = 46 in humans (Jauch et al. 1992).
Karyotype analysis relies on the identification of individual chromosomes and has been a challenge for most nonmodel plant and animal species, especially those with polyploidy and/or those with a large number of small chromosomes. Chromosome banding and fluorescence in situ hybridization (FISH) were two milestone techniques in the history of chromosome identification and karyotype analysis. Unfortunately, only a few plant species with large chromosomes have benefited from the chromosome banding techniques (Friebe et al. 1996). G-banding, which is commonly used in karyotyping in mammalian species, does not generate bands on chromosomes from most plants (Greilhuber 1977; Anderson et al. 1982); while FISH can be universally applied in plant species (Schwarzacher et al. 1989; Lim et al. 2000; Mandakova et al. 2010; Szinay et al. 2012; Weiss-Schneeweiss and Schneeweiss 2013). Various types of DNA probes can be used in FISH, including repetitive DNA sequences (Mukai et al. 1993; Fransz et al. 1998; Kato et al. 2004) and large-insert genomic DNA clones (Jiang et al. 1995; Dong et al. 2000; Kulikova et al. 2001; Kim et al. 2002). However, it is often a major challenge to establish a FISH-based chromosome identification system in a nonmodel species because of the lack of chromosome-specific DNA probes. Although karyotypes have been described in many plant species, individual chromosomes were not identified in most of these reported karyotypes. Such karyotypes, therefore, are not comparable among related species and cannot be used for evolutionary studies.
The Solanaceae is an important plant family comprising >3000 species. One of the genus, Solanum, contains several major food crops, including potato, tomato, and eggplant. Solanaceae species were derived ∼40 MYA from an ancestral diploid species with 2n = 24 chromosomes. Nearly all diploid family members have maintained this chromosome number (Wu et al. 2006). However, this identical basic chromosome number does not indicate maintenance of genomic synteny of the 12 homeologous chromosomes among the solanaceous species. Although both potato and tomato genomes have been sequenced (The Potato Genome Sequencing Consortium 2011; The Tomato Genome Consortium 2012), the karyotypes, genomes, and their evolution in other solanaceous species are largely unknown.We developed a novel chromosome identification system using solanaceous species as a model. We selected a set of 54,672 oligonucleotides (oligos) from the single copy sequences associated with 26 specific chromosome regions in the potato genome. These oligos were massively synthesized de novo in parallel and were labeled as FISH probes (Han et al. 2015). The pooled oligos produced 26 distinct FISH signals, which can be used as a “bar code” or a “banding pattern” to identify all 12 potato chromosomes. Strikingly, this bar code has been maintained among distantly related Solanum species, including tomato and eggplant, which diverged from potato ∼5–8 and 15 MYA, respectively (Y. Wang et al. 2008; Wu and Tanksley 2010; Sarkinen et al. 2013). Modifications to this bar code in different species can be inferred as potential rearrangements of the associated chromosome(s) during evolution. We demonstrate that the oligo-FISH-based techniques are powerful new tools for chromosome identification and karyotyping research in nonmodel species.
Materials and Methods
Plant materials
Seven diploid species were used in FISH mapping, including the doubled monoploid Solanum tuberosum Group Phureja clone DM1-3 516 R44 [doubled monoploid (DM)], S. bulbocastanum (PI 498223; Oaxaca, Mexico), tomato (S. lycopersicum) variety Micro Tom, S. etuberosum (E genome, PI 558306; O’Higgins, Chile), S. melongena (eggplant) (PI 665010, cultivar Black Beauty), S. caripense (PI 243342, Costa Rica), and pepper (Capsicum annuum var. annuum ACE F1). Tetraploid potato cultivar “Katahdin” and hexaploid species S. demissum (PI 225711; Boyaca, Colombia) were also used in FISH mapping.
Oligo-FISH probe design
The oligo probes were designed using Chorus software (https://github.com/forrestzhang/Chorus) with only minor modifications (Han et al. 2015). Briefly, the repetitive sequences in the potato genome (The Potato Genome Sequencing Consortium 2011; Hardigan et al. 2016) were filtered and remaining sequences were then divided into oligos (45 nt) in a step size of 5 nt. Each oligo was aligned to the potato genome to filter out those with duplicates in the genome (>75% similarity over all 45 nt). Oligos within the centromeric regions (Gong et al. 2012) were also excluded. Oligos with dTm >10 [dTm = melting temperature (Tm) − hairpin Tm] were kept to build a probe database. Oligo sequences that were homologous to the tomato genome were preferentially selected for chromosome painting probes. We adjusted the number of oligos across the chromosomes to ensure that the painting probes produce uniform signals on the entire chromosomes. For the bar code oligo probes, we first selected target regions with a relatively high density of oligos based on the density distribution profile on the entire chromosome. We then selected oligos that show >90% homology with tomato sequences. The oligos were synthesized by Arbor Biosciences (Ann Arbor, MI) were labeled following published protocols (Han et al. 2015).
Oligo-FISH
To prepare mitotic metaphase chromosomes, root tips were harvested from greenhouse-grown plants and treated with nitrous oxide at a pressure of 160 psi (∼10.9 atm) for 20–50 min. The root tips were then fixed in fixative solution (3 ethanol:1 acetic acid) and kept at −20°. An enzymatic solution with 3% cellulase (Yakult Pharmaceutical, Tokyo, Japan), 1.5% pectinase (Plant Media), and 1% pectolyase (Sigma Chemical, St. Louis, MO) was used to digest the root tips for 50 min at 37°, and slides were prepared using a stirring method. Briefly, root tips were put on a microscope slide and macerated with a needle in 20 µl of 45% acetic acid. Then, the suspension was spread with a needle on a hot plate at 50° for 2 min. Chromosomes were fixed by adding 200 µl of ethanol:acetic acid (3:1) fixative solution on a hot plate at 50° for 10 sec. Afterward, an additional 200 µl of ethanol:acetic acid (3:1) fixative solution was dropped on the tilted slide, which was dried at room temperature. Slides were also prepared using the dropping method (Kato et al. 2004) for chromosome painting experiments.
FISH was performed following published protocols (Dong et al. 2000). The hybridization mixture (500 ng of each labeled probe of single-stranded DNA, 50% formamide, 10% dextran sulfate, 2× SSC) was applied directly to denaturated chromosome slides and incubated for 2 days at 37°. Approximately 2000 ng of sheared genomic DNA (with average size of 100 bp) prepared from S. etuberosum and S. caripense was used as blocking DNA in chromosome painting experiments. The hybridization mixture for chromosome painting was denatured at 95° for 8 min and incubated at 37° for 2 hr before being applied to denatured chromosome slides. Biotin- and digoxigenin-labeled probes were detected by anti-biotin fluorescein (Vector Laboratories, Burlingame, CA) and anti-digoxigenin rhodamine (Roche Diagnostics, Indianapolis, Indiana), respectively. Chromosomes were counterstained with DAPI in VectaShield antifade solution (Vector Laboratories). FISH images were captured using a QImaging Retiga EXi Fast 1394 CCD camera attached to an Olympus BX51 epifluorescence microscope. Images were processed with Meta Imaging Series 7.5 software. The final contrast of the images was processed using Adobe Photoshop CS3 software.
Karyotyping
The short (S) and long (L) arms of individual chromosomes were measured from 10 complete metaphase cells for each species using the computer application MicroMeasure version 3.3 (Reeves and Tear 2000). The chromosomal arm measurements were used to calculate the total length of each chromosome (tl = S + L), total length of entire set of chromosomes (TL = ∑tl), arm ratio (AR = L/S) of each chromosome, and relative length of each chromosome (RL = tl/TL × 100).
Synteny analysis of potato and tomato DNA sequence
Potato genome (V404) (The Potato Genome Sequencing Consortium 2011) and tomato genome (SL3.0) (The Tomato Genome Consortium 2012) were aligned using MUMmer 3 (Kurtz et al. 2004). The parameters used for mummer were “-mum -n -c -b -l 30” and the parameters used for gaps were “-l 60 -f .12 -s 1000.” Synteny blocks between potato and tomato genome were identified using DAGchainer (Haas et al. 2004) with parameters “-o -0f -e-2f -A 10.” The positions of potato and tomato centromeres were determined as the major peaks of CENH3 chromatin immunoprecipitation-sequencing reads for each chromosome. For chromosomes with unassembled centromeric/pericentromeric sequences, the centromere positions were determined by analyzing the distribution of centromeric repeats, transposable elements, and sequencing gaps in the chromosomes.
Data availability
Supplemental Material, Table S1 in File S1 contains all information about the number and locations of oligos associated with each of the 26 individual FISH signals generated by the two bar code FISH probes. The Chorus software used for oligo-FISH probe design is freely available (https://github.com/forrestzhang/Chorus).
Results
Development of oligo-based FISH probes for chromosome identification in Solanum species
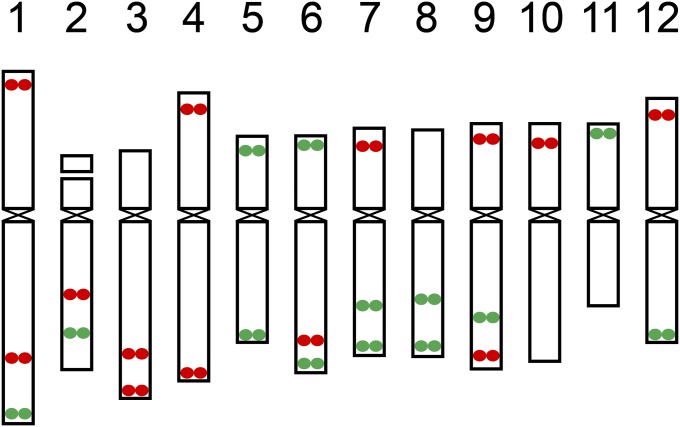
We developed two oligo-FISH probes: PB9446 (green) and PB8495 (red). These two probes contain 27,306 and 27,366 oligos (45 nt), respectively, and are derived from 26 different regions on the 12 potato chromosomes (Table S1 in File S1). These two probes were designed to produce 26 distinct FISH signals, which can be used as a bar code or banding pattern to uniquely label each of the 12 potato chromosomes (Figure 1). Each chromosomal region is covered by 2000–2250 oligos (Table S1 in File S1) that were selected using our oligo-FISH probe development pipeline (Han et al. 2015). The oligos were selected from single copy sequences in the potato genome (The Potato Genome Sequencing Consortium 2011; Hardigan et al. 2016). The oligos associated with each of 26 FISH signals spanned a genomic region ranging from 184 to 707 kb (Table S1 in File S1). Some chromosomal arms contained two signals, which were separated by at least 7 Mb (Table S1 in File S1) to ensure the separation of the two signals on the same arm.
Figure 1.
Predicted locations of the oligo-FISH signals on 12 potato chromosomes. Oligos were selected from a total of 26 chromosomal regions (13 red regions and 13 green regions). The 12 chromosomes can be distinguished from each other based on number and location of the red/green signals. The centromere positions on the 12 chromosomes in the potato reference genome were based on the locations of sequences associated with CENH3 nucleosomes (Gong et al. 2012).
A total of 54,672 oligos were included in the two probes. Sequence analysis showed that 33,911 oligos (62%) are associated with annotated potato genes, including 16,489 with coding sequences, 13,354 with introns, and 4068 with 5′ and 3′ UTRs. The remaining oligos were derived from intergenic regions. We analyzed the sequence similarity of these potato oligos with the tomato genome sequence (The Tomato Genome Consortium 2012). Only 3023 oligos (11%) were identical to the corresponding tomato sequences. In addition, 19,033 oligos (35%) showed one to four mismatches (>90% homology) with the tomato sequences.
Chromosome identification in diploid and polyploid potato species
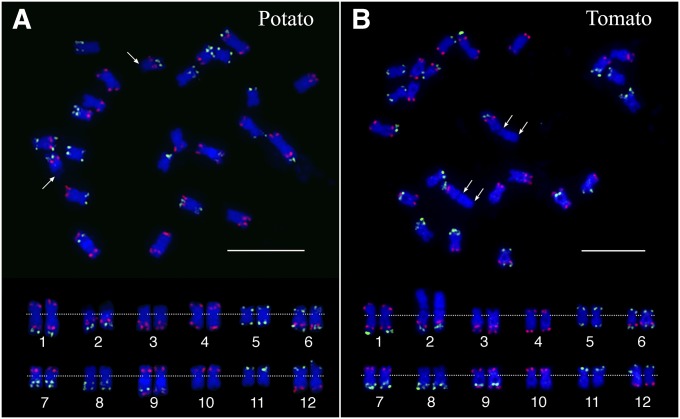
The two oligo-FISH probes were labeled and hybridized to the somatic metaphase chromosomes prepared from S. tuberosum Group Phureja clone DM1-3 516 R44 (2n = 2x = 24) (DM), which is a homozygous clone and has been fully sequenced (The Potato Genome Sequencing Consortium 2011). The green and red FISH signals derived from the two probes (Figure 2A) matched to the predicted patterns (Figure 1). The signals formed a bar code that uniquely labels the 12 chromosomes. Chromosome 2 is the only nucleolus organizer (Nor) chromosome in the potato genome (Dong et al. 2000). The 45S ribosomal RNA genes were located at the distal end of the short arm, which is distinctly decondensed and stained faintly by DAPI (Figure S1 in File S1). Karyotyping analysis revealed that most potato chromosomes are metacentric or submetacentric (except for chromosome 2) with an arm ratio ranging from 2.67 to 1.19 (Table 1). Chromosomes 1 and 2 (without including the 45S rDNA region) represent the largest and smallest chromosomes, respectively (Table S2 in File S1).
Figure 2.
FISH mapping of potato and tomato chromosomes using two oligo-FISH probes. (A) FISH mapping of DM potato. Arrows point to the 45S rDNA regions associated with chromosome 2 (FISH mapping of the 45S rDNA on the same metaphase cell is shown in Figure S1 in File S1). The rDNA region is distinctly decondensed compared to the rest of the chromosome. (B) FISH mapping of tomato. The double arrows indicate the extent of the 45S rDNA regions (FISH mapping of the 45S rDNA on the same metaphase cell is shown in Figure S1 in File S1). The rDNA region is similarly condensed compared with the rest of the chromosome. The top panels show a complete metaphase cell from potato and tomato, respectively. Homologous chromosomes in the bottom panel were digitally excised from the same cells and paired. The centromeres of the chromosomes are aligned by a dotted line. Bar, 10 µm.
Table 1. Arm ratio of individual chromosomes in six Solanum species.
| Chromosome | S. tuberosum (potato) | S. bulbocastanum | S. lycopersicum (tomato) | S. etuberosum | S. caripense (tzimbalo) | S. melongena (eggplant) |
|---|---|---|---|---|---|---|
| 1 | 1.80 ± 0.46 | 2.20 ± 0.38 | 1.57 ± 0.25 | 1.71 ± 0.63 | 2.56 ± 0.60 | 1.44 ± 0.27 |
| 2a | 3.63 ± 0.61 | 3.94 ± 0.73 | 3.31 ± 1.37 | 2.89 ± 0.60 | 3.32 ± 1.13 | 2.58 ± 0.76 |
| 3 | 2.67 ± 0.49 | 2.29 ± 0.64 | 2.96 ± 0.49 | 1.76 ± 0.31 | 2.83 ± 0.68 | 1.37 ± 0.18 |
| 4 | 1.50 ± 0.22 | 1.64 ± 0.25 | 2.21 ± 0.37 | 1.21 ± 0.14 | 1.43 ± 0.43 | 1.46 ± 0.24 |
| 5 | 1.30 ± 0.13 | 1.32 ± 0.22 | 1.17 ± 0.12 | 1.25 ± 0.14 | 1.23 ± 0.24 | 1.35 ± 0.21 |
| 6 | 1.98 ± 0.29 | 1.78 ± 0.49 | 2.11 ± 0.34 | 1.63 ± 0.27 | 2.53 ± 0.64 | 1.63 ± 0.25 |
| 7 | 1.85 ± 0.32 | 1.75 ± 0.21 | 1.67 ± 0.32 | 1.27 ± 0.28 | 2.40 ± 0.69 | 1.20 ± 0.13 |
| 8 | 1.90 ± 0.25 | 2.04 ± 0.49 | 1.84 ± 0.38 | 2.37 ± 0.54 | 2.61 ± 0.55 | 1.14 ± 0.11 |
| 9 | 1.96 ± 0.29 | 1.55 ± 0.18 | 1.81 ± 0.29 | 1.26 ± 0.22 | 1.58 ± 0.48 | 1.47 ± 0.20 |
| 10 | 1.38 ± 0.18 | 1.50 ± 0.33 | 1.44 ± 0.20 | 1.51 ± 0.19 | 1.52 ± 0.32 | 1.48 ± 0.20 |
| 11 | 1.19 ± 0.15 | 1.16 ± 0.10 | 1.55 ± 0.17 | 1.17 ± 0.11 | 1.50 ± 0.34 | 1.22 ± 0.28 |
| 12 | 1.43 ± 0.27 | 1.28 ± 0.21 | 1.32 ± 0.21 | 1.53 ± 0.32 | 1.49 ± 0.65 | 1.12 ± 0.09 |
Measurement was conducted on each chromosomal arm in 10 metaphase cells.
The 45S rDNA on the short arm of chromosome 2 was not included in the measurement.
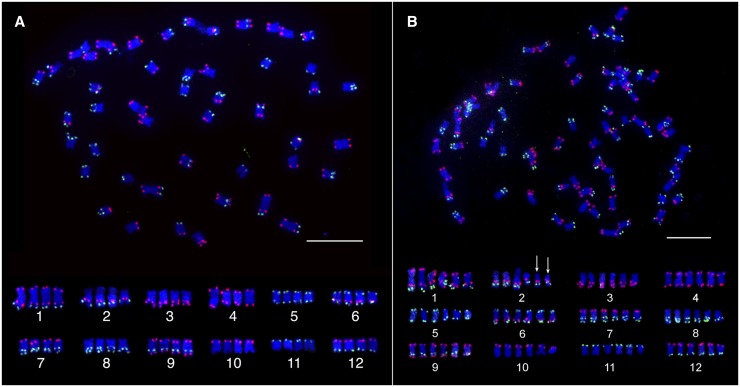
We then performed FISH on cultivated potato (S. tuberosum, 2n = 4x = 48), an autotetraploid species. We observed four identical copies of each of the 12 chromosomes from potato cultivar Katahdin (Figure 3A). S. demissum (2n = 6x = 72) was recognized as an allohexaploid species based on traditional chromosome pairing analyses of hybrids between S. demissum and various Solanum species (Matsubayashi 1991). The consensus conclusion from traditional cytogenetic studies was that S. demissum contains two similar genomes that differ from the third genome (Matsubayashi 1991). We identified 6 copies of each of the 12 potato chromosomes in S. demissum (Figure 3B). The FISH signal patterns from the six homologous/homeologous chromosomes were identical to those from DM potato. Interestingly, two of the six copies of chromosome 2 lack the 45S ribosomal gene arrays (Figure S2 in File S1).
Figure 3.
Chromosome identification in polyploid Solanum species. (A) Chromosome identification of potato cultivar Katahdin. The top panel shows a complete metaphase cell hybridized with two oligo-FISH probes. The bottom panel shows the 4 homologous chromosomes of each of the 12 potato chromosomes digitally excised from the same cell. (B) Chromosome identification in the hexaploid species S. demissum. The top panel shows a complete metaphase cell hybridized with two oligo-FISH probes. The bottom panel shows the 6 homologous chromosomes of each of the 12 potato chromosomes digitally excised from the same cell. The two arrows indicate the two copies of chromosome 2 that are not associated with 45S rDNA (FISH mapping of the 45S rDNA is showed in Figure S2 in File S1). Bar, 10 µm.
Comparative karyotyping of potato and tomato
DNA sequence-based analysis suggested that tomato and potato have diverged for ∼5–8 MY (Y. Wang et al. 2008; Sarkinen et al. 2013). Chromosome synteny between the potato and tomato has been well maintained based on comparative genetic linkage mapping and comparative cytogenetic mapping (Tanksley et al. 1992; Iovene et al. 2008; Tang et al. 2008; Gaiero et al. 2017). We conducted DNA sequence-based synteny analyses between the 12 pairs of pseudomolecules from potato and tomato genomes. Multiple inversions in different sizes were found to be associated with all 12 homeologous chromosome pairs (Figure S3 in File S1), which revealed abundant intrachromosomal rearrangements, but no interchromosomal arrangement, occurred during the divergence of these two species.
The two oligo-FISH probes generated an identical signal bar code on tomato and potato chromosomes (Figure 2). Two tomato chromosomes showed distinct morphology compared to the potato homeologues. The tomato 45S ribosomal RNA genes were also located at the distal region of the short arm of chromosome 2 (Figure 2B and Figure S1 in File S1). However, the 45S ribosomal DNA (rDNA) region was as condensed as the rest of the tomato chromosome 2 (Figure S1 in File S1), which was consistently observed in all metaphase cells. This unique condensation pattern of the 45S rDNA region makes chromosome 2 the longest chromosome in tomato (Figure 2B). Chromosome 4 from the two species showed a distinct difference in arm ratios. Potato chromosome 4 is a submetacentric chromosome with an arm ratio of 1.50; while tomato chromosome 4 appeared to be a subtelocentric (or acrocentric) chromosome with an arm ratio of 2.21 (Figure 2 and Table 1). At least two inversions in the long arms, each spanning several megabases of DNA, distinguished the two chromosomes (Figure S3 in File S1). By contrast, no inversion was detected in the short arms of the two chromosomes. It is not clear whether the different arm ratios of these two chromosomes were caused by an inversion that spanned the centromere of the chromosome in one species or by some other chromosomal rearrangement events.
Comparative karyotyping of Solanum species that are distantly related to potato
To reveal the karyotype evolution of the Solanum species, we performed comparative oligo-FISH in five additional species using the two probes developed in potato. These species have diverged variously from potato, including S. bulbocastanum (a wild species closely related to potato), S. etuberosum, S. caripense (tzimbalo), S. melongena (eggplant), and C. annuum (pepper), which are more distantly related to potato than tomato is to potato (Lou et al. 2010).
S. bulbocastanum:
The FISH signals generated on S. bulbocastanum chromosomes were identical to those from potato (Figure 4). The arm ratio (Table 1) and relative length (Table S2 in File S1) of individual S. bulbocastanum chromosomes were also highly similar to the homeologous potato chromosomes.
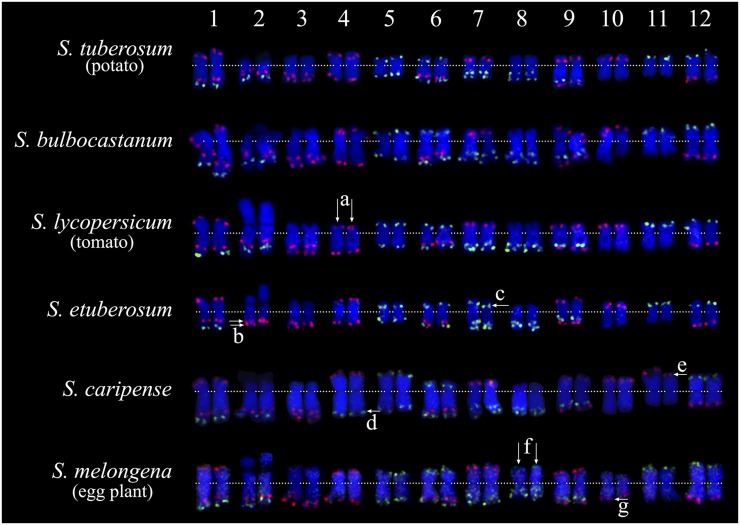
Figure 4.
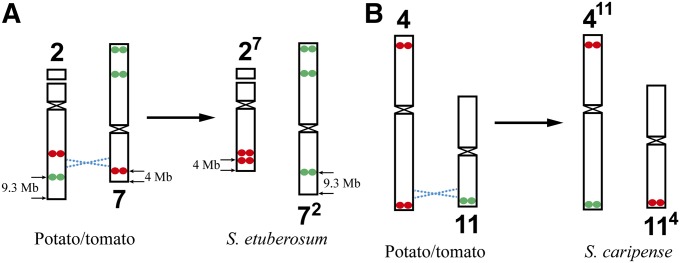
Comparative karyotyping of six diploid Solanum species. Chromosomes 1–12 from each species are arranged from left to right. Karyotypes of potato and tomato were developed from the same metaphase cells in Figure 2. Karyotypes of the remaining four species are developed from the same metaphase cells in Figure S4 in File S1. (a) Double arrows point to the two copies of tomato chromosome 4, which have a distinct arm ratio compared to chromosome 4 from other species. (b) Double arrows point to two closely linked red signals on S. etuberosum chromosome 2, the bottom red signal is predicted to be derived from the short arm of chromosome 7. For comparison, we used the karyotype of potato as our reference, see switches between red and green signals among these two species. (c) Arrow indicates the green signal on the short arm of S. etuberosum chromosome 7, which is predicted to be derived from the long arm of chromosome 2. (d) Arrow points to the green signal on the long arm of S. caripense chromosome 4, which is predicted to be derived from the short arm of chromosome 11. (e) Arrow points to the red signal on S. caripense chromosome 11, which is predicted to be derived from the long arm of chromosome 4. (f) Double arrows point to the two copies of eggplant chromosome 8, which have a distinctly large short arm compared to chromosome 8 from other species. (g) Arrow indicates the location of the red signal on the long arm of eggplant chromosome 10. This signal is located at the short arm of chromosome 10 from other species.
S. etuberosum:
S. etuberosum is a nontuberizing wild species that has been used in potato breeding due to its resistance to various potato diseases (Dong et al. 1999; Novy et al. 2002, 2007). Phylogenetically, S. etuberosum is more distantly related to potato than tomato is to potato (Lou et al. 2010). The FISH signals on most S. etuberosum chromosomes were identical to those on potato chromosomes. Chromosome 2 is the sole Nor chromosome (Figure S4 in File S1). However, signal modifications were observed on chromosomes 2 and 7. The long arm of chromosome 2 lost its distal green signal and gained an additional red signal (“b” in Figure 4). By contrast, the short arm of chromosome 7 lost its distal red signal but gained a green signal (“c” in Figure 4). A reciprocal translocation between chromosomes 2 and 7 would explain the observed FISH signal pattern changes (Figure 5A). The distal red signal on S. etuberosum chromosome 2 is more close to the end of the chromosome compared to the distal green signal on potato/tomato chromosome 2 (Figure 4). This can be explained by the fact that the green signal on chromosome 2 is 9.3 Mb away from the end, while red signal on chromosome 7 is only 4 Mb away from the end (Figure 5A). The other 10 S. etuberosum chromosomes showed a similar arm ratio and relative length to the homeologous potato chromosomes (Table 1 and Table S2 in File S1).
Figure 5.
Predicted reciprocal chromosomal translocations identified in Solanum species. (A) A reciprocal translocation between chromosomes 2 and 7 in S. etuberosum. Chromosomes 2 and 7 from potato/tomato are hypothesized to be the ancestral types. A reciprocal translocation (dashed blue lines) is predicted based on the modifications to the oligo-FISH bar code, which result in the two translocation chromosomes 27 and 72, respectively, in S. etuberosum. (B) A reciprocal translocation between chromosomes 4 and 11 in S. caripense. The chromosomes 4 and 11 from potato/tomato are hypothesized to be the ancestral types. A reciprocal translocation (dashed blue lines) is predicted based on the modifications of the oligo-FISH bar code, which result in the two translocation chromosomes 411 and 114, respectively, in S. caripense.
S. caripense:
S. caripense, also known as tzimbalo, is an evergreen shrub native to South America and is grown for its edible fruit. The S. caripense chromosomes were visibly larger than potato chromosomes. Phylogenetically, S. caripense is more distantly related to potato than S. etuberosum is to potato (Lou et al. 2010). Overall S. caripense showed a similar karyotype as potato and tomato. However, we observed distinct FISH signal patterns on chromosomes 4 and 11, respectively. The red signal on the long arm of chromosome 4 was replaced by a green signal (“d” in Figure 4). On the other hand, the green signal on the short arm of chromosome 11 was replaced by a red signal (“e” in Figure 4). A reciprocal translocation between chromosomes 4 and 11 would explain this signal pattern change (Figure 5B). The rest of the S. caripense chromosomes showed a similar arm ratio and relative length to the homeologous potato chromosomes (Table 1 and Table S2 in File S1).
Eggplant (S. melongena):
Eggplant diverged from a common ancestor of potato/tomato ∼15.5 MYA (Wu and Tanksley 2010). The two oligo-FISH probes generated uniform but generally weak background signals on all eggplant chromosomes. Surprisingly, the patterns derived from the major FISH signals matched those from potato and tomato chromosomes (Figure 4). Eggplant chromosome 8 is a metacentric chromosome with an arm ratio of 1.14. However, chromosome 8 from the other five Solanum species have subtelocentric morphology with an arm ratio ranging from 1.84 to 2.61 (Figure 4 and Table 1). Since the two green signals on the long arm of chromosome 8 of S. melongena were clearly closer to the centromere than those on chromosome 8 of other Solanum species (“f” in Figure 4), chromosome 8 of S. melongena likely resulted from an inversion spanning the centromere, and a large fragment from the long arm was moved to the short arm due to the inversion. Similarly, a pericentric inversion is also likely involved in chromosome 10, which would explain the red signal at the distal region on the long arm (“g” in Figure 4), which is located on the short arms of chromosome 10 in other species (Figure 4).
Pepper (C. annuum):
Pepper diverged from a common ancestor of potato/tomato ∼19.6 MYA (Wu and Tanksley 2010). The two oligo-FISH probes produced massive background signals on pepper chromosomes (Figure S5 in File S1). Punctuated major signals were observed on every chromosome. However, most of the pepper chromosomes cannot be unambiguously identified based on the signal patterns on potato chromosomes, suggesting that major structural arrangements have occurred between most potato and pepper chromosomes. The sizes of the pepper chromosomes appeared to be at least twice that of potato chromosomes. The current sequence assemblies estimate 3000 Mb for the pepper genome (https://www.ncbi.nlm.nih.gov/assembly, GCA_000512255.1), which is significantly larger than the potato genome (∼800 Mb). These results suggest that the pepper genome has undergone major expansion and rearrangements during evolution.
Confirmation of interchromosomal translocation by oligo-based chromosome painting
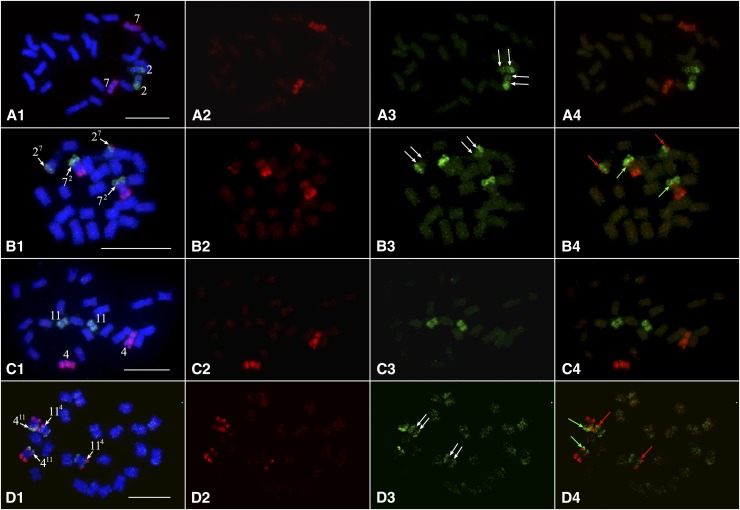
We developed oligo-based chromosome painting probes to validate the interchromosomal translocations in S. etuberosum and S. caripense, which were predicted based on bar code FISH signal modifications relative to potato chromosomes (Figure 5). Oligos unique to a single potato chromosome were computationally identified and synthesized in parallel (Han et al. 2015). We selected 27,392 oligos for both potato chromosomes 2 and 7. The chromosome 7 probe generated uniform FISH signals on DM chromosome 7 (Figure 6A2). However, the chromosome 2 probe generated weak signals on the short arm and the proximal region on the long arm of DM chromosome 2 (Figure 6A3). These two probes, especially that for chromosome 2, generated very weak signals in the pericentromeric regions of chromosome 2 and 7 of S. etuberosum (Figure 6B3). This is likely caused by divergence of the DNA sequences located in the pericentromeric regions. Nevertheless, chromosome painting clearly showed that a small chromosome 7 segment was translocated to chromosome 2 (27). In contrast, a relatively large chromosome 2 segment was translocated to chromosome 7 (72) (Figure 6, B1 and B4). Thus, the chromosomal painting results matched to the predicted reciprocal translocation based on the modification to the bar code (Figure 5A).
Figure 6.
Validation of chromosomal translocations by chromosome painting. (A1–A4) Painting of chromosomes 2 (green) and 7 (red) of DM potato. Red (A2), green (A3), and both red and green (A4) fluorescence signals were digitally separated from (A1). Double white arrows in (A3) indicate relatively weak FISH signals that span the short arm and proximal region of the long arm of chromosome 2. (B1–B4) Painting of chromosomes 2 (green) and 7 (red) in S. etuberosum. Red (B2), green (B3), and both red and green (B4) fluorescence signals were digitally separated from (B1). Double white arrows in (B3) indicate very weak or background level FISH signals that span the short arm and proximal region of the long arm of chromosome 2. Red arrows in (B4) point to the breakpoint where a small chromosome 7 fragment attached to chromosome 2 (27). Green arrows in B4 point to the breakpoint where a large chromosome 2 fragment attached to chromosome 7 (72). (C1–C4) Painting of chromosomes 4 (red) and 11 (green) of DM potato. Red (C2), green (C3), and both red and green (C4) fluorescence signals were digitally separated from (C1). (D1–D4) Painting of chromosomes 4 (red) and 11 (green) in S. caripense. Red (D2), green (D3), and both red and green (D4) fluorescence signals were digitally separated from (D1). Double white arrows indicate background level FISH signals that span pericentromeric region of chromosome 11. Red arrows in (D4) point to the breakpoint where a chromosome 4 fragment attached to chromosome 11 (114). Green arrows in (B4) point to the breakpoint where a chromosome11 fragment attached to chromosome 4 (411). Bar, 10 µm.
Similarly, we developed painting probes for potato chromosomes 4 and 11, each containing 27,392 oligos. Both probes generated uniform FISH signals on DM chromosomes with only limited hybridization background (Figure 6, C2 and C3). The painting probes, however, produced unambiguous hybridization signals only at the distal ends of chromosomes 4 and 11 of S. caripense (Figure 6, D1 and D4). Only background-level FISH signals were detected in the pericentromeric regions of the homeologous chromosomes in S. caripense (Figure 6D3). Nevertheless, chromosome painting in S. caripense clearly revealed the reciprocal translocation between chromosome 4 and 11, resulting in chromosomes 411 and 114, respectively (Figure 6, D1 and D4). The exchanged chromosomal segments from the two chromosomes showed a similar size (Figure 6D4). Thus, the chromosomal painting results in S. caripense also matched the predicted reciprocal translocation based on the modifications to the oligo-FISH bar code (Figure 5B).
Discussion
Oligo-FISH bar code: a new chromosome identification methodology
FISH is the most important technique for chromosome identification in plants (Jiang and Gill 1994, 2006). Repetitive DNA sequences were commonly used as probes in FISH-based chromosome identification (Mukai et al. 1993; Kato et al. 2004). However, it is often challenging to find a repeat that would produce distinct FISH signals on individual chromosomes in a plant species. More importantly, the FISH signals from repetitive DNA probes can potentially be highly polymorphic among different varieties and accessions, which may prevent consistent identification of individual chromosomes (Jiang and Gill 2006). Alternatively, large-insert genomic DNA clones, such as bacterial artificial chromosome (BAC) clones, can be used as FISH probes for chromosome identification (Jiang et al. 1995). However, this approach is dependent on the availability of a large-insert genomic DNA library as well as a major effort to isolate clones specific to every chromosome (Dong et al. 2000; Cheng et al. 2001; Kulikova et al. 2001; Kim et al. 2002; K. Wang et al. 2008). In addition, BACs from plant species with large and complex genomes often contain high proportions of repetitive DNA sequences and do not produce chromosome-specific FISH signals (Zhang et al. 2004; Janda et al. 2006).
We demonstrate that oligo-FISH bar codes provide a powerful and efficient technique for plant chromosome identification. It has several major advantages compared to the repeat- or BAC-based FISH probes: (1) Oligo-based FISH probes can be designed in any species with a sequenced genome, which has been demonstrated in several animal and plant species (Boyle et al. 2011; Yamada et al. 2011; Beliveau et al. 2012; Han et al. 2015). Thus, a single or few oligo pools can be designed to identify all chromosomes in a plant species with a sequenced genome. If the majority of oligos are associated with genic sequences, the same bar code can be expected from different varieties and accessions in the same species. (2) We demonstrate that a bar code probe can potentially be used to identify homeologous chromosomes among distantly related species, which allow for evolutionary studies. (3) Oligos can be selected from multiple regions from the same chromosome. Such a cocktail oligo probe will generate a unique hybridization pattern that resembles FISH signal patterns generated from multiple BACs derived from a single chromosome (Iovene et al. 2008; Szinay et al. 2008, 2012; Tang et al. 2008). An unlimited number of possible patterns can be designed for each chromosome. (4) Each oligo-based probe can be used for nearly 1,000,000 FISH experiments (Han et al. 2015). Thus, such bar code oligo-FISH probes are cost effective and can be maintained as a permanent resource.
The total number of FISH signals will be the most important factor in designing an oligo-FISH bar code. Oligos spanning 30–50 kb of single copy sequences can generate a strong FISH signal on metaphase chromosomes. However, it may be difficult to identify such long stretches of single copy sequences in some plant genomes. If multiple signals are designed on a single chromosome arm, the groups of oligos should be separated by a sufficient distance to ensure separate FISH signals. We demonstrate that 7 Mb is sufficient to consistently separate two FISH signals on potato metaphase chromosomes. However, a longer distance (>10 Mb) should be considered for plant species with chromosomes much larger than those of potato.
Chromosomal inversion and translocations in Solanum species
Chromosomal evolution of the solanaceous species has been investigated traditionally using pairwise comparative genetic linkage mapping (Wu and Tanksley 2010). Since genetic linkage maps and DNA markers were best developed in tomato (Tanksley et al. 1992), most of the pairwise mapping was performed between tomato and other solanaceous species, including potato, eggplant, pepper, and Nicotiana species (Bonierbale et al. 1988; Tanksley et al. 1992; Livingstone et al. 1999; Doganlar et al. 2002; Wu et al. 2009, 2010). Comparative FISH mapping has also been conducted among Solanum species using common sets of BACs isolated from potato or tomato (Iovene et al. 2008; Tang et al. 2008; Lou et al. 2010; Szinay et al. 2012; Gaiero et al. 2017). These comparative studies showed that inversions were the most common cause of chromosomal rearrangements among the solanaceous species. Translocations were also reported in some comparisons, for example, tomato and eggplant were found to differ by 24 inversions and 5 translocations based on eggplant linkage mapping using a set of 232 tomato-derived DNA markers (Wu et al. 2009).
The resolution of linkage mapping is restricted by the number of markers used. Genotyping or mapping errors, caused by wrong marker order or population size, may result in misidentified chromosomal rearrangements, such as inversion. In addition, population-based linkage mapping is an expensive and time-consuming approach; it has mostly been conducted in crops or economically important plant species. Although translocations were reported in some of the comparative mapping investigations among Solanum species, no cytological evidence was provided for any of the predicted translocations. For example, linkage mapping suggested that eggplant chromosome 5 is an equivalent of a fusion of the short arm of chromosome 5 with the long arm of chromosome 12 in tomato. Similarly, eggplant chromosome 11 is an equivalent of a fusion of the short arm of chromosome 11 with the short arm of chromosome 4 in tomato (Wu et al. 2009). However, our comparative oligo-FISH does not indicate whole-arm translocations associated with eggplant chromosomes 4, 5, 11, and 12 (Figure 4). We cannot exclude the possibility that the interchromosomal translocations are specific to the eggplant accession used by Wu et al. (2009). Thus, application of additional eggplant genotypes in oligo-FISH mapping may explain the discrepancy of results based on genetic linkage mapping and comparative oligo-FISH mapping.
It is intriguing that chromosomal inversions are highly common among the Solanum species (Figure S3 in File S1). By contrast, chromosomal translocations are relatively rare. Interestingly, we discovered reciprocal translocations in S. etuberosum and S. caripense, and both are wild species. Strikingly, the oligo-FISH probes generated nearly identical signal patterns on chromosomes from potato and eggplant (Figure 4), which have diverged for ∼15.5 MY (Wu and Tanksley 2010). A recent study in humans showed that a translocation can change the spatial position of the translocated chromosome fragment in the nucleus and, thus, alter the expression of the associated genes (Harewood et al. 2010). Since potato, tomato, and eggplant are crop species, selection in breeding practice may have eliminated chromosomal variants that may have negatively affected the fitness of the species due to the altered gene expression associated with the chromosomal rearrangement. Translocations have previously been reported to be rare in wheat cultivars but common in their wild ancestors (Badaeva et al. 1995). Analysis of the presence of the translocations in multiple populations of S. etuberosum and S. caripense will reveal if these chromosomal variants have been fixed in these wild species.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300344/-/DC1.
Acknowledgments
We thank John Bamberg and Robert Jarret for providing the germplasm used in this study. We thank two anonymous reviewers for their valuable comments and edits on the manuscript. This research was supported partially by National Science Foundation grant ISO-1237969 to J.J. A scholarship to G.T.B. and funds to G.A.T. were provided by CAPES (Brazilian Ministry of Education). J.-M.R. and K.S. are employees of Arbor Biosciences and J.-M.R. is an owner of Arbor Biosciences.
Author contributions: J.J. conceived the research, G.T.B. and L.H. conducted FISH experiments. T.Z. and H.Z. designed oligo-FISH probes. K.S. and J.-M.R. synthesized probes and provided reagents. G.T.B., L.H., G.A.T., and J.J. analyzed data. J.J. wrote the article.
Footnotes
Communicating editor: A. Paterson
Literature Cited
- Anderson L. K., Stack S. M., Mitchell J. B., 1982. An investigation of the basis of a current hypothesis for the lack of G-banding in plant chromosomes. Exp. Cell Res. 138: 433–436. [DOI] [PubMed] [Google Scholar]
- Badaeva E. D., Jiang J. M., Gill B. S., 1995. Detection of intergenomic translocations with centromeric and noncentromeric breakpoints in Triticum araraticum - mechanism of origin and adaptive significance. Genome 38: 976–981. [DOI] [PubMed] [Google Scholar]
- Beliveau B. J., Joyce E. F., Apostolopoulos N., Yilmaz F., Fonseka C. Y., et al. , 2012. Versatile design and synthesis platform for visualizing genomes with oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 109: 21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale M. W., Plaisted R. L., Tanksley S. D., 1988. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120: 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S., Rodesch M. J., Halvensleben H. A., Jeddeloh J. A., Bickmore W. A., 2011. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 19: 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. K., Buell C. R., Wing R. A., Gu M., Jiang J. M., 2001. Toward a cytological characterization of the rice genome. Genome Res. 11: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganlar S., Frary A., Daunay M. C., Lester R. N., Tanksley S. D., 2002. A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161: 1697–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Novy R. G., Helgeson J. P., Jiang J., 1999. Cytological characterization of potato - Solanum etuberosum somatic hybrids and their backcross progenies by genomic in situ hybridization. Genome 42: 987–992. [Google Scholar]
- Dong F. G., Song J. Q., Naess S. K., Helgeson J. P., Gebhardt C., et al. , 2000. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101: 1001–1007. [Google Scholar]
- Fransz P., Armstrong S., Alonso-Blanco C., Fischer T. C., Torres-Ruiz R. A., et al. , 1998. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13: 867–876. [DOI] [PubMed] [Google Scholar]
- Friebe B., Endo T. R., Gill B. S., 1996. Chromosome banding methods, pp. 123–153 in Plant Chromosomes: Laboratory Methods, edited by Fukui K., Nakayama S. CRC Press, Boca Raton, FL. [Google Scholar]
- Gaiero P., van de Belt J., Vilaro F., Schranz M. E., Speranza P., et al. , 2017. Collinearity between potato (Solanum tuberosum L.) and wild relatives assessed by comparative cytogenetic mapping. Genome 60: 228–240. [DOI] [PubMed] [Google Scholar]
- Gong Z. Y., Wu Y. F., Koblizkova A., Torres G. A., Wang K., et al. , 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J., 1977. Why plant chromosomes do not show G-bands. Theor. Appl. Genet. 50: 121–124. [DOI] [PubMed] [Google Scholar]
- Haas B. J., Delcher A. L., Wortman J. R., Salzberg S. L., 2004. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646. [DOI] [PubMed] [Google Scholar]
- Han Y. H., Zhang T., Thammapichai P., Weng Y. Q., Jiang J. M., 2015. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan M. A., Crisovan E., Hamilton J. P., Kim J., Laimbeer P., et al. , 2016. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 28: 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L., Schutz F., Boyle S., Perry P., Delorenzi M., et al. , 2010. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 20: 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovene M., Wielgus S. M., Simon P. W., Buell C. R., Jiang J. M., 2008. Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 180: 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J., Safar J., Kubalakova M., Bartos J., Kovarova P., et al. , 2006. Advanced resources for plant genomics: a BAC library specific for the short arm of wheat chromosome 1B. Plant J. 47: 977–986. [DOI] [PubMed] [Google Scholar]
- Jauch A., Wienberg J., Stanyon R., Arnold N., Tofanelli S., et al. , 1992. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl. Acad. Sci. USA 89: 8611–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. M., Gill B. S., 1994. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37: 717–725. [DOI] [PubMed] [Google Scholar]
- Jiang J. M., Gill B. S., 2006. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49: 1057–1068. [DOI] [PubMed] [Google Scholar]
- Jiang J. M., Gill B. S., Wang G. L., Ronald P. C., Ward D. C., 1995. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92: 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Lamb J. C., Birchler J. A., 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Childs K. L., Islam-Faridi M. N., Menz M. A., Klein R. R., et al. , 2002. Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45: 402–412. [DOI] [PubMed] [Google Scholar]
- Kulikova O., Gualtieri G., Geurts R., Kim D. J., Cook D., et al. , 2001. Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J. 27: 49–58. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., et al. , 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langergraber K. E., Prufer K., Rowney C., Boesch C., Crockford C., et al. , 2012. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. USA 109: 15716–15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. Y., Matyasek R., Lichtenstein C. P., Leitch A. R., 2000. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma 109: 245–258. [DOI] [PubMed] [Google Scholar]
- Livingstone K. D., Lackney V. K., Blauth J. R., van Wijk R., Jahn M. K., 1999. Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152: 1183–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Q. F., Iovene M., Spooner D. M., Buell C. R., Jiang J. M., 2010. Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 119: 435–442. [DOI] [PubMed] [Google Scholar]
- Mandakova T., Joly S., Krzywinski M., Mummenhoff K., Lysak M. A., 2010. Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22: 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi M., 1991. Phylogenetic relationships in the potato and its related species, pp. 93–118 in Chromosome Engineering in Plants: Genetics, Breeding, Evolution (Vol. 2), edited by Tsuchiya T., Gupta P. Elsevier, Amsterdam. [Google Scholar]
- Mukai Y., Nakahara Y., Yamamoto M., 1993. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence insitu hybridization using total genomic and highly repeated DNA probes. Genome 36: 489–494. [DOI] [PubMed] [Google Scholar]
- Novy R. G., Nasruddin A., Ragsdale D. W., Radcliffe E. B., 2002. Genetic resistances to potato leafroll virus, potato virus Y, and green peach aphid in progeny of Solanum etuberosum. Am. J. Potato Res. 79: 9–18. [Google Scholar]
- Novy R. G., Gillen A. M., Whitworth J. L., 2007. Characterization of the expression and inheritance of potato leafroll virus (PLRV) and potato virus Y (PVY) resistance in three generations of germplasm derived from Solanum etuberosum. Theor. Appl. Genet. 114: 1161–1172. [DOI] [PubMed] [Google Scholar]
- Reeves A., Tear J., 2000. MicroMeasure version 3.3. http://www.colostate.edu/Depts/Biology/MicroMeasure.
- Sarkinen T., Bohs L., Olmstead R. G., Knapp S., 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol. Biol. 13: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T., Leitch A. R., Bennett M. D., Heslopharrison J. S., 1989. In situ localization of parental genomes in a wide hybrid. Ann. Bot. 64: 315–324. [Google Scholar]
- Szinay D., Chang S. B., Khrustaleva L., Peters S., Schijlen E., et al. , 2008. High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J. 56: 627–637. [DOI] [PubMed] [Google Scholar]
- Szinay D., Wijnker E., van den Berg R., Visser R. G. F., de Jong H., et al. , 2012. Chromosome evolution in Solanum traced by cross-species BAC-FISH. New Phytol. 195: 688–698. [DOI] [PubMed] [Google Scholar]
- Tang X. M., Szinay D., Lang C., Ramanna M. S., van der Vossen E. A. G., et al. , 2008. Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics 180: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S. D., Ganal M. W., Prince J. P., de Vicente M. C., Bonierbale M. W., et al. , 1992. High-density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Potato Genome Sequencing Consortium , 2011. Genome sequence and analysis of the tuber crop potato. Nature 475: 189–194. [DOI] [PubMed] [Google Scholar]
- The Tomato Genome Consortium , 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Guan B., Guo W. Z., Zhou B. L., Hu Y., et al. , 2008. Completely distinguishing individual A-genome chromosomes and their karyotyping analysis by multiple bacterial artificial chromosome - fluorescence in situ hybridization. Genetics 178: 1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Diehl A., Wu F. N., Vrebalov J., Giovannoni J., et al. , 2008. Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics 180: 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H., Schneeweiss G. M., 2013. Karyotype diversity and evolutionary trends in angiosperms, pp. 209–230 in Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes, edited by Greilhuber J., Dolezel J., Wendel J. F. Springer Vienna, Vienna. [Google Scholar]
- Wu F. N., Tanksley S. D., 2010. Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. N., Mueller L. A., Crouzillat D., Petiard V., Tanksley S. D., 2006. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. N., Eannetta N. T., Xu Y. M., Tanksley S. D., 2009. A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor. Appl. Genet. 118: 927–935. [DOI] [PubMed] [Google Scholar]
- Wu F. N., Eannetta N. T., Xu Y. M., Plieske J., Ganal M., et al. , 2010. COSII genetic maps of two diploid Nicotiana species provide a detailed picture of synteny with tomato and insights into chromosome evolution in tetraploid N. tabacum. Theor. Appl. Genet. 120: 809–827. [DOI] [PubMed] [Google Scholar]
- Yamada N. A., Rector L. S., Tsang P., Carr E., Scheffer A., et al. , 2011. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. 132: 248–254. [DOI] [PubMed] [Google Scholar]
- Zhang P., Li W. L., Fellers J., Friebe B., Gill B. S., 2004BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112: 288–299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental Material, Table S1 in File S1 contains all information about the number and locations of oligos associated with each of the 26 individual FISH signals generated by the two bar code FISH probes. The Chorus software used for oligo-FISH probe design is freely available (https://github.com/forrestzhang/Chorus).