Abstract
How the brain makes trillions of synaptic connections using a genome of only 20,000 genes is a major question in modern neuroscience. Alternative splicing is one mechanism that can increase the number of proteins produced by each gene, but its role in regulating synapse formation is poorly understood. In Drosophila, photoreceptors form a synapse with multiple postsynaptic elements including lamina neurons L1 and L2. L1 and L2 express distinct isoforms of the homophilic repulsive protein Dscam2, and since these isoforms cannot bind to each other, cell-specific expression has been proposed to be necessary for preventing repulsive interactions that could disrupt the synapse. Here, we show that the number of synapses are reduced in flies that express only one isoform, and L1 and L2 dendritic morphology is perturbed. We propose that these defects result from inappropriate interactions between L1 and L2 dendrites. We conclude that regulated Dscam2 alternative splicing is necessary for the proper assembly of photoreceptor synapses.
Keywords: splicing, Dscam2, synapse, isoform, Drosophila
THE genome has evolved many mechanisms to allow a finite number of genes to perform a plethora of biological processes. One way that this can be accomplished is by increasing the diversity of the proteome. During neurodevelopment, the stochastic expression of distinct isoforms is necessary for receptive field elaboration and spacing, as has been shown for Drosophila Dscam1 (Hughes et al. 2007; Matthews et al. 2007; Soba et al. 2007) and the mouse protocadherin γ-subcluster (Lefebvre et al. 2012). The prominence of probabilistic expression of unique isoforms may have evolved as a strategy to establish distinct neuronal identities for ubiquitous processes such as self-avoidance.
To contribute to proteome diversity, alternative isoforms need to have distinct biochemical functions or to be expressed in unique spatiotemporal patterns. Alternative splicing is most prevalent in the nervous system, presumably so that it can contribute to the functional complexity of the brain (Merkin et al. 2012; Raj and Blencowe 2015). Although examples of regulated isoform expression between neuronal subtypes in Caenorhabditis elegans (Norris et al. 2014; Tomioka et al. 2016), Drosophila (Lah et al. 2014), and the mammalian nervous system (Iijima et al. 2014; Schreiner et al. 2014; Fuccillo et al. 2015; Zhang et al. 2016; Chen et al. 2017) exist, whether distinct isoforms play unique roles in synapse formation is largely unexplored (Nguyen et al. 2016). For the purpose of neuronal connectivity, both the mammalian neurexins and Drosophila Dscam2 produce isoforms with distinct binding specificities in a cell type-specific manner. Neurexins are transsynaptic cell recognition molecules known to modulate synaptic potentiation through extensive alternative splicing of up to 11,520 isoforms (Schreiner et al. 2014). The cell type-specific binding specificity and regulated expression of different neurexins to different extracellular ligands, such as neuroligins and cerebellins (Ko et al. 2009; Siddiqui et al. 2010; Uemura et al. 2010; Matsuda and Yuzaki 2011; Boucard et al. 2012), is crucial for postsynaptic long-term potentiation (Aoto et al. 2013). In Drosophila, Dscam2 is a cell recognition molecule that is important for cell type-specific avoidance (tiling) and self-avoidance (Millard et al. 2007, 2010). Alternative splicing of Dscam2 produces two isoforms with different extracellular domains that confer isoform-specific homophilic recognition (Millard et al. 2007). Previously, we showed that Dscam1 and Dscam2 redundantly promote self-avoidance of sister neurites in the overlapping dendritic arrays of lamina neurons, L1 and L2 (Millard et al. 2010). This prevents neurites from the same cell from incorporating onto the same presynaptic site (synaptic exclusion), ensuring the invariant pairing of L1 and L2 neurites. What remained puzzling was how Dscam2 could distinguish between self (L1/L1 and L2/L2) and nonself (L1/L2) pairing. A model was proposed where cell-specific Dscam2 isoform expression could mediate self-avoidance and avoid inappropriate interactions between L1 and L2 through isoform-specific homophilic binding. In our subsequent study, we demonstrated that L1 and L2 cells indeed express distinct Dscam2 isoforms (Lah et al. 2014; Li et al. 2015). However, what has not been tested is whether cell-specific expression of Dscam2 is required for photoreceptor (R cell) synapse formation. If L1 and L2 expressed the same Dscam2 isoform, repulsion between their dendrites at R cell synapses might be expected to prevent synapses from forming entirely. However, R cell synaptogenesis is a robust process and, to our knowledge, there are few examples in the literature of mutations in genes that change the number of R cell synapses (Hiesinger et al. 2006; Schwabe et al. 2014). Here, we test whether regulated alternative splicing of Dscam2 is necessary for the assembly of R cell synapses. Using both light and electron microscopy (EM), we found that R cell synapses are reduced in flies expressing a single Dscam2 isoform. We also found morphological defects in the dendrites of L1 and L2, suggesting that inappropriate interactions between these cells decrease dendritic elaboration. We conclude that regulated Dscam2 alternative splicing permits proper synaptic organization by preventing inappropriate interactions between L1 and L2 dendrites.
Materials and Methods
Fly genetics and transgenic lines
Flies were cultured on standard yeast–agar media at 25° in a room with windows that exposed them to natural day/night cycles. The following Gal4 driver lines were used to restrict expression to specific cells: svp-Gal4 (L1neurons; Kyoto#103727; Tan et al. 2015), bab1-Gal4 (L2 neurons; Bloomington#47736; Tan et al. 2015), L1L2-Gal4 (L1 and L2 neurons; Kyoto#105191; Rister et al. 2007), C202a-Gal4 (Rister et al. 2007), and 8-18-Gal4 (L2 neurons). The following transgenes were used for labeling or genetic analysis: Dscam210A-A and Dscam210B-D (Lah et al. 2014), brp-SNAP knock-in (Kohl et al. 2014), UAS-Syt4GYB (Singari et al. 2014), UAS(FRT.stop)myr::smGdP-V5-THS-UAS(FRT.stop)myr::smGdP-cMyc (Nern et al. 2015), 27G05-FLP (Pecot et al. 2013), and UAS-mCD8GFP, FRT79D, and tub-Gal80 (Lee and Luo 2001). Specific genotypes used in each experiment can be found in Supplemental Material, Tables S1 and S2 in File S1.
Immunohistochemistry
Flies were maintained on cornmeal/agar media at 25° and were dissected for analysis within 1 day of eclosion. Immunohistochemistry was conducted as described previously (Lee and Luo 2001; Lah et al. 2014). Fly brains were fixed for 45 min in [4% paraformaldehyde (PFA), 2% for Cysteine string protein (Csp), and 50% phosphate-buffered lysine (PBL) in 0.0025% Triton X-100], and blocked in PBS containing 10% goat serum and 0.5% Triton X-100 for 60 min (Hamanaka and Meinertzhagen 2010). Primary and secondary antibodies were incubated overnight and washed 3× with PBS and 0.5% Triton X-100. SNAP labeling was conducted essentially as described in Kohl et al. (2014). Brains were permeabilized in PBS with 0.3% Triton X-100 for 10 min and incubated with chemical tag for 60 min at room temperature. Antibody dilutions used were as follows: rabbit anti-GFP (1:1000; Invitrogen, Carlsbad, CA), mouse mAb24B10 (1:20; DSHB), mouse anti-svp 6F7 [1:20; Developmental Studies Hybridoma BankDSHB (DSHB)], mouse anti-Csp (1:10; DSHB), mouse anti-dlg (1:20; DSHB), mouse anti-synapsin (1:10; DSHB), mouse anti-nc82 (1:10; DSHB), rat anti-mCherry (1:500; Invitrogen), DyLight anti-mouse Cy3 (1:1000; Jackson Laboratory), DyLight anti-rabbit 488 (1:1000; Jackson Laboratory), DyLight anti-mouse 647 (1:1000; Jackson Laboratory), V5-tag:DyLight anti-mouse 405 (1:200; AbD Serotec), V5-tag:DyLight anti-mouse 550 (1:500; AbD Serotec), and SNAP-surface 549 (4 μM; New England Biolabs, Beverly, MA) (File S2) .
EM
Fly heads dissected in a sagittal plane were fixed in 4% PFA and 2.5% glutaraldehyde overnight. EM cross-sections of the lamina neuropil were processed by PELCO BioWave for tissue preparation. After buffer rinse (0.1 M cacodylate buffer), osmium tetroxide fixation (1% osmium tetroxide in cacodylate buffer), water rinse, dehydration (50–100% ethanol), and resin infiltration (epon), the samples were embedded and polymerized. After resin polymerization, the samples were thin-sectioned by Leica EM UC6 ultramicrotome and stained by uranyl acetate (5% in 50% ethanol) and lead citrate, and lamina cartridges were imaged using a Hitachi 7700 120 kV Transmission electron microscope. Electron micrographs were then deidentified and analyzed by two counters. R cells were defined by the presence of capitate projections and/or electron-dense synaptic vesicles. Individual lamina neuron identities could not be elucidated and were designated as non-R cells. Using ImageJ, counters noted the number of T-bars (associated and not associated with R cells), capitate projections, and multiple membrane-containing structures defined by the criteria of at least a double membranous structure (could be more), and a cluster of such structures were classified as one unit. Upon reidentification of files, the data were sorted for statistical analysis (see below in Statistical analyses). The statistical differences between the different genotypes were similar between the two counters.
Image acquisition and deconvolution
Imaging was performed at the Queensland Brain Institute’s Advanced Microscopy Facility and School of Biomedical Sciences Imaging Facility. Synaptic data and MultiColor FlpOut (MCFO) was imaged on a spinning-disk confocal system (Marianas; 3I), consisting of a Axio Observer Z1 (Zeiss [Carl Zeiss], Thornwood, NY) equipped with a CSU-W1 spinning-disk head (Yokogawa Corporation of America), ORCA-Flash4.0 v2 sCMOS camera (Hamamatsu Photonics), and 63× 1.4 NA P-Apo and 100× 1.4 NA P-Apo objectives. Image acquisition was performed using SlideBook 6.0 (3I). Huygens software (Scientific Volume Imaging) was used to deconvolve each channel of raw image stacks individually using a theoretical point spread function, automatic background estimation, 40 iterations, signal-to-noise ratio of 20, optimized iteration mode, quality change threshold of 0.1%, and a one-brick layout. After manual inspection, images that contained artifacts or the borders of individual puncta that were not sharply defined were removed from analysis. Imaris (Bitplane, St. Paul, MN) was used to quantify synapse number (shown as puncta), after manual baseline subtraction using the surfaces tool to segregate the region of interest (lamina cartridge), and the spot tool to quantify with puncta specifications as x,y diameter of 0.18 μm and z projection of 0.36 μm (Mosca and Luo 2014). All images were visually inspected in three dimensions during the spot tool function to verify quantification. MARCM (mosaic analysis with a repressible cell marker) data were imaged on a Leica SP8 laser scanning confocal system with a 63× Glycerol NA 1.3 objective.
Quantification of dendrite width
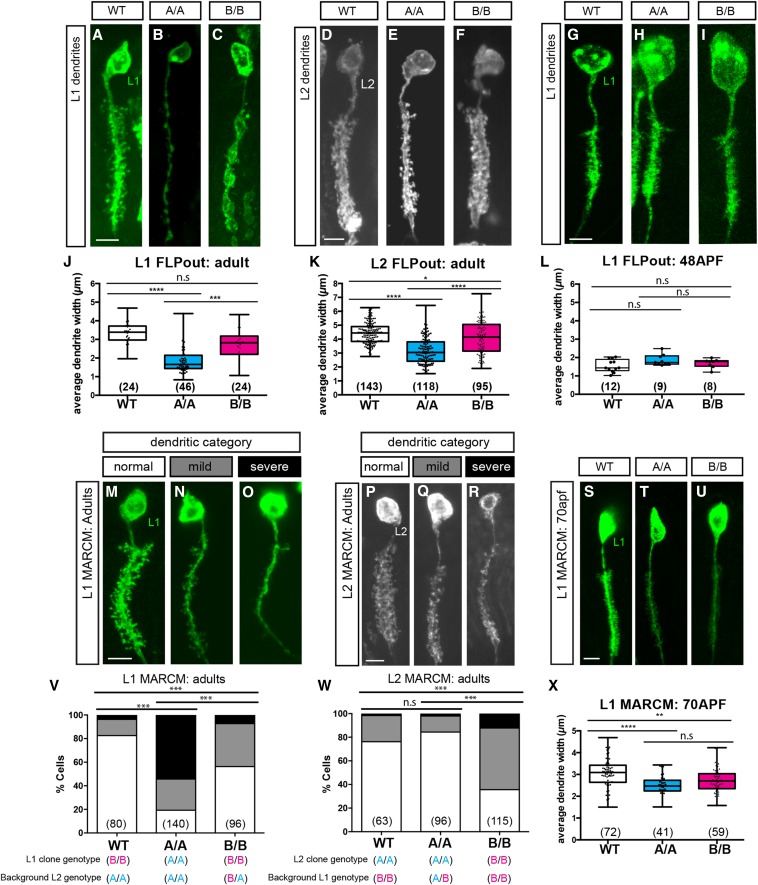
Individual L1 or L2 cells were isolated for quantification of dendrites. The width of the proximal and distal sections of the array, and the point halfway between the two, were measured using ImageJ. An average was calculated with these three measurements to give the mean width of the array. This average was then plotted to give results in Figure 4.
Figure 4.
Reduced dendrite complexity in Dscam2 single-isoform lines. (A–C) Representative confocal images of L1 dendrites generated using lamina neuron-specific FLP (27G05-FLP) and MultiColor FLPout transgenes. (A) Wild-type (WT), (B) Dscam2A (A/A), and (C) Dscam2B (B/B). (D–F) Representative confocal images of L2 dendrites generated as in (A–C). (D) WT, (E) A/A, and (F) B/B. Bar, 5 μM (A–F). (G–I) Representative confocal images of dendrites generated as in (A–C) at 48 hr APF (after puparium formation). (G) WT, (H) A/A, and (I) B/B. Bar, 5 μm. (J–L) Quantification of L1 and L2 dendritic width in WT, A/A, and B/B animals (see Materials and Methods). (G) L1 dendritic width in adults. (I) L2 dendritic width in adults. (L) L1 dendritic width at 48 hr APF. Boxplot format (middle line, median; error bars = min and max, box = 25–75% quartiles, and each data point is a different cell). Clone number in parentheses. Kruskal–Wallis test with Dunn’s multiple comparison test; n.s (not significant) P > 0.05, * P < 0.05, *** P < 0.001, and **** P < 0.0001. (M–R) MARCM in L1 and L2 cells with Dscam2 single-isoform alleles. (M–O) Representative confocal images of L1 clones with different severities of dendritic loss. (P–R) Representative confocal images of L2 dendrites with different severities of dendritic loss. (S–U) Representative confocal images of L1 MARCM clones at 70 hr APF. (S) WT, (T) A/A, and (U) B/B. Bar, 5 μm. (V and W) Categorization and quantification of L1 (V) and L2 (W) Dscam2 single-isoform MARCM. Data in stacked bars (white, normal; gray, mild; and black, severe). Fisher’s exact test; n.s P > 0.05 and *** P < 0.001. Clone number in parentheses. (X) Quantification of dendritic width in L1 MARCM clones at 70 hr APF. Boxplot format (middle line, median; error bars = min and max, box = 25–75% quartiles, and each data point in a different cell). Clone numbers in parentheses. Kruskal–Wallis test with Dunn’s multiple comparison test; n.s P > 0.05 and ** P < 0.01. ANOVA test with Tukey’s multiple comparison test was used where data followed a Gaussian distribution.
Statistical analyses
Statistical analyses were performed using Prism 7.0 software (GraphPad). D’Agostino and Pearson omnibus normality tests were conducted to determine whether all data sets followed a Gaussian (normal) distribution. The significance for normality test was set at P < 0.05, where statistical significance equates to a data set that does not follow a normal distribution. Normally distributed data sets were subject to a parametric ANOVA test and a Tukey’s multiple comparison tests. Data sets containing non-Gaussian distributions were subjected to a Kruskal–Wallis test (nonparametric ANOVA) and a Dunn’s multiple comparisons test. For categorical data, we conducted Fischer’s exact test. Statistical significances in figures are depicted with asterisks as follows: not significant P > 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001. See Supplemental Information for a full list of tests for all data sets obtained in this study (Table S2 in File S1).
Data availability
Protocols and reagents are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
The size of lamina cartridges is reduced in Dscam2 single-isoform mutants
To assess whether Dscam2 alternative splicing is required for lamina cartridge formation, we used two previously described fly lines that express a single isoform of Dscam2 from its endogenous locus (Lah et al. 2014). In these flies, the spatial and temporal patterns of Dscam2 expression are unperturbed, but all Dscam2-positive cells express only one of the possible two extracellular domains of Dscam2. The synaptic modules that contain R cell synapses, called lamina cartridges, consist of six presynaptic R cell terminals surrounding postsynaptic L1 and L2 lamina neurons. L1 and L2 dendrites project radially to form en passant synaptic connections with the R cells (Figure 1A). Although lamina cartridges in the single-isoform lines visualized with an antibody against chaoptin (mAb24B10) had distinct boundaries between each another, cartridges from the single-isoform lines were smaller than control cartridges (Figure S1, B, D, H, and I). This suggested that Dscam2 isoform diversity is important in maintaining proper lamina cartridge size.
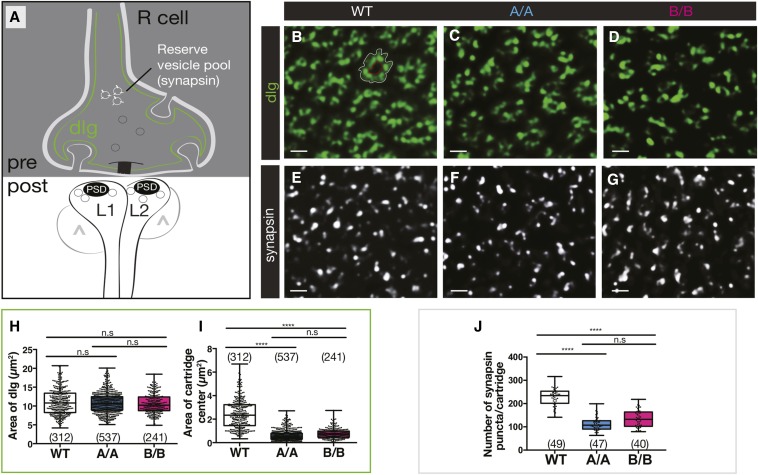
Figure 1.
Dscam2 single-isoform mutants have a reduction in presynaptic markers and reduced lamina cartridge size. (A) Schematic representation of an R cell tetrad synapse showing localization of dlg (green) and synapsin (white) on the presynaptic membrane. The postsynaptic side of the synapse includes L1-expressing Dscam2B, L2-expressing Dscam2A, and amacrine/L3/glial cells (^) that represent the variable components of the postsynaptic complex. PSD, postsynaptic density. (B–G) Representative deconvolved optical cross-sections of lamina cartridges from wild-type (WT), Dscam2A (A/A), and Dscam2B (B/B) single-isoform lines. (B–D) dlg labeling (green) and (E–G) anti-synapsin (white). (H and I) Quantification of anti-dlg-labeled area (between white outline and red outline) (H) and the center region of each cartridge devoid of labeling (within red outline) (I). (J) Quantification of synapsin puncta per lamina cartridge using “spot tool” in Imaris. As the cartridge is difficult to define with synapsin labeling alone, costaining with synaptotagmin 4 (see Figure 2) was performed to demarcate cartridges. Boxplot format: middle line = median, range bars = min and max, box = 25–75% quartiles, and each data point = single cartridge. Cartridge number for each genotype is shown in parentheses. Kruskal–Wallis test with Dunn’s multiple comparison test. n.s (not significant) P > 0.05 and **** P < 0.0001. Bar, 2 μm.
Presynaptic defects in Dscam2 single-isoform lines
To determine what contributes to the reduction in lamina cartridge size, we first analyzed the expression of presynaptic-associated protein Discs large (dlg). A member of the membrane-associated guanylate kinase family, dlg is a cytoplasmic scaffolding protein associated with the presynaptic membrane of Drosophila photoreceptors and invaginations of glia called capitate projections. We therefore used dlg as marker for the structural integrity of the presynaptic terminal (Woods and Bryant 1993; Hamanaka and Meinertzhagen 2010) (Figure 1A). To determine if the organization of the terminal was disrupted, two measurements were taken: the area demarcated by dlg and the area devoid of the marker in the center of each cartridge, where lamina neurons L1 and L2 reside. The area demarcated by dlg was similar between Dscam2 single-isoform lines and controls (Figure 1, B, D, and H), whereas the area occupied by L1 and L2 within the cartridge was reduced (Figure 1I). Since dlg within the presynaptic membrane was unperturbed, the reduction in cartridge size (Figure 1H) that we observed was likely due to changes in postsynaptic contributions to the cartridge.
Although dlg expression appeared normal, we wondered whether other presynaptic proteins more intimately involved in synaptic release were perturbed. Synapsin is a conserved phosphoprotein that bridges vesicles and the cytoskeleton, and is known to be a marker for the reserve synaptic vesicle pool (Greengard et al. 1993; Pieribone et al. 1995) (Figure 1A). The number of synapsin puncta per cartridge was reduced in Dscam2A single-isoform lines and to a lesser extent in Dscam2B single-isoform lines compared to controls (Figure 1, E, G, and J). We also analyzed Csp, another conserved presynaptic protein that is essential for regulated neurotransmission (Zinsmaier et al. 1994). It acts as a chaperone to traffic the synaptic vesicle and coordinate neurotransmitter release by binding to two of the major components of the vesicular exocytosis machinery (syntaxin and synaptotagmin 1) while promoting the activity of presynaptic calcium channels (Figure S1A) (Nie et al. 1999; Wu et al. 1999). In control cartridges, Csp highlighted regions of the presynaptic membrane that come into contact with the postsynaptic lamina neuron dendrites. In Dscam2 single-isoform lines, the areas demarcated by Csp and occupied by L1 and L2 were reduced compared to controls (Figure S1, E, G, J, and K). This confirmed our synapsin results and suggested that although the structural integrity of the presynaptic terminals was intact, the synaptic release machinery was perturbed in Dscam2 single-isoform lines.
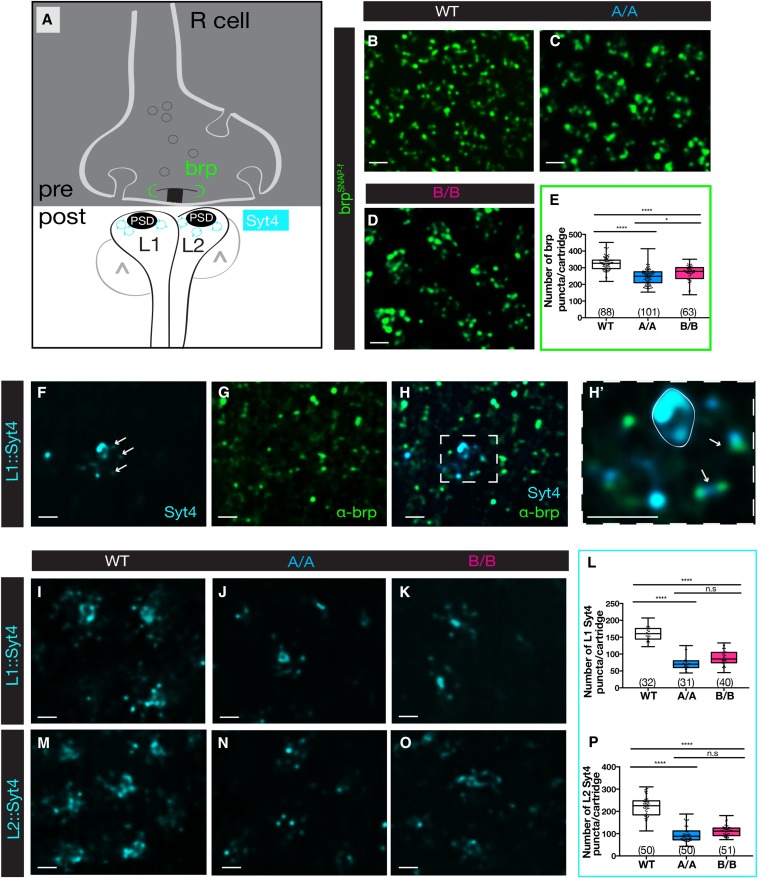
The reduction in presynaptic machinery suggested that the number of synapses could be reduced in single-isoform lines. We therefore analyzed the structural T-bar ribbon-associated protein, Bruchpilot (brp), an ortholog of protein rich in amino acids glutamate (E), leucine (L), lysine (K) and serine (S) (ELKS) family synaptic proteins in vertebrates (Wagh et al. 2006). Comparative studies using EM and light microscopy have shown that there is a one-to-one relationship between brp puncta and synapses defined as an active zone apposed to a postsynaptic density (Chen et al. 2014; Mosca and Luo 2014; Sugie et al. 2015). Thus, brp is widely accepted as the most reliable marker to quantify the number of synapses in flies using light microscopy (Figure 2A). Visualization of brp was achieved using a SNAP-tagged knock-in construct inserted into an endogenous brp locus (Kohl et al. 2014). Brp-SNAP animals were crossed into a Dscam2 single-isoform background and subsequently labeled with the chemical substrate that detects the SNAP transgene. Brp was quantified as the number of puncta per cartridge in a semiautomated fashion using Imaris software. All brp puncta within a cartridge were included in the analysis, as previous studies have determined that the majority of synapses occur between R cells and L1 and L2 in tetrads; however, reciprocal connections between L2 and L4 are also minimally represented (Meinertzhagen and O’Neil 1991). In comparison to controls, a decrease in the number of brp puncta per cartridge was observed in Dscam2 single-isoform lines (Figure 2, B and E). Interestingly, the reduction in brp puncta was more dramatic in animals expressing isoform A compared to isoform B, suggesting that the two isoforms are not functionally identical. These brp data are consistent with the reduction in Csp area and synapsin numbers, and indicate that synapse formation is reduced in Dscam2 single-isoform lines. While a reduction in synapses is apparent, disrupting Dscam2 isoform expression does not exclude the absolute formation of synapses. This suggests that regulated Dscam2 alternative splicing is necessary to achieve normal levels of R cell synapses.
Figure 2.
Synapses marked by brp and postsynaptic Syt4 are reduced in Dscam2 single-isoform mutants. (A) Schematic as in Figure 1, showing localization of brp (green) to the T-bar and localization of Syt4 (magenta) to the postsynaptic density (PSD). (B–D) Representative deconvolved confocal slices of lamina cartridges expressing the chemical tag brp-SNAP (green) and labeled with SNAP-surface 549 in wild-type controls (WT), Dscam2A (A/A), and Dscam2B (B/B) single-isoform lines. (E) Quantification of brp puncta per lamina cartridge using “spot tool” in Imaris. (F) Representative deconvolved confocal slice of a lamina cartridge expressing svp-Gal4 (L1)::Syt4-mCherry. Syt4 postsynaptic puncta are marked with arrows. The strong labeling in the middle of the cartridge is the primary L1 neurite. (G) Anti-brp (nc82) labeling of the confocal slice shown in (F). (H and H’) Merged images showing that Syt4 puncta are apposed to brp puncta (arrows in (H’). The prominent expression in the primary neurite is outlined in white. (I–O) Representative deconvolved confocal slices of Syt4 expression in L1 and L2 cells in WT, A/A, and B/B single-isoform lines. (I–K) L1 expression (svp-Gal4::UAS Syt4-mCherry). (M–O) L2 expression (bab1-Gal4::UAS Syt4-mCherry). (L and P) Quantification of Syt4 puncta in L1 (L) and L2 (P). Boxplot format: middle line = median, error bars = min and max, box = 25–75% quartiles, and each data point = single cartridge. Cartridge number for each genotype is shown in parentheses. Kruskal–Wallis test with Dunn’s multiple comparison test. n.s (not significant) P > 0.05 and **** P < 0.0001. Bar, 2 μm.
Postsynaptic defects in Dscam2 single-isoform lines
To investigate the postsynaptic integrity of R cell synapses in single-isoform lines, we used synaptotagmin 4 (Syt4) to label the postsynaptic L1 and L2 neurons. Syt4 is localized in a punctate fashion to postsynaptic membranes (Lloyd et al. 2000; Adolfsen and Littleton 2001) and is proposed to associate with postsynaptic vesicles. Like other synaptotagmin proteins, it acts as a Ca2+ sensor, but Syt4 specializes in retrograde vesicle release to the presynaptic terminal, rather than presynaptic vesicle fusion (Adolfsen et al. 2004; Yoshihara et al. 2005). We expressed Syt4 in either L1 or L2 neurons using specific Gal4 drivers (seven-up and bab1, respectively) in conjunction with a tagged UAS Syt4 transgene (Singari et al. 2014). Not every cartridge was labeled using this method, most likely due to mosaic expression of the Gal4 lines, but we only imaged and quantified the cartridges that had clear Syt4 labeling. In controls, Syt4 puncta localized to synaptic regions of the lamina cartridge (Figure 2A), but it was also detected on the membrane of the primary neurite in the middle of each cartridge, likely due to the trafficking of the ectopically expressed Syt4 to the dendritic compartment (Figure 2, F–H and H’). Colabeling with a brp antibody (nc82) demonstrated that brp and Syt4 puncta were apposed to each other, suggesting that Syt4 localizes to the postsynaptic density, as previously reported (Adolfsen et al. 2004). In single-isoform lines, the pattern of Syt4 puncta was similar to controls, but the number of puncta was reduced by 50% in both L1 and L2 (Figure 2, I–P). The 20–30% reduction in presynaptic brp puncta compared to the 50% reduction in postsynaptic Syt4 suggests that pre- and postsynaptic defects are not equal in single-isoform lines. The increased severity on the postsynaptic side of the synapse is likely due to changes in the complexity of lamina neuron dendrites (see below).
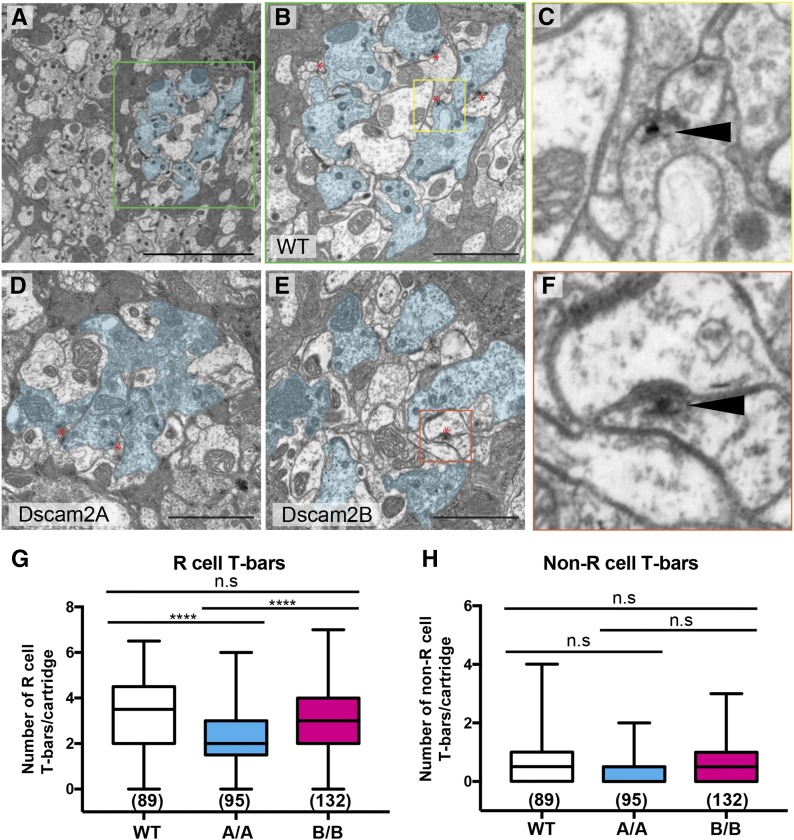
Reduced R cell T-bars and capitate projections in Dscam2 single-isoform lines
To validate the light microscopy results observed in Dscam2 single-isoform mutants, we examined the lamina cartridges using EM. Uniform random cross-sections were obtained in ∼2 μm intervals from the most superficial part of the lamina neuropil. To quantify ultrastructural characteristics of control and single-isoform cartridges, we deidentified the EM images and had two counters analyze T-bars and capitate projections in a double-blinded fashion (see Materials and Methods). During the course of this analysis, we observed unidentified multiple-membrane-containing structures in many of the EM sections, so these were quantified as well. In cross-sections of the WT lamina neuropil, R cell profiles line the perimeter of each cartridge surrounding L1 and an L2 in the center (Figure 3, A and B). In single-isoform mutants, disruptions in the positions of R cell and lamina neuron profiles were observed (Figure 3, D and E), consistent with our light microscopy analysis. The multiple-membrane-containing structures were observed at a much higher frequency in single-isoform cartridges compared to controls (Figure S2, D–G, K, and O), but since their origin was unknown, these were not analyzed further. Consistent with our brp results, we observed a significant decrease in R cell-associated T-bars (Figure 3, B–E and Figure S2, H and L) and capitate projections (Figure S2, C, J, and N) in Dscam2A animals when compared to controls, whereas Dscam2B mutants showed a reduction that was not statistically significant. The number of T-bars not associated with R cells was similar between Dscam2 single-isoform mutants and control animals (Figure 3H and Figure S2, I and M). Since the number of R cells and their associated rhabdomeres were not different between single-isoform mutants and control animals (Figure S1L and Figure S2A), we ruled out the possibility that the decrease in T-bars was due to a loss of R cells. Together, our EM results suggest that regulated Dscam2 isoform expression is necessary for obtaining normal T-bar and capitate projection numbers in lamina cartridges.
Figure 3.
Reduced R cell T-bars in Dscam2A single-isoform lines. (A–F) Electron micrographs of representative lamina sections from wild-type (WT), Dscam2A, and Dscam2B animals imaged using transmission electron microscopy. (A) A WT section at low magnification. The R cells of a single cartridge are outlined in cyan. The green box outlines a single-lamina cartridge. (B) A single WT lamina cartridge with R cells lining the perimeter and presumptive lamina neurons L1 and L2 in the center. The yellow box outlines an example of a T-bar. (C) An example of a T-bar (black solid arrow head) in WT animals. The T-shaped platform is closely associated with a dense region of presynaptic vesicles opposing the postsynaptic density. (D) A Dscam2A single-isoform (A/A) lamina cartridge. (E) A Dscam2B single-isoform (B/B) lamina cartridge. The orange box outlines an example of a non-R cell-associated T-bar. Bar, 5 μM (A) or 2 μM (B, D, and E). Red asterisks denote T-bars (B, D, and E). (F) Non-R cell-associated T-bar (black solid arrow head) in B/B animals. (G and H) Quantification of T-bars associated and not associated with R cells. Shown is the average of two counters (see Materials and Methods). Boxplot format: middle line = median, error bars = min and max, box = 25–75% quartiles, and each data point = T-bar. Cartridge number for each genotype is shown in parentheses. Kruskal–Wallis test with Dunn’s multiple comparison test. n.s (not significant) P > 0.05 and **** P < 0.0001.
Reduced dendrite complexity in Dscam2 single-isoform lines
Our analysis of synaptic proteins in the lamina cartridges revealed that the area occupied by L1 and L2 in each cartridge was reduced compared to controls, while R cells remained intact. This suggested that the dendrites of these cells could be reduced in complexity, presumably due to inappropriate Dscam2 interactions. To test this, we used a MCFO approach by coupling a Gal4 driver expressed in both L1 and L2 with LN-FLP (lamina neuron flip) and UAS>stop>epitope-myr-smGFP to visualize L1 and L2 dendrites at the single-cell level (Nern et al. 2015). L1 and L2 cells were distinguished by their unique axon arbors, which target distinct layers in the medulla. We quantified the average width of L1 and L2 dendritic arrays (see Materials and Methods) expressing different Dscam2 isoforms. We found that the width of both L1 and L2 dendritic arrays was decreased in single-isoform lines (Figure 4, A–F, J, and K). Similar to what we observed for synapses, the Dscam2A single-isoform line produced more severe phenotypes compared to the Dscam2B line. To distinguish whether phenotypes result from the incorrect initiation of dendrites or the inappropriate interactions between dendrites after elaboration, we visualized L1 neurons at 48 hr after puparium formation (APF). L1 dendritic arrays were indistinguishable between Dscam2 single-isoform lines and controls at this time point (Figure 4, G, I, and L), arguing that initial dendritogenesis is not impaired. These data suggest that expression of distinct Dscam2 isoforms in L1 and L2 prevents inappropriate (presumably repulsive) interactions between these cells that lead to defects in dendrite morphogenesis.
Dscam2 cell type-specific alternative splicing is essential for dendrite morphogenesis
To gain insight into whether these dendritic phenotypes are autonomous to L1 and L2, we generated L1 and L2 cells homozygous for a single isoform in an otherwise heterozygous background using mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo 2001). Although the single-isoform lines represent gain-of-function alleles, we could compare L1 and L2 clones expressing either the correct or incorrect isoforms in the identical heterozygous background. This would allow us to correlate the severity of dendritic defects with the similarity of isoforms between L1 and L2. Similar to the FLPout experiments, L1 MARCM clones expressing a single Dscam2 isoform exhibited reductions in dendritic complexity with varying severities. This was categorized into phenotypic classes: normal, dendrites have a stereotypical bottle brush-like dendritic array; mild, dendrites are present but reduced in number; and severe, dendrites appear to be absent with the primary neurite exposed (Figure 4, M, R, V, and W). Using these parameters, we categorized individual L1 clones in a blind fashion. L1 clones expressing the incorrect isoform (Dscam2A) displayed the highest penetrance of dendritic loss (Figure 4V, 81%) when compared to wild-type and L1 clones expressing the correct isoform (Figure 4V, wild-type: 18% and Dscam2B: 44%). The width of the L1 dendritic arrays was also compromised (Figure S3A). L1 clones expressing Dscam2A had the most severe reduction in dendritic width and this was observed as early as 70 hr APF (Figure 4, S–U and X). We also analyzed L2 single-isoform clones and a reciprocal trend was observed. L2 clones expressing the wrong isoform (Dscam2B) exhibited the highest penetrance of dendritic loss (Figure 4W; 64%) when compared to wild-type and L2 clones expressing the correct isoform (Figure 4W; wild-type: 24% and Dscam2A: 16%). Similar to the FLPout experiments, the L2 dendritic phenotypes were less severe than those in L1. However, the MARCM experiments exhibited the expected trends in terms of L1–L2 repulsion. Phenotypes were stronger when both cells expressed the same isoform compared to when they shared one isoform between them (Figure 4, V–X). This is consistent with our previous data in L1 and L2 axon terminals (Lah et al. 2014), and argues that expression of the same isoform in L1 and L2 causes the dendritic phenotypes.
Discussion
In this study, we show that regulated Dscam2 alternative splicing is necessary for R cell synaptogenesis and for the elaboration of postsynaptic dendrites. In fly lines expressing only one Dscam2 isoform, we observed a reduction in both pre- and postsynaptic markers using light microscopy and a reduction in R cell T-bars using EM. Lastly, we observed a reduction in the complexity of L1 and L2 dendrites in single-isoform lines. Our data suggest that these phenotypes are caused by repulsion between L1 and L2 when these cells express identical Dscam2 isoforms.
Unexpectedly, we found that the Dscam2A single-isoform lines exhibited a stronger phenotype than their Dscam2B counterpart. This was evident in both the analysis of synapse numbers and in the quantification of L1 and L2 dendritic arrays. Although there was a trend for a reduction in T-bars in Dscam2B lines, it did not reach statistical significance by EM. Analysis of brp puncta did demonstrate a significant reduction in Dscam2B animals by light microscopy, likely due to the numbers involved (∼17,000 puncta over 63 cartridges vs. 500 T-bars counted over 140 random sections). The differences in dendritic phenotypes are more difficult to explain. One possibility is that Dscam2A interactions have an increased affinity or avidity compared to Dscam2B interactions and that this leads to an increase in repulsive signaling. However, obvious differences in homophilic binding were not observed in previous studies using cell aggregation and pull-down assays (Millard et al. 2007). Alternatively, changes in the magnitudes of these phenotypes could reflect diverse functions for the two isoforms that become unmasked when regulated isoform expression is eliminated. For example, isoform B may function differently from isoform A when expressed in A-expressing cells.
A comparison of the pre- and postsynaptic defects in the single-isoform lines reveals more dramatic phenotypes on the postsynaptic side. For example, brp was reduced by 20–30%, but Syt4 was reduced by 50% in both lines. The reduction in the complexity of L1 and L2 dendrites also suggested a stronger disruption of the postsynaptic compartment. We propose that the tightly regulated postsynaptic composition of the R cell synapse may be altered when cell-specific Dscam2 alternative splicing is eliminated. With fewer dendrites available to participate in synapse formation and inappropriate interactions between L1 and L2 processes, there are likely synapses that contain only one process from either L1 or L2 as opposed to the invariant pairing of L1 and L2 observed at wild-type synapses. These synapses would presumably still contain a presynaptic T-bar, but the Syt4 contribution from the absent postsynaptic cell would be missing. However, even if we had expressed Syt4 in both L1 and L2 simultaneously instead of individually, the resolution of our light microscopy would not have been able to distinguish between one vs. two Syt4 puncta. Thus, one explanation for the greater reduction in postsynaptic compared to presynaptic puncta is that some synapses form in the absence of a canonical L1–L2 postsynaptic pair.
During the course of our EM study, we observed multiple-membrane-containing structures in both single-isoform lines at a much higher frequency than in wild-type (Figure S1). Although we have not identified these structures, they show similarity to both autophagosomes (Kishi-Itakura et al. 2014) and multi-lamellar bodies (Weiss and Minke 2015). This could indicate that neurons within these single-isoform cartridges are not as healthy as their wild-type counterparts, presumably because their postsynaptic composition is incorrect. Although we did not observe a reduction in R cell numbers at either the light microscropy or the EM level, we always analyzed animals that were < 1 day old. Older single-isoform animals may exhibit photoreceptor degeneration.
Our data demonstrating a reduction in R cell synapses in Dscam2 single-isoform lines contrasts with previous work that has proposed that R cell synapse formation proceeds irrespective of disruptions to the postsynaptic compartment or cartridge organization (Brandstätter et al. 1992; Hiesinger et al. 2006). However, these studies were carried out in flies with surgically injured lamina neurons or with loss-of-function mutations in candidate genes that lead to changes in the activity or the organization of the lamina cartridge. Our Dscam2 single-isoform lines are gain-of-function mutants with respect to L1 and L2, as these cells have acquired the ability to interact in a Dscam2-dependent manner. Since Dscam2 functions as a repulsive cue in these cells, we propose that repulsion between L1 and L2 dendrites reduces synapse number and dendrite complexity. However, R cells and other lamina neurons also express Dscam2 and could be contributing to the phenotypes observed in single-isoform lines. L3 is a variable component of the postsynaptic tetrad and is estimated to be present in about ∼30% of all photoreceptor synapses (Meinertzhagen and O’Neil 1991). L3 expresses isoform A, like L2. Photoreceptors express isoform B mRNA (Lah et al. 2014), but they do not require Dscam2 for R cell targeting (Millard et al. 2007; Tadros et al. 2016), and whether they express Dscam2 protein at all is still unclear. Using an antibody that recognizes the cytoplasmic domain of Dscam2, protein cannot be detected in the eye disc or optic stalk of third instar larvae (S. S. Millard and S. L. Zipursky, unpublished data). Dscam2 protein is also absent from R7 and R8 terminals during pupal development (Millard et al. 2007). The caveat to these expression studies is that there may be secreted forms of Dscam2 protein that this antibody would not recognize (Graveley et al. 2011). Although secreted proteins would not be able to mediate contact-dependent repulsion like membrane-bound forms, they could modulate Dscam2B interactions. Regardless of the cells involved, disrupting the regulated expression of the two Dscam2 extracellular isoforms clearly disrupts synapse formation.
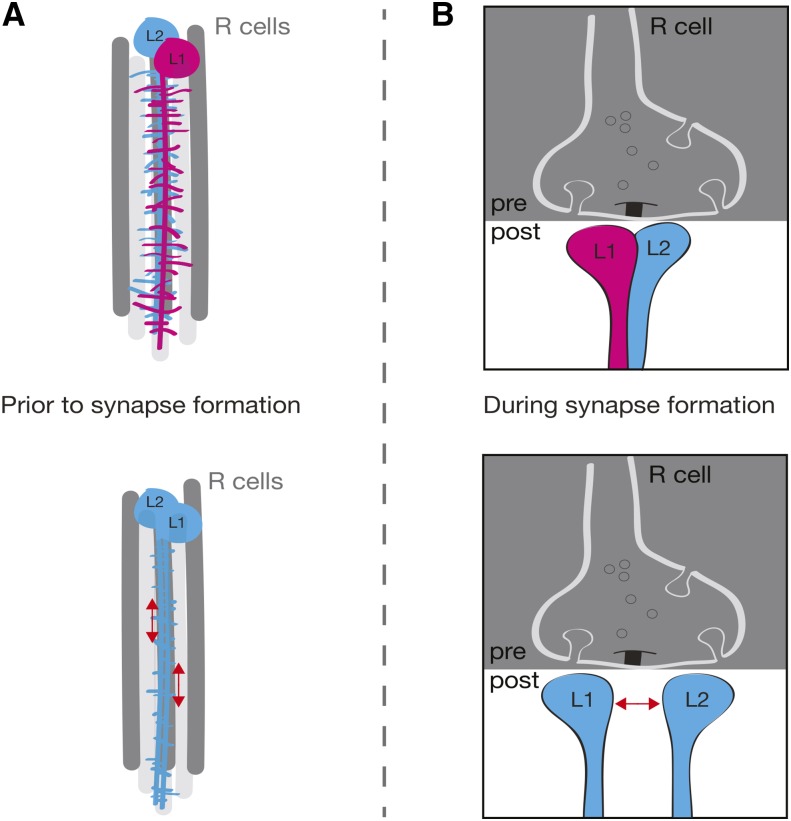
Our data using the Dscam2 single-isoform lines provide conclusive evidence for the model that Dscam2 cell-specific isoform expression is necessary for synaptic exclusion at R cell synapses. When L1 and L2 express the same isoform, the number of synapses and the complexity of the postsynaptic dendritic array is reduced. The reduction in synapses is likely the result of two aberrant processes dependent on repulsion: (1) a decrease in the number postsynaptic elements available for forming synapses and (2) the failure of L1 and L2 postsynaptic pairing at R cell terminals (Figure 5).
Figure 5.
Model: cell-specific isoform expression is necessary for R cell synapse formation. (A) Side view of the developing cartridge. When L1 and L2 express different Dscam2 isoforms, dendrites from these cells elaborate extensive branches that meet to form synapses at the R cell terminal (top). In animals expressing a single isoform, L1 and L2 dendritogenesis is normal, but dendritic growth is stunted due to inappropriate repulsion between these cells. This results in a reduction of postsynaptic dendrites prior to synapse formation (bottom). (B) Cross-sectional view of the developing synapse. When L1 and L2 express different Dscam2 isoforms, nascent synapses incorporate one dendritic element from each of these cells. Multiple elements from the same cells are excluded through Dscam1/Dscam2 self-avoidance (Millard et al. 2010) (top). In animals expressing a single isoform, nascent synapses that incorporate L1 and L2 elements are subject to inappropriate repulsion at the forming synapse. This could lead to several scenarios including: normal synapses (presumably due to compensatory mechanisms that overcome inappropriate repulsion), synapses that lack an L1 or an L2 element, and synapses that lack both L1 and L2 (bottom). Red arrows indicate inappropriate repulsion between L1 and L2. Dscam2A = blue, Dscam2B = pink.
Thus, both probabilistic (Dscam1) and regulated (Dscam2) alternative splicing play crucial roles in the formation of photoreceptor synapses. Identifying the splicing regulator of Dscam2 may reveal other target transcripts that also contribute to synaptogenesis through cell-specific alternative splicing.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300432/-/DC1.
Acknowledgments
We thank Shaun Walters and Luke Hammond for technical assistance with microscopy; the Millard laboratory for manuscript feedback; Ning Liu for technical assistance with electron microscopy; and Wei Jun Tan for technical assistance with stock building. S.K.K. and J.S.S.L. were supported by an Australian Postgraduate Award (Research Training Scheme). This study was supported by an Australian National Health and Medical Research Council grant (APP1105792).
Author contributions: S.S.M. and G.J.-e.S. conceived study and experimental designs. S.K.K., G.J.-e.S. and J.S.S.L. carried out the synaptic protein imaging studies. S.K.K. and J.S.S.L. carried out the lamina neuron FLPout experiments. J.S.S.L. and G.J.-e.S. carried out the lamina neuron MARCM dendrite analysis. S.K.K. and J.S.S.L. quantified the electron microscopy data, which was sectioned and imaged by Ning Liu in P.G.N.’s laboratory. J.S.S.L., S.K.K., and S.S.M. wrote the paper.
Footnotes
These authors contributed equally to this work.
Communicating editor: L. Luo
Literature Cited
- Adolfsen B., Littleton J. T., 2001. Genetic and molecular analysis of the synaptotagmin family. Cell. Mol. Life Sci. 58: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolfsen B., Saraswati S., Yoshihara M., Littleton J. T., 2004. Synaptotagmins are trafficked to distinct subcellular domains including the postsynaptic compartment. J. Cell Biol. 166: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J., Martinelli D. C., Malenka R. C., Tabuchi K., Sudhof T. C., 2013. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154: 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard A. A., Ko J., Sudhof T. C., 2012. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J. Biol. Chem. 287: 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstätter J. H., Seyan H. S., Meinertzhagen I. A., 1992. The effects of the loss of target cells upon photoreceptor inputs in the fly’s optic lobe. J. Neurocytol. 21: 693–705. [DOI] [PubMed] [Google Scholar]
- Chen W. V., Nwakeze C. L., Denny C. A., O’Keeffe S., Rieger M. A., et al. , 2017. Pcdhalphac2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science 356: 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Akin O., Nern A., Tsui C. Y., Pecot M. Y., et al. , 2014. Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo M. V., Foldy C., Gokce O., Rothwell P. E., Sun G. L., et al. , 2015. Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms. Neuron 87: 326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard P., Valtorta F., Czernik A. J., Benfenati F., 1993. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science 259: 780–785. [DOI] [PubMed] [Google Scholar]
- Hamanaka Y., Meinertzhagen I. A., 2010. Immunocytochemical localization of synaptic proteins to photoreceptor synapses of Drosophila melanogaster. J. Comp. Neurol. 518: 1133–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesinger P. R., Zhai R. G., Zhou Y., Koh T.-W., Mehta S. Q., et al. , 2006. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Curr. Biol. 16: 1835–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. E., Bortnick R., Tsubouchi A., Baumer P., Kondo M., et al. , 2007. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 54: 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T., Iijima Y., Witte H., Scheiffele P., 2014. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. J. Cell Biol. 204: 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J., Zhang C., Arac D., Boucard A. A., Brunger A. T., et al. , 2009. Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 28: 3244–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J., Ng J., Cachero S., Ciabatti E., Dolan M.-J., et al. , 2014. Ultrafast tissue staining with chemical tags. Proc. Natl. Acad. Sci. USA 111: E3805–E3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah G. J., Li J. S., Millard S. S., 2014. Cell-specific alternative splicing of Drosophila Dscam2 is crucial for proper neuronal wiring. Neuron 83: 1376–1388. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L., 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Lefebvre J. L., Kostadinov D., Chen W. V., Maniatis T., Sanes J. R., 2012. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488: 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. S. S., Shin G. J.-e., Millard S. S., 2015. Neuronal cell-type-specific alternative splicing: a mechanism for specifying connections in the brain? Neurogenesis (Austin) 2: e1122699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd T. E., Verstreken P., Ostrin E. J., Phillippi A., Lichtarge O., et al. , 2000. A genome-wide search for synaptic vesicle cycle proteins in Drosophila. Neuron 26: 45–50. [DOI] [PubMed] [Google Scholar]
- Matsuda K., Yuzaki M., 2011. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur. J. Neurosci. 33: 1447–1461. [DOI] [PubMed] [Google Scholar]
- Matthews B. J., Kim M. E., Flanagan J. J., Hattori D., Clemens J. C., et al. , 2007. Dendrite self-avoidance is controlled by Dscam. Cell 129: 593–604. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen I. A., O’Neil S. D., 1991. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 305: 232–263. [DOI] [PubMed] [Google Scholar]
- Merkin J., Russell C., Chen P., Burge C. B., 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard S. S., Flanagan J. J., Pappu K. S., Wu W., Zipursky S. L., 2007. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature 447: 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard S. S., Lu Z., Zipursky S. L., Meinertzhagen I. A., 2010. Drosophila dscam proteins regulate postsynaptic specificity at multiple-contact synapses. Neuron 67: 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca T. J., Luo L., 2014. Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. Elife 3: e03726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D., Rubin G. M., 2015. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112: E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. M., Schreiner D., Xiao L., Traunmuller L., Bornmann C., et al. , 2016. An alternative splicing switch shapes neurexin repertoires in principal neurons vs. interneurons in the mouse hippocampus. Elife 5: e22757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Ranjan R., Wenniger J. J., Hong S. N., Bronk P., et al. , 1999. Overexpression of cysteine-string proteins in Drosophila reveals interactions with syntaxin. J. Neurosci. 19: 10270–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Gao S., Norris M. L., Ray D., Ramani A. K., et al. , 2014. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell 54: 946–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot M. Y., Tadros W., Nern A., Bader M., Chen Y., et al. , 2013. Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron 77: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieribone V. A., Shupliakov O., Brodin L., Hilfiker-Rothenfluh S., Czernik A. J., et al. , 1995. Distinct pools of synaptic vesicles in neurotransmitter release. Nature 375: 493–497. [DOI] [PubMed] [Google Scholar]
- Raj B., Blencowe B. J., 2015. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron 87: 14–27. [DOI] [PubMed] [Google Scholar]
- Rister J., Pauls D., Schnell B., Ting C.-Y., Lee C.-H., et al. , 2007. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron 56: 155–170. [DOI] [PubMed] [Google Scholar]
- Schreiner D., Nguyen T. M., Russo G., Heber S., Patrignani A., et al. , 2014. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 84: 386–398. [DOI] [PubMed] [Google Scholar]
- Schwabe T., Borycz J. A., Meinertzhagen I. A., Clandinin T. R., 2014. Differential adhesion determines the organization of synaptic fascicles in the Drosophila visual system. Curr. Biol. 24: 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui T. J., Pancaroglu R., Kang Y., Rooyakkers A., Craig A. M., 2010. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 30: 7495–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singari S., Javeed N., Tardi N. J., Marada S., Carlson J. C., et al. , 2014. Inducible protein traps with dominant phenotypes for functional analysis of the Drosophila genome. Genetics 196: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P., Zhu S., Emoto K., Younger S., Yang S. J., et al. , 2007. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugie A., Hakeda-Suzuki S., Suzuki E., Silies M., Shimozono M., et al. , 2015. Molecular remodeling of the presynaptic active zone of Drosophila photoreceptors via activity-dependent feedback. Neuron 86: 711–725. [DOI] [PubMed] [Google Scholar]
- Tadros W., Xu S., Akin O., Yi C. H., Shin G. J., et al. , 2016. Dscam proteins direct dendritic targeting through adhesion. Neuron 89: 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Zhang K. X., Pecot M. Y., Nagarkar-Jaiswal S., Lee P. T., et al. , 2015. Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163: 1756–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M., Naito Y., Kuroyanagi H., Iino Y., 2016. Splicing factors control C. elegans behavioural learning in a single neuron by producing DAF-2c receptor. Nat. Commun. 7: 11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Lee S. J., Yasumura M., Takeuchi T., Yoshida T., et al. , 2010. Trans-synaptic interaction of GluRdelta2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141: 1068–1079. [DOI] [PubMed] [Google Scholar]
- Wagh D. A., Rasse T. M., Asan E., Hofbauer A., Schwenkert I., et al. , 2006. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49: 833–844. [DOI] [PubMed] [Google Scholar]
- Weiss S., Minke B., 2015. A new genetic model for calcium induced autophagy and ER-stress in Drosophila photoreceptor cells. Channels 9: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. F., Bryant P. J., 1993. ZO-1, DlgA and PSD-95/SAP90: homologous proteins in tight, septate and synaptic cell junctions. Mech. Dev. 44: 85–89. [DOI] [PubMed] [Google Scholar]
- Wu M. N., Fergestad T., Lloyd T. E., He Y., Broadie K., et al. , 1999. Syntaxin 1A interacts with multiple exocytic proteins to regulate neurotransmitter release in vivo. Neuron 23: 593–605. [DOI] [PubMed] [Google Scholar]
- Yoshihara M., Adolfsen B., Galle K. T., Littleton J. T., 2005. Retrograde signaling by Syt 4 induces presynaptic release and synapse-specific growth. Science 310: 858–863. [DOI] [PubMed] [Google Scholar]
- Zhang X., Chen M. H., Wu X., Kodani A., Fan J., et al. , 2016. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166: 1147–1162.e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier K. E., Eberle K. K., Buchner E., Walter N., Benzer S., 1994. Paralysis and early death in cysteine string protein mutants of Drosophila. Science 263: 977–980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protocols and reagents are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.