Peischl et al. explore the way evolutionary forces shape genetic variability in expanding human populations. Over a few generations of separate evolution...
Keywords: range expansion, Quebec, mutation load, genetic drift
Abstract
Humans have colonized the planet through a series of range expansions, which deeply impacted genetic diversity in newly settled areas and potentially increased the frequency of deleterious mutations on expanding wave fronts. To test this prediction, we studied the genomic diversity of French Canadians who colonized Quebec in the 17th century. We used historical information and records from ∼4000 ascending genealogies to select individuals whose ancestors lived mostly on the colonizing wave front and individuals whose ancestors remained in the core of the settlement. Comparison of exomic diversity reveals that: (i) both new and low-frequency variants are significantly more deleterious in front than in core individuals, (ii) equally deleterious mutations are at higher frequencies in front individuals, and (iii) front individuals are two times more likely to be homozygous for rare very deleterious mutations present in Europeans. These differences have emerged in the past six to nine generations and cannot be explained by differential inbreeding, but are consistent with relaxed selection mainly due to higher rates of genetic drift on the wave front. Demographic inference and modeling of the evolution of rare variants suggest lower effective size on the front, and lead to an estimation of selection coefficients that increase with conservation scores. Even though range expansions have had a relatively limited impact on the overall fitness of French Canadians, they could explain the higher prevalence of recessive genetic diseases in recently settled regions of Quebec.
THE impact of recent demographic changes or single bottlenecks on the overall fitness of populations is still highly debated (Lohmueller et al. 2008; Fu et al. 2014; Lohmueller 2014; Simons et al. 2014; Do et al. 2015; Henn et al. 2015, 2016; Gravel 2016). Simulation and theoretical approaches suggest that populations on expanding wave fronts experience strong genetic drift (Peischl et al. 2013, 2015), leading to relaxed selection (Gravel 2016) resulting in the build-up of expansion load (Peischl et al. 2013). The main reason for this expansion load is that low population densities and strong genetic drift at the wave front promote the genetic surfing of both neutral and selected variants (Peischl et al. 2013). This relatively inefficient selection on the wave front leads to the preservation of many new mutations, unless very deleterious (Peischl et al. 2013), and it shifts the site frequency spectrum (SFS) of standing variants, thus increasing the contribution of (partially) recessive variants to mutation load (Peischl and Excoffier 2015). After a range expansion, both a decrease of diversity and an increase in the recessive mutation load with distance from the source is expected (Kirkpatrick and Jarne 2000; Peischl and Excoffier 2015). This pattern has recently been shown to occur in non-African human populations, where a gradient of recessive load has been observed between North Africa and the Americas (Henn et al. 2016). Whereas the bottleneck out of Africa that started ∼50,000 years ago (e.g., Gravel et al. 2011) must have created a mutation load, the exact dynamics of this load increase due to the expansion process are still unknown. It is also unclear if a much more recent expansion could have had a significant impact on the genetic load of populations.
The settlement of Quebec can be considered as a series of demographic and spatial expansions following initial bottlenecks. The majority of the 6.5 million French Canadians living in Quebec are the descendants of ∼8500 founder immigrants of mostly French origin (Charbonneau et al. 2000; Laberge et al. 2005). This French immigration started with the founding of a few settlements along the Saint Laurence river at the beginning of the 17th century (Charbonneau et al. 2000). Most new settlements were restricted to the Saint Laurence Valley until the 19th century, after which remote territories began to be colonized. Bottlenecks and serial founder effects occurring during range expansions are thought to have profoundly affected patterns of genetic diversity, leading to large frequency differences when compared to the French source population (Laberge et al. 2005). Even though the French Canadian population has expanded 700-fold in ∼300 years, its genetic diversity is actually not what is expected in a single panmictic, exponentially growing population, as allele frequencies have drifted much more than expected in a fast-growing population (Heyer 1995, 1999). It has been shown that genetic surfing (Klopfstein et al. 2006; Peischl et al. 2016) has occurred during the recent colonization of the Saguenay-Lac St-Jean area (Moreau et al. 2011), and that the fertility of women on the wave front was 25% higher than of those living in the core of the settlement, giving them more opportunity to transmit their genes to later generations. In addition, female fertility was found to be heritable on the front but not in the core (Moreau et al. 2011), a property that further contributes to lowering of the effective size of the front population (Austerlitz and Heyer 1998; Sibert et al. 2002) and the enhancement of drift on the wave front. Social transmission of fertility (Austerlitz and Heyer 1998) and genetic surfing during range expansions, or a combination of both (Moreau et al. 2011), have been proposed to explain a rapid increase of some low-frequency variants. Thus, it seems that differences in allele frequencies between French Canadians and continental Europeans are due to a mixture of the random sampling of initial immigrants (founder effect) and of strong genetic drift having occurred in Quebec after the initial settlement, resulting in a genetically and geographically structured population of French Canadians (Bherer et al. 2011).
The demographic history of Quebec has not only affected patterns of neutral diversity, but also the prevalence of some genetic diseases, independently from inbreeding (De Braekeleer 1991; Heyer 1995; Laberge et al. 2005; Yotova et al. 2005), as well as the average selective effect of segregating variants (Casals et al. 2013). Even though French Canadians have fewer mutations segregating in the population than the French, these mutations are found at loci which are, on average, much more conserved, and thus are potentially more deleterious than those segregating in the French population (Casals et al. 2013). Recurrent founder effects, low densities, and intergenerational correlation in reproductive success could all contribute to increase drift and reduce the efficacy of selection on expanding wave fronts, and thus lead to the development of a stronger mutation load (Peischl et al. 2013). Furthermore, even short periods of increased drift and relaxed selection can affect the efficacy of selection in future generations (Gravel 2016), resulting in differential rates of purging in front and core populations. It is therefore likely that the excess of low-frequency deleterious variants observed in French Canadian individuals (e.g., Casals et al. 2013) could be at least partly due to the expansion process rather than to the sole initial bottleneck.
To better understand and quantify the effect of a recent expansion process on the amount and pattern of deleterious genetic variation, we screened the ascending genealogies of 3916 individuals from the CARTaGENE cohort (Awadalla et al. 2013) that were linked to the BALSAC genealogical database (http://balsac.uqac.ca/). Using stringent criteria on the quality of genealogical information (see Materials and Methods), we selected 51 (front) individuals whose ancestors were as close as possible to the front of the colonization of Quebec, and 51 (core) individuals whose ancestors were as far as possible from the front (see Materials and Methods, Figure 1, and Supplemental Material, File S2 and Animation S2 in File S3). We then sequenced these 102 individuals at very high coverage (mean 89.5× and range 67–128×) for ∼106.5 Mb of exomic and UTR regions, and contrasted their genomic diversity to detect if sites with various degrees of conservation and deleteriousness have been differentially impacted by selection.
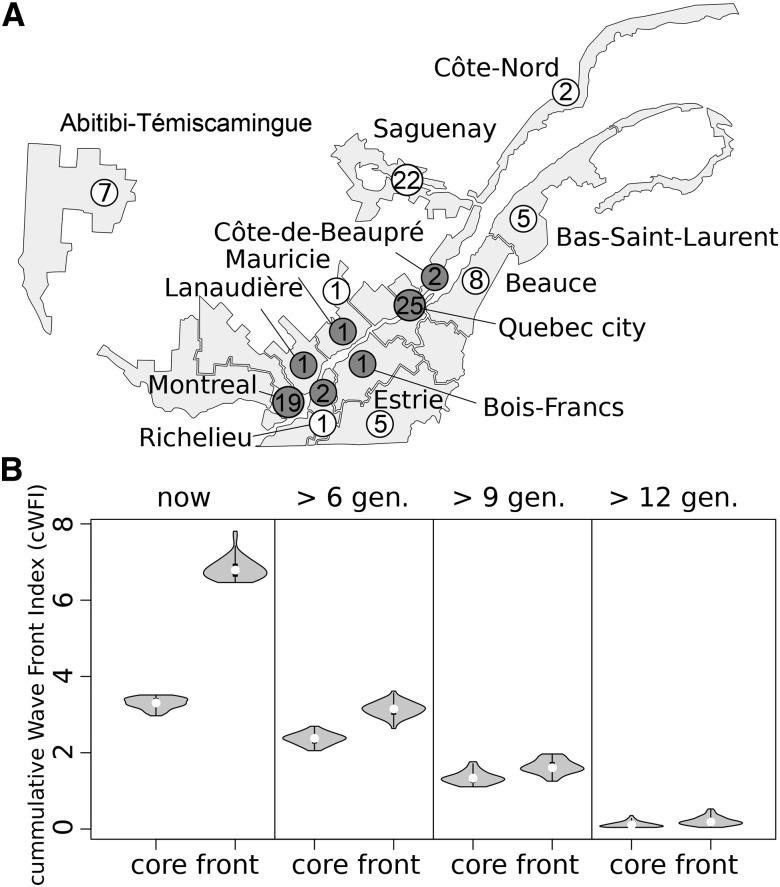
Figure 1.
Location and number of sampled individuals and distribution of the cumulative wave front indices (cWFI). (A) Front and core sampled individuals are shown in white and gray, respectively. The numbers inside circles indicate the sample size for each location. (B) The leftmost panel shows the distribution of cWFI among sampled individuals. The other three panels display the cWFI of the ancestors of the sampled individuals that lived 6, 9, or 12 gen. (generations) ago, which shows that observed differences in cWFI between current samples have mostly emerged in the six most recent generations.
Materials and Methods
Selection of individuals to sequence
We selected individuals to be sequenced by screening the genealogy of 3916 individuals of the CARTaGENE biobank (Awadalla et al. 2013), who could be connected to the BALSAC genealogical database (http://balsac.uqac.ca) thanks to the information they provided on their parents and grandparents. The BALSAC database includes records from all Catholic marriages in Quebec from 1621 to 1965, totaling more than three million records (five million individuals). The ascending genealogies of the 3916 CARTaGENE individuals were assessed for their maximum generation depth, their completeness defined as the fraction of ancestors that are traced back in an individual’s genealogy at generation g relative to the maximum number of ancestors () at that generation (Jetté 1991), as well as our ability to assess the front or core status of the ancestors. Thus, we first eliminated 420 genealogies that spanned <12 generations (maximum generation depth <12 generations); we also eliminated 537 genealogies that had a mean depth of less than eight generations, 578 genealogies whose completeness (Jetté 1991) computed over the last six generations was <95%, and 97 additional genealogies whose completeness computed over the 12 generations was <30%. Genealogies were also filtered based on the quantity of information available for the computation of a cumulative wave front index (cWFI), defined as where the summation is over all ancestors in the genealogy, is the genetic contribution of the i-th ancestor, is the wave front index of the i-th ancestor, defined as (Moreau et al. 2011), and g is the number of generations elapsed since the foundation of the location where the ancestor reproduced [see Moreau et al. (2011) for more details]. A value of 1 would imply that all the ancestors of the focal individual reproduced on the wave front. To ensure that differences in between individuals are not due to a lack of information on the core front status of individuals in the genealogy, we eliminated 717 genealogies for which a single was missing for any individual belonging to the six most recent generations ( completeness < 1 for the six most recent generations) and 15 additional genealogies for which the completeness until generation 12 was <0.5. We also excluded genealogies for which the total number of individuals with computable WFI until generation 12 was either too small or too large from the analysis, so that the was computed on genealogies of comparable total sizes. The 10% smallest and the 15% largest genealogies were thus eliminated (389 genealogies) from further analyses. The 1163 remaining individuals were ranked according to their and we then selected individuals with the 10% smallest and 10% highest We also eliminated from these two groups those individuals that were too closely related. The kinship coefficient (Wright 1922) was thus computed between all members of these groups to determine their relatedness. For 41 pairs of individuals more related than second cousins ( > 1/64), one of the two individuals was removed at random. Furthermore, we removed individuals with inbreeding coefficients >0.05. Finally, the 60 individuals with the lowest and the 60 individuals with the largest were selected for further DNA analyses. Among these, 51 individuals of each category for which peripheral blood samples were available in the CARTaGENE biobank were further considered for DNA extraction and sequencing. The geographic location of the marriage place of 102 individuals’ parents is reported in Figure 1, and examples of the location of the ancestors of front and core individuals at various periods are shown in Animation S1 in File S2 and Animation S2 in File S3. In our final sample, only three individuals had inbreeding coefficients >0.02 (Figure S28 in File S1).
DNA extraction, library preparation, and sequencing
Peripheral blood samples preserved in EDTA tubes from 102 selected individuals from the CARTaGENE cohort were processed for DNA extraction using the FlexiGene DNA kit as recommended by the supplier (QIAGEN, Valencia, CA). Total DNA was quantified by measurement with a NanoDrop 8000 spectrophotometer (Thermo Scientific), followed by double-stranded DNA (dsDNA) quantification with a QUBIT 2.0 fluorometer (Life Technologies). DNA libraries were prepared for each sample following the standard protocol of the KAPA Library Preparation Kit for Illumina sequencing platforms. A Covaris S2 fragmentation (duty cycle: 10%, intensity: 5.0, cycles per burst: 200, duration: 120 sec, mode frequency: sweeping, displayed power Covaris S2: 23W) was performed on 1 µg dsDNA input (50 µl total volume) for each sample to generate 180–200 bp average size fragments. The resulting 3′ and 5′ overhangs were end-repaired, 3′-adenylated, and ligated to specific indexed adaptors. After a dual solid phase reversible immobilization (SPRI) size selection of 250–450 bp adapter-ligated fragments, final precapture library enrichment was performed by ligation-mediated polymerase chain reaction (LM-PCR) followed by a library amplification cleanup with magnetic beads (AMpure XP, Agencourt). Following the protocol for whole-exome capture with the Roche NimbleGen SeqCap EZ Exome + UTR Library kit (User’s Guide v4.2, http://www.nimblegen.com/products/seqcap/ez/exome-utr/index.html), the size distribution of the enriched fragments was then checked using a DNA 1000 chip on an Agilent 2100 Bioanalyzer for whole-exome capture validation. The 102 uniquely indexed amplified DNA samples were mixed into 34 pool libraries of three different indexed DNAs each, and were then hybridized to specific SeqCap EZ Hybridization Enhancing oligos at +47° for 72 hr. After a washing step followed by a SeqCap EZ Pure Capture Beads recovery of the targeted sequences (here whole exome + UTRs), the multiplex DNA samples were amplified by a postcapture LM-PCR, cleaned with AMpure XP magnetic beads, and bioanalyzed with a DNA 1000 chip to quantify and qualify the amplified captured multiplexed DNA samples. Prior to sequencing, a final validation by quantitative PCR assays was carried out on the DNA samples to assess the relative fold enrichment in precaptured sequences vs. postcaptured ones. Finally, these 34 DNA pools (one pool per lane) were paired-end (2 × 100 bp) sequenced on an Illumina HiSequation 2500 System.
Alignment and variant calling
Before mapping reads, a quality control was done using FASTQC, and trimming of the adapters and of poor-quality read ends was done using Trim Galore (≥ Q20). The reads were then mapped to the hg19 reference genome using BWA v 0.5.9r16 using the default parameters. PCR duplicates were removed using Picard-tools v1.56 (http://broadinstitute.github.io/picard/). We kept properly paired and uniquely mapped reads using Samtools v0.1.19-44428cd. After these steps, we estimated the mean sequence coverage per individual, across the targeted exomic and UTR regions of cumulative length ∼106.5 Mb, to be between 67 and 128× (Figure S41 in File S1). Realignment around indels (insertion/deletions) and variants recalibration were performed with GATK v3.2-2. GATK v3.2-2 was also used to call variants using the workflow recommended by the Broad Institute (https://www.broadinstitute.org/gatk/guide/best-practices?bpm=DNAseq). We performed first step using HaplotypeCaller, reporting the calls in genome variant call format (GVCF) mode. Then the joint genotyping calls were performed using the GenotypeGVCFs subprogram of GATK, to get the raw SNP and indel calls. The last step of recalibrating and filtering the genotype calls was done with VQSR, using the recommended options separately on the SNP and indel calls.
Sequence analysis
We removed all variants associated with a quality score <30. We kept 426,301 SNPs and 43,081 indels, and used ANNOVAR to functionally characterize these variants. Table S5 in File S1 gives the number of variants in each ANNOVAR functional class. Individual genotypes associated with low read depth (i.e., <10) and low genotype quality (i.e., <20) were marked as missing genotypes. We also collected polymorphism data for 305 individuals from three European populations [British from England and Scotland (GBR), Spanish from Spain (IBS), and Italian from Tuscany, Italy (TSI), Table S6 in File S1] from the 1000 Genomes phase 3 panel (1000 Genomes Project Consortium et al. 2015). Note that the 1000 Genomes phase 3 panel set of variants consists of polymorphisms called from a combination of both low- and high-coverage data (between 8 and 30×). Our comparison of French Canadians and individuals from European populations to the 1000 Genomes phase 3 panel was restricted to the genomic regions that intersected between the targeted regions sequenced in the present study and the high-coverage target of the 1000 Genomes phase 3 panel, which amounts to ∼46.4 Mb. Since the 1000 Genomes data had lower coverage (∼65×) than our French Canadian data (∼89.5×), which could lead to differences in genotype frequencies other than those due to population history, we performed a downsampling of French Canadian individual reads using Samtools v1.1, option “-s,” to randomly select reads and reduce the coverage to 65×.
This step was then followed by the SNP calling and recalibration steps described in the previous section. We defined shared SNPs between French Canadians and individuals from the 1000 Genomes phase 3 panel as SNPs found in both data sets. Differences in number of various types of sites were obtained by a permutation test consisting of randomly permuting individuals between front and core, reestimating the desired statistics on the permuted samples, and estimating the P-value of the observed statistics in the generated empirical null distribution.
Assessment of mutation effects
The ancestral state of all mutations was characterized, following the 1000 Genomes Project Consortium et al. (2015), using the human ancestor genome inferred from the alignment of six primate genomes (Homo sapiens, Pan troglodytes, Gorilla gorilla, Pongo abelii, Macaca mulatta, and Callithrix jacchus) (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/). The biological impact of SNPs was assessed via Genomic Evolutionary Rate Profiling Rejected Substitution (GERP RS) scores (Cooper et al. 2005; Davydov et al. 2010), which measure, at a given genomic location, the difference between the expected and the observed number of mutations at a given position occurring across a phylogeny of 35 mammals. GERP RS scores were obtained from the University of California, Santa Cruz genome browser (http://hgdownload.cse.ucsc.edu/gbdb/hg19/bbi/All_hg19_RS.bw). Note that the human sequence was not included in the calculation of GERP RS scores. The human reference sequence was indeed excluded from the alignment for the calculation of both the neutral rate and site-specific “observed” rate for the RS score to prevent any bias in the estimates. Mutations were classified as being “neutral,” “moderate,” “large,” or “extreme” for GERP RS scores with ranges [-2,2[, [2,4[, [4,6[ and [6,∞[, respectively. GERP RS scores of 0 indicated that the alignment of mammalian sequences was too shallow at that position to get a meaningful estimate of constraint (Goode et al. 2010), and sites with such scores were removed from all analyses involving GERP RS scores.
We also used additional methods to assess the functional effect of SNPs and to characterize short indels. We used PolyPhen-2 (Adzhubei et al. 2010) to predict the damaging effect of missense mutations. PolyPhen-2 predicts the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations. We also used PhyloP scores, which measure evolutionary conservation at individual alignment sites to identify putatively deleterious variants (Pollard et al. 2010). PhyloP scores were computed from the alignments of 36 eutherian mammal sequences, without the human reference sequence, and denoted as PhyloPNH (Fu et al. 2013). Finally, we used combined annotation dependent depletion (CADD) scores, which integrate several annotations including conservation metrics, regulatory information, transcript information, and protein-level scores into a single measure (C-score) for each variant (Kircher et al. 2014). We used scaled C-scores, phred-like scores ranging from 0.001 to 99, in our analyses, as these scores are easily interpretable. A scaled C-score > 10 indicates that the corresponding variant is predicted to be in the 10% most deleterious classes of variants. A scaled C-score > 20 indicates that the corresponding variant is predicted to be in the 1% most deleterious class of variants. Mutations were classified as being neutral, moderate, large, or extreme for CADD scores, with ranges [0,10[, [10,20[, [20,30[ and [30,∞ [, respectively.
Assessment of relaxation of selection
Assessing mutation load from genomic data is an inherently difficult problem [for further discussion see Lohmueller (2014)]. Instead, we used GERP RS scores as a proxy for selection intensity and calculated the average GERP RS score for each individual across all sites at which the focal individual carried a derived allele. The average GERP RS score has a straightforward interpretation: it measures the average degree of conservation of segregating sites. We expect that selection keeps strongly conserved sites (large GERP RS scores) at low frequency, whereas weakly conserved sites should be found at higher frequency. A relaxation of selection, for instance due to strong genetics drift, should therefore lead to an increase of the average GERP RS score. Here, we focus on the average RS score per site. The average GERP RS score per site is simply the average of GERP RS scores calculated over all sites at which an individual carries at least one copy of a derived mutation: where n is the number of segregating sites per individual and is the GERP RS score of site Note that this measure does not distinguish between heterozygous sites and derived homozygous sites. To account for the frequency of derived alleles, we also calculated the average GERP RS score across sites that have a given derived allele frequency (DAF).
Detection of outlier SNPs and gene ontology analysis
To detect potential outlier SNPs based on levels of genetic differentiation, we used the outlier FST method proposed by Beaumont and Nichols (1996) and implemented in the Arlequin software (Excoffier and Lischer 2010). In brief, this test uses coalescent simulations to generate the joint distribution of FST and heterozygosity between populations expected under a finite-island model, having the same average FST value as that observed. This null distribution is then used to compute the P-value of each SNP based on its observed FST and heterozygosity levels. SNPs with FST values outside the 99% quantile based on the simulations were considered as outliers. These SNPs were then annotated to Ensembl gene identifiers with the R package BiomaRt (Durinck et al. 2009). SNPs were mapped to a gene if they were located in the gene transcript or within 10 kb of it. If a SNP was allocated to more than one gene with this method, we uniquely allocated it to its closest gene. If more than one SNP was assigned to a given gene, we only kept the SNP with the highest FST value.
We conducted a gene ontology (GO) enrichment analysis on the list of significant SNPs using the topGO R package (Alexa et al. 2006). We applied the default algorithm using a Kolmogorov–Smirnov test to detect highly differentiated biological processes and obtain their P-values. This approach integrates information about relationships between the GO terms and the different scores of the genes (here, the P-values) into the calculation of the statistical significance. We kept only GO terms that included >10 genes in this analysis.
Maximum likelihood estimation of past demography and selection coefficients
To infer demography and selection coefficients, we considered only sites that are found as private singletons in the European 1000 Genomes populations and that are found to be polymorphic in Quebec. We used the current frequency of these variants in Europe as a proxy for their frequency during the foundation of Quebec. This allowed us to directly estimate front and core effective population sizes without having to estimate additional parameters for the European population.
We modeled the evolution of allele frequencies at independent sites under random genetic drift and natural selection in two panmictic populations, denoted the core and the front. Variables describing properties of the front and core are denoted with sub- or superscript and respectively. For simplicity, we only present calculations for the front; the core can be treated analogously. Then denotes the number of sites with a DAF of Let the SFS on the front be denoted by where is the effective population size at the front and denotes the time (in generations) since the founding of Quebec. Assuming a Wright–Fisher model of drift and genic selection (that is, no dominance or epistasis), the SFS then evolves according to
| (1) |
where denotes the binomial distribution and is the strength of selection against the derived allele. We calculated the current allele frequency distribution (16 generations after the onset of the settlement) with the initial condition where is the expected allele frequency distribution in the bottlenecked population and is the sample size in Europe. We then obtained the expected allele frequency distribution for a sample of individuals by
| (2) |
Let be the relative frequency of sites with a DAF of To account for the fact that we only considered sites shared between Europe and Quebec, we corrected the allele frequency distribution by multiplying the proportion of sites that are not found to be polymorphic at the front, by i.e., we count only the proportion of sites where the derived allele is lost in the front but is polymorphic in the core, and then renormalize such that Note that this renormalization accounts for the loss of variants in both the front and core. Thus, we account for the loss of variants in both the front and core in the estimation process, since the loss of variants is reflected in the shape of the renormalized SFS. We can then calculated the likelihood from our data as
where and denote the observed DAFs in front and core, respectively. The likelihood was then maximized numerically via a grid search in the parameter space.
Contribution of expansion to variance in allele frequencies
To estimate the relative contribution of the expansion to the total variation in allele frequencies, we first iterated Equation (1) with the maximum likelihood estimates (MLE) parameters to obtain expected allele frequency distributions after the bottleneck () and at the end of the expansion We then calculated the variance in allele frequencies after the bottleneck () and at the end of the expansion () from the allele frequency distributions. Using the decomposition the proportion of variance explained by the expansion process is given by
Demographic and selection inference from the SFS
We used a total of 43,407,133 neutral sites (with GERP RS cores between −2 and 2) to estimate the past demography of French Canadians from the joint (front and core) SFS using fastsimcoal2 (Excoffier et al. 2013). The demographic scenario is shown in Figure S31 in File S1. We performed 50 runs of fastsimcoal2, each with 50 cycles, and we used 200,000 simulations per likelihood estimations. The estimated parameters obtained from the run maximizing the likelihood are shown in Table S4 in File S1.
Based on the demography inferred from neutral sites, we then used Fitdadi (Kim et al. 2017) to estimate the Distribution of Fitness Effects (DFE) from the SFS computed on 40,341,124 sites assumed to be under selection (with GERP RS scores ≥ 2). This estimation was done independently in the core and the front population, assuming that the DFE followed a γ distribution and using a multinomial likelihood. In each case, we fixed the shared and population-specific demographic parameters shown in Table S4 in File S1, and searched for the γ distribution parameters maximizing the likelihood computed from the SFS inferred from the deleterious sites. The resulting DFEs are reported in Figure S32 in File S1.
Inference of DFE with DFE-α
We used DFE-α (Eyre-Walker and Keightley 2007; Schneider et al. 2011) to estimate the DFE of deleterious mutations independently for front and core populations. We first estimated the demographic and mutational parameters under all implemented models (1-, 2-, or 3-epoch demographic history with piecewise constant population sizes) using predicted neutral sites. We then used the SFS of predicted deleterious sites to infer the DFE, as modeled by a γ distribution. The resulting demographic histories are reported in Table S7 in File S1 and the inferred DFEs are shown in Figure S33 in File S1.
Individual-based simulations
We performed individual-based simulations of a range expansion in a two-dimensional habitat consisting of a lattice of 11 × 11 discrete demes (stepping-stone model). Generations are discrete and nonoverlapping, and mating within each deme is random. Migration is homogeneous and isotropic, except that the boundaries of the habitat are reflecting, i.e., individuals cannot migrate out of the habitat. Population size grows logistically within demes. Our simulations start from a single panmictic ancestral population, representing France. After a burn-in phase that ensures that the ancestral population are at mutation–selection–drift balance, a propagule of founders is placed on the deme with coordinates (3,6) on the 11 × 11 grid representing French Canada (see Figure S42 in File S1). During the next six generations, the population expands along a one deme-wide corridor in the middle of the habitat (representing the Saint Laurence River corridor). During these six generations, all colonized demes in French Canada receive migrants from the ancestral populations in equal proportions. The number of migrants was chosen to roughly match historical records (Charbonneau et al. 2000). In particular, we chose 1000, 2000, 1000, 1000, 1000, and 2000 pioneer immigrants from the ancestral population for the first six generations, respectively. After that, the expansion continues into the remaining habitat for 11 generations. See Figure S42 in File S1 for a sketch of the model.
We chose a carrying capacity of K = 1000 diploid individuals and the size of the ancestral population was 10,000. Migration rate was set to m = 0.2 and the within-deme growth rate was R = 2 [that is, at low densities the population doubles within one generation, reflecting the average absolute fitness of around four to five surviving children getting married per woman (Moreau et al. 2011)]. We simulated a set of 10,000 independent diallelic loci per individual. The genome-wide mutation rate was set to U = 0.1. Mutations occur only in one direction and back mutations were ignored. We performed two types of simulation: (i) evolution of neutral mutations and (ii) evolution of sites under purifying selection. In the latter case, we assumed that all sites had the same selection coefficient s. Mutations were considered to be either codominant or completely recessive and interact multiplicatively across loci, that is, there was no epistasis. We also simulated and recorded the cWFI of each individual. The simulation code can be downloaded from: https://github.com/CMPG/ADMRE.
Data availability
The data will be submitted to the European Genome-phenome Archive repository. The accession number is EGAS00001001957.
Results
French Canadians vs. Europeans
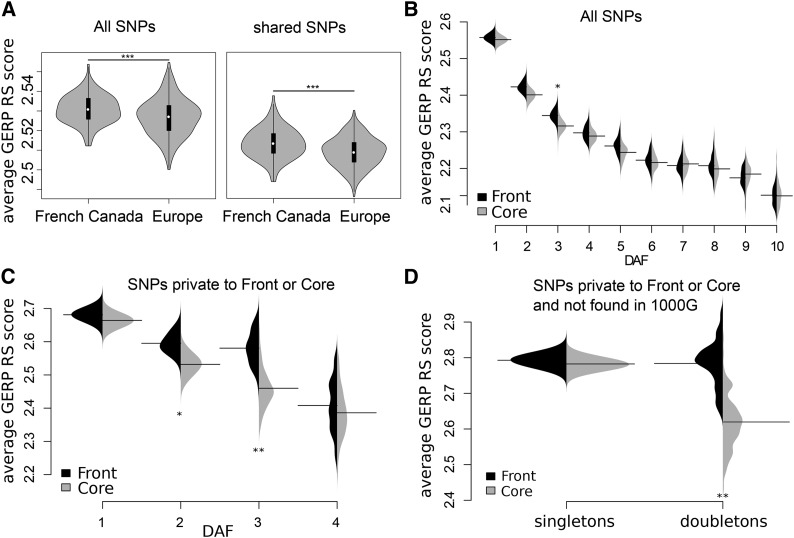
French Canadians are genetically very divergent from three European populations of the 1000 Genome phase 3 panel (1000 Genomes Project Consortium et al. 2015) (Great Britain, Spain, and Italy, Figure S1 in File S1), as expected after a strong bottleneck. When focusing on SNPs shared between French Canadians and Europeans, and thus on relatively high-frequency variants, core individuals are found genetically closer to European samples than front individuals in the first two principal component analysis (PCA) axes (Figure S1B in File S1), in keeping with stronger drift having occurred on the wave front. The qualitative outcome of the PCA does not change if we remove inbred individuals (Figures S2 and S3 in File S1) or if we account for linkage among SNPs (Figure S3 in File S1). Furthermore, we checked that these differences between Europeans and French Canadians were not due to the lower coverage of the 1000 Genomes exomes (65× on average) by downsampling our data from 89.5 to 65× and repeating the PCA (Figure S1, C and D in File S1). If we assess the functional impact of point mutations with GERP RS scores (Davydov et al. 2010), sites polymorphic in French Canadians are on average more conserved than sites polymorphic in Europeans (Figure 2A). Again, downsampling to account for differences in coverage does not affect this result (Figure S4 in File S1). Thus, even though French Canadians have fewer polymorphic sites than 1000 Genomes populations from Europe, their variants are on average potentially more deleterious than those found in European samples (Figure 2A), in line with previous results (Casals et al. 2013). We find the same pattern if we calculate the average GERP RS score per derived allele rather than per polymorphic site (Figure S5 in File S1). Note that these results still hold if we focus only on SNPs that are shared between 1000 Genomes and Quebec samples, even though the distributions are slightly more overlapping (Figure 2A and Figure S5B in File S1).
Figure 2.
(A) Distributions of average GERP RS scores per site per individual in three European 1000 Genomes populations, as well as in core and front individuals. Left: all sites. Right: sites shared between 1000 Genomes samples and Quebec (Student’s t-test P-values = 10−7 and 10−5, respectively). (B) Average GERP RS score per site having different DAFs. The solid horizontal lines show the average GERP RS score per site. The violin plots show the average GERP RS score distribution obtained by bootstrap (1000 replicates). (C) Like (B), but for mutations private to the front or to the core. (D) Like (B), but for singletons and doubletons that are private to front or core and not found in the 1000 Genomes phase 3 panel. For the sake of clarity, higher DAF classes are not shown in (B–D). Only SNPs with GERP RS scores > 0 were used for the calculations of GERP RS scores in all panels. Asterisks indicate significance levels obtained by permutation tests: * P < 0.05, ** P < 0.01, and *** P < 0.001. 1000G, 1000 Genomes; DAF, derived allele frequency; GERP RS, GERP Rejected Substitution.
Genomic diversity of front and core individuals
In French Canadians, front individuals have a significantly smaller number of variants than core individuals (Table 1), consistent with higher rates of drift. The allele frequencies in front and core individuals are overall very similar (Figure S6 in File S1), but there is a significant deficit of singletons on the front as compared to the core (Pperm < 0.001, Figure S7 in File S1 and Table S1 in File S1), which is balanced by an excess of doubletons on the front (Pperm < 0.001). Note that this pattern is consistently found for all GERP RS score categories (Figures S7 and S8 in File S1) and is consistent with simulations of range expansions (Figure S9 in File S1). We then looked at whether genes containing SNPs with large frequency differences between front and core (i.e., those with FST P-value < 0.01) were overly represented in some GO categories. Among these genes, allele frequency differences between the front and the core ranged from 0.069 to 0.294 (see Table S2 in File S1 for a list of strongly differentiated genes). The top 25 significantly enriched GO categories (Table S3 in File S1) are generally involved in very conserved processes like gene expression, development, and cell growth (see supplemental notes and Figure S10 in File S1), suggestive of a relaxation of selection rather than specific adaptations to the front environment.
Table 1. Summary of genetic diversity in front and core samples.
| Type and no. of polymorphism | Core (n = 51) | Front (n = 51) | Total (n = 102) | |
|---|---|---|---|---|
| Total no. of SNPs | 426,301 | |||
| No. of SNPs with inferred ancestral/derived state | 314,483 | > | 308,396 | 396,424 |
| No. of SNPs without missing data | 266,547 | > | 261,355 | 328,372 |
| No. of exonic SNPs | 83,653 | > | 81,763 | 107,525 |
| No. of nonsynonymous SNPs | 40,750 | > | 39,595 | 55,133 |
| No. of predicted deleterious SNPs | 66,300 | > | 64,763 | 85,748 |
| No. of SNPs private to one of the two groups of individuals | 78,310 | > | 72,353 | 150,663 |
| No. of SNPs without missing data and not seen in 1000 Genomes phase 3 panel | 31,608 | > | 29,811 | 56,669 |
| No. of SNPs without missing data, private to one of the two groups, and not seen in 1000 Genomes phase 3 panel | 26,858 | > | 25,061 | 51,919 |
| Average number of derived predicted neutral alleles per individual (−2 GERP RS ) | 50,983.94 | ≈ | 50,996.51 | |
| Average number of derived predicted deleterious alleles per individual (GERP RS ) | 22,017.57 | ≈ | 22,034.16 | |
| Average number of derived homozygous predicted deleterious sites per individual (GERP RS ) | 6,037.53 | ≈ | 6,057.75 | |
| No. of indels | 33,789 | > | 33,297 | 43,081 |
| Heterozygosity | ||||
| All sites | 0.0588 | ≈ | 0.0586 | |
| Exons | 0.0548 | ≈ | 0.0547 | |
| Introns | 0.0632 | ≈ | 0.0630 | |
| 5′ UTR | 0.0489 | ≈ | 0.0487 | |
| 3′ UTR | 0.0623 | ≈ | 0.0623 |
Significant differences between front and core are indicated by “>’’ (permutation test, Pperm < 0.001), and nonsignificant differences are indicated by “≈’’ (P > 0.05). No., number; GERP RS, GERP Rejected Substitution.
Similar number of derived alleles in front and core individuals
We first computed the total number of predicted neutral alleles per individual (−2 GERP RS score < 2) and, as expected, we did not find any differences between front and core (Figure S11 in File S1 and Table 1). We then computed the total number of predicted deleterious alleles (GERP RS score 2) in front and core individuals. This measure has previously been used as proxy for mutation load (Simons et al. 2014; Do et al. 2015), implicitly assuming that mutations interact additively and that all mutations have equal effects. In line with theoretical predictions (Kirkpatrick and Jarne 2000; Peischl and Excoffier 2015), we found virtually no difference between front and core individuals (Figures S12A and S13 in File S1 and Table 1), with a slight but nonsignificant excess of derived alleles in front individuals as compared to individuals from the core (). When we consider the cumulative GERP RS score per individual, considering all variants irrespective of their frequencies, we find a similarly slight and nonsignificant excess in the front as compared to the core (Figure S12B in File S1). However, we note that the relationship between GERP RS scores and selection coefficients is highly nonlinear (Racimo and Schraiber 2014), which means that both the total number of derived alleles and the cumulative GERP RS scores do not necessarily reflect the mutation load, even if we assume additive effects of mutations (see also ref Henn et al. 2015). Furthermore, if (some) mutations are (partially) recessive, these two measures are insensitive to a change in mutation load that would be caused by increased genetic drift (Kirkpatrick and Jarne 2000; Peischl and Excoffier 2015). If we only count derived homozygous sites, the difference in the cumulative GERP RS score between front and core is more pronounced as compared to the total additive measures (Figure S12C in File S1 and see Table 1 for total number of derived homozygous sites), but it remains statistically insignificant (). Note that the lack of significant differences between front and core is not surprising, given their short divergence time (Figure 1B). However, we find a significantly higher ( < 0.001) cumulative GERP RS score in the core for singletons, which is compensated by a significantly lower statistic for doubletons, tripletons, and quadrupletons ( < 0.05, Figure S14 in File S1). We confirmed these patterns using alternative deleteriousness scoring systems (Figures S15–S18 in File S1). Since it appears difficult to assess mutation load from sequence data in human populations (see e.g., Lohmueller 2014), in the following we shall focus on statistics other than genetic load that can measure a relaxation of selection via changes in the allele frequencies of deleterious mutations, and compare them to analytical results and individual-based simulations.
Low frequency variants in front individuals are more conserved
In a population that experiences high rates of drift, and hence less-efficient selection (Gravel 2016), we expect that the average (negative) selection coefficient associated to segregating variants increases as compared to a population that experiences less drift (Peischl and Excoffier 2015). Therefore, we considered the average GERP RS score per site as a proxy for the average deleteriousness of a segregating site, since strongly conserved sites are expected to be under strong purifying selection. The examination of low-frequency variants that are enriched for deleterious mutations (Boyko et al. 2008; Nelson et al. 2012; Kiezun et al. 2013) should allow us to better evidence the presence of differential selection between front and core, and we therefore stratified the average GERP RS score according to its DAF. We actually found a negative relationship between the frequency of mutations and their average GERP RS scores (Figure 2B), and low-frequency variants (DAF < 5%) had significantly larger GERP RS scores (and are thus potentially more deleterious) on the front than in the core (Pperm = 0.038). Since new variants should also be enriched for deleterious mutations (Boyko et al. 2008; Keinan and Clark 2012), we then focused on mutations private to front or to core individuals. With this additional filtering, the differences in GERP RS scores between front and core for low-frequency mutations were much more pronounced (Figure 2C), with significant differences for both doubletons and tripletons (Pperm = 0.03 and Pperm = 0.0025, respectively). We checked that these results were not due to our use of GERP RS scores by repeating our analyses using alternative proxies for the damaging effects of mutations. Overall, we found very similar evidence of reduced selection in front populations (Figures S17–S27 in File S1) for point mutations (and for indels) identified as under selection by PolyPhen-2 (Adzhubei et al. 2010), PhyloPNH (Fu et al. 2013), and CADD (Kircher et al. 2014), suggesting that our results are robust to alternative scoring systems for deleteriousness.
New deleterious mutations are at higher frequencies on the front
We further enriched our data for new mutations that occurred during the colonization of Quebec by focusing only on French Canadian mutations that are not observed in the entire 1000 Genomes phase 3 panel, and which are private either to the core or to the front samples. In this filtered data set, we found a significant excess of predicted deleterious (GERP RS score ≤ 2) singletons in the core (Pperm < 0.001), and an excess of doubletons in the front (Pperm < 0.001, Table S1 in File S1). Interestingly, the doubletons on the front were as conserved as singletons in both core and front samples, suggesting that doubletons on the front are variants that would be singletons in the core (Figure 2D). We can test the claim that differential selection has allowed mutations at more-conserved sites to reach or be maintained at higher frequencies on the front by comparing the cumulative GERP RS score of neutral and predicted deleterious doubletons. If the increase in the average degree of conservation of doubletons at the front is due to a purely neutral process, such as inbreeding or demography, we should see similar differences in the cumulative GERP RS score of doubletons between front and core for neutral and deleterious variants. However, we find that the cumulative GERP RS scores for these doubletons are similar in front and core individuals for neutral sites (−2 ≤ GERP RS < 2), but significantly larger in front individuals for nonneutral GERP RS score categories (GERP RS ≥ 2) (Figure 3). To see if inbreeding could explain the observed excess of deleterious doubletons in the front, we compared samples from the region of Saguenay, where remote inbreeding is higher than in the rest of Quebec (Figure S28 in File S1), with front samples coming from other regions of Quebec. We find that doubletons in less-inbred non-Saguenay individuals are at loci that are on average more conserved than those of Saguenay individuals (Figure S29 in File S1), showing that inbreeding cannot explain the increase in frequency of rare deleterious variants.
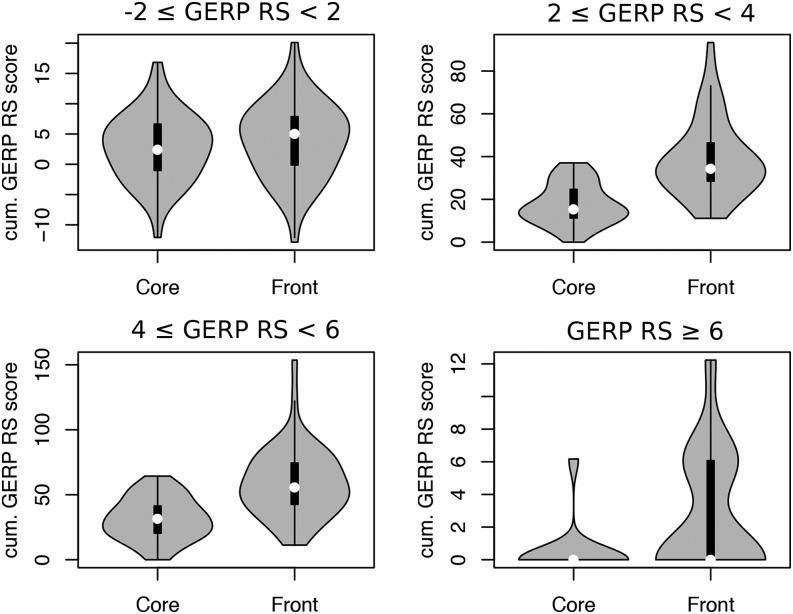
Figure 3.
Distribution of the cumulative additive GERP RS scores of doubletons in front and core individuals for different GERP RS categories. Sites were considered if they were not seen in derived states in 1000 Genomes samples and if they were private to the core or to the front. Differences between front and core are significant for the three categories of sites potentially under selection (P = 10−11, 10−9, and 10−4 for mildly, strongly, and extremely deleterious sites, respectively), but not for the neutral sites (−2 GERP RS score < 2, P = 0.34). GERP RS, GERP Rejected Substitution.
Likelihood-based demographic and selection coefficient inference
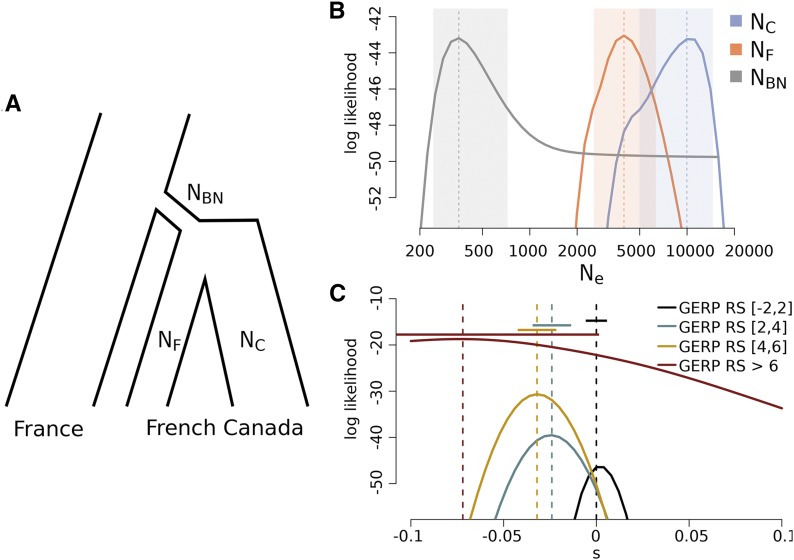
We used the allele frequency distributions of mutations that are singletons in European 1000 Genomes populations and that are still seen in Quebec to estimate the parameters of a simple demographic model for the settlement of French Canada, as well as selection coefficients of sites belonging to different GERP RS categories. In this model, a small founding population splits off from the ancestral population, and then further splits into two subpopulations: the front and the core (Figure 4A). We estimate the effective population size of the founding population (NBN), the front (NF), and the core (NC) under a maximum-likelihood framework based on intergenerational allele frequency transition matrices (see Materials and Methods for details). We report here results for a model in which we fix the duration of the initial bottleneck to one generation, but the analysis of a model with a seven-generation bottleneck yields qualitatively similar results, which can be found in the Supplemental Material (Figure S30 in File S1). We infer that French Canadians passed through a bottleneck equivalent to = 354 effective diploid individuals, and that the front population was ∼2.5 smaller ( = 3972) than the core population ( = 9977) (Figure 4B). We then used these MLEs to estimate the contribution of the range expansion to the total variance in allele frequencies on the front as where is the variance in allele frequencies after the bottleneck and is the remaining variance due to the expansion process. We found that explains ∼20% of the total variance in allele frequencies that occurred since the initial settlement at the expansion front. Therefore, we estimate that under our simple model, 20% of the genetic divergence between Europe and the front has been generated by the expansion process, whereas the remaining 80% is due to the initial bottleneck shared by the core. We also estimated the strength of selection associated with rare variants under our estimated demographic model. In agreement with predictions, the MLE for the selection coefficient associated with predicted neutral variants is centered around zero, whereas the selection coefficients associated with predicted deleterious sites become more negative with increasing GERP RS score (Figure 4C, maximum likelihood estimates and 95% C.I.s: ). Note that the most negative selection coefficient for GERP RS scores 6 is not significantly different from zero due to the small number of sites belonging to this category. We note that these estimates are based on the evolution of rare (0.1%), segregating variants and that these alleles are significantly enriched for deleterious alleles (Boyko et al. 2008) (irrespective of their associated GERP RS scores estimated in mammals). Therefore, the inferred selection coefficients do not correspond to the DFE of all variants, which should have considerably lower selection coefficients. Nevertheless, our results suggest a clear monotonic relationship between GERP RS scores and deleteriousness.
Figure 4.
MLE of effective population sizes and selection coefficients of rare variants. (A) Sketch of the model for the demographic history of core and front populations. Likelihoods were calculated based on the expectation of the change in allele frequency distribution of rare variants (that is, singletons in the European sample). Marginal likelihoods and MLE for effective population sizes of bottleneck, and in front and core (B), and selection coefficients for different GERP RS categories (C). Shaded areas indicate 95% C.I.s in (B), and horizonal bars indicate 95% C.I.s in (C). GERP RS, GERP Rejected Substitution; MLE, maximum likelihood estimation; NBN, effective population size of the founding population; NC, effective population size of the core; NF, effective population size of the front.
Whereas the previous approach concentrates on the fate of sites that are singletons in Europe, we used an alternative approach to estimate demographic parameters and the DFE effects based on the joint SFS computed between core and front samples. Using only assumed neutral sites (with GERP RS between −2 and +2), we first estimated the demographic parameters of a more realistic demographic model (see Figure S31 in File S1), including recent exponential growth in both core and front populations using the program fastsimcoal2 (Excoffier et al. 2013). In this approach, we obtain results (reported in Table S4 in File S1) congruent with the previous (and analytically more tractable) model, in the sense that just after divergence, the initial core population was ∼1.3 times larger than the initial front population, supporting the view of a larger extent of drift in the front after divergence. This demographic scenario was then used to estimate the parameters of a γ-distributed DFE from the SFS of sites assumed to be under selection (GERP RS 2) with Fitdadi (Kim et al. 2017), in the front and in the core populations separately. We found that the average selection coefficient for these potentially deleterious sites is smaller in the core than in the front thus suggesting that mutations tend to have stronger deleterious effects in front populations as compared to the core. If we assume a monotonic relationship between GERP RS scores and selection coefficients, this difference seems especially due to the most deleterious sites, since the average selection coefficient for GERP RS [2,4[ is almost identical at the front ( and in the core ( whereas selection coefficients are larger in front than in core for GERP RS [4,6[ () and GERP RS 6 () (Figure S32 in File S1). Likelihood comparisons suggest that the DFEs estimated in the two population are actually different (see legend of Figure S32 in File S1), which could imply that both differential demography and differential selection have shaped the SFS of selected sites. This interpretation is supported by inference of the DFE under simpler demographic models using the software DFE-α (Eyre-Walker and Keightley 2007; Schneider et al. 2011, Figure S33 in File S1). When comparing the inferred DFEs to previous studies, we find that the average strength of selection that we infer is lower than what has been reported previously (Boyko et al. 2008; Kim et al. 2017). However, it should be noted that previous studies were done on nonsynonymous sites, whereas we have included synonymous mutations in our approach. The fact that synonymous sites tend to have lower GERP RS scores could therefore explain why our approach yields an overall more-neutral DFE compared to previous studies. An alternative reason why our estimates of the DFE differ from previous ones could be the use of the multinomial likelihood function in Fitdadi (Kim et al. 2017), which only uses the proportional SFS, rather than absolute counts and the number of monomorphic positions. Indeed, Kim et al. (2017) have shown that estimates for the scale parameter can be less exact when using the multinomial likelihood, which could explain the observed discrepancy of the estimated DFEs.
Variants with low frequency in Europe have been more impacted by selection in the core
Because neutral sites should only be affected by drift and not by selection, stronger drift at the front should increase the variance of neutral allele frequencies (Gravel 2016), but should not affect their average frequency. In contrast, if we follow a set of deleterious variants that were present at a given frequency in the ancestral population, the frequency of these variants should be smaller in the core as compared to the front if the purging of deleterious variants was more efficient. To test these predictions, we again used mutations that are singletons in European 1000 Genomes populations and that are still seen in Quebec, and contrasted empirical patterns of allele frequencies with predictions from the maximum likelihood model described above (Figure 4A). In agreement with theoretical predictions (Figure S34 in File S1), we found no significant difference in the average DAFs () of European singletons predicted to be neutral (GERP RS score between −2 and 2) (= 0.00720 vs. 0.00717 in front and core, respectively, Pperm = 0.34, Figure S35 in File S1), and a slightly larger variance of DAFs on the front (SD (): 0.0163 vs. 0.0159, Pperm = 0.072, Figure S36 in File S1). Contrastingly, predicted deleterious sites have significantly higher DAFs on the front than in the core (Pperm = 0.0146 for sites with GERP RS score > 4), and the difference between front and core is increasing with increasing GERP RS score (Figure S35 in File S1), in keeping with higher rates of purging in the ancestry of core individuals. In line with neutral evolution of predicted deleterious alleles, the average DAF of European singletons seems to be independent of the GERP RS score at the front, whereas DAF clearly decreases with increasing GERP RS score in the core, indicative of faster purging of more-deleterious mutations (Figure S35 in File S1). We also checked the distribution of GERP RS scores among those sites and found no notable differences between front and core (Figure S37 in File S1), showing that the larger frequency of these deleterious variants at the front is not compensated by overall lower GERP RS scores of these variants as compared to the core.
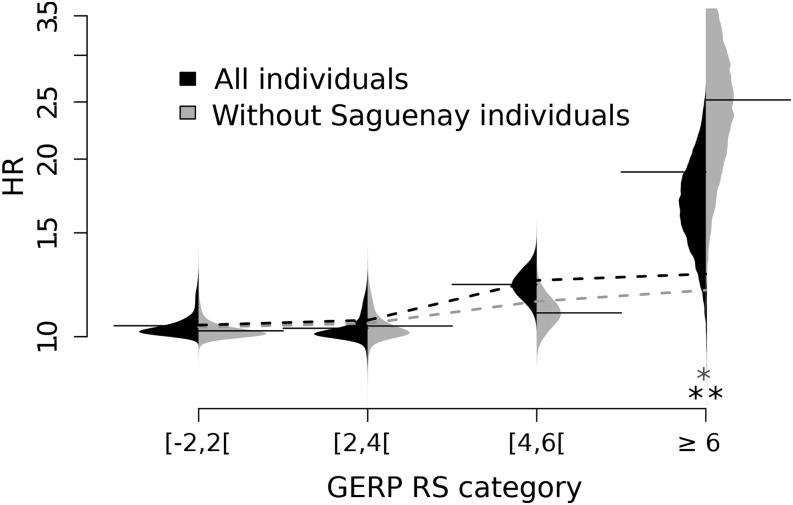
Since differences between core and front individuals are strongest for rare alleles, these differences may have an impact on the homozygosity of recessive deleterious alleles and thus influence disease incidence. We used the ClinVar database (Landrum et al. 2014) to identify pathogenic variants [causing Mendelian disorders, see Richards et al. (2015)] in the set of SNPs segregating in French Canadians. The distribution of GERP RS scores for pathogenic variants is clearly shifted toward higher GERP RS scores as compared to the distribution for all SNP loci (Figure S38 in File S1), confirming that GERP RS is a valid deleteriousness scoring system. We found that front individuals carry more known pathogenic variants than core individuals (8.96 vs. 7.82, respectively, Figure S40 in File S1, ) and have an 11.8% higher probability of being homozygotes for these pathogenic variants than core individuals (), suggesting that the expansion process has also affected disease-causing mutations. This is medically relevant, since among the 92 pathogenic variants for which we could find information on their dominance/recessivity status (out of 116), a large majority (75, or 81.5%) were considered as fully recessive. For rare deleterious variants (i.e., derived singletons in Europe with GERP RS score 2), this excess in homozygosity is 9.5%. Of importance, this excess increases with GERP RS scores and reaches ∼90% (Pperm = 0.021) for sites with a GERP RS score > 6 (Figure 5). The fact that the variance in allele frequencies becomes more similar between front and core with increasing GERP RS score (Figure S36A in File S1) shows that the excess in homozygosity on the front is not due to an increase in the variance of allele frequencies in the front alone. Note also that the increase of homozygosity cannot be explained by the higher inbreeding level prevailing on the front, and that the differences in homozygosity between front and core become even more pronounced if one removes more-inbred Saguenay individuals (Pperm = 0.008, Figure 5). This last result shows that stronger purifying selection in the core, rather than higher inbreeding on the front, is directly responsible for the lower frequencies of deleterious mutations in the core.
Figure 5.
Ratio of expected homozygosity (HR) for variants that are singletons in European 1000 Genomes populations. , where and are derived allele frequencies in front and core individuals, respectively. The horizontal solid lines indicate HR for different GERP Rejected Substitution (GERP RS) score categories. The dashed lines indicate the expected HR values that would be due to differences in estimated inbreeding levels between front and core, calculated as where Violin plots show the distribution of 5000 bootstrap replicates. We find significant differences between the expected values for GERP RS scores > 6 (all individuals: P = 0.021 and without Saguenay individuals: P = 0.008, obtained by bootstrap).
Simulations can reproduce observed differences between front and core
Whereas it seems difficult to perform demographic inferences under a complex spatially explicit model, we have used forward simulations to see how well a model of range expansion can explain our observations (see Materials and Methods for details on the simulations). Our simulations reveal that the observed excess of singletons in core populations, as well as the excess of doubletons in front populations, is consistent with a model of range expansion (Figure S9 in File S1). Even though previous theoretical work showed that range expansions lead to a flattening of the whole SFS (Sousa et al. 2014), our current results are in line with theory since we are considering a much shorter evolutionary period here [16 generations instead of, e.g., 150 in Peischl and Excoffier (2015)]. Indeed, during the onset of the expansion, drift should have more impact on the low-frequency entries of the SFS (e.g., singletons and doubletons), because these are the entries for which the number of sites is largest and because they are close to an absorbing state (loss). Importantly, simulations also confirm these features (i.e., a deficiency of singletons and excess of doubletons on the front as compared to the core) of the SFS for negatively selected (recessive or codominant) mutations (Figure S9 in File S1). Finally, our simulations confirm that an excess of homozygosity should rapidly develop on the front and that it should increase with the deleteriousness of mutations (Figure S39 in File S1), in keeping with the observed patterns in Quebec (Figure 5). Altogether, our simulation results show that a model of range expansion can explain most of the observed differences between front and core individuals in Quebec.
Discussion
The interaction between demography and selection has been a central theme in population genetics. A particularly hotly debated topic is whether and to what extent recent demography has affected the efficacy of selection in modern humans (Lohmueller et al. 2008; Gazave et al. 2013; Fu et al. 2014; Lohmueller 2014; Simons et al. 2014; Do et al. 2015; Henn et al. 2015, 2016; Gravel 2016). The original conclusion that European populations show a larger proportion of predicted deleterious variants when compared to African populations (Lohmueller et al. 2008) has been recently revisited in a series of studies that reached different and apparently opposite conclusions [reviewed in Lohmueller (2014)]. However, this controversy might have arisen because different studies focused on different patterns or processes. First, people focused either on measures of the efficacy of selection (defined as the amount of change in load per generation) or on measures of the mutation load [e.g., see Gravel (2016), for a detailed study of this distinction). Second, people either measured the load as being due to codominant (Simons et al. 2014; Do et al. 2015) or partially recessive (Henn et al. 2016) mutations, which can lead to drastically different conclusions about the consequences of demographic change on mutation load (Henn et al. 2015, 2016). Finally, most theoretical and empirical work has focused on the effects of bottlenecks and recent population growth, but ignored the out-of-Africa expansion process and the spatial structure of human populations (Sousa et al. 2014). While it has now been shown that old human expansions could lead to the buildup of a recessive mutation load (Henn et al. 2016), it is still unclear whether very recent or ongoing expansions could also affect patterns of diversity in genomic regions under selection, and what the exact genomic signatures of these recent expansions are.
Our current study uses a unique combination of historical records, detailed genealogical information, and genomic data to assess the impact of such a recent range expansion on functional genetic diversity, and to disentangle the effects of genetic drift, purifying selection, and inbreeding during an expansion. As expected, given the short divergence time of front and core, we find that the allele frequency distributions in front and core populations are very similar across all GERP RS categories (Figures S6 and S7 in File S1), resulting in overall balanced numbers of predicted deleterious alleles or cumulative GERP RS scores per individual (Figures S12 and S13 in File S1 and Table 1, see also Figures S15 and S16 in File S1). However, a closer look reveals significant differences between front and core, particularly for low-frequency variants (Figure 2 and Figures S14 and S22–S24 in File S1), which should be enriched for deleterious mutations and thus be more sensitive to differential selection. The significant differences that we have detected between front and core individuals all suggest that larger frequencies of deleterious mutations on the front are due to relaxed purifying selection on the front. The fact that front and core individuals mainly diverged six generations ago with respect to the position of their ancestors to the colonization front (Figure 1B) suggests that the relaxation of natural selection can affect modern populations remarkably quickly. The recent divergence between front and core populations (∼1780, Figure S28 in File S1) has left traces in the genomic diversity of French Canadians that are of two kinds. First, front individuals show increased genetic drift relative to core individuals, as attested by their overall lower levels of diversity (Table 1), their larger genetic divergence from Europeans (Figure S1 in File S1), and their lower estimated effective size (Figure 4B and Figure S31, Table S4, and Table S7 in File S1). This result confirms the genetic surfing effect previously identified in the Saguenay Lac St-Jean region (Moreau et al. 2011), but it is not driven by samples from the Saguenay area (e.g., Figure S29 in File S1). Rather, it is a property shared by all individuals with ancestors having lived on the front, and presently found in the most peripheral regions of Quebec (Figure 1). Second, potentially relaxed selection on the front as compared to the core is supported by several lines of evidence. The main evidence comes from the fact that sites targeted by mutations tend to be more conserved in front than in core individuals (Figure 2, B–D), and that rare, putatively deleterious derived alleles, have a higher probability of being homozygous at the front (Figure 5). The relaxed selection hypothesis is especially obvious when one considers deleterious mutations that are at low frequencies (singletons) in Europe and that are present at lower frequencies in core than in front individuals, or mutations that are now at low frequencies in Quebec and that are occurring at more conserved (and thus potentially more deleterious) sites in front than in core individuals (e.g., private doubletons and tripletons in Figure 2, C and D).
At first sight, the increased frequency of rare and potentially deleterious alleles (i.e., doubletons) in front individuals could be attributed to their higher inbreeding levels. However, there are several lines of argument against this interpretation. First, we note that there are ∼5% more doubletons on the front than in the core (21,332 vs. 20,284, Figure S7 and Table S1 in File S1), which cannot be explained by a difference in inbreeding level of only 0.3% (Figure S28 in File S1). Instead, individual-based simulations show that the excess of doubletons at the front is consistent with a model of range expansion (Figure S7 and S9 in File S1). Second, the proportion of doubleton sites where both derived alleles are in the same individual is smaller than expected (1/101 = 0.99%) in both front (0.651%) and core (0.646%) individuals, which is indicative of similar (Pperm = 0.898) levels of selection against derived homozygotes in both samples. Third, if higher inbreeding (and not relaxed selection) on the front had increased the frequency of all rare mutations irrespective of their deleterious effect, more-deleterious mutations should have been better purged by selection than less-deleterious mutations, provided that a part of the mutations is (partially) recessive. The observed doubletons on the front should then be on average less conserved due to the more-efficient purging in the core. However, we find the opposite, with doubletons at the front being more conserved than in the core (Figure 2), which means that the number of doubletons at highly conserved sites has increased proportionally more than at neutral sites. Fourth, we find that less-inbred individuals from the front tend to have rare variants that are more deleterious than more-inbred individuals from the Saguenay area (Figure S28 and S29 in File S1). Finally, the difference in inbreeding level between front and core individuals cannot explain the twofold increased expected homozygosity for extremely deleterious variants on the front (Figure 5), and removing Saguenay individuals from the analysis amplifies the excess of derived homozygotes on the front (Figure 5). Contrastingly, a model of range expansion can explain the increase in derived homozygosity at the expansion front (Figure S39 in File S1). Taken together, these results suggest that differences between front and core individuals are mainly driven by increased drift at the expansion front and more-efficient selection against deleterious mutations in the core.
In line with previous results (Casals et al. 2013), we find that all French Canadians carry, on average, more-strongly conserved mutations than Europeans (Figure 2A). Even though it has been proposed that this is the result of a mere founder effect (Casals et al. 2013), current French Canadians descend from ∼8500 French founders (Laberge et al. 2005), which implies a relatively mild founder effect that would take hundreds to thousands of generations to increase load to such an extent (Lohmueller et al. 2008; Peischl et al. 2013). More likely, this load could have been created during the initial settlement and range expansion that occurred in Quebec along the Saint-Laurence valley. A major loss of diversity and an increase in the frequency of rare deleterious variants might indeed have occurred during the first nine generations of the settlement of Quebec, until the middle of the 18th century, before current front and core individuals actually diverged (Figure 1B). The importance of these early generations is supported by genealogical analyses of the genetic contributions of the founders having lived at different periods. Early settlers have indeed contributed 45–90% to the current French Canadian gene pool (Heyer 1995; Bherer et al. 2011), depending on the regions of Quebec, and early founders contributed proportionally more than later individuals to the current French Canadian gene pool (Heyer 1995; Bherer et al. 2011; Moreau et al. 2011). Overall, our modeling of the evolution of European singletons suggests that the bottleneck shared between core and front populations explains ∼80% of the variance in allele frequencies at the expansion front, whereas only 20% of this variance can be attributed to the separate expansion of the ancestors of front individuals (Figure 4). Note that this latter value should be considered as a lower bound for the total contribution of the expansion, because front and core samples have a shared history of being on the expansion front in the first few generations in Quebec, and this shared expansion is absorbed into the estimate of the bottleneck population size in our estimation procedure.
Overall, our results clearly suggest that the low effective size prevailing on the wave front of the colonization has made selection overall less efficient at the population level than in the core. Consequently, over a very short time (nine generations or less, see Figure 1 and Figure S28 in File S1), deleterious variants have been more-efficiently purged in the core and maintained at lower frequencies than in front individuals. This excess of deleterious mutations in front individuals has probably had only a minor effect on the total mutation load and on the fitness of most individuals, because these mutations are still at very low frequencies. Nevertheless, this wave front effect might be medically relevant as rare deleterious variants have a higher probability of being homozygous on the front than in the core, suggesting that rare recessive diseases should be more common in individuals whose ancestors lived on the front. In agreement with this prediction, we find that front individuals have, on average, one more known pathogenic variant than core individuals (8.96 vs. 7.82, respectively, Figure S40 in File S1, ). Since most (85%) of these pathogenic variants are recessive, front individuals are more likely to be derived homozygous and thus to develop some disease. Importantly, this effect is noticeably stronger than the relative risk of developing a rare disease due to inbreeding. In addition, the evidence of relaxed selection on recent wave fronts suggests that prolonged periods of range expansions over hundreds of generations should have promoted the spread of deleterious mutations in newly settled territories, which could have contributed to global variation in mutation load and in a burden of genetic diseases in expanding modern human populations. This hypothesis should to be testable when ancient DNA becomes available in early pioneer populations.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300551/-/DC1.
Acknowledgments
We thank Claude Bhérer for detailed comments on the article, the CARTaGENE participants and team for data collection and assistance, Marc Tremblay for help in connecting CARTaGENE individuals to the Balsac genealogical database, Remy Brugmann for Bioinformatic analyses, Ryan Gutenkunst and Bernard Kim for help with dadi and Fitdadi computations, and the Ubelix High Performance Computing cluster of the University of Bern. We confirm that informed consent was obtained from all subjects. This work has been made possible by Swiss National Science Foundation grants 31003A-143393 and 310030B-166605 to L.E. A.H. is a Fonds de la Recherche en Santé du Québec Research Fellow and holds a Medical Research Council eMedLab Medical Bioinformatics Career Development Fellowship, funded from award MR/L016311/1. P.A. is supported by the Ministry of Research of Ontario. K.J.G. was funded by a European Molecular Biology Organization long-term fellowship (ALTF 2-2016). The authors declare that they have no conflicts of interest.
Footnotes
Communicating editor: J. Akey
Literature Cited
- 1000 Genomes Project Consortium. Auton A., Brooks L. D., Durbin R. M., Garrison E. P., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J., Lengauer T., 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607. [DOI] [PubMed] [Google Scholar]
- Austerlitz F., Heyer E., 1998. Social transmission of reproductive behavior increases frequency of inherited disorders in a young-expanding population. Proc. Natl. Acad. Sci. USA 95: 15140–15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla P., Boileau C., Payette Y., Idaghdour Y., Goulet J. P., et al. , 2013. Cohort profile of the CARTaGENE study: Quebec’s population-based biobank for public health and personalized genomics. Int. J. Epidemiol. 42: 1285–1299. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Nichols R. A., 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. Biol. Sci. 263: 1619–1626. [Google Scholar]
- Bherer C., Labuda D., Roy-Gagnon M. H., Houde L., Tremblay M., et al. , 2011. Admixed ancestry and stratification of Quebec regional populations. Am. J. Phys. Anthropol. 144: 432–441. [DOI] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F., Hodgkinson A., Hussin J., Idaghdour Y., Bruat V., et al. , 2013. Whole-exome sequencing reveals a rapid change in the frequency of rare functional variants in a founding population of humans. PLoS Genet. 9: e1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau H., Desjardins B., Légaré J., Denis H., 2000. The population of the St. Lawrence Valley, 1608–1760, pp. 99–142 in A Population History of North America, edited by Haines M. R., Steckel R. H. Cambridge University Press, New York. [Google Scholar]
- Cooper G. M., Stone E. A., Asimenos G., Program N. C. S., Green E. D., et al. , 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15: 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov E. V., Goode D. L., Sirota M., Cooper G. M., Sidow A., et al. , 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Braekeleer M., 1991. Hereditary disorders in Saguenay-Lac-St-Jean (Quebec, Canada). Hum. Hered. 41: 141–146. [DOI] [PubMed] [Google Scholar]
- Do R., Balick D., Li H., Adzhubei I., Sunyaev S., et al. , 2015. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 47: 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., Huber W., 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4: 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Lischer H. E., 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V. C., Foll M., 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Fu W., O’Connor T. D., Jun G., Kang H. M., Abecasis G., et al. , 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Gittelman R. M., Bamshad M. J., Akey J. M., 2014. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am. J. Hum. Genet. 95: 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E., Chang D., Clark A. G., Keinan A., 2013. Population growth inflates the per-individual number of deleterious mutations and reduces their mean effect. Genetics 195: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode D. L., Cooper G. M., Schmutz J., Dickson M., Gonzales E., et al. , 2010. Evolutionary constraint facilitates interpretation of genetic variation in resequenced human genomes. Genome Res. 20: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., 2016. When is selection effective? Genetics 203: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Henn B. M., Gutenkunst R. N., Indap A. R., Marth G. T., et al. , 2011. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA 108: 11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B. M., Botigue L. R., Bustamante C. D., Clark A. G., Gravel S., 2015. Estimating the mutation load in human genomes. Nat. Rev. Genet. 16: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B. M., Botigue L. R., Peischl S., Dupanloup I., Lipatov M., et al. , 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA 113: E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E., 1995. Genetic consequences of differential demographic behavior in the Saguenay region, Quebec. Am. J. Phys. Anthropol. 98: 1–11. [DOI] [PubMed] [Google Scholar]
- Heyer E., 1999. One founder/one gene hypothesis in a new expanding population: Saguenay (Quebec, Canada). Hum. Biol. 71: 99–109. [PubMed] [Google Scholar]
- Jetté R., 1991. Traité de Généalogie. Les Presses de l’Université de Montréal, Montréal. [Google Scholar]
- Keinan A., Clark A. G., 2012. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336: 740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A., Pulit S. L., Francioli L. C., van Dijk F., Swertz M., et al. , 2013. Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 9: e1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Y., Huber C. D., Lohmueller K. E., 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics 206: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., et al. , 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Jarne P., 2000. The effects of a bottleneck on inbreeding depression and the genetic load. Am. Nat. 155: 154–167. [DOI] [PubMed] [Google Scholar]
- Klopfstein S., Currat M., Excoffier L., 2006. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23: 482–490. [DOI] [PubMed] [Google Scholar]
- Laberge A. M., Michaud J., Richter A., Lemyre E., Lambert M., et al. , 2005. Population history and its impact on medical genetics in Quebec. Clin. Genet. 68: 287–301. [DOI] [PubMed] [Google Scholar]
- Landrum M. J., Lee J. M., Riley G. R., Jang W., Rubinstein W. S., et al. , 2014. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42: D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., 2014. The distribution of deleterious genetic variation in human populations. Curr. Opin. Genet. Dev. 29: 139–146. [DOI] [PubMed] [Google Scholar]
- Lohmueller K. E., Indap A. R., Schmidt S., Boyko A. R., Hernandez R. D., et al. , 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau C., Bherer C., Vezina H., Jomphe M., Labuda D., et al. , 2011. Deep human genealogies reveal a selective advantage to be on an expanding wave front. Science 334: 1148–1150. [DOI] [PubMed] [Google Scholar]
- Nelson M. R., Wegmann D., Ehm M. G., Kessner D., St Jean P., et al. , 2012. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peischl S., Excoffier L., 2015. Expansion load: recessive mutations and the role of standing genetic variation. Mol. Ecol. 24: 2084–2094. [DOI] [PubMed] [Google Scholar]
- Peischl S., Dupanloup I., Kirkpatrick M., Excoffier L., 2013. On the accumulation of deleterious mutations during range expansions. Mol. Ecol. 22: 5972–5982. [DOI] [PubMed] [Google Scholar]
- Peischl S., Kirkpatrick M., Excoffier L., 2015. Expansion load and the evolutionary dynamics of a species range. Am. Nat. 185: E81–E93. [DOI] [PubMed] [Google Scholar]
- Peischl S., Dupanloup I., Bosshard L., Excoffier L., 2016. Genetic surfing in human populations: from genes to genomes. Curr. Opin. Genet. Dev. 41: 53–61. [DOI] [PubMed] [Google Scholar]
- Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F., Schraiber J. G., 2014. Approximation to the distribution of fitness effects across functional categories in human segregating polymorphisms. PLoS Genet. 10: e1004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., et al. , 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Charlesworth B., Eyre-Walker A., Keightley P. D., 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189: 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibert A., Austerlitz F., Heyer E., 2002. Wright-Fisher revisited: the case of fertility correlation. Theor. Popul. Biol. 62: 181–197. [DOI] [PubMed] [Google Scholar]
- Simons Y. B., Turchin M. C., Pritchard J. K., Sella G., 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa V., Peischl S., Excoffier L., 2014. Impact of range expansions on current human genomic diversity. Curr. Opin. Genet. Dev. 29: 22–30. [DOI] [PubMed] [Google Scholar]
- Wright S., 1922. Coefficients of inbreeding and relationship. Am. Nat. 56: 330–338. [Google Scholar]
- Yotova V., Labuda D., Zietkiewicz E., Gehl D., Lovell A., et al. , 2005. Anatomy of a founder effect: myotonic dystrophy in Northeastern Quebec. Hum. Genet. 117: 177–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be submitted to the European Genome-phenome Archive repository. The accession number is EGAS00001001957.