ABSTRACT
Protein misfolding is the common theme for neurodegenerative disorders including Huntington disease (HD), which is mainly caused by cytotoxicity of the mutant HTT (huntingtin) protein (mHTT). The soluble mHTT has an expanded polyglutamine (polyQ) stretch that may adopt multiple conformations, among which the one recognized by the polyQ antibody 3B5H10 is the most toxic due to unknown mechanisms. In a recent study, we showed that the 3B5H10-recognized mHTT species has a slower degradation rate due to its resistance to selective macroautophagy/autophagy. In HD mouse brain tissues as well as HD patient fibroblasts and post-mortem brain tissues, the 3B5H10-recognized mHTT species lacks Lys63-polyubiquitination and SQSTM1/p62 interaction, which are essential for cargo recognition by selective autophagy. Collectively, we discovered that the mHTT protein is subject to conformation-dependent recognition by selective autophagy, which is more selective than what we perceived: the process can be selective among different conformations of the same protein, leading to conformation-dependent differences in protein degradation and toxicity.
KEYWORDS: Huntington disease, polyQ, neurodegeneration, protein aggregates, p62
Autophagy is a key cellular process that provides important quality control for misfolded and aggregation-prone proteins, which are engulfed into phagophores, the precursors to autophagosomes, and delivered to lysosomes for protein degradation. Growing evidence has shown that autophagy-mediated degradation is not limited to the process of engulfing bulk cytosol into phagophores, and can occur relatively selectively through cargo recruitment, a process referred to as “selective autophagy.” During cargo recruitment, autophagy receptors such as SQSTM1/p62 and NBR1 function as adaptor proteins tethering cargo in phagophores for subsequent clearance. Thus, selective autophagy offers additional selectivity for degradation of the protein cargos by SQSTM1 or NBR1 recognition, which is dependent on certain protein modification signals such as Lys63-poly-ubiquitination. Meanwhile, it is still unclear how selective is “selective autophagy.” More specifically, while selective autophagy is able to distinguish misfolded aggregated proteins from normal proteins, it is unknown whether selective autophagy can selectively recognize certain misfolded soluble conformations from others of the same protein. This question is especially important for studies of neurodegenerative disorders, which are usually caused by misfolded disease-relevant proteins degraded by autophagy pathways including selective autophagy.
Our recent discovery revealed that selective autophagy is actually selective for different soluble conformations of the same protein. Protein misfolding is a common hallmark for most neurodegenerative disorders. Among them, Huntington disease is a monogenetic disorder caused by the mutant HTT gene that encodes the mutant HTT protein (mHTT) with an expanded polyglutamine (polyQ) stretch. There are currently 9 discovered neurodegenerative disorders caused by different proteins with polyQ expansion. Autophagy-mediated degradation is the major pathway that clears mHTT, and its degradation rate can be used to predict the timing of the onset of neurodegeneration. Previous studies suggest that different polyQ conformations in mHTT may lead to difference in toxicity, but there is a lack of mechanistic explanation regarding why a certain conformation is more toxic.
We hypothesized that the expanded polyQ in mHTT may have multiple conformations, which may lead to differences in mHTT degradation rates. The mHTT species with the slowest degradation rate might cause the highest toxicity. To test this hypothesis, we established a click-chemistry- and homogeneous time resolved fluorescence-based pulse-chase assay and measured the degradation rates of different mHTT species with different polyQ conformations recognized by different polyQ antibodies. We discovered that the most toxic mHTT species recognized by monoclonal polyQ antibody 3B5H10 has the slowest degradation rate. Compared to the other mHTT species recognized by another polyQ antibody, MW1, the 3B5H10-recognized mHTT species is insensitive to autophagy-mediated degradation and completely resistant to selective autophagy (Fig. 1). This result is pretty striking because the 2 species are considered to be the same except for their recognition by different polyQ antibodies. Thus, the mHTT protein with expanded polyQ exhibits a conformation-dependent recognition by selective autophagy (CORSA). We further discovered that the CORSA of mHTT is due to a lack of Lys63-polyubiquination and SQSTM1-binding of the 3B5H10-recognized mHTT species, possibly due to its distinct conformation. We confirmed this in Huntington disease patient cells, mouse brains and human patient post-mortem brain tissues, while the mechanisms remain to be further clarified.
Figure 1.
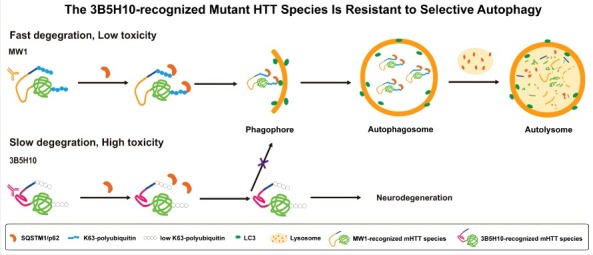
A schematic model explaining the slower degradation and higher toxicity of the 3B5H10-recognized mHTT species. The 3B5H10-recognized mHTT species lacks the K63-polyubiquitination and fails to be recognized by the selective autophagy receptor SQSTM1. As a result, this species fails to be sequestered by phagophores for degradation mediated by selective autophagy, leading to higher toxicity.
In summary, we discovered that the mHTT protein is subject to CORSA, indicating that selective autophagy is able to distinguish different soluble conformations of the same protein. It is possible that CORSA also occurs in other proteins that cause neurodegenerative disorders, especially different polyQ proteins that cause different types of spinocerebellar ataxia. Revealing the mechanistic details of CORSA may provide novel insights of disease mechanisms and therapeutic targets.
Acknowledgments
The authors are supported by the National Key Research and Development Program of China (2016YFC0905100) and National Natural Science Foundation of China (91649105) and for funding. Authors have no competing interests that might be perceived to influence the results and/or discussion reported in this paper.


