ABSTRACT
Meiosis is a specialized cellular division occurring in organisms capable of sexual reproduction that leads to the formation of gametes containing half of the original chromosome number. During the earliest stage of meiosis, prophase I, pairing of homologous chromosomes is achieved in preparation for their proper distribution in the coming divisions. An important question is how do homologous chromosomes find each other and establish pairing interactions. Early studies demonstrated that chromosomes are dynamic in nature and move during this early stage of meiosis. More recently, there have been several studies across different models showing the conserved nature and importance of this chromosome movement, as well as the key components involved in chromosome movement. This review will cover these major findings and also introduce unexamined areas of regulation in meiotic prophase I chromosome movement.
KEYWORDS: LINC, meiosis, recombination, synapsis
Chromosome movement is a key aspect of many biologic processes. It is well-established that chromosome movement plays an important role in chromosome segregation (Reviewed in1). In addition, chromosomal movement is important for basic cellular processes such as transcription and DNA damage repair (Reviewed in2,3). Chromosome movement is also a conserved feature of the meiotic program and is required for the timely and/or successful completion of all meiotic events. A common feature of this movement is linking chromosomes to the cytoskeleton via nuclear transmembrane protein complexes, where movement forces are generated in the cytoplasm. Here we will focus on this, one of the many forms of chromosome movement: we will review how chromosome movement is generated and the way it affects chromosome pairing and synapsis in meiosis.
Overview of meiosis
Meiosis involves 2 cell divisions, the first of which segregates homologous chromosomes to opposite poles, reducing ploidy by half. In most organisms, this segregation requires crossovers, the exchange of DNA sequences between homologous chromosomes, which in turn, is dependent on stable associations of homologs in meiotic prophase I (reviewed in4). In early meiotic prophase I, chromosomes form pairing interactions that bring chromosomes to close physical associations. The processes of synapsis then stabilizes these pairing interactions throughout the homolog pair, and is mediated by the synaptonemal complex (SC), a meiosis-specific protein complex. Absent or misregulated assembly of the SC prevents the stabilization of pairing interactions that are essential for meiosis, leading to chromosome missegregation.
Meiotic prophase I is divided into 5 stages that are defined by the behavior of chromosomes; early stages (leptotene to zygotene) show low levels of chromosome pairing and initiation of SC assembly, while the following stage (pachytene) is characterized by full synapsis and stable pairing (Reviewed in4). In late meiotic prophase I, the SC disassembles (diplotene) and condensed bivalents with chiasmata (pairs of homologs connected by crossover(s)) are visible in diakinesis (these stages may be of short duration in some organisms). Recombination also proceeds sequentially between these stages;4 formation of meiotically induced DNA double strand breaks (DSBs) in leptotene, their processing in zygotene, and their repair during pachytene to form crossovers. For all of these recombination events to proceed properly, pairing promoted by chromosomal movement is necessary. Although the process of meiotic recombination is conserved among species, there are variations in the recombination program (we will not discuss them here). For the rest of this review, we will be discussing what is known about meiotic prophase I chromosomal movement in a variety of organisms as well as a recent finding on how this chromosomal movement is regulated.
Chromosome movement in meiotic prophase I
Meiotic prophase I chromosomes are frequently described as having ongoing rapid prophase movement (RPM), due to their observably rigorous movement pattern. Analysis of chromosome movement across organisms showed that chromosomes can move in a coordinated manner and/or a solitary manner (Fig. 1A). In the case of coordinated movement, all chromosomes move in the same direction, which can be detected with movement of the whole nuclear envelope. Solitary movement is found when one or few chromosomes move in one direction while the others obtain a different trajectory.
Figure 1.
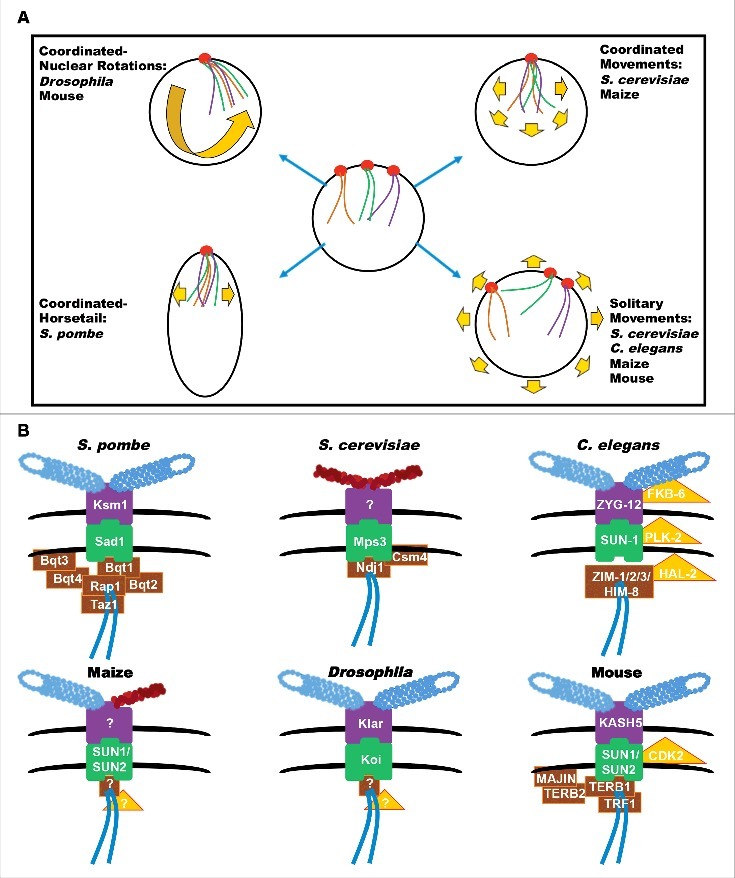
Chromosome Movement Patterns and Protein Components of Chromosome Movement. (A) Examples of the different chromosomal movement patterns and with which organism each is associated. Red circles: LINC complex; Yellow arrows: possible directions of chromosome movement. (B) Six different model organisms with their meiotic LINC complexes and associated- structural/regulatory protein components described in the text. Blue tubes: microtubules; Red tubes: actin; Black lines: nuclear envelope; Purple boxes: KASH proteins; Green boxes: SUN proteins; Brown boxes: structural-associated proteins; Yellow triangles: regulation-associated proteins.
The dynamic nature of meiotic prophase nuclei was first noted ∼50 y ago in a study performed in hamster spermatogonia.5 More recently, 2 studies in mice examined these movements in a manner that allowed for a more in depth analysis.6,7 These studies showed that mouse chromosomes are extremely dynamic and exhibit both independent and coordinated movement (the latter associated with nuclear rotation). Maize, like mice, show both coordinated and solitary movements of chromosomes,8 and this is also true of S. cerevisiae.9,10 However, not all organisms show both coordinated and solitary movements of chromosomes. In Drosophila, the entire nuclear membrane and associated chromosomes rotate as one unit;11 coordinated chromosome movement is also observed in S. pombe, where nuclei and chromosomes move synchronously, acquiring a characteristic horse-tail shape.12 In C. elegans, movement is not coordinated and each chromosome cluster (conglomerate of chromosomes) shows a distinct trajectory of movement.13,14 Chromosome movements were also detected in other non-model organisms; for example, one of the first studies to show quantitative analysis of meiotic chromosome movement was done in house crickets, in which chromosome movements near the nuclear envelope were reported.15
Chromosome movement is more common in early meiotic prophase in mice, plants, and worms, compared with yeast. Studies in rat and mouse spermatocytes, and in maize, indicate that chromosome oscillations occur from late leptotene to early pachytene with peak movement in zygotene.6,8,16 In C. elegans, similar movement is found in the leptotene/zygotene stage and is greatly reduced in pachytene.13,14 In terms of movement duration, Drosophila and yeasts represent 2 extremes. In Drosophila, movement is restricted to nuclei at the last mitotic division before meiotic entry (the 8-cell cyst).11 Whereas the most extended relative period of movement is found in S. pombe and S. cerevisiae where chromosomes move vigorously throughout all or most of meiotic prophase I, respectively.9,10,12
Chromosome movement velocity in meiotic prophase I is higher than what is found in pre-meiotic or mitotic cells. The average velocity of chromosomes varies widely between species (∼80–500 nm/sec) and occasionally different velocities have been reported in separate studies of the same organism. Moreover, some chromosomes show RPMs that double or even quadruple the average velocity (for methods of detection see Table 1). In S. pombe, overall chromosome movement is relatively slow (83nm/sec).11,12 Measurement of movement rates in mouse zygotene estimate an average of 109–120nm/sec6, which is comparable to C. elegans (average 125nm/sec).13,14 Movement velocity of S. cerevisiae meiotic chromosomes varies between studies, from lower estimates (average 208–272 nm/sec) to higher values (average 300–500nm/sec).9,10,17 Similar velocities are also reported for maize (average 400nm/sec)8 and Drosophila (300nm/sec11 and H.J.R personal communication).
Table 1.
Meiotic chromosome movement by species.
| Movement by | Type of Movement | Timing | Average Velocity | Mode of detection | |
|---|---|---|---|---|---|
| S. pombe | Microtubules45 | Coordinated (horse-tail) 12 | Throughout prophase I 9,10,12 | ∼83 nm/sec12 | Leading edge of the stained DNA by Hoechst 33342 staining12 |
| S. cerevisiae | Actin 9,10 | Solitary and coordinated 5,6,7 |
Throughout prophase I 9,10,12 | 200–500 nm/sec9,10 | Rap1-GFP9 ZIP1::GFP 10 |
| Maize | Microtubules and actin8 | Solitary and coordinated 8 | Leptotene-zygotene transition and zygotene 6,8,16 | ∼400 nm/sec8 | Heterochromatin knobs or chromosome ends by DAPI staining8 |
| C. elegans | Microtubules47 | Solitary13,14 | Leptotene-zygotene13,14 | 125–400 nm/sec13,14 | ZYG-12::GFP13 SUN-1::GFP14 |
| Drosophila | Microtubules11 | Coordinated (nuclear rotations) 11 | Pre-meiotic (8- cell cyst)11 | 300 nm/sec11 | CID-RFP11 |
| Mouse | Microtubules39,57 | Solitary and coordinated (nuclear rotations)5,6,7 | Leptotene-zygotene6,8,16 | 109–120 nm/sec6 | Pericentric heterochromatin by Hoechst 33342 staining6 and GFPTRF17 |
Overall, patterns of movements vary between organisms, some with rapid (maize) or slower (mouse and C. elegans) movement velocities for a restricted duration, while others demonstrate high (S. cerevisiae) or low (S. pombe) velocities for most of prophase I. An interesting question that remains, is why the velocities, duration, and overall movement pathways differ from organism to organism. The pattern of movement could be an outcome of the number and size of chromosomes relative to the available forces to move them (i.e., dynein molecules, see below). It could also be that a different system of chromosome movement regulation is in place in different organisms (regulation reviewed below).
Telomere clustering in meiotic prophase I
In most organisms, chromosome ends (telomeres or telomere-proximal regions) are connected to the nuclear envelope. Meiotic prophase I involves a phase in which the ends of chromosomes cluster at the nuclear envelope: chromosomes ends are more frequently found close together rather than apart. In mammals, plants, and budding yeast, this clustering is tight and pronounced. In these organisms, the clustered configuration is defined as a bouquet. In mouse and humans, bouquet formation is correlated with the initiation of synapsis (between leptotene and zygotene) and is relatively short: ∼0.1% of the time in prophase I (15 min of the 11 d in prophase I).6,18 Transient bouquet formation is also found in S. cerevisiae (∼30 sec/4 hours = ∼0.1%), in which bouquet structures form and fall apart repeatedly in early prophase I9. In maize, the bouquet is formed at the end of leptotene and persists longer, throughout zygotene.19 C. elegans chromosomes that enter meiosis cluster to one side of the nucleus, but do not form a bouquet, as only a single chromosome end is attached to the nuclear envelope.20 Moreover, although several chromosomal ends may be found in close proximity, they do not all coalesce.14 Clusters of telomeres can be formed that contain a subset of chromosomes. The clustered arrangement of chromosomes in C. elegans is maintained for a relatively long time, throughout leptotene and zygotene (7 hours/∼3 d = ∼10%). Drosophila and S. pombe show 2 extremes of clustering behaviors; in S. pombe, telomere clustering is maintained throughout prophase I12, while in Drosophila, telomeres do not attach to the nuclear envelope.21 It has been proposed that centromere clustering can replace telomere clustering/bouquet formation in Drosophila.22,23 Together, these studies indicate that although clustering is an early-prophase phenomenon correlating with the establishment of pairing interactions, it exhibits a variable duration.
Clustering/bouquet formation is found in stages during which chromosomes move, suggesting that movement is required for clustering. In S. pombe and C. elegans, chromosomes move with the highest velocities when they are clustered.12-14 However, the bouquet stage is not necessarily the stage with the highest velocity in all organisms; in mouse, telomeres that cluster move less compared with stages preceding or following bouquet formation,7 and in S. cerevisiae, chromosomes move faster after bouquet formation.9 Bouquet formation was shown to be dependent on chromosome movement in S. pombe24 and in S. cerevisiae.9,10 Telomere clustering can occur only if telomeres are recruited to the nuclear envelope, as preventing telomere attachment to the nuclear envelope in S. pombe, S. cerevisiae, and mice leads to defects in telomeric clustering.25-29 However, little is known about the mechanisms of forming and resolving chromosome clustering once chromosomes are attached. These mechanisms may involve post-translational modification of proteins connecting chromosomes to the nuclear membrane (see below). In C. elegans, the kinases PLK-2 and CHK-2 are required for the clustering of chromosomes at early prophase I.30,31 Studies in S. cerevisiae showed that resolution of clustering requires sister-chromatid cohesion.9 Interestingly, in mice, the complex that recruits chromosomes to the nuclear membrane also binds a sister-chromatid cohesion protein.32
Although telomere clustering is not conserved across model organisms in meiosis, chromosome clustering is conserved. It is reasonable to hypothesize that the purpose of this clustering is to promote pairing interactions. The driving force behind chromosome movement and clustering are similar (cytoskeletal anchoring and movement forces, as will be discussed below). However, clustering requires either coordinated movement of chromosomes toward each other and/or a mechanism that ensures that chromosomes that encountered each other will remain in close proximity. The precise nature of these regulating mechanisms is still unclear.
Linking chromosomes to the cytoskeleton
The ubiquitous LINC (Linker of Nucleoskeleton and Cytoskeleton) complex plays an important and conserved role in connecting chromosomes to the cytoskeleton. The LINC complex is composed of 2 classes of proteins: Sad1p/Unc84 (SUN) and Klarsicht/Anc1/Syne1 homology (KASH) domain proteins (for meiotic LINC see Figure 1A). The SUN domain proteins span the inner nuclear envelope, interacting with proteins that recruit chromosomes to the LINC complex. The KASH domain proteins are embedded in the outer nuclear envelope and interact with the cytoskeleton of meiotic cells. SUN domain proteins homotrimerize and interact with 3 subunits of KASH domain proteins in the perinuclear space.33,34 LINC complexes can interact with each other to form macro-complexes (observed as “patches”), however, the mechanism for such interaction is still unclear and may involve a variety of interaction surfaces [reviewed in35].
Perturbing the function of the LINC complex leads to defects in chromosome movement and clustering. Deletion of the S. pombe SUN domain protein, Sad1, prevents formation of a functional spindle, and removal of the KASH domain protein Ksm1 leads to elimination of telomere-led chromosome movement, as well as telomere clustering.36,37 In mice, the LINC complex is composed of SUN1 and the meiotic specific KASH5.38,39 SUN2, another SUN domain protein, is able to partially substitute for SUN1s function in mouse Sun1 null mutants.29 As expected, telomere movement is eliminated in Kash5 mutants and severely impaired in Sun1 mutant mice.6 A central role for the LINC complex in meiotic chromosome movement is also present in C. elegans where chromosome movement is eliminated in sun-1 mutants.14 In Drosophila, KASH/Klar and SUN/koi mutants reduce nuclear oscillations.11 It is not yet clear how deletion of maize LINC complex affects movement, but Arabidopsis Sun1 and Sun2 double mutants have reduced bouquet formation.40 S. cerevisiae SUN/Mps3 mutants also reduce chromosome movement and bouquet formation;41 no KASH domain protein has been yet identified in this system. The conserved role of the LINC complex in chromosome movements is attributed to linking chromosomes to the cytoskeleton. However, it is possible that the LINC complex also plays a direct role in microtubule dynamics, such as organizing microtubules, as was proposed by studies in S. pombe.42 Furthermore, meiotic LINC complex proteins are not always meiosis-specific and can have additional functions during meiosis (e.g., DNA damage repair43). Therefore, elucidating the functions of the LINC complex, and its many components, will give us a better understanding of not only how the LINC complex affects different parts of the cell at different stages of the cell cycle, but also how these different functions may overlap and affect each other.
The role of the cytoskeleton in chromosome movements
Chromosome movements in meiotic prophase I are driven by the cytoskeleton that is connected to chromosomes through the LINC complex (Table 1). Microtubules are necessary for meiotic chromosome movement in animals and in S. pombe. Inhibition of microtubule polymerization in zygotene of rat spermatocytes prevents chromosome movements.44 Similar experiments in mouse highlighted the importance of microtubules and indicated that actin plays no role in meiotic chromosome movements.6,7,39 The 2 very distinct types of chromosome movement; the horse-tail meiotic chromosome movement in S. pombe,45,46 and the nuclear/chromosomal rotations in Drosophila, are also dependent upon microtubules.11 In C. elegans, inhibition of microtubule polymerization or dynein, but not actin, lead to arrest in chromosome movements.47 Despite the predominance of microtubule-dependent mechanisms, actin can play an important role in chromosome movements in some organisms; inhibiting actin polymerization reduces or eliminates chromosome movement in S. cerevisiae,9,10 and both microtubules and actin are required for movements in maize.8 All of these studies indicate that both coordinated and solitary chromosome movements utilize the cytoskeleton and can use either the actin and/or the microtubule network.
Microtubule organization in mitotic cells is orchestrated by the centrosome, which acts as the microtubule organizing center (MTOC), from which astral microtubules emanate. In yeast, the equivalent structure to the centrosomal MTOC is the Spindle Pole Body (SPB). In S. pombe, meiotic chromosome movement is driven by astral microtubules emanating from the SPB and anchored at the cell membrane, generating a pulling force.45,48-50 Drosophila meiotic microtubules nucleate mainly from an ER structure (fusome), but also from the nuclear envelope and the centrosome.11 Microtubule nucleation events are driven by γTubulin, which localizes to the centrosome/SPB in mitotic cells and therefore is used as a marker for MTOC. In mouse, γTubulin maintains stable localization and is not dynamic as found for the leading edges of chromosome movements.7 Studies in C. elegans and mouse agree that γTubulin can be found close to the nuclear envelope but is not present at all sites of microtubule-nuclear envelope associations.6,51 Altogether, this suggests that chromosome movement in animals is not coordinated by pulling or pushing forces (microtubule polymerization) but by a “walking” mechanism. In agreement, dynein inhibition has similar effects on chromosome movement as microtubule depolymerization.7,47 Additional evidence for this mechanism comes from studies in mouse where several chromosomes can move on the same path, and chromosomes can move in a back and forth manner.6 This movement is not passive, as studies in C. elegans have shown it to be an energy-dependent mechanism.52 Thus, in metazoans, the cytoskeletal network forms paths on which chromosome movement is actively directed. How the meiotic cytoskeleton is regulated to ensure these specialized functions is still an open question.
Mechanisms connecting the LINC complex to chromosomes
The proteins connecting the LINC complex to chromosomes are highly divergent (Fig. 1B). The common feature is that the site of connection is near or at the termini of chromosomes. The typical site of connection is the telomeric repeats, but in C. elegans chromosomal binding sites contain repetitive sequences on one or 2 chromosomes, termed pairing centers.53 These pairing centers are found in proximity to telomeres, and therefore are functionally similar to the canonical telomeric attachment.53
S. pombe telomeres are bound by the protein Rap1, which interacts with the meiosis-specific proteins Taz1, Bqt1, and Bqt2.54 Bqt1 serves as a bridge between Bqt2-Taz1 and the LINC complex.54 In S. cerevisiae, SUN/Mps3 interacts with Ndj1, a telomere-associated protein, which is required for Mps3 localization to telomeres.55 In mice, the manner by which telomeres are recruited to LINC changes throughout prophase. In early prophase I, TERB1 links SUN1 and the telomere binding protein TRF1,32,56 while in mid-prophase I, the TERB2-MAJIN complex binds to telomeres via DNA binding domains found in MAJIN, making TRF1-TERB1 binding dispensable.57 The C. elegans pairing-center binding proteins (ZIM-1/2/3 and HIM-8) associate with the nuclear envelope, and absence of these proteins results in an extended period of chromosome polarization.58 It is not yet clear if pairing center proteins bind to SUN-1 directly or not. As expected, proteins that are involved in connecting telomeres to the LINC complex were shown to be required for telomere clustering and/or chromosome movements in S. pombe,54 S. cerevisiae,28 and mice.32
LINC complexes localize to the nuclear envelope in both meiotic and mitotic cells. In most organisms, localization of LINC complexes in mitotic cells is uniform, whereas meiotic LINC localization is restricted. In mouse meiosis, Sun1 and Sun2 form foci at the nuclear envelope, throughout meiotic prophase I, which move to one side of the nucleus in zygotene.59 Kash5 also forms foci and localizes next to telomeres in a ring-like structure.39 Similarly, C. elegans SUN-1 and KASH/ZYG-12 co-localize with markers of chromosome ends, but these LINC proteins form patches and can therefore accommodate more than one chromosome end.60 Maize Sun2 localization in zygotene is diffuse, but concentrated to one half of the nuclear envelope, close to where telomeres are localized.61 In S. cerevisiae, LINC behaves differently; mitotic Sun/Mps3 forms a single focus, while its meiotic localization is extended to form patches.55 The localization of the LINC complex has yet to be examined in Drosophila meiotic nuclei. Overall, meiotic localization of LINC forms a pattern by which localization is restricted at only a few sites to the nuclear envelope, in agreement with it interacting only with chromosomal ends.
The connection between LINC and chromosomes may be assisted by other proteins that stabilize the complex at the nuclear membrane. In mice, TERB2-MAJIN are important for TERB1 recruitment to LINC.57 Interestingly, MAJIN has a trans-membrane domain (in addition to its DNA binding domain) that may help in stabilizing the LINC-TERB1 complex at the nuclear membrane.57 Similarly, S. pombe Bqt3 and Bqt4 connects telomeres to the nuclear envelope, where Bqt4 interacts with the telomeric binding protein Rap1.62 This complex may cooperate with LINC in meiotic attachment of telomeres to the nuclear envelope. The absence of the meiotic KASH domain protein in S. cerevisiae, led to the speculation that the trans-membrane protein, Csm4, could serve a similar function. However, Csm4 physically interacts with Ndj1, which puts Csm4 on the nuclear side of the membrane and therefore it is likely to act as a LINC-accessory protein, similarly to Bqt3 or MAJIN.63,64 In C. elegans, the Caenorhabditis-specific protein HAL-2 regulates the recruitment of the autosomal pairing center proteins to the nuclear envelope.30,65
LINC also can be affected by post-translational modifications. In C. elegans, PLK-2, a polo kinase, and the kinase CHK-2 are both required for SUN-1 phosphorylation on S12.30,31,66 This phosphorylation acts as a signal for telomere clustering in C. elegans and may be part of a surveillance mechanism for meiotic progression; once DNA damage repair reaches a certain point of progression SUN-1 aggregates are dissolved and chromosome movement relaxes.67 Another kinase, CDK, may be responsible for LINC regulation in mice by targeting TERB1,57 and Cdk2 may regulate its interaction with SUN1.68 SpeedyA, a noncanonical activator of CDKs, was shown to be important for Cdk2 localization to telomeres in mouse spermatocytes, suggesting a telomeric pre-complex of SpeedyA and Cdk2.68,69 Despite the advances in understanding the regulation of the LINC complex by post-translational modifications in mouse and C. elegans, further elucidation of signaling pathways await discovery in other organisms.
The role of chromosomal movements in meiotic prophase I
Meiotic chromosome movements are required for pairing and synapsis, which consequently, is required for the formation of all obligatory crossover events. How much each organism depends upon chromosome movements to achieve these events varies. Synapsis defects in LINC mutants range from the inability to processively load SC proteins to aggregation of SC in a structure termed the polycomplex. The most drastic effect of depleting the LINC complex in meiosis is revealed in mice, where a Sun1 knockout leads to pachytene arrest and massive apoptosis.38 In these LINC complex mutants, chromosomes fail to completely synapse and are unable to complete recombination. Meiosis in other organisms still progresses in the absence of the LINC complex, but may be severely impaired. The double mutant of Sun1 and Sun2 in Arabidopsis shows partial synapsis, reduced pairing, and severe reduction in crossovers that leads to elimination of almost all seed production.40 In C. elegans, dynein inhibition leads to delayed pairing and polycomplex formation, and knock down of all pairing center proteins leads to failure to form crossovers as well.30,47 Elimination of telomere-led chromosome movement in S. pombe results in a large reduction in pairing, recombination frequency, and spore viability (∼70% dead).36 The role of chromosome movements in S. cerevisiae may be limited: meiotic events, such as recombination, are delayed but not severely impaired when chromosome movements halt, leading to a modest reduction in spore viability (∼10%). While the effect on SC polymerization is notable, leading to polycomplex formation, the effect on crossover formation is mild or undetectable in S. cerevisiae.27,41,55,70 It is not surprising that Drosophila meiosis, which shows chromosome movements just before meiotic entry, has a limited dependence upon chromosome movements: Drosophila LINC mutants show only a delay in pairing, accompanied by some polycomplex formation.11 The role of chromosome movements in crossover formation has not yet been examined in Drosophila. Chromosome movement's effect on crossovers is likely limited to its governance of chromosome pairing which then controls proper recombination progression. Why some organisms rely less on pairing facilitated by chromosome movement to complete recombination is still unclear, but, speculatively, this may be due to more robust DNA repair.
Why is chromosome movement essential for pairing and synapsis?
Chromosome pairing and synapsis appear to be the most direct targets of meiotic chromosome movements. One straight-forward explanation for this requirement is that chromosomes need to be brought in close proximity to pair and allow for the stabilization of such interactions by the processive assembly of the SC. This explains why chromosomes are not only linked to the nuclear envelope but also brought together forming a cluster/bouquet in early prophase I. Another function of chromosome movement is to perturb interactions between non-homologous chromosomes that may lead to non-homologous synapsis. Clear evidence for this comes from studies in C. elegans in which attenuation, but not abolishment, of chromosome movements leads to non-homologous synapsis.47,60 Defects in chromosome movements results in an increase of ectopic, non-allelic, recombination in S. pombe.71 This was further corroborated by a study in S. pombe showing that when oscillations are inhibited by microtubule depolymerization for a short amount of time, non-homologous pairing interactions are stabilized.72 Lastly, when synapsis proceeds from more than one site on a chromosome, interlocks can be formed. Interlocks are 2 pairs (or more) of synapsing chromosomes that are entangled when one chromosome is locked between 2 other synapsing chromosomes. Such interlocks can be observed in Sun mutants in Arabidopsis.40 Chromosome movement helps to “shake off” these entanglements and release the interlocked chromosomes and thus facilitate proper synapsis. Despite the prevalence of this model in the literature, little direct evidence supports interlock formation in mutants with bona fide defects in movements. It could be that interlock resolution by chromosome movements is essential only for organisms with fairly long chromosomes.
The absence of chromosome movements, in the presence of SC proteins, frequently leads to polycomplex formation [in S. cerevisiae, Drosophila, and C. elegans28,30,47]. In C. elegans, if movement of one chromosome is perturbed, SC components will form a functional SC only on the other moving chromosomes and no polycomplex will be formed.58 This indicates that quiescent chromosomes are not preferred sites of SC assembly, but in the absence of an alternative, they can nucleate multi-directional SC assembly (polycomplex), as opposed to linear assembly (SC). Others have proposed that the chromosome movements leads to stretching of chromosomes which may be a prerequisite for linear SC assembly.13 Thus, chromosome movement directly affects pairing and SC assembly, and loss of movement allows for proper and improper interactions to form; movement helps stabilize the proper interactions during this process.
Can there be too much movement?
Chromosome movements do not occur continuously in meiosis, chromosomes move in one direction, stop and then resume or change direction. Live-imaging studies of chromosome movements record frequent directional changes [mouse,6 maize,8 S. cerevisiae,9,10 and C. elegans13,14]. In some organisms, directional changes and directional movements were shown to be interrupted with long pauses.6 It is possible that pauses reflect the time required to switch LINC-dynein to a new microtubule filament (i.e., the time required to establish new interactions between KASH domain proteins to a microtubule via dynein). However, it is also possible that these pauses can serve as a point of regulation. It is reasonable to suggest that once pairing is initiated by bringing 2 homologous chromosomes together, movement needs to be attenuated to allow processive and uninterrupted synapsis. Our studies suggest that such negative regulation of chromosome movement is a prerequisite for timely pairing and synapsis.73 FKBP52, an HSP90 co-chaperone, was shown to depolymerize microtubules in vitro and interacts with microtubules in mitotic rat cell culture.74 We have shown that the FKBP52 homolog in C. elegans, fkb-6, is essential for timely pairing and synapsis, and fkb-6 mutants have a similar phenotype to mutants of dynein or KASH/zyg-12.47 However, while dynein and KASH/zyg-12 mutants reduce chromosome movements, fkb-6 mutants make it more frequent. As expected, KASH/zyg-12 mutants antagonize fkb-6 function in epistasis experiments. This study suggests that negative regulation of chromosome movement is also key to proper chromosome pairing and that the resting phase of chromosome movement may be an important hub for regulation. Whether regulation of chromosome movement is active in other organisms remains to be explored, as well as what other cell processes may be involved with its regulation.
Concluding remarks
Seventy years of research on meiotic chromosome movement has led to a detailed analysis of these movements, establishing their necessity for proper pairing, synapsis, and therefore, the downstream events that are required for proper chromosome segregation in meiosis. These studies have also identified the key proteins involved in chromosome movement and show that the LINC complex is a key component for this movement. Despite this significant progress, many gaps of knowledge still remain. We know little about the mechanisms that fine-tune chromosome movement. Since chromosome movement, in terms of speed and directionality, varies during meiotic prophase I it is important to identify the mechanisms that regulate such movement. One important aspect of that will be to fully identify the modes of negative regulation, a poorly studied aspect of chromosome movement. Regulated chromosome movement prevents the SC from aggregating to form polycomplexes. It is still not clear what it is about chromosome movement that actively blocks aberrant SC polymerization. Moreover, it is still not clear what signals for bouquet formation and dissolution once chromosomes are attached to the LINC complex. Studies in mice and S. pombe have begun to shed light on the complexity of the mechanisms for anchoring telomeres to the nuclear envelope. The roles of other protein complexes, besides LINC, in connecting the chromosomes to the nuclear envelope in other organisms should also be addressed. Lastly, intra-nuclear forces, such as ones mediated by nuclear actin, may also be involved in some cases of chromosomal movement in the nucleus, which awaits exploration. These questions, and others, allow for the continuing research of this key process in meiosis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Robert Malone and members of the Smolikove Lab for critical reading of this manuscript. We apologize to colleagues whose work has not been cited due to space limitation.
Funding
S. S. is funded by National Science Foundation research grant 1515551.
References
- [1].Duro E, Marston AL. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes & Development. 2015;29:109-22. doi: 10.1101/gad.255554.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Osorio DS, Gomes ER. Connecting the nucleus to the cytoskeleton for nuclear positioning and cell migration. Adv Exp Med Biol. 2014;773:505-20. doi: 10.1007/978-1-4899-8032-8_23. PMID:24563363 [DOI] [PubMed] [Google Scholar]
- [3].Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355-64. doi: 10.1016/j.cell.2013.02.010. PMID:23498942 [DOI] [PubMed] [Google Scholar]
- [4].Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor Perspectives in Biology. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. PMID:25986558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yao KT, Ellingson DJ. Observations on nuclear rotation and oscillation in Chinese hamster germinal cells in vitro. Experimental Cell Research. 1969;55:39-42. doi: 10.1016/0014-4827(69)90451-0. PMID:4888994 [DOI] [PubMed] [Google Scholar]
- [6].Lee C-Y, Horn HF, Stewart CL, Burke B, Bolcun-Filas E, Schimenti JC, Dresser ME, Pezza RJ. Mechanism and regulation of rapid telomere prophase movements in mouse meiotic chromosomes. CellReports. 2015;11:551-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shibuya H, Morimoto A, Watanabe Y. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 2014;10:e1004821. doi: 10.1371/journal.pgen.1004821. PMID:25502938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sheehan MJ, Pawlowski WP. Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proceedings of the National Academy of Sciences. 2009;106:20989-94. doi: 10.1073/pnas.0906498106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. The Journal of Cell Biology. 2005;170:213-23. doi: 10.1083/jcb.200501042. PMID:16027219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:16934-9. doi: 10.1073/pnas.0704860104. PMID:17939997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Christophorou N, Rubin T, Bonnet I, Piolot T, Arnaud M, Huynh J-R. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nature Cell Biology. 2015;17:1388-400. doi: 10.1038/ncb3249. PMID:26458247 [DOI] [PubMed] [Google Scholar]
- [12].Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270-3. doi: 10.1126/science.8146661. PMID:8146661 [DOI] [PubMed] [Google Scholar]
- [13].Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. The Journal of Cell Biology. 2012;196:47-64. doi: 10.1083/jcb.201106022. PMID:22232701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, et al.. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001219. doi: 10.1371/journal.pgen.1001219. PMID:21124819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rickards GK. Prophase chromosome movements in living house cricket spermatocytes and their relationship to prometaphase, anaphase and granule movements. Chromosoma. 1975;49:407-55. doi: 10.1007/BF00285133. PMID:1132283 [DOI] [PubMed] [Google Scholar]
- [16].Parvinen M, Söderström KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534-5. doi: 10.1038/260534a0. PMID:1264213 [DOI] [PubMed] [Google Scholar]
- [17].Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188-201. doi: 10.1016/j.cell.2008.04.050. PMID:18585353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Scherthan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. The Journal of Cell Biology. 1996;134:1109-25. doi: 10.1083/jcb.134.5.1109. PMID:8794855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. The Journal of Cell Biology. 1997;137:5-18. doi: 10.1083/jcb.137.1.5. PMID:9105032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Goldstein P, Slaton DE. The synaptonemal complexes of caenorhabditis elegans: comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma. 1982;84:585-97. doi: 10.1007/BF00292857. PMID:7075356 [DOI] [PubMed] [Google Scholar]
- [21].Carpenter AT. Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma. 1975;51:157-82. doi: 10.1007/BF00319833. PMID:806439 [DOI] [PubMed] [Google Scholar]
- [22].Tanneti NS, Landy K, Joyce EF, McKim KS. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol. 2011;21:1852-7. doi: 10.1016/j.cub.2011.10.005. PMID:22036181 [DOI] [PubMed] [Google Scholar]
- [23].Takeo S, Lake CM, Morais-de-Sá E, Sunkel CE, Hawley RS. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol. 2011;21:1845-51. doi: 10.1016/j.cub.2011.09.044. PMID:22036182 [DOI] [PubMed] [Google Scholar]
- [24].Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. The Journal of Cell Biology. 1999;145:1233-49. doi: 10.1083/jcb.145.6.1233. PMID:10366596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828-31. doi: 10.1038/33947. PMID:9572143 [DOI] [PubMed] [Google Scholar]
- [26].Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825-8. doi: 10.1038/33941. PMID:9572142 [DOI] [PubMed] [Google Scholar]
- [27].Trelles-Sticken E, Dresser ME, Scherthan H. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. The Journal of Cell Biology. 2000;151:95-106. doi: 10.1083/jcb.151.1.95. PMID:11018056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252-5. doi: 10.1126/science.276.5316.1252. PMID:9157883 [DOI] [PubMed] [Google Scholar]
- [29].Link J, Leubner M, Schmitt J, Göb E, Benavente R, Jeang K-T, Xu R, Alsheimer M. Analysis of meiosis in SUN1 deficient mice reveals a distinct role of SUN2 in mammalian meiotic LINC complex formation and function. PLoS Genet. 2014;10:e1004099. doi: 10.1371/journal.pgen.1004099. PMID:24586178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Labella S, Woglar A, Jantsch V, Zetka M. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Developmental Cell. 2011;21:948-58. doi: 10.1016/j.devcel.2011.07.011. PMID:22018921 [DOI] [PubMed] [Google Scholar]
- [31].Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y, et al.. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139:920-33. doi: 10.1016/j.cell.2009.10.045. PMID:19913286 [DOI] [PubMed] [Google Scholar]
- [32].Shibuya H, Ishiguro K-I, Watanabe Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nature Cell Biology. 2014;16:145-56. doi: 10.1038/ncb2896. PMID:24413433 [DOI] [PubMed] [Google Scholar]
- [33].Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035-47. doi: 10.1016/j.cell.2012.03.046. PMID:22632968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, Zhou Z. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Research. 2012;22:1440-52. doi: 10.1038/cr.2012.126. PMID:22945352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hieda M. Implications for diverse functions of the LINC complexes based on the structure. Cells. 2017;6:3. doi: 10.3390/cells6010003. PMID:28134781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Molecular and General Genetics MGG. 1997;254:238-49. doi: 10.1007/s004380050412. PMID:9150257 [DOI] [PubMed] [Google Scholar]
- [37].Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. The Journal of Cell Biology. 1995;129:1033-47. doi: 10.1083/jcb.129.4.1033. PMID:7744953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Developmental Cell. 2007;12:863-72. doi: 10.1016/j.devcel.2007.03.018. PMID:17543860 [DOI] [PubMed] [Google Scholar]
- [39].Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K-I, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. The Journal of Cell Biology. 2012;198:165-72. doi: 10.1083/jcb.201204085. PMID:22826121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Varas J, Graumann K, Osman K, Pradillo M, Evans DE, Santos JL, Armstrong SJ. Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 2015;81:329-46. doi: 10.1111/tpj.12730. PMID:25412930 [DOI] [PubMed] [Google Scholar]
- [41].Lee C-Y, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012;8:e1002730. doi: 10.1371/journal.pgen.1002730. PMID:22654677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yoshida M, Katsuyama S, Tateho K, Nakamura H, Miyoshi J, Ohba T, Matsuhara H, Miki F, Okazaki K, Haraguchi T, et al.. Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. The Journal of Cell Biology. 2013;200:385-95. doi: 10.1083/jcb.201207168. PMID:23401002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lawrence KS, Tapley EC, Cruz VE, Li Q, Aung K, Hart KC, Schwartz TU, Starr DA, Engebrecht J. LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. The Journal of Cell Biology. 2016;215:801-21. doi: 10.1083/jcb.201604112. PMID:27956467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Salonen K, Paranko J, Parvinen M. A colcemid-sensitive mechanism involved in regulation of chromosome movements during meiotic pairing. Chromosoma. 1982;85:611-8. doi: 10.1007/BF00330775. PMID:7128279 [DOI] [PubMed] [Google Scholar]
- [45].Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. Journal of Cell Science. 1998;111(Pt 6):701-12. PMID:9471999 [DOI] [PubMed] [Google Scholar]
- [46].Svoboda A, Bähler J, Kohli J. Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma. 1995;104:203-14. doi: 10.1007/BF00352185. PMID:8529460 [DOI] [PubMed] [Google Scholar]
- [47].Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907-19. doi: 10.1016/j.cell.2009.10.039. PMID:19913287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Molecular Biology of the Cell. 2001;12:3933-46. doi: 10.1091/mbc.12.12.3933. PMID:11739791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Niccoli T, Yamashita A, Nurse P, Yamamoto M. The p150-Glued Ssm4p regulates microtubular dynamics and nuclear movement in fission yeast. Journal of Cell Science. 2004;117:5543-56. doi: 10.1242/jcs.01475. PMID:15509865 [DOI] [PubMed] [Google Scholar]
- [50].Saito TT, Okuzaki D, Nojima H. Mcp5, a meiotic cell cortex protein, is required for nuclear movement mediated by dynein and microtubules in fission yeast. The Journal of Cell Biology. 2006;173:27-33. doi: 10.1083/jcb.200512129. PMID:16585273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. The Journal of Cell Biology. 2009;186:229-41. doi: 10.1083/jcb.200902101. PMID:19635841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Labrador L, Barroso C, Lightfoot J, Müller-Reichert T, Flibotte S, Taylor J, Moerman DG, Villeneuve AM, Martinez-Perez E. Chromosome movements promoted by the mitochondrial protein SPD-3 are required for homology search during caenorhabditis elegans meiosis. PLoS Genet. 2013;9:e1003497. doi: 10.1371/journal.pgen.1003497. PMID:23671424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nature Cell Biology. 2009;11:934-42. doi: 10.1038/ncb1904. PMID:19620970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59-69. doi: 10.1016/j.cell.2006.01.048. PMID:16615890 [DOI] [PubMed] [Google Scholar]
- [55].Conrad MN, Lee C-Y, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA [Internet]. 2007;104:8863-8. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=17495028&retmode=ref&cmd=prlinks. doi: 10.1073/pnas.0606165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Daniel K, Tränkner D, Wojtasz L, Shibuya H, Watanabe Y, Alsheimer M, Tóth A. Mouse CCDC79 (TERB1) is a meiosis-specific telomere associated protein. BMC Cell Biology. 2014;15:17. doi: 10.1186/1471-2121-15-17. PMID:24885367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shibuya H, Hernandez-Hernandez A, Morimoto A, Negishi L, Hoog C, Watanabe Y. MAJIN Links Telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell. 2015;163:1252-66. doi: 10.1016/j.cell.2015.10.030. PMID:26548954 [DOI] [PubMed] [Google Scholar]
- [58].Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Developmental Cell. 2006;11:817-29. doi: 10.1016/j.devcel.2006.09.020. PMID:17141157 [DOI] [PubMed] [Google Scholar]
- [59].Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci USA. 2007;104:7426-31. doi: 10.1073/pnas.0609198104. PMID:17452644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Developmental Cell. 2007;12:873-85. doi: 10.1016/j.devcel.2007.05.004. PMID:17543861 [DOI] [PubMed] [Google Scholar]
- [61].Murphy SP, Gumber HK, Mao Y, Bass HW. A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.). Front Plant Sci. 2014;5:314. doi: 10.3389/fpls.2014.00314. PMID:25071797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. The Journal of Cell Biology. 2009;187:413-27. doi: 10.1083/jcb.200902122. PMID:19948484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kosaka H, Shinohara M, Shinohara A. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 2008;4:e1000196. doi: 10.1371/journal.pgen.1000196. PMID:18818742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008;4:e1000188. doi: 10.1371/journal.pgen.1000188. PMID:18818741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang W, Miley N, Zastrow MS, MacQueen AJ, Sato A, Nabeshima K, Martinez-Perez E, Mlynarczyk-Evans S, Carlton PM, Villeneuve AM. HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet. 2012;8:e1002880. doi: 10.1371/journal.pgen.1002880. PMID:22912597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Harper NC, Rillo R, Jover-Gil S, Assaf ZJ, Bhalla N, Dernburg AF. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Developmental Cell. 2011;21:934-47. doi: 10.1016/j.devcel.2011.09.001. PMID:22018922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, La Volpe A, Jantsch V. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 2013;9:e1003335. doi: 10.1371/journal.pgen.1003335. PMID:23505384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mikolcevic P, Isoda M, Shibuya H, del Barco Barrantes I, Igea A, Suja JA, Shackleton S, Watanabe Y, Nebreda AR. Essential role of the Cdk2 activator RingoA in meiotic telomere tethering to the nuclear envelope. Nat Comms. 2016;7:11084 Available from: http://www.nature.com/doifinder/10.1038/ncomms11084. doi: 10.1038/ncomms11084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tu Z, Bayazit MB, Liu H, Zhang J, Busayavalasa K, Risal S, Shao J, Satyanarayana A, Coppola V, Tessarollo L, et al.. Speedy A-Cdk2 binding mediates initial telomere-nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proceedings of the National Academy of Sciences. 2017;114:592-7. doi: 10.1073/pnas.1618465114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chua PR, Roeder GS. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes & Development. 1997;11:1786-800. doi: 10.1101/gad.11.14.1786 [DOI] [PubMed] [Google Scholar]
- [71].Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. The EMBO Journal. 2000;19:3831-40. doi: 10.1093/emboj/19.14.3831. PMID:10899136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chacón MR, Delivani P, Tolić IM. Meiotic nuclear oscillations are necessary to avoid excessive chromosome associations. CellReports. 2016;17:1632-45 [DOI] [PubMed] [Google Scholar]
- [73].Alleva B, Balukoff N, Peiper A, Smolikove S. Regulating chromosomal movement by the cochaperone FKB-6 ensures timely pairing and synapsis. The Journal of Cell Biology. 2017;216(2):393-408, 155:jcb.201606126–22. doi: 10.1083/jcb.201606126. PMID:28077446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chambraud B, Belabes H, Fontaine-Lenoir V, Fellous A, Baulieu EE. The immunophilin FKBP52 specifically binds to tubulin and prevents microtubule formation. The FASEB Journal. 2007;21:2787-97. doi: 10.1096/fj.06-7667com. PMID:17435176 [DOI] [PubMed] [Google Scholar]


