Abstract
The gut microbiota has diverse and essential roles in host metabolism, development of the immune system and as resistance to pathogen colonization. Perturbations of the gut microbiota, termed gut dysbiosis, are commonly observed in diseases involving inflammation in the gut, including inflammatory bowel disease, infection, colorectal cancer and food allergies. Importantly, the inflamed microenvironment in the gut is particularly conducive to blooms of Enterobacteriaceae, which acquire fitness benefits while other families of symbiotic bacteria succumb to environmental changes inflicted by inflammation. Here we summarize studies that examined factors in the inflamed gut that contribute to blooms of Enterobacterieaceae, and highlight potential approaches to restrict Enterobacterial blooms in treating diseases that are otherwise complicated by overgrowth of virulent Enterobacterial species in the gut.
INTRODUCTION
The symbiosis between symbiotic microbes and animals is the result of at least 500 million years of co-evolution,1,2 giving rise to an intricately balanced but yet complex and diverse microbial community of as many as 1014 bacteria in the healthy human gastrointestinal tract, which are collectively referred to as the gut microbiota. The human gut microbiota is composed of more than 500 species within 4 dominant phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria.3,4 Firmicutes and Bacteroidetes account for more than 90% of the bacterial population in the colon, whereas Actinobacteria and Proteobacteria are scarce in the colon but relatively more enriched in the ileum. The advent of new technologies in the past decade has substantially advanced our understanding of the complexity and functional diversity of the gut microbiota. Symbiotic bacteria produce enzymes that digest complex carbohydrates and regulate bile acid metabolism.5 Synthesis of certain vitamins as well as nutrients, such as short-chain fatty acids (SCFAs), requires gut symbiotic bacteria.6 SCFAs, produced by bacterial fermentation of undigested carbohydrates in the colonic lumen,7 are absorbed by host cells, and oxidation of SCFAs releases energy that can be utilized both locally in the colon as well as systemically when transported to the liver via the portal bloodstream.8,9 The SCFAs acetate, proprionate and butyrate have been shown to contribute to the development of colonic regulatory T (Treg) cells that limit local inflammation in the gut, as well as engage the G protein-coupled receptor GPR43 on neutrophils to diminish their infiltration into tissues and hence mitigate inflammation.10,11 Another major function of the gut microbiota is to provide microbial signals for proper development and function of the host immune system. For example, Bacteroides fragilis and Clostridial species exhibit the unique capacity to induce Treg responses.12–15 Development of local T-cell-dependent and -independent immunoglobulin A response, as well as the gut-associated lymphoid tissue, are dependent on the presence of symbiotic bacteria in the intestine.16–19
What is gut dysbiosis?
Given the diverse functions of the gut microbiota, maintenance of a balanced gut microbiota is essential for intestinal homeostasis and human health. Over a century ago, Nobel Prize laureate Elie Metchnikoff was the first one to introduce the term dysbiosis to describe disrupted symbiosis, and to theorize that human health could be enhanced by introducing beneficial bacteria found in yogurt.20 Gut dysbiosis refers to altered composition of the gut microbiota that is associated with functional changes in the microbial transcriptome, proteome or metabolome. A myriad of both intestinal and extra-intestinal disorders are linked to gut dysbiosis, including inflammatory bowel disease (IBD), infection, food allergies, asthma, diabetes, obesity, multiple sclerosis, autism, periodontitis, and colorectal cancer.21–26 Gut dysbiosis is thought to originate from shifts in relative bacterial abundances, caused by dietary changes, inflammation, immune deficiency, infection or exposure to antibiotics or toxins.21 Recent studies have suggested that perturbations of the gut microbiota may foster “blooms” of otherwise low-abundance and harmful bacteria that contribute to disease (Table 1). In particular, Enterobacterial blooms are commonly observed in gut dysbiosis in various contexts involving inflammation in the gut.27–36
Table 1.
Enterobacterial blooms in human diseases and mouse disease models
| Species | Source of inflammation | Impact of Blooms | References |
|---|---|---|---|
| Mucosa-associated symbiotic E. coli strains | Patients with inflammatory bowel disease | Monocolonization increased incidence of invasive carcinoma in the IL-10-deficient colorectal cancer mouse model | 27 |
| Symbiotic E. coli | C. jejuni-colonized infant mice | Reduced colonization resistance against C. jejuni | 35 |
| Enterobacterial species | Mouse oral C. rodentium infection | Unknown | 34 |
| Proteobacterial species | Toll-like receptor 5-deficient mice | Transient instability of the gut microbiota | 28 |
| Adherent and invasive E. coli (AIEC) | Ileitis in patients with Crohn’s disease | Unknown | 30 |
| E. coli isolates with heightened virulence | Isolated from patients with Crohn’s disease or ulcerative colitis | In vitro, enhanced capability to activate NLRP3 inflammation and resistance to macrophage killing. | 42 |
| Blooms of Klebsiella pneumoniae and Proteus mirabilis | T-bet−/− ×Rag2−/−ulcerative colitis (TRUC) mouse model | Oral infection with these strains elicited colitis in Rag2−/− and WT adult mice | 41 |
| Salmonella and E. coli | Mouse oral Salmonella model | High densities of Salmonella and E. coli lead to horizontal gene transfer of the colicin-plasmid p2 from Salmonella to E. coli | 98 |
| E. coli pathobiont | Ampicillin and neomycin-treated mice | Multidrug-resistant E. coli capable of inducing lethal NAIP5–NLRC4 inflammasome in systemic infection | 36 |
| Enterobacterial species | Clindamycin-treated mice | Enhanced susceptibility to Clostridium difficile-induced colitis | 29 |
Abbrevations: C. jejuni, Campylobacter jejuni; E. coli, Escherichia coli; IL-10, interleukin-10; WT, wild type.
Blooms of Enterobacteriaceae
Enterobacteriaceae, including symbionts Escherichia coli, Klebsiella spp., and Proteus spp., are a large family of gram-negative facultative bacteria within the class Gammaproteo-bacteria and the phylum Proteobacteria. Enterobacteriaceae reside in the gut at low levels and are localized in close proximity to the mucosal epithelium due to their relative higher tolerance of oxygen diffused from the epithelium. In the newborn intestine, the first colonizers in the aerobic gastrointestinal tract are facultative anaerobes such as Enterobacteriaceae, which deplete oxygen to create a new environment suitable for colonization of strict anaerobes, such as Bacteroides, Clostridium, and Bifidobacterium just a few days after birth.37 During the first few months, breast milk allows oligosaccharide fermenters such as Bifidobacterium to thrive. Subsequent weaning and introduction of solid foods rich in polysaccharides not digestible by host enzymes gives rise to the expansion of polysaccharide fermenters Bacteroides, Clostridium, Ruminococcus, and simultaneously a decrease in Bifidobacterium and Enterobacteriaceae.38,39 Interestingly, Enterobacteriaceae are among the most commonly overgrown symbionts in many conditions involving inflammation, such as IBD, obesity, colorectal cancer, celiac disease, and antibiotic treatment. Both environmental and nutritional changes as a result of inflammation in the gut may confer a growth advantage to Enterobacteriaceae. The inflamed gut in various contexts, either infection by a pathogen,34 chemically induced colitis,40 or deficiencies in host immunity,28,41 appears to provide a favorable environment for expansion of Enterobacteriaceae. For example, an increased prevalence of Enterobacteriaceae, including adherent-invasive E. coli (AIEC), is also found in patients with Crohn’s disease or ulcerative colitis, the two forms of IBD.42–46 Despite the increased abundance of E. coli including AIEC in patience with IBD, there is insufficient evidence to substantiate the hypothesis of infectious causes of IBD by AIEC in the gut. On the other hand, the bloom of E. coli appears to be a consequence rather than a cause of inflammation in IBD, likely due to the unique ability of E. coli to thrive in the inflamed gut. Furthermore, the bloom of E. coli in the inflamed gut is likely not exclusive to AIEC. Enter-obacterial blooms often negatively impact host defense against pathogens or injury. For example, Enterobacterial blooms also enhance the susceptibility to Clostridium difficile-induced colitis in mice treated with one dose of clindamycin.29 E. coli pathobionts that had emerged in antibiotics or dextran sulfate sodium (DSS)-treated mice caused bacteremia and ultimately mouse mortality.36,40 In a study of human fecal microbiota, higher abundance of Bacteroides and Escherichia species was associated with higher susceptibility to Campylobacter infection.47 In addition, a recent study of humanized mice reported that expansion of Proteobacteria due to antibiotic treatment, or transplantation of E. coli isolated from a patient with Crohn’s disease, exacerbated gluten-induced enteropathy.48 Potent inflammatory PAMPs in Enterobacteriaceae such as lipopolysaccharides are thought to promote disease. Lipopolysaccharide from Enterobacteriaceae has been shown to exacerbate NSAID-induced intestinal injury and increase intestinal permeability in celiac disease.49,50
Inflammation-driven blooms of Enterobacteriaceae are associated in various diseases (Table 1) and could potentially contribute to the pathogenesis of disease development. A deeper understanding of the mechanisms underlying the blooms of Enterobacteriaceae in the context of inflammation would shed light on approaches to mitigating Enterobacteriaceae-mediated pathogenesis. Here we provide a comprehensive review of recent literature elucidating both host and microbial factors in the inflamed gut that give rise to blooms of Enterobacterial species.
Inflammation-induced environmental changes that drive dysbiosis
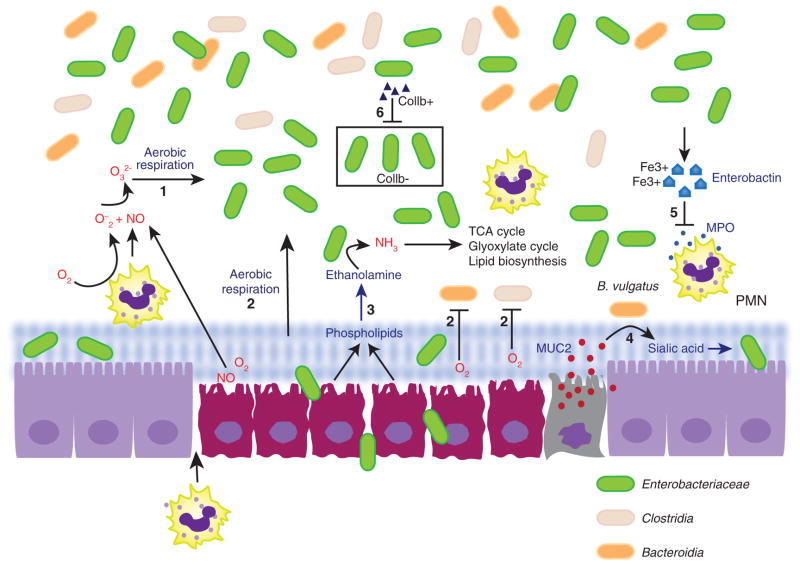
Inflammatory host response in the gut can be initiated by insults such as infection, injury and even antibiotic treatment. Several studies have provided evidence that an inflammatory tissue environment is conducive to perturbations of the gut microbiota often characterized by blooms of particular bacterial species that are genetically endowed with the capability of utilizing nutrients more abundantly found in the inflamed gut. One of the initial observations of infection-driven reduced diversity of the gut microbiota was in a mouse model of Helicobacter hepaticus, suggesting that invasion by a pathogen causes significant disturbances in the gut microbial community structure.51 However, colonization of a pathogen per se is not always associated with altered gut microbiota, as infection with another pathogen, Campylobacter jejuni, established gut colonization in mice but failed to trigger a robust inflammatory response or alterations of the composition of the gut microbiota.34 Of note, some pathogens within the family Enterobacteriaceae, namely Citrobacter rodentium and Salmonella, initially utilize virulence factors to induce intestinal inflammation, which subsequently confers a growth advantage for these pathogens in the intestinal lumen.32,34,52 In addition, oral infection with Citrobacter rodentium elicits robust inflammatory response and significantly reduced the microbial diversity of colonic microbiota while promoting the growth of facultative Enterobacteriaceae.34 Several mechanisms have been proposed to account for blooms of Enterobacteriaceae in the inflamed gut (Figure 1), which include: (1) nutritional changes, (2) anaerobic respiration, (3) aerobic respiration, (4) mucin utilization, (5) metal acquisition, (6) production of antimicrobials, (7) horizontal gene transfer.
Figure 1.
Mechanisms of inflammation-driven blooms of Enterobacteriaceae in the gut. (1) Superoxide radicals and nitric oxide react to give rise to nitrate (NO3−), which can be reduced by nitrate reductase-expressing Enterobacteriaceae in anaerobic nitrate respiration. (2) Higher levels of oxygen in the inflamed gut permit aerobic respiration by Enterobacteriaceae, while inhibiting growth of obligate anaerobes Bacteroidia and Clostridia. (3) Phospholipids from dying epithelial cells are broken down to give rise to ethanolamine, which is converted to ammonia that can be utilized in tricarboxylic acid cycle, glyoxylate cycle and lipid biosynthesis. (4) The release of sialic acid from MUC2 is mediated by cecal sialidase activity from Bacteroides vulgatus, and Enterobacteriaceae can take up sialic acid for synthesis of bacterial capsule and lipooligosaccharides. (5) Bacterial siderophore Enterobactin released by Escherichia coli inhibits the bacteriocidal effect of myeloperoxidase from neutrophils. (6) Colicin Ib (Collb)-expressing Enterobacterial species kill closely related but Collb-sensitive Enterobacterial species by releasing Collb, which forms pores in Collb-sensitive bacteria and disrupts cell wall synthesis.
Nutritional changes
Diet has a pivotal role in determining the composition of the gut microbiota.53 For example, the abundance of oligosaccharides in human breast milk favors the proliferation of bacteria equipped with carbohydrate processing enzymes, such as Bifidobacterium and Bacteroides spp., thus giving rise to distinct gut microbiota in breast milk-fed infants, in contrast to formula-fed infants that have higher levels of Clostidium spp.54,55 Mice fed on a “Western diet” with high sugar and fat display a reduction in Bacteroidetes but an overgrowth of Firmicutes.56 A vegetarian diet in humans is associated with lower intestinal pH and increased SCFA that might have an inhibitive effect on the growth of E. coli and other members of Enterobacteriaceae.57 Hence, nutrient sources have a profound role in shaping the composition of the gut microbiota. In the inflamed gut, collateral damage of the mucosal epithelium due to inflammatory response and increased shedding of dead epithelial cells result in an elevation of phospholipids such as phosphatidylcholine and phosphatidylethanolamine, which are derived from lipids of dead epithelial cell membrane. Ethanolamine can be used as a source of carbon and/or nitrogen by a variety of species in the Firmicutes, Actinobacteria and Proteobacteria phyla, as well as pathogenic species such as Salmonella and Pseudomonas.58 Utilization of ethanolamine involves breaking ethanolamine into ammonia and acetaldehyde by an ethanolamine ammonia lyase in a vitamin-B12-dependent manner.58,59 The ammonia can be used as a cellular supply of reduced nitrogen, and the acetaldehyde is converted to the metabolically useful compound acetyl-CoA, which is absorbed in various metabolic cycles, such as the tricarboxylic acid cycle, the glyoxylate cycle, or lipid biosynthesis.58
Anaerobic respiration
Under inflammatory conditions, gut luminal oxygen levels rise partly due to the elevated blood flow and hemoglobin. In addition, host proinflammatory cytokine signaling leads to activation of intestinal epithelial reactive oxygen species-generating enzymes such as Nox-1 and Duox2,60,61 while neutrophils transmigrating into the gut lumen to generate copious reactive oxygen species via the phagocyte NADPH oxidase.62 Furthermore, reactive nitrogen species are also produced in the gut lumen by both epithelial cells and transmigrating neutrophils. Inflammatory host response results in respiratory electron acceptors that favor bacterial growth by anaerobic respiration, including nitrate respiration, for bacterial growth. A previous study showed that the amounts of nitrate in inflamed colon rise rapidly when a superoxide radical reacts with nitric oxide (NO) to form peroxynitrite (ONOO−), a potent antimicrobial that is rapidly converted to nitrate (NO3−). The nitrate-rich tissue environment confers a growth advantage for Enterobacteriaceae such as E. coli through nitrate respiration, since genes encoding nitrate reductase are found within the genomes of Enterobacteriaceae but largely absent in obligate anaerobic bacteria belonging to the classes Bacteroidia and Clostridia.63,64 In a similar fashion, Salmonella enterica (Salmonella) infection leads to generation of nitrate in the inflamed intestine that selectively supports the growth of Salmonella by nitrate respiration.65 In addition, neutrophil respiratory burst leads to oxidation of thiosulfate to tetrathionate ( ), which can be used by Salmonella as a respiratory electron acceptor, thereby boosting the growth of Salmonella in the inflamed gut.66 Furthermore, reactive oxygen species and reactive nitrogen species can oxidize organic sulfides, such as methionine, or tertiary amines, such as trimethylamine (TMA), to form S-oxides and N-oxides, respectively, that are used as electron acceptors for anaerobic respiration to fuel the growth of E. coli in the inflamed gut.66
Aerobic respiration
Successful colonization of Enterobacteriaceae in the mammalian intestine attributes to their respiratory flexibility. Enterobacteriaceae are facultative anaerobes that are able to respond to different oxygen availability in the intestine, switch between aerobic and anaerobic respiration, and obtain energy from fermentation instead of respiration when exogenous electron acceptors are not available. In the absence of oxygen, E. coli can resort to anaerobic respiration using nitrate, nitrite, trimethylamine-N-oxide (TMAO), dimethyl sulfoxide (DMSO) and fumarate, in the order of preference, as electron acceptors.67 Inflammation or infection has been shown to lead to mucosal hypoxia. For instance, a recent study demonstrated a significant reduction in gut tissue oxygen levels in mice infected orally with Salmonella.68 Higher blood flow and hemoglobulin within the luminal environment in an inflamed intestine is thought to result in a more aerobic microenvironment in the lumen that favors the bloom of facultative anaerobes such as Enterobacteriaceae.59 However, it remains challenging to measure luminal oxygen levels. In response to an aerobic environment, E. coli expresses terminal oxidases that mediate aerobic respiration using oxygen as a terminal electron acceptor.69 Expression of terminal oxidases is regulated at the transcriptional level by the O2 and NO3− sensing regulators FNR, ArcA/B, NarX/L, and NarP/Q.59 Aerobic (oxidative) respiration is thermodynamically more favorable than anaerobic respiration or fermentation. The higher concentration of oxygen derived from higher blood flow and hemogloblin within the luminal environment in an inflamed intestine favors the growth of facultative anaerobes such as Enterobacteriaceae while inhibiting the growth of obligate anaerobes including mainly Bacteroidia and Clostridia. In fact, the importance of oxygen intake in blooms of Enterobacteriaceae is highlighted by multiples studies. Blooms of Enterobacteriaceae were observed in patients following small bowel transplantation due to increased oxygen intake in the bowel.70 Bacterial respiration of butyrate, a SCFA, was shown to render epithelial cells in the colon hypoxic.71 In addition, a recent study illustrated that when butyrate-producing Clostridia are depleted by streptomycin treatment, epithelial oxygenation is elevated, leading to luminal expansion of Salmonella via aerobic growth mediated by Salmonella type III cytochrome bd-II oxidase and synergized with nitrate reductases.72 A previous study showed that aerobic respiration is required for commensal and pathogenic E. coli to colonize the mouse intestine, as E. coli mutants lacking ATP synthase, which is required for all respiratory energy-conserving metabolism, were eliminated by competition with respiratory-competent wild-type strains. Likewise, E. coli mutants, which lacked the high-affinity cytochrome bd oxidase that is required for aerobic respiration when oxygen tensions are low, also failed to colonize the mouse intestine.73 These results suggest that even in the steady state, a low threshold of oxygen in the intestinal tissue may be pivotal for Enterobacteriaceae to establish stable colonization in the intestine, and an increase in local oxygen availability, as seen in the inflamed gut, further drives the blooms of Enterobacteriaceae. It remains technically challenging, however, to accurately measure the concentration of oxygen in the gut lumen. Data on oxygen levels in the intestine are difficult to compare between different laboratories due to technical limitations of the available techniques. Hence, the role of oxygen levels in blooms of Enterobacteriaceae in the inflamed gut remains a subject of active investigation. Up to date, spectral electron paramagnetic resonance imaging in living mice to measure oxygenation and Pd-porphyrin used to quantify luminal oxygen have been used, but not in the context of chronic or acute inflammation in the gut.59 Accurate measurement of oxygen concentrations in the inflamed gut and its impact on Enterobacterial blooms will be insightful for developing approaches to dampen Enterobacterial blooms and restore healthy gut microbiota.
Mucin utilization
The mucus layer coating the gut epithelium is a critical first line of host defense against ingested food or potential invading commensal or pathogenic bacteria in the gut lumen. Colonization of commensal bacteria is limited to the outer mucus layer, but the inner mucus layer is largely devoid of bacteria. Secretory mucin glycoprotein (MUC2) and bioactive epithelial membrane-bound mucins (MUC1, MUC3, and MUC17) are produced by intestinal goblet cells. Lack of MUC2 mucin in mice is associated with increased bacterial adhesion to the surface epithelium, increased intestinal permeability and enhanced susceptibility to colitis induced by DSS.74,75
N-acetyl-D-galactosamine, N-acetyl-D-glucosamine, N-acetylneuraminic acid (sialic acid), L-fucose and D-galactose are five major mucin-derived sugars that can be potential nutrient sources for bacteria. Increased production and secretion of mucin is a hallmark of intestinal inflammation triggered by enteric infection, likely as a mechanism to facilitate expulsion of pathogens and maintain the integrity of the mucus layer. Secretory MUC2, a major colonic mucin in both humans and mice, has been shown to be upregulated by the transcription factor nuclear factor (NF) κB during inflammation via NFκB-binding sites in MUC2. Increased numbers of mucosa-associated E. coli are observed in human IBD.44 Conversely, a recent study reported enrichment of E. coli in the colons of mice with DSS-induced colonic damage and inflammation, and the growth advantage of E. coli is conferred by elevated mucin-derived sialic acid during inflammation.76 Sialic acid is one of the major carbohydrates in mucins and can be taken up by bacteria lacking de novo biosynthetic pathways for these sugars, such as E. coli, and incorporated into bacterial capsule and lipooligosaccharides.77 During DSS-induced inflammation, the release of sialic acid from mucin was mediated by cecal sialidase activity from Bacteroides vulgatus, which also expanded in the DSS-induced inflamed gut. Treatment with a sialidase inhibitor was effective in mitigating DSS-induced colitis in mice.76 Higher levels of sialic acid contribute to expansion of both S. Typhimurium and Clostridium difficile in mice treated with antibiotics.78 In addition, increased fecal glycosidases were observed in patients with Crohn’s disease, consistent with enhanced mucin-degrading activity in the inflamed gut. Mucin-degrading bacteria, Mucispirillum schaedleri, Akkermansia musiniphila and B. acidifaciens, expand during intestinal inflammation and mediate release of less complex sugars (lactose, melibiose, raffinose, and galactinol) from mucins.59 Accumulation of these sugars and metabolites may either directly or indirectly lead to depletion of commensal bacteria mainly from the Bacteroidetes and Clostridia phyla in the inflamed gut, but on the other hand confer a growth advantage to Enterobacteriaceae as well as pathogens such as S. Typhimurium and Clostridium difficile.59,78
Metal acquisition
Iron is a vital nutrient for both the host and pathogens. Iron in vertebrates is mostly sequestered intracellularly by the iron storage protein ferritin or by heme as a co-factor of hemoglobin or myoglobin. Furthermore, iron is insoluble in the extracelluar neutral pH of serum and hence inaccessible to pathogens.79 Therefore, free iron in the circulation is limited to a low concentration, 10−24 M, as a mechanism to restrict growth of pathogens. To circumvent host iron restriction, many bacteria produce and release iron-chelating molecules, termed side-rophores, with high affinities for Fe3+ to compete against host-mediated sequestration of iron via heme acquisition and transferrin/lactoferrin receptors.79,80 Enterobacteriaceae are equipped with an arsenal of catecholate-type and hydroxamate-type siderophores that are critical for their survival in low iron environments, either in the circulation or in inflamed tissues.81,82 In the inflamed gut, a variety of antimicrobial proteins are produced and released by host cells to limit the availability of iron for bacterial growth. Lipocalin-2, for instance, is produced by both mucosal epithelial cells and infiltrating neutrophils in response to inflammation, as a strategy to prevent bacterial iron acquisition by binding and sequestering the siderophore enterobactin.83,84 A recent study showed that enterobactin, a catecholate siderophore released by E. coli, inhibits the activity of the neutrophil bacteriocidal enzyme myeloperoxidase both in vitro and in the inflamed gut, hence promoting the survival of E. coli in the inflamed gut.85 A study that sequenced the genomes of AIEC from intestines of patients with Crohn’s disease, dogs with granulomatous colitis, and mice with acute ileitis demonstrated common over-expression of genes encoding iron acquisition (yerinabactin, chu operon) in these more virulent bacteria relative to nonpathogenic E. coli.86 The ability of Enterobacteriaceae to acquire iron can be advantageous in the response to a pathogen that is dependent on iron for intestinal colonization. For instance, E. coli strain Nissle 1917 inoculated into mice limited Salmonella intestinal colonization by competing for iron using siderophores.87 Therefore, these studies underscore the importance of iron acquisition in promoting blooms of more virulent Enterobacteriaceae in the inflamed gut, which often give rise to disease.
Another metal that is sequestered by host response in the inflamed gut is zinc, which is an essential element for the growth of most organisms including bacteria. The intracellular concentration of zinc is tightly regulated, as too little zinc does not support growth but too much is toxic. Therefore, many bacteria have evolved high-affinity zinc-binding and transport systems.88 Calprotectin, a heterodimer composed of S100A8 and S100A9, is an antimicrobial protein that chelates zinc or manganese for its activity and is upregulated by IL-17 and IL-22 in response to infection.89,90 Elevation of Calprotectin in the extracellular fluids of patients with inflammatory disorders, such as rheumatoid arthritis and vasculitis, renders calprotectin a marker for inflammation.91 In response to inflammation in the gut, transmigration of neutrophils into the intestinal lumen leads to massive release of Calprotectin by dead neutrophils in the intestinal lumen, thereby drastically reducing the availability of zinc to bacteria in the intestinal lumen. Salmonella overcomes Calprotectin-mediated zinc sequestration by neutrophils by expressing a high-affinity zinc transporter (ZnuABC) while a znuA mutant Salmonella exhibits impaired growth in the inflamed gut.92 In addition, a high-affinity transporter for zinc is essential for Campylobacter jejuni to replicate and colonize in the gut.88 Hence, these studies underline the acquisition of the micronutrient zinc in the inflamed gut as an important means for Salmonella and likely resident Enterobacteriaceae to bloom while other members of the gut microbiota succumb to the low-zinc microenvironment. However, it remains unclear whether resident Enterobactericeae in the gut express high-affinity transporters for zinc akin to ZnuABC in Salmonella in response to inflammation in the gut.
Inflammation-driven production of antimicrobials
In addition to competition for limited nutrients in the gut, bacteria can bloom at the expense of other bacteria by producing antimicrobials. For example, bacteriocins termed colicins produced by Enterobacteriaceae can kill phylogenetically close relatives by pore formation in the inner membrane, nuclease activity or disruption of cellwall synthesis. A recent study revealed that colicin lb (Collb) production in Salmonella confers a growth advantage to Salmonella over colicin lb-sensitive resident E. coli in the inflamed gut.93 A Salmonella strain that is unable to induce intestinal inflammation, however, does not exhibit Colicin lb-mediated outcompetition of resident E. coli.93 Expression of Collb (cib) is in fact driven by low iron availability and the SOS response (which refers to the bacterial response to DNA damage) in a Fur- and LexA-dependent manner, respectively, within an inflammatory environment.94,95 Neutrophils recruited to the inflamed tissue release copious iron-sequestering lactoferrin upon degranulation,96 and iron availability is further restricted by lipocalin-2 abundantly produced by both neutrophils and epithelial cells to bind to and inhibit the bacterial siderophore enterobactin.97 Moreover, SOS response is instigated in the inflamed gut when bacteria sense DNA damage inflicted by oxidative stress in the inflamed gut.95 Therefore, the inflammatory environment in the gut creates unique conditions that potentiate the effects of colicins, which as a fitness factor contribute to blooms of colicin-expressing Enterobacteriaceae in the inflamed gut.
Horizontal gene transfer
Horizontal gene transfer could be another strategy for Enterobacterieacea to acquire virulent factors essential for thriving in the inflamed gut despite perturbations caused by environmental changes.98,99 Recent studies have suggested that inflammation-inflicted blooms of Enterobacteriaceae may be a consequence of enhanced fitness of Enterobacteriaceae due to horizontal gene transfer. The mammalian intestine is predominantly colonized by obligate anaerobes within the Firmicutes and Bacteroidetes phyla, which is theorized to suppress and maintain facultative anaerobes such as Enter-obacteriaceae at low densities (far less than 108 c.f.u./g) in the normal gut. The low densities of Enterobacteriaceae result in low frequencies of efficient conjugative plasmid transfer, or horizontal gene transfer (HGT).98,100 The genomes of Enter-obacteriaceae in general contain a large portion of horizontally acquired DNA, which in fact makes up more than 20% of an E. coli genome. In addition, the relative small size of genome and short doubling time of E. coli likely contributes to the high adaptability and propensity of E. coli to bloom in the presence of perturbations of the microbiota in the inflamed gut.101 One hypothesis is that inflammation-driven blooms of Enterobac-teriaceae foster HGT due to high densities of Enterobacteriaceae in the inflamed gut.98,99 Stecher et al.98 showed that in a mouse model of Salmonella oral infection, Salmonella-induced inflammation parallels blooms of both Salmonella and resident commensal E. coli, which favors conjugative HGT between Salmonella and E. coli. Thus, the adaptation of Enterobacteriaceae to growth in the inflamed gut, inflicted by either pathogen infection or impaired host immunity, may enhance selection of strains with higher pathogenic potential and ultimately the emergence of pathobionts due to acquisition of virulence factors and antibiotic resistance.99,102 In fact, expansion of pathobioints, a hallmark of dysbiosis, is associated with chronic inflammatory conditions observed in immunocompromised mice.41,103 Of note, HGT is more common but not exclusive to Enterobacteriaceae; an open pangenome shaped by HGT is found in Enterococcus spp. as well.59,104 Clinically, a recent study confirmed in vivo HGT of OXA-48, a highly prevalent carbapenemase worldwide, from OXA-48-harboring K. pneumoniae to E. coli in the gut of an infected patient. Therefore, HGT might, at least partially, contribute to the emergence of carbapenem-resistant Enterobacteriaceae that poses an urgent threat to health care in developed countries.102
CONCLUSIONS AND FUTURE DIRECTIONS
Inflammation in the gastrointestinal tract, due to infection, antibiotic use or dietary changes, result in a plethora of stressors that favor the blooming of Enterobacterial species with acquired inflammation-driven fitness factors to overcome environmental and nutritional limitations. We are still at the infancy of understanding the impact of Enterobacterial blooms in gut dysbiosis on host metabolism and adaptation to future insults such as infection, antibiotic use or dietary changes. The effort to understand mechanisms underlying gut dysbiosis has been hindered by technical difficulties to culture many obligate or facultative anaerobic gut symbiotic bacteria in an in vitro system. The recent successful isolation and culture of segmented filamentous bacteria,105 an important gut symbiotic bacterium that drives gut homeostatic Th17 and immunoglobulin A response,106,107 will likely affect many studies on dysbiosis and its relation to mucosal immune response. One conceivable consequence of overgrowth of Enterobacterial species with heightened resistance to environmental stressors is systemic translocation of these Enterobacterial species that may ultimately lead to bacteremia and even sepsis. Indeed, a growing number of outbreaks by bacteria from the family Enterobacteriaceae have emerged in the hospital setting.108,109 Bacteremia due to Gramnegative symbiotic E. coli accounts for 25–50% of all bloodstream infections globally.110 Therefore, understanding factors in the inflamed gut that confer a growth advantage to Enterobactericeae will shed light on therapeutics to prevent flourished Enterobacterial species in the gut from altering host homeostasis and contributing to disease development. Inflammation in the gut arises from various conditions, but the fundamental environmental changes in the inflamed gut are consistent in different disease settings and commonly foster the emergence of virulent species of Enterobacteriaceae. Dampening inflammation would be an ideal approach to restricting blooms of Enterobacterieaceae as well as many detrimental effects of inflammation, but it remains a challenge to tone down inflammatory response, as seen in the treatment for rheumatoid arthritis and cancer.111,112 Thus, maneuvering nutritional changes, oxidative stress, or iron/zinc availability commonly observed in the inflamed gut might be a better approach to either diminish blooms of Enterobacteriaceae or decelerate the outcompetition of symbionts within other families, thereby restoring a healthy gut microbiota and host homeostasis.
Acknowledgments
This work was supported by NIH grants T32HL007517 and 5P30DK034933 (to MYZ), DK091191 and DK095782 (to GN).
Footnotes
DISCLOSURE
The authors declared no conflict of interest.
References
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faust K, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Besten G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 10.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Round JL, Mazmanian SK. Inducible Foxp3 + regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Macpherson AJ, et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 18.Hirota K, et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 20.Mackowiak PA. Recycling metchnikoff: probiotics, the intestinal micro-biome and the quest for long life. Front Public Health. 2013;1:52. doi: 10.3389/fpubh.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 22.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butto LF, Haller D. Dysbiosis in intestinal inflammation: Cause or consequence. Int J Med Microbiol. 2016 doi: 10.1016/j.ijmm.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnich N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli and Crohn’s disease. Curr Opin Gastroenterol. 2007;23:16–20. doi: 10.1097/MOG.0b013e3280105a38. [DOI] [PubMed] [Google Scholar]
- 31.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barman M, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa M, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 2014;41:620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Haag LM, et al. Campylobacter jejuni induces acute enterocolitis in gnotobiotic IL-10−/− mice via Toll-like-receptor-2 and -4 signaling. PLoS One. 2012;7:e40761. doi: 10.1371/journal.pone.0040761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fallani M, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 40.Zeng MY, et al. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De la Fuente M, et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int J Med Microbiol. 2014;304:384–392. doi: 10.1016/j.ijmm.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 46.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 47.Dicksved J, Ellstrom P, Engstrand L, Rautelin H. Susceptibility to Campylobacter infection is associated with the species composition of the human fecal microbiota. Mbio. 2014;5:e01212–e01214. doi: 10.1128/mBio.01212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galipeau HJ, et al. Intestinal microbiota modulates gluten-induced immunopathology inhumanizedmice. AmJ Pathol. 2015;185:2969–2982. doi: 10.1016/j.ajpath.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace JL, et al. Proton pump inhibitors exacerbate NSAID-induced small intestinal injury by inducing dysbiosis. Gastroenterology. 2011;141:1314–1322. e1311–e1315. doi: 10.1053/j.gastro.2011.06.075. [DOI] [PubMed] [Google Scholar]
- 50.Cinova J, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS One. 2011;6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuehl CJ, Wood HD, Marsh TL, Schmidt TM, Young VB. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect Immun. 2005;73:6952–6961. doi: 10.1128/IAI.73.10.6852-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fallani M, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 55.Marcobal A, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmer J, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012;66:53–60. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- 58.Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stecher B. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MBP-0008-2014. doi:101128/microbiolspec.MBP-0008-2014. [DOI] [PubMed] [Google Scholar]
- 60.Rokutan K, et al. NADPH oxidases in the gastrointestinal tract: a potential role of Nox1 in innate immune response and carcinogenesis. Antioxid Redox Signal. 2006;8:1573–1582. doi: 10.1089/ars.2006.8.1573. [DOI] [PubMed] [Google Scholar]
- 61.Harper RW, et al. Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett. 2005;579:4911–4917. doi: 10.1016/j.febslet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 62.Chin AC, Parkos CA. Neutrophil transepithelial migration and epithelial barrier function in IBD: potential targets for inhibiting neutrophil trafficking. Ann NY Acad Sci. 2006;1072:276–287. doi: 10.1196/annals.1326.018. [DOI] [PubMed] [Google Scholar]
- 63.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spees AM, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. Mbio. 2013;4 doi: 10.1128/mBio.00430-13. doi:101128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivera-Chavez F, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 68.Jennewein J, et al. Low-oxygen tensions found in Salmonella-infected gut tissue boost Salmonella replication in macrophages by impairing antimicrobial activity and augmenting Salmonella virulence. Cell Microbiol. 2015;17:1833–1847. doi: 10.1111/cmi.12476. [DOI] [PubMed] [Google Scholar]
- 69.Karhausen J, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartman AL, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelly CJ, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivera-Chavez F, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones SA, et al. Respiration of Escherichia coli in the mouse intestine. Infect Immun. 2007;75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 76.Huang YL, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bouchet V, et al. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc Natl Acad Sci USA. 2003;100:8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 81.Reissbrodt R, Rabsch W. Further differentiation of Enterobacteriaceae by means of siderophore-pattern analysis. Zentralbl Bakteriol Mikrobiol Hyg A. 1988;268:306–317. doi: 10.1016/s0176-6724(88)80015-4. [DOI] [PubMed] [Google Scholar]
- 82.Faber F, Baumler AJ. The impact of intestinal inflammation on the nutritional environment of the gut microbiota. Immunol Lett. 2014;162:48–53. doi: 10.1016/j.imlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goetz DH, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 84.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 85.Singh V, et al. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat Commun. 2015;6:7113. doi: 10.1038/ncomms8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dogan B, et al. Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm Bowel Dis. 2014;20:1919–1932. doi: 10.1097/MIB.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 87.Deriu E, et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gielda LM, DiRita VJ. Zinc competition among the intestinal microbiota. Mbio. 2012;3:e00171–00112. doi: 10.1128/mBio.00171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 92.Liu JZ, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nedialkova LP, et al. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS Pathog. 2014;10:e1003844. doi: 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cascales E, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Butala M, Zgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ward PP, Uribe-Luna S, Conneely OM. Lactoferrin and host defense. Biochem Cell Biol. 2002;80:95–102. doi: 10.1139/o01-214. [DOI] [PubMed] [Google Scholar]
- 97.Bachman MA, Miller VL, Weiser JN. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009;5:e1000622. doi: 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stecher B, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA. 2012;109:1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 100.Scott KP. The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell Mol Life Sci. 2002;59:2071–2082. doi: 10.1007/s000180200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mason TG, Richardson G. Escherichia coli and the human gut: some ecological considerations. J Appl Bacteriol. 1981;51:1–16. doi: 10.1111/j.1365-2672.1981.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 102.Gottig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VA. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis. 2015;60:1808–1815. doi: 10.1093/cid/civ191. [DOI] [PubMed] [Google Scholar]
- 103.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qin X, et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012;12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schnupf P, et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 108.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 109.Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27:128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kang CI, et al. Bloodstream infections due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for mortality and treatment outcome, with special emphasis on antimicrobial therapy. Antimicrob Agents Chemother. 2004;48:4574–4581. doi: 10.1128/AAC.48.12.4574-4581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 112.Brennan FM, McInnes IB. Evidence that cytokines play a role in rheumatoid arthritis. J Clin Invest. 2008;118:3537–3545. doi: 10.1172/JCI36389. [DOI] [PMC free article] [PubMed] [Google Scholar]