Abstract
Long non-coding RNAs (lncRNAs) have been reported to perform significant roles in cancer development and progression. Our research has found that a novel lncRNA n326322 was higher in nasopharyngeal carcinoma (NPC) cells. Moreover, the gain and loss of functional approaches revealed that the overexpression of lncRNA-n326322 promoted NPC cell proliferation and invasion, whereas the downregulation of lncRNA-n326322 suppressed cell proliferation and invasion. Further experiments demonstrated that potential mechanism may be associated with the activation of PI3K/AKT and ERK/MAPK pathways. Taken together, these results indicate that lncRNA-n326322 is associated with tumorigenesis of NPC.
Keywords: nasopharyngeal carcinoma, lncRNA-n326322, invasion
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a common aggressive carcinoma arising from the nasopharynx epithelium [1]. It is vastly prevalent in Southeast China and some other Asian countries [1]. Thanks to the development of radiation therapy and adjuvant chemotherapy, the average 5-year survival rate for NPC patients has been greatly improved [1–4]. However, tumor recurrence and distant metastases are still the main obstacles for patient survival [5–6]. Elucidating the molecular mechanisms of pathogenesis to find notable therapeutic strategies for NPC patients is urgently needed.
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides in length with limited or no protein-coding functions [7]. Accumulating evidence indicates that dysregulated lncRNA expression levels are correlated with cancer progression and development by modulating gene expression in transcriptional or posttranscriptional levels [8–13]. For example, lncRNA urothelial cancer-associated 1 (UCA1) has been shown to overexpress in bladder cancer, and the high expression of UCA1 frequently enhanced the invasive and migratory capacity of bladder cancer cells [13]. Recent studies have also reported that lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) could promote epithelial-to-mesenchymal transition (EMT) and metastasis in many cancers, including renal cell carcinoma, oral squamous cell carcinoma, and endometrioid endometrial carcinoma [14–16]. However, only a few studies exist, focusing on dysregulated lncRNAs in NPC and their functions that remain largely unknown [17–19].
In this study, we have identified a new lncRNA-n326322 (Noncode V3 ID: n326322), which was upregulated in NPC cell lines. We also found that lncRNA-n326322 could regulate cell proliferation through PI3K/AKT pathway and promote cell invasion through ERK/MAPK pathway in NPC.
RESULTS
lncRNA-n326322 is upregulated and associated with tumorigenesis of NPC
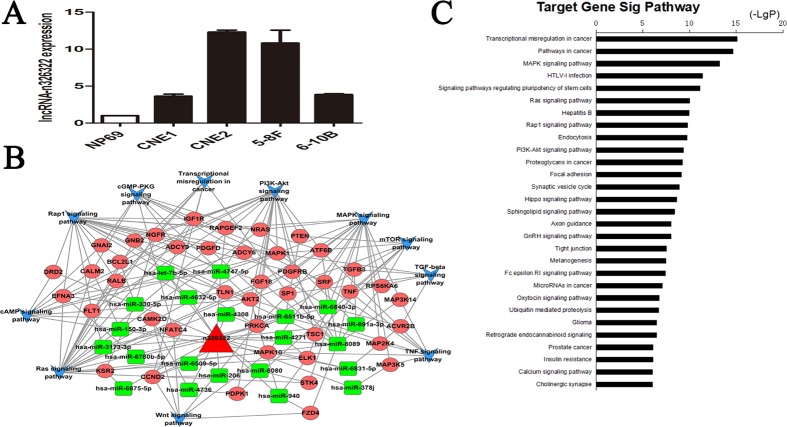
The expression levels of lncRNA-n326322 in NPC cell lines were analyzed by qRT-PCR, and the results showed that lncRNA-n326322 expression was significantly higher in NPC cells than in immortalized NP69 cells (Figure 1A). To predict the potential biological functions of the novel lncRNA-n326322 in tumor formation, we employed bioinformatic analyses based on some emerging themes. One of these themes is that lncRNAs perform their regulatory functions partly through interaction with RNA-binding proteins (RBPs). Moreover, with the rapid development of high-throughput sequencing of RNAs, it has provided powerful ways to map the interaction of lncRNAs and RBPs [20, 21]. We used RBPDB (http://rbpdb.ccbr.utoronto.ca/), a database of RNA-binding specificities, to search for the possible RBPs that could be sequence complementary with lncRNA-n326322. As shown in Table 1, we found a total of 16 RBPs that could match to lncRNA-n326322, including RBMY1A1, EIF4B, and Pum2, which have been reported to have an important influence on tumor progression [22–24]. In addition, accumulating studies have demonstrated a competitive endogenous RNA (ceRNA) hypothesis stating that lncRNA might serve as a molecular sponge to modulate miRNA expression [25]. We employed database software to determine conceivable miRNAs that could bind to lncRNA-n326322 and eventually found a total of 30 paired miRNAs. Then, we performed computational methods to confirm these possible miRNA targets. Here, we integrated lncRNA-n326322, its paired miRNAs, and miRNA targets to construct a global ceRNA network (Figure 1B). To further explore this interaction network, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to analyze these targets. Interestingly, we found that these lncRNA-associated ceRNA targets are involved in some KEGG pathways including cancer pathways and some other signaling pathways (Figure 1C). Among these enriched signaling pathways, such as the MAPK and Ras signaling pathways, as well as the PI3K–Akt signaling pathway, have been identified to contribute to the tumorigenesis of NPC [26–28].
Figure 1. lncRNA-n326322 is upregulated in NPC cell lines.
(A) Expression of lncRNA-n326322 in NPC cell lines. (B) Global view of ceRNA network consists of the miRNAs, miRNA targets and lncRNA-n326322. (C) Significantly enriched KEGG pathway of miRNA targets in the ceRNA network.
As discussed above, lncRNA-n326322 is upregulated and associated with tumorigenesis of NPC.
lncRNA-n326322 promotes NPC cell proliferation
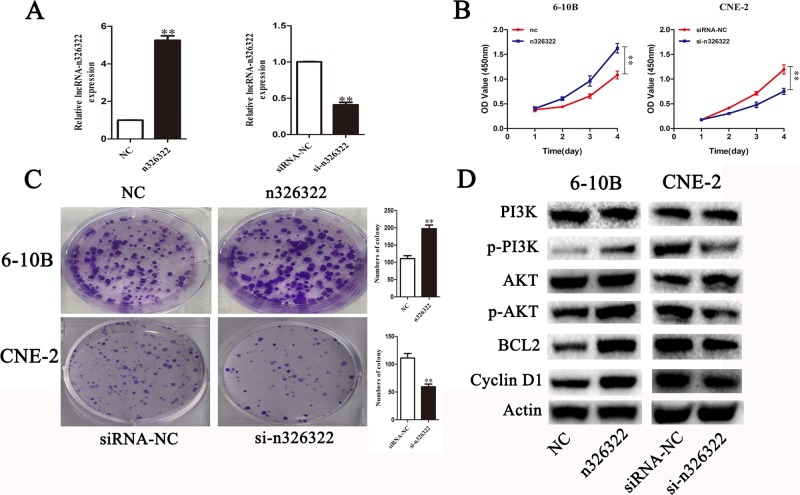
To investigate the effects of lncRNA-n326322 on the proliferation of NPC cells, gain and loss of functional approaches were performed. As shown in Figure 1A, NPC cell lines of CNE-2 expressed the highest levels of n326322, whereas 6-10B showed the lowest levels of n326322 among these tested NPC cell lines. Thus, we selected the CNE-2 and 6-10B cells for further studies. The expression levels of lncRNA-n326322 in 6-10B cells or CNE-2 cells were analyzed by qRT-PCR after transfection with pcDNA-n326322 or n326322-siRNA. In comparison with the negative control, the 6-10B cells transfected with n326322 overexpression vector evidently exhibited an increased n326322 mRNA level, whereas the knockdown efficiency of lncRNA-n326322 in CNE-2 cells was nearly 60%. Based on the growth curves determined by Cell Counting Kit-8 (CCK-8) assays, the forced expression of lncRNA-n326322 dramatically improved cell viability, whereas suppression of n326322 had the opposite effect in CNE-2 cells (Figure 2B). We also used colony formation assays to confirm our findings. As shown in Figure 2C, lncRNA-n326322 substantially increased the colony formation efficiency in NPC cell lines.
Figure 2. LncRNA-n326322 promotes NPC cell proliferation.
(A) The efficiency of siRNA-n326322 and n326322 overexpression was determined by qRT-PCR. **P < 0.05 compared with control. (B) CCK8 assays indicated that the upregulation of lncRNA-n326322 promoted 6-10B cell viability, and the downregulation of lncRNA-n326322 inhibited the proliferation of CNE2 cells. **P < 0.05 compared with control. (C) Colony formation was used to analyze cell viability of transfected 6-10B and CNE-2 cells. **P < 0.05 compared with control. (D) The Western blot analysis showed the expression of the PI3K/AKT signaling pathways controlled by lncRNA-n326322.
To understand the mechanism of n326322 in promoting cell proliferation, Western blot analysis was performed. Based on our previous bioinformatic analyses, we believed that PI3K/Akt may be involved in NPC progression. The PI3K/Akt signaling pathway has been reported to possess important roles in cell growth and proliferation [28, 29]. Interestingly, dozens of studies have demonstrated that the PI3K/Akt signaling pathway is one of the downstream signals activated by lncRNAs [30, 31]. Jing et al. illustrated that the knockdown of HOTAIR suppressed the cell proliferation by serving as a molecular sponge of miR-326 via the PI3K/Akt signaling pathway in human glioma cells [30]. Additionally, Zhu et al. showed that lncRNA HULC, an oncogene in gliomas, promoted angiogenesis by up-regulating ESM-1 expression through the PI3K/Akt signaling pathway [31]. In this present study, overexpression of lncRNA-n326322 significantly improved the levels of phosphorylated PI3K and Akt, whereas downregulation of lncRNA-n326322 suppressed these protein levels. However, the expression levels of PI3K and Akt were stable (Figure 2D). The expression levels of BCL2 and Cycling D1, known as important downstream components of PI3K/Akt signaling pathway, were also detected by the Western blot analysis [31]. The levels of BCL2 and Cycling D1 were improved due to the restoration of lncRNA-n326322 and depressed owing to the knockdown of lncRNA-n326322. All these results reveal that n326322 regulates NPC cell proliferation via PI3K/Akt signaling pathways.
lncRNA-n326322 promotes NPC cell invasion and migration
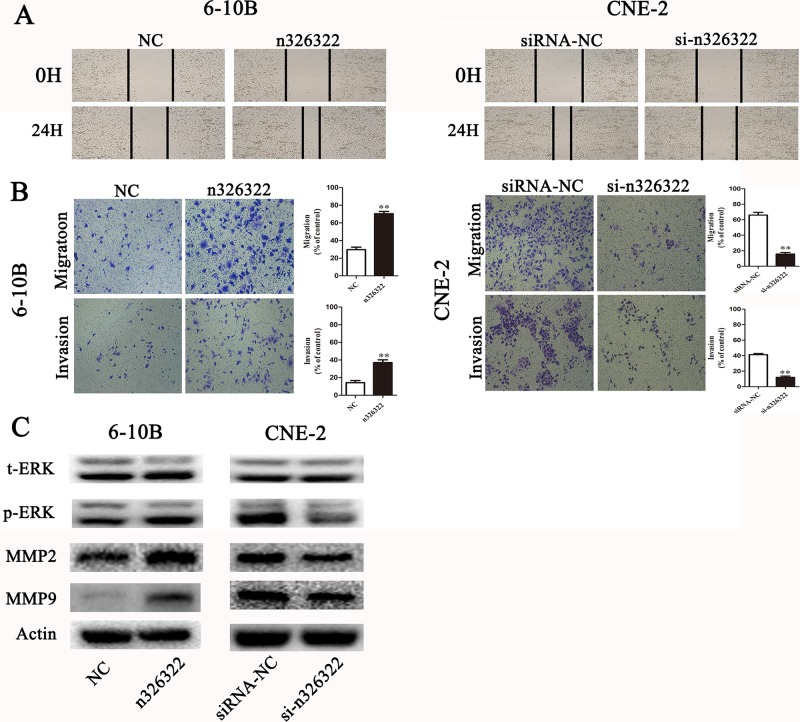
To examine the promotion of the invasive behavior of NPC cells by lncRNA-n326322, wound-healing and transwell assays were used. Wound-healing assays demonstrated that exogenous expression of lncRNA-n326322 simulated the migratory potential of 6-10B cells, but knock-down of lncRNA-n326322 expression significantly delayed scratch healing in CNE-2 cells (Figure 3A). The transwell assay without matrigel also confirmed that upregulated lncRNA-n326322 improved cell migration, whereas downregulated lncRNA-n326322 inhibited this ability (Figure 3B). Furthermore, the transwell assay with matrigel showed that the number of invasive cells increased in n326322 overexpressing cells compared with the negative control cells, whereas the number of invasive cells with downregulated lncRNA-n326322 was decreased (Figure 3B).
Figure 3. lncRNA-n326322 promotes NPC cell invasion and migration.
(A) The migration capability of NPC cells regulated by lncRNA-n326322 was determined by wound-healing assays. (B) Transwell assay with or without matrigel showed that upregulation of lncRNA-n326322 increased cell invasion and migration in 6-10B cells, whereas downregulation of lncRNA-n326322 suppressed cell invasion and migration capability in CNE-2 cells. **P < 0.05 compared with control. (C) Western blot results demonstrated that the overexpression of lncRNA-n326322 induced the expression of p-ERK, MMP2 and MMP9, and the knockdown of lncRNA-n326322 inhibited the expression of p-ERK, MMP2 and MMP9.
According to our bioinformatic analyses, the ERK/MAPK pathway, which was often aberrantly activated in human cancers and led to enhanced cell invasiveness [32, 33], might take part in n326322-driven tumor invasion. We performed the Western blot analysis to examine the expression levels of upregulated or downregulated lncRNA-n326322 on the ERK/MAPK signaling pathway. As shown in Figure 3C, the protein levels of phosphorylated ERK were higher in lncRNA-n326322 overexpressing cells, and the opposite result was observed in lncRNA-n326322 knock-down cells, whereas no detectable changes were found in the total levels of ERK. It is widely acknowledged that matrix metalloproteinases (MMPs) are the vital downstream signaling components of the ERK/MAPK pathway [34, 35]. Particularly, upregulations of MMP2 and MMP9 are associated with NPC tumor progression and enhance the migration and invasion of NPC cells [36]. Here, we identified that the expression levels of MMP2 and MMP9 in lncRNA-n326322 overexpressing cells were reduced and increased in lncRNA-n326322 knock-down cells compared with those in the control cells. Hence, lncRNA-n326322 might promote the invasive and migratory capacities of NPC cells through the ERK/MAPK signaling pathways.
DISCUSSION
Accumulating evidence has revealed that lncRNAs play important biological roles in human cancers, and that dysregulated lncRNAs are significantly related to the occurrence and development of tumors [37–40]. Several lncRNAs have also been identified in NPC development, indicating a poor prognosis [41–45]. In this present study, lncRNA-n326322 was overexpressed more in the NPC cell lines compared with NP69, and the bioinformatic analyses showed that lncRNA-n326322 might be associated with tumorigenesis of NPC.
lncRNA-n326322 (Noncode V3 ID: n326322; Noncode Gene ID: NONHSAG056983 and Noncode TRANSCRIPT ID: NONHSAT150079) is located in chromosome 1 (chr1: 175618208–175632895), and the mature lncRNA-n326322 transcript is 491 bp in length. In the present study, we used CCK8 and colony formation assays to confirm that the overexpression of lncRNA-n326322 in 6-10B cell lines promoted cell proliferation, whereas the downregulation of lncRNA-n326322 in CNE-2 cells significantly reduced cell growth. Then, we performed the Western blot analysis to investigate the possible cancer-related pathways that could increase cell viability. Many lncRNAs have been reported to promote tumorigenesis through the PI3K/Akt signaling pathway [30, 41]. We test the expression of p-PI3K and p-Akt and surprisingly found that they were markedly enhanced by exogenous expression of lncRNA-n32632 and reduced by downregulated expression of lncRNA-n326322, whereas no changes were detected in the expression of PI3K and Akt. Meanwhile, the expression levels of BCL2 and Cycling D1, the downstream molecules of PI3K/Akt signaling pathway, were also increased due to the restoration of lncRNA-n326322 and depressed owing to the knockdown of lncRNA-n326322. These lines of evidence indicated that lncRNA-n326326 promoted cell proliferation by activating the PI3K/Akt signaling pathway. However, the mechanism of how lncRNA-n326322 sensitized this pathway remains unclear. Interaction with RBP and CeRNA hypothesis are important mechanisms for lncRNAs to perform biological functions [20, 25]. On the other hand, through our established online software analysis, we uncovered some potential RBPs for lncRNA-n326322 targets. EIF4B, one of these RBPs, has been considered as an oncogene in tumor progression, and Shahbazian et al. showed that the mTOR/PI3K pathway converged on eIF4B [46, 47]. In this present study, lncRNA-n326322 might facilitate PI3K/Akt signaling by regulating the expression of EIF4B. On the other hand, through the bioinformatic analysis, we found that lncRNA-n326322 may improve the expression of FGF18 by serving as a molecular sponge to modulate miR-8089. FGF18 is a positive regulator of PI3K/Akt signaling pathways [48].
lncRNA-n326322 could activate invasion and migration of NPC cells. Western blot results revealed that the elevated invasion and migration by overexpression of lncRNA-n326322 was possibly linked to the activated ERK/MAPK pathway, which has been widely confirmed in cancers [49, 50]. MMP2 and MMP9, downstream of the ERK/MAPK pathway, are popular key enzymes for invasion and migration [51]. During our study, the data showed that upregulated lncRNA-n326322 induced the expression of p-ERK, MMP2, and MMP9; on the contrary, downregulated lncRNA-n326322 reduced the expression of p-ERK, MMP2, and MMP9. Although the precise mechanisms on how lncRNA-n326322 activated the ERK/MAPK signaling pathway are still unknown, a growing number of reports have thrown light on lncRNA-mediated regulation on this pathway [52, 53]. A recent study has reported that a new lncRNA HCG11-derived oncogenic growth through interaction with the RNA-binding protein IGF2BP1, contributes in facilitating the ERK/MAPK pathway in hepatocellular carcinoma [52]. Wang et al. also found that lncRNA UCA1 could function as a molecular sponge of miR-216b and activate the FGFR1/ERK signaling pathway in hepatocellular carcinoma [53]. In this present study, the underlying mechanisms that lncRNA-n326322 facilitate the ERK/MAPK signaling pathway may be also associated with RBPs and miRNAs. Our bioinformatic analysis provided some RBPs or miRNAs, such as eIF4B and miR-150, which have been proved to interact with the ERK/MAPK pathway [46, 47, 54]. However, further studies need to be performed to confirm whether these molecules are involved in lncRNA-mediated regulation on the ERK/MAPK signaling pathway.
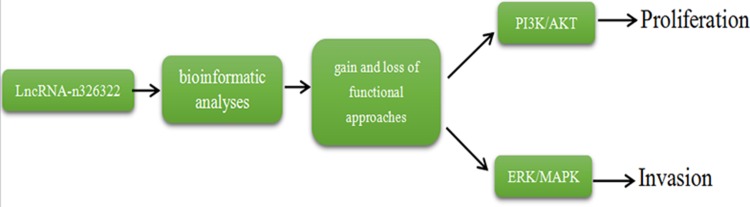
In summary, we identified novel lncRNA-n326322 as an important mediator of proliferation and invasion of NPC cells by PI3K/Akt and ERK/MAPK signaling pathways (Figure 4) and provided a new insight into the mechanism of invasion and metastasis of NPC.
Figure 4. lncRNA-n326322 promotes the proliferation and invasion of NPC cells.
MATERIALS AND METHODS
Cell culture
A human immortalized nasopharyngeal epithelial cell line (NP69) and four NPC cell lines (CNE1, CNE2, 5–8F, 6–10B) originated from the Research Center of Clinical Oncology of the Affiliated Jiangsu Cancer Hospital, Nanjing Medical University, Nanjing, China. Cells were cultured in RPMI 1640 medium (Gibco, Grand Island, NY) containing 5% fetal bovine serum (Gibco, USA) and incubated at 37°C in the presence of 5% CO2.
RNA extraction and quantitative real-time PCR assays
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions. Then, 2 μg of total RNA was reversely transcribed into cDNA using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). Real-time PCR was performed using SYBR Green PCR master mix on the ABI7300 real-time PCR machine (Applied Bio-systems). The expression of n326322 and β-actin was examined using the following specific primers: 5′-TCATTCCACATCAAGCCCCG-3′ and 5′-TGGATATCGGGCTACAGGT-3′ and 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′. Fold changes for n326322 expression levels were calculated using the 2ΔΔCt method
Cell transfection
siRNA-NC and siRNA-n326322 were synthesized by RiboBio (Guangzhou, China). The full length of n326322 was subcloned into the pcDNA3.1 plasmid, and the pcDNA3.1 empty vector was used as the control plasmid. CNE-2 and 6-10B cells were cultured in six-well plates for 24 h and transfected with siRNA-n326322 or n326322 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. To determine the efficiency of siRNA-n326322 or n326322, the expression of n326322 was assessed with real-time PCR.
Cell viability assay
Growth curves were determined by the CCK-8 according to the manufacturer's instructions. After the 48 h transfection, 3 × 103 cells/well were seeded in 96-well plates with five replicate wells for each group. Cells were cultured for 1, 2, 3, or 4 days before 10 ml CCK-8 was added to the 100 μL of culture medium for each well and incubated at 37°C for 1 h according to the manufacturer's instructions. The optical density in each well was determined at a wavelength of 450 nm by a microplate reader. The experiment was repeated three times, and the data shown are the average of these three repeated measurements.
Colony forming assay
To measure the colony-forming activity, CNE-2 and 6-10B cells were trypsinized and placed into a six-well plate (1 × 102 cells/well) after transfection and cultured for 10 days. The colonies were fixed with paraformaldehyde and stained with 0.1% crystal violet. Then, the numbers of colonies were counted under an inverted microscope. All steps were performed in triplicate.
Cell wound-healing assay
Wound-healing assay was conducted to examine the capacity of cell migration. CNE2 and 6-10B cells were cultured in six-well plates and transfected with siRNA or n326322, and then maintained for 48 h until confluency. A wound was created by manually scratching the surface of the plates with a 200-μL pipette tube, and the floated cells were removed by washing with PBS twice. Cell migration was monitored under an optical microscope with a magnification of 100×. Each experiment was performed in triplicate.
Cell invasion assay
Matrigel-coated (BD Biosciences) membrane and migration assay with no matrigel membrane were used to assess cell invasion or migration ability according to the manufacturer's instructions. Transfected CNE-2 and 6-10B cells were cultured for 48 h. The cells were resuspended (2 × 104 cells/well) in 200 μL of serum-free media and placed in the upper compartment of a chamber (Corning, NY, USA). Meanwhile, the bottom chamber was filled with 500 μL of RPMI-1640 and 20% fetal bovine serum. The cells that migrated or invaded the lower surface of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet after 24 h of incubation. Then, five random fields of cells were counted under a light microscope at a magnification of 100×. All experiments were performed three times.
Western blot analysis
Total cellular proteins were extracted from the transfected cells using modified RIPA buffer (Beyotime, Shanghai, China), and protein concentration was quantified using a BCA protein assay kit (Beyotime, Shanghai, China). A total of 15 mg of protein from each sample was separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking with BSA in Tris-buffered saline/Tween-20, The membranes were incubated with the indicated primary antibodies: anti-PI3K/p-PI3K (1:1000; Cell Signaling Technology, USA), anti-Akt/p-Akt (1:1000; Cell Signaling Technology, USA), anti-ERK/p-ERK (1:1000; Cell Signaling Technology, USA), anti-MMP9 (1:1000; Cell Signaling Technology, USA), and anti-β-actin(1:2000;Cell Signaling Technology, USA) at 4°C overnight and then incubated with the corresponding secondary antibodies at room temperature for 2 h. Immunoreactive bands were visualized using the ECL detection reagent (Millipore, Billerica, MA, USA). All data analyses were repeated thrice independently.
Statistical analysis
All statistical analyses were conducted using Student's t-test and one-way analysis of variance with Graphpad 5.0 and SPSS 13.0. Data were expressed as mean ± standard deviation in at least three separate experiments. P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS AND FUNDING
We thank the members of the Research Center of Clinical Oncology of of Jiangsu province for their technical assistance. Funding: This work was supported by the Jiangsu Provincial Commission of Health and Family Planning Young Scholars Award [grant numbers: Q201501]; the Jiangsu Clinical Medicine Science and Technology Special Fund [grant number: BL2014091]; the National Natural Science Foundation of China [grant number: 81672989]; and the Medical Young Talent Foundation of Jiangsu Provincial Health Department [grant number: QNRC2016648].
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Lee AW, Ng WT, Chan YH, Sze H, Chan C, Lam TH. The battle against nasopharyngeal cancer. Radiother Oncol. 2012;104:272–8. doi: 10.1016/j.radonc.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Kim YS, Kim BS, Jung SL, Lee YS, Kim MS, Sun DI, Yoo EJ, Mun SK, Yoon SC, Chung SM, Kim HK, Jo SH, Kang JH. Radiation therapy combined with (or without) cisplatin-based chemotherapy for patients with nasopharyngeal cancer: 15-years experience of a single institution in Korea. Cancer Res. Treat. 2008;40:155–163. doi: 10.4143/crt.2008.40.4.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong RL, Ting LL, Ko JY, Hsu MM, Sheen TS, Lou PJ, Wang CC, Chung NN, Lui LT. Induction chemotherapy with mitomycin, epirubicin, cisplatin, fluorouracil, and leucovorin followed by radiotherapy in the treatment of locoregionally advanced nasopharyngeal carcinoma. J Clin Oncol. 2001;19:4305–4313. doi: 10.1200/JCO.2001.19.23.4305. [DOI] [PubMed] [Google Scholar]
- 4.Lee AW, Yau TK, Wong DH, Chan EW, Yeung RM, Ng WT, Tong M, Soong IS, Sze WM. Treatment of stage IV(A-B) nasopharyngeal carcinoma by induction-concurrent chemoradiotherapy and accelerated fractionation. Int J Radiat Oncol Biol Phys. 2005;63:1331–1338. doi: 10.1016/j.ijrobp.2005.05.061. [DOI] [PubMed] [Google Scholar]
- 5.Razak AR, Siu LL, Liu FF, Ito E, O'Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46:1967–78. doi: 10.1016/j.ejca.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Huang T, Chen MH, Wu MY, Wu XY. Correlation between expression of extracellular matrix metalloproteinase inducer and matrix metalloproteinase-2 and cervical lymph node metastasis of nasopharyngeal carcinoma. Ann Otol Rhinol Laryngol. 2013;122:210–215. doi: 10.1177/000348941312200311. [DOI] [PubMed] [Google Scholar]
- 7.Chen XM, Zhang DD, Luo JJ. Research Progress on Long Noncoding RNAs. Progress in Biochemistry and Biophysics. 2014;41:997–1009. [Google Scholar]
- 8.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Serviss JT, Johnsson P, Grandér D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;18:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim Biophys Sin (Shanghai) 2014;46:1011–5. doi: 10.1093/abbs/gmu104. [DOI] [PubMed] [Google Scholar]
- 12.Chen YN, Xiong XD. Long Noncoding RNA and Epigenetic Regulation. Progress in Biochemistry and Biophysics. 2014;41:723–730. [Google Scholar]
- 13.Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 2016;107:18–27. doi: 10.1111/cas.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Zhang C, Chen R, Xiong H, Qiu F, Liu S, Zhang M, Wang F, Wang Y, Zhou X, Xiao G, Wang X, Jiang Q. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016;383:28–40. doi: 10.1016/j.canlet.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Gao W, Chan JY, Wong TS. Differential expression of long noncoding RNA in primary and recurrent nasopharyngeal carcinoma. Biomed Res Int. 2014;2014:404567. doi: 10.1155/2014/404567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–64. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng A, Tang JP, Goh CH, Hui KM. Regulation of the H19 imprinting gene expression in human nasopharyngeal carcinoma by methylation. Int J Cancer. 2013;104:179–87. doi: 10.1002/ijc.10926. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Wei Y, Yan Z, Wu C, Chang Z, Zhu Y, Li K, Xu Y. The characteristic landscape of lncRNAs classified by RBP-lncRNA interactions across 10 cancers. Mol Biosyst. 2017;13:1142–1151. doi: 10.1039/c7mb00144d. [DOI] [PubMed] [Google Scholar]
- 21.Yoon JH, Gorospe M. Cross-Linking Immunoprecipitation and qPCR (CLIP-qPCR) Analysis to Map Interactions Between Long Noncoding RNAs and RNA-Binding Proteins. Methods Mol Biol. 2016;1402:11–17. doi: 10.1007/978-1-4939-3378-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Yang F, Wu X, Huang JF, Yuan JH, Tao QF, Jiang GH, Zheng JS, Sun SH. Long non-coding RNA RBMY2FP promotes proliferation of male hepatocellular carcinoma by directing DNA methylation and activating RBMY1A1 via DNMT1. Oncotarget. 2016;30 Jul. https://doi.org/10.18632/oncotarget.10952 [Epub ahead of print] [Google Scholar]
- 23.Madden JM, Mueller KL, Bollig-Fischer A, Stemmer P, Mattingly RR, Boerner JL. Abrogating phosphorylation of eIF4B is required for EGFR and mTOR inhibitor synergy in triple-negative breast cancer. Breast Cancer Res Treat. 2014;147:283–93. doi: 10.1007/s10549-014-3102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, Condorelli G, Banfi S, Verde P. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34:3240–50. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Sun B, Zhu J, Zhou N, Yang Z, Gu J. Expression of RKIP in chronic myelogenous leukemia K562 cell and inhibits cell proliferation by regulating the ERK/MAPK pathway. Tumour Biol. 2014;35:10057–10066. doi: 10.1007/s13277-014-2312-3. [DOI] [PubMed] [Google Scholar]
- 27.Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, Chu GC, Jiang S, Hu J, Chang K, Vilar E, Song X, Zhang J, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017;31:370–382. doi: 10.1101/gad.293449.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng HW, Chen YF, Wong JM, Weng CW, Chen HY, Yu SL, Chen HW, Yuan A, Chen JJ. Cancer cells increase endothelial cell tube formation and survival by activating the PI3K/Akt signalling pathway. J Exp Clin Cancer Res. 2017;36:27. doi: 10.1186/s13046-017-0495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J, Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene. 2017;607:41–46. doi: 10.1016/j.gene.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, Xue YX. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–49. doi: 10.18632/oncotarget.4290. https://doi.org/10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan G, Jiang Y. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7:14429–40. doi: 10.18632/oncotarget.7418. https://doi.org/10.18632/oncotarget.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, Cao Y, Bao RF, Mu JS, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohsaka S, Hinohara K, Wang L, Nishimura T, Urushido M, Yachi K, Tsuda M, Tanino M, Kimura T, Nishihara H, Gotoh N, Tanaka S. Epiregulin enhances tumorigenicity by activating the ERK/MAPK pathway in glioblastoma. Neurooncology. 2014;16:960–970. doi: 10.1093/neuonc/not315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deryugina E, Quigley J. Matrix metalloproteinases and tumor metastasis. Cancer Metast Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 35.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–2. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Lv X, Chen J, Xie C, Xia W, Jiang C, Zeng T, Ye Y, Ke L, Yu Y, Liang H, Guan XY, Guo X, et al. CCL2-CCR2 axis promotes metastasis of nasopharyngeal carcinoma by activating ERK1/2-MMP2/9 pathway. Oncotarget. 2016;7:15632–47. doi: 10.18632/oncotarget.6695. https://doi.org/10.18632/oncotarget.6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 38.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 40.Lin Z, Hu Y, Lai S, Xue M, Lin J, Qian Y, Zhuo W, Chen S, Si J, Wang L. Long Noncoding RNA: its partners and their roles in cancer. Neoplasma. 2015;62:846–54. doi: 10.4149/neo_2015_103. [DOI] [PubMed] [Google Scholar]
- 41.Zhang W, Huang C, Gong Z, Zhao Y, Tang K, Li X, Fan S, Shi L, Li X, Zhang P, Zhou Y, Huang D, Liang F, et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545–54. doi: 10.1007/s10735-013-9503-x. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Lin Y, Yang X, Wu X, He X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473:913–9. doi: 10.1016/j.bbrc.2016.03.150. [DOI] [PubMed] [Google Scholar]
- 43.Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang HD, Li G, Zhang JF. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7:4712–23. doi: 10.18632/oncotarget.6731. https://doi.org/10.18632/oncotarget.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, Chen P, Shi L, Lian Y, Jing Y, Tang K, Li Z, Zhou Y, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–18. doi: 10.18632/oncotarget.4057. https://doi.org/10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37:4025–33. doi: 10.1007/s13277-015-4227-z. [DOI] [PubMed] [Google Scholar]
- 46.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5’cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 47.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–64. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 49.Xu WH, Zhang JB, Dang Z, Li X, Zhou T, Liu J, Wang DS, Song WJ, Dou KF. Long non-coding RNA URHC regulates cell proliferation and apoptosis via ZAK through the ERK/MAPK signaling pathway in hepatocellular carcinoma. Int J Biol Sci. 2014;10:664–676. doi: 10.7150/ijbs.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchegger K, Silva R, López J, Ili C, Araya JC, Leal P, Brebi P, Riquelme I, Roa JC. The ERK/MAPK pathway is overexpressed and activated in gallbladder cancer. Pathol Res Pract. 2017;213:476–482. doi: 10.1016/j.prp.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 51.Forsyth PA, Wong H, Laing TD, Rewcastle NB, Morris DG, Muzik H, Leco KJ, Johnston RN, Brasher PM, Sutherland G, Edwards DR. Gelatinase-A (MMP-2), gelatinase-B (MMP-9) and membrane type matrix metalloproteinase-1 (MT1- MMP) are involved in different aspects of the pathophysiology of malignant gliomas. Br J Cancer. 1999;79:1828–1835. doi: 10.1038/sj.bjc.6990291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Zheng Y, Liu H, Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int J Oncol. 2017;51:791–800. doi: 10.3892/ijo.2017.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–917. doi: 10.18632/oncotarget.3219. https://doi.org/10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun W, Zhang Z, Wang J, Shang R, Zhou L, Wang X, Duan J, Ruan B, Gao Y, Dai B, Qu S, Liu W, Ding R, et al. MicroRNA-150 suppresses cell proliferation and metastasis in hepatocellular carcinoma by inhibiting the GAB1-ERK axis. Oncotarget. 2016;7:11595–608. doi: 10.18632/oncotarget.7292. https://doi.org/10.18632/oncotarget.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]