Abstract
Objective
Recent evidence from genetic, cell biology and model animal studies have suggested a pivotal role of autophagy in mediating systemic lupus erythematosus (SLE). However, the genetic basis has not yet been thoroughly examined. The aim of the present study was therefore to identify additional susceptibility variants in autophagy-related genes, and their functional significance.
Methods
First, we performed a gene family-based genetic association analysis in patients with SLE using ImmunoChip, and selected the top-associated polymorphisms for replication in additional cohorts. To identify regulatory clues, we analyzed publicly available blood expression quantitative trait locus data and Encyclopedia of DNA Elements data on transcription factor binding sites and cell type-specific differential expression. The functional effects were tested by luciferase reporter assays, electrophoretic mobility gel shift assays (EMSA) and differential gene expression assays.
Results
In 14,474 samples, we observed that the rare Chinese variant rs933717T was associated with susceptibility to SLE (case 0.11% vs. control 0.87%, p = 2.36 × 10−10, OR = 0.13). The rs933717 risk allele C correlated with increased MAP1LC3B expression: increased MAP1LC3B mRNA was observed in patients with SLE and in lupus-prone mice. In reporter gene constructs, the risk allele increased luciferase activity up to 2.7~3.8-fold in both HEK 293T and Jurkat cell lines, and the binding of HEK293T and Jurkat nuclear extracts to the risk allele was also increased.
Conclusion
We observed a likely genetic association between LC3B, a widely-used marker for autophagy, and susceptibility to SLE.
Keywords: SLE, genetic association, autophagy, MAP1LC3B
INTRODUCTION
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with diverse clinical phenotypes and outcomes (1). Although the exact pathogenesis of SLE remains unclear, genetic factors are known to play key roles. To date, hypothesis-free genome-wide association studies (GWAS) have identified over 80 susceptibility loci (2). However, all these variants account for no more than 30% of the total genetic risk in SLE, and our knowledge of the genetic background is still limited (2). Further pathway testing analysis might help to improve our understanding of the pathogenic processes implicated in SLE.
Autophagy is an evolutionarily-conserved homeostatic process by which cytoplasmic materials are delivered to lysosomes for degradation (3). The autophagic process is implemented by autophagy related proteins which also play a part in regulating immune responses, including the direct elimination of microorganisms, inflammation, antigen presentation, lymphocyte homeostasis and secretion of immune mediators (4). Recent studies suggest that autophagy is deregulated in peripheral T and B cells in lupus-prone mice and SLE patients (5–7). Strikingly, autophagosomes in CD19+ B cells of NZB/W significantly increased before the disease had developed (7). In addition, more recent data indicated that a related process, LC3-associated phagocytosis (LAP), controlled immune responses to dying cells: its inhibition led to the development of a SLE-like disease (8–10). Inhibition of autophagy is likely to ameliorate murine lupus via reduction of proinflammatory cytokines. All these studies suggest that autophagy plays a highly important role in SLE (11). Thus it would be useful to discover the extent to which disease-associated genetic variants in LAP-related genes affect autophagy in humans.
Genetic advances resulting from by genome-wide association studies (GWASs) and follow-up studies found variants in genes related to autophagy including ATG5, ATG7, IRGM, DRAM1, CDKN1B, APOL1, MTMR3, CLEC16A, LRRK2 and ATG16L2, especially in Asian populations (12–20). Most of these genetic associations (ATG5, ATG7, IRGM, MTMR3, LRRK2 and ATG16L2) could be replicated in our previous reports (2, 13, 14 and 20). However, only a few of these genetic associations were reported in Caucasians (ATG5, IRGM and CLEC16A) or Africans (APOL1). This could suggest a genetic heterogeneity, and that the autophagy gene has a major genetic impact in Asian populations (18, 21). Thus there is a need for a systematic evaluation of genetic associations in human SLE patients from a pathway perspective, especially in Asians. In addition, the question remains as to whether the associated variant might have functional significance.
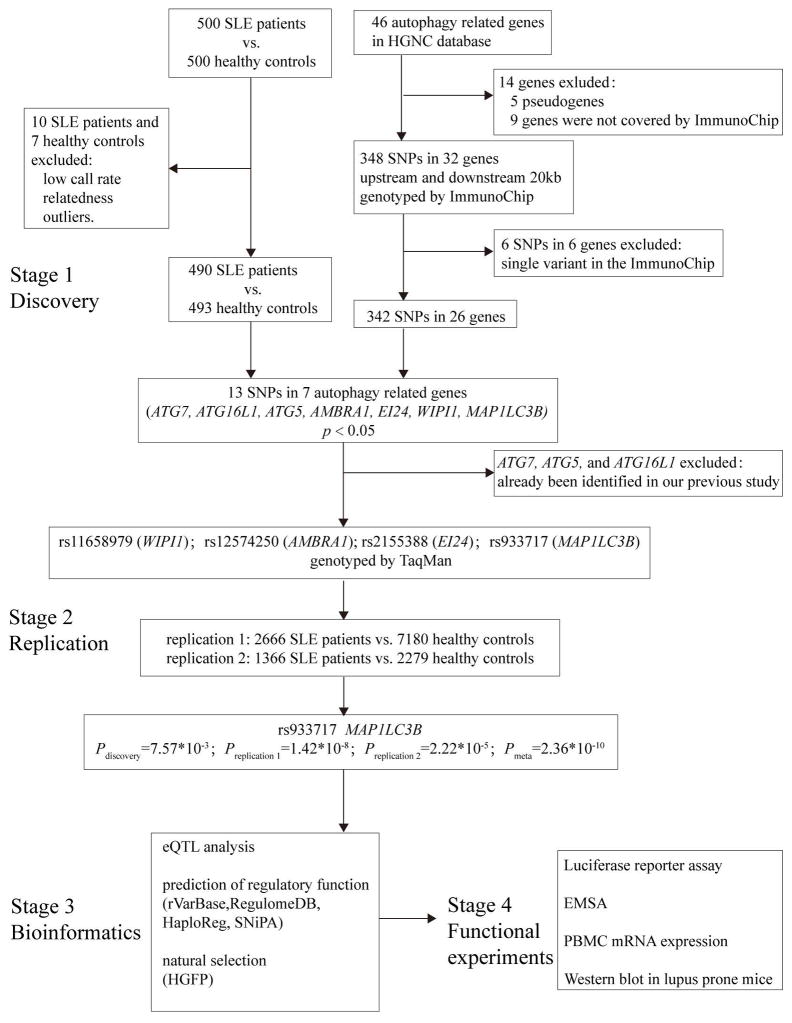
To find additional autophagy gene variants in Chinese SLE patients, in our present study, we included genes involved in the autophagic process according to HUGO gene nomenclature committee, and conducted a genetic association studyin more than 10,000 Han Chinese using a four-stage strategy (Figure 1), including genetic discovery, genetic replication, bioinformatic analysis and experimental validation.
Figure 1. Flow chart of our study design.
In stage 1, we systematically selected autophagy-related genes and conducted a tentative genetic discovery study using ImmunoChip. Stage 2 aimed to replicate novel loci discovered in stage 1 using independent populations. After combined analysis with discovery and replication 1 cohorts, SNPs with p < 2.6 × 10−7 were further replicated using the replication 2 cohort. After the two-stage genetic association analysis, bioinformatics analysis was performed to annotate the susceptibility loci in stage 3. Given the possible functional annotations for the novel loci by stage 3, in-house experimental validations were performed in vitro in stage 4.
MATERIAL AND METHODS
Sample description
The discovery cohort consisted of 500 healthy donors (32.0 ± 8.6 years; female:male = 3:1) and 500 SLE patients (Northern Han Chinese) (31.9 ± 11.2 years; female: male = 6:1). Replication cohort 1 consisted of 7180 healthy donors (37.6 ± 15.3 years; female: male = 4:1) and 2666 SLE patients (34.6 ± 13.4 years; female: male = 9:1) (Northern Han Chinese). To further replicate our genetic finding, 1,366 SLE patients and 2,279 controls (replication cohort 2) were enrolled (all Southern Han Chinese). All patients were diagnosed by local rheumatologists, and the diagnosis of SLE met the American College of Rheumatology (ACR) revised criteria for the classification of SLE (22). The study was approved by the medical ethics committee of Peking University and all participants provided informed consent (IRB: 2016[1139]).
Genes and SNP selection
A total of 46 autophagy-related genes were found by searching the HUGO Gene Nomenclature Committee database (www.genenames.org).
As shown in Figure 1, 5 genes were excluded as they were pseudogenes (BECN1P1, ATG3P1, ATG12P1, ATG12P2 and ATG4AP1), and 9 genes were not covered by ImmunoChip (MAP1LC3C, ATG4B, SNX4, WIPI2, RB1CC1, ATG2A, DRAM1, ATG2B and GABARAPL2). This left a total of 32 genes for enrollment into further analysis, namely ATG4C, DRAM2, ATG9A, ATG16L1, ATG7, ATG3, ATG10, ATG12, ATG5, ATG9B, SNX30, AMBRA1, ATG13, ATG16L2, EI24, GABARAPL1, ATG101, MAP1LC3B2, ULK1, ATG14, MAP1LC3B, GABARAP, ULK2, BECN1, VMP1, WIPI1, EPG5, ATG4D, MAP1LC3A, SOGA1, ATG4A and VMA21. Single nucleotide polymorphisms (SNP) in autophagy-related genes were included, along with 20 kb upstream and downstream. The results for 6 genes (ATG9A, ATG3, ATG13, GABARAP, BECN1 and SOGA1) should be interpreted with caution due to single variant involvement in the ImmunoChip (23) (Figure 1). We analyzed a total of 342 SNPs, mapped to 26 loci associated with autophagy-related genes (> 2 SNPs per gene).
Genotyping and quality control
During the discovery stage, 1000 participants (500 SLE patients and 500 healthy controls) were genotyped using ImmunoChip, and quality control was undertaken as previously reported (23). Of these, 10 SLE patients and 7 healthy controls were excluded due to a low call rate, relatedness or the presence of outliers (Figure 1). During the replication stage, genotyping was performed by TaqMan allele discrimination assays (replication cohort 1) using a TaqMan universal PCR master mix and a pre-designed SNP genotyping assay mix. These mixes contained PCR primers and probes purchased from ABI (Applied Biosystems, Foster City, CA). Analysis was performed using a 7500 Sequence Detection System (Applied Biosystems) or directly extracted from a HumanOmniZhongHua-8 BeadChip (replication cohort 2, unpublished).
Bioinformatic analysis
Variants were annotated using the Encyclopedia of DNA Elements (ENCODE) and blood expression quantitative trait locus (eQTL) data. Regulatory features were annotated using the following databases: rVarBase (http://rv.psych.ac.cn), Ensemble (http://asia.ensembl.org/index.html), rSNPBase (http://rsnp.psych.ac.cn), SNiPA (http://snipa.helmholtz-muenchen.de/snipa/index.php), RegulomeDB (http://regulome.stanford.edu) and HaploReg v4.1 (http://archive.broadinstitute.org/mammals/haploreg). Genetic signatures of natural selection in the human genome were analyzed using the HGDP Selection Browser (http://hgdp.uchicago.edu).
Allele-dependent gene expression regulations/eQTLs were queried using the Gene Expression Variation (GENEVAR) database (http://www.sanger.ac.uk/humgen/genevar) and the Genome-Wide Repository of Associations between SNPs and Phenotypes (GRASP) database (https://grasp.nhlbi.nih.gov/overview.aspx). Differential mRNA expression data were obtained from Array Express (http://www.ebi.ac.uk/arrayexpress).
Luciferase reporter assay
Sequences of 101bp flanking rs933717 were synthesized and subcloned into pGL4.23 (luc2/minP, Promega, USA) using the KpnI and BglII restriction sites (sequences are given in supplementary Table 1). HEK 293T (1.5 × 105 cells/well) and Jurkat (2 × 105 cells/well) were transfected with 0.8 μg pGL4.23 DNA containing rs933717 and 0.08 μg pRL-TK vector (as transfection control) using lipofectamine 2000 (Thermo, USA). After 48 h, cells were lysed and analyzed for luciferase activity using the Dual-Luciferase Assay System (Promega, USA). Each experiment was repeated three times.
Electrophoretic mobility shift assay (EMSA)
Nuclear proteins from HEK293T and Jurkat cells were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo, USA). EMSA was performed according to the instructions for the DIG Gel Shift Kit (Roche, 3353591910). The single-stranded oligonucleotides used to synthesize the double-stranded ones are given in supplementary table 2. For supershift assays, 10 μl of anti–EBF-1 (catalog no. H00001879-D01P; Novus Biologicals), anti-ROAZ (catalog no. ab169096; Abcam), or anti-ZIC4 (catalog no. sc-101202; Santa Cruz Biotechnology) antibody was incubated with nuclear proteins from HEK 293T or Jurkat cells for 1 hour before adding the relevant labeled probe. Electrophoresis took place in 5% polyacrylamide gels (in 0.5 × Tris/Borate/EDTA TBE buffer) and the dye was run 2/3 of the way to the bottom of the plate in 0.5 × TBE buffer at 50 V. Then, the gel was transferred to nylon membranes (LC2003, Thermo Fisher) at 200 mA for 30 min and cross-linked at 120 × 100 uJ/cm2. After incubation for 30 min in blocking solution, the membrane was incubated with antibody solution for 1 h and washed twice in washing buffer for 15 min. The membrane was then exposed and scanned using a LAS-3000 Imaging system (GE Healthcare Bioscience). The experiments were carried out in triplicate.
LC3B-II western blot in MRLlpr/lpr and C57BL mice
The lymph nodes from 10 MRLlpr/lpr mice (weight 23.1 ± 0.8g) and 10 C57BL mice (weight 22.4 ± 0.9g) were obtained for western blot. The antibodies used were specific for ACTB/Beta-Actin (8457, Cell Signaling Technology) and LC3 (L7543, Sigma-Aldrich). Tissue proteins from the inguinal lymph nodes were extracted and lysed in RIPA buffer (100 mM Tris-HCl, pH 8; 150 mM NaCl; 1% Triton X-100; 1 mM MgCl2; and 25 mM Na3VO4) in the presence of complete protease inhibitor mixture (P8340, Sigma-Aldrich). The proteins were separated by 15% SDS-PAGE and then transferred to a PVDF membrane (Millipore, Billerica, MA, USA). The antibody dilutions were anti-LC3 (1:1000) and ACTB (actin beta) (1:1000). The signal was detected using enhanced chemiluminescence detection reagents, and images were visualized using a LAS-3000 Imaging system (GE Healthcare Bioscience). LC3B-II levels were normalized by densitometry to ACTB levels using ImageJ Software.
All the mice were purchased from the Model Animal Research Center of Nanjing University and maintained in pathogen-free housing conditions. Assay procedures were performed according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals (Ministry of Health, People’s Republic of China, 1998). The study was approved by the Ethics Committee of Peking University.
Statistical analysis
Allelic and genotypic associations were assessed using PLINK (https://www.cog-genomics.org/plink) to give the odds ratio (OR) with a 95% confidence interval (95% CI). Quantitative variables with a normal distribution were expressed as means and standard deviations, and a Student’s t-test was performed. In eQTL analysis, the Spearman’s coefficient was calculated. Statistical analysis was performed with the SPSS 13.0 software package (SPSS Inc., USA).
RESULTS
Novel genetic associations of MAP1LC3B rs933717 in SLE
The discovery stage of the genetic analysis found 13 SNPs in 7 autophagy-related genes (ATG7, ATG16L1, ATG5, AMBRA1, EI24, WIPI1 MAP1LC3B) with p < 0.05. Of these genes, ATG5 showed the most associations, consistent with previous reports on GWAS and candidate genes (Supplementary Table 3) (12 13).
Since the aim was to identify novel gene associations, we excluded ATG7, ATG5 and ATG16L1 from further study because they had been associated in our previous paper (13). We therefore studied 4 other SNPs in promising loci in an additional replication cohort (replication cohort 1). From these, we identified 2 novel loci that were significantly associated with SLE (p < 0.05); the strongest signal was from rs933717 of MAP1LC3B (Pdiscovery = 7.57 × 10−3, Preplication1 = 1.42 × 10−8), which approached genome-wide significance (5 × 10−8, Table 1). Therefore, rs933717 was replicated in a second independent cohort (Preplication2 = 2.22 × 10−5), and reached genome-wide significance when the cohorts were combined (P = 2.36 ×10−10) (Table 1).
Table 1.
Associations of MAP1LC3B rs933717 with susceptibility to SLE in a Chinese population
| SNP | Chr | Minor allele | Stage (n: case/control) | MAF (Case/Control) | p | Odds ratio (95%CI) |
|---|---|---|---|---|---|---|
| rs933717 | 16 | T | Discovery (490/493) | 0.20/1.22 | 7.57 × 10−3 | 0.17 (0.04–0.74) |
| Replication1 (2666/7180) | 0.13/0.88 | 1.42 × 10−8 | 0.15 (0.07–0.32) | |||
| Replication2 (1366/2279) | 0.02/0.75 | 2.22 × 10−5 | 0.05 (0.01–0.36) | |||
| Combined (4522/9952) | 0.11/0.87 | 2.36 × 10−10 | 0.13 (0.07–0.24) | |||
| rs12574250 | 11 | A | Discovery (490/493) | 5.20/7.71 | 2.39 × 10−2 | 0.66 (0.46–0.95) |
| Replication1 (2666/7180) | 5.95/7.49 | 1.94 × 10−4 | 0.78 (0.69–0.89) | |||
| Combined (3156/7673) | 5.83/7.51 | 2.24 × 10−5 | 0.77 (0.68–0.87) | |||
| rs11658979 | 17 | G | Discovery (490/493) | 10.82/14.20 | 2.34 × 10−2 | 0.73 (0.56–0.96) |
| Replication1 (2666/7180) | 11.74/12.22 | 0.36 | 0.96 (0.87–1.05) | |||
| Combined (3156/7673) | 11.60/12.35 | 0.10 | 0.93 (0.85–1.02) | |||
| rs2155388 | 11 | A | Discovery (490/493) | 7.55/10.75 | 1.39 × 10−2 | 0.68 (0.50–0.93) |
| Replication1 (2666/7180) | 8.08/8.67 | 0.19 | 0.93 (0.83–1.04) | |||
| Combined (3156/7673) | 8.00/8.80 | 0.04 | 0.89 (0.80–0.99) |
CI, confidence interval; MAF, minor allele frequency; SNP, single-nucleotide polymorphism. Combined p-values from Cochran’s and Mantel-Haenszel statistics. A Hardy–Weinberg equilibrium test was performed in the controls of all three stages. For ImmunoChip, the p threshold with Bonferroni correction was < 2.6 × 10−7 when considering 192,194 variants.
Bioinformatic functional annotations of MAP1LC3B rs933717
eQTL analysis
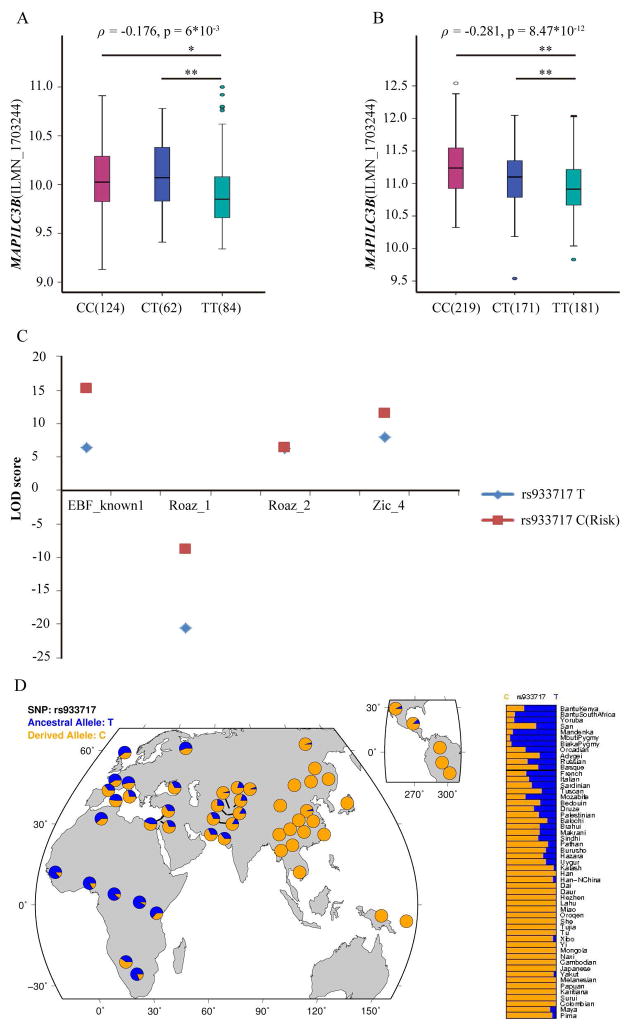
MAP1LC3B rs933717 was annotated to be eSNPs in ENCODE, as it showed that rs933717CC (risk) correlated with increased MAP1LC3B expression (p=6*10−3) in Epstein-Barr virus-transformed lymphoblastoid cell lines (EBV-LCL) from 270 HapMap samples in GENEVAR database (Figure 2A). With a larger sample size, this association may be even stronger: array quantification of 618 individuals from 7 HapMap3 populations (p = 8.47 × 10−12, Figure 2B), GTEx2015_v6project (p = 1.55 × 10−7, 24) as well as a genome-wide eQTL analysis in whole peripheral blood from 1,469 unrelated individuals (p = 9.7 × 10−9, 25). In addition, risk genotypes of rs933717 were significant correlated with higher expression of several autophagy-related genes, suggesting trans-QTL effects (Supplementary Table 4).
Figure 2. Series of bioinformatics analyses supporting the functional role of MAP1LC3B rs933717.
P < 0.01 is marked as * and p < 0.001 is marked as **.
(A) eQTL analysis in the GENVAR database.
The expression data in transformed B-cell lines were a pool of 4 populations from healthy HapMap samples including US individuals with European ancestry (CEU), Han Chinese individuals from Beijing (CHB), Yoruba individuals in Ibadan, Nigeria (YRI) and Japanese individuals in Tokyo, Japan (JPT).
(B) eQTL analysis in HapMap3 individuals.
The lymphoblast expression data consisted of a pool of 618 individuals from 7 HapMap3 populations including 80 CHB, 82 JPT, 108 YRI, 82 Gujarati Indians in Houston (GIH), 83 Luhya in Webuye, Kenya (LWK), 45 Mexican ancestry in Los Angeles (MEX) and 138 Maasai in Kinyawa (MKK). A total of 47 individuals were not successfully genotyped at rs933717, leaving 571 individuals for further analysis.
(C) Transcription factors prediction
Altering the rs933717 allele from T to C increased the binding affinity for transcription factors EBF_known1, Roaz_1, Roaz_2 and Zic_4.
(D) Genetic signatures of natural selection in the human genome.
Detailed global allele frequency distribution of rs933717 in 53 world populations.
y-axes of Figures 2A and B denotes MAP1LC3B expression in gene expression chips. β = partial regression coefficient. Numbers in parentheses in the x-axis labels denotes the sample size of peoples with the genotype. ILMN_1703244 denotes the probe in gene expression chip (Illumina Inc).
Prediction of regulatory function
The in-silico bioinformatics analysis with ENCODE data demonstrated that rs933717 resided in a transcription factor (TF) binding region. Four binding site motifs span the rs933717 region for binding by the following TFs: EBF_known1, Roaz_1, Roaz_2 and Zic_4. The differences between the LOD scores for the alleles T and C (risk) were 8.8, 11.9, 0.2 and 3.7 for EBF_known1, Roaz_1, Roaz_2 and Zic_4, respectively (Figure 2C). The data showed that rs933717 could affect transcriptional enhancer activity, suggesting that rs933717 might reside in an important region for gene transcriptional regulation (Supplementary Table 5, 6 and 7).
Genetic signatures of natural selection in the human genome
We further investigated the detailed global allele frequency distribution of rs933717 in 53 populations. Intriguingly, the risk allele rs933717C showed regional enrichment: the highest frequencies (~100%) were found in East Asia, South Asia and Southeast Asia, followed by the Middle East and Europe and the lowest were found in Africa (Figure 2D). Population selection analysis indicated that standardized integrated haplotype scores (iHS) were 1.781 in northern and western European (CEU) and 1.477 in Yoruba (YRI); and Fay and Wu’s H+ was −13.663 in CEU and −3.449 in YRI. Neither the iHS nor Fay and Wu’s H+ were available in an East Asian population (26, 27).
The risk allele ‘C’ at rs933717 showed increased reporter gene activity
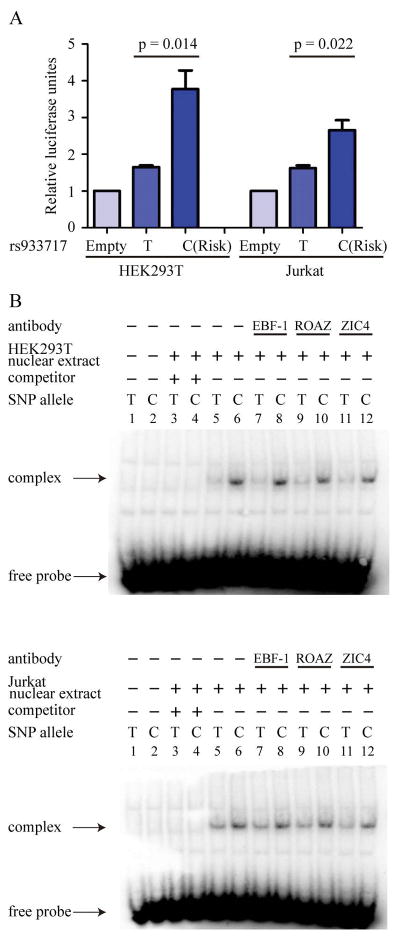
The effects of the variant-containing sequences on gene expression were assessed by luciferase reporter assay, in which 110pb around rs933717 were cloned into pGL4.23 (luc2/minP) reporter vectors and transfected into HEK293T and Jurkat cells. We observed that the presence of the risk allele ‘C’ of rs933717 significantly increased luciferase activity from 1.7-fold to 3.8-fold (p = 0.014) in HEK293T cells and from 1.6-fold to 2.7-fold (p = 0.022) in Jurkat cells (Figure 3A).
Figure 3. Experimental validation of bioinformatics analysis.
(A) Luciferase reporter assay
Luciferase reporter assays of the transcriptional activity of rs933717 in HEK293T and Jurkat cell lines, presented as means ± SEM. The luciferase activity of rs933717C was significantly greater than of the rs933717T protective allele in both cell lines (p < 0.05).
(B) EMSA
Alleles of rs933717 showed different binding affinities for HEK293T and Jurkat cell lines. EMSA showed complex formation after addition of HEK293 and Jurkat nuclear extract (lanes 5 and 6). After competition with a 200-fold excess of unlabeled probes, the complex was eliminated (lanes 3 and 4). ROAZ, EBF1 and ZIC4-specific antibodies were not observed to super-shift the complex formation using either the C or T probe (lanes 7–12). The DIG-labeled C probe showed a higher affinity for nuclear protein-DNA complex (lanes 5–12).
Differential binding of nuclear extract at rs933717
Evidence of enhancer activity was provided by EMSA: risk allele C of rs933717 containing oligonucleotides showed a greater affinity for nuclear protein-DNA complex with HEK293T and Jurkat cell nuclear extract than protective allele T of rs933717 containing oligonucleotides (Figure 3B, lanes 5 and 6). The nuclear protein-DNA complex was abolished by addition of excessive unlabeled competitor probes (Figure 3B, lanes 3 and 4). In silico analysis using the JASPAR, TransFac, and MAPPER databases predicted binding at the transcription factor binding sites for ROAZ, EBF-1, and ZIC4. However, in the supershift assay, ROAZ-, EBF-1–, and ZIC4-specific antibodies were not observed to supershift the complex formation with either an rs933717 C probe or an rs933717 T probe (Figure 3B, lanes 7–12). In contrast, in the same 2 EMSA experiments performed with HEK293T and Jurkat cell-derived nuclear samples, the DIG-labeled C probe showed a higher affinity for the nuclear protein-DNA complex (Figure 3B, lanes 5–12). This also indicated that rs933717C differed from rs933717T in formation of the nuclear protein-DNA complex.
Autophagy is increased in SLE patients and in the lymph nodes of lupus-prone mice
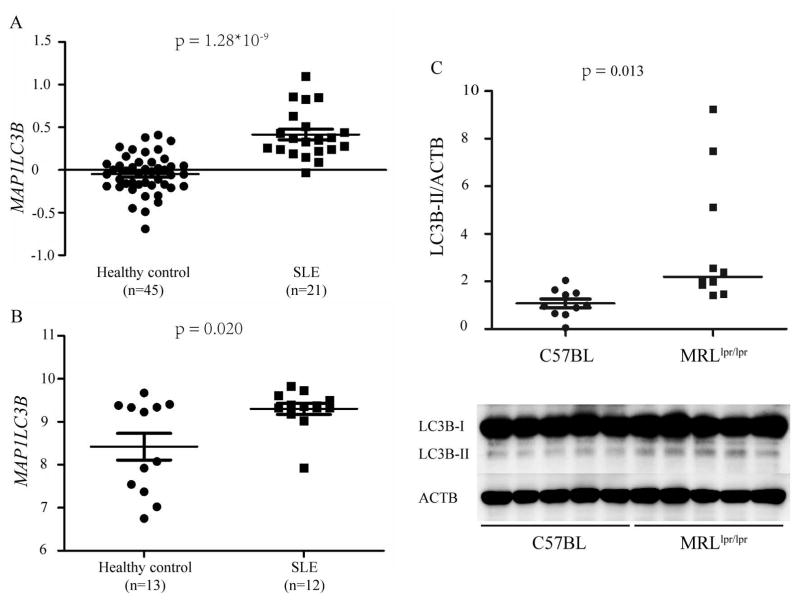
Consistent with the observation that risk alleles are associated with higher levels of gene expression, increased MAP1LC3B expression was also observed in patients with SLE. We used mRNA expression data from a comparatively large project focusing on the gene expression profiling of PBMC (E-GEOD-20864) from 21 SLE vs. 45 healthy individuals in a GEO database. This study found significantly elevated levels of MAP1LC3B expression in SLE patients compared to normal controls (p = 1.28 × 10−9, Figure 4A). The result was successfully replicated (in an independent cohort enrolled in our own center) by expression microarray conducted using an Illumina HT-12 v4 Expression BeadChip. Compared with the 12 healthy controls, the level of MAP1LC3B expression was significantly higher in the 13 SLE patients (p =2.00×10−2) (Figure 4B).
Figure 4. Autophagy was increased in SLE patients and in lymph nodes from MRLlpr/lpr mice.
(A) MAP1LC3B was significantly greater in SLE patients according to GEO data MAP1LC3B mRNA expression in the PBMC of 21 SLE patients and 45 healthy individuals was analyzed based on E-GEOD-20864 from Array Express. E-GEOD-20864 is a comparatively large project focusing on the gene expression profiling of PBMC from SLE vs. healthy individuals.
(B) MAP1LC3B was significantly greater in SLE patients in our own cohort 12 healthy controls and 13 SLE patients were enrolled and the expression microarray conducted by Illumina HT-12 v4 Expression BeadChip.
(C) Autophagy was significantly greater in lymph nodes taken from MRLlpr/lpr mice. Blots shown were representative of independent experiments performed in 10 C57BL mice and 10 MRLlpr/lpr mice. The densitometry analysis of LC3B-II levels relative to ACTB is shown in the scatterplot with the line indicating the median level. LC3B-II was significantly greater in the lymph nodes from MRLlpr/lpr mice than the C57BL control mice (p = 0.013).
Autophagy was quantified in lymph nodes obtained from MRLlpr/lpr lupus-prone mice and C57BL healthy control mice by LC3 immunoblot. The cellular autophagy response was assessed using LC3-II normalized to ACTB, the most widely used index. The level of autophagy was significantly higher in the lymph nodes from MRLlpr/lpr mice than healthy control mice (p = 0.013, Figure 4C).
DISCUSSION
In the current study, we conducted various analyses. First, we carried out a gene family-based genetic association involving 26 autophagy-related genes. Although ImmunoChip was designed for dense mapping of immune-related genes instead of autophagy pathway analysis, an increasing number of studies highlighted its power for detecting previously-overlooked susceptibility loci (23). Also, the ImmunoChip design included several rare variants considered to have significant functional effects which might have been previously overlooked.
We observed that rs933717 (MAP1LC3B) was associated with susceptibility to SLE in more than 10,000 Chinese individuals with genome-wide significance. Of special note, rs933717 was found to be associated with susceptibility to age-related cataracts in a GWAS using 530,101 SNPs from the Illumina 660W-Quad in a total of 7,397 individualsin eMERGE (electronic medical records and genomics) network study. However, the p value was 0.000041, suggesting that a large sample size may be critically necessary. We found data to suggest that the T allele has a protective function, consistent with this previous report (28). In contrast, expression analysis suggested a likely pathogenic role of increased MAP1LC3B expression in SLE patients, which might be because of an allele-dependent effect of risk allele rs933717C. To identify possible molecular mechanisms, functional annotations were performed using ENCODE data, which supported a likely regulatory feature of these variants. To confirm these bioinformatic clues, an in-house functional luciferase reporter assay and EMSA were carried out. Both supported an enhancer role of the rs933717 region, and LC3B-II was higher in lupus-prone mice than control mice. Taken together, the results from genetic association, expression association and functional validation tests strongly suggest that MAP1LC3B rs933717 is involved in SLE etiology.
Microtubule-associated protein LC3B is encoded by the MAP1LC3B gene. LC3B is a central protein in autophagy since it is involved in cargo recruitment into, biogenesis and maturation of autophagosomes. More importantly, LC3B is the most widely-used autophagosome marker (29). In our present study, expression analysis suggested a likely pathogenic role of increased LC3B in SLE patients. Further animal experiments also showed that LC3B was higher in lupus-prone mice than in healthy control mice, corroborating the current observation that higher levels of LC3B were consistently observed in lupus-prone mice models and SLE patients (5–7). Following these observations, it further suggested that autophagy is genetically associated with SLE. Our data suggested that rs933717 of MAP1LC3B could increase the expression of LC3B in immune cells by both cis- and trans-eQTL effects. Genetic association, expression association and functional validation highlight the likely important role of rs933717 in Chinese SLE patients. However, the associations were not able to determine whether rs933717 was the single causal variant or whether it was just in linkage disequilibrium with a nearby variant. Thus, further fine-mapping analysis is needed.
SLE is an autoimmune disease and characterized as the abnormality of immune system including antigen presentation by antigen presenting cells (APCs), the survival of autoreactive T/B cells which lead to the production of large amount of autoantibodies. Recent studies indicated that autophagy was involved in the presentation of intracellular antigens to major histocompatibility complex (MHC) class II molecules (30, 31). Higher rates of autophagy led to a more active MHC presentation process which might lead to the breakdown of immune tolerance and an increase of the immune response. In addition, increased autophagy would promote the survival of autoimmune T/B cells, which in turn would lead to high levels of autoantibody and cytokine production and multiple organ/system involvement (32–39). Thus, high levels of autophagy might promote the initiation of autoimmunity in SLE by influencing multiple aspects of the immune system. More importantly, it was recently reported that LC3-associated phagocytosis controls immune responses to dying cells and its inhibition leads to the development of SLE-like disease. Our data reinforced the notion that LC3 might be genetically involved in SLE (9).
We included SNP mapping to autophagy-related genes along with 20kb upstream and downstream. Although rs933717 was annotated to be an intronic variant of FBXO31, its associated, rSNP-related, genes included MAP1LC3B (proximal transcriptional regulation) and FBXO31 (distal transcriptional regulation and RNA binding protein-mediated regulation) (Ensemble: http://asia.ensembl.org/index.html; and rSNPBase: http://rsnp.psych.ac.cn/). It is hard to determine the exact causal variant and gene in this kind of strategy of forward genetic association. In most cases, GWAS implicated disease-susceptible loci rather than directly identifying disease-associated genes. This is because many of the disease-associated SNPs were located in an intergenic region with the susceptible gene, or the functional locus was located within a region near to the SNP, which often contained multiple genes. We thus also analyzed the differential expressions of FBXO31 between SLE patients and healthy controls using data from GEO and from our own cohort. We found no significant difference in expressions of FBXO31 (GEO GSE20864 data p = 0.092; data from our center p = 0.343). Thus, rs933717 was more likely to be involved in the pathogenesis of SLE by influencing MAP1LC3B. F-box proteins, in conjunction with Skp1, Cul1 and Rbx1, generate F-box containing complex (or SCF complex) that are responsible for the ubiquitination of proteins, leading to their activation or degradation. The F-box protein FBXO31 is required for normal mitotic progression and genome stability due to its role in regulating FOXM1 levels during the G2/M transition. Therefore, future study targeting FBXO31 is also of interest.
Genetic associations between autophagy gene variants was pronounced in Asian populations, as most autophagy genetic associations could be replicated in our previous reports (2, 13, 14, 20). However, few of these genetic associations were reported in Caucasians or Africans, suggesting genetic heterogeneity and that the autophagy gene has a major genetic impact in Asian populations (18, 21). The rs933717 variant is seen in Asians but is more common in Caucasians, which might be because of natural selection. However, the mechanisms behind how this kind of natural selection affects autophagy and SLE epidemiology needs to be further investigated, including independent replications from other populations (23).
In the current study, differential effects of nuclear protein binding with the rs933717 C/T allele were observed, which may support the notion that rs933717 influences the expression of MAP1LC3B. However, the exact transcription factor was not clearly determined as it was not validated by ROAZ or EBF1 antibody-specific super-shift assay. Further exploration to seek the binding factor, such as CHIP-on-chip or ChIP-seq, or in other cell lines, is still warranted.
In summary, we systemically evaluated genes involved in autophagy in a large sample by a four-stage strategy, including genetic discovery, genetic replication, bioinformatic analysis and experimental validation. We observed likely novel genetic associations between LC3B (encoded by MAP1LC3B), an excellent marker for autophagic structures, and susceptibility to SLE. Our work was committed toward a more complete understanding of SLE pathogenesis, especially focusing on autophagy. A deeper knowledge of the relationship between autophagy and SLE is likely to lead the discovery of new therapeutic targets.
Supplementary Material
Acknowledgments
We thank all the members of our laboratory for their technical assistance. We also thank the patients, their families and healthy donors for their cooperation and for giving consent to participate in this study.
FUNDINGS
This work was supported by the National Science Foundation of China [grant number 81570629]; the National Key Research and Development Program of China [grant number 2016YFC0904102]; the Training Program of the Major Research Plan of the National Natural Science Foundation of China [grant number 91642120]; the Natural Science Foundation for Innovation Research Group of China [grant number 81321064]; the Capital of Clinical Characteristics and the Applied Research Fund [grant number Z141107002514037]; the Beijing Natural Science Foundation [grant numbers 7152148, 7131016], the Chinese Society of Nephrology [grant number 15020030591], the Beijing Nova Program [grant number 2017019], and the US National Institutes of Health [grant numbers AR060366, MD007909]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICT OF INTEREST STATEMENT
None declared.
References
- 1.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;9:929–39. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 2.Molineros JE, Yang W, Zhou XJ, Sun C, Okada Y, Zhang H, et al. Confirmation of five novel susceptibility loci for Systemic Lupus Erythematosus (SLE) and integrated network analysis of 82 SLE susceptibility loci. Hum Mol Genet. 2017;26:1205–16. doi: 10.1093/hmg/ddx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;7330:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;10:722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alessandri C, Barbati C, Vacirca D, Piscopo P, Confaloni A, Sanchez M, et al. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J. 2012;11:4722–32. doi: 10.1096/fj.12-206060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gros F, Arnold J, Page N, Decossas M, Korganow AS, Martin T, et al. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy. 2012;7:1113–23. doi: 10.4161/auto.20275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis. 2015;5:912–20. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandyopadhyay U, Overholtzer M. LAP: the protector against autoimmunity. Cell Res. 2016;8:865–6. doi: 10.1038/cr.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;7601:115–9. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;42:17396–401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Yue Y, Dong C, Shi Y, Xiong S. Blockade of macrophage autophagy ameliorates activated lymphocytes-derived DNA induced murine lupus possibly via inhibition of proinflammatory cytokine production. Clin Exp Rheum. 2014;5:705–14. [PubMed] [Google Scholar]
- 12.International Consortium for Systemic Lupus Erythematosus G. Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;2:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH, et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheumatic Dis. 2011;7:1330–7. doi: 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- 14.Zhou XJ, Nath SK, Qi YY, Cheng FJ, Yang HZ, Zhang Y, et al. Brief Report: identification of MTMR3 as a novel susceptibility gene for lupus nephritis in northern Han Chinese by shared-gene analysis with IgA nephropathy. Arthritis Rheumatol. 2014;10:2842–8. doi: 10.1002/art.38749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YM, Cheng FJ, Zhou XJ, Qi YY, Zhao MH, Zhang H. Rare Variants of ATG5 Are Likely to Be Associated With Chinese Patients With Systemic Lupus Erythematosus. Medicine (Baltimore) 2015;22:e939. doi: 10.1097/MD.0000000000000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Tang H, Zhang Y, Tang X, Zhang J, Sun L, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet. 2013;1:41–51. doi: 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;7:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;2:390–6. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, et al. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity. 2015;5:942–52. doi: 10.1016/j.immuni.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YM, Zhou XJ, Cheng FJ, Qi YY, Hou P, Zhao MH, et al. Autophagy-related gene LRRK2 is likely a susceptibility gene for systemic lupus erythematosus in northern Han Chinese. Oncotarget. 2017;8:13754–61. doi: 10.18632/oncotarget.14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;11:1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1997;9:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 23.Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet. 2016;3:323–30. doi: 10.1038/ng.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;6235:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;8:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;3:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Scott LJ, Bonnycastle LL, Jackson AU, Chines P, Pruim R, et al. Tag SNP selection for Finnish individuals based on the CEPH Utah HapMap database. Genet Epidemiol. 2006;2:180–90. doi: 10.1002/gepi.20131. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie MD, Verma SS, Hall MA, Goodloe RJ, Berg RL, Carrell DS, et al. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol Vis. 2014:1281–95. [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;1:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwart W, Griekspoor A, Kuijl C, Marsman M, van Rheenen J, Janssen H, et al. Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity. 2005;2:221–33. doi: 10.1016/j.immuni.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;5:1250–9. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 32.Zhou XJ, Zhang H. Autophagy in immunity: implications in etiology of autoimmune/autoinflammatory diseases. Autophagy. 2012;9:1286–99. doi: 10.4161/auto.21212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou XJ, Cheng FJ, Zhang H. Emerging view of autophagy in systemic lupus erythematosus. Int Rev Immunol. 2015;3:280–92. doi: 10.3109/08830185.2013.879711. [DOI] [PubMed] [Google Scholar]
- 34.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;1:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arsov I, Li X, Matthews G, Coradin J, Hartmann B, Simon AK, et al. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death Differ. 2008;9:1385–95. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy. 2007;3:266–7. doi: 10.4161/auto.3908. [DOI] [PubMed] [Google Scholar]
- 37.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;7:4046–55. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 38.Arsov I, Adebayo A, Kucerova-Levisohn M, Haye J, MacNeil M, Papavasiliou FN, et al. A role for autophagic protein beclin 1 early in lymphocyte development. J Immunol. 2011;4:2201–9. doi: 10.4049/jimmunol.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;6:986–97. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.