Abstract
Obesity is an excess accumulation of adipose tissue mass, and, together with its sequelae, in particular type II diabetes and metabolic syndrome, obesity presents a major health crisis. Although obesity is simply caused by increased adipose mass, the heterogeneity of adipose tissue in humans means that the response to increased energy balance is highly complex. Individual subjects with similar phenotypes may respond very differently to the same treatments; therefore, obesity may benefit from a personalized precision medicine approach. The variability in the development of obesity is indeed driven by differences in sex, genetics, and environment, but also by the various types of adipose tissue as well as the different cell types that compose it. By describing the distinct cell populations that reside in different fat depots, we can interpret the complex effect of these various players in the maintenance of whole-body energy homeostasis. To further understand adipose tissue, adipogenic differentiation and the transcriptional program of lipid accumulation must be investigated. As the cell- and depot-specific functions are described, they can be placed in the context of energy excess to understand how the heterogeneity of adipose tissue shapes individual metabolic status and condition.
Keywords: adipose tissue, cellular heterogeneity, adipocyte, preadipoctye
Introduction
Currently, the U.S. Centers for Disease Control and Prevention estimate that more than half of the U.S. population is overweight or obese and almost one in every five children is obese.1,2 This epidemic is occurring worldwide, as more than 1 billion adults worldwide are overweight and over 300 million people rank as obese. Obesity leads to a broad spectrum of other sequelae including insulin resistance, type 2 diabetes, and atherosclerosis, which collectively form the metabolic syndrome.3 The diversity of pathologies that arise with increased adiposity underscores the many different roles that adipose tissue plays in normal physiology and, as we will highlight, the different cell types that reside in fat. Strikingly, almost 20 million Americans have type 2 diabetes, and over 40 million people in the United States have metabolic syndrome, sharing a collection of abnormalities that each generate risk for the others and present clinically as many of our most common medical disorders, including dyslipidemias, non-alcoholic fatty liver, cardiovascular disease, renal failure, and even some cancers.4,5
To characterize obesity as simply an increase in adipose tissue mass is an oversimplification. In humans, obesity-associated morbidity and mortality are linked to both fat accumulation and fat distribution. Increased fat mass can be caused by increased adipocyte size (i.e., hypertrophy), increased adipocyte number (i.e., hyperplasia), or both. Individuals with adult-onset obesity commonly display a hypertrophy phenotype, whereas individuals with early-onset obesity exhibit both adipocyte hypertrophy and hyperplasia.6 Fat distribution also contributes to the intersubject variability of obesity and plays an important role in metabolic risk. Central obesity (i.e., apple-shape obesity), characterized by excessive intra-abdominal/visceral fat accumulation, is associated with high risk for the development of insulin resistance, diabetes, and metabolic syndrome. By contrast, increased subcutaneous fat accumulation (i.e., pear-shape obesity) exhibits low risk for metabolic disorders.7 Recently, large-scale meta-analyses have started to map out the genetic loci and suggest potential pathways that are associated with fat accumulation and distribution.8–10
In addition to interindividual differences, diverse cellular compositions within fat tissue also contribute to the heterogeneity of human adipose tissue and linkage with the complex functions. In many tissues, heterogeneous populations of cell types interact to maintain homeostasis and perform the physiologic role of the tissue. Some cellular functions and processes can be shared among different cell types; however, others are exclusive to a single type of cell within the organ. All tissues, for example, contain blood vessels composed of endothelial and smooth muscle cells. It is well recognized that different subpopulations of neurons exist in the brain,11 and different fiber types constitute the skeletal muscle.12 Different cell types in the pancreas are specialized to secrete specific hormones, and subpopulations of these distinct cell types are thought to exist.13,14 Adipose tissue is no exception, and the heterogeneous nature of adipose tissue and a variety of roles for the cells that compose this tissue in its various depots throughout the body have been reported.15,16 In rodents, in addition to the heterogeneity of cell types within the adipose depot, cell-intrinsic features, such as cell size and level of insulin signaling, contribute to depot-specific endocrine and metabolic functions.17–20. Understanding the interplay among different cells types within adipose tissue will be key to defining the specific roles of individual depots in human health and disease.
Heterogeneity of adipose depots: functions and locations
Functional differences among adipose depots
While the key feature of adipose tissue is energy is stored in the form of lipid droplets that aggregate inside cells,21–23 not all fat depots are created equal. Metabolic syndrome may also stem from specific adipose tissue dysfunction.16 There are two functionally different types of adipose tissues: white adipose tissue (WAT), which is the main site of energy storage, and brown adipose tissue (BAT), which is dedicated to thermogenesis and energy expenditure24 by virtue of the expression of a mitochondrial protein called uncoupling protein 1 (UCP1),27 which acts as a proton-leak channel in the inner mitochondrial membrane through which the proton gradient generated by the electron transport chain can be dissipated as heat rather than used to drive adenosine triphosphate (ATP) synthesis.28 These two types of adipocytes differ in cellular lipid morphology. Energy-storing WAT depots are dominated by adipocytes that contain a single, unilocular droplet, while energy-expending BAT contains cells with many smaller droplets in a multilocular pattern.25 By storing lipid droplets with higher surface area, access for water-soluble enzymes to their substrates is increased, allowing increasing lipolytic rates,26 for example during cold exposure.
In addition to the classical brown adipocytes, a second cell type called brown-like beige/brite adipocytes or recruited adipocytes (referred to as beige adipocytes hereafter) arise in WAT depots during prolonged cold exposure or β3 adrenergic stimulation. They are distinguished from white adipocytes by their multilocular lipid droplets and share many features with the classical brown adipocytes, such as high mitochondrial content and expression of UCP1. In rodents, brown and beige adipocytes arise from distinct cellular lineages,29 and several studies have shown transcriptional differences between them that might drive differential impacts on metabolism,30 certainly quantitatively but also qualitatively. For example, UCP1 expression is highly inducible in beige adipocytes, whereas it is expressed at constitutively high levels in brown adipocytes.31,32 In humans, the adipose tissue commonly referred to as BAT seems to be a mix of these two cells types;33 however, since they both share the function of thermogenesis and therefore have high metabolic rates, discrimination between them by current techniques to measure substrate uptake is not possible. What is known is that, in humans, BAT mass is negatively correlated with body mass index, and BAT activation can improve glucose homeostasis,34,35 while in rodent models BAT activation can improve lipid metabolism and ameliorate atherosclerosis.36,37
Depot-specific heterogeneity
Recent data have suggested that different fat depots located in different anatomical locations of body appear to have distinct cellular compositions and diverse functions.16,38,39 In humans, fat depot–specific differences in function are clinically relevant owing to the observation that increased abdominal white fat is associated with insulin resistance,40 while subcutaneous white adipose tissue exerts a protective effect against metabolic syndrome.41–43 BAT is mainly located in the interscapular region in rodents, while, in adult humans, brown/beige fat is mainly clustered around the neck, clavicle, spinal cord, and perirenal regions.44 Clearly, the health and size of the tissues in which the body chooses to store energy as lipids have an impact on whole-body metabolism and health.
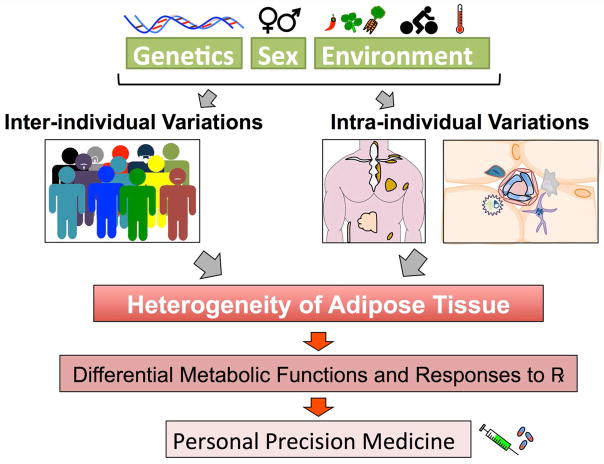
To begin to understand the differences between different depots of adipose tissue in humans, transcriptional profiling of whole adipose tissue has been used. Before even considering the differences in gene expression, it is important to remember that many factors, such as genetic background of the individual and the depots sampled, can contribute to the gene expression patterns observed in whole-tissue lysates of human tissues (Fig. 1). With these considerations in mind, all comparisons of different depots seem to identify a signature of developmental genes45–47 that are differentially regulated among different fat depots. For example, expression of the genes Short Stature Homeobox 2 (Shox2), Engrailed 1 (En1), T-Box 15 (Tbx15), Homeobox 5 (Hoxa5), Hoxc8, Hoxc9 and Glypican 4 (Gpc4) is depot specific in mice48 and humans,45,49 while the expression of general adipogenic markers proliferator-activated receptor γ (PPARγ) and the CCAAT/enhancer-binding proteins α, β, and δ (C/EBPs) is not. Furthermore, Tbx15, Hoxa5 and Gpc4 expression in adipose tissue correlate with body mass index,45 which means that the developmental signature in each individual fat depot may play a role in controlling its growth and function. It is not hard to imagine, given the broad distribution of adipose tissue throughout the body, that these gene signatures are a relic of the patterning programs that direct development of progenitor cells to the sites of the different fat pads. Underscoring the idea that different fat depots are derived from distinct pools of progenitors, studies in mice to trace progenitor cells have shown that BAT but not WAT is largely composed of adipocytes that arise from a myogenic factor 5 (MYF5)+ early progenitor cell shared with skeletal muscle.29 Guertin’s group went on to carefully quantify the number of adipocytes arising from this lineage in each depot and found that no two depots were similar; each adipose tissue depot had a differently sized population of MYF5+ and MYF5− adipocytes.50 All of this information serves to highlight the fact that interpretation of whole-tissue transcriptional profiling is limited by one main issue in adipose tissue: cellular heterogeneity.
Figure 1.
The heterogeneity of adipose tissue has different drivers. The genetic background of different individuals can synergize with differences in cellular heterogeneity as well as variation in adipose tissue depot size and function. A person’s adipose tissue would then drive a distinct metabolic response to physiologic stimuli, such as diet, but also to therapeutic intervention (Px). This may require a personalized medicine approach to treat metabolic disease and indeed instruct decisions in the diet.
Cellular heterogeneity of adipose tissue
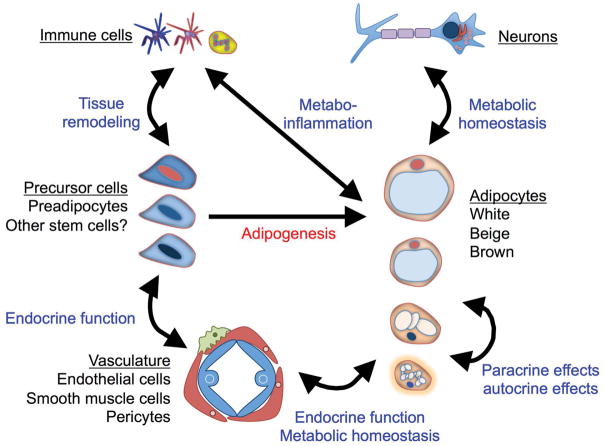
Adipocytes are mature, lipid-laden cells that are found in individual, discrete depots distributed throughout the body; however, this is by no means always the case, as adipocytes can also be found in non-adipose tissue, such as bone, liver, skin, muscle, and heart.51–54 In each adipose tissue depot, many other different cell types comingle with adipocytes, including cells from the immune system, vascular system, and nervous system (Fig. 2). Although other cells make a significant contribution to the heterogeneity of individual adipose tissue depots,55 in terms of maintaining energy balance and adiposity, the mature adipocytes are most important. Adipose tissue also plays a critical role in the endocrine system; however, this can also be complex, as certain hormones are specifically secreted by mature adipocytes from certain depots, which further highlights the diverse functions of adipose tissue.38,56–59 Since the majority of adipose tissue by volume is adipocytes, their biology, especially as it relates to energy storage, has been a prime focus. Adipocytes also have a major influence on the expression profile of whole adipose tissue, and, since tissue lysates are often used as a sample, especially in human studies, the transcriptional regulation of these cells is critical to understand.
Figure 2.
Cellular heterogeneity of adipose tissue. Different cell types interact in adipose tissue to maintain homeostasis. To generate and maintain a pool of mature adipocytes, different precursor populations (depicted as slightly different shades with different color nuclei) are maintained in a stem cell niche, where they can react to different cues that regulate their adipogenic differentiation. The stem cell niche is located in close proximity to the vasculature, where it can respond to endocrine signals that can also modulate the function of mature adipocytes. Cells from the immune system are critical for tissue remodeling and also regulate the inflammatory milieu of adipose tissue response to energy balance in a process called metabo-inflammation. Finally, different kinds of mature adipocytes arise from the distinct progenitor cell pools and can be acutely activated by neurons to control thermogenesis and lipolysis.
Currently, three different adipocyte cell types have been described (white, brown and beige); however, there may be others. It is possible that there are subgroups of each type of adipocyte that are further specialized for certain functions such that the overall contribution of the depot is a reflection of the population of different adipocytes.
Preadipocytes serve as a pool for replenishing mature adipocytes during normal tissue turnover or in response to stimuli. In humans, it has been estimated that approximately 10% of the mature adipocyte population turn over each year60 and must be supported by a population of preadipocytes, supported in turn by a niche. Adipocyte turnover in rodents is considerably faster, with as many as 5% of adipocytes replaced daily.61 Generally speaking, preadipocytes are described as committed adipocyte progenitor cells; however, it is clear that there is more than one stage to this commitment.62,63 Whether distinct subclasses of preadipocytes are specifically responsible for secretion of specific hormones is unknown, as the markers that define specific substages of differentiation have only begun to be defined. In addition to cells at different stages of differentiation, preadipocyte subpopulations that preferentially differentiate into white, beige, or brown adipocytes result in a broad diversity of immature cells.62,64 Recent studies have identified cell surface markers that delineate between human brown, white, and beige adipocytes,65 and significant effort has focused on identifying specific markers that identify preadipocyte populations predisposed to differentiate into each of these cell types. Work from our lab identified the cell surface marker integrin β1 (CD29) as a predictor of thermogenic differentiation and, by sorting preadipocytes with high CD29 expression, we effectively enriched for a population of cells that differentiated into mature adipocytes with high UCP1 expression.33 The exact numbers and cell-intrinsic functions of these different subpopulations seems to be depot specific, and they have been studied extensively.66–70 Several putative preadipocyte populations share a characteristic close proximity to vascular cells,71–73 which will be discussed further below. Preadipocytes also interact with cells from the immune system during times of tissue remodeling, which in adult life occurs during energy imbalance, which will also be addressed later in this review. Finally, preadipocytes are known to be the major source of certain adipose tissue derived secreted factors, such as plasminogen activator inhibitor 1 (PAI-1)74 and tumor necrosis factor α(TNF-α),75 and thus the number and function of preadipocytes is critical to endocrine function of the adipose tissue depot.
The presence of immune cells in adipose tissue has been long appreciated, but the links between adipose tissue function and the immune system are complicated, and our understanding is constantly evolving. Adipose tissue resident macrophages were the first immune cells observed in adipose tissue and have been associated with obesity for more than a decade, although the interplay between specific subpopulations of macrophages has recently become more thoroughly appreciated.76–78 Considerable evidence supports the presence of at least two distinct macrophage populations: classically activated, proinflammatory M1 macrophages and alternatively activated, anti-inflammatory M2 macrophages.79,80 Natural Killer (NK) cells are also recruited to adipose tissue in obese states and drive insulin resistance.81 Presumably, this switch to a proinflammatory cell population is largely responsible for the increased cytokines and inflammatory gene expression profile from obese adipose tissue that characterizes metabo-inflammation.82–84 Macrophages are not the only immune cell in adipose tissue,85 and regulation of inflammation is not the only role the immune system plays in adipose tissue. Tissue remodeling, for example during prolonged cold exposure or β-adrenergic stimulation, recruits a subset of innate immune cells, including M2 macrophages, eosinophils, and the type 2 cytokines interleukin-4 and interleukin-13 (IL-4 and IL-13), which are required for activation of beige adipogenesis.86–88 Other noncanonical type 2 cytokines, such as IL-33, are required for brown adipogenesis; however, the involvement of immune cells in these contexts may be even more complex.89
One of the major functions of adipose tissue is lipolysis, which increases the availability of free fatty acids. Metabolic challenges, such as a cold environment, stimulate sympathetic neural efferent activity to WAT, driving lipolysis to release stored energy for use in other tissues and thereby increasing the availability of free fatty acids as one source of fuel for BAT thermogenesis. Lipolysis is largely driven by increased sympathetic input, so it is not difficult to imagine neurons as part of the adipose tissue milieu.9. Intriguingly, recent work has defined a neural circuit that also senses these fatty acids and feeds back to increase BAT thermogenesis.91 This circuit may reinforce the effect of cold-sensing neurons that activate BAT and increase energy expenditure.90 WAT is also innervated by sensory nerve fibers that form networks with metabolic brain areas; moreover, activation of these afferents is reported to increase sympathetic nervous system outflow.92 However, the endogenous stimuli sufficient to drive WAT afferents during metabolic challenges, as well as their functional relation to BAT thermogenesis, remain unknown.
Adipose tissue must also be vascularized, meaning endothelial cells and smooth muscle cells are also part of the architecture of adipose tissue. As previously mentioned, putative preadipocyte populations are located in close physical proximity to the vasculature of adipose tissue.93 Interestingly, this phenotype is recapitulated in adipose tissue cultured ex vivo, where over time vascular proliferation and extension of blood vessel structures from the tissue explant is accompanied by the emergence of a population of preadipocytes along these structures.72 Taken together, these data imply that the adipose tissue vasculature forms an important part of the local stem cell niche and that circulating signals may control stem cell turnover and differentiation, in addition to local signals. Adipose tissue must respond to lipolytic stimuli and efficiently release or take up stored energy, such that proper vascularization is critical to tissue health.72 In BAT, vascularization is high to deliver nutrients into BAT and distribute heat throughout the body by efficiently moving blood through the tissue. This also facilitates endocrine function, and WAT must also be vascularized for similar reasons, but to a lesser extent than BAT. The dynamics of WAT vascularization have been more thoroughly investigated than those of BAT, and a large body of literature exists on vascular dysfunction in adipose tissue that arises during metabolic syndrome. Hypoxia exerts a proinflammatory effect in WAT via hypoxia-inducible factor (HIF1a), with an opposing anti-inflammatory effect of the HIF2a isoform.94 In addition to the impact of vascular dysfunction on adipocyte function, adipocytes can also affect vascular function by modulating vascular stretch, with healthy adipose tissue surrounding the vasculature exerting an anticontractile effect.95
Together, the many different cell types in adipose tissue work together to function in whole-body energy metabolism. When there are changes to metabolic homeostasis, different cell types are activated and recruited to respond.
Adipogenesis
Mature adipocytes are terminally differentiated, presumably from precursor cells that are already committed to the adipocyte lineage (i.e., preadipocytes).7 The process of adipocyte differentiation from preadipocytes has been extensively studied in vitro using two-dimensional models and, more recently, co-culture models to examine the effects of one cell type on the differentiation of preadipocytes in vitro.96,97 Studies have begun to shed light on in vivo adipocyte differentiation using three-dimensional cultures that mimic in vivo microenvironments98–100 or genetic marker systems to label specific cell types and track their differentiation in vivo.101–103 Many different cell models have been used to study differentiation of adipocytes in vitro, with the goal mainly to understand the mechanisms that underlie the development of the lipid droplet and the partitioning of energy to and from it. As previously mentioned, not all mature adipocytes are created for energy storage. In addition to white adipocytes, brown adipocytes and brown-like beige adipocytes are specialized in expending energy to increase heat production. Brown and beige fat cells are known to exist in anatomically defined depots in mice and humans.44,104–106 Though these cells have different functions, they share a general adipogenic differentiation process.
In vitro adipocyte differentiation
In vitro adipocyte differentiation occurs when preadipocytes, which can be isolated because they are not lipid laden and therefore pellet during centrifugation (mature adipocytes float), are isolated and either immortalized or simply differentiated from primary cells using induction cocktails.107 A commonly accepted induction cocktail contains insulin, triiodothyronine (T3), and compounds that raise cAMP and inhibit cycloxygenase-2, such as indomethacin, 3-isobutyl-1-methylxanthine (IBMX), and dexamethasone.64 These signals synergize to activate an adipogenic differentiation program characterized by suppression of adipogenic inhibitors genes, such as preadipocyte factor-1 (Pref-1), necdin, and wingless-type MMTV integration site family member 10A (Wnt10a) and activation of adipogenic activators, such as PPARγ and C/EBPs.108 By activating the adipogenic gene expression program, preadipocytes begin a maturation process characterized by the development of an advanced metabolite-handling system, which can be most obviously recognized visually by lipid accumulation.109
While similar to white adipocytes in terms of adipogenesis, brown and beige adipocytes are different from white fat cells in several features, as discussed above. UCP1 is expressed uniquely in thermogenically competent cells and serves to identify brown and beige adipocytes. Expression of UCP1 is induced in response to cold and diet, as well as hormones and growth factors.110 UCP1 is more highly expressed in brown/beige adipocytes than white adipocytes, and, importantly, differentiation of primary preadipocytes isolated from WAT and BAT results in expression of UCP1 only in brown adipocytes. Clearly, brown adipocytes are able to access a program of differentiation that is distinct from that available to white adipocytes, and differences in chromatin structure (addressed below) may play a role in allowing brown but not white preadipocytes to express UCP1. Though brown preadipocytes differentiated in vitro seem to phenotypically recapitulate mature brown adipocytes in vivo, white adipocytes are unable to fully mature into unilocular cells in vitro33,111 (Fig. 3). There are many factors, such as availability of extracellular matrix and cytoskeletal proteins, that contribute to the differences between in vitro and in vivo differentiation. It is also probable that the distribution of anabolic and catabolic signals in vitro is different from that in vivo. Adding to the complexity, distinct size classes of lipid droplets exist in each cell that differ in their composition, ability to recruit proteins to their surface, and ultimately, function.112
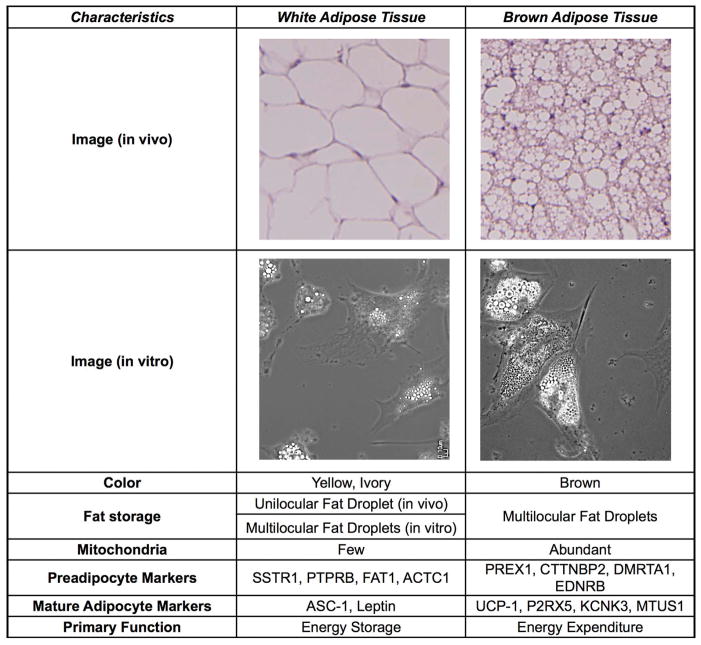
Figure 3.
Characteristics of white and brown adipose tissue. Abundant mitochondria and a high level of vascularization make brown adipose tissue distinct from white adipose tissue in vivo; however, in vitro–differentiated adipocytes from brown and white precursors are morphologically indistinguishable. In vivo, white adipose tissue stores lipid in a unilocular fat droplet, whereas, in vitro, white adipocytes share a multilocular morphology with brown adipose tissue in vivo and in vitro. On a molecular level, the genes that mark cells from these two different depots are different both at the preadipocyte stage and in mature adipocytes. Human white preadipocytes express high levels of SSTR1, PTPRB, FAT1, and ACTC1 and after differentiation can be discriminated from brown adipocytes by increased expression of ASC-1 and leptin, which allow them to perform their primary function of energy storage. Human brown preadipocytes, on the other hand, express PREX1, CTTNBP2, DMRTA1, and EDNRB. Differentiation leads to expression of UCP1, which facilitates energy expenditure, and human brown adipocytes can be further discriminated from white adipocytes by expression of P2RX5, KCNK3, and MTUS1.
Because white adipocytes do not become unilocular in vitro, it is difficult to use this feature to define in vitro differentiated white adipocytes. Further, lipid droplet accumulation in vitro can be rather dynamic, with lipids appearing and then disappearing from the same cell within 8 h (Fig. 3). On the other hand, lipid droplet morphology is related to metabolic capacity,113 and one study using droplet morphology to screen candidate drugs succeeded in uncovering the antithermogenic effect of Janus kinase (Jak) signaling, which could be blocked to activate mitochondrial biogenesis and UCP1 expression. Regardless of the distinctions between adipocytes derived from in vitro versus in vivo differentiation, in vitro cellular models can recapitulate most of the metabolic capacity that occurs in vivo, such as glucose and fatty acid utilization in response to insulin, and have offered great tools in the studies aiming to understand the molecular pathways of adipogenesis and drug discovery.33
While many murine adipocyte models have been studied in vitro for many years, it was not until recently that some groups generated human preadipocyte models to better understand the similarities to and differences from mouse cells.33,72,111 Critically, the groups generating these cells were able to produce paired cell lines isolated from different adipose tissue depots from the same subject, which allows the genetic heterogeneity of human population studies to be eliminated.
In vivo adipocyte differentiation
Most of our understanding of in vivo adipogenesis is derived from lineage-tracing experiments using transgenic mice to indelibly mark preadipocyte cells and then allow those cells to differentiate into mature adipocytes. These studies are of course limited to murine adipogenesis and require the resource-intense step of generating transgenic mice, where the gene is used to label preadipocytes. One drawback is that, to generate transgenic mice, the genetic identity of the preadipocytes must be known a priori. Though limited logistically, these studies provide compelling evidence about the lineage of adipocytes, clearly demonstrating that most of the adipocytes are derived from a platelet-derived growth factor receptor α (PDGFRα)–expressing precursor cell.73,114 Expression of zinc-finger protein 423 (ZFP423) begins a commitment to adipogenic differentiation,71,115 and ZFP423 plays an essential role in maintaining white fat identity.116 PDGFRβ, on the other hand, marks a population of bipotent preadipocytes capable of white and beige adipocyte differentiation depending on the physiologic stimulation.117 This may indicate that the PDGFRβ population is heterogeneous itself or may represent a unique preadipocyte population.
As mentioned previously, the interscapular brown fat depot arises developmentally from a lineage of cells in the dermomyotome that is shared with skeletal muscle and expresses the transcription factors En1, paired box protein 7 (PAX7), and MYF5,29,118,119 while the majority of white and beige adipocyte precursors do not express MYF5.29,50,120,121 Brown adipogenesis includes activation of a thermogenic differentiation program, and, in many ways, this program is shared between brown and beige adipocytes; however, the two cell types have distinct gene expression profiles and developmental origins in mice.29,122 Thermogenic gene expression is driven by PRD1–BF1–RIZ1 homologous domain containing 16 (PRDM16) and the transcriptional co-activator proliferator-activated receptor γ coactivator 1α (PGC1α), which co-activates mitochondrial biogenesis along with mitochondrial transcription factor A (TFAM).123–125 Complete differentiation of brown adipocytes requires PRDM16, which acts as a regulatory switch in preadipocytes to allow brown adipocyte differentiation and can be overexpressed in non-adipogenic cells to activate this process.126 In mice, adipocyte-specific deletion of PRDM16 has little impact on differentiation and function of interscapular brown adipocytes, but beige adipocyte formation in response to cold or β3-adrenergic stimulation is impaired.127
Since the DNA-binding domain of PRDM16 is dispensable for its function in brown adipogenesis, the search for transcriptional co-activators that may mediate PRDM16’s effect on gene transcription has resulted in several hits.126 Interestingly, these proteins include chromatin-modifying enzymes, such as euchromatic histone-lysine N-methyltransferase 1 (EHMT1) and C-terminal–binding protein 1 (CTBP1) and CTBP2,128 which recruit histone deacetylase enzymes to chromatin.129 It is reasonable to assume that the protein landscape of the UCP1 promoter could exists in three different states, depending on the cell context. Some cells may close the chromatin structure of the UCP1 promoter, rendering it inaccessible. This may be distinct from cells where expression of UCP1 could be induced, where low and high expression levels could correlate with different transcription factor occupancy. Supporting the notion that thermogenic competency is a function of chromatin state, PRDM16 in human embryonic stem cells has specific histone 3 lysine 9 (H3K9) methyltransferase activity. The H3K9 methylation state is a critical determinant of gene activity, and preadipocytes that do not express PRDM16 may fail to properly modify chromatin associated with the thermogenic differentiation program to allow access to UCP1.130,131
Another PRDM16 co-activator, zinc-finger protein 516 (ZFP516), can directly interact with the UCP1 promoter by binding it to drive expression132. ZFP516 binds to the proximal UCP1 promoter only 70 bp from the transcription start site to regulate UCP1 gene expression. This is in contrast to other regulatory elements, which are known to activate UCP1 expression via an enhancer element approximately 2.2 kb from the transcription start site.133,134 Other transcription factors, such as early B cell factor 2 (EBF2), are also thought to act as a co-regulators of gene expression with PRDM16; however. whether EBF2 regulates UCP1 expression directly is unknown.135 EBF2 was shown to mark cells from both white and brown adipose tissue that were committed to the thermogenic fate, and EBF2-expressing cells give rise to brown and beige adipocytes, suggesting that EBF2 and its targets poise these cells to express UCP1 after differentiation.136 The white fat–determining factor ZFP423 distinguishes between white and beige adipogenesis by suppressing EBF2 and PRDM16 activation.116
Thermogenic gene expression is further driven by the transcriptional co-activator PGC1, which activates mitochondrial biogenesis with mitochondrial transcription factor TFAM.137 Cellular tyrosine kinases in the Jak, spleen tyrosine kinase (Syk) and proto-oncogene tyrosine protein kinase Src families that negatively regulate thermogenic differentiation have also been identified.113,138 During the later stages of differentiation, both sympathetic dependent and independent pathways could further enhance the thermogenic program. The canonical pathway that activates mature BAT metabolic activity in response to cold relies on sympathetic input resulting in the release of the catecholamine neurotransmitter norepinephrine (NE) to activate G protein–coupled adrenergic receptors and increase intracellular cAMP. This process could be mimicked using pharmacological compounds, such as the β3-adrenergic receptor agonist CL 316,243. These stimuli enhance BAT thermogenic activity and induce beige adipocyte formation in white adipose tissue.68 The latter is mediated by increased differentiation of precursor cells into thermogenically competent cells, as well as direct transdifferentiation.69 In addition to the canonical catecholamine pathway, several alternate pathways capable of increasing BAT thermogenic activity and/or brown fat differentiation have been described.12,70 These include classical hormones, such as thyroid hormone and insulin, and newly identified endocrine factors, such as bile acid, natriuretic peptides, fibroblast growth factor 21 (FGF21), irisin, and members of the bone morphogenetic protein family (BMPs).139
Factors that regulate adipose heterogeneity
Genetics influence adipose heterogeneity
To identify potential candidate genes controlling obesity, recent studies have combined large clinical cohorts with publically available data to perform meta-analysis of single nucleotide polymorphism (SNP) variation in populations with sample sizes reaching almost a quarter of a million subjects. In a result that betrays its own complexity, fewer than 100 loci associate with obesity.8,9,10 In some cases, determining the causal gene is relatively straightforward, as the SNP is located near a gene with known functions congruent with a role in energy metabolism or a gene, such as melanocortin 4 receptor (MC4R), brain-derived neurotrophic factor (BDNF), or pro-opiomelanocortin (POMC), that causes monogenic obesity in mice. In association studies, variation in these genes has drastic effects on adipose tissue in some way, making their relation to obesity obvious. In other cases, assigning a causal gene is less straightforward: for example when a polymorphism in a distal regulatory region affects expression of a gene that is not necessarily the closest gene to the SNP. Recent evidence suggests that this may be the case for SNPs located near the fat mass and obesity–associated (FTO) locus, which actually regulate expression of the Iroquois homeobox protein 3 (IRX3) gene140 rather than the FTO gene that was originally implicated in the SNPs’ association with obesity.141 This example serves to highlight the importance of pairing an understanding of the functional importance of genes with phenotypic association data, and, by using gene ontology to categorize the genes implicated by SNPs, pathways identified include synaptic function, cell–cell adhesion, glutamate signaling, RNA binding/processing, lipid metabolism, and endocrine function.9,10 Additionally, more general pathways, for example mitogen-activated protein kinase (MAPK) signaling, are also upregulated as part of a mitogenic signature of cell survival that could be expected in an environment of energy excess.8,9 Still, one frustrating result of the impressive effort to generate genetic associations with human obesity has been the ability to explain a mere 3% of the phenotypic variation with the loci identified thus far, suggesting that the individual contributions of genetic variants to phenotype may be subtle.8
It is important to remember that these genome-wide association studies (GWAS) identify loci that associate with obesity and its sequelae that have known mutant alleles that can change that gene’s effect on obesity. This means that heterogeneity among the population in the DNA sequence in and around this gene may alter the accumulation of adipose tissue in some way. The minor alleles that are unknown and the alleles in genes with only moderate effects remain elusive, adding to the overall heterogeneity of adipose tissue that must be accounted for.
Gender-specific adipose heterogeneity
Gender clearly plays a fundamental role in fat mass and distribution.142 Women tend to accumulate more subcutaneous fat in gluteal and femoral regions, while men store fat in upper body and visceral areas.143 Sexual dimorphism in mice actually extends to the specific lipid content of adipose tissue, as the BAT of female mice contains a distinctly different lipidomic signature than that of male mice.144 Clearly, gender-specific hormones regulate aspects of adipogenesis, and castration of male mice can feminize aspects of adipocyte function in vitro.20 With that in mind, it is also critical to remember that, in addition to sex-specific aspects of adipose tissue biology presumably driven by differences in sex hormones, some hormones are actually secreted from adipose tissue itself.38,145–148 The secretion of these putative adipokines can itself be sex specific,149 leading to a complicated model wherein hormone levels and sensitivity are both sexually dimorphic. Intriguingly, some of the differences between adipocytes from male and female mice, including lipogenic and lipolytic rate as well as insulin sensitivity, are cell intrinsic.20 Sexual dimorphism can also be detected in the proclivity for spontaneous differentiation, which is higher in cells from human females compared with males.70 Understanding the mechanisms regulating depot-specific fat expansion and function in physiological and pathophysiological conditions may facilitate developing strategies for modulating whole-body metabolism.
Energy demand regulates adipose tissue heterogeneity
Dynamic energy demand requires changes to tissue morphologies, function, and cellular compositions to maintain homeostasis. Given the major role of adipose tissue in energy homeostasis, it is key to understand how this tissue copes with changes in energy homoeostasis to store, dispense, and use fuel as needed. A positive energy balance triggers an overall increase in pathways of energy storage, which causes expansion of adipose tissue to an extent that varies by depot; however, adipocytes undergo hypertrophy characterized by increased lipid droplet size (Fig. 4). In addition to hypertrophy of existing adipocytes, extended periods of energy excess lead to WAT expansion by increased preadipocyte proliferation and differentiation, and in mice this process occurs rapidly and is dependent on the phosphoinositide 3-kinase (PI3K)–protein kinase B (AKT2) pathway.150 Owing to increased cell death of hypertrophic adipocytes, hyperplasia seems to affect tissue size less than hypertrophy, although the local microenvironments of each depot, as well as cell-intrinsic properties of preadipocytes from each depot, mean that the extent of hyperplasia and hypertrophy varies between depots. In humans, lower body adipose tissue located in subcutaneous depots of the buttocks and thighs seems to expand by hypertrophy, while abdominal subcutaneous adipose tissue in the visceral cavity expands by hyperplasia.151
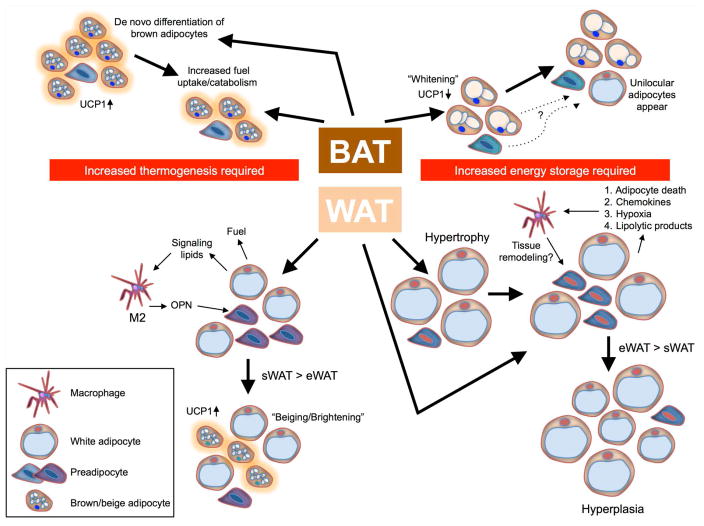
Figure 4.
Energy balance drives changes in BAT and WAT that can activate preadipocyte differentiation. In BAT, increased thermogenic demand activates energy expenditure by catabolizing fuel, and a long-term imbalance can stimulate differentiation of new brown adipocytes and increased tissue size. Excess energy can be stored in BAT and is usually characterized by a decrease in number and increase in size of lipid droplets, as well as decreased expression of UCP1. In some mouse models, such as chronic high fat–diet feeding, BAT adipocytes can even appear unilocular, although it is unclear if these cells are truly white adipocytes or if this unilocular lipid droplet is a further adaptation of brown adipocytes to energy excess. WAT participates in energy expenditure during thermogenesis, first as a fuel source that can release stored fuel to thermogenic BAT, but, during chronic cold exposure, these lipolysis products are also able to recruit macrophages that secrete molecules, such as OPN, to stimulate differentiation of UCP1+ beige/brite adipocytes in situ. Energy excess causes expansion of WAT, first by hypertrophy of adipocytes but then, through a process similarly associated with macrophage recruitment, through the differentiation of adipocytes into new mature adipocytes. Hypertrophy and hyperplasia occur at different rates in different adipose tissue depots.
In mice, however, it is visceral adipose tissue that expands via hyperplasia, at least in response to high-fat diet.18 To support this process, macrophages are recruited to adipose tissue during chronic energy excess by adipocyte death, chemokine release, hypoxia, or lipolytic products;76,77 however, the ratio of M1 to M2 polarization varies among depots. Since subcutaneous adipose tissue is considered “healthy adipose tissue” and visceral adipose tissue is thought to be “unhealthy adipose tissue,” the relative rates of hypertrophy and hyperplasia in these two depots during times of energy excess are thought to underlie their different contributions to the metabolic syndrome. In addition to high-fat diet, caloric restriction has also been used to measure the effect of energy balance on adipose tissue composition and function and has some interesting effects beyond what one might expect with simple negative energy balance. One well-known phenotype of caloric restriction is extended life span,152,153 but underlying this are delays in the onset of many diseases, including metabolic syndrome, and an improvement in tissue metabolism that can be measured by increased sensitivity of energy-sensing pathways.154–156 In broad terms, caloric restriction decreases adipocyte size, increases the number of M2-polarized tissue-resident macrophages, and can promote the recruitment of beige adipocytes; however, consistent with the theme of this review, the effect size varies by depot.157 A better understanding of the cellular and molecular mechanisms that underlie WAT energy storage and trigger hypertrophy or hyperplasia is key to developing treatments for metabolic syndrome, obesity, and diabetes.
Another environmental challenge that modulates energy homeostasis is cold exposure. During periods of prolonged cold exposure, in addition to the hyperplasia that occurs in BAT, a population of adipocytes that express UCP1 termed beige/brite adipocytes arise in WAT from preadipocytes that are activated to differentiate in situ in a process called browning or beiging.158 In transgenic mice lacking classical BAT, these cells are even able to fully compensate and maintain normal thermogenesis during cold exposure, demonstrating their ability to catabolize fuel and generate heat.159 These cells arise from progenitor cells residing in WAT that maintain thermogenic competency, allowing them to activate both general adipogenic lipid accumulation and the gene network that drives thermogenesis and ultimately converges on UCP1.
A final common modulator of energy balance is exercise and, indeed as many who exercise hope, this has profound effects on adipose tissue. In addition to the well-recognized calorie-burning effect of exercise, one other major effect of exercise is induction of lipolysis in WAT to liberate stored energy for use as fuel, and the extent to which lipolysis is activated by exercise varies among adipose tissue depots.160,161 In addition to direct effects on the tissue itself, adipose tissue from exercised animals can improve glucose homeostasis, potentially by secreting factors that promote insulin action or secretion in distal tissues; however, the effect on glucose homeostasis is also depot dependent.162,163
Conclusions
Adipose tissue contains many different cell types, and only by understanding the interplay among them can the contribution of each adipose depot and the tissue as a whole be understood in the control of energy balance. By studying the mechanisms that govern preadipocyte differentiation, it may be possible to direct the differentiation of these cells either in vivo or ex vivo toward a desired phenotype to treat metabolic syndrome. Clearly, owing to the current obesity epidemic, the most desirable phenotype is one that is biased toward energy expenditure and against energy storage, so the comparison of brown and white adipocyte differentiation is especially relevant. This process is undoubtedly similar in both cell types; however, the energy-dispensing properties and morphological phenotype of BAT cells is remarkably well maintained in vitro and can be used to identify new targets that promote or inhibit thermogenic potential.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein EA, et al. Obesity and severe obesity forecasts through 2030. American journal of preventive medicine. 2012;42:563–570. doi: 10.1016/j.amepre.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Craft S. Insulin resistance and Alzheimer’s disease pathogenesis: potential mechanisms and implications for treatment. Curr Alzheimer Res. 2007;4:147–152. doi: 10.2174/156720507780362137. [DOI] [PubMed] [Google Scholar]
- 4.Hsu IR, Kim SP, Kabir M, Bergman RN. Metabolic syndrome, hyperinsulinemia, and cancer. Am J Clin Nutr. 2007;86:s867–s871. doi: 10.1093/ajcn/86.3.867S. [DOI] [PubMed] [Google Scholar]
- 5.Cornier MA, et al. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch J, Batchelor B. Adipose tissue cellularity in human obesity. Clin Endocrinol Metab. 1976;5:299–311. doi: 10.1016/s0300-595x(76)80023-0. [DOI] [PubMed] [Google Scholar]
- 7.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Fu J, Hofker M, Wijmenga C. Apple or pear: size and shape matter. Cell Metab. 2015;21:507–508. doi: 10.1016/j.cmet.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nature reviews Neuroscience. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott W, Stevens J, Binder-Macleod SA. Human skeletal muscle fiber type classifications. Physical therapy. 2001;81:1810–1816. [PubMed] [Google Scholar]
- 13.Katsuta H, et al. Subpopulations of GFP-marked mouse pancreatic beta-cells differ in size, granularity, and insulin secretion. Endocrinology. 2012;153:5180–5187. doi: 10.1210/en.2012-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roscioni SS, Migliorini A, Gegg M, Lickert H. Impact of islet architecture on beta-cell heterogeneity, plasticity and function. Nat Rev Endocrinol. 2016 doi: 10.1038/nrendo.2016.147. [DOI] [PubMed] [Google Scholar]
- 15.Cohen P, Spiegelman BM. Cell biology of fat storage. Mol Biol Cell. 2016;27:2523–2527. doi: 10.1091/mbc.E15-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrinelli V, Carobbio S, Vidal-Puig A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia. 2016;59:1075–1088. doi: 10.1007/s00125-016-3933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joe AW, Yi L, Even Y, Vogl AW, Rossi FM. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 2009;27:2563–2570. doi: 10.1002/stem.190. [DOI] [PubMed] [Google Scholar]
- 18.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macotela Y, et al. Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macotela Y, Boucher J, Tran TT, Kahn CR. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes. 2009;58:803–812. doi: 10.2337/db08-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaffney EF, Vellios F, Hargreaves HK. Lipoblastomatosis: ultrastructure of two cases and relationship to human fetal white adipose tissue. Pediatric pathology. 1986;5:207–216. doi: 10.3109/15513818609041202. [DOI] [PubMed] [Google Scholar]
- 22.Kindblom LG, Save-Soderbergh J. The ultrastructure of liposarcoma. A study of 10 cases. Acta pathologica et microbiologica Scandinavica Section A, Pathology. 1979;87a:109–121. doi: 10.1111/j.1699-0463.1979.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napolitano L, Fawcett D. The fine structure of brown adipose tissue in the newborn mouse and rat. The Journal of biophysical and biochemical cytology. 1958;4:685–692. doi: 10.1083/jcb.4.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricquier D, Nechad M, Mory G. Ultrastructural and biochemical characterization of human brown adipose tissue in pheochromocytoma. J Clin Endocrinol Metab. 1982;54:803–807. doi: 10.1210/jcem-54-4-803. [DOI] [PubMed] [Google Scholar]
- 26.D’Andrea S. Lipid droplet mobilization: The different ways to loosen the purse strings. Biochimie. 2016;120:17–27. doi: 10.1016/j.biochi.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Porter C, et al. Human and Mouse Brown Adipose Tissue Mitochondria Have Comparable UCP1 Function. Cell Metab. 2016;24:246–255. doi: 10.1016/j.cmet.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rousset S, et al. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53(Suppl 1):S130–S135. doi: 10.2337/diabetes.53.2007.s130. [DOI] [PubMed] [Google Scholar]
- 29.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shabalina IG, et al. UCP1 in Brite/Beige Adipose Tissue Mitochondria Is Functionally Thermogenic. Cell Rep. 2013;5:1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 32.Kalinovich AV, de Jong JM, Cannon B, Nedergaard J. UCP1 in adipose tissues: two steps to full browning. Biochimie. 2017;134:127–137. doi: 10.1016/j.biochi.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Xue R, et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med. 2015;21:760–768. doi: 10.1038/nm.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanssen MJ, et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat Med. 2015;21:863–865. doi: 10.1038/nm.3891. [DOI] [PubMed] [Google Scholar]
- 35.Hanssen MJ, et al. Short-term Cold Acclimation Recruits Brown Adipose Tissue in Obese Humans. Diabetes. 2016;65:1179–1189. doi: 10.2337/db15-1372. [DOI] [PubMed] [Google Scholar]
- 36.Berbee JF, et al. Brown fat activation reduces hypercholesterolaemia and protects from atherosclerosis development. Nature communications. 2015;6:6356. doi: 10.1038/ncomms7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khedoe PP, et al. Brown adipose tissue takes up plasma triglycerides mostly after lipolysis. J Lipid Res. 2015;56:51–59. doi: 10.1194/jlr.M052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villarroya F, Cereijo R, Villarroya J, Giralt M. Brown adipose tissue as a secretory organ. Nat Rev Endocrinol. 2017;13:26–35. doi: 10.1038/nrendo.2016.136. [DOI] [PubMed] [Google Scholar]
- 39.Kwok KH, Lam KS, Xu A. Heterogeneity of white adipose tissue: molecular basis and clinical implications. Exp Mol Med. 2016;48:e215. doi: 10.1038/emm.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reaven GM. Banting lecture: Role of insulin resistance in human disease. Diabetes. 1988;37:1595–1607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 41.Snijder MB, et al. Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. Am J Clin Nutr. 2003;77:1192–1197. doi: 10.1093/ajcn/77.5.1192. [DOI] [PubMed] [Google Scholar]
- 42.Snijder MB, et al. Trunk fat and leg fat have independent and opposite associations with fasting and postload glucose levels: the Hoorn study. Diabetes Care. 2004;27:372–377. doi: 10.2337/diacare.27.2.372. [DOI] [PubMed] [Google Scholar]
- 43.Van Pelt RE, Jankowski CM, Gozansky WS, Schwartz RS, Kohrt WM. Lower-body adiposity and metabolic protection in postmenopausal women. J Clin Endocrinol Metab. 2005;90:4573–4578. doi: 10.1210/jc.2004-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen KY, et al. Brown Adipose Reporting Criteria in Imaging STudies (BARCIST 1.0): Recommendations for Standardized FDG-PET/CT Experiments in Humans. Cell Metab. 2016;24:210–222. doi: 10.1016/j.cmet.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gesta S, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gesta S, et al. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc Natl Acad Sci U S A. 2011;108:2771–2776. doi: 10.1073/pnas.1019704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee KY, et al. Shox2 is a molecular determinant of depot-specific adipocyte function. Proc Natl Acad Sci U S A. 2013;110:11409–11414. doi: 10.1073/pnas.1310331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto Y, et al. Adipose Depots Possess Unique Developmental Gene Signatures. Obesity (Silver Spring ) 2010;18:872–878. doi: 10.1038/oby.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tchkonia T, et al. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am J Physiol Endocrinol Metab. 2007;292:E298–E307. doi: 10.1152/ajpendo.00202.2006. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Gurmaches J, Guertin DA. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat Commun. 2014;5:4099. doi: 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheller EL, Rosen CJ. What’s the matter with MAT? Marrow adipose tissue, metabolism, and skeletal health. Ann N Y Acad Sci. 2014;1311:14–30. doi: 10.1111/nyas.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lecka-Czernik B, Rosen CJ. Skeletal integration of energy homeostasis: Translational implications. Bone. 2016;82:35–41. doi: 10.1016/j.bone.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaggini M, Saponaro C, Gastaldelli A. Not all fats are created equal: adipose vs. ectopic fat, implication in cardiometabolic diseases. Hormone molecular biology and clinical investigation. 2015;22:7–18. doi: 10.1515/hmbci-2015-0006. [DOI] [PubMed] [Google Scholar]
- 54.Rivera-Gonzalez GC, et al. Skin Adipocyte Stem Cell Self-Renewal Is Regulated by a PDGFA/AKT-Signaling Axis. Cell Stem Cell. 2016;19:738–751. doi: 10.1016/j.stem.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villaret A, et al. Adipose tissue endothelial cells from obese human subjects: differences among depots in angiogenic, metabolic, and inflammatory gene expression and cellular senescence. Diabetes. 2010;59:2755–2763. doi: 10.2337/db10-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villarroya J, Cereijo R, Villarroya F. An endocrine role for brown adipose tissue? Am J Physiol Endocrinol Metab. 2013;305:E567–E572. doi: 10.1152/ajpendo.00250.2013. [DOI] [PubMed] [Google Scholar]
- 57.Li F, et al. Myokines and adipokines: Involvement in the crosstalk between skeletal muscle and adipose tissue. Cytokine Growth Factor Rev. 2016;33:73–82. doi: 10.1016/j.cytogfr.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Mancuso P. The role of adipokines in chronic inflammation. ImmunoTargets and therapy. 2016;5:47–56. doi: 10.2147/ITT.S73223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawicka K, Krasowska D. Adipokines in connective tissue diseases. Clinical and experimental rheumatology. 2016;34:1101–1112. [PubMed] [Google Scholar]
- 60.Spalding KL, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 61.Rigamonti A, Brennand K, Lau F, Cowan CA. Rapid cellular turnover in adipose tissue. PLoS One. 2011;6:e17637. doi: 10.1371/journal.pone.0017637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 64.Schulz TJ, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ussar S, et al. ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Sci Transl Med. 2014;6:247ra103. doi: 10.1126/scitranslmed.3008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cartwright MJ, Tchkonia T, Kirkland JL. Aging in adipocytes: potential impact of inherent, depot-specific mechanisms. Exp Gerontol. 2007;42:463–471. doi: 10.1016/j.exger.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirkland JL, Hollenberg CH, Gillon WS. Age, anatomic site, and the replication and differentiation of adipocyte precursors. Am J Physiol. 1990;258:C206–C210. doi: 10.1152/ajpcell.1990.258.2.C206. [DOI] [PubMed] [Google Scholar]
- 68.Kirkland JL, Hollenberg CH, Gillon WS. Effects of fat depot site on differentiation-dependent gene expression in rat preadipocytes. Int J Obes Relat Metab Disord. 1996;20(Suppl 3):S102–S107. [PubMed] [Google Scholar]
- 69.Tchkonia T, et al. Fat depot origin affects adipogenesis in primary cultured and cloned human preadipocytes. Am J Physiol Regul Integr Comp Physiol. 2002;282:R1286–R1296. doi: 10.1152/ajpregu.00653.2001. [DOI] [PubMed] [Google Scholar]
- 70.Tchoukalova YD, et al. Sex- and depot-dependent differences in adipogenesis in normal-weight humans. Obesity (Silver Spring, Md) 2010;18:1875–1880. doi: 10.1038/oby.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta RK, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Min SY, et al. Human ‘brite/beige’ adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice. Nat Med. 2016;22:312–318. doi: 10.1038/nm.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YH, Petkova AP, Granneman JG. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab. 2013;18:355–367. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu XJ, et al. Insulin sensitive and resistant obesity in humans: AMPK activity, oxidative stress, and depot-specific changes in gene expression in adipose tissue. J Lipid Res. 2012;53:792–801. doi: 10.1194/jlr.P022905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cartier A, et al. Visceral obesity and plasma glucose-insulin homeostasis: contributions of interleukin-6 and tumor necrosis factor-alpha in men. J Clin Endocrinol Metab. 2008;93:1931–1938. doi: 10.1210/jc.2007-2191. [DOI] [PubMed] [Google Scholar]
- 76.Weisberg SP, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ortega Martinez de Victoria E, et al. Macrophage content in subcutaneous adipose tissue: associations with adiposity, age, inflammatory markers, and whole-body insulin action in healthy Pima Indians. Diabetes. 2009;58:385–393. doi: 10.2337/db08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lumeng CN, Delproposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee BC, et al. Adipose Natural Killer Cells Regulate Adipose Tissue Macrophages to Promote Insulin Resistance in Obesity. Cell Metab. 2016;23:685–698. doi: 10.1016/j.cmet.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased adipose tissue expression of tumor necrosis factor-a in human obesity and insulin resistance. J Clin Invest. 1995;95:2409–2415. doi: 10.1172/JCI117936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-a: Direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 84.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 85.Ferrante AW., Jr The immune cells in adipose tissue. Diabetes Obes Metab. 2013;15(Suppl 3):34–38. doi: 10.1111/dom.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee MW, et al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 2015;160:74–87. doi: 10.1016/j.cell.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rao RR, et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell. 2014;157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brestoff JR, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 2015;519:242–246. doi: 10.1038/nature14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Odegaard JI, et al. Perinatal Licensing of Thermogenesis by IL-33 and ST2. Cell. 2016;166:841–854. doi: 10.1016/j.cell.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartness TJ, Ryu V. Neural control of white, beige and brown adipocytes. International journal of obesity supplements. 2015;5:S35–39. doi: 10.1038/ijosup.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garretson JT, et al. Lipolysis sensation by white fat afferent nerves triggers brown fat thermogenesis. Molecular metabolism. 2016;5:626–634. doi: 10.1016/j.molmet.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bartness TJ, Liu Y, Shrestha YB, Ryu V. Neural innervation of white adipose tissue and the control of lipolysis. Front Neuroendocrinol. 2014;35:473–493. doi: 10.1016/j.yfrne.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang W, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trayhurn P, Wang B, Wood IS. Hypoxia in adipose tissue: a basis for the dysregulation of tissue function in obesity? Br J Nutr. 2008;100:227–235. doi: 10.1017/S0007114508971282. [DOI] [PubMed] [Google Scholar]
- 95.Aghamohammadzadeh R, Heagerty AM. Obesity-related hypertension: epidemiology, pathophysiology, treatments, and the contribution of perivascular adipose tissue. Ann Med. 2012;44(Suppl 1):S74–84. doi: 10.3109/07853890.2012.663928. [DOI] [PubMed] [Google Scholar]
- 96.Miki Y, et al. The advantages of co-culture over mono cell culture in simulating in vivo environment. The Journal of steroid biochemistry and molecular biology. 2012;131:68–75. doi: 10.1016/j.jsbmb.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 97.Pandurangan M, Hwang I. Application of cell co-culture system to study fat and muscle cells. Applied microbiology and biotechnology. 2014;98:7359–7364. doi: 10.1007/s00253-014-5935-9. [DOI] [PubMed] [Google Scholar]
- 98.Pope BD, Warren CR, Parker KK, Cowan CA. Microenvironmental Control of Adipocyte Fate and Function. Trends Cell Biol. 2016;26:745–755. doi: 10.1016/j.tcb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Unser AM, Tian Y, Xie Y. Opportunities and challenges in three-dimensional brown adipogenesis of stem cells. Biotechnology advances. 2015;33:962–979. doi: 10.1016/j.biotechadv.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chun TH, et al. A pericellular collagenase directs the 3-dimensional development of white adipose tissue. Cell. 2006;125:577–591. doi: 10.1016/j.cell.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 101.Jeffery E, et al. The Adipose Tissue Microenvironment Regulates Depot-Specific Adipogenesis in Obesity. Cell Metab. 2016;24:142–150. doi: 10.1016/j.cmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeffery E, et al. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Berry R, Rodeheffer MS, Rosen CJ, Horowitz MC. Adipose Tissue Residing Progenitors (Adipocyte Lineage Progenitors and Adipose Derived Stem Cells (ADSC) Current molecular biology reports. 2015;1:101–109. doi: 10.1007/s40610-015-0018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Virtanen KA, Nuutila P. Brown adipose tissue in humans. Curr Opin Lipidol. 2011;22:49–54. doi: 10.1097/MOL.0b013e3283425243. [DOI] [PubMed] [Google Scholar]
- 105.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 107.Van RL, Bayliss CE, Roncari DA. Cytological and enzymological characterization of adult human adipocyte precursors in culture. J Clin Invest. 1976;58:699–704. doi: 10.1172/JCI108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo L, Li X, Tang QQ. Transcriptional regulation of adipocyte differentiation: a central role for CCAAT/enhancer-binding protein (C/EBP) beta. J Biol Chem. 2015;290:755–761. doi: 10.1074/jbc.R114.619957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chapman AB, Knight DM, Dieckmann BS, Ringold GM. Analysis of gene expression during differentiation of adipogenic cells in culture and hormonal control of the developmental program. J Biol Chem. 1984;259:15548–15555. [PubMed] [Google Scholar]
- 110.Villarroya F, Peyrou M, Giralt M. Transcriptional regulation of the uncoupling protein-1 gene. Biochimie. 2016;134:86–92. doi: 10.1016/j.biochi.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 111.Shinoda K, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21:389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang S, et al. Morphologically and Functionally Distinct Lipid Droplet Subpopulations. Sci Rep. 2016;6:29539. doi: 10.1038/srep29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moisan A, et al. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat Cell Biol. 2015;17:57–67. doi: 10.1038/ncb3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gupta RK, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shao M, et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab. 2016;23:1167–1184. doi: 10.1016/j.cmet.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vishvanath L, et al. Pdgfrbeta(+) Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab. 2016;23:350–359. doi: 10.1016/j.cmet.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang C, Weng Y, Shi F, Jin W. The Engrailed-1 Gene Stimulates Brown Adipogenesis. Stem cells international. 2016;2016:7369491. doi: 10.1155/2016/7369491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanchez-Gurmaches J, Guertin DA. Adipocyte lineages: Tracing back the origins of fat. Biochim Biophys Acta. 2013;1842:340–51. doi: 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shan T, et al. Distinct populations of adipogenic and myogenic Myf5-lineage progenitors in white adipose tissues. J Lipid Res. 2013;54:2214–2224. doi: 10.1194/jlr.M038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes Dev. 2008;22:1269–1275. doi: 10.1101/gad.1681308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harms MJ, et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 2014;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kajimura S, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rosenfeld JA, et al. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143–110. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 132.Dempersmier J, et al. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol Cell. 2015;57:235–246. doi: 10.1016/j.molcel.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cassard-Doulcier AM, Gelly C, Bouillaud F, Ricquier D. A 211-bp enhancer of the rat uncoupling protein-1 (UCP-1) gene controls specific and regulated expression in brown adipose tissue. Biochem J. 1998;333( Pt 2):243–246. doi: 10.1042/bj3330243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kozak UC, et al. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol. 1994;14:59–67. doi: 10.1128/mcb.14.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang W, et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A. 2014;111:14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vernochet C, et al. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 2012;16:765–776. doi: 10.1016/j.cmet.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Usui M, Uno M, Nishida E. Src family kinases suppress differentiation of brown adipocytes and browning of white adipocytes. Genes Cells. 2016;21:302–310. doi: 10.1111/gtc.12340. [DOI] [PubMed] [Google Scholar]
- 139.Townsend KL, Tseng YH. Brown adipose tissue: recent insights into development, metabolic function, and therapeutic potential. Adipocyte. 2012;1:13–24. doi: 10.4161/adip.18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Palmer BF, Clegg DJ. The sexual dimorphism of obesity. Mol Cell Endocrinol. 2015;402:113–119. doi: 10.1016/j.mce.2014.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jensen MD. Adipose tissue and fatty acid metabolism in humans. Journal of the Royal Society of Medicine. 2002;95(Suppl 42):3–7. [PMC free article] [PubMed] [Google Scholar]
- 144.Hoene M, et al. The lipid profile of brown adipose tissue is sex-specific in mice. Biochim Biophys Acta. 2014;1842:1563–1570. doi: 10.1016/j.bbalip.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 145.Villarroya J, Cereijo R, Villarroya F. An endocrine role for brown adipose tissue? Am J Physiol Endocrinol Metab. 2013;305:E567–572. doi: 10.1152/ajpendo.00250.2013. [DOI] [PubMed] [Google Scholar]
- 146.Gunawardana SC. Adipose tissue, hormones, and treatment of type 1 diabetes. Curr Diab Rep. 2012;12:542–550. doi: 10.1007/s11892-012-0300-9. [DOI] [PubMed] [Google Scholar]
- 147.Karastergiou K, Mohamed-Ali V. The autocrine and paracrine roles of adipokines. Mol Cell Endocrinol. 2009 doi: 10.1016/j.mce.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 148.Loomba-Albrecht LA, Styne DM. Effect of puberty on body composition. Curr Opin Endocrinol Diabetes Obes. 2009;16:10–15. doi: 10.1097/med.0b013e328320d54c. [DOI] [PubMed] [Google Scholar]
- 149.Combs TP, et al. Sexual differentiation, pregnancy, calorie restriction, and aging affect the adipocyte-specific secretory protein adiponectin. Diabetes. 2003;52:268–276. doi: 10.2337/diabetes.52.2.268. [DOI] [PubMed] [Google Scholar]
- 150.Jeffery E, Church CD, Holtrup B, Colman L, Rodeheffer MS. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol. 2015;17:376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tchoukalova YD, et al. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proc Natl Acad Sci U S A. 2010;107:18226–18231. doi: 10.1073/pnas.1005259107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Colman RJ, et al. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nature communications. 2014;5:3557. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wei M, et al. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS genetics. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Canto C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda, Md) 2011;26:214–224. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.De Guzman JM, et al. Chronic caloric restriction partially protects against age-related alteration in serum metabolome. Age (Dordrecht, Netherlands) 2013;35:1091–1104. doi: 10.1007/s11357-012-9430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fabbiano S, et al. Caloric Restriction Leads to Browning of White Adipose Tissue through Type 2 Immune Signaling. Cell Metab. 2016;24:434–446. doi: 10.1016/j.cmet.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 158.Petrovic N, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Thermogenically competent nonadrenergic recruitment in brown preadipocytes by a PPARgamma agonist. Am J Physiol Endocrinol Metab. 2008;295:E287–E296. doi: 10.1152/ajpendo.00035.2008. [DOI] [PubMed] [Google Scholar]
- 159.Schulz TJ, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Savard R, Smith LJ, Palmer JE, Greenwood MR. Site specific effects of acute exercise on muscle and adipose tissue metabolism in sedentary female rats. Physiol Behav. 1988;43:65–71. doi: 10.1016/0031-9384(88)90099-6. [DOI] [PubMed] [Google Scholar]
- 161.Chen N, et al. Depot-specific effects of treadmill running and rutin on white adipose tissue function in diet-induced obese mice. J Physiol Biochem. 2016;72:453–467. doi: 10.1007/s13105-016-0493-5. [DOI] [PubMed] [Google Scholar]
- 162.Stanford KI, et al. A novel role for subcutaneous adipose tissue in exercise-induced improvements in glucose homeostasis. Diabetes. 2015;64:2002–2014. doi: 10.2337/db14-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stanford KI, Middelbeek RJ, Goodyear LJ. Exercise Effects on White Adipose Tissue: Beiging and Metabolic Adaptations. Diabetes. 2015;64:2361–2368. doi: 10.2337/db15-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]