Summary
The cohesin complex is built upon the SMC1/SMC3 heterodimer, and mammalian meiocytes feature two variants of SMC1 named SMC1α and SMC1β. It is unclear why these two SMC1 variants have evolved. To determine unique versus redundant functions of SMC1β, we asked which of the known functions of SMC1β can be fulfilled by SMC1α. Smc1α was expressed under control of the Smc1β promoter in either wild-type or SMC1β-deficient mice. No effect was seen in the former. However, several major phenotypes of SMC1β-deficient spermatocytes were rescued by SMC1α. We observed extended development before apoptosis and restoration of axial element and synaptonemal complex lengths, chromosome synapsis, sex body formation, processing of DNA double-strand breaks, and formation of MLH1 recombination foci. This supports the concept that the quantity rather than the specific quality of cohesin complexes is decisive for meiotic chromosome architecture. It also suggests plasticity in complex composition, because to replace SMC1β in many functions, SMC1α has to more extensively associate with other cohesins. The cells did not complete meiosis but died to the latest at the pachytene-to-diplotene transition. Telomere aberrations known from Smc1β−/− mice persisted, and DNA damage response and repair proteins accumulated there regardless of expression of SMC1α. Thus, whereas SMC1α can substitute for SMC1β in many functions, the protection of telomere integrity requires SMC1β.
Keywords: meiosis, spermatocytes, chromosomes, cohesin, DNA repair, DNA recombination, telomeres
Highlights
-
•
SMC1α can substitute for SMC1β in many meiotic functions
-
•
Key for many meiotic functions is the quantity of cohesin, not the specific type
-
•
Telomere integrity can be preserved only by SMC1β
-
•
In the absence of SMC1β, a DNA damage response is triggered at telomeres
Why are there two SMC1 variants in meiocytes? Biswas et al. asked which of the functions of SMC1β can be fulfilled by SMC1α. Several major phenotypes of SMC1β-deficient spermatocytes were rescued by SMC1α, but telomere deficiencies were not. Thus, whereas SMC1α can substitute for SMC1β in many functions, telomere integrity requires SMC1β.
Introduction
Meiosis poses particular challenges to chromosome structure and dynamics (for recent reviews, see [1, 2, 3, 4]). In prophase of meiosis I, the two sister chromatids generated from each chromosome during premeiotic replication compact into axial elements (AEs) characterized by meiosis-specific proteins like SYCP2 and SYCP3. In pachytene of prophase I, the two homologous AEs have paired and formed the synaptonemal complex (SC), featuring SYCP1 often used as a marker for synapsis. In diplotene, the homologs desynapse but remain linked to each other via chiasmata, characteristic meiotic recombination structures. To initiate recombination, programmed DNA double-strand breaks (DSBs) are introduced early in prophase I. These DSBs become repaired through homologous recombination, some leading to chiasmata. Meiosis II is similar to mitotic cell division, where sister chromatids are separated.
Cohesins are essential supporters of meiotic chromosome structure and dynamics (recently reviewed in [5, 6, 7, 8, 9]). Three cohesin proteins form the ring-like core cohesin complex: SMC1; SMC3; and kleisin. Besides the canonical proteins SMC1α, SMC3, and RAD21 present in all nucleus-bearing somatic cells and in mammalian meiocytes, the latter express additional cohesins, which are largely meiosis specific: the SMC1 variant SMC1β and the kleisins RAD21L and REC8. As a fourth cohesin protein, an SA-type protein associates with the tripartite complex. SA1 and SA2 are found in somatic and some meiotic cells, SA3 (STAG3) exists almost exclusively in meiocytes. Thus, mammalian meiocytes form several different cohesin complexes.
Studies using mouse strains deficient in one or two meiotic cohesins have shed substantial light on their specific roles, although many questions remain open [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21]. Cohesin complexes determine the length of chromosome axes, i.e., of AEs and SCs, by restricting chromosome compaction that is exerted by AE/SC proteins like SYCP3 [22]. Less cohesin on these axes results in shortened axes, and in some instances, such as absence of STAG3 or combined REC8/RAD21L deficiency, the axes vanished almost completely. Synapsis is impaired to different degrees by the absence of specific cohesins, and the pairing of the X and Y chromosomes is abolished in all known cohesin deficiency models [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21]. These sex chromosomes pair at a short ca. 700-kbp region called PAR (pseudoautosomal region) and thus remain unsynapsed along most of their length. Specific proteins associate with the sex chromosomes, such as the phosphorylated H2AX (γH2AX), and a distinct chromatin domain named “sex body” forms, within which gene expression is silenced. Because in many cohesin mutants, synapsis fails on several autosomes, silencing factors are recruited to those autosomes and thus are reduced at the sex chromosomes, where some genes, such as Zfy1/2, become expressed with lethal consequences for the spermatocytes [23]. The cells die at the early/mid-pachytene stage, corresponding to stage IV of seminiferous tubule development. Besides synapsis, meiotic sister chromatid cohesion requires cohesins, which also support meiotic recombination and maintenance of proper telomere structures.
One of the major unsolved questions in meiotic cohesin biology is why mammalian meiocytes express two variants of SMC1. Are there functions specific for SMC1α versus SMC1β? In meiosis, SMC1β appears to be the more prominent SMC1 variant. At meiotic entry in preleptotene, SMC1β becomes expressed while there is plenty of SMC1α on the chromatin. SMC1α then slowly disappears, and there is not much seen on chromosomes anymore after prophase I. SMC1β remains present on the centromeres until the metaphase/anaphase II transition [11, 24]. In the absence of SMC1β, AEs and SCs are shortened in length by about half and the surrounding chromatin extends approximately (app.) twice as much from these axes. These spermatocytes show incomplete chromosome synapsis, suffer telomere damage, and loose some sister chromatid cohesion. They die in early/mid-pachytene (tubular stage IV) [24]. It was evident though that some sister chromatid cohesion remained intact in prophase I of Smc1β−/− mice, that the majority of homologs still synapsed, and that AEs and SCs formed [13, 24, 25]. Such remaining functions ought to be supported by SMC1α complexes. However, would SMC1α complexes be able to fulfill all cohesin functions in prophase I if there would be sufficient and timely expression of SMC1α? In the present study, we asked whether there are functions specific for SMC1β or whether its functions are redundant between the two SMC1 variants. The analysis of spermatocytes from a strain expressing SMC1α under control of the Smc1β gene’s expression regulatory elements in either wild-type or Smc1β−/− background showed that SMC1β can be replaced by SMC1α for many, but not all, functions. No rescue was seen in protection of telomeres, and SMC1β appears to prevent an aberrant DNA damage response at telomeres.
Results
Smc1α Transgene Expression and Spermatocyte Development
To determine whether SMC1α can substitute for SMC1β and fulfill some or all of its functions in male and female meiosis, we placed the mouse Smc1α cDNA within a bacterial artificial chromosome (BAC) containing the Smc1β gene regulatory regions (Figures 1A and S1A). Thus, Smc1α gene expression was driven from the Smc1β promoter elements. These transgenic mice were bred with SMC1β-deficient mice described before [24]. The genotype of the resulting mouse strain was named Smc1β−/−Smc1βpromSmc1α, in short, Smc1β−/−1α. We obtained five independent founders, three of which were analyzed in detail. All data presented here represent all three lines. This strain was compared to wild-type (WT) Smc1β+/+ and Smc1β−/− strains, and for some phenotypes, WT mice expressing the Smc1βpromSmc1α transgene (Smc1β+/+1α) were also investigated.
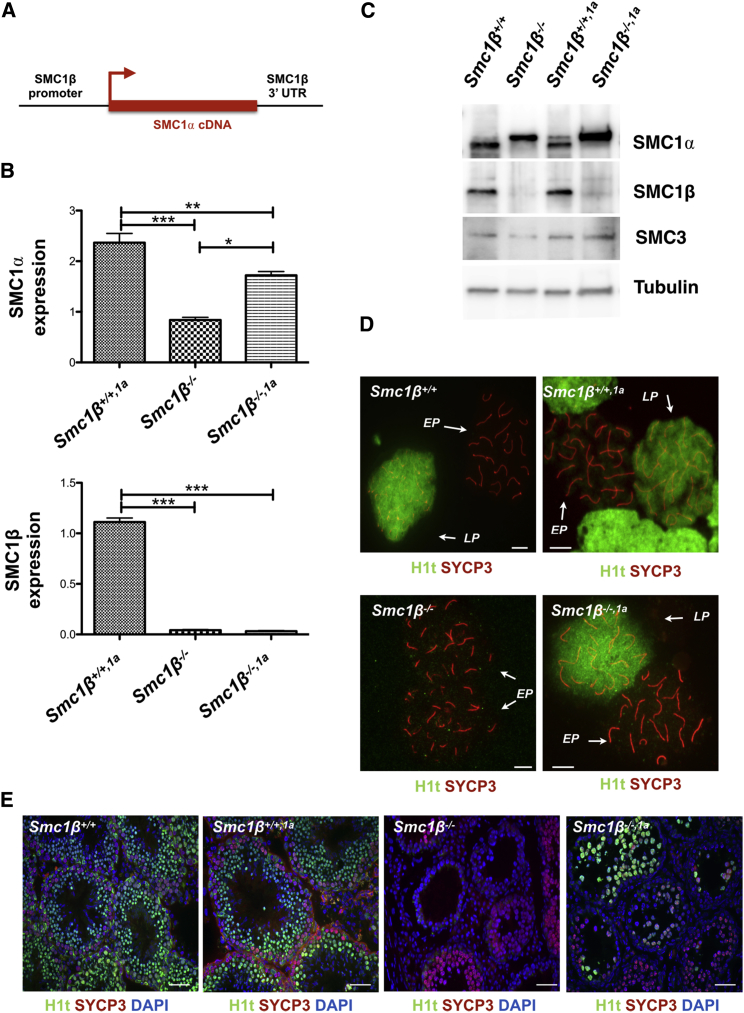
Figure 1.
Experimental Approach and SMC1α Expression
(A) Schematic diagram for SMC1α expression under control of the Smc1β promoter.
(B) Relative expression of Smc1α and Smc1β in FACS-sorted 4N spermatocytes analyzed by RT-PCR. SEM was calculated using three biological and three technical repeats for each genotype. Relative expression of Smc1α was normalized to Smc1β+/+ levels. According to Bonferroni’s multiple comparison test, the mean values for Smc1α expression (Smc1β+/+,1a: 2.365 ± 0.1826; Smc1β−/−: 0.8375 ± 0.0584; and Smc1β−/−,1a: 1.720 ± 0.0767) are all significantly different (p < 0.05). The mean values for Smc1β expression in all genotypes (Smc1β+/+,1a: 1.113 ± 0.039; Smc1β−/−: 0.0415 ± 0.00724; and Smc1β−/−,1a: 0.03 ± 0.0057) are significantly different (p < 0.0001) except between Smc1β−/−,1a and Smc1β−/− (p = 0.3441).
(C) Protein levels of SMC1α, SMC1β, SMC3, and tubulin in different genotypes.
(D and E) H1t-positive spermatocyte spreads (scale bar, 5 μm; D) and cross sections (scale bar, 50 μm; E) of all genotypes.
See also Figures S1 and S7.
First, expression of the Smc1α transgene in meiocytes was analyzed. Levels of Smc1α mRNA (Figures 1B and S1B) in fluorescence-activated cell sorting (FACS)-sorted 4N spermatocytes were about 2.4-fold increased in Smc1β+/+1α mice compared to WT and app. 2-fold higher in Smc1β−/−1α than in Smc1β−/− spermatocytes. The levels of Smc1β mRNA in WT were not affected by expression of the Smc1α transgene and were at background levels in Smc1β−/− spermatocytes. These results were reflected on the protein level (Figures 1C and S1C). In extracts from FACS-sorted 4N spermatocytes (Figure S1D), we observed two bands close to each other for SMC1α, with the upper band very prominent in the Smc1β−/− background (Figure 1C). This was independent of the Smc1α transgene. The distinct features of the two SMC1α bands are therefore subject to a separate study. In the WT background, SMC1β protein levels were not affected by Smc1α transgene expression. We also compared RNA expression in sorted Smc1β+/+1α spermatocytes of Smc1α (endogenous and transgene), Smc1α transgene, and Smc1β and observed app. the same levels of expression of the Smc1α transgene and of Smc1β (Figure S1E). This shows that, in the BAC, the Smc1β promoter drives Smc1α transgene as expected at Smc1β-like levels or slightly higher.
Next, we analyzed spermatogenesis (Figures 1D, 1E, S1D, and S1F), which was unperturbed in WT mice expressing the Smc1α transgene. Spermatogenesis in Smc1β−/− mice proceeds maximally to early/mid-pachytene, corresponding to seminiferous tubule stage IV [24]. In Smc1β−/−1α mice, however, spermatocytes reached the end of pachytene. This was evident from positive staining for the histone variant H1t (Figures 1D and 1E). Starting in mid-pachytene, expression of H1t serves as a marker for progression beyond the stage IV early/mid-pachytene checkpoint [26, 27]. In WT, 67% of pachytene chromosome spreads were strongly positive for H1t, which associates with the entire chromatin. H1t-positive cells are almost entirely absent from tubules of Smc1β−/− mice (<<1% of pachytene-like Smc1β−/− spermatocytes) [24]. In contrast, 43% of Smc1β−/−1α pachytene spermatocytes were strongly H1t positive. However, no diplotene cells were observed in Smc1β−/−1α mice. FACS analysis of total testis cell suspensions (Figure S1F) confirmed that there is no effect of transgene expression in the WT and there are no haploid cells in the Smc1β−/−1α strain, which shows about the same profile for 4N (prophase I) and 2N (somatic) cells as the Smc1β−/− mice; the small increase through late pachytene cells is not visible by this FACS analysis. There were no cells positive for pericentromeric pH3, a marker for mid-diplotene, and no cells showed desynapsis. Thus, all Smc1β−/−1α spermatocytes have died until the very end of pachytene, i.e., between tubular stage IV and stage X to the latest.
Whereas expression was readily detectable in spermatocytes, we did not obtain significant expression of the transgene in oocytes (Figures S1G–S1I). Neither in the WT nor in the Smc1β−/− background did we detect increased levels of Smc1α (Figure S1G). Phenotypes of Smc1β−/− oocytes reported earlier were not rescued. Like in the Smc1β−/− strain, there were no oocytes at the germinal vesicle stage in aged Smc1β−/−1α mice, and the oocyte number was also not altered in WT mice carrying the Smc1α transgene (Figure S1H). Similarly, metaphase I chromosome spreads of Smc1β−/−1α oocytes showed some univalents, i.e., pairs of sister chromatids that have lost their linkage to the corresponding homologous pair, like in the Smc1β−/− strain (Figure S1I). Thus, this study focused on spermatocytes.
Expression of the Smc1α transgene in WT spermatocytes did not cause any phenotype, which was confirmed by further analysis (see below). The Smc1β+/+1α mice were fully fertile, as were Smc1β+/−1α mice. In line with our central question for the ability of SMC1α to compensate for loss of SMC1β, most of the subsequent studies therefore focused on Smc1β−/−1α mice.
Chromosome Axes and Synapsis
The perhaps most prominent chromosomal phenotype of Smc1β−/− spermatocytes is the length reduction of AEs and SCs by about half [24]. Together with data from other meiosis-specific cohesin mutants, which also showed axes length reduction to varying degrees (for a table providing a recent overview, see [28]), it was proposed that cohesin complexes restrict the axes compaction that is mediated by non-cohesin proteins, such as AE protein SYCP3. This hypothesis was supported by the reconstitution of nearly normal axes length in Smc1β−/− Sycp3−/− double-deficient mice [22].
We asked whether increased expression of SMC1α rescues axes length in Smc1β−/− meiocytes. Early Smc1β−/−1α pachytene spermatocytes showed an average SC length approaching WT dimensions, i.e., 5.99 μm length compared to 7.11 μm in the WT and 3.2 μm in the Smc1β−/− samples (Figures 2A–2C), indicating a high degree of rescue of axes length. This rescue equally affected all chromosomes. Thus, SMC1α can substitute for SMC1β, suggesting that the quantity rather than the specific quality of the cohesin complexes determines axis length.
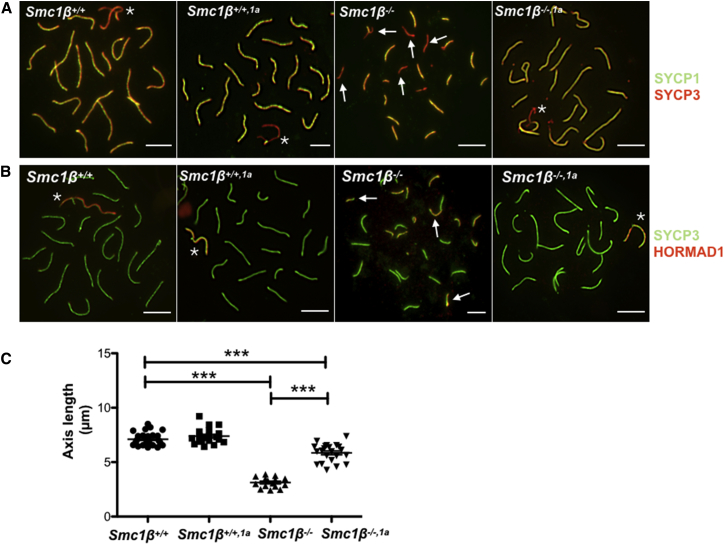
Figure 2.
Synapsis and Axis Length
(A) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β+/+,1a, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (red) for AEs/lateral elements (LEs) and with anti-SYCP1 (green) for SCs. Arrows indicate unsynapsed regions.
(B) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β+/+,1a, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (green) for AEs/LEs and anti-HORMAD1 (red) for unsynapsed region. Arrows indicate unsynapsed regions.
(C) Graphical representation of chromosome length (SYCP-stained) of spermatocyte spreads measured using ImageJ software. (n = 27, 7.111 μm [mean length; ±0.1153 μm SEM] Smc1β+/+; n = 20, 7.388 μm [±0.1561 μm] Smc1β+/+,1a; n = 15, 3.130 μm [±0.1235 μm] Smc1β−/−; n = 22, 5.852 μm [±0.1756 μm] Smc1β−/−,1a). According to Bonferroni’s multiple comparison test, all pairwise differences were statistically relevant (p < 0.05) except for the comparison of Smc1β+/+ versus Smc1β+/+,1a; scale bar, 5 μm; asterisks mark the XY chromosomes.
See also Figures S2, S3, and S7.
Synapsis of the AEs is also impaired in Smc1β−/− meiocytes, where on average 7.3 (34.7%) of the autosomes partially or entirely fail to synapse [24, 25]. The sex chromosomes never synapse in this mutant. Synapsis can be monitored by staining pachytene spermatocytes for the SC-specific SYCP1 or for HORMAD1, which associates with unsynapsed regions of chromosomes. Figures 2A and 2B show both types of analysis and demonstrates regions of asynapsis in the Smc1β−/−, but neither in the WT control nor in the Smc1β−/−1α chromosomes. Smc1β−/−1α autosomes synapse along their entire length, and like in WT, the sex chromosomes synapse at the PAR, although they display some morphological abnormalities (see below). This shows that synapsis was rescued in the Smc1β−/−1α cells.
Cohesin Association with Chromosomes
Would Smc1α transgene expression in the SMC1β-deficient spermatocytes affect the presence of other cohesins on the AEs or SCs? In Smc1β−/− spermatocytes, SMC3, SMC1α, RAD21, REC8, RAD21L, and STAG3 are still present on the axes, although at reduced levels and often in a more punctuate pattern [24].
Figures S2 and S3 show cohesins on mid-pachytene chromosomes in control samples (WT; Smc1β−/−) and in Smc1β−/−1α cells. The higher compaction of Smc1β−/− chromosomes tends to yield more intense signals, and thus, intensity comparisons to Smc1β−/− chromosomes are not accurately possible. Taking this limitation into consideration, the signals for several cohesins seem to be slightly more intense on Smc1β−/−1α chromosomes. REC8 and RAD21L signals appear more intense on Smc1β−/−1α chromosomes, similar to WT, possibly indicating that SMC1α formed axes-associated complexes with these kleisins. Whether this applies also to SMC3 and STAG3 cannot be determined with certainty from these chromosome spreads. Loss of STAG3 confers reduction of axis length even more dramatic than that of Smc1β−/− cells. This suggested that SMC1α/STAG3 complexes contribute very significantly to axis length [21]. In Smc1β−/−1α cells, the additionally expressed SMC1α may recruit STAG3 to chromosome axes, suggesting that the increase in axis length happens in a STAG3 complex. Overall, the cohesins are present on Smc1β−/−1α chromosomes and tend to appear moderately enhanced.
DNA Double-Strand Break Repair and Sex Chromosome Morphology
Smc1β−/− spermatocytes properly generate SPO11-dependent DSBs but do not repair them as efficiently as WT cells [25]. DMC1 and RAD51 foci, which form at DSBs as part of the repair process, persist longer. Analyzing RAD51 and DMC1 foci in early pachytene cells of WT, Smc1β−/−, and Smc1β−/−1α spermatocytes showed that expression of SMC1α rescues the processing of RAD51 and DMC1 foci, which are processed at WT levels (Figures 3A–3D).
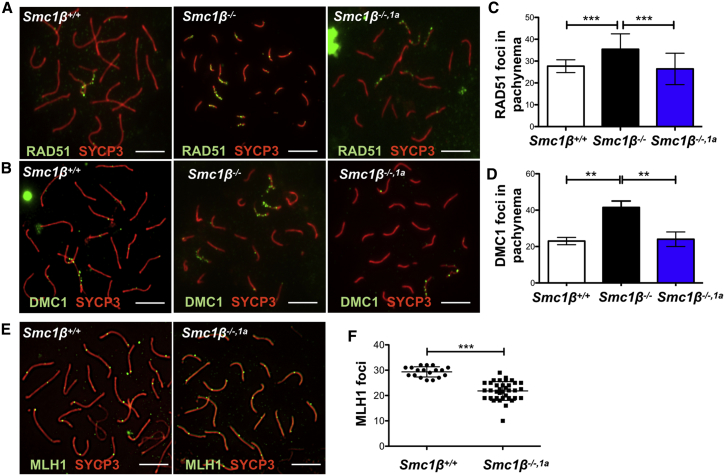
Figure 3.
Double-Strand Break Processing
(A and B) Immunofluorescence staining of spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, probed with anti-SYCP3 (red) for AEs/LEs and anti-RAD51 (A) or anti-DMC1 (green; B) as indicated for DNA double-strand break repair foci.
(C) Graph showing numbers of RAD51 foci in pachytene stage cells (n = 25, 27 RAD51 foci ± 2.949 SD, Smc1β+/+; n = 56, 35 RAD51 foci ± 7.045 SD, Smc1β−/−; n = 26, 26 RAD51 foci ± 7.184 SD, Smc1β−/−,1a). The p values between all genotypes are <0.0001 except between Smc1β+/+ and Smc1β−/−,1a (p = 0.429).
(D) Graph showing numbers of DMC1 foci in pachytene stage cells. (n = 140, 23 DMC1 foci, ±1.89 SD, Smc1β+/+; n = 100, 42 DMC1 foci ± 5.89, Smc1β−/−; n = 150, 25 DMC1 foci ± 6.184, Smc1β−/−,1a). The p values between all genotypes are <0.0001 except between Smc1β+/+ and Smc1β−/−,1a (p = 0.258).
(E) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, probed with anti-SYCP3 (red) for AEs/LEs and anti-MLH1 for late recombinational nodule.
(F) Graphical representation of number of MLH1 foci in Smc1β+/+ and Smc1β−/−,1a spermatocytes. (Smc1β+/+: n = 18, 29 MLH1 foci ± 2.029 SD; Smc1β−/−,1a: n = 36, 22 MLH1 foci ± 3.713; p < 0.0001 according to unpaired t test; scale bar, 5 μm).
See also Figures S4 and S7.
Processed DSBs may turn into recombination foci. To test this, we analyzed the occurrence of MLH1 foci (Figures 3D and 3E), which from mid-pachynema onward mark sites of future chiasmata. There are no MLH1 foci in Smc1β−/− spermatocytes, which fail to efficiently process DSBs and do not reach mid-to-late pachynema [24]. Smc1β−/−1α spermatocytes, however, showed MLH1 on each chromosome. The number of MLH1 foci was about 22 per cell compared to 30 foci per cell in WT. This confirms not only the extended progression of Smc1β−/−1α spermatogenesis but also shows that SMC1β is neither specifically required to process DSBs nor to form MLH1 foci. An additional marker to visualize recombination foci is CDK2, which localizes to some of the MLH1 foci and is required for meiotic recombination [29, 30]. Staining for CDK2 in spermatocytes of the three genotypes revealed that the WT control shows app. 9.36 (average foci number; ±3.128 SD) CDK2 foci, separate from signals at the telomeres (Figures S4A and S4B). Smc1β−/− spermatocytes do not show any CDK2 foci. In Smc1β−/−1α spermatocytes, 3.33 (±1.12) CDK2 foci appear, one-third of WT. CDK2 much more prominently localizes to telomeres [31]. We observed a similar absence of CDK2 signals on some telomeres of Smc1β−/− and Smc1β−/−1α spermatocytes, consistent with our analysis of telomeres (see below).
Rescue of DSB repair is also indicated by the WT-like behavior of γH2AX, which associates with DSBs. In Smc1β−/− spermatocytes, γH2AX persists on many autosomes and thus is diffusely distributed in clouds within the nuclei (Figure 4A) as opposed to associating like in WT with the sex body, the specifically structured chromatin at the sex chromosomes. Smc1β−/−1α displayed a proper sex body, intensely stained for γH2AX, which was not distributed on autosomes.
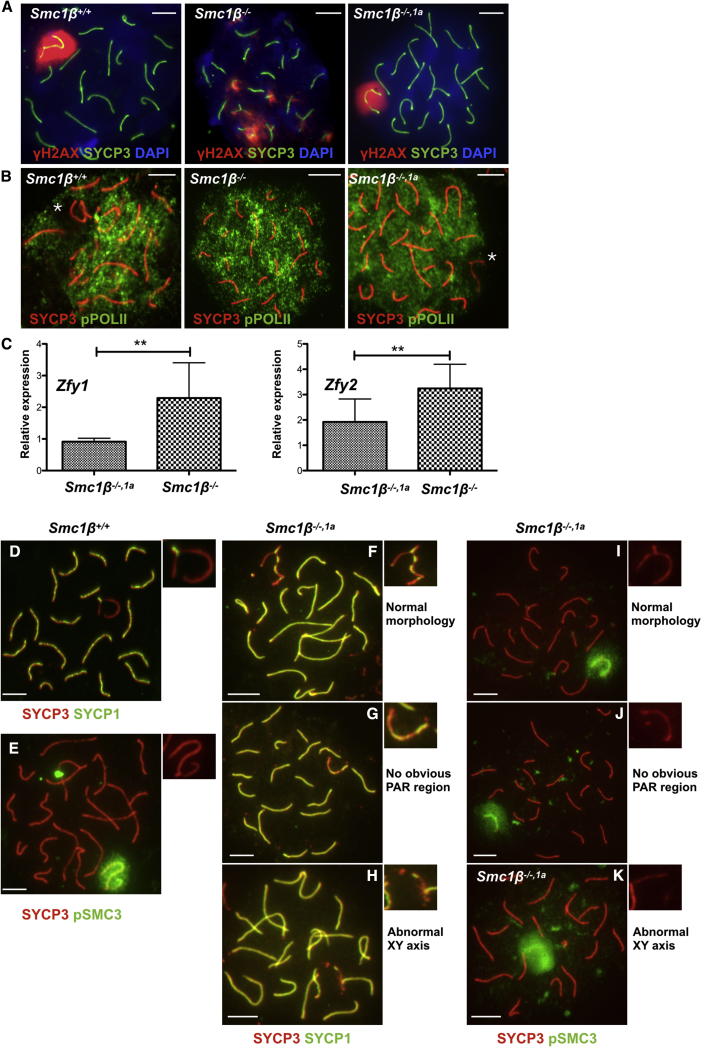
Figure 4.
Sex Body Formation
(A) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (green) for AEs/LEs and with anti-phosphorylated H2AX (γH2AX) for DNA damage.
(B) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (red) for AEs/LEs and with anti-phosphorylated RNA POLII (green). Asterisk indicates sex chromosomes.
(C) Graphical representation of relative expression of ZFY1 (Smc1β−/−,1a 0.9158 ± 0.10584, Smc1β−/−: 2.292 ± 1.212 SD; n = 3; p < 0.0001) and ZFY2 (Smc1β−/−,1a 1.924 ± 0.904, Smc1β−/− 3.243 ± 1.052 SD; [n = 3; p = 0.0023]). Relative expression was normalized to Smc1β+/+ levels.
(D–K) Spermatocyte chromosome spreads of Smc1β+/+ and Smc1β−/−,1a mice, stained with anti-SYCP3 (red) for AEs/LEs and with anti-SYCP1 (green) (D and F–H) and with anti-phosphorylated SMC3 (E and I–K).
(D, E, F, and I) Normal sex chromosome morphology.
(G and J) No obvious sex chromosome PAR region.
(H and K) Abnormal XY axis morphology.
See also Figure S7.
The sex chromosomes are transcriptionally repressed, which can be seen in WT cells by absence of signals for phosphorylated RNA polymerase II (Figure 4B). No such distinct region (“black hole”) can be seen in Smc1β−/− spermatocytes, but in Smc1β−/−1α spermatocytes, the sex chromosomes are devoid of piwiRNA (pRNA) PolII signals. This supports the notion that, in Smc1β−/−1α spermatocytes, a functional, transcriptionally silenced sex body is formed. Smc1β−/− spermatocytes fail to silence gene expression on the sex chromosomes and express the Zfy1/2 gene pair known to trigger apoptosis if expressed in early/mid-pachytene [25, 32]. Because silencing of the sex chromosomes appeared normal in Smc1β−/−1α spermatocytes, their extended survival may be supported by repressed expression of Zfy1/2. RT-PCR revealed that there is indeed significantly less Zfy1/2 expressed in Smc1β−/−1α spermatocytes (Figure 4C). This confirms that SMC1α can substitute for SMC1β in forming sex body chromatin and silencing sex chromosomes.
Localization of H3K9me3, which marks heterochromatin [33] and is aberrantly diffuse in Smc1β−/− spermatocytes, appeared normal in Smc1β−/−1α spermatocytes, indicating that the distribution of heterochromatin, particularly at the centromeres, is also rescued (Figure S4C).
Further detailed analysis showed though that sex chromosome pairing is not perfect. Staining WT pachytene cells for SYCP1 and SYCP3 yielded sex chromosomes with the expected morphology that were double positive only at the synapsed PAR (Figure 4D). In the Smc1β−/−1α spermatocytes, 38.6% (n = 417) of the sex chromosomes showed a WT-like morphology and staining, but there were cells (52.9%) where we did not detect a PAR and some that showed an irregular sex chromosome axes morphology (8.4%; Figures 4E–4K). Similarly, when the cells were stained for SYCP3 and the phosphorylated form of SMC3, which stains the sex body in late pachytene [25], most of the Smc1β−/−1α cells showed a normal morphology of the sex chromosomes, but some sex chromosomes (40%; n = 300) appeared more stretched and/or the two sex chromosomes were slightly apart within the sex body. Thus, whereas most of the sex chromosome features lost in the Smc1β−/− spermatocytes are rescued by SMC1α transgene expression, the rescue is not complete.
Centromere and Telomere Aberrations
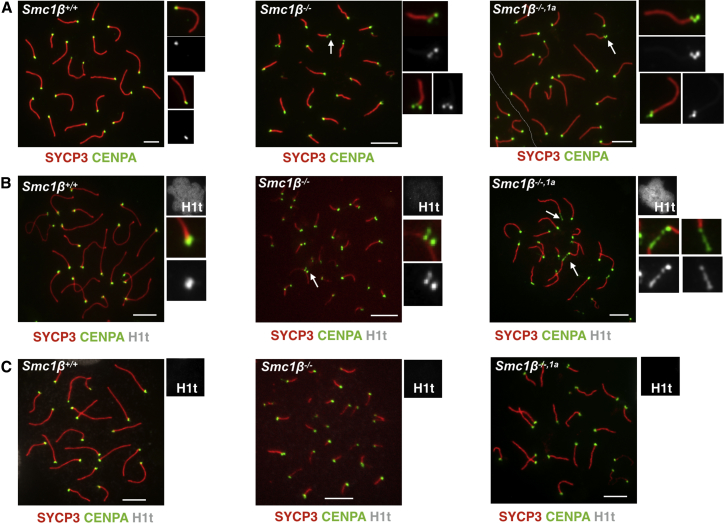
In mice, the centromeres are located in the subtelomeric region, and their cohesion is affected by SMC1β deficiency, which does not affect much of cohesion along chromosome arms [25]. Centromeric cohesion can be assessed by staining for CENP-A. Fully synapsed four sister chromatids in full cohesion display one CENP-A signal per chromosome. Loss of centromeric cohesion yields two separate signals. However, loss of synapsis at the centromeres has the same effect. If there are three or four signals, loss of cohesion and of synapsis must have occurred.
In WT, there was one CENP-A signal per chromosome (Figure 5A). In the Smc1β−/− spermatocytes, there were at least four chromosomes per cell that show 2 clearly separate CENP-A spots and at least one chromosome that shows 3 spots (marked by an arrow). A similar number of chromosomes carrying two or three separate signals was observed in Smc1β−/−1α cells (Figure 5A). Anti-centromere antibody (ACA) stains centromeric/pericentromeric heterochromatin and produces a more diffuse signal compared to CENP-A but confirmed the continued presence of at least 2 signals on several chromosomes per cell, and in some instances, the loss of cohesion at the centromere was seen clearly on sex chromosomes (Figure S5).
Figure 5.
Centromeres
(A) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (red) for AEs/LEs and with anti-CENP-A (green) for centromere.
(B and C) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (green) for AEs/LEs, anti-CENPA (red), and anti-H1t (gray) showing H1t-positive and H1t-negative spermatocytes, respectively. AEs/LEs stained with anti-SYCP3 and centromere with anti-CENP-A. White arrows indicate abnormal centromere structure (scale bar, 5 μm).
See also Figures S5 and S7.
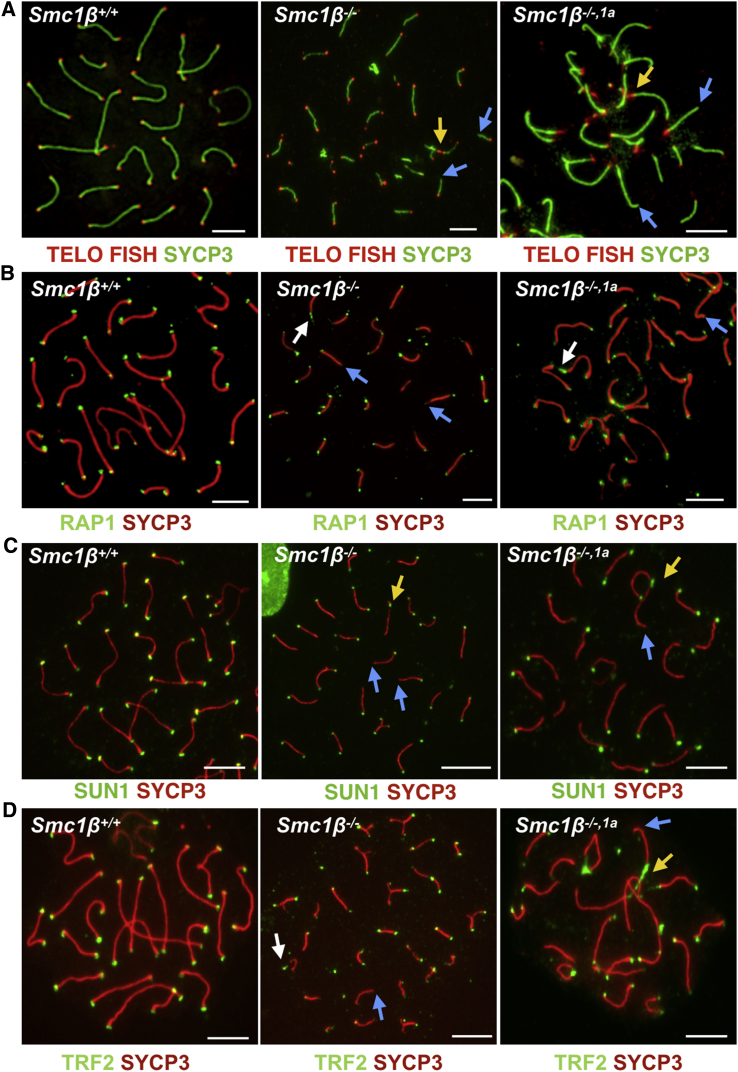
To distinguish between early pachytene and late pachytene cells, we co-stained H1t together with SYCP3 and CENP-A (Figures 5B and 5C). There were no or only very weakly H1t-positive Smc1β−/− spermatocytes, but the late pachytene Smc1β−/−1α chromosomes in 96% of these cells showed several grossly aberrant CENP-A signals, which appeared as long stretches or bridges between chromosome ends. Some other signals showed as 2 dots, some as three. The stretches and bridges were not apparent or very small in H1t-negative cells. Thus, the incidence of centromeric aberrations, which included loss of centromeric cohesion and synapsis, increased toward the end of pachytene and was not rescued in Smc1β−/−1α cells. The centromeric stretches and bridges seen in late pachytene Smc1β−/−1α spermatocytes were reminiscent of telomere aberrations that we reported earlier for Smc1β−/− mice [13]. There were chromosomes without telomeres, stretches of telomeres separated from the main axes, as well as telomere bridges and fusions.
To test whether expression of SMC1α protects telomeres from such damage, we stained telomeres on chromosome spreads by telo-fluorescence in situ hybridization (FISH) and for RAP1 (Figures 6A and 6B). As expected, the Smc1β−/−, but not the WT, spermatocytes showed telomere aberrations. However, the Smc1β−/−1α cells also showed these telomere aberrations in both types of staining. This was further confirmed by staining for the telomere protein TRF2 and for the telomere-membrane anchor protein SUN1 (Figures 6C and 6D). The average number of telomere aberrations per cell was 11 to 14 whereas, in the WT control, there were none (app. 500 telomeres analyzed per genotype). Overall, there was no statistically significant difference between Smc1β−/− and Smc1β−/−1α cells (Figure S6A). These aberrations included solitary telomere signals and axes lacking telomeres as the most frequent types of aberrations (70%–80%), as well as axes featuring long stretches of telomeres and some (<5% of all aberrations) end-to-end associations leading to chromosome fusions or occasionally a circular chromosome.
Figure 6.
Telomeres
(A) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (green) for AEs/LEs and with telomere FISH probe (red).
(B–D) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 (red) for AEs/LEs and for telomeres with anti-RAP1 (green; B), anti-SUN1 (green; C), and anti-TRF2 (green; D). Yellow arrow indicates fused telomeres, blue arrow indicates chromosome ends without telomere, and white arrow indicates telomere stretches (scale bar, 5 μm).
See also Figures S6 and S7.
Quantification of the telo-FISH signal provides an indication for the presence of telomeric sequences, i.e., for telomere length and the length distribution profile. Figures S6B–S6E show that there is a large decrease in telomere length from WT to Smc1β−/− spermatocytes as reported earlier [13]. The Smc1β−/−1α cells show a similar length reduction with mostly no statistical difference to Smc1β−/−, although there appears to be a mild increase in area. We cannot exclude a very minor rescue of the telomere length phenotype, possibly an effect of the overall increase in axis length.
DNA Damage Response Activity at Spermatocyte Chromosome Ends
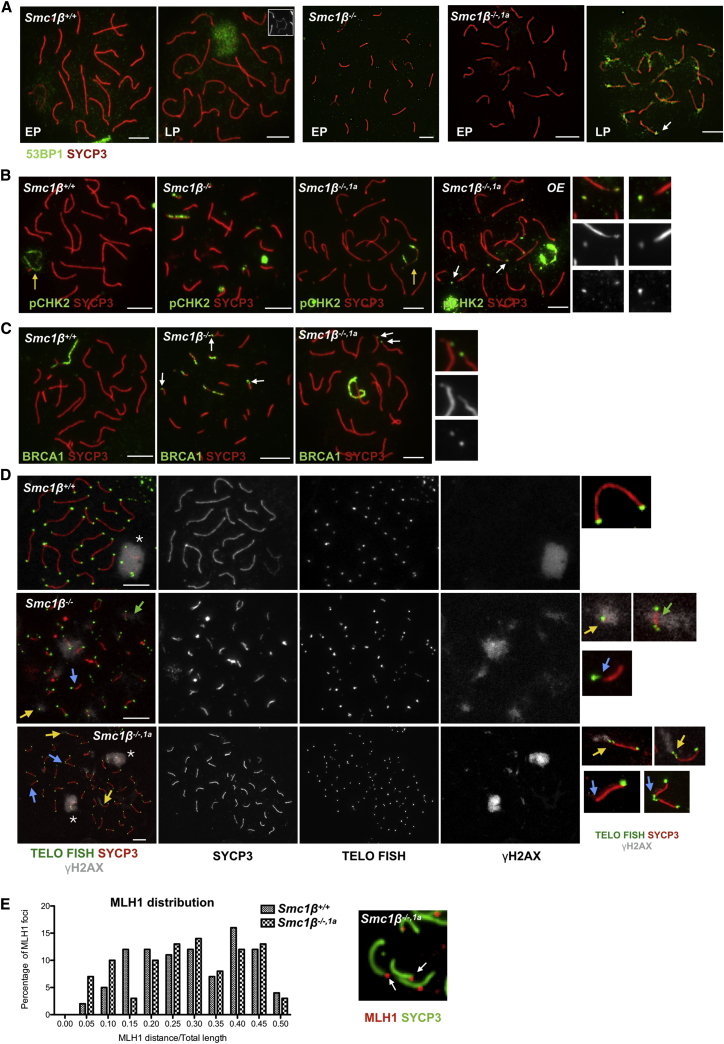
Thus, the centromere and telomere phenotypes were not rescued by SMC1α. Both reflect processes that occur at the chromosomal ends, and we asked whether these ends suffered damage that would evoke a DNA damage response (DDR). We analyzed DDR proteins on these chromosomes. Figure 7 shows staining for the end-joining protein 53BP1, which in WT pachytene cells localizes to the sex chromosomes only from mid-pachytene onward [34, 35]. In Smc1β−/− cells, which die in early/mid-pachytene, accumulation of 53BP1 on sex chromosome axes was not seen as expected, and it was also absent of WT and Smc1β−/−1α early/mid-pachytene cells (Figure 7A). In late pachytene, staining of the WT sex body with anti-53BP1 was readily detectable, whereas in the Smc1β−/−1α cells, the staining was much more widespread and particularly heavy at the ends of many chromosomes. Often, one end per chromosome was 53BP1 positive. Together, this indicated that a DDR occurred at ends of many chromosomes.
Figure 7.
Recombination at Dysfunctional Chromosome Ends
(A) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 for AEs/LEs and with anti-53BP1 for DNA repair protein (EP, early pachynema; LP, late pachynema). White arrow shows accumulation of 53BP1 at the end of the chromosome.
(B) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 for AEs/LEs and with anti-pCHK2. OE indicates over-exposed image of Smc1β−/−,1a spermatocyte. White arrow shows accumulation of phosphorylated CHK2 at the end of the chromosome. Yellow arrow indicates sex chromosomes. Magnified images show phosphorylated CHK2-positive chromosome ends.
(C) Spermatocyte chromosome spreads of Smc1β+/+, Smc1β−/−, and Smc1β−/−,1a mice, stained with anti-SYCP3 for AEs/LEs and with anti-BRCA1 for DNA repair protein. White arrow shows accumulation of BRCA1 at the end of the chromosome. Magnified images show BRCA1-positive chromosome ends.
(D) Chromosome spreads stained for telomere by telo-FISH (green), with anti-SYCP3 for AEs/LEs and with anti- phosphorylated H2AX (γH2AX) for DNA damage. Magnified images show normal and dysfunctional telomeres. Yellow arrow indicates γH2AX-positive dysfunctional telomere. Blue arrow indicates γH2AX-negative dysfunctional telomere. Green arrow shows γH2AX-positive asynapsed chromosome.
(E) Graphical representation of MLH1 distribution along the chromosome of Smc1β+/+ and Smc1β−/−,1a spermatocytes. White arrows in the magnified image of Smc1β−/−,1a spermatocyte show MLH1 at the end of chromosome. According to Chi-square analysis, the histograms of Smc1β+/+ and Smc1β−/−,1a spermatocytes are not statistically significantly different; p = 0.2644 (scale bar, 5 μm).
See also Figure S7.
To further investigate this apparent DDR, the ATM (ataxia telangiectasia mutated) effector kinase CHK2 was examined, which becomes phosphorylated when activated and localizes to the sex chromosome axes [36]. In WT, anti-pCHK2 produced signals only on the sex chromosomes (Figure 7B), but in the Smc1β−/− spermatocytes and in the Smc1β−/−1α cells, signals were also detected on some autosome ends or near ends, indicating broken-off telomeres (Figure 6 and for Smc1β−/−) [13]. In Smc1β−/−1α cells, staining was also observed sometimes at the sex chromosomes like in the WT, consistent with rescue of sex chromosome pairing.
The DDR protein BRCA1, which functions as a recruiter of DDR kinases and downstream effectors mostly in DSB repair by homologous recombination [37], was reported to localize to sex chromosome axes in pachytene spermatocytes [38] and suggested to promote meiotic DSB repair [39]. The presence of BRCA1 on sex chromosomes was confirmed for the WT and seen also for Smc1β−/−1α cells (Figure 7C). In Smc1β−/− cells, the individualized sex chromosomes and unsynapsed autosomes also carried some BRCA1. In addition, several ends of Smc1β−/− and of Smc1β−/−1α chromosomes showed distinct BRCA1 signals, further supporting the hypothesis of a DDR activity at these ends.
To further assess this activity and to test the overlap with telomeres, we stained the chromosomes for γH2AX as a DSB repair marker and for telomeres. Except the expected staining of the sex body, there was no γH2AX signal in WT cells (Figure 7D). The Smc1β−/− cells showed rather widespread signals, showing asynapsis of many chromosomes, but often also overlapped with particular intensity with the telomere FISH signal. A very similar pattern was observed for Smc1β−/−1α chromosomes, where, besides the expected sex body staining, γH2AX signals were accumulating at telomeres, including those that showed aberrant structures like stretches. Some chromosomes that lacked telomere sequences entirely or appeared fused did not display γH2AX signals. Here, the DDR response was presumably finished or never initiated. About five chromosomes per cell showed γH2AX signals at the chromosome ends, which, considering that there were app. 11 telomere aberrations per Smc1β−/−1α cell, indicates ongoing DDR responses at about half of the defective telomeres. Slightly lower numbers of BRCA1 (3.3), pCHK2 (3.6), and 53BP1 (3.2) signals were found at chromosome ends, which may not be as persistent as γH2AX signals (between 1,558 and 2,993 telomeres were analyzed).
Because there is DDR activity at the ends of many Smc1β−/− spermatocyte chromosomes, would this involve the homologous recombination machinery active in spermatocytes? Analyzing the spatial distribution of MLH1 foci, we observed a shift toward the ends of chromosomes (Figure 7E). There were significantly more MLH1 foci at close distance to the chromosomal ends on the Smc1β−/−1α chromosomes than on WT chromosomes. Within the terminal 10 μm of the chromosomes, there were 7% of MLH1 foci in WT and 18% of MLH1 foci in Smc1β−/−1α spermatocytes (p < 0.0001). This chromosome-terminal localization of MLH1, together with the HR promoter BRCA1, suggests a DDR-activated homologous recombination activity at telomeres. Prevention of this activity requires SMC1β. Crossover interference appears still to work as there were no MLH1 foci in the vicinity of the terminal foci.
Discussion
Why do mammalian meiocytes express two SMC1 variants? Which functions of the two SMC1 proteins are unique to SMC1β, and which are redundant? One approach to get initial insights into this problem was to identify those functions that depend on the presence of one SMC1 type. SMC1α deficiency models are very difficult to generate, because the essential Smc1α gene resides on the X chromosome and its expression is very sensitive to manipulation (unpublished data). In any case, such a strain would not reveal whether expression of SMC1α in meiocytes instead of SMC1β would rescue deficiencies observed in Smc1β−/− mice. SMC1β deficiency was extensively studied [13, 24, 25, 40], yet the division of functions between SMC1α and SMC1β remained unclear. Therefore, we expressed SMC1α driven by the Smc1β promoter elements to determine whether SMC1α can rescue SMC1β-deficiency phenotypes. Several phenotypes were indeed complemented, but certain phenotypes were not, suggesting that SMC1α and SMC1β are functionally redundant in many, but not all, respects. Figure S7 summarizes our findings in a model. Whereas axis length, synapsis, sex body formation, and processing of programmed DSBs were restored to near WT levels, telomere integrity was not. This also showed that the telomere defects observed in absence of SMC1β are not a consequence of reduced axis length of Smc1β−/− spermatocytes.
The data further suggest that spermatocyte death observed in early/mid-pachytene in the Smc1β−/− strain is not a consequence of telomere aberrations. These aberrations persist in the Smc1β−/−1α strain, and yet the cells develop further to the very end of pachytene. Our earlier notion that death of Smc1β−/− spermatocytes is likely triggered by expression of the derepressed sex chromosome genes Zfy1/2, whose expression we now observed to be repressed in Smc1β−/−1α spermatocytes, is thus further supported.
Expression of the Smc1α transgene was clearly seen at the level of mRNA. Immuno-probing of FACS-sorted 4N spermatocyte extracts for SMC1α showed two bands close together. The lower band was most prominent in the WT background, whereas in Smc1β−/− cells, the upper band was strongest. This was independent of the Smc1α transgene, suggesting that it has no relevance for this study. Whereas the precise nature of the two bands and their distinct representation is not known, they may, for example, reflect different posttranslational modifications (PTMs) of SMC1α if SMC1β is absent. The unavoidable differences in cell composition of the FACS-sorted populations (the WT background showed more fetal calf serum [FSC]-low cells) may have also contributed, as the lower band may be the prominent form in the WT background.
The functional redundancy of both SMC1 variants suggests that, for some roles of cohesin, the total amount of cohesin in spermatocytes matters much more than the specific type of SMC1. Indications that cohesin dosage is important came from studies on heterozygote cohesin-deficient mice, which are fertile but show subtle changes in meiotic chromosome appearance and recombination frequencies [40]. Axis length, repair of DSBs, and synapsis are intimately linked in prophase I, and it is likely that the contribution of cohesin to axis length is key not only for a proper axis: loop ratio but also for efficient DSB repair and synapsis. Earlier, we showed that the antagonistic interplay of SMC1β and SYCP3 determines axis length whereby SMC1β limits the compaction mediated by SYCP3. This is consistent with the hypothesis that the amount of cohesin available to be loaded onto the axis is a length-determining factor.
In many Smc1β−/−1α spermatocytes, we did not detect PAR synapsis and some cells showed an irregular sex chromosome axes morphology. This may indicate that SMC1α cannot fully restore sex chromosome pairing in all cells, but the morphological differences are often small, depend on staining intensities, and may be rather a result of slightly suboptimal levels of SMC1α protein in these cells. The interpretation of rather small deviations from the WT is difficult. Unlike in Smc1β−/− spermatocytes, both Smc1β−/−1α sex chromosomes resided in the sex body, even if slightly apart. Overall, most of the sex chromosome features lost in the Smc1β−/− spermatocytes, particularly sex body formation and gene silencing, are rescued by Smc1α transgene expression.
For most roles of cohesin in early meiosis, the two SMC1 variants seem to be redundant. There is no specific requirement for SMC1β in preserving axis length, supporting efficient repair of Spo11-introduced DSBs, promoting synapsis and enabling sex chromosome pairing, sex body formation, and silencing of sex chromosome genes. This suggests that both SMC1 variants can form cohesin complexes with similar or identical activities and likely the same cohesin protein partners. This is consistent with the above concept that, for some roles, cohesin quantity is more important than cohesin quality. Earlier data suggested the formation of six or more distinct cohesin complexes in mammalian meiocytes, with certain preferential cohesin subunit associations for either SMC1α or SMC1β. Both SMC1 variants were found in either co-precipitation studies or by concluding from cohesin deficiency models to associate with any of the three kleisins and any of the three STAG (SA) proteins, although to a different extent. Some associations were reported only in specific cohesin mutant backgrounds, leaving it uncertain whether these complexes also exist in WT spermatocytes [16, 17, 18, 19, 20, 21, 24, 28, 41, 42]. The data reported here suggest that there is more flexibility in cohesin complex formation than perhaps previously appreciated, for SMC1α replaces SMC1β in many functions. Variable associations of either one SMC1/SMC3 heterodimer with a kleisin and STAG protein may depend primarily on the relative concentration of the SMC1 variants. There is certainly an upper limit of how many cohesin complexes can be loaded onto a chromosome, and thus, there may be competition between the variants as long as they are both present in early prophase I.
As reported earlier, SMC1β contributes to centromeric cohesion [25], and unlike many other phenotypes, the loss of centromeric cohesion in Smc1β−/− spermatocytes is not rescued in Smc1β−/−1α spermatocytes. Nevertheless, from earlier studies, it was clear that not all centromeric cohesion depends on SMC1β as some is preserved in Smc1β−/− spermatocytes and thus can only be provided by SMC1α. Why then does overexpression of SMC1α not rescue loss centromeric cohesion? Murine centromeres are telocentric, and because meiotic telomere integrity requires SMC1β, the structural aberrations of telomeres in absence of SMC1β may also affect centromeres. These telomere deficiencies cannot be rescued by SMC1α and thus may impair centromeric cohesion. This speculation is supported by the observation that about a fifth or fourth of telomeres are defective and the same number of centromeres display multiple centromere signals. DDR and DNA repair proteins accumulate at many telomeres in absence of SMC1β, indicating widespread telomere damage. The full extent of telomere damage may thus be underestimated by microscopic telo-FISH analysis. Were expression levels of the SMC1α transgene too low to rescue telomere damage? Considering the extensive rescue of many other phenotypes, including axis length that requires decorating with cohesin all along the chromosomes, it is unlikely that expression levels were not sufficiently high to rescue phenotypes at the rather short telomeres.
How is SMC1α different from SMC1β so that SMC1α cannot protect telomere integrity? The amino acid sequences differ mainly in the C terminus, where only SMC1β carries a stretch of basic amino acids, which we showed earlier binds DNA and promotes reannealing of cDNA single strands [11]. This C-terminal tail may either serve as an interaction module specifically for telomere-relevant factors and/or it may help promote a particular telomere DNA structure, such as t-loops. We observed that DDR proteins accumulate at the telomeres and that MLH1 foci are biased toward the telomeres in absence of SMC1β. This may suggest that SMC1β protects telomeres from an alternative-lengthening-of-telomeres (ALT)-like process. It may also suggest that SMC1β preserves damaged telomere DNA structures that would trigger the DDR. Both explanations are not mutually exclusive. Given the ability of SMC1α to restore MLH1 foci and given the well-documented structural role of cohesins on meiotic chromosomes, we think it likely that SMC1β supports a protective telomere DNA structure. These questions may be addressed in future studies now that we know of the specific role of SMC1β at telomeres.
STAR★Methods
Key Resources Table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SMC1α (IF: 1:100; IB: 1:2000) | Bethyl | Cat# A300-055A; RRID: AB_2192467 |
| Rabbit anti-RAD51 (IF: 1:100) | Santa Cruz | Cat# SC-8349; RRID: AB_2253533 |
| Rabbit anti-DMC1 (IF: 1:100) | Santa Cruz | Cat# SC-22768; RRID: AB_2277191 |
| Rabbit anti-RAD21 (IF: 1:200) | Bethyl | Cat# A300-080A; RRID: AB_2176615 |
| Rabbit anti-RAD21 (IF: 1:100) | Abcam | Cat# ab154769 |
| Rabbit anti-RAD21L (IF: 1:500) | Dr. A. Pendas | N/A |
| Rabbit anti-STAG3 (IF: 1:100) | Jessberger Lab | N/A |
| Rabbit anti-SYCP3 (IF: 1:500) | Novus Biologicals | Cat# NB300-230; RRID: AB_10001748 |
| Human anti-ACA (IF: 1:5) | Antibodies Inc. | Cat# 15-235-0001 |
| Rabbit anti-antiSMC1b-N (5048) (IF: 1:100) | Jessberger Lab | N/A |
| Rabbit anti-SMC3 (IF: 1:100; IB: 1:2000) | Bethyl | Cat# A300-060A; RRID: AB_67579 |
| Mouse anti-γH2AX Ser 139 (IF: 1:700) | Upstate | Cat# 05-636; RRID: AB_309864 |
| Mouse anti-SYCP3 (clone 60C10) (undil. supernatant) | Dr. C. Heyting | N/A |
| Guinea pig anti-HORMAD1 (IF: 1:700) | Dr. A. Toth | N/A |
| Rabbit anti-SYCP1 (IF: 1:500) | Abcam | Cat# ab15090; RRID: AB_301636 |
| Rabbit anti-CENPA (IF: 1:100) | Cell Signaling | Cat# 2048S; RRID: AB_1147629 |
| Guinea pig anti-REC8 (IF: 1:100) | Dr. C. Höög | N/A |
| Guinea pig anti-SUN1 (IF: 1:500) | Dr. M. Alsheimer | N/A |
| Rabbit anti-pChk2 (Thr68) (IF: 1:100) | Cell Signaling | Cat# 2661T; RRID: AB_331479 |
| Mouse anti-MLH1 (IF: 1:50) | Cell Signaling | Cat# 3515S; RRID: AB_2145615 |
| Rabbit anti-TRF2 (IF: 1:100) | Novus Biologicals | Cat# NB110-57130; RRID: AB_844199 |
| Rabbit anti-RAP1 (IF: 1:50) | IMGENEX | Cat# IMG-289; RRID: AB_317310 |
| Rabbit anti-53BP1 (IF: 1:200) | Novus Biologicals | Cat# NB100-904; RRID: AB_10002714 |
| Mouse anti-CDK2 (IF: 1:100) | Santa Cruz | Cat# SC-6248; RRID: AB_627238 |
| Rabbit anti-H3K9me3 (IF: 1:500) | Upstate | Cat# 07-442; RRID: AB_310620 |
| Goat Anti-mouse IgG-Cy3 (IF: 1:500) | Biolegends Inc. | Cat# 405309; RRID: AB_893530 |
| Goat Anti-Rabbit IgG-Alexa Fluor 488 (IF: 1:500) | Invitrogen | Cat# A11034; RRID: AB_2576217 |
| Goat Anti-mouse IgG-Alexa Fluor 488 (IF: 1:500) | Invitrogen | Cat# A11001; RRID: AB_2534069 |
| Goat Anti-Mouse IgG-FITC (IF: 1:500) | Bio-Rad/AbD Serotec | Cat# 101002; RRID: AB_609711 |
| Goat Anti-Guinea pig-Alexa Fluor 568 (IF: 1:500) | Invitrogen | Cat# A11075; RRID: AB_141954 |
| Goat Anti- Human-Alexa Fluor 568 (IF: 1:500) | Invitrogen | Cat# A21090; RRID: AB_1500627 |
| Goat Anti-Rabbit-HRP (IF: 1:5000) | Jackson Lab | Cat# 111-035-003; RRID: AB_2313567 |
| Oligonucleotides | ||
| Smc1α Primer: (F: 5′-CCCAATGGCTCTGGTAAGTCA-3′; R: 5′-GCTCCATGTATCAGGTCCCG-3′) | this study | N/A |
| Smc1β Primer:(F: 5′-GCATGGATTGCTTGGAAGAT-3′; R: 5′-CTGACGTTTTCCCTCATGGT-3′) | this study | N/A |
| Smc1α-tg F: CCAAGTACCCAGATGCCAAC | this study | N/A |
| Smc1β- 3′UTR R: TAGCTGGTGGCTGTTCAAGA | this study | N/A |
| Zfy1 Primer: (F: 5′-CAGATTGTGTTTCTGAAGCAGTCT-3′; R: 5′-TCCTGACTCTGCATTCATGG-3′) | [25] | N/A |
| Zfy2 Primer: (F: 5′-TCTGGAGCAGCAAGATGATG-3′; R: 5′-TGCACACCTTGATAACTTCTGG-3′ | [25] | N/A |
| Gapdh Primer: (F: 5′-GGAGTTGCTATGAAGTTGC-3′; R: 5′-GAGAAACCTGCCAAGTATGAC-3′) | [25] | N/A |
| Rsp16 Primer: (F: 5′-AATGGGCTCATCAAGGTGAACGGA-3′; R: 5′-TTCACACGGACCCGAATATCCACA-3′) | this study | N/A |
| Experimental Models: Organisms/Strains | ||
| Smc1β−/− | [24] | N/A |
| Smc1β−/−,1a | This paper | N/A |
| Software and Algorithms | ||
| Prism v5.0 | GraphPad Software | http://www.graphpad.com |
| Fiji | https://fiji.sc | https://fiji.sc |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rolf Jessberger (rolf.jessberger@tu-dresden.de).
Experimental Model and Subject Details
Mice
Smc1β−/− mice and Smc1βprom-EGFP mice were previously described [13, 24]. To generate mice expressing SMC1α in an SMC1β-like fashion, an Smc1βprom-SMC1⍺ BAC was generated by recombineering and insertion of the Smc1⍺ cDNA (pCR-BluntII-TOPO-Smc1⍺-cDNA-3876) into an Smc1β BAC (BAC RP23-451I21), replacing the SMC1β coding region region keeping the Smc1β promoter and 3` UTR regions unaltered. This BAC was linearized and injected into zygotes. Mice were bred and maintained in the animal facility of the Medical Faculty, Technische Universität Dresden (Dresden, Germany), according to approved institutional guidelines. All experiments, which involved euthanasia and organ removal only, were approved by the State animal welfare office. The mice were of 6 to 16 weeks of age unless otherwise specified and of male sex except for experiments show in Figure S2, where female mice were used.
Method Details
Analysis of Hoechst 33342-stained cells by flow cytometry
Testes were taken from mice and the tunica albuginea was removed to isolate seminiferous tubules. The tubules were digested for 20’ at 32°C in 1 mL of digestion buffer (HBSS supplemented with 20 mM HEPES (pH 7.2), 1.2 mM MgSO4.7H2O, 1.3 mM CaCl2.2H2O, 4 μg/ml DNase, 1 μg/ml of collagenase type I, 0.05% lactate). Tubules were then rinsed with PBS and again digested for 20’ at 32°C in 1 mL of digestion buffer. The cell suspension was then filtered through a 40 μm mesh to remove cell clumps and the cells were pelleted by centrifugation. Cells were resuspended in FACS incubation buffer (HBSS supplemented with 20 mM HEPES (pH 7.2), 1.2 mM MgSO4, 1.3 mM CaCl2, 6.6 mM sodium pyruvate, 0.05% lactate, glutamine, and 1% fetal calf serum) at a density of 2 million cells/ml. 5 μg/ml of Hoechst 33342 was added and the cells were incubated at 32°C for 1 h. 2 μg/ml of PI was added to exclude dead cells. An LSRII flow cytometer (BD Biosciences) was used to analyze the cells with FACSDiva software (BD Biosciences, version 6.1.3). The Hoechst dye was excited with the 355 nm UV laser and emission filters Emerald (585/42 nm, LP) and Alexa 350 (505 nm) were applied. PI was excited with the 488 nm blue laser and emission was filtered using PE (685 nm).
Cell sorting
Testis single cell suspensions were prepared by the method described above. Sorting of meiocytes was performed with the 85 μm nozzle in an ARIA II (BD Biosciences). GFP positive cells were excited by 488 nm laser and sorted using filters of 502 LP and 530/30 BP. Cells with Hoechst and PI staining were excited by 355 nm and 488 nm laser respectively. PI signal was detected by using 685 LP and 710/40 BP filter. Hoechst blue signal was detected using 600 LP and 620/10 BP filters in front of first detector and Hoechst red was detected with 440/40 BP filter in front of the second detector. Cells were collected in DMEM with FCS and pooled by centrifugation and frozen at −80°C for subsequent qPCR and protein extraction. FlowJo (Tree Star Inc.) and FACSDiva software were used for cell sorting and data analysis.
Single cell suspension and chromosome spreads
Surface-spread chromosomes were prepared by detergent spreading adapted from Wojtasz et al. [43]. Testis was taken from the sacrificed mice and the tunica albuginea was removed. Tubules were digested in 1ml of 1μg/ml of collagenase type I - PBS buffer for 10’ at 32°C with slight agitation. The tubules and cells were pelleted by centrifugation, the supernatant removed, and resuspended in 500μl of 0.025% trypsin and incubated for 5′ at 32°C. Then 200 μl of media with FCS was added to the cell suspension, which was then filtered througha 40 μm mesh and centrifuged. The pellet was resuspended in 300μl of PBS, and the single cell suspension was used for the chromosome spreads. 1.5 μl of the single cell suspension were dropped on 7 μl of 0.25% of NP40 on slides. Cells were allowed to lyse for 2 mins and then fixed by adding 24 μl of S fix (1% paraformaldehyde, 10 mM sodium borate buffer pH 9.2). Samples were incubated for 1 hr at room temperature in a humid chamber. Slides were dried under a hood and washed two times for 1 min each time with 0.4% Agepon (AgfaPhoto) and three times for 1 min each time with water. Slides were used immediately or kept at −20°C until staining.
Testis cryosection
Isolated testes were placed in 2% (v/v) of formaldehyde/PBS for 40’ at RT for fixation before incubation in 30% (w/v) sucrose/PBS overnight. Subsequently, testes were mounted in O.C.T (Sakura Finetek Europe), shock-frozen on dry ice and stored at −80°C. 8 μm thick sections were made from frozen testis, placed on the slides and dried for at-least 30’ at RT. Slides were treated with ice cold methanol for 10’ and 1’ with ice cold acetone. After completely drying, the slides were kept at –80°C or used immediately for staining.
Immunofluorescence staining and FISH
Chromosome spreads and sections were treated in the same way. Slides were blocked with either blocking buffer (2% BSA, 0.1% Triton X in PBS) or 10% goat serum for at least 1 h at RT before applying the primary antibody. Slides were incubated with primary antibodies for at least 3 h at 37°C. Then slides were washed with blocking buffer and incubated with secondary antibodies for at least 1 h. After the secondary antibody treatment slides were washed with blocking buffer and mounted with Vectashield containing 1μg/ml of DAPI.
FISH was performed according to the manufacturer’s protocol (Metasystems’ GmbH). Ten μl of probe mixture was put on slides and covered with a coverslip. Both sample and probe were denatured simultaneously by heating on a hotplate at 75°C for 2 min, followed by incubation of slides in a humidified chamber at 37°C overnight for hybridization. Post hybridization washes were performed with 0.4x SSC at 72°C for 2 min and 2x SSC, 0.05% Tween-20 at room temperature for 30 s. Slides were incubated with DAPI and signals were analyzed.
Microscopy and image analysis
Fluorescence was visualized with Zeiss Axiophot fluorescence microscope and analysis of images was performed using ImageJ version 1.43u.
Protein extraction and western blotting
FACS sorted cells were treated with RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS, 5 mM NaOV4, 10 mM NaF, protease inhibitors (Roche)) and kept for 10 min on ice. After centrifugation for 10 min at 14000 rpm at 4°C, the supernatant was collected and used as total cell extract.
Proteins extracted by the protocol described above were separated on a 6% SDS gel. Gels were then blotted to a Hybond-C nitrocellulose membrane using the Mini Trans-Blot Electrophoretic Transfer cell (Bio-Rad) for 2h at 100V. Membranes were then blocked with 1% (w/v) milk/PBS-T for 1hr and after three washes with PBS-T membranes were incubated with the primary antibody overnight at 4°C. After three washes in PBS-T, secondary antibody incubation was performed for 1h at room temperature. Following three washes, protein signals were visualized by chemoluminescence with the Pierce ECL western blotting system (Thermo Fisher Scientific).
RNA Isolation
Total testis RNA was extracted from sorted cells or unsorted cells of Smc1β−/− and WT control littermate mice with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, testes were isolated and the tunica albuginea was detached. Testes were homogenized in TRIzol reagent prior to phenol-chloroform extraction of RNA. The amount of the RNA solubilized in water was confirmed by use of the BioAnalyzer (Bio-Rad).
Quantitative reverse-transcription PCR analysis
Testis RNA was reverse transcribed using Superscript II Reverse Transcriptase (Invitrogen). For PCR amplification, primers were designed for 180 to 200 bp long intron-spanning PCR products using the web-based tool Primer blast. For each reaction, 2 μl of diluted cDNA generated from 80 ng of RNA was amplified in a 20 μl reaction volume using the QuantiTect SYBR Green PCR kit (QIAGEN) in a Rotorgene 3000 thermal cycler (Corbett Research). Reactions were performed in duplicates for each gene. Gapdh was used to normalize the signal of the gene of interest.
Oocyte qPCR
Oocytes were extracted in M2 media (Sigma; M7167) plus 2.5 mM Milrinone (Sigma; M4659) to maintain GV arrest. The surrounding somatic cells were removed by pipetting. The oocytes were treated briefly with Tyrode’s solution (Sigma; T1788) to remove the zona pellucida. Between 50 to 150 denuded GV oocytes were added to 350 μL RLT buffer from the QIAGEN RNAeasy kit (QIAGEN; 74134). The samples were processed according to the QIAGEN protocol and the RNA sample was eluted in 30 μL H2O. Half of the sample was used to produce cDNA using the Promega cDNA kit (Promega; M3682), together with oligo(dT)15 (Promega; C1101). To normalize the samples, the volume of cDNA was diluted according to the initial cell number. The qPCR was carried out in duplicates using SYBR Green (Thermo Scientific; 4309155). Rsp16 was used as a house-keeping gene to normalize the data [44]. The Smc1βprom-smc1α transgene negative sample was set to the value of 1 in each case. Primers were:
Oocyte chromosome spreads
Oocytes were extracted in M2 media and matured at 37°C until the desired stage (MI 6 hr). The zona pellucida was removed used Tyrode’s solution (Sigma; T1788). Cells were added to a well containing 15 μL of “oocyte spread solution” (1% PFA, 0.2% Triton X-100, 3mM DTT, pH 9.2 with Boric Acid). Cells were incubated for 20 mins then allowed to air dry. Slides were washed in PBS and mounted using Vectashield mounting media (Vector; H-1000) plus DAPI. Slides were imaged on a Zeiss widefield microscope. Images show one representative Z slice.
Quantification and Statistical Analysis
Statistical details for each figure can be found in the figure legends. Statistical tests were performed using GraphPad Prism version 5.0. The comparisons between different genotypes were done using the non-parametric one-way ANOVA algorithm and post test analyses were done using either Tukey or Bonferonni’s multiple comparison test as indicated.
Data and Software Availability
Data presented in this paper have been deposited at Mendeley Data and are available at https://doi.org/10.17632/4yts42t464.1.
Acknowledgments
We thank Dr. Attila Toth for helpful discussion and reagents and Drs. C. Höög, A. Pendas, and M. Alsheimer for reagents. This study was funded by grants to R.J. from the Deutsche Forschungsgemeinschaft (DFG) through project SPP1384 (grant number JE150/10-2) and from the European Union Horizon 2020 program GermAge (grant number 634113).
Author Contributions
U.B. performed and designed experiments and co-wrote the manuscript. M.S. performed and designed experiments. R.J. designed experiments and co-wrote the manuscript. All authors designed figures and tables and read and edited the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: January 11, 2018
Footnotes
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.cub.2017.12.020.
Supplemental Information
References
- 1.Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D., Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahoon C.K., Hawley R.S. Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 2016;23:369–377. doi: 10.1038/nsmb.3208. [DOI] [PubMed] [Google Scholar]
- 4.Cooper T.J., Garcia V., Neale M.J. Meiotic DSB patterning: a multifaceted process. Cell Cycle. 2016;15:13–21. doi: 10.1080/15384101.2015.1093709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J. Roles of cohesin and condensin in chromosome dynamics during mammalian meiosis. J. Reprod. Dev. 2013;59:431–436. doi: 10.1262/jrd.2013-068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNicoll F., Stevense M., Jessberger R. Cohesin in gametogenesis. Curr. Top. Dev. Biol. 2013;102:1–34. doi: 10.1016/B978-0-12-416024-8.00001-5. [DOI] [PubMed] [Google Scholar]
- 7.Remeseiro S., Cuadrado A., Losada A. Cohesin in development and disease. Development. 2013;140:3715–3718. doi: 10.1242/dev.090605. [DOI] [PubMed] [Google Scholar]
- 8.Rankin S. Complex elaboration: making sense of meiotic cohesin dynamics. FEBS J. 2015;282:2426–2443. doi: 10.1111/febs.13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding D.Q., Haraguchi T., Hiraoka Y. A cohesin-based structural platform supporting homologous chromosome pairing in meiosis. Curr. Genet. 2016;62:499–502. doi: 10.1007/s00294-016-0570-x. [DOI] [PubMed] [Google Scholar]
- 10.Prieto I., Suja J.A., Pezzi N., Kremer L., Martínez-A C., Rufas J.S., Barbero J.L. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat. Cell Biol. 2001;3:761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- 11.Revenkova E., Eijpe M., Heyting C., Gross B., Jessberger R. Novel meiosis-specific isoform of mammalian SMC1. Mol. Cell. Biol. 2001;21:6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eijpe M., Offenberg H., Jessberger R., Revenkova E., Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J. Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adelfalk C., Janschek J., Revenkova E., Blei C., Liebe B., Göb E., Alsheimer M., Benavente R., de Boer E., Novak I. Cohesin SMC1beta protects telomeres in meiocytes. J. Cell Biol. 2009;187:185–199. doi: 10.1083/jcb.200808016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutiérrez-Caballero C., Herrán Y., Sánchez-Martín M., Suja J.A., Barbero J.L., Llano E., Pendás A.M. Identification and molecular characterization of the mammalian α-kleisin RAD21L. Cell Cycle. 2011;10:1477–1487. doi: 10.4161/cc.10.9.15515. [DOI] [PubMed] [Google Scholar]
- 15.Ishiguro K., Kim J., Fujiyama-Nakamura S., Kato S., Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011;12:267–275. doi: 10.1038/embor.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Hirano T. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J. Cell Biol. 2011;192:263–276. doi: 10.1083/jcb.201008005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llano E., Herrán Y., García-Tuñón I., Gutiérrez-Caballero C., de Álava E., Barbero J.L., Schimenti J., de Rooij D.G., Sánchez-Martín M., Pendás A.M. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J. Cell Biol. 2012;197:877–885. doi: 10.1083/jcb.201201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda T., Fukuda N., Agostinho A., Hernández-Hernández A., Kouznetsova A., Höög C. STAG3-mediated stabilization of REC8 cohesin complexes promotes chromosome synapsis during meiosis. EMBO J. 2014;33:1243–1255. doi: 10.1002/embj.201387329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopkins J., Hwang G., Jacob J., Sapp N., Bedigian R., Oka K., Overbeek P., Murray S., Jordan P.W. Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet. 2014;10:e1004413. doi: 10.1371/journal.pgen.1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llano E., Gomez-H L., García-Tuñón I., Sánchez-Martín M., Caburet S., Barbero J.L., Schimenti J.C., Veitia R.A., Pendas A.M. STAG3 is a strong candidate gene for male infertility. Hum. Mol. Genet. 2014;23:3421–3431. doi: 10.1093/hmg/ddu051. [DOI] [PubMed] [Google Scholar]
- 21.Winters T., McNicoll F., Jessberger R. Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J. 2014;33:1256–1270. doi: 10.1002/embj.201387330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak I., Wang H., Revenkova E., Jessberger R., Scherthan H., Höög C. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J. Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahadevaiah S.K., Bourc’his D., de Rooij D.G., Bestor T.H., Turner J.M., Burgoyne P.S. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J. Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revenkova E., Eijpe M., Heyting C., Hodges C.A., Hunt P.A., Liebe B., Scherthan H., Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 25.Biswas U., Wetzker C., Lange J., Christodoulou E.G., Seifert M., Beyer A., Jessberger R. Meiotic cohesin SMC1β provides prophase I centromeric cohesion and is required for multiple synapsis-associated functions. PLoS Genet. 2013;9:e1003985. doi: 10.1371/journal.pgen.1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albig W., Drabent B., Kunz J., Kalff-Suske M., Grzeschik K.H., Doenecke D. All known human H1 histone genes except the H1(0) gene are clustered on chromosome 6. Genomics. 1993;16:649–654. doi: 10.1006/geno.1993.1243. [DOI] [PubMed] [Google Scholar]
- 27.Drabent B., Bode C., Bramlage B., Doenecke D. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem. Cell Biol. 1996;106:247–251. doi: 10.1007/BF02484408. [DOI] [PubMed] [Google Scholar]
- 28.Biswas U., Hempel K., Llano E., Pendas A., Jessberger R. Distinct roles of meiosis-specific cohesin complexes in mammalian spermatogenesis. PLoS Genet. 2016;12:e1006389. doi: 10.1371/journal.pgen.1006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickerson H.D., Joshi A., Wolgemuth D.J. Cyclin A1-deficient mice lack histone H3 serine 10 phosphorylation and exhibit altered aurora B dynamics in late prophase of male meiosis. Dev. Biol. 2007;306:725–735. doi: 10.1016/j.ydbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viera A., Rufas J.S., Martínez I., Barbero J.L., Ortega S., Suja J.A. CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J. Cell Sci. 2009;122:2149–2159. doi: 10.1242/jcs.046706. [DOI] [PubMed] [Google Scholar]
- 31.Viera A., Alsheimer M., Gómez R., Berenguer I., Ortega S., Symonds C.E., Santamaría D., Benavente R., Suja J.A. CDK2 regulates nuclear envelope protein dynamics and telomere attachment in mouse meiotic prophase. J. Cell Sci. 2015;128:88–99. doi: 10.1242/jcs.154922. [DOI] [PubMed] [Google Scholar]
- 32.Royo H., Polikiewicz G., Mahadevaiah S.K., Prosser H., Mitchell M., Bradley A., de Rooij D.G., Burgoyne P.S., Turner J.M. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Takada Y., Naruse C., Costa Y., Shirakawa T., Tachibana M., Sharif J., Kezuka-Shiotani F., Kakiuchi D., Masumoto H., Shinkai Y. HP1γ links histone methylation marks to meiotic synapsis in mice. Development. 2011;138:4207–4217. doi: 10.1242/dev.064444. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed E.A., van der Vaart A., Barten A., Kal H.B., Chen J., Lou Z., Minter-Dykhouse K., Bartkova J., Bartek J., de Boer P., de Rooij D.G. Differences in DNA double strand breaks repair in male germ cell types: lessons learned from a differential expression of Mdc1 and 53BP1. DNA Repair (Amst.) 2007;6:1243–1254. doi: 10.1016/j.dnarep.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Broering T.J., Alavattam K.G., Sadreyev R.I., Ichijima Y., Kato Y., Hasegawa K., Camerini-Otero R.D., Lee J.T., Andreassen P.R., Namekawa S.H. BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. J. Cell Biol. 2014;205:663–675. doi: 10.1083/jcb.201311050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modzelewski A.J., Holmes R.J., Hilz S., Grimson A., Cohen P.E. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev. Cell. 2012;23:251–264. doi: 10.1016/j.devcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 38.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D.M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 39.Xu X., Aprelikova O., Moens P., Deng C.X., Furth P.A. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 2003;130:2001–2012. doi: 10.1242/dev.00410. [DOI] [PubMed] [Google Scholar]
- 40.Murdoch B., Owen N., Stevense M., Smith H., Nagaoka S., Hassold T., McKay M., Xu H., Fu J., Revenkova E. Altered cohesin gene dosage affects Mammalian meiotic chromosome structure and behavior. PLoS Genet. 2013;9:e1003241. doi: 10.1371/journal.pgen.1003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bannister L.A., Reinholdt L.G., Munroe R.J., Schimenti J.C. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- 42.Herrán Y., Gutiérrez-Caballero C., Sánchez-Martín M., Hernández T., Viera A., Barbero J.L., de Álava E., de Rooij D.G., Suja J.A., Llano E., Pendás A.M. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 2011;30:3091–3105. doi: 10.1038/emboj.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C.R., Cooke H.J., Jasin M., Keeney S. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hached K., Xie S.Z., Buffin E., Cladière D., Rachez C., Sacras M., Sorger P.K., Wassmann K. Mps1 at kinetochores is essential for female mouse meiosis I. Development. 2011;138:2261–2271. doi: 10.1242/dev.061317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.