Abstract
Electronic medical record (EMR) systems provide easy access to radiology reports and offer great potential to support quality improvement efforts and clinical research. Harnessing the full potential of the EMR requires scalable approaches such as natural language processing (NLP) to convert text into variables used for evaluation or analysis. Our goal was to determine the feasibility of using NLP to identify patients with Type 1 Modic endplate changes using clinical reports of magnetic resonance (MR) imaging examinations of the spine. Identifying patients with Type 1 Modic change who may be eligible for clinical trials is important as these findings may be important targets for intervention. Four annotators identified all reports that contained Type 1 Modic change, using N = 458 randomly selected lumbar spine MR reports. We then implemented a rule-based NLP algorithm in Java using regular expressions. The prevalence of Type 1 Modic change in the annotated dataset was 10%. Results were recall (sensitivity) 35/50 = 0.70 (95% confidence interval (C.I.) 0.52–0.82), specificity 404/408 = 0.99 (0.97–1.0), precision (positive predictive value) 35/39 = 0.90 (0.75–0.97), negative predictive value 404/419 = 0.96 (0.94–0.98), and F1-score 0.79 (0.43–1.0). Our evaluation shows the efficacy of rule-based NLP approach for identifying patients with Type 1 Modic change if the emphasis is on identifying only relevant cases with low concern regarding false negatives. As expected, our results show that specificity is higher than recall. This is due to the inherent difficulty of eliciting all possible keywords given the enormous variability of lumbar spine reporting, which decreases recall, while availability of good negation algorithms improves specificity.
Keywords: Natural language processing, Radiology reporting, Lumbar spine imaging, Modic classification
Introduction
Electronic medical record (EMR) systems provide ready access to radiology reports and hold significant promise to communicate radiology results, as well as for use in quality improvement projects and clinical research [1]. However, while there has been some movement towards structured radiology reporting [2] (e.g., the Breast Imaging Reporting and Data System [3]) as well as increased use of templates, the vast majority of radiology reports use unstructured free text [1]. Harnessing the full potential of the EMR to gather information on large numbers of patients requires scalable, inexpensive approaches such as natural language processing (NLP).
NLP, or computational linguistics, is a subfield of computer science that uses computational techniques to learn, understand, and produce human language content [4]. The overall goal is to translate natural human language into a structured format or discrete representation suitable for processing by computer algorithms. NLP can be thought of as a framework or pipeline with multiple steps.
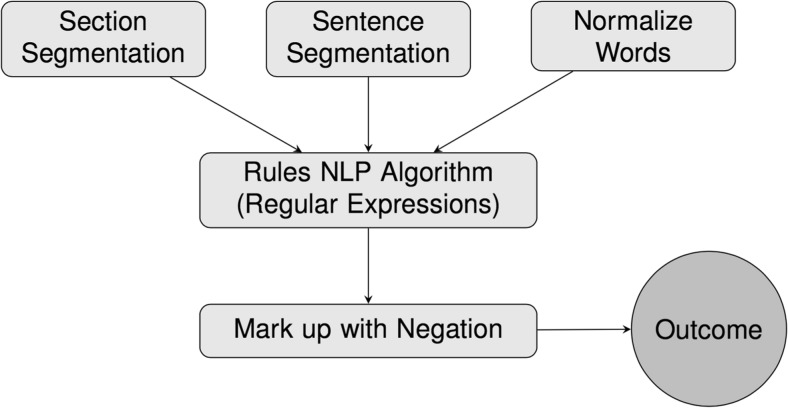
Our goal was to develop and evaluate a rule-based NLP algorithm, using logical classification rules constructed by human medical experts, to determine presence of specific findings in free-text radiology reports (Fig. 1).
Fig. 1.
Our natural language processing (NLP) pipeline
There are several advantages to rule-based approaches as opposed to machine learning methods, one of which is the minimal requirement for set-up, as only a list of keywords is needed to implement algorithms based on regular expression matching. Additionally, excellent off-the-shelf packages are available so that phrase identification can accurately identify both positive and negative identification of clinical findings reported in text reports [5, 6]. Lastly, less annotation (labeling) is required, as labels are only needed for evaluation of fixed processing algorithms, whereas machine learning methods require both a large development data set and a separate evaluation data set.
Our overall goal was to explore the feasibility of using NLP to identify targeted subsets of patients who would be candidate participants for select clinical trials. In this paper, we present our results regarding the ability of an NLP algorithm to identify patients with Type 1 Modic endplate changes [7] found on magnetic resonance (MR) imaging of the spine. These changes are characterized by low T1 and high T2 signals within the endplate of a vertebral body, commonly encountered in clinical MRI of the spine, both in patients with low back pain (LBP), but also in patients without LBP [8]. Modic Type 1 changes are believed to result from endplate fissuring with subsequent development of vascular granulation tissue which results in bone marrow edema [7].
Identifying patients with Type 1 Modic endplate changes who may be eligible for clinical trials is important as these findings are potential targets for intervention.
Materials and Methods
Radiology Reports and Annotation
We randomly sampled 200 lumbar spine MR radiology reports from a previous study based on a prospectively assembled cohort of older adults with back pain, the Back pain Outcomes using Longitudinal Data (BOLD) cohort [9]. Two annotators (a board-certified radiologist and a physical therapist) read and extracted the document level reported presence of Type 1 Modic endplate change. Based on annotation of these reports, we identified eight reports which documented presence of Type 1 Modic change (sample prevalence 4%), for which a simple string-matching regular expressions approach resulted in 0.90 recall and 0.99 negative recall in case identification.
These initial results led to designing and evaluating a rigorous and reproducible rule-based NLP algorithm aimed at identifying Type 1 Modic change among MR reports of adult individuals undergoing lumbar spine imaging. For this second phase, we randomly selected 458 lumbar spine MR radiology reports obtained as part of a prospective, multi-institutional pragmatic randomized trial, the Lumbar Imaging with Reporting of Epidemiology (LIRE) study [10]. The LIRE trial collects data from four participating integrated health care delivery systems (Kaiser Permanente of Northern California, Group Health Cooperative, Mayo Clinic Health System, and Henry Ford Health System).
Our validation sample size of N = 458 encompassed annotated MR reports from the LIRE cohort. To test the null hypothesis that PPV < 0.90 using a one-sample superiority test with 80% power at the 0.05 level, a validation sample of N = 426 was required, if the true PPV were 0.96 and we assumed a typical rule-based model with moderately high sensitivity (0.60) and high specificity (0.90) to detect Type 1 Modic Changes with 11% finding prevalence. Our post hoc sample size calculation indicated that our annotated sample was sufficiently large to detect such an effect.
We recruited four annotators, all with expertise in spine disorder diagnosis and treatment. These annotators included two board-certified radiologists (JG and HH), a spine physiatrist (PS), and a physical therapist (SR). To streamline the annotation process, we first elicited from the annotators a list of synonyms, or keywords related to Type 1 Modic endplate change. These keywords were supplemented with online searches of medical databases such as MEDLINE/PUBmed and other NLM databases. The four annotators then proceeded to identify all radiology reports that reported presence of Type 1 Modic endplate change using rules derived from these synonyms and keywords, as described in detail below.
The annotation interface was designed in REDCap (Research Electronic Data Capture) [11], an electronic data capture system used to administer surveys and collect data for clinical trials. The data capture system had a functionality for the annotators to flag a record if there was an ambiguity in the current set of annotation rules. Figure 2 provides a screenshot of the annotation user interface. Subsequently, annotators adjudicated disagreements by consensus between each annotator pair. When consensus could not be reached, a senior radiologist provided adjudication. We then amended the annotation rules for the next round. Three rounds were conducted, with 58 reports annotated in the first round, 100 in the second, and 300 in the third round. This process resulted in the set of 11 general rules listed in Table 1.
Fig. 2.
Annotation user interface. Text from the anonymized radiology report is displayed. Below, findings are listed with checkboxes to indicate present. There is an option to flag the report for discussion if it is ambiguous and requires further review or adjudication. An annotator can also add a comment to be reviewed during consensus meetings and suggest additional synonyms. The red “(Adj)” flags were used in adjudication discussions to signal the findings where annotators disagreed on the presence of the finding
Table 1.
Overall annotation rules
| 1. If there is a conflict between a finding noted in the body of the report vs. the impression section of the report, code what is in the impression. |
| 2. If a finding is more specific in one section of the report than in another, code both findings. |
| 3. If a report notes a possible finding, for example using the words “possible,” “probable,” or “minimal” finding, code the finding as present. |
| 4. When a finding is described as “not excluded” (or a similar type of “hedging” phrase) consider the finding to be possible and therefore present unless the finding cannot be diagnosed with the modality. |
| 5. Primarily interested in the lumbar spine. Ignore cervical and upper thoracic (T1–T6) findings (including scout images) but include lower thoracic (T7–T12) findings or findings where the thoracic level is not specified. |
| 6. We ignore transcription errors in the report unless there is not enough context to interpret the phrase or sentence. |
| 7. Code only findings that are explicitly described rather than inferring. |
The outcome of the annotation process was considered as the reference standard in this project as detailed by the annotation guidelines document. The reference standard data set consists of the set of 458 lumbar spine MR radiology reports labeled for whether Type 1 endplate change was present in each report. These labels were obtained either through agreement between independent annotators, discussion through consensus meetings, or adjudication by the senior neuroradiologist.
As an example for an NLP rule, Table 2 contains a structured specification for Type 1 Modic endplate change.
Table 2.
Specification for Type 1 Modic endplate change
| Examples with keywords in italics: |
| • Edematous endplate changes |
| • Endplate edema |
| • Endplate signal Modic type 1–2 changes |
| • For where reactive endplate changes are present, particularly edema along the left superior L5 endplate. |
| • Minimal edema in the superior L5 endplate with more chronic appearance |
| • Modic type 1 degenerative endplate change |
| • Type 1 Modic degenerative endplate change |
| Edema synonyms: |
| • Acute phase signal change |
| • Edema |
| • (High OR increased) AND (signal OR STIR or T2) |
| • T2 hyperintensity |
| • Type 1 Modic (do not need to be associated with “endplate”) |
| Endplate synonyms: |
| • Endplate |
| Exclusion criteria: |
| • Type 2 endplate changes and synonyms |
| ○ Synonyms: fatty endplate change, fat transformation of endplate |
| • Type 3 endplate changes and synonyms |
| ○ Synonyms: endplate sclerosis, sclerotic endplate, endplate irregularity (unless edema also present) |
| Note: The following should be included as “endplate edema” only if the report indicates that the abnormal signal involves only the endplate |
| • (High OR increased) AND (signal OR STIR OR T2) |
| • Bone marrow edema |
| • T2 hyperintensity |
Implementation of a Rule-Based NLP Framework
A rule-based NLP framework in Java (v 4.6.0) (Oracle Corporation, Redwood Shores, CA) was implemented based on regular expressions (REGEX), incorporating the Apache Lucene (v 6.1.0) Application Program Interface (API) (The Apache Software Foundation, Forest Hill MD), and an implementation of the NegEx algorithm [5]. All utilized software used was open source. Our algorithm incorporated a standard NLP pipeline, including text pre-processing, section segmentation, sentence segmentation, and concept identification. Concept identification refers to semantic analysis where meaning is assigned to the words and phrases by linking them to semantic types, such as symptom, disease, and procedure, and concepts [12]. In Fig. 1, we outline the steps used in our specific implementation of NLP algorithm. For pre-processing, we cleaned and normalized text with spell checking, lower casing, stop word removal, and reducing inflected/derived words to their word stem (stemming); using the standard tokenizer in Apache Lucene. For section segmentation, we implemented a deterministic string split algorithm to separate the impression section from the body of the report text. For sentence segmentation, we used the default tokenizer in Apache Lucene.
We performed concept identification at the document level, using the same keyword list as for annotation, accounting for possible spelling variations using appropriate regular expressions. To incorporate negation and exclusion criteria as described, we adapted the NegEx algorithm with its default settings, applied at the sentence level [5].
Our algorithm implemented an interpretation of Type 1 Modic endplate change as detailed in Table 2. A key component of Type 1 Modic endplate change is “endplate edema,” which is a composite finding, comprised of two entities: “endplate” (or synonyms) and “edema” (or synonyms). Our algorithm allowed a maximum word distance of 3 when endplate precedes edema (e.g., endplate edema), and maximum word distance of 10 when endplate follows edema (e.g., edema seen at the endplate). If this composite finding was present, our algorithm proceeded to check if any of the exclusion criteria applied (Table 2). In such a manner, “endplate edema” was identified as present in the report only if there was at least one sentence in the report text, for which a keyword was identified and not negated and not excluded by exclusion criteria. Table 3 provides more details on the NLP algorithm.
Table 3.
Pseudocode with REGEX
| FOR each report: |
| Initialize REGEX: = 0 |
| Initialize (REP_POS, REP_NEG): = (0,0) |
| FOR each of BODY and IMPRESSION sections: |
| Initialize (SEC_POS, SEC_NEG): = (0,0) |
| FOR each sentence: |
| Search for base KEYWORD |
| FOR each KEYWORD: |
| Search for EXCLUSION surrounding KEYWORD |
| IF at least one KEYWORD = 1 AND EXCLUSION = 1: |
| (SEN_POS, SEN_NEG) = (1,1) |
| IF at least one KEYWORD = TRUE AND EXCLUSION = 0: |
| (SEN_POS, SEN_NEG) = (1,0) |
| FOR all sentences in section: |
| IF at least one (SEN_POS, SEN_NEG) = (1,1): |
| (SEC_POS, SEC_NEG) = (1,1) |
| IF at least one (SEN_POS, SENE_NEG) = (1,0): |
| (SEC_POS, SEC_NEG) = (1,0) |
| (REP_POS, REP_NEG):= (REP_POS, REP_NEG) of IMPRESSION |
| IF (SEC_POS, SEC_NEG) of IMPRESSION = (0,0): |
| (REP_POS, REP_NEG):= (SEC_POS, SEC_NEG) of BODY |
| IF (REP_POS, REP_NEG) = (1,0) |
| REGEX: = 1 |
A novel aspect of our algorithm was that it utilized the “impression trumps body” rule, whereby the algorithm considered the evidence in both the body and impression sections of the radiographic report texts, and decided in favor of the impression section whenever there is a conflict.
To describe the overall effort to develop the NLP rule-based approach detailed here, it is helpful to consider it in four parts: (1) project planning and support, (2) development of annotation database and interface, (3) annotation work, and (4) NLP algorithm development and validation.
Project planning and support, including defining project aims and documentation, took approximately 30 h of effort. The development of annotation database and interface took approximately 15 h of effort. The total effort of annotation work for all four annotators was estimated to have taken approximately 30 h. Lastly, developing and validating a rule-based algorithm consumed on the order of 15 h. Therefore, the total initial development effort was estimated on the order of 90 h.
Statistical Analysis
For the proposed NLP algorithm, recall (sensitivity), specificity, precision (positive predictive value (PPV)), negative predictive value (NPV), and the F1 score were calculated. The F1 score, defined as = 2 × (recall × precision) / (precision + recall), is the harmonic mean of recall and precision, and provides a single, overall measure of NLP classification performance. Given that F1 score is a combined summary of both precision and recall, it is a relative term with no absolute ranges of poor, fair, good, or excellent. For all these error metrics, corresponding 95% confidence intervals using a normal approximation to binomial proportions were reported. For the F1 score, 95% confidence intervals using a delta method approximation were reported. Cohen’s Kappa coefficient on the 458 reports to assess inter-annotator agreement of our annotation process was calculated. All analyses were performed using R (v 3.3.0) [13].
This study was Health Insurance Portability and Accountability Act (HIPAA) compliant and approved by University of Washington Institutional Review Board Protocol # 39009 for the BOLD study and Group Health Cooperative Institutional Review Board Protocol # 476829 for the LIRE pragmatic trial.
Results
The inter-annotator agreement data between the four annotators was analyzed with Cohen’s Kappa coefficient. The inter-annotator agreement was 0.88 (95% C.I. 0.70, 1.0). This was calculated from the point estimates for all 6 annotator pairs (four annotators, choose two). Based on the Landis and Koch magnitude guidelines [14], 0.88 falls in the range of 0.81–1.0 which is considered “almost perfect” agreement.
In our study, inter-annotator disagreements fell into two main categories: (1) when an alternative phrase was used to denote Type 1 Modic endplate change, for example “acute phase degenerative change” or “edema in the L2 through L5 vertebral bodies probably related to disc degeneration”; and (2) when endplate edema was associated with a fracture, for example “…endplate edema at T11 level superiorly which may suggest an acute or subacute wedge-compression deformity”.
Regarding the “impression trumps body” rule, no direct contradictions between Findings and Impression were observed specifically for Type 1 Modic endplate change in our set of MRI reports; however, there were two MRI reports were endplate edema was only mentioned in the Impression. These two reports were therefore considered positive for this finding.
The results from running the NLP algorithm are summarized in Table 4. The prevalence of Type 1 Modic endplate changes was 50/458 = 0.11 (95% C.I. 0.07–014). The NLP recall (sensitivity) and specificity were 35/50 = 0.70 (95% C.I. 0.52–0.82) and 404/408 = 0.99 (95% C.I. 0.97–1.0), respectively. The precision (PPV) and the NPV were 35/39 = 0.90 (95% C.I. 0.75–0.97) and 404/419 = 0.96 (95% C.I. 0.94–0.98), respectively. The F1 score was 0.79 (95% C.I. 0.43–1.0).
Table 4.
Comparison of NLP with reference standard annotation for Type 1 Modic change with MR reports (N = 458)
| Reference standard annotation | |||||
|---|---|---|---|---|---|
| Rule-based NLP | Present | Absent | Total | ||
| Present | 35 | 4 | 39 | ||
| Absent | 15 | 404 | 419 | ||
| Total | 50 | 408 | 458 | ||
| Prevalence | Recall | Specificity | Precision | NPV | F1 score |
| 0.10 (0.07, 0.14) | 0.70 (0.52, 0.82) | 0.99 (0.97, 1) | 0.90 (0.75, 0.97) | 0.96 (0.94, 0.98) | 0.79 (0.43, 1) |
Overall, 15 false negatives and 4 false positives were observed in our results. The four false positives and reasons for them are detailed in Table 5. Reasons for false negatives included instances where the findings were discussed in a complex way, possibly combined with a typographical error. For example, “…interval L5 superior Schmorl’s node is seen with mild type I discogenic sigl.” or “…reactive edema identified within the opposing L5-S1 and plates…”, and cases where mixed types of Modic enplate changes were present “mild mixed edema and fatty type endplate changes…”.
Table 5.
False positives (n = 4) and reasons for them
| Report text | Issue resulting in false positive |
|---|---|
| ... The endplate edema and anterior endplate enhancement seen at the L5 interspace on 1/20/2014 has resolved… | The distance between “endplate edema” and “has resolved” was too long for the negation detection algorithm |
| …There is endplate edema at T11 level superiorly which may suggest an acute to subacute wedge compression deformity superimposed upon old wedge compression deformity of T11… | The distance between “endplate edema” and “wedge compression deformity” was too long for the algorithm |
| …Subacute phase degenerative change at the anterior endplates… | Not properly accounting for “subacute phase” |
| …Acute phase degenerative change along the superior endplate of L4… | Not properly accounting for “acute phase” |
Discussion
In this study, we describe development of a rule-based NLP algorithm for identifying Type 1 Modic endplate changes in our sample of lumbar spine MR radiology reports. We are not aware of prior published NLP algorithms in the specific domain of MR spine reporting. We have demonstrated that NLP provides a feasible and scalable approach to abstract useful information from radiology report text in the EMR for clinical or research purposes. Specifically, our NLP algorithm identified reports with documented Type 1 Modic changes with high PPV (precision).
After the initial development of our algorithm, it can be run in seconds on thousands reports for minimal cost.
The reference standard developed as part of this effort for evaluation of our NLP application appears to demonstrate high integrity and reliability. First, a relative large number of reports were randomly selected from multiple institutions with different styles of reporting. Four different annotators were part of the effort to annotate each of these reports, and consensus was achieved for each finding in each report. Also, as discussed above, the average Cohen’s kappa coefficient among the six annotator pairs was high, 0.88.
Our results show that for our NLP algorithm, specificity is significantly higher than recall. The main reasons for this include difficulty of eliciting all possible keywords given the enormous variability of how lumbar spine findings are reported, which decreases recall. Conversely, availability of good negation algorithms to rule out the presence of a given finding improves specificity.
Despite efforts to standardize spine reporting [15], enormous variability in the reporting of lumbar spine findings remains, which was the main challenge in our work. Somewhat low prevalence of Type 1 Modic endplate changes in our sample is another limitation of this study.
Performance of rule-based NLP can be limited by several factors, including limited number of reports for a specific finding, findings that are complex to identify, ambiguity in reports, and feature sets which are not sufficiently rich. We observed appropriate pre-processing steps, including spell checking, word stemming, and removal of “nonsense” words are helpful.
As mentioned above, F1 scores are frequently used to compare different NLP systems, allowing combination of two measures into a single figure of merit. However, this measure is limited to being a relative term with no absolute range or ranges of poor, fair, good, or excellent. Despite the challenges of our particular domain, our F1 score, 0.79, is comparable to the work of Cheng et al. [16] who used an NLP algorithm to extract tumor status (stable/progression/regression) from MR reports, a relatively straightforward task with established vocabulary, and obtained an F1 score of 0.81. However, when their same algorithm was used to extract significance of the findings from the reports, a question dealing with much less standardized vocabulary, the F1 score decreased to 0.69.
In contrast to the complex domain of radiology findings reporting, Lakhani et al. [17] developed a rule-based algorithm to automate detection of non-routine communication of results in radiology reports, which is likely less complex and variable. That study demonstrated precision of 0.97 and recall of 0.98 for identifying radiology reports containing documentation of non-routine communication, and an F1 score of 0.976 [17].
One issue in developing rule-based algorithms is determining how far apart key words can be in a sentence to be considered together. In the work of Lakhani et al. [17], for example, they chose a maximum separation of 14 words. In our work, we allowed a maximum word distance of 3 when endplate precedes edema (e.g., endplate edema), and maximum word distance of 10 when endplate follows edema (e.g., edema seen at the endplate), restricted to the same sentence. Lakhani et al. also repeated the iterative refinement process of the algorithm until the precision and recall changed by no more than 0.5% between iterations.
Our work demonstrates an application for which rule-based NLP is feasible. Type 1 Modic endplate change is a relatively rare finding. In our annotated dataset, we observed a prevalence of about 10%. With such extreme outcome class imbalance, it is expected a machine learning system trained on raw data will result in low precision [18].
Conclusions
Rule-based NLP is a feasible approach for identifying patients with Type 1 Modic endplate change if the emphasis is on identifying only relevant cases with low concern regarding false negatives. As expected, our results show that for this particular rule-based NLP application, specificity is significantly higher than recall. The main reasons for this include difficulty eliciting all possible keywords given the enormous variability of how lumbar spine findings are reported, which decreasing recall, while availability of good negation algorithms to rule out presence of a given finding improves specificity.
Acknowledgements
This work is supported by the National Institutes of Health (NIH) Common Fund, through a cooperative agreement (5UH3AR06679) from the Office of Strategic Coordination within the Office of the NIH Director. The views presented here are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
BOLD funding through AHRQ grant no. 1R01HS022972.
References
- 1.Cai T, et al. Natural Language Processing Technologies in Radiology Research and Clinical Applications. Radiographics. 2016;36:176–191. doi: 10.1148/rg.2016150080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langlotz CP. Structured radiology reporting: are we there yet? Radiology. 2009;253:23–25. doi: 10.1148/radiol.2531091088. [DOI] [PubMed] [Google Scholar]
- 3.Burnside ES, et al. The ACR BI-RADS experience: learning from history. J Am Coll Radiol. 2009;6:851–860. doi: 10.1016/j.jacr.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirschberg J, Manning CD. Advances in natural language processing. Science. 2015;349:261–266. doi: 10.1126/science.aaa8685. [DOI] [PubMed] [Google Scholar]
- 5.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 6.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42:839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modic MT, Steinberg PM, Ross JS, Masaryk TJ, Carter JR. Degenerative disk disease: assessment of changes in vertebral body marrow with MR imaging. Radiology. 1988;166:193–199. doi: 10.1148/radiology.166.1.3336678. [DOI] [PubMed] [Google Scholar]
- 8.Jensen TS, Karppinen J, Sorensen JS, Niinimäki J, Leboeuf-Yde C. Vertebral endplate signal changes (Modic change): a systematic literature review of prevalence and association with non-specific low back pain. Eur Spine J. 2008;17:1407–1422. doi: 10.1007/s00586-008-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvik JG, et al. Back pain in seniors: the back pain outcomes using longitudinal data (BOLD) cohort baseline data. BMC Musculoskelet Disord. 2014;15:134. doi: 10.1186/1471-2474-15-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvik JG, et al. Lumbar imaging with reporting of epidemiology (LIRE)—protocol for a pragmatic cluster randomized trial. Contemp Clin Trials. 2015;45:157–163. doi: 10.1016/j.cct.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pons E, Braun LM, Hunink MG, Kors JA. Natural language processing in radiology: a systematic review. Radiology. 2016;279:329–343. doi: 10.1148/radiol.16142770. [DOI] [PubMed] [Google Scholar]
- 13.R Core Team . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 14.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 15.Fardon DF, Williams AL, Dohring EJ, Murtagh FR, Gabriel Rothman SL, Sze GK. Lumbar disc nomenclature: version 2.0: Recommendations of the combined task forces of the North American Spine Society, the American Society of Spine Radiology and the American Society of Neuroradiology. Spine J. 2014;14:2525–2545. doi: 10.1016/j.spinee.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Cheng LT, Zheng J, Savova GK, Erickson BJ. Discerning tumor status from unstructured MRI reports—completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging. 2010;23:119–132. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakhani P, Kim W, Langlotz CP. Automated detection of critical results in radiology reports. J Digit Imaging. 2012;25:30–36. doi: 10.1007/s10278-011-9426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Q, Dunbrack RL. The role of balanced training and testing data sets for binary classifiers in bioinformatics. PLoS One. 2013;8:e67863. doi: 10.1371/journal.pone.0067863. [DOI] [PMC free article] [PubMed] [Google Scholar]