Abstract
The global escalation in antibiotic resistance cases means alternative antimicrobials are essential. The aim of this study was to investigate the antimicrobial capacity of apple cider vinegar (ACV) against E. coli, S. aureus and C. albicans. The minimum dilution of ACV required for growth inhibition varied for each microbial species. For C. albicans, a 1/2 ACV had the strongest effect, S. aureus, a 1/25 dilution ACV was required, whereas for E-coli cultures, a 1/50 ACV dilution was required (p < 0.05). Monocyte co-culture with microbes alongside ACV resulted in dose dependent downregulation of inflammatory cytokines (TNFα, IL-6). Results are expressed as percentage decreases in cytokine secretion comparing ACV treated with non-ACV treated monocytes cultured with E-coli (TNFα, 99.2%; IL-6, 98%), S. aureus (TNFα, 90%; IL-6, 83%) and C. albicans (TNFα, 83.3%; IL-6, 90.1%) respectively. Proteomic analyses of microbes demonstrated that ACV impaired cell integrity, organelles and protein expression. ACV treatment resulted in an absence in expression of DNA starvation protein, citrate synthase, isocitrate and malate dehydrogenases in E-coli; chaperone protein DNak and ftsz in S. aureus and pyruvate kinase, 6-phosphogluconate dehydrogenase, fructose bisphosphate were among the enzymes absent in C.albican cultures. The results demonstrate ACV has multiple antimicrobial potential with clinical therapeutic implications.
Introduction
Antibiotic resistance is rapidly becoming a major worldwide problem. There has been a steady increase in the number of pathogens that show multiple drug resistance. In fact the World Health Organization predicts that infections involving antibiotic resistant pathogens will pose major patient care management issues in the future1.This will inevitably lead to an increase in hospital stays, cost, patient morbidity and mortality. In the immunocompromised and at risk patients severe microbial infections can result in sepsis. Sepsis can rapidly lead to systemic inflammation and organ failure2. In response to microbial invasion, the innate immune system reacts by triggering tissue damage. Mononuclear cells recognize pathogens associated with molecular patterns (PAMPs) present on the microbial surface. This results in intracellular signaling cascades which initiate pro-inflammatory cytokine and chemokine release into the blood circulation. Unchecked, the chemokines will continue to recruit more immune cells to the site of infection which release further pro-inflammatory cytokines enhancing inflammation in a continuous feedback loop3. Essentially, antibiotics are antimicrobials but can also act as immune modulators reducing the release of pro-inflammatory cytokines such as IL-1β, IL-6, TNFα, IL-8, and interferon-gamma (INF-gamma). Antibiotics can also affect mononuclear phagocytic function and modulate the activity of nuclear transcription factors such as NF-ĸB and activator proteins4–7. Microorganisms such as E-coli, S. aureus and C. albicans form part of the human microbiota. However pathogenic forms of these microbes have been implicated in blood or urinary tract infections, gastroenterititis, endocarditis, soft tissue infections and organ malfunction8–10.
The anti-microbial agents used to treat gram negative infections such as β-lactams, fluroquinolones, sulfamethaoxathoxazole and trimethroprin are becoming increasingly ineffective. Strains of S. aureus have emerged with reduced susceptibility to vancomycin and methicillin6,11,12. Furthermore, antibiotic action itself can be problematic in terms of cell membrane permeability, intracellular inactivation and the inability to reach intracellular structures in which organisms can hide. Alternative supplementation that can combat a plethora of microbes without concurrent side effects would be of significant healthcare interest as the discovery of effective new antibiotic has been slow but should be a global priority.
The Old Testament and Hippocrates reported on the use of ACV in combination with honey to combat infection and protect open skin wounds. Historically, vinegar has been produced and sold as a commercial commodity for over 5000 years. In fact up until the sixth century BC, the Babylonians were making vinegars for consumption as well as for use in healing13. Vinegar is the resultant product when ethyl alcohol is converted to acetic acid by Acetobacter. It can be produced by different methods and a variety of raw materials such as wine, malted barley, alcohol, fruits and cider14. ACV is produced from cider that has undergone acetous bioconversion and has relatively low acidity (5% acetic acid). It also contains organic acids, flavonoids, polyphenols, vitamins and minerals15. ACV has been hailed as a supplement aiding weight loss, hyperlipademia, hypercholesterliaemia, nutritional support, antioxidant defence and lowering blood pressure. Utilising organic acids as nutritional supplements has been regarded as safe and can eliminate harmful intestinal bacteria16–18. The positive impact of dietary ACV supplementation has been highlighted in vivo. ACV decreased the serum lipid profile in mice fed a high cholesterol diet over 28 days. Intragrastric ACV addition induced a protective effect against erythrocyte, kidney and liver oxidative injury as well as lowering cholesterol levels16. ACV also decreased blood triglyceride and very low density lipoprotein levels in rats which had induced cholesterol induced hepatic steatosis18. Despite the known health benefits of dietary organic acid supplementation, to the best of our knowledge the direct effect of ACV on microbes and mononuclear leucocytes has not been examined. The aim of the present study was to investigate the antimicrobial activity of ACV on microbes and associated inflammatory pathways.
Results
The antibacterial and antifungal activity of ACV against E. coli, S. aureus and C. albicans
In order to determine the anti-microbial activity of ACV, E. coli, S. aureus and C. albicans were directly cultured with different concentrations of ACV. Figure 1 represents the experimental results. The minimum dose required to restrict growth for C. albicans was neat, undiluted ACV (5% acidity), for S. aureus it was a 1/2 dilution (2.5% acidity) and for E-coli, growth was restricted at a significantly lower dilution of 1/50 (equivalent to 0.1% acidity). We also measured the equivalent zones of inhibition (in mm) for each of the microbes at varying dilutions of ACV which is depicted in the photographs of the culture plates (Fig. 2). To translate the MIC into supplementary tablet dosages required, we tested concentration ranges from 400 µg/ml in doubling microdilutions to the lowest of 3.1 µg/ml against each microbe. The MIC for ACV tablets at which no growth was visible was 62 µg/ml for E. coli; 125 µg/ml for S. aureus and 250 µg/ml for C. albicans respectively. Both sets of results were also confirmed further by microdilution. We used the Braggs ACV for all future experiments at the minimum inhibitory dilution required for each organism.
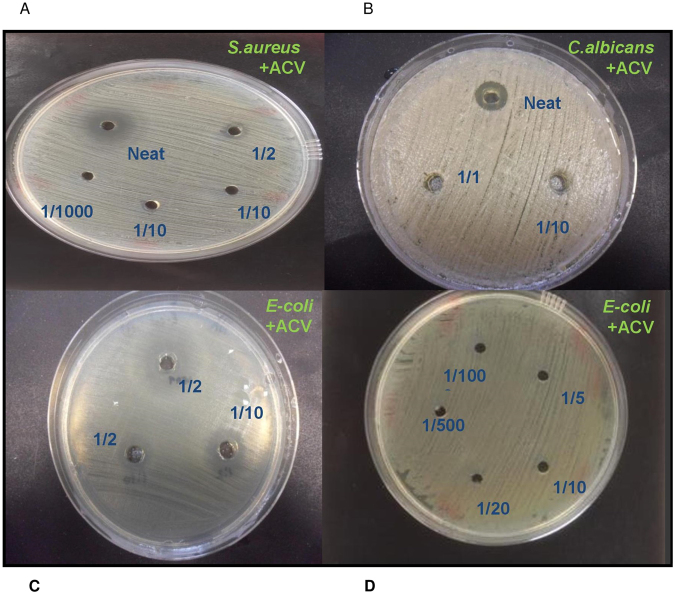
Figure 1.
Effect of varying concentrations of ACV on microbial growth after incubation at 37 °C for 24 h. (a) S. aureus; (b) C. albicans, (c) E-coli (d) E-coli. ACV was either applied neat or diluted 1:2 or 1:10 v/v in distilled water. Zones of microbial growth inhibition are indicated by clear zones and vary with ACV dilutions for each microbe. Photographs were taken using a 20 Mega pixel Samsung camera.
Figure 2.
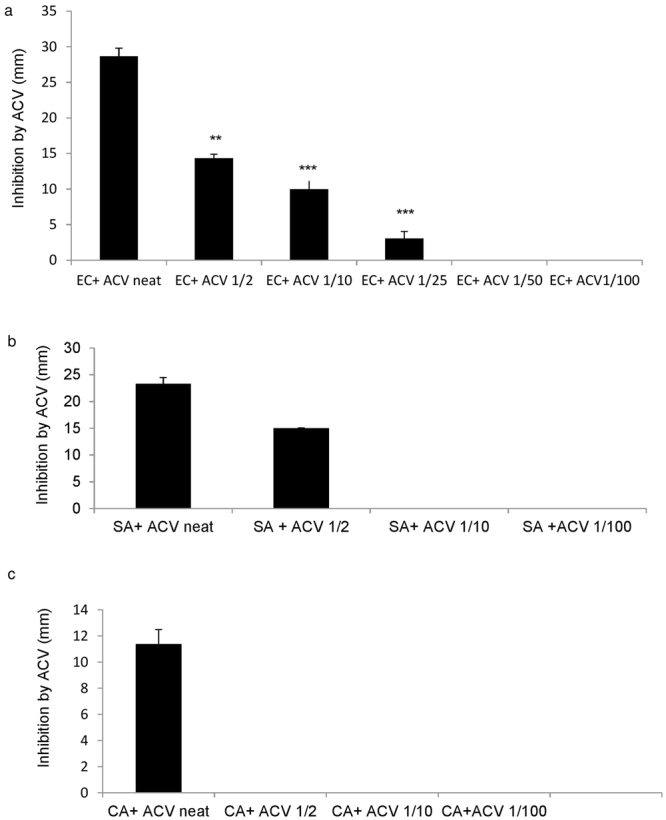
Inhibition of microbe growth by ACV after incubation for 24 h at 37 °C. (a) E-coli; (b) S. aureus (c) C. albicans. Zone of inhibition was measured in mm. These experiments represent data from three repeats. EC = E-coli, SA = S. aureus, CA = C. albicans, ACV = Apple cider vinegar.
Downregulation of pro-inflammatory cytokine secretion by ACV in monocytes exposed to microbes
The microbes utilised in our study have been extensively studied and are known to cause inflammation through their capacity to stimulate the leucocyte pro-inflammatory cytokine cascades19,20. Hence, we proceeded to measure mononuclear derived TNF-α and IL-6 cytokines as indicators of inflammation which are also the markers of choice for clinical diagnosis of septic infections21.
Figure 3 depicts the effects of a dose dependent reduction in TNFα and IL-6 release from monocytes which have been co-cultured with ACV, together with either C. albicans, E-coli or S. aureus for 24 h.
Figure 3.
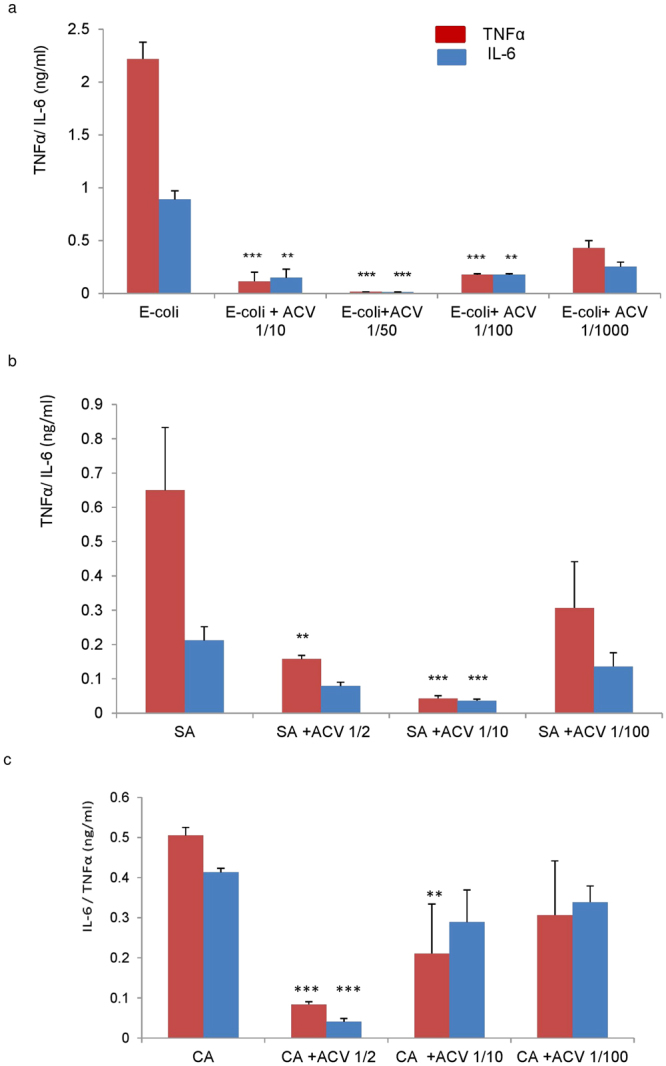
Effect of ACV on pro-inflammatory cytokine secretion from human monocytes infected with (a) E-coli; (b) C. albicans and (c) S. aureus after incubation for 24 h at 37 °C. ACV was added at dilutions of 1/10, 1/25, 1/50, 1/100 or 1/1000. EC = E-coli, SA = S. aureus, CA = C. albicans. The minimum inhibitory dilution of ACV required for significant pro-inflammatory downregulation varied with each microbe. For TNFα, a 1/50 ACV minimum inhibitory dilution was required for EC, p = 0.0008; 1/10 for SA, p = 0.01; 1/2 for CA, p = 0.0003 respectively. For IL-6, 1/50 ACV dilution was required for EC, p = 0.0008; 1/10 for SA, p = 0.03; 1/2 for CA, p = 0.008 respectively. Results represented are mean +/− SD of 3 experiments. We used student’s paired t-tests for statistical evaluation (Excel 2017) with statistical significance taken when p < 0.05. EC = E-coli, SA = S. aureus, CA = C. albicans, ACV = Apple cider vinegar.
Consistent with the microbial growth inhibition data depicted in Fig. 2, the effect of ACV at the minimum inhibitory concentration of 1/50 resulted in a significant reduction in monocyte derived TNFα (p = 0.008) and IL-6 release (P = 0.001) in monocytes cultured with E-coli. For S. aureus the minimum inhibitory concentration for ACV was found to be 1/10 in terms of reduction of TNFα (p = 0.011) and IL-6 (p = 0.03). For C.albicans the minimum inhibitory dilution was lower at 1/2 dilution, for TNFα (p = 0.003) and IL-6 (p = 0.008). It was imperative to ascertain whether the monocytes were alive during inoculation with the various microbes especially after incubation for 24 h at 37 °C. We added Trypan blue directly to monocytes which had been co-cultured with microbes after 2, 6 and 24 h. Light microscopy revealed that greater than 90% of cells were alive after 24 h in all co-cultures as demonstrated in (Fig. 4a,b,c) which represents the light microscopic images of monocytes and the microbes in co-cultures.
Figure 4.
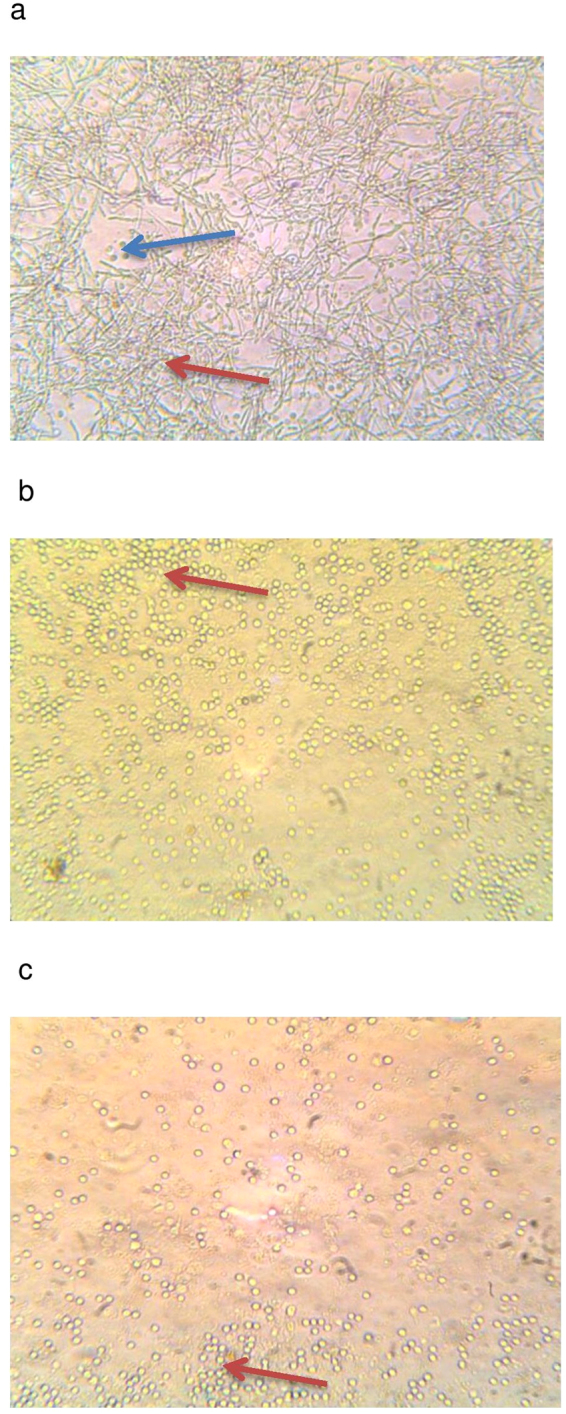
(a–c) Photos of monocytes in co-culture with microbes. Monocytes were cultured with the microbes and ACV. Trypan blue addition indicated over 95% viability. Red arrows indicate microbes and the blue arrow shows monocytes in Fig. 4a which are not visible in Fig. 4b and c since they have been covered by the microbes. Photographs were taken after 24 h incubation at 37 °C under × 100 magnification using a light microscope indicated over 90% viability with monocytes at 2, 4, 6 and 24 h of co-culture (24 h photos shown). A = C. albicans, B = E-coli and C = S. aureus respectively.
Upregulation of phagocytic capacity
We also investigated whether ACV could have an effect on the phagocytic function of monocytes alone and also after a 4 h exposure to microbes with or without ACV treatment. A 14.2, 13.7 and 20.4% increase in monocyte phagocytic capacity was observed after E. coli, S. aureus and C. albicans co-culture with ACV respectively and in comparison to the resting unstimulated monocytes. Results are expressed as the mean and SD of 3 similar experiments (Table 1). This suggests that ACV can increase phagocytic potential in monocytes which is significant as microbial phagocytosis is a key effector function of innate immunity22.
Table 1.
Effect of ACV on human monocyte phagocytic capacity.
| Monocyte co-culture conditions | Monocyte phagocytic capacity expressed as % change increase in side scatter. (mean ± SD) |
|---|---|
| Medium | 6.1 ± 1.4 |
| E-coli | 9.0 ± 2.4 |
| S. aureus | 22.3 ± 2.2 |
| C albicans | 31.9 ± 2.4 |
| E-coli + ACV | 23.2 ± 3.7 |
| S. aureus + ACV | 36.0 ± 5.2 |
| C albicans + ACV | 52.3 ± 4.1 |
In vitro differentiated monocytes were incubated with microbes for 4 h at 37 °C. Cells cultured with microbes with or without ACV were washed and processed for detection on the Beckton Dickinson flow cytometer. An analysis of changes in regional gated profiles and % shift in side scatter was measured. Data is presented as mean ± SD of 3 similar experiments.
Proteomic results of E. coli, S. aureus and C. albicans after exposure to ACV
The bottom-up proteomic study of ACV treated E-coli cultures revealed the absence in detection of key enzymes; citrate synthase, isocitrate dehydrogenase, deoxyribose–phosphate idolase, malate dehydrogenase, aminomethyltransferase and formate acetyltransferasesuccinyl-CoA ligase (Table 2). An absence in acyl carrier protein, DNA protein including DNA protection during starvation protein, integration host factor subunit alpha and ribosome associated inhibitor A was evident. Following ACV treatment S. aureus cultures failed to express 50 s ribosomal proteins L2, L15, L23, L24, enzymes; alcohol dehydrogenase, catalase, formate acetyltransferase, L-lactate dehydrogenase-2, ornithine aminotransferase and serine hydroxymethyl transferase (Table 3). Cell division protein ftsZ and chaperone protein Dnak were also not detected. However an important pentose phosphate pathway enzyme: 6 phosphogluconate dehydrogenase decarboxylating was displayed. Table 4 demonstrates that key enzymes required for glycolysis and candida immunogenicity were undetected after 24 h of exposure to ACV in C. albicans. These incorporated fructose bisphosphate aldolase, phosphogluconate dehydrogenase, pyruvate kinase and peptidyl-propyl cis-trans isomerase.
Table 2.
List of E-coli proteins identified following ACV treatment. E-coli were cultured with 1/50 dilution of ACV or alone in broth for 24 hours at 37 °C in a shaking incubator. After which mass spectroscopy analysis was carried out.
| Protein Name | Mass (Da) | Control E-coli culture | ACV Treated E-coli culture |
|---|---|---|---|
| 30S ribosomal protein S1 | — | * | |
| 30S ribosomal protein S11 | 13950 | * | |
| 30S ribosomal protein S4 | 23512 | * | |
| 30S ribosomal protein S6 | 15177 | * | |
| 30S ribosomal protein S7 | 17593 | * | |
| 30S ribosomal protein S8 | 14175 | * | |
| 50S ribosomal protein | 14923 | * | * |
| 50S ribosomal protein L1 | 24714 | * | |
| 50S ribosomal protein L17 | 14413 | * | * |
| 50S ribosomal protein L2 | 29956 | * | |
| 50S ribosomal protein L6 | 18949 | * | |
| 60 kDa chaperonin | 57464 | * | * |
| Acyl carrier protein | 8693 | * | |
| Aminomethyltransferase | 40235 | * | |
| Autonomous glycyl radical cofactor | — | * | |
| Citrate synthase | 48383 | * | |
| Cytidine deaminase | 31805 | * | |
| Deoxyribose-phosphate aldolase | 27958 | * | |
| DNA protection during starvation protein | 18684 | * | |
| DNA-binding protein H-NS | 15587 | * | |
| DNA-binding protein HU-alpha | 9529 | * | |
| DNA-binding protein HU-beta | 9220 | * | |
| Elongation factor Tu 1 | 43427 | * | * |
| Enolase | 45683 | * | * |
| Formate acetyltransferase 1 | 85588 | * | |
| Glutamate/aspartate periplasmic-binding protein | 33513 | * | |
| Glyceraldehyde-3-phosphate dehydrogenase | 35681 | * | * |
| Integration host factor subunit alpha | 11347 | * | |
| Isocitrate dehydrogenase [NADP] | 46070 | * | |
| Major outer membrane lipoprotein Lpp | 8375 | * | |
| Malate dehydrogenase | 32488 | * | |
| Outer membrane protein A | 37292 | * | * |
| Ribosome-associated inhibitor A | 12777 | * | |
| Succinate dehydrogenase flavoprotein subunit | 65008 | * | * |
| Succinyl-CoA ligase [ADP-forming] subunit beta | 42244 | * | |
| Transaldolase B | 35368 | * | |
| Uridine phosphorylase | 27313 | * | * |
The “*” indicates the presence of protein whilst the blank region denotes no detection of that particular protein.
Table 3.
List of S. aureus proteins identified following ACV treatment.
| Protein name | Mass (Da) | Control S. aureus culture | ACV- Treated S. aureus culture |
|---|---|---|---|
| 30S ribosomal protein S1 | 43250 | * | * |
| 30S ribosomal protein S12 | 15334 | * | |
| 30S ribosomal protein S2 | 29133 | * | |
| 30S ribosomal protein S3 | 24085 | * | * |
| 30S ribosomal protein S4 | 22999 | * | |
| 30S ribosomal protein S5 | 17732 | * | |
| 30S ribosomal protein S6 | 11588 | * | |
| 30S ribosomal protein S7 | 17783 | * | * |
| 30S ribosomal protein S8 | 14822 | * | |
| 50S ribosomal protein L1 | 24693 | * | |
| 50S ribosomal protein L13 | 16323 | * | * |
| 50S ribosomal protein L14 | 13241 | * | * |
| 50S ribosomal protein L15 | 15587 | * | |
| 50S ribosomal protein L2 | 30194 | * | |
| 50S ribosomal protein L21 | 11326 | * | * |
| 50S ribosomal protein L23 | 10599 | * | |
| 50S ribosomal protein L24 | 11529 | * | |
| 50S ribosomal protein L25 | 23773 | * | * |
| 50S ribosomal protein L29 | 8085 | * | |
| 50S ribosomal protein L4 | 22451 | * | * |
| 50S ribosomal protein L6 | 19774 | * | |
| 6-phosphogluconate dehydrogenase, decarboxylating | 51941 | * | |
| Alcohol dehydrogenase | 36424 | * | |
| Arginine deiminase | 47113 | * | * |
| Bacterial non-heme ferritin | 23773 | * | * |
| Catalase | 58457 | * | |
| Cell division protein FtsZ | 41012 | * | |
| Chaperone protein DnaK | 66338 | * | |
| DNA-binding protein HU | 9620 | * | * |
| Elongation factor Tu | 43134 | * | * |
| Enolase | 47145 | * | * |
| ESAT-6 secretion system extracellular protein A | 11029 | * | |
| Formate acetyltransferase | 85264 | * | |
| Fructose-bisphosphate aldolase class 1 | 32907 | * | |
| Isocitrate dehydrogenase [NADP] | 46451 | * | |
| L-lactate dehydrogenase 2 | 34468 | * | |
| Ornithine aminotransferase 2 | 43675 | * | |
| Ornithine carbamoyltransferase, catabolic | 37853 | * | * |
| Probable malate:quinone oxidoreductase | 56135 | * | * |
| Putative universal stress protein SA1532 | 18521 | * | * |
| Pyruvate dehydrogenase E1 component subunit beta | 35194 | * | * |
| Serine hydroxymethyltransferase | 45384 | * |
S. aureus were cultured with 1/10 dilution of ACV or alone in broth for 24 hours at 37 °C in a shaking incubator. After which mass spectroscopy analysis was carried out. The “*” indicates the presence of protein whilst the blank region denotes no detection of that particular protein.
Table 4.
List of C. albicans proteins identified following ACV treatment.
| Protein name | Mass (Da) | Control C.albicans culture | ACV- Treated C.albicans culture |
|---|---|---|---|
| 40S ribosomal protein S1 | 29083 | * | * |
| 6-phosphogluconate dehydrogenase | 71270 | * | |
| Alcohol dehydrogenase 1 | 37255 | * | * |
| Elongation factor 1-alpha 1 | 50436 | * | * |
| Elongation factor 2 | 93865 | * | * |
| Enolase 1 | 47202 | * | * |
| Fructose-bisphosphate aldolase | 39362 | * | |
| Glucose-6-phosphate isomerase | 61148 | * | |
| Glyceraldehyde-3-phosphate dehydrogenase | 35925 | * | * |
| Heat shock protein SSA1 | 70452 | * | * |
| Mitochondrial outer membrane protein porin | 29748 | * | |
| Peptidyl-prolyl cis-trans isomerase | 17678 | * | |
| Phosphoglycerate kinase | 45266 | * | * |
| Phosphoglycerate mutase | 27437 | * | * |
| Plasma membrane ATPase 1 | 98083 | * | * |
| Pyruvate decarboxylase | 62744 | * | * |
| Pyruvate kinase | 55752 | * | |
| Small heat shock protein 21 | 21482 | * | * |
| Triosephosphate isomerase | 26880 | * | * |
| White colony protein WHS11 | 6991 | * |
C. albicans were cultured with 1/2 dilution of ACV or alone in broth for 24 hours at 37 °C in a shaking incubator. After which mass spectroscopy analysis was carried out. The “*” indicates the presence of protein whilst the blank region denotes no detection of that particular protein.
Discussion
ACV has multiple antimicrobial properties on different microbial species, affecting microbe growth, suppressing mononuclear cytokine and phagocytic responses. The tandem mass spectroscopy results are in cohesion with these observations. The microbes underwent significant impairment following ACV addition which damaged cell integrity, structural and metabolic proteins as well as nuclear material. Indeed the enzymes citrate synthase, isocitrate dehydrogenase, malate dehydrogenase, aminomethyltransferase and formate acetyltransferasesuccinyl-CoA ligase are crucial for E-coli growth, gene regulation and central, intracellular carbon metabolism. An absence of these enzymes would affect glycolytic, tricarboxylic acid cycle, pentose phosphate, glycoxlate shunt and oxidative phosphorylation pathways in E. coli23. Furthermore, the absence of DNA binding protein from starved cells, is significant as it protects E-coli and functions to control gene regulation during cell starvation24. With respect to other proteins, we observed the presence of ribosomal proteins (L1, L6 and 30 s ribosomal proteins (S1, S4, S6, S7, S8, S11) in ACV treated E. coli compared to control E. coli. These are mostly RNA binding proteins hence their presence could have been due to partial disintegration of 50 s and 30 s ribosomal breakdown whereas these subunits might remain intact in untreated E. coli cultures. The absence of ribosome associated inhibitor A could interrupt E. coli growth cycles as it serves to minimise translational errors25. Collectively these results support the cytotoxic effects of ACV we observed on E-coli. ACV treated S. aureus cultures did not express the chaperone protein Dnak and the cell division protein ftsz. This is significant as previous studies have shown that a non-functional DnaK system can cause a reduced tolerance to heat, oxidative, antibiotic stresses and lowered carotenoid production26. There was also an absence of gateway enzymes involved in multiple pathways such as ornithine aminotransferase27. 6 phosphogluconate dehydrogenase decarboxylating was expressed which is not surprising as it plays a critical role in protecting cells from oxidative stress28. The effect of ACV on C.albican protein moieties was less dramatic, nevertheless we did detect the absence of key enzymes which are fundamental in maintaining cell integrity and biosynthetic pathways29.
There could be a strong possibility that ACV acts like other anti-pathogenic compounds in diverting monocyte responses through toll receptor signalling pathways. There is evidence that E-coli in particular can induce a typical M1 monocyte profile through mechanisms involving NF-kB activation. This results in upregulation of inflammatory cytokines TNF-α and PI3 kinase stimulation30. The in vitro M1 monocyte phenotype is also prominent in severe sepsis. This was shown in a study using baboons where a substantial mortality rate correlated with high serum levels of TNFα and IL-6 following induced sepsis infection31. Unchecked high levels of circulating M1 cytokines can rapidly lead to cardiac arrest and death hence any factor capable of lowering pro-inflammatory cytokine concentrations is essential in therapy32. It has been reported that ACV consists of acetic acid, flavonoids such as gallic acid, tyrosol catechin, epicatechin, benzoic acid, vaninilin, caftaric acid, coutaric acid, caffeic acid, acid and ferrulic acid These constituents have been reported to affect immune defence and oxidative responses18,33.
Furthermore, the mechanism of ACV activity could be attributed in part to the apple polyphenol content. Yang et al. (2010) reported on the cellular protective effects of apple polyphenols on induced liver damage whereby histopathological tissue destruction was limited and liver activity maintained in mice that received the polyphenols34. The mechanisms involved were free radical scavenger action, lipid peroxidation modulation and the antioxidant upregulation capacity of ACV. Interestingly, a study by Denis et al. demonstrated the anti-inflammatory potential of apple phenols on gastrointestinal cell inflammation which involved downregulation of TNF and IL-6 cytokines35. Another means of action could involve the acetic acid component of ACV which is able to reduce the cell hydrogen potential hence could potentially facilitate diffusion across the plasma membrane of microbes. Furthermore, there is evidence that organic acids can alter immune responses by binding to GPR3, a G protein coupled receptor which is mostly expressed on inflammatory leukocytes36. Also, an investigation reported on upregulated blood and plasma antioxidant enzyme release after apple consumption which would encourage immune protection16.
The positive benefits of dietary ACV supplementation have been highlighted in vivo. ACV decreased the serum lipid profile in mice fed a high cholesterol diet over 28 days. Intragastric ACV addition induced a protective effect against erythrocyte, kidney and liver oxidative injury as well as lowering cholesterol levels18. ACV also decreased blood triglyceride and very low density lipoprotein levels in rats which had induced cholesterol induced hepatic steatosis33. In an infection induced model of denture stomatitis, ACV addition resulted in anti-fungal activity against Candida Spp which was comparable to nystatin in terms of reducing microbial adherence and destruction37.
Severe infections, autoimmunity or transplantation can inevitably lead to ineffective immunity in patients. An analysis of macrophages from Crohn’s disease patients revealed that they had defective responses to E-coli due to Crohn’s related systemic immunosuppression38. A recent report showed that co-administration of ACV with L-casei boosted systemic and mucosal immune responses, antioxidant enzyme and growth genes in fish39. Equally, perhaps additive dietary supplementation with ACV could be of benefit in acute infections, autoimmune induced immune dysregulation or antibiotic redundancy in humans. Future studies would establish whether ACV could be used as a potential therapeutic, In vivo models of infection could be induced by infusing microbes systemically into mice followed by treatment with or without intraperitoneal ACV. Intragastric ACV has been fed to animals used as models of obesity and infection in the past18,37,39. ACV efficacy could be evaluated by measuring microbial burden, serum cytokine levels, leukocyte counts and tissue pathology. Side effects could include acid reflux, nausea or delayed digestion as ACV has a pH of 4.2. However, the acidity could be neutralised by the addition of sodium bicarbonate to preparations. The results of this study could have clinical implications as ACV could be used as an additive component of an antimicrobial therapeutic regimen especially in immunocompromised patients presenting with infections of the aforementioned microbes.
We conclude that ACV can have multiple antimicrobial effects directly on E-coli, S. aureus and C. albicans. ACV addition can also decrease induced inflammatory cytokine release during mononuclear leukocyte infection and increases monocyte phagocytic capacity. Mechanisms include alteration of the microbial protein physiology destroying structural pathogenic proteins and metabolic enzymes. Collectively our results highlight the potent antimicrobial and therefore beneficial actions of ACV. This preliminary study encourages further work on dietary ACV supplementation investigating its antimicrobial role and the constituents which could be responsible for this activity.
Materials and Methods
Chemical reagents, microorganisms, media and culture conditions
A selection of microbial specimens which represented a typical gram positive, a gram negative and a yeast species were chosen for initial investigation. Microbial strains: E-coli strain NCTC 10418 and S. aureus strains NCTC 6571 were purchased from Health Protection Agency (Colindale, U.K.). C. albicans strain 90828 was purchased from the American Type Culture Collection (LGC Promochem).
Reagents
Dulbecco’s modified media, dimethyl-ethyl-sulphonyl-oxide, HANKS balanced salt solution, histopaque, ethanol, phosphate buffered saline, paraformaldehyde, acetone, dithiothreitol, iodoacetamide, trypsin from porcine pancreas of proteomics grade, formic acid, acetonitrile, HPLC-grade water, methanol and Whatman Mini-UniPrep syringeless filter devices (pore size 0.45 µm) were purchased from Sigma Aldrich (Poole, U.K.). TNF-alpha, interleukin-6 (IL-6) enzyme linked immunosorbent assays (ELISA’s) were purchased from Research and Development Systems (Abingdon, U.K.). Mueller hinton agar was purchased from Oxoid, UK. Braag’s Apple Cider Vinegar and apple cider vinegar tablets (500 mg, Troo healthcare) were purchased from commercial sources.
Inoculum preparation and measurement of anti-microbial activity of ACV
Cultures of E. coli and S. aureus were grown in nutrient media whereas C. albicans was grown in Sabourand media. All cultures were cultivated in a shaking incubator at 37 °C for 24 h overnight prior to use. Mueller hinton agar (MHA) was prepared by dissolving 38 g in 1 litre of distilled water, boiling the mixture for 1 min, after cooling and autoclaving, the media was poured into petri dishes. The plates were left to dry and subsequently stored at 37 °C. All microbial cultures were adjusted to 0.5 McFarland’s standard 1.5 × 108 CFU/ml and 4 × 106 CFU/ml of each organism used in experiments. Each microbe was swabbed evenly onto plates containing MHA. For sample addition, 100 µL of ACV at varying concentrations was added to the wells which were punched into the agar. The plates were then incubated at 37 °C for 24 h. Zones of inhibition surrounding samples were identified, photographed and measured in mm40. Experiments were repeated at least 5 times.
Ethical Approval and Informed Consent
All experimental protocols were approved by the Middlesex University Natural Sciences Ethics Committee number 2323. Further the methods were carried out in accordance to the relevant guidelines and regulations. Informed consent was received when applicable.
Human mononuclear cell isolation procedure from whole peripheral blood
Human leucocyte rich cones and serum were obtained from volunteer donors collected from NHS Cord blood and transplant bank at Colindale, London and treated as described previously41. Briefly the cones were washed with phosphate buffer saline to harvest the leucocyte rich cells. These were then spun on histopaque density gradient at 1200 RPM for 20 min. Monocytes were purified using the CD14 positive mononuclear portion which was isolated according to manufacturer’s instruction provided with the pan monocyte isolation kit. Cells washed with HANKS balanced solution, counted and cultured into 24 well plates at 4 × 105 cells per mL. Cells were allowed to adhere for an hour at after which non-adherent cells were washed away and full media replenished with Dulbecco’s media containing 10% human serum. Monocytes were determined using light microscopy and flow cytometry phenotypic analysis of differentiation markers as described previously using CD1441. Freshly isolated monocytes were cultured with varying concentrations of ACV and either C. albicans, E. coli or S. aureus at counts of 4 × 106 CFU/ml respectively for 24 h at 37 °C and 5% CO2 after which supernatants were collected and analysed for TNF-α or IL-6 secretion using ELISA kits following manufacturer’s protocols.
Monocyte phagocytic capacity measurement by flow cytometry
Isolated human mononuclear cells were cultured at 4 × 105/mL in 24 well plates over a period of two days after which they were incubated with microbes (4 × 106 CFU/ml) for 4 h at 37 °C and 5 CO2. Cells were then scraped replenished in ice cold PBS containing 1 mM EDTA, washed and removed from plates. The resultant pellets were fixed in 400 µL of 4% paraformaldehyde and analysed using a FACS Calibur flow cytometer (Beckton Dickinson Immunocytometry Systems, UK and Cell Quest software).
Preparation of microbial tryptic digests for mass spectroscopy analysis
An aliquot of microbial suspension was collected and treated over 24 h with 1/100 ACV, washed with PBS, resuspended in PBS and then treated with 1 mL of ice-cold acetone. The bacterial cells were harvested after centrifugation 13,000 g for 5 min. The pellet was dried and then reconstituted in 50 mM ammonium bicarbonate. Cells were lysed using a Soniprep 150 Plus (MSE, U.K.) for 10 s with the amplitude set at 13. Subsequently proteins were denatured and reduced with 3 μL of 100 mM dithiothreitol in 50 mM ammonium bicarbonate at 95 °C for 5 min followed by alkylation with 6 μL of 100 mM iodoacetamide in the dark at room temperature for 20 min. Proteins were then digested with 2 μL of trypsin (0.1 μg/μL dissolved in 50 mM ammonium bicarbonate) at 37 °C for 3 h. A further 2 μL of trypsin was added to the sample and the mixture was incubated at 37 °C for an additional 2 h. The samples were diluted in 150 µL 50 mM ammonium bicarbonate and passed through Mini-Uni Prep filter devices.
Liquid Chromatography-Electrospray Ionisation Tandem Mass Spectrometry
The tryptic peptides were analysed using a Shimadzu Prominence HPLC system hyphenated to an electrospray ionisation hybrid ion-trap time-of-flight (IT-TOF) mass spectrometer (Shimadzu, U.K.) operated in tandem mass spectrometry mode. Peptides were separated using an Ascentis Express 150 × 2.1 mm, 2.7 µm C18 column (Sigma-Aldrich, Poole, U.K.) using a flow rate of 0.21 mL/min. The column oven temperature was set to 40 °C. Data was acquired and processed using LabSolutions®software (version 3.50.348, Shimadzu, UK). A linear gradient elution profile composed of ‘A’−0.1% formic acid in water and ‘B’- 0.1% formic acid in acetonitrile was used. The gradient profile was 0–40% B, 70 min; 40–90% B, 1 min; maintained at 90% B, for 3 min; 90–0% B, 1 min; and 15 min re-equilibration at 0% B. An injection volume of 40 µL was used. Samples were kept in the auto sampler set to 4 °C. Mass spectrometry analysis was performed in MS/MS mode using positive ions electrospray. The precursors’ acquisition range was set to 400–1,800 m/z while the fragments acquisition range was set to 200–1,500 m/z. For both precursors and fragments the ion accumulation time was set to 30 msec. The other instrument conditions were set as follows: detector voltage 1.6 kV, CID energy 70%, nebulising gas flow 1.5 L/min, CDL temperature 200 °C, heat block temperature 200 °C, interface voltage 4.5 V, detector voltage 2 kV. The data acquisition was performed in a 37.5 min interval.
LC-MS/MS Data Processing
MS/MS data were extracted from the resulting instrument files using Mascot Distiller software (version 2.5.1.0, Matrix Science, London, UK). For precursors peak picking the following parameters were used: correlation threshold – 0.7, minimum signal to noise ratio – 5, minimum peak m/z – 50, maximum peak m/z – 100,000, minimum peak width – 0.02 Da, expected peak width – 0.2 Da, maximum peak width – 2 Da. The MS/MS ion list was searched using Mascot search engine against all entries in Swiss-Prot database (2016_2). For database search the following parameters were used: two missed cleavages, carbamidomethylation of cysteine (as fixed modification) and oxidation of methionine (as variable modification). The tolerance for precursor peptides was set to 10 ppm and for fragments to 0.3 Da. Peptide charges used for peak picking was +2, +3 and +4.
Statistical analysis
All experimental results are expressed as the mean ± standard deviation (SD). Statistical analyses was carried out using one way ANOVA or students t-test, outcomes were considered significant where p < 0.05 (when comparing apple cider vinegar treated microbes to the untreated groups in all experiments). All experiments were repeated at least 3–5 times. Analysis was carried out using Excel software version 2016.
Data availability
The datasets generated and analyzed during the current study reside with the corresponding author and can be made available upon request.
Acknowledgements
We would like to thank Manika Choudhury and Alejandra Gonzalez Baez for their technical assistance.
Author Contributions
D.Y. conceived and performed the experiments, data analysis and wrote the manuscript. V.S. contributed to the mass spectroscopy experiments and data analysis. A.S. contributed to manuscript writing and data review.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization (WHO). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: Switzerland (2017).
- 2.Mora-Rillo M, et al. Impact of virulence genes on sepsis severity and survival in Escherichia coli bacteraemia. Virulence. 2015;6:93–100. doi: 10.4161/21505594.2014.991234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rittirsch D, Flierl PA, Ward MA. Harmful molecular mechanisms in sepsis. Nature Reviews Immunology. 2008;8:776–787. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cigana C, Assaei BM, Melotti P. Azithromycin selectively reduces tumour necrosis factor alpha levels in cystic fibrosis airway epithelial cells. Antimicrob agents Chemother. 2007;51:975–81. doi: 10.1128/AAC.01142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culic O, et al. Azithromycin modulates neutrophil function and circulating inflammatory mediators in healthy human subjects. Eur J Pharmacol. 2002;450:277–89. doi: 10.1016/S0014-2999(02)02042-3. [DOI] [PubMed] [Google Scholar]
- 6.Marbach H, et al. Identification of a distinctive phenotype for endocarditis-associated clonal complex 22 MRSA isolates with reduced vancomycin susceptibility. J Med Microbiol. 2017;66:584–591. doi: 10.1099/jmm.0.000470. [DOI] [PubMed] [Google Scholar]
- 7.Aghai ZH, et al. Azithromycin suppresses activation of nuclear factor kappa B and synthesis of pro-inflammatory cytokines in tracheal aspirate cells from premature infants. Pediatr Res. 2007;62:483–8. doi: 10.1203/PDR.0b013e318142582d. [DOI] [PubMed] [Google Scholar]
- 8.Nurjadi D, Klein S, Zimmermann S, Heeg K, Zanger P. Transmission of ST8-USA300 Latin American Variant Methicillin-Resistant Staphylococcus aureus on a Neonatal Intensive Care Unit: Recurrent Skin and Soft- Tissue Infections as a Marker for Epidemic Community-Associated-MRSA Colonization. Infect Control Hosp Epidemiol. 2017;18:1–3. doi: 10.1017/ice.2017.88. [DOI] [PubMed] [Google Scholar]
- 9.Ward DV, et al. Metagenomic Sequencing with Strain-Level Resolution Implicates Uropathogenic E. coli in Necrotizing Enterocolitis and Mortality in Preterm Infants. Cell Rep. 2016;14:2912–24. doi: 10.1016/j.celrep.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–84. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Çek M, et al. Antibiotic prophylaxis in urology departments. global prevalence study of infections in urology investigators. Eur Urol. 2013;63:386–94. doi: 10.1016/j.eururo.2012.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan, S. B. et al. Reduced vancomycin susceptibility of methicillin-susceptible Staphylococcus aureus: no significant impact on mortality but increase in complicated infection. Antimicrob Agents Chemother. AAC.00316–17, 10.1128/AAC.00316-17 (2017). [DOI] [PMC free article] [PubMed]
- 13.Hippocrates: The Genuine Works of Hippocrates. Volume I and II. Edited by: Adams F. New York: William Wood and Company (1886).
- 14.Ebner, H., Farnworth, H. & Sellemer, S. Vinegar in biotechnology: A multi-volume comprehensive treatise, 2nd edition, 579–91 (1995).
- 15.del Campo G, Berregi I, Santos JI, Dueñas M, Irastorza A. Development of alcoholic and malolactic fermentations in highly acidic and phenolic apple musts. Bioresour Technol. 2008;99:2857–63. doi: 10.1016/j.biortech.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Avci A, et al. Effects of apple consumption on plasma and erythrocyte antioxidant parameters in elderly subjects. Exp Aging Res. 2007;33:429–37. doi: 10.1080/03610730701525352. [DOI] [PubMed] [Google Scholar]
- 17.Pourmouzaffar S, Hajimardloo A, Mindare HK. Dietary effect of apple cider vinegar and propionic acid on immune related transcriptional response and growth performance in white shrimp. Litopenaeus vannamei. Fish Shellfish Immunology. 2017;60:60–71. doi: 10.1016/j.fsi.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Nazıroğlu M, et al. Apple cider vinegar modulates serum lipid profile, erythrocyte, kidney, and liver membrane oxidative stress in ovariectomized mice fed high cholesterol. J Membr Biol. 2014;24:667–73. doi: 10.1007/s00232-014-9685-5. [DOI] [PubMed] [Google Scholar]
- 19.Palaska I, Gagari E, Theoharides TC. The effects of P. gingivalis and E.coli LPS on the expression of proinflammatory mediators in human mast cells and their relevance to periodontal disease. J. Biol Regul Homeost agents. 2016;30:655–664. [PubMed] [Google Scholar]
- 20.Swan ZD, Bouwer AL, Wonderlich ER, Barrat-Boyes SM. Persistent accumulation of gut macrophages with impaired phagocytic function correlates with SIV disease progression in macaques. Eur J Immunol. 2017 doi: 10.1002/eji.201646904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walborn, A., Hoppensteadt, D., Syed, D., Mosier, M. & Fareed, J. Biomarker Profile of Sepsis-Associated Coagulopathy Using Biochip Assay for Inflammatory Cytokines. Clin Appl Thromb Hemost., 10.1177/1076029617709084 (2017). [DOI] [PMC free article] [PubMed]
- 22.Procenca JT, Barral DC, Gordo I. Commensal to pathogen: One single transposon insertion results in two pathoadaptive traits to Escherichia coli-macrophage interaction. Sci Reports. 2017;7(1):4504. doi: 10.1038/s41598-017-04081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahan, N., Maeda, K., Matsuoka, Y., Sugimoto, Y. & Kurata, H. Development of an accurate kinetic model for the central carbon metabolism of Escherichia coli. Microb Cell Fact. 21–15(1), 112, 10.1186/s12934-016-0511-x (2016). [DOI] [PMC free article] [PubMed]
- 24.Calhoun LN, Kwon YM. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol. 2011;110(2):375–86. doi: 10.1111/j.1365-2672.2010.04890.x. [DOI] [PubMed] [Google Scholar]
- 25.Kawashima SA, et al. Potent reversible and potent chemical inhibitors of eukaryotic ribosome biogenesis. Cell. 2016;30:1573–88. doi: 10.1016/j.cell.2016.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh VK, et al. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153:3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 27.Ginguay, A., Cynober, L., Curis, E. & Nicolis, I. Ornithine Aminotransferase, an Important Glutamate-Metabolizing Enzyme at the Crossroads of Multiple Metabolic Pathways. Biology. 7, 6(1). pii: E18. 10.3390/biology6010018 (2017). [DOI] [PMC free article] [PubMed]
- 28.Martins RN, Harper CG, Stokes GB, Masters CL. Increased cerebral glucose-6-phosphate dehydrogenase activity in Alzheimer’s disease may reflect oxidative stress. J Neurochem. 1986;46:1042–1045. doi: 10.1111/j.1471-4159.1986.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 29.Kelly J, Kavanagh K. Proteomic analysis of protein released from growth arrested candida albicans following exposure to caspofungin. Med Mycol. 2010;48:598–605. doi: 10.3109/13693780903405782. [DOI] [PubMed] [Google Scholar]
- 30.Ni W, et al. Escherichia coli maltose-binding protein activates mouse peritoneal macrophages and induces M1 polarization via TLR2/4 in vivo and in vitro. Int Immunopharmacol. 2014;21:171–80. doi: 10.1016/j.intimp.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Bengtsson A, et al. Anti-TNF treatment of baboons with sepsis reduces TNF-alpha, IL-6 and IL-8, but not the degree of complement activation. Scand J Immunol. 1998;48:509–14. doi: 10.1046/j.1365-3083.1998.00433.x. [DOI] [PubMed] [Google Scholar]
- 32.Simisopoulou M, et al. Differential expression of cytokines and chemokines in human monocytes induced by lipid formulations of amphotericin B. Antimicrob Agents Chemother. 2005;49:1397–1403. doi: 10.1128/AAC.49.4.1397-1403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budak NH, et al. Effects of apple cider vinegars produced with different techniques on blood lipids in high-cholesterol-fed rats. J Agric Food Chem. 2011;22–59(12):6638–44. doi: 10.1021/jf104912h. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Li Y, Wang F, Wu C. Hepatoprotective effects of apple polyphenols on CCI4-induced acute liver damage in mice. J Agric Food Chem. 2010;26–58(10):6525–31. doi: 10.1021/jf903070a. [DOI] [PubMed] [Google Scholar]
- 35.Denis, M. C. et al. Apple peel polyphenols and their beneficial actions on oxidative stress and inflammation. Furtos A, Dudonne S, Montoudis A, Garofalo C, Desjardins Y, Delvin E, Levy E. PLoS One8(1) (2013). [DOI] [PMC free article] [PubMed] [Retracted]
- 36.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 37.Mota AC, et al. Antifungal Activity of Apple Cider Vinegar on Candida Species Involved in Denture Stomatitis. J Prosthodont. 2015;24(4):296–302. doi: 10.1111/jopr.12207. [DOI] [PubMed] [Google Scholar]
- 38.Vazeille E, et al. Monocyte-derived macrophages from Crohn’s disease patients are impaired in the ability to control intracellular adherent-invasive Escherichia coli and exhibit disordered cytokine secretion profile. J Crohns Colitis. 2015;9(5):410–20. doi: 10.1093/ecco-jcc/jjv053. [DOI] [PubMed] [Google Scholar]
- 39.Safari R, Hoseinifar SH, Nejadmoghadam S, Khalili M. Apple cider vinegar boosted immunomodulatory and health promoting effects of Lactobacillus casei in common carp (Cyprinus carpio) Fish Shellfish Immunol. 2017;7(67):441–448. doi: 10.1016/j.fsi.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Ostrovsky EA, et al. Methods for the antimicrobial activity and determination of minimum inhibitory concentration of planrt extracts. Brazilian J Pharmacogn. 2008;18:301–7. doi: 10.1590/S0102-695X2008000200026. [DOI] [Google Scholar]
- 41.Yagnik, D. Macrophage derived platelet activating factor implicated in the resolution phase of gouty inflammation. Int J Inflam. 526496, 10.1155/2014/526496 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study reside with the corresponding author and can be made available upon request.