ABSTRACT
Rpc34 is a subunit of the Rpc82/34/31 subcomplex residing on the DNA-binding cleft of RNA polymerase (Pol) III. Rpc34 contains a structurally flexible N-terminal tandem winged-helix (tWH) domain related to the TFIIE transcription factor. While the second WH (WH2) fold of the tWH domain is known to function in DNA melting activity during transcription initiation, the functional role of the WH1 fold is unknown. In this study, we generated a series of new Rpc34 tWH mutants conferring a cold-sensitive growth phenotype. We found that the tWH mutations severely compromised in vitro transcription activity due to destabilization of the preinitiation complex (PIC). Site-specific protein photo-cross-linking analysis indicated that the tWH domain persistently interacts with protein subunits of the Pol III cleft in the PIC and the ternary elongation complex (TEC). Furthermore, purified Pol III proteins with tWH mutations also showed reduced efficiency in RNA elongation. Our study results suggest that the tWH domain is an important protein module above the Pol III cleft that integrates protein and nucleic acid interactions for initiation and elongation.
KEYWORDS: transcription, RNA polymerase III, Rpc34, winged-helix domain
INTRODUCTION
In eukaryotic cells, transcription of genomic DNA is conducted by three different RNA polymerases (Pol), namely, Pol I, Pol II, and Pol III. While Pol I and Pol II are responsible for rRNA and mRNA transcription, respectively, Pol III transcribes short untranslated RNAs such as tRNA, 5S rRNA, U6 snRNA, and 7SL RNA (1, 2). The transcription process is divided into different stages, including initiation, elongation, termination, and reinitiation. During Pol III transcription initiation, transcription factors TFIIIA and TFIIIC function in recognizing the promoter elements inside the transcribed region, and TFIIIB is subsequently recruited to the upstream region of the transcribed gene. Finally, TFIIIB recruits Pol III to the transcription start site to complete the formation of a preinitiation complex (PIC) (3). The Pol III PIC spontaneously melts DNA to load the single-stranded DNA bubble into the Pol III cleft for subsequent initiation of RNA synthesis (4).
Pol III is the largest RNA Pol, comprising 17 subunits with a total molecular mass of ∼700 kDa (2). The Pol III complex contains a Pol II-like 12-subunit core structure and two specific subcomplexes, i.e., the Rpc37/53 heterodimer and Rpc82/34/31 heterotrimer, respectively, homologous to the TFIIF and TFIIE transcription factors in the Pol II system. Recent high-resolution cryo-electron microscopy (cryo-EM) studies have revealed that these two Pol III-specific subcomplexes are in positions adjacent to the Pol III active site cleft formed by the two largest subunits, Rpc160 and Rpc128 (5–7). The Rpc37/53 subcomplex is anchored at the lobe domain of Rpc128 and participates in all steps of the Pol III transcription cycle. In particular, Rpc37 has been proposed to regulate transcription termination through an interaction with the transcription bubble (8–11). Structural resolution of the Rpc82 subunit has revealed four extended winged-helix (eWH1 to eWH14) folds followed by a C-terminal coiled-coil domain, with the subunit utilizing its eWH1 and eWH14 domains to anchor the Rpc82/34/31 subcomplex on the clamp domain of Rpc160 (6, 7, 12, 13). The positioning of the Rpc82/34/31 subcomplex explains its functional contribution to transcription initiation (14–17).
Saccharomyces cerevisiae Rpc34 is composed of 317 amino acids (Fig. 1A), and its N-terminal half contains two WH folds (for WH1, amino acids [aa] 11 to 75; for WH2, aa 87 to 150) (Fig. 1B), jointly referred to as the tandem WH (tWH) domain. The tWH domain is also found in the Rpa49 subunit of Pol I and the Tfa2 (TFIIEβ in human) subunit of the TFIIE transcription factor in the Pol II machinery (2). The C-terminal half of Rpc34 contains a third WH fold (WH3; aa 170 to 270) and an acidic terminus. According to cryo-EM structural analysis (5), the WH3 fold and the acidic C terminus are bound, respectively, to the eWH4 and coiled-coil domains of Rpc82. Due to structural flexibility, the tWH domain has not been resolved by cryo-EM (6, 7, 13, 18) but, based on a chemical cross-linking study (19), it has been proposed that the domain is positioned directly above the cleft to interact with the transcription bubble. Functional insights into the tWH domain have come from a mutational analysis in which mutations of charged residues in the WH2 fold were found to severely compromise DNA melting in transcription initiation (17). Supporting its DNA melting activity, an electrophoretic gel mobility shift (EMSA) analysis indicated that the WH2 fold of human Rpc39 protein (an Rpc34 ortholog) strongly binds double-stranded DNA (12). However, the WH1 fold inhibits DNA binding. To date, the functional role of WH1 has not been characterized.
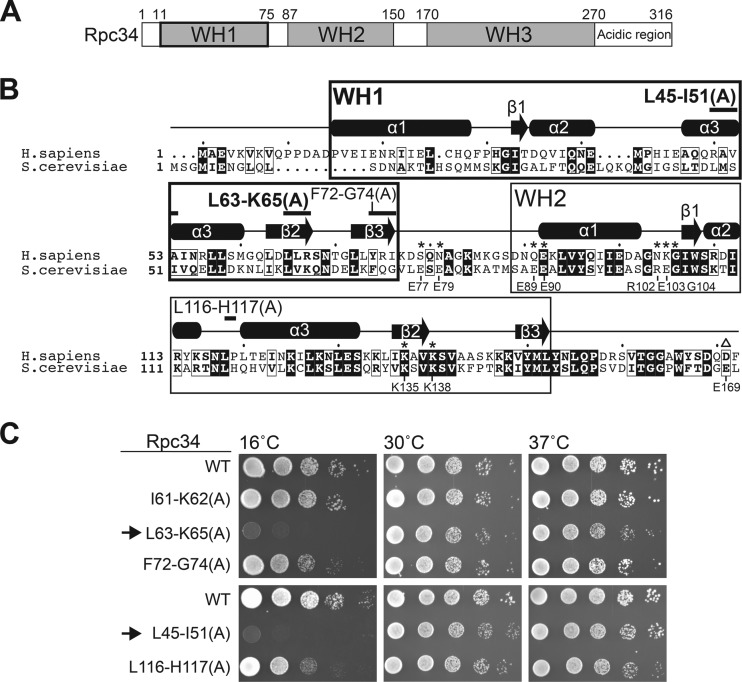
FIG 1.
New functionally important regions in the tWH domain. (A) Structural domains in Rpc34. (B) The sequence alignment between human (Homo sapiens) Rpc39 (Rpc6; PORL3F) and yeast (S. cerevisiae) Rpc34 proteins was generated by the Clustal Omega and ESPript programs. The WH1 and WH2 domains are boxed. Secondary structures were predicted by ESPript (cylinders, α-helices; arrows, β-strands). The new alanine replacement mutations are indicated by black bars above the sequence, and alanine mutations analyzed in a previous publication (17) are labeled with asterisks. UniProt accession numbers are as follows: for H. sapiens hRpc39, Q9H1D9; for S. cerevisiae Rpc34, P32910. (C) Cell growth analysis of the Rpc34 mutations by serial-dilution spot assay. Arrows point to two Rpc34 mutation strains with alanine replacements in L45-I51 and L63-K65 within the WH1 fold. These two strains conferred a severe cold-sensitive cell growth phenotype at 16°C.
To further understand the functional role of the tWH domain in Pol III transcription, we analyzed functional phenotypes of a series of new Rpc34 mutants with a cold-sensitive cell growth defect. We found that mutations in both tWH folds conferred an in vitro transcription defect attributable to destabilization of the PIC. Site-specific cross-linking analysis indicates that the tWH domain interfaces with the Pol III cleft in both the transcription initiation and elongation complexes. Consistent with the positioning, further analysis indicates that the tWH mutants reduce transcription elongation efficiency. Our data provide new insights into the functional requirements of the tWH domain in the Pol III transcription cycle.
RESULTS
Important new functional regions of the tWH domain.
To understand the functional importance of the tWH domain of Rpc34, we first generated a series of S. cerevisiae mutant strains. In contrast to a previous mutational study targeting charged residues (17), we replaced stretches of amino acid sequences by alanines throughout the domain. As demonstrated in Fig. 1B and C, two Rpc34 mutant strains with alanine replacing all residues in sequence blocks L45-I51 and L63-K65 of the WH1 fold conferred a severe cold-sensitive cell growth phenotype (assayed at 16°C). The two alanine replacement mutants highlight the functional importance of the α3 and β2 secondary structures of WH1. In addition, we obtained two mutant strains, namely, F72-G74(A) and L116-H117(A), that conferred a slightly cold-sensitive cell growth phenotype (Fig. 1B and C). All of our new tWH mutations are distinct from the residues described in a previous analysis by Brun et al. (17) (see Fig. 1B here; residues marked with asterisks). Our newly obtained mutant strains allowed us to conduct a functional analysis of previously uncharacterized structural regions in the tWH domain.
The tWH mutations affect Pol III transcription activity.
In our subsequent analysis of the Rpc34 tWH mutants, we applied immunoprecipitation (IP) analysis to address whether the mutations affect structural integrity. In pulldown experiments, we utilized anti-Flag antibody beads to immobilize Flag-epitope-tagged Rpc82 from yeast whole-cell extracts (WCE). As shown in Fig. 2A, Rpc34 and Rpc128 were also coimmunoprecipitated, and the precipitated protein levels were not affected by Rpc34 mutations. We concluded that stable subunit association of the Pol III complex is not compromised by mutations in the Rpc34 tWH domain.
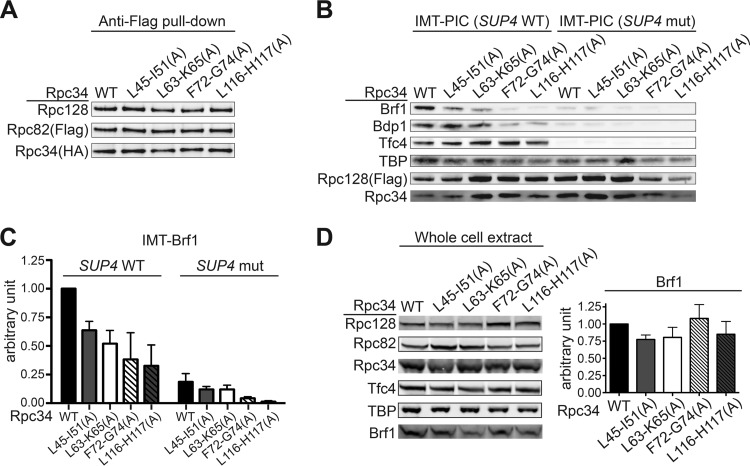
FIG 2.
PIC formation analysis of the Rpc34 tWH mutants. (A) Coimmunoprecipitation with Rpc34 mutants. The Pol III complex was immunoprecipitated through binding of Flag epitope-tagged Rpc82 with anti-Flag–agarose beads. Coimmunoprecipitated Rpc128 and Rpc34 (HA epitope tagged) were revealed by the respective antibodies. (B) Western analysis of proteins in the Pol III PIC from the immobilized template assay. The DNA template contained the SUP4 tRNA gene (SUP4 WT) or the mutant SUP4 sequence (SUP4 mut) with mutations in the A and B boxes of the internal promoters. (C) Quantitation of Brf1 in the IMT assays. Error bars indicate standard deviations (SD) of results from three independent IMT experiments performed using separately prepared whole-cell extracts (WCEs). (D) Proteins in the whole-cell extracts. Results of Western analysis of proteins in WCEs are displayed on the left. (Right panel) Quantitation of Brf1 in the WCEs; error bars indicate SD of results from three independently prepared WCEs.
We next assayed Pol III transcriptional activity of the Rpc34 mutants. We first analyzed PIC formation by the immobilized template (IMT) assay with WCEs from mutant strains. Whereas the protein levels of Pol III subunits (Rpc128, Rpc82, and Rpc34) and Tfc4 (a subunit of TFIIIC), as well as of TBP (a DNA-binding subunit of TFIIIB), were the same or slightly higher on the wild-type (WT) SUP4 template, the levels of Brf1 and Bdp1 (subunits of TFIIIB) were reduced due to the tWH mutations (Fig. 2B and C). The lower levels of Brf1 and Bdp1 in the Rpc34 mutant extracts are specific since the association of these factors and Tfc4 with the DNA were promoter dependent (Fig. 2B and C). Based on the levels of Brf1 in different WCEs, the PIC formation phenotype was unlikely to have been caused by variations in Brf1 content (Fig. 2D). As Rpc34 is known to interact with Brf1 (17, 20, 21), the mutations likely affect stable associations of Brf1 and Bdp1 in the PIC, resulting in the losses of both proteins after stringent washes in the IMT assay. This in vitro phenotype is surprising as the TFIIIB complex is known to bind very tightly to promoter DNA (22, 23). However, our IMT analysis suggests that the tWH domain of Rpc34 provides an important form of protein support for Brf1 and Bdp1 in the higher-order PIC.
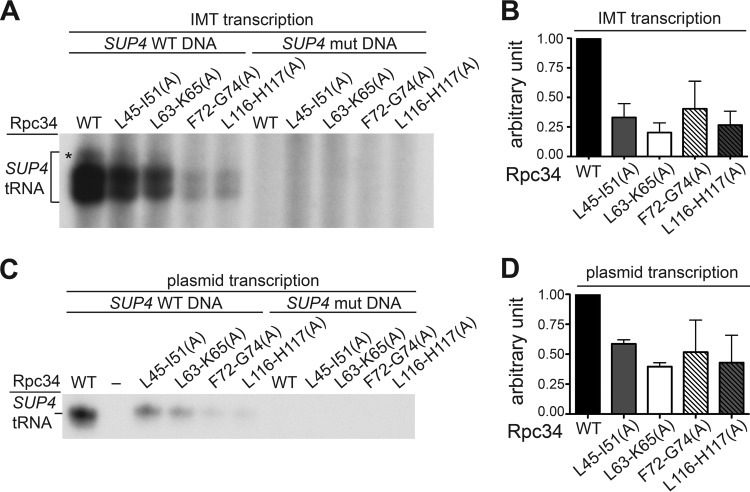
We further conducted in vitro transcription analysis with the isolated PICs. As demonstrated in Fig. 3A and B, the tWH mutants exhibited significantly reduced in vitro SUP4 tRNA synthesis. This reduced tRNA synthesis correlates with the reductions of Brf1 and Bdp1 in the IMT assays. Additional analyses via plasmid transcription assays were consistent with the IMT analysis, with tWH mutants also exhibiting reduced tRNA synthesis in those analyses (Fig. 3C and D). In summary, our study data suggest a functional role for the tWH domain in PIC stabilization for transcription.
FIG 3.
Transcription activity of the Rpc34 tWH mutants. (A) In vitro transcription activity of Pol III with Rpc34 mutations. The SUP4 tRNA transcripts were generated by addition of NTPs to the isolated PICs from the IMT assays. An asterisk highlights the read-through RNA transcripts that resulted from transcription termination at the second terminator sequence of the SUP4 tRNA gene. (B) Quantitation of the transcriptional activity of the isolated PICs. The RNA transcripts from the wild-type SUP4 (SUP4 WT) template in the experiment described for panel A were quantified. Error bars indicate SD of results from three independent IMT transcription experiments performed with separately prepared WCEs. (C) In vitro transcription activity with plasmid DNA and WCEs of Rpc34 mutants. (D) Quantitation of the transcriptional activity from the plasmid transcription assay. Error bars indicate SD of results from three independent plasmid transcription experiments performed with separately prepared WCEs.
The tWH domain interacts with Rpc37 and Rpc82 in Pol III initiation and elongation.
According to cryo-EM structural analysis (5), Rpc34 WH3 and the acidic C terminus interact with the eWH4 and coiled-coil domains of Rpc82, respectively. Consistent with the cryo-EM structure, a previous cross-linking analysis using a lysine-specific cross-linker revealed Rpc82 binding sites within the WH3 fold of Rpc34 (summarized in Fig. 4A) (19). Lysine-specific cross-linking analysis also mapped a number of amino acid positions in the Rpc34 tWH domain having interactions with Rpc128, Rpc82, and Rpc37, all of which indicated positioning of the tWH domain above the Pol III cleft (19). In particular, the WH2 fold was modeled as connecting with the upstream edge of the DNA bubble based on multiple intersubunit lysine links. Since the previous cross-linking study was conducted with a purified Pol III sample, we further conducted a cross-linking analysis of Rpc34 within the PIC. In this new analysis, we site-specifically incorporated the photo-cross-linker p-benzoyl-l-phenylalanine (BPA) as a nonnatural amino acid into Rpc34 in yeast. We employed WCEs with BPA-containing Rpc34 variants to assemble the PIC using an immobilized DNA template containing the SUP4 tRNA gene. The BPA cross-linking analysis was also extended to the transcribing Pol III complex that is also referred to as the ternary elongation complex (TEC; Pol III-DNA-RNA). To isolate the TECs, we further added ATP, UTP, and CTP to the preassembled PICs to allow Pol III stalling on synthesis of the 17-nucleotide-long RNA (Fig. 4B). The isolated complexes were subjected to UV irradiation to generate protein cross-links, revealing specific protein interactions for BPA-substituted positions in Rpc34.
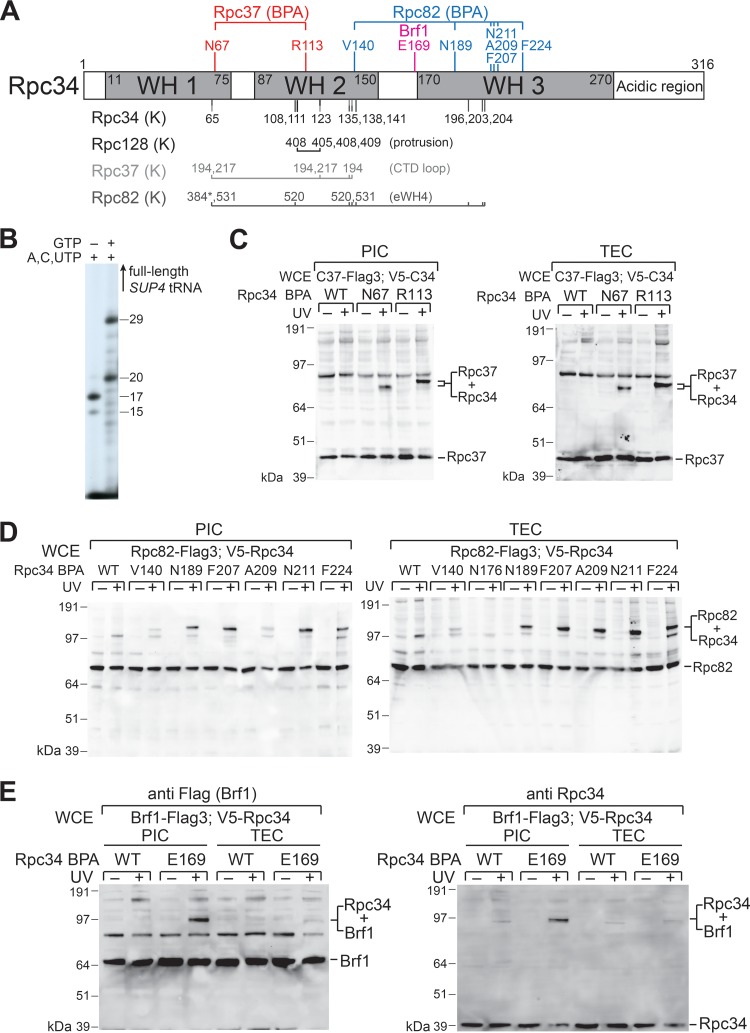
FIG 4.
Protein-protein interactions of the tWH domain in initiation and elongation. (A) Summary of protein interaction mapping by BPA and lysine-specific cross-linking. Positions of BPA cross-links are shown above the schematic of Rpc34. A BPA substitution at E169 immediately N terminal to the WH3 fold generated a cross-link with Brf1 (21). Below the schematic, lysine residues of Rpc34 involved in lysine-specific cross-linking with Rpc128, Rpc37, and Rpc82 are listed. Also indicated are the structural domains in which the cross-linked lysines are located. K384 of Rpc82 (marked with an asterisk) is located within an unstructured loop of the eWH3 domain. Other lysine residues of Rpc82 are positioned in the eWH4 domain. CTD, C-terminal domain. (B) RNA synthesis. The autoradiogram shows RNA transcripts from the addition of ATP, CTP, and UTP (lane 1) or the complete set of NTPs (lane 2) to the preassembled PIC on the SUP4 tRNA gene. The major RNA gel band in lane 1 corresponds to the 17-nucleotide RNA from the stalled Pol III ternary elongation complex (TEC). Upper and lower RNA gel bands represent RNA products resulting from misincorporation and exonuclease activities of Pol III, respectively. In lane 2, the two lower (20- and 29-nt) RNA gel bands correspond to the major pausing sites of Pol III elongation on the SUP4 tRNA gene reported in the literature (25). (C) Rpc34-Rpc37 BPA cross-linking in the TEC and PIC. In the Western analysis of BPA cross-linking, the anti-Flag antibody was utilized to reveal C-terminally Flag epitope-tagged Rpc37 (C37-Flag3), as well as the Rpc37-Rpc34 cross-links. The Rpc37-Rpc34 cross-links were also validated in a parallel analysis using anti-V5 antibody to probe N-terminally V5 epitope-tagged Rpc34 (V5-C34) (data not shown). WCE, whole-cell extract. (D) Rpc34-Rpc82 BPA cross-linking in the TEC and PIC. The anti-Flag antibody was utilized to reveal the Flag epitope-tagged Rpc82 as well as the cross-links. (E) Rpc34-Brf1 BPA cross-linking. BPA substitution at the E169 residue of Rpc34 generated a cross-link with Brf1 in the PIC. The intensity of Rpc34-Brf1 cross-linking was reduced significantly in the TEC. Both anti-Flag and anti-Rpc34 antibodies were applied in the Western analysis.
As indicated in Fig. 4C and D and summarized in Fig. 4A, the Rpc37 cross-linking sites are found in the WH1 and WH2 folds, whereas the Rpc82 cross-linking sites are localized in the WH3 fold (with one exception in the WH2 fold). Overall, our BPA cross-linking results correlate well with a previous lysine-specific cross-linking analysis (19). However, we did not detect Rpc128 cross-linking (Fig. 4A). We presume that this reflects differences in the chemistry of their cross-linking reactions and/or the length of the cross-linker. Upon photolysis, the benzophenone functional group of BPA preferentially reacts with CH bonds of neighboring amino acids, linking amino acids within distances of approximately 10 Å (Cα to Cα). The DSS cross-linker used in the lysine-specific cross-linking study is more flexible, with a reaction distance of up to 20 Å (Cα to Cα) (24). Summarizing the cross-linking analyses, the tWH domain maintains close contacts with Rpc82 and Rpc37 within the PIC and the TEC. In contrast, a previously discovered interaction between Rpc34 and the transcription initiation factor Brf1 occurs only in the PIC (Fig. 4E) (21).
The tWH domain functions in RNA elongation.
Our cross-linking analyses indicate that the tWH domain likely maintains a network of protein-nucleic interactions above the Pol III cleft in both initiation and elongation. To address whether the new tWH mutations affect the elongation process, we analyzed RNA extension by chasing the isolated TECs to longer RNA products with further addition of GTP. Based on the RNA extension profiles shown in Fig. 5, the L45-I51(A) mutation apparently increased Pol III pausing at the previously reported early pausing sites of the SUP4 tDNA (25), suggesting a role in elongation for this tWH region.
FIG 5.
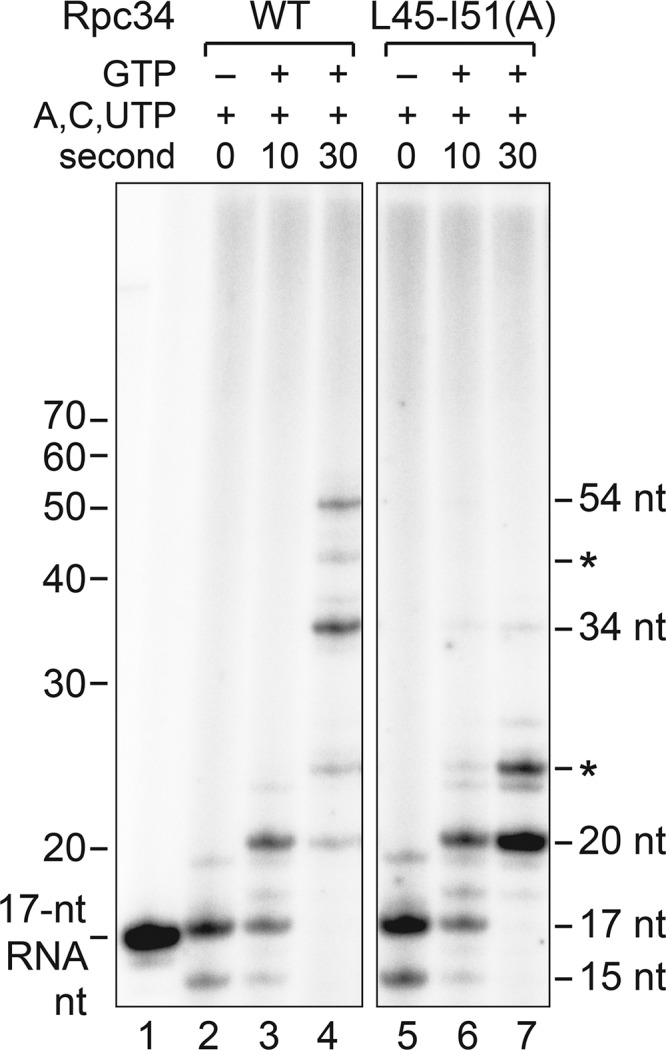
Elongation analysis of the Rpc34 tWH mutant. The autoradiogram shows RNA transcripts from the TECs stalled on the immobilized SUP4 tDNA with a subset of ribonucleotides (A, U, and C) (lanes 2 and 5) or chased TECs with further addition of GTP (lanes 3, 4, 6, and 7). The band in lane 1 represents a 32P-labeled 17-nucleotide RNA. RNA gel bands (20, 34, and 54 nt) corresponding to the major pausing sites of Pol III elongation on the SUP4 tRNA gene are indicated. Asterisks highlight the RNA products from unidentified pausing sites.
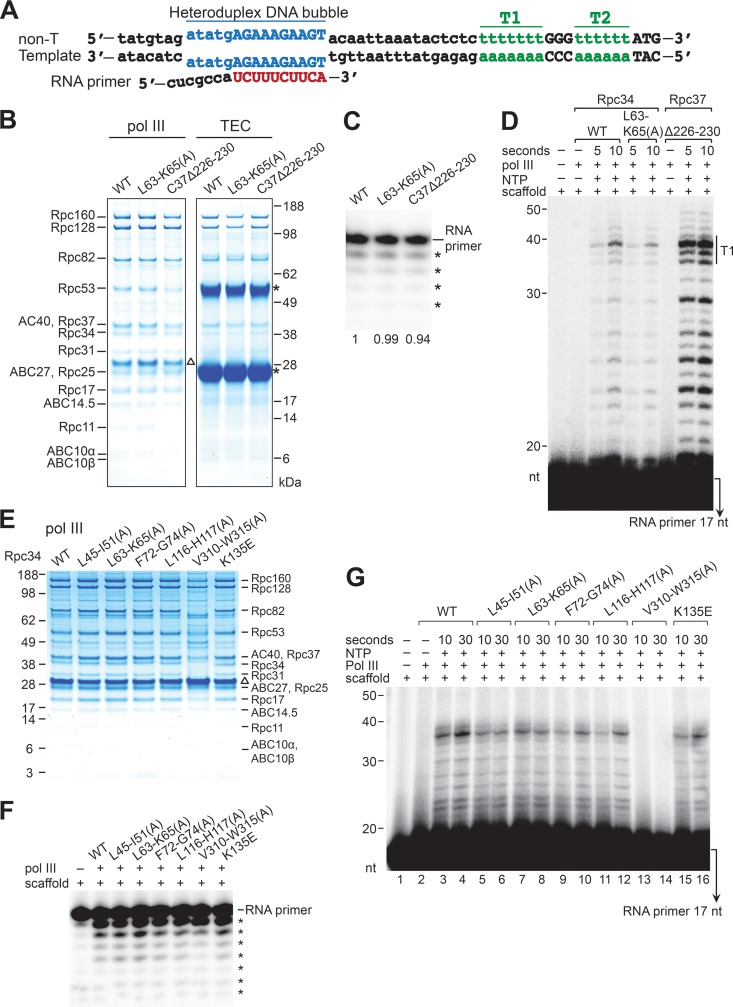
To further analyze Pol III elongation, we reconstituted the TEC with purified Pol III and a nucleic acid scaffold for RNA elongation. As demonstrated in Fig. 6A, the nucleic acid scaffold was designed based on the previous EM study of the transcribing Pol III to contain a heteroduplex DNA bubble, DNA:RNA hybrid, and downstream duplex DNA (5). The purified Pol III complexes are shown in Fig. 6B (left panel). The composition of the purified Pol III complex generated from the immunoaffinity purification process was not altered by either the L63-K65(A) mutation of Rpc34 or an internal deletion mutation in the C-terminal region of Rpc37. We subsequently assembled the TECs and utilized anti-Flag antibody beads to pull down Flag-epitope-tagged Pol III and the bound nucleic acid scaffold (Fig. 6B, right panel). Comparing the amounts of Pol III subunits and RNA primer (Fig. 6B and C), similar levels of TECs were isolated with the wild-type and mutant Pol IIIs. However, based on their RNA extension products (Fig. 6D), elongation by Pol III with the Rpc34 L63-K65(A) mutation was found to be less efficient. In contrast, Pol III with the Rpc37 internal deletion exhibited increased elongation efficiency (Fig. 6D), which is consistent with previous studies indicating that mutations in the Rpc37 C-terminal region alter Pol III properties in elongation and termination (8–10). We further applied the scaffold elongation assay to analyze other Pol III mutants containing Rpc34 tWH mutations (Fig. 6E and F). All tWH mutations resulted in reduced Pol III elongation efficiency (Fig. 6G). Therefore, we conclude that both WH folds of the tWH domain function in Pol III elongation. Supporting this specific functional activity, an Rpc34 C-terminal mutation that causes dissociation of the Rpc82/34/31 subcomplex [Fig. 6E, V310-W315(A)] completely abolished Pol III elongation (Fig. 6G, lanes 13 and 14).
FIG 6.
Elongation efficiency analysis of the Rpc34 WH1 mutant. (A) The DNA/RNA scaffold. The scaffold contains a 15-nucleotide bubble with mismatched bases in blue and two terminator sequences (T1 and T2) in green. The RNA primer (red) contains 10 nucleotides complementary to the template strand. non-T, nontemplate strand. (B) The Pol III complex. (Left panel) Purified Pol III subunits in the Coomassie blue-stained gel are indicated. (Right panel) Pol III subunits in the TEC. The DNA/RNA scaffold shown in panel A was utilized to form the TEC with the purified Pol III, and the TEC was immunoprecipitated and analyzed by SDS-PAGE and Coomassie staining. As indicated above the gels, the Pol III complexes derived from wild-type Rpc34, the Rpc34 L63-K65(A) mutant, and the Rpc37 internal deletion Δ226-230 mutant. The triangle indicates the TEV protease used in Pol III purification. Asterisks indicate the immunoglobulin heavy and light chains of the anti-Flag–agarose beads. (C) RNA primer in the TEC. Asterisks indicate the truncated RNA primers derived from Pol III backtracking activity. The relative intensities of RNA primers are indicated below the lanes. (D) RNA elongation. RNA primer extension from the TEC was conducted for 5 and 10 s after the addition of NTPs. T1, terminated RNAs from the T1 terminator. The RNA peak represents 37 nt (terminated at the fourth nucleotide position in the T1 terminator). (E) The purified Pol III complexes. Mutations in Rpc34 are indicated above the lanes. Mutations in the Rpc34 tWH domain are as indicated in Fig. 1B and C. The Pol III sample purified from the Rpc34 V310-W315(A) mutant strain lacks Rpc82, Rpc34, and Rpc31 subunits. (F) RNA primer in the TEC. Asterisks indicate the truncated RNA primers derived from Pol III backtracking (exonuclease) activity. The relative intensities determined for the RNA primers were as follows: WT, 1.00; L45-I51(A), 1.01; L63-K65(A), 1.01; F72-G74(A), 0.89; L116-H117(A), 0.90; V310-W315(A), 0.90; K135E, 0.91. (G) RNA elongation activity. RNA primer extension from the TEC was conducted for 10 and 30 s after the addition of NTPs. No RNA extension product was observed for the Rpc34 V310-W315(A) mutant (lanes 13 and 14), indicating that the Rpc82/34/31 subcomplex is required for elongation.
DISCUSSION
The TFIIE-related subunit Rpc34 is important for Pol III recruitment and DNA melting in transcription initiation (14, 17). Biochemical analyses have indicated that Rpc34 establishes a protein network with Rpc160, Rpc128, Rpc82, and Rpc37 within the Pol III complex. Rpc34 is also known to interact with transcription initiation factors Brf1, Bdp1, and TBP (14, 17, 20, 21). Recently available cryo-EM structures of free and transcribing Pol III (5) revealed that the C-terminal half of Rpc34 binds stably with Rpc82 and that the N-terminal tWH domain is structurally flexible. The tWH domain thus acts as a mobile structural module, as evidenced by a previous study showing a requirement for the tWH domain in the dynamic DNA melting process. In this study, we further identified new structural regions in the tWH domain that are functionally important for initiation and elongation. In addition, our site-specific cross-linking analysis supports a position for the tWH domain above the Pol III cleft in both transcription steps.
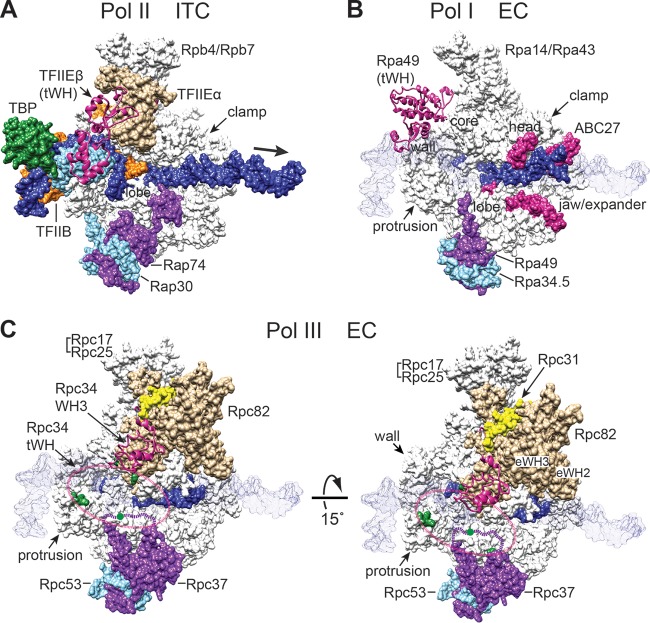
The tWH domain is a common structural module in all eukaryotic Pol systems (2). The tWH domain resides, respectively, in the C-terminal region of the Rpa49 subunit of Pol I and in the N-terminal region of the transcription factor TFIIEβ of the Pol II machinery. The structural position of tWH has been resolved in the Pol II closed, open, and initially transcribing complexes by cryo-EM analyses (Fig. 7A) (26–28). Based on these cryo-EM structures, the α-subunit of TFIIE (TFIIEα) and the Rap30 subunit of TFIIF directly interact with DNA around the upstream edge of the transcription bubble (Fig. 7A), suggesting a functional contribution to DNA melting. The TFIIEβ tWH domain likely acts as a secondary structural factor in DNA melting, with its WH1 and WH2 folds binding to Rap30 and TFIIEα, respectively. In comparison to TFIIE, the positions of the tWH domains in the Pol I and Pol III systems are not well defined. The tWH domain of the Rpa49 subunit was not resolved from high-resolution X-ray crystal structures (29, 30) but was proposed to be located at different positions above the DNA-binding cleft of Pol I. In an EM analysis of elongating Pol I (31), the tWH domain was inferred to be positioned near the wall and clamp regions of the cleft (Fig. 7B). In another EM study of free Pol I (32), tWH was suggested to interact with the clamp head, lobe, and jaw/expander domains, as well as with the ABC27 subunit. This localization of the tWH domain in the free Pol I complex is also consistent with an earlier cross-linking analysis (24). Thus, it is likely that the tWH domain switches to different positions to fulfill its functional roles in multiple stages of the Pol I transcription cycle (32, 33).
FIG 7.
Different structural positions of the tWH domain in the Pol I, II, and III transcription complexes. (A) Structure of the Pol II initially transcribing complex (ITC). The human Pol II ITC model is displayed based on the structural coordinate file from the Protein Data Base (PDB identifier 5iy9) (26). The molecular surfaces of Pol II (white), TBP (green), TFIIB (orange), TFIIEα (brown), Rap74 (the large subunit of TFIIF; purple), and Rap30 (the small subunit of TFIIF; pale blue) are labeled. The open promoter DNA is colored in blue, and an arrow points to the downstream DNA. The backbone trace of TFIIEβ is represented by the pink ribbon model. The clamp and lobe domains, as well as the Rpb4/7 dimer, are indicated. (B) The elongation complex (EC) of Pol I. The yeast Pol I elongation complex is displayed based on PDB coordinate file 5m64 (31). Protein surfaces are colored as described for the respective homologous proteins in the Pol II ITC model shown in panel A. The tWH domain of Rpa49 is displayed with the pink ribbon model for comparison with TFIIEβ. Structural surfaces highlighted in pink are the binding regions proposed by Pilsl et al. (32) for the Rpa49 tWH domain, including regions in the clamp head, jaw/expander, and lobe domains, as well as the ABC27 subunit. The blue molecular surface represents the DNA molecule. For comparison, the open promoter DNA in the Pol II ITC structure (A) is shown as a semitransparent blue DNA surface in the background. (C) The Pol III EC structure. (Left panel) The yeast Pol III EC is oriented in a view similar to the Pol II ITC view in panel A, and all Pol III protein subunits are colored as described for their respective homologous proteins in the Pol II ITC (A). The coordinate file is 5fj8 (PDB) (5). Structural information for an Rpc37 C-terminal loop (amino acids 197 to 224) is not available in the coordinate file, and a purple dashed line denotes this missing sequence block. Amino acids colored in dark green represent residues in Rpc128, Rpc82, and Rpc37 involved in lysine-lysine cross-linking with the Rpc34 tWH domain (also see Fig. 5A). A pink dashed oval indicates the likely position of the Rpc34 tWH domain. The blue molecular surface represents the DNA molecule. The WH3 domain of Rpc34 is displayed with the pink ribbon model. For comparison, the open promoter DNA in the Pol II ITC structure (A) is shown as a semitransparent blue DNA surface in the background. (Right panel) A 15° rotation of the structural model. Positions of the Rpc82 eWH2 and eWH3 domains are indicated.
Although the structure of the Rpc34 tWH domain in the Pol III complex was not resolved in the recent high-resolution EM analysis (5), a possible localization has been derived based on multiple protein and DNA cross-linking analyses. According to the protein cross-linking analysis (19), the tWH domain interacts with the clamp domain of Rpc160, the protrusion domain of Rpc128, the C-terminal loop of Rpc37, and the eWH4 domain of Rpc82 (Fig. 7C). This positioning analysis is further supported by our current BPA cross-linking study, which indicates that the tWH domain maintains contacts with Rpc37 and Rpc82 in the PIC and TEC (Fig. 4). Additionally, a previous DNA photoaffinity analysis indicated that Rpc34 interacts with DNA bases of the transcription bubble (34, 35) (Fig. 7C). The tWH domain thus serves as a central protein module to integrate a dynamic protein-nucleic acid network above the Pol III cleft. Supporting the functional importance of the tWH domain, our IMT analysis indicates that the tWH mutations affect stable associations of Brf1 and Bdp1 in the PIC. Although these two TFIIIB components together with TBP are known to bind stably to the TFIIIC-bound promoter for subsequent recruitment of Pol III (22, 23, 36), our results suggest that Brf1 and Bdp1 become less stably bound in the resulting higher-order PIC. We speculate that a conformational change likely occurs within the PIC, with the tWH domain playing an important role in stabilizing the postrecruitment complex on the promoter. Furthermore, as the Rpc37 subunit has been found to function in the facilitated reinitiation and termination processes (10), Rpc34 likely regulates steps following RNA elongation, with the interaction being between the tWH domain and the Rpc37 C-terminal loop. However, a more complete understanding of the specific functional roles of the tWH domain requires further structural analyses of the Pol III PIC and TEC, as well as additional functional studies of Rpc34 at all steps of the Pol III transcription cycle.
MATERIALS AND METHODS
Plasmids, yeast strains, and cell growth.
The gene sequence encoding Rpc34 with a hemagglutinin (HA) epitope tag on the N terminus was cloned into the pRS315 (cen ars Leu2+) vector to generate the plasmid pYYU-1 (HA-Rpc34/pRS315). Multiple alanine replacements were introduced into the Rpc34 coding sequence of pYYU-1 by in vitro mutagenesis. The plasmids containing the wild-type and mutant Rpc34 genes were transfected into the YLy3 strain [MATα ade2::his3G his3Δ200 leu2Δ met15Δ lys2Δ trp1Δ63 ura3Δ (rpc34::KanMX4) Rpc34/pRS316 (Ura3+)] originally derived from the BY4705 S. cerevisiae strain (21, 37). Rpc34 mutant yeast strains were then generated through the 5FOA dropout method (plasmid shuffling), replacing the wild-type plasmid (Ura3+) with the mutant one (Leu2+) (38). Rpc34 mutant strains were also generated with the genotypic background containing either a Flag epitope tag on the C terminus of Rpc82 or a tandem affinity purification (TAP; Flag3-His6-tobacco etch virus [TEV] protease cleavage sequence-protein A) tag on the Rpc128 C terminus. The Rpc82-Flag and Rpc128-TAP tagged strains were employed in immunoprecipitation (IP) assays and affinity purification of Pol III, respectively. To generate yeast strains for BPA (Bachem) incorporation in Rpc34, we cloned the Rpc34 coding sequence into 2μm vector pRS425 (LEU2+) (39). The Rpc34 gene was driven by the yeast ADH1 promoter, and Rpc34 contained a V5 epitope tag on the N terminus. The TAG (amber) nonsense codon was introduced to replace a specific amino acid codon in Rpc34, resulting in a series of Rpc34 amber mutant/pRS425 plasmids. The Rpc34 amber plasmids were cotransfected with plasmid pLH157 (containing coding sequence of the suppressor tRNACUA and a BPA-tRNA synthetase) into the YLy3 strain to generate individual yeast strains for BPA incorporation in Rpc34. More detailed information about the genetic approach was provided previously (8, 21).
For cell growth analysis, the Rpc34 mutant and wild-type (WT) strains were grown in yeast extract-peptone-dextrose (YPD) medium and harvested at an optical density (OD) at 600 nm of 1.0. The cell cultures were then diluted to concentrations of 10−1, 10−2, 10−3, and 10−4. Each diluted cell culture (5 μl) was spotted onto either YPD plates or synthetic complete-glucose plates (lacking leucine). Plates were incubated at temperatures of 16, 25, 30, 35, and 37°C.
Immunoprecipitation.
To prepare the yeast whole-cell extract (WCE) for IP assay, a 1-liter cell culture of the WT strain or the Rpc34 mutant strain was grown in YPD medium to an OD of 2.0. The harvested cells were lysed and processed for WCE preparation following a previously described protocol (8, 19). WCE (1 mg) was mixed with 50 μl of anti-Flag antibody–agarose beads (M2; Sigma-Aldrich) and incubated at 4°C for 2 h in WCE buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 5 mM MgCl2, 1 mM EDTA, 20% glycerol). The bound proteins were washed three times with 200 μl of WCE buffer, and the proteins were extracted by boiling with NuPAGE sample buffer (Invitrogen) for subsequent SDS-PAGE and Western blot analysis. Immunostained protein bands were visualized by using an Odyssey infrared imaging system (Li-COR Biosciences).
The immobilized template assay for PIC/TEC formation and BPA photo-cross-linking assays.
To isolate the Pol III PIC with the immobilized template (IMT) assay, yeast WCEs from the 1-liter cell culture were incubated with the 5′-end biotin-conjugated DNA fragment containing the following S. cerevisiae SUP4 tRNA gene sequence: AACAATTAAATACTCTCGGtagccaagttgGTTTAAGGCGCAAGACTTTAATTTATCACTACGAAATCTTGAGATCGGGcgttcgactccCCCCCGGGAGATTTTTTT. The A and B boxes of the promoter elements are in lowercase. The details of the procedure were described previously (8). Briefly, the biotinylated DNA (603 bp) was amplified by PCR and subsequently immobilized on streptavidin magnetic beads (Dynal) in transcription buffer containing 20 mM HEPES (pH 7.9), 80 mM KCl, 5 mM MgCl2, 1 mM EDTA, 2% glycerol, 0.01% Tween 20, and 1 mM dithiothreitol (DTT). Each WCE of 800 μg was mixed with 2 μg of immobilized DNA in a final volume of 100 μl transcription buffer for 30 min of incubation at 30°C. The isolated PICs were washed three times using the transcription buffer for Western blotting. A mutant SUP4 tDNA template was used as a negative control for analyzing specific proteins binding to the immobilized DNA template. In the mutant SUP4 tDNA, the A and B boxes were changed to the following sequences: for the mutant A box, TTAATAGTGGA; for the mutant B box, CTTGAAGTGGT.
For the BPA photo-cross-linking experiment, the isolated PICs from the IMT assay were resuspended in a volume of 200 μl transcription buffer and treated with a total energy amount of 8,000 μJ cm−2 UV (UVB) irradiation in a Spectrolinker XL-1500 UV oven (Spectronics). After UV treatment, the reaction mixture was further washed three times, separated by SDS-PAGE, and analyzed by Western blotting.
To isolate the TEC with the IMT assay, the above-described isolated PICs were resuspended in a volume of 20 μl transcription buffer additionally containing ATP, CTP, and UTP (each 500 μM) to allow stalling of the TEC downstream of the transcription start site after synthesis of the 17-nucleotide (nt) RNA. After incubation at 30°C for 15 min, the volume of reaction mixture was increased to 200 μl by adding transcription buffer, and UV irradiation was subsequently applied. After three washes with the transcription buffer, the TECs were analyzed by Western blotting.
In vitro transcription assay.
In vitro transcription activity analysis was conducted with the isolated PICs from the IMT assays. The isolated PICs were resuspended in 17 μl of transcription buffer containing 200 ng α-amanitin, 4 units of RNase inhibitor (Promega), and 1 mM DTT. A mixture of nucleoside triphosphates (NTPs) (3 μl) was subsequently added, and the resulting reaction mixture contained 500 μM (each) ATP, UTP, and CTP; 50 μM GTP; and 0.16 μM [α-32P]GTP (3,000 Ci/mmol). After allowing the reaction to proceed at 30°C for 30 min, transcription was quenched by adding 180 μl of 0.1 M sodium acetate, 10 mM EDTA, 0.5% SDS, and 200 μg/ml glycogen. The transcripts were extracted by phenol-chloroform and ethanol precipitation before being separated on 6% (wt/vol) denaturing urea polyacrylamide gels and visualized by autoradiography.
To conduct the in vitro transcription assay with plasmid DNA, 30 μg of WCE was preincubated with 150 ng plasmid DNA containing the SUP4 tRNA gene in a final volume of 17 μl transcription buffer additionally with 200 ng α-amanitin, 4 units of RNase inhibitor (Roche), and 1 mM dithiothreitol (DTT). Transcription was started with further addition of 3 μl NTPs (ATP [500 μM], UTP [500 μM], CTP [500 μM], GTP [50 μM], and [α-32P]GTP [0.08 μM {3,000 Ci/mmol}]) to reach a final volume of 20 μl. After reaction at 30°C for 30 min, RNA products were extracted and analyzed by denaturing polyacrylamide (6%) gel electrophoresis. The SUP4 pre-tRNA transcripts were visualized by autoradiography and quantified with the ImageQuant TL program (GE).
To analyze RNA products of the TECs from the IMT assays, ATP (125 μM), CTP (125 μM), and radiolabeled UTP (α-32P; 0.42 μM [3,000 Ci/mmol]) were incubated with the isolated PICs for 20 min. In comparison with the above-described transcription assays, the nucleotide concentration was reduced to limit downstream stalling caused by misincorporation. The RNA transcripts in the isolated TECs were separated by denaturing polyacrylamide (16%) gel electrophoresis and visualized by autoradiography. To analyze the RNA extension products from the GTP-chased TECs, GTP (125 μM) and additional UTP (500 μM) were added to the preassembled TECs. RNAs were harvested and analyzed by the procedure described above.
Pol III purification.
To purify Pol III with an affinity purification procedure, a total of 5 liters of yeast cell culture was grown in YPD medium to an OD of 2, and the harvested cells were resuspended in the TAP-tag purification buffer containing 40 mM HEPES (pH 7.5), 10% glycerol, 350 mM NaCl, 0.1% Tween 20, 1 mM DTT, and protease inhibitors phenylmethylsulfonyl fluoride (PMSF; 1 mM), benzamidine (2.58 mM), leupeptin (0.7 mM), and pepstatin A (1 μM). The yeast cells were lysed by glass bead beating and centrifuged in a JLA-8.1000 rotor (Beckman Coulter) at 5,000 rpm for 10 min to collect the clarified lysate. The lysate was further centrifuged in a type 45 Ti rotor (Beckman Coulter) at 35,000 rpm for 1 h. The resulting lysate was subsequently incubated with 1 ml IgG-Sepharose beads (GE Healthcare Life Sciences) for 3 h at 4°C. The bound proteins on beads were washed once with 50 ml TAP-tag purification buffer, followed by a wash with 2 ml TEV protease cleavage buffer containing 10 mM Tris (pH 8), 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, and 10% glycerol. Subsequently, the beads were resuspended with 1 ml TEV protease buffer containing 50 μg TEV protease, and protease digestion was conducted overnight at 4°C. After digestion, Pol III in the supernatant was removed for later use in TEC formation with the DNA/RNA scaffold. Protein subunits of the purified Pol III were separated by SDS-PAGE, followed by Coomassie blue staining. Pol III subunits on the Coomassie blue-stained gel were confirmed by mass spectrometry. The total protein concentration was assayed by the Bradford colorimetric method (Bio-Rad protein assay), and the yield was ∼0.1 mg.
TEC formation with purified Pol III and the DNA/RNA scaffold and RNA extension.
A DNA/RNA scaffold was designed with the following oligonucleotides: (i) template strand (5′-CATAAAAAACCCAAAAAAAGAGAGTATTTAATTGTTGAAGAAAGAGTATACTACATA); (ii) nontemplate strand (5′-TATGTAGATATGAGAAAGAAGTACAATTAAATACTCTCTTTTTTTGGGTTTTTTATG); and (iii) RNA primer (5′-CUCGCCAUCUUUCUUCA). As also depicted in Fig. 5A, the DNA/RNA scaffold contained a 15-nt bubble with mismatched single-stranded DNA bases and terminator sequences (T1 and T2) in the downstream duplex DNA. The 5′ end of the RNA primer was subjected to radioisotope labeling using T4 polynucleotide kinase and [γ-32P]ATP. The RNA primer was initially annealed with the template DNA strand to form a DNA/RNA hybrid with a final quantity of 0.7 pmol. A 1-μg volume of purified Pol III was added to the DNA/RNA hybrid mixture for incubation at room temperature for 10 min, and the nontemplate DNA strand was subsequently added for another 10 min of incubation. The assembled TECs were precipitated using anti-Flag–agarose beads (M2; Sigma-Aldrich) to pull down Pol III with Flag epitope-tagged Rpc128. RNA extension was initiated with the addition of an NTP mixture (500 μM). The synthesized RNAs were separated by denaturing polyacrylamide (15%) gels and visualized by using a phosphorimaging plate (Fuji Film) for subsequent scanning with a Typhoon FLA 9000 imaging scanner (GE).
ACKNOWLEDGMENTS
We thank Seok-Kooi Khoo, Chih-Chien Wu, Yu-Chun Lin, Yue-Chang Chou, and Chih-Syuan Liu for technical assistance throughout the study. We thank John O'Brien for English editing. We express our sincere appreciation to an anonymous reviewer who suggested performing the control experiment whose results are shown in Fig. 2 and the RNA extension analysis of the stalled elongation complex whose results are shown in Fig. 5.
This work was supported by grants 103-2311-B-001-021-MY3 and 105-2627-M-001-009 from the Ministry of Science and Technology, Taiwan, Republic of China, and a Career Development Award to H.-T.C. from Academia Sinica, Taiwan, Republic of China.
REFERENCES
- 1.Schramm L, Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 2.Vannini A, Cramer P. 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Geiduschek EP, Kassavetis GA. 2001. The RNA polymerase III transcription apparatus. J Mol Biol 310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 4.Kassavetis GA, Blanco JA, Johnson TE, Geiduschek EP. 1992. Formation of open and elongating transcription complexes by RNA polymerase III. J Mol Biol 226:47–58. doi: 10.1016/0022-2836(92)90123-2. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Muller CW. 2015. Molecular structures of unbound and transcribing RNA polymerase III. Nature 528:231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. 2010. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Lindner D, Muller CW. 2010. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J 29:3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CC, Lin YC, Chen HT. 2011. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol 31:2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arimbasseri AG, Maraia RJ. 2015. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol Cell 58:1124–1132. doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. 2006. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J 25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassavetis GA, Prakash P, Shim E. 2010. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem 285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lefèvre S, Dumay-Odelot H, El-Ayoubi L, Budd A, Legrand P, Pinaud N, Teichmann M, Fribourg S. 2011. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat Struct Mol Biol 18:352–358. doi: 10.1038/nsmb.1996. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann NA, Jakobi AJ, Vorlander MK, Sachse C, Muller CW. 2016. Transcribing RNA polymerase III observed by electron cryo-microscopy. FEBS J 283:2811–2819. doi: 10.1111/febs.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Roeder RG. 1997. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev 11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 15.Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J 14:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiannilkulchai N, Stalder R, Riva M, Carles C, Werner M, Sentenac A. 1992. RPC82 encodes the highly conserved, third-largest subunit of RNA polymerase C (III) from Saccharomyces cerevisiae. Mol Cell Biol 12:4433–4440. doi: 10.1128/MCB.12.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brun I, Sentenac A, Werner M. 1997. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J 16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, Sentenac A, Muller CW, Schoehn G. 2007. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol Cell 25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Wu CC, Herzog F, Jennebach S, Lin YC, Pai CY, Aebersold R, Cramer P, Chen HT. 2012. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc Natl Acad Sci U S A 109:19232–19237. doi: 10.1073/pnas.1211665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoo B, Brophy B, Jackson SP. 1994. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev 8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 21.Khoo SK, Wu CC, Lin YC, Lee JC, Chen HT. 2014. Mapping the protein interaction network for TFIIB-related factor Brf1 in the RNA polymerase III preinitiation complex. Mol Cell Biol 34:551–559. doi: 10.1128/MCB.00910-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassavetis GA, Joazeiro CA, Pisano M, Geiduschek EP, Colbert T, Hahn S, Blanco JA. 1992. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell 71:1055–1064. doi: 10.1016/0092-8674(92)90399-W. [DOI] [PubMed] [Google Scholar]
- 23.Kassavetis GA, Nguyen ST, Kobayashi R, Kumar A, Geiduschek EP, Pisano M. 1995. Cloning, expression, and function of TFC5, the gene encoding the B″ component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci U S A 92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jennebach S, Herzog F, Aebersold R, Cramer P. 2012. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res 40:5591–5601. doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaaban SA, Bobkova EV, Chudzik DM, Hall BD. 1996. In vitro analysis of elongation and termination by mutant RNA polymerases with altered termination behavior. Mol Cell Biol 16:6468–6476. doi: 10.1128/MCB.16.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. 2016. Near-atomic resolution visualization of human transcription promoter opening. Nature 533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P. 2016. Transcription initiation complex structures elucidate DNA opening. Nature 533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 28.Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD. 2016. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166:1411–1422.e1416. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. 2013. RNA polymerase I structure and transcription regulation. Nature 502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NM, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Muller CW. 2013. Crystal structure of the 14-subunit RNA polymerase I. Nature 502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 31.Tafur L, Sadian Y, Hoffmann NA, Jakobi AJ, Wetzel R, Hagen WJ, Sachse C, Muller CW. 2016. Molecular structures of transcribing RNA polymerase I. Mol Cell 64:1135–1143. doi: 10.1016/j.molcel.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilsl M, Crucifix C, Papai G, Krupp F, Steinbauer R, Griesenbeck J, Milkereit P, Tschochner H, Schultz P. 2016. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat Commun 7:12126. doi: 10.1038/ncomms12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P. 2010. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell 39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 34.Lannutti BJ, Persinger J, Bartholomew B. 1996. Probing the protein-DNA contacts of a yeast RNA polymerase III transcription complex in a crude extract: solid phase synthesis of DNA photoaffinity probes containing a novel photoreactive deoxycytidine analog. Biochemistry 35:9821–9831. doi: 10.1021/bi960525x. [DOI] [PubMed] [Google Scholar]
- 35.Bartholomew B, Durkovich D, Kassavetis GA, Geiduschek EP. 1993. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol 13:942–952. doi: 10.1128/MCB.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB. 2003. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature 422:534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 37.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]