Abstract
Horizontal gene transfer via plasmid conjugation enables antimicrobial resistance (AMR) to spread among bacteria and is a major health concern. The range of potential transfer hosts of a particular conjugative plasmid is characterised by its mobility (MOB) group, which is currently determined based on the amino acid sequence of the plasmid-encoded relaxase. To facilitate prediction of plasmid MOB groups, we have developed a bioinformatic procedure based on analysis of the origin-of-transfer (oriT), a merely 230 bp long non-coding plasmid DNA region that is the enzymatic substrate for the relaxase. By computationally interpreting conformational and physicochemical properties of the oriT region, which facilitate relaxase-oriT recognition and initiation of nicking, MOB groups can be resolved with over 99% accuracy. We have shown that oriT structural properties are highly conserved and can be used to discriminate among MOB groups more efficiently than the oriT nucleotide sequence. The procedure for prediction of MOB groups and potential transfer range of plasmids was implemented using published data and is available at http://dnatools.eu/MOB/plasmid.html.
Introduction
Antimicrobial resistance (AMR) is a pressing global issue, as it diminishes the activity of 29 antibiotics and consequently leads to over 25,000 deaths each year in Europe alone1,2. The development of AMR in microbial communities is facilitated by horizontal gene transfer (HGT) of conjugative elements (including plasmids and integrative elements)3 carrying antibiotic resistance genes along with virulence genes4,5. It is therefore important to determine the routes of plasmid transfer among bacteria6,7, based on determining their host range8.
It is currently known that each of the 6 established mobility superclasses of conjugative elements have limited transfer host range8. Conjugation systems of each of these MOB groups are classified according to the conservation of the amino acid sequences of relaxase, the central enzyme that enables relaxation and transfer of elements from donor to recipient cells9,10. Besides relaxases, the relative conservative nature of MOB groups can be detected among other protein components of conjugation systems, which are comprised of (i) auxiliary proteins that take part in formation of the relaxation complex (relaxosome) in the origin of transfer (oriT) DNA region11, (ii) coupling protein (type IV)12,13, which connects the relaxosome with (iii) the mating complex (type IV secretion system, T4SS) that forms the transfer channel between donor and recipient cells14. These protein components were shown to coevolve to a large extent within their respective MOB groups12,13,15. In addition to the conservative nature of proteins involved in DNA transfer, it has also been observed that a relaxase from a certain MOB group enables the most efficient transfer only of plasmids belonging to that same group16. Therefore, one can expect that the substrate for relaxases, the bare noncoding sites in oriT, should also possess some MOB-specific properties that enable their cognate relaxases to initiate the conjugation process most efficiently (Fig. 1, Table 1).
Figure 1.
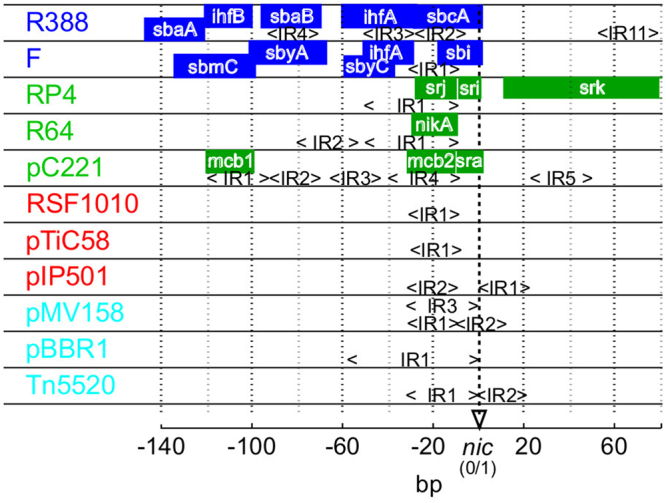
Schematic representation of available experimental data on oriT regions from four MOB groups. oriT data from MOB F (blue), P (green), Q (red) and V (cyan) supports that the conservation of structural properties within each MOB group is greater than between groups. Known binding sites for auxiliary proteins and relaxases are marked (colored squares) and are frequently characterized by inverted repeats (<IR>). Relaxase binding sites are nearest to the nic site (between 0 and 1 bp). General characteristics of MOB groups are: (F) system of multiple auxiliary proteins including the DNA-bending protein IHF44,55, (P) up to 5 proteins including relaxase involved in relaxation (RP4)47,54,61 (Q) a shorter oriT region of only 38 bp that covers besides relaxase 2 auxiliary proteins without clear binding sites (RSF1010)48,49,62, (V) no known auxiliary proteins50,51,63.
Table 1.
oriT structural properties that enable relaxasome formation and nicking of DNA to initiate transfer of conjugative elements. Shown are experimentally determined oriT structural features and predicted structural properties that were used to interpret them.
| Experimental oriT structural features | Reference | Predicted properties |
|---|---|---|
| Direct and inverted repeats of DNA sequence around nic that define extensive secondary structures (e.g. hairpins) and act as protein-DNA recognition regions | 44,64 | Deformability SDef |
| Duplex stability SStab | ||
| DNA melting bubbles and destabilizations that facilitate relaxase nicking, aided by lower duplex stability around nic and by DNA thermal dynamics | 21,33,65,66 | Thermally induced duplex destabilizations STIDD |
| Intrinsically curved or flexible regions that facilitate binding and changes in oriT structure around nic (e.g. IHF protein binding in MOB F) | 44,55,67 | Bending propensity SBend |
| Persistence length SPer | ||
| Differences in DNA spacing and orientation between binding sites and nic | 68 | Helical repeats SHel |
The specific conservation of oriT properties within MOB groups can also be expected, since DNA binding proteins recognize a particular site on DNA by a physicochemical interaction with the DNA. Prior to binding, proteins slide on DNA in controlled 1D diffusion processes in search of their active binding sites17,18. Therefore, some of the essential features of DNA recognition that optimize the protein-DNA indirect readout process are the conformational and physicochemical DNA structural properties at the specific binding sites and around them19,20. In the case of initiation of conjugation, the oriT region is a recognition site and it is also an enzymatic substrate, since the relaxase recognizes specific DNA as well as makes a nick in the DNA to initiate conjugation21,22. However, contrary to the conserved amino or nucleic acid sequences of relaxases and auxiliary proteins, the oriT is a noncoding region and low conservation of nucleotide sequence is expected9,11.
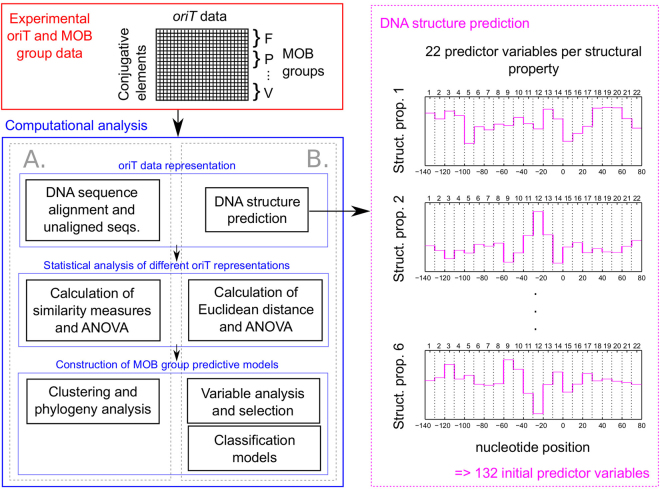
Therefore, in order to pinpoint the specific properties in oriT that are conserved within plasmids of a particular MOB group, the conventional approach based on clustering of similar DNA sequences is unlikely to be successful. A more advanced approach is required to classify MOB groups based on the analysis of oriT structural properties. The aims of the present study were to (i) analyze the DNA structural properties of oriT regions from different MOB groups (Fig. 1, Table 1), (ii) determine if DNA structural properties are conserved within MOB groups and can be used to discriminate among them and (iii) implement the classification procedure as a webtool available to the wider research community.
Methods
oriT datasets
To construct and analyze statistical and predictive models a training and a testing dataset were used. The training dataset comprised nucleotide sequences of oriT regions of 64 elements that were obtained from the Genbank database. In these sequences the oriT regions were identified and aligned according to published experimental information on nic sites (Supp. Table S1). Despite the scarce amount of published data, which limited the amount of MOB groups used and the size of the training dataset, the dataset was balanced, with approximately 16 elements from each MOB and contained oriTs from all known MOB subgroups10. For the testing dataset we obtained 136 oriT regions from plasmids, for which the only previously available information was that of their MOB groups, determined on the basis of amino acid sequences of relaxases10. The locations of nic sites in these plasmids were determined by finding the minimal Euclidean distance between structural properties of training oriTs and the testing dataset. The positions of resulting oriT regions were verified using experimental data and relaxase locations10 (Supp. Table S2). By combining the training and testing datasets, the expanded dataset of 200 elements was of an appropriate size to support a statistical and machine learning analysis (Supp. Fig. S1: see learning curves). The testing dataset was thus used for cross validations as well as training of predictive models. In both datasets the part of the oriT regions with relevant protein binding features from −140 bp to +80 bp according to the nic site were analysed (Fig. 1: see references).
Nucleotide sequence analysis
The oriT dataset of 200 elements was aligned using the ClustalW algorithm23 and grouped based on the following distances between DNA sequences: (I) the p-distance: the ratio of the amount of different sequence positions to sequence size, and (ii) the 2-parameter Kimura distance: models transitional and transversional nucleotide substitution rates24 (Fig. 2). Clustering of similar sequences was performed with the Neighbor Joining method using p-distance, and with the Maximum Likelihood method using the Kimura distance. The topology of constructed trees was tested with the bootstrap25. The classification accuracy of condensed trees was estimated as the average ratio of branches that contained elements from a specific MOB group to all elements in that group. Mega version 6.06 software26 was used for all calculations with default settings. The bootstrap parameter was set to 1000 repetitions and cutoff values of 50% and 80% were used for positioning of branches within a constructed tree. DNA sequence conservation per basepair was evaluated using information content analysis based on Shannon’s entropy, where the maximum information content of 2 bits reflected maximum sequence conservation and vice-versa27,28.
Figure 2.
Overview of the performed computational analysis. DNA sequences of oriT regions and their MOB groups were used to compare (A) the conventional approach based on analysis of primary sequences with (B) our new approach based on DNA structure prediction. oriT regions were ligned up to the nic functional site.
Prediction of structural variables
In contrast to the conventional sequence-based analysis, an alternative representation of oriT regions was developed based on computed DNA structural properties (Fig. 2). Parametric models were used to predict conformational and physicochemical properties. Conformational properties included (i) DNA deformability, which affects DNA-protein interactions, as given by volumes of conformation space (SDef) with the model based on data of DNA-protein crystal complexes29, (ii) DNA bending propensity (SBend) with the model based on DNaseI enzyme digestion data30 and (iii) DNA persistence length (SPer, proportional to stiffness) and DNA helical repeats (SHel, equal to number of bps per helix turn) with the model based on cyclization experiments of short DNA fragments31. Physicochemical properties included (i) relative DNA duplex stability (SStab) with the thermodynamic nearest neighbor (NN) model using the unified NN parameters at 37 °C32 and (ii) thermally induced duplex destabilization (TIDD, STIDD) with our recently developed method based on machine learning algorithms using 6 bp of neighboring regions at a threshold of 0.1 Å33. Predicted structural properties spanned 10 bp using a sliding window approach, due to its potential to detect the conserved regions among similar MOB groups with higher accuracy and solve the problem of leftover nucleotides at the end of the sequence. To increase the ratio of signal to noise, the predictions were averaged in windows of 10 consecutive basepairs (Fig. 2). This also decreased the number of variables used in the analysis per DNA structural property and per oriT region from an initial 220 to 22. For calculation of the DNA sequence and structural properties Matlab software (Mathworks, MA, USA) was used.
Statistical analysis
A central measure of the conservation of data within groups is the ratio of the variability of the data between groups versus the average variability of data in each group, which is given as the F statistic and can be statistically evaluated with analysis of variance (ANOVA). Since our data did not follow a normal distribution (Supp. methods S1), a non-parametric multivariate ANOVA34 was used (Supp. Methods S2). In this procedure the variability of the data was evaluated based on an inter-point geometric approach that enabled the use of different distance measures including: (i) the p-distance with nucleotide sequences and (ii) the Euclidean distance with structural variables. The same non-parametric procedure was used to analyze the conservation of (i) individual structural variables and (ii) nucleotide sequences at specific oriT positions in windows of 10 bp (Fig. 2: comparison at 22 positions). To avoid Type I errors due to multiple comparisons the Bonferroni correction was applied35. Differences between means of groups of data were tested with the Mann-Whitney-Wilcoxon test36. Input data was standardized to zero mean and unit variance. All analyses were performed in Matlab, except distribution analysis for which SPSS ver. 22 (IBM, NY, USA) was used.
Variable analysis and selection
Subsets of the most informative structural variables for predicting MOB groups were obtained using a backward variable selection procedure. The procedure included (i) ranking of variables according to one of three criteria of relative variable importance, and (ii) performing backward selection with classification tests, to select the optimal subset that led to highest classification measures (see ‘Construction of predictive models’ below). The initial criteria for ranking of variables were based on p-values of the F statistic. However, since the ANOVA procedure that was used did not enable analysis of potential interactions between variables, which were presumed to play an important role in discrimination between groups, two of the most efficient and frequently used variable selection algorithms37 were applied to detect interactions between variable. These were (i) Correlation-based feature selection (CFS) Subset Evaluator algorithm38 with the Greedy Stepwise search method to detect moderate levels of interaction and (ii) ReliefF Attribute Evaluator algorithm39 with the Ranker search method used to detect higher order interactions.
Construction of predictive models
Two types of classification tests were performed using either (i) different subsets of predictor variables or (ii) different subsets of data. In the backward variable selection procedure, the influence of the number of ranked variables on MOB prediction was evaluated by stepwise removal of variables with the lowest ranks. With each subset, 10 repetitions of classification tests were performed. To evaluate the effect of removing elements with low classification frequency (the ratio of correct classifications to number of classifications) from the training dataset, 100 repetitions were performed. The classification tests comprised (i) 10-fold cross validations (CVs) using the training dataset (CV_64), (ii) 10-fold CVs using the full set of 200 elements (CV_200) and (iii) testing the trained models with the testing dataset (Test). The classification tests were evaluated with six of the most relevant classification performance measures for multi-group classification (Supp. Methods S3)40–43, including Precision (Pre) and Recall (Rec). The Multilayer perceptron algorithm with default settings was used for construction and testing of predictive models. Matlab was used to run the algorithms and to analyze the data. Algorithm implementations in Weka software43 version 3.7.9 were used.
Results
Structure prediction improves discrimination of MOB groups
The conventional phylogenetic sequence analysis of the dataset of oriT regions (Fig. 2, Supp. Tables S1 and S2) led to an inaccurate discrimination of MOB groups. Dendrograms of aligned oriT sequences based on calculated sequence distances, either p-distance or Kimura, contained large numbers of clusters (up to 48 per MOB group) from which elements could not be sorted into their respective MOB groups (Supp. Fig. S2A–D: estimated class. accuracy did not exceed 0.110 ± 0.104; 95% confidence bounds given) Therefore, a different sequence alignment approach was used, in which oriT sequences were lined up according to the nic site (see Table 1, Fig. 1). However, the results again indicated that MOB groups could not be correctly resolved (Supp. Fig. S2E–H: estimated class. accuracy did not exceed 0.082 ± 0.045). The oriT region also showed low information content, i.e. low sequence conservation in individual MOB groups (Supp. Fig. S3: below 0.518 bits) and even lower among all MOB groups (below 0.152 bits) both in sequence and nic based alignments. However, the F statistic obtained from the analysis of variance of MOB groups by comparing the overall variance of data between groups with the variance of data within groups was shown to be statistically significant with the aligned sequences at an alpha level of 0.05 (F = 0.728, p = 0.029), contrary to the nic based alignment (F = 0.525, p = 0.475).
Since the oriT region contains many structural features that were presumed to be crucial for achieving better MOB discrimination, we predicted 6 known structural properties as an alternative representation of oriT data (see Table 1 and Fig. 2). Using the structural variables a significantly larger F statistic was obtained than with unaligned and aligned sequences (p < 0.001 and p = 0.047, respectively), showing significantly higher conservation of structural properties within MOB groups (F = 1.000, p < 0.001; Supp. Table S3).
Predicted structural properties distinguish functionally important sites in oriT
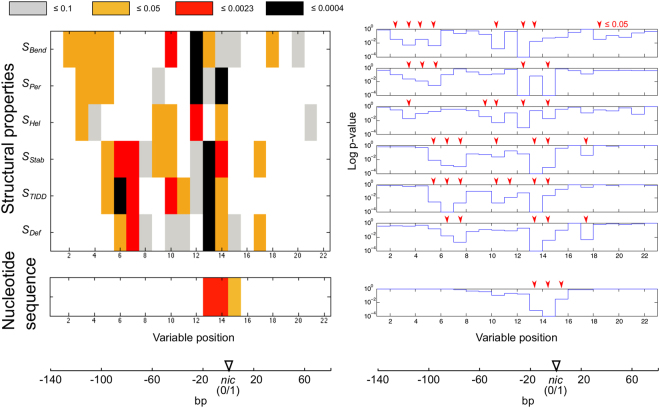
Analysis of variance of nucleotide sequence and structural properties at the 22 variable positions in oriT showed that structural properties were significantly conserved at multiple oriT positions (Fig. 3: 1 to 2 significant positions with the most stringent corrections for multiple testing, except with property SHel). However, nucleotide sequences were conserved only around the nic site (1 significant position; see Supp. Fig. S3). Up to a two fold increase of conserved positions was thus obtained with the structural variables compared to the nucleotide sequences (Fig. 3: 28% vs. 14% of positions, respectively, with uncorrected p).
Figure 3.
Conservation of structural variables and nucleotide sequences according to analysis of variance. Variables of 6 structural properties and nucleotide sequences in windows of 10 bp were compared at 22 positions in oriT regions (labeled ‘Variable position’ on the x axis). P values of the F statistic (y axis) are given at levels of significance that are (i) uncorrected (0.05) and (ii) corrected for multiple comparisons within a particular structural property or nucleotide sequence spanning 22 variables (0.0023) or (iii) whole set of 6 structural properties (0.0004).
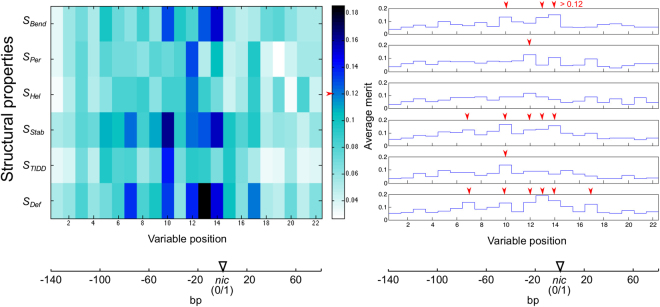
When structural variables were ranked according to their relative importance of discrimination of MOB groups using machine learning algorithms (Supp. Table S4: ReliefF and CFS algorithms), the highest measures of classification performance were obtained with a subset of 16 highest ranked variables using the ReliefF algorithm (Fig. 4: testing models built with training dataset using testing dataset; Supp. Fig. S4 and Table S5). This was a significant improvement to using the full set of 132 variables (p < 0.002) as well as to the classification performance measures obtained with subsets of variables ranked according to p-values or the CFS algorithm (p < 0.006). The most informative structural properties according to the variable subset obtained with the ReliefF algorithm were DNA deformability SDef, duplex stability SStab and bending propensity SBend (Fig. 4: 6, 5 and 3 highest ranked variables, respectively), whereas thermally induced duplex destabilization STIDD and persistence length SPer were less informative (1 variable each). No variables from helical repeats SHel were present among the highest ranked variables, though SHel12 was the 17th highest ranked according to ReliefF (see Supp. Table S3).
Figure 4.
Variable analysis using the ReliefF algorithm. Relative importance (ReliefF Average merit on the y axis) of the structural variables of 6 structural properties (labeled ‘Variable position’) in the oriT regions is shown. The cutoff level of relative importance (Average merit) for the subset of 16 highest ranked variables and the positions of these variables are marked with red arrows.
The majority of the 16 highest ranked structural variables were upstream from nic (Figs 4 and 5: 15 out of 16) and over half of these (Figs 4 and 5: 9 of 16) were less than 30 bp away from nic. In group MOB F, in the region from −100 to −40 bp the mean stability SStab7,10, destabilizations STIDD10 and deformability SDef7,10 showed largest deviations from other groups (Supp. Fig. S5; differences were significant p < 0.006) and coincided with inverted repeats and auxilliary protein binding sites (Fig. 1: eg. sbaB and sbyA)44. Similarly, in the interval from approximately −50 to −10 bp the mean bending propensity was lower in MOB F than elsewhere (SBend10,13, see Supp. Fig. S6; p < 0.001) and SBend10 coincided with an IHF binding site (Fig. 1: ihfA)44,45. In MOB P, significant increases in bending propensity SBend2–5 from −130 to −90 bp and a decrease in deformability SDef6,7 from −90 to −70 bp coincided with binding site mcb1 and inverted repeats, respectively (p < 0.006). The region downstream from nic also showed relevance for MOB P discrimination, since mean deformability SDef17 and DNA stability (Supp Table S4: SStab17 is ranked just below the 16 subset) were lower and bending propensity SBend18 was higher compared to other groups (Fig. 1: positions correspond to IR5 in pC221 and TraK binding site srk in RP446; see Supp. Fig. S7; p < 0.002)46,47. In MOB Q, mean persistence length SPer12, stability SStab12 and deformability SDef12,13,14 as well as the significantly conserved amount of helical repeats SHel12 showed large deviations from other groups at around −20 bp, corresponding to locations of IRs involved in relaxase binding (Fig. 1; p < 0.002)48,49. Similarly, MOB V displayed a low mean stability SStab12,13,14 and high amount of destabilizations around −10 bp (Supp. Table S4: STIDD12,15 are ranked immediately below the 16 variable subset; all p < 0.001), coinciding with IRs50,51.
Figure 5.
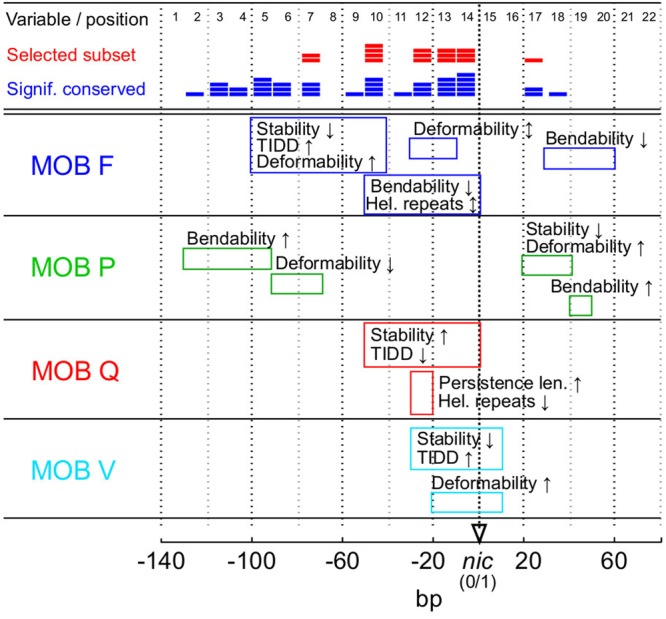
Overview of structural properties and variable analysis in oriT regions from four MOB groups. Shown are the most prominent structural properties that separated a particular MOB group from the other groups (see details in Supp. Fig. S5). Also depicted at specific positions are the amount of variables from the selected subset (Fig. 4, red color) and the amount of variables with significant conservation (Fig. 3, blue color).
Structure based approach enables prediction of transfer range
Using machine learning algorithms with the selected structural variables, predictive models were built that could classify input oriT regions into their corresponding MOB groups with high precision and recall (Supp. Table S6: PreTest = 0.975 ± 0.001, RecTest = 0.973 ± 0.001, PreCV_200 = 0.958 ± 0.001, RecCV_200 = 0.949 ± 0.002). Since certain elements in the training dataset were frequently inaccurately classified, we examined how their removal from the dataset affected classification performance. Results showed that removal of any elements from the training dataset negatively affected the performance of the models. Although removal of the first nine elements (see Supp. Table S6) with a classification frequency below 0.2 led to improved results of cross validations (PreCV_64 increasing to 0.842 ± 0.008, RecCV_64 to 0.790 ± 0.006 to PreCV_64 to 0.988 ± 0.003 and RecCV_64 to 0.979 ± 0.004, P < 0.001), testing with the 140 element dataset showed a decrease in predictive performance (PreTest = 0.975 ± 0.001, RecTest = 0.973 ± 0.001 to PreTest = 0.789 ± 0.001, RecTest = 0.763 ± 0.001, P < 0.001).
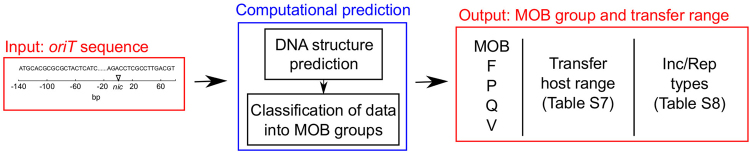
In order to facilitate the prediction of the plasmid transfer range using our models, we collected all currently available data into two tables8,10,52 (Supp. Tables S7 and S8), which link the MOB classification of plasmids with known transfer hosts and Inc/Rep types. The predictive classification models based either on the set of 64 experimentally obtained elements or the whole set of 200 elements were implemented as a webtool available at http://dnatools.eu/MOB/plasmid.html (Fig. 6). The input is a DNA sequence, which is a 230 bp long oriT region with the nic site located between positions 140 and 141. The output consists of (i) the predicted MOB group of the particular oriT and plasmid as well as (ii) the range of potential transfer hosts (Supp. Table S7) and Inc/Rep types (Supp. Table S8) in the MOB group, according to the data available for the training elements.
Figure 6.
Overview of the oriT structure-based prediction procedure. Based on an input oriT sequence, the computational procedure predicts (i) the MOB group of the particular oriT and plasmid as well as (ii) the range of potential transfer hosts and Inc/Rep types (see Discussion). Two types of predictive classification models are available to the user, based the training sets of either 64 or 200 elements.
Discussion
The approach that is currently used to classify a particular plasmid is based on analysis of amino acid sequences of relaxases and accessory proteins. Here however, we showed for the first time that plasmids can be correctly classified into MOB groups based on predicted structural properties of noncoding oriT sequences, without any information about the relaxase. The oriT regions act as relaxase recognition sites as well as enzymatic substrates for nicking. Accordingly, we can conclude that oriT structural properties have co-evolved with the relaxases and accessory proteins involved in the DNA recognition, nicking and transfer reactions within their particular MOB group, as theory and experimental evidence suggested16–19.
This is supported by the analysis of variance, which showed that within the MOB groups oriT regions contained significantly conserved structural properties (Fig. 3). However, the statistical procedure did not account for any possible interactions between the structural properties and structural variables, which were presumed to be important in oriT due to latent structural connections. We therefore performed additional analysis and selection of variables using machine learning algorithms (Fig. 4, Supp. Table S4). Ranking the variables based on their importance in discrimination of MOB groups helped us to identify the structurally informative oriT regions. The subset of 16 highest ranked variables (see Fig. 4, Supp. Table S4) thus included 12 variables that were determined to be significantly conserved with the analysis of variance (Fig. 3: p < 0.05). Of these 12 variables, 3 variables were below the corrected significance level of p < 0.0004 and 5 were below p < 0.0023 (see Fig. 3). With 3 of the 4 additional variables included in the subset of 16 highest ranked variables (Fig. 4) and not determined to be significantly conserved, p was below 0.1, showing a moderate degree of conservation (Fig. 3: bending propensity SBend14, stability SStab12 and deformability SDef10). These variables were probably included due to variable interactions, which were also likely the reason that some of the most significant variables (4 of 7 with p < 0.0004) were not included in the selected subset.
The selected structural variables that enabled the most accurate classification of MOB groups were the most informative, since they coincided with experimentally determined oriT structural properties. By comparing the variables with oriT protein binding sites we observed a higher conservation of structural properties at or around specific protein binding sites than at other positions (Figs 1 and 5, Supp. Fig. S5). The region in the immediate vicinity of nic was the most relevant for analysis of oriT regions and their classification (Fig. 5: over half of the selected variables), since it is the most important for DNA relaxation. This region contains inverted repeats and well characterized binding sites in all MOB groups (Fig. 1)11. The structural variables around nic reflected specific relaxase binding and nicking properties in the particular groups of elements. For instance, formation of DNA melting bubbles and hairpins involved in relaxation separated MOB groups Q and V50,51 from other MOB groups (Fig. 5). As expected according to experimental data, most of the selected attributes were upstream from nic, since this region has a greater role in the control of relaxation than the downstream region. This was most prominent in groups MOB F and P, since they have more auxiliary protein binding sites and span farther upstream than other groups (Fig. 1)11,53. The downstream region also showed relevance for classification, since certain elements in MOB F and P contain downstream binding sites for auxiliary proteins (Fig. 1: RP4 and pC221 in MOB P, R388 in MOB F: deviations in mean stability SStab20, deformability SDef20 and bendability SBend20 corresponded with IR11, p < 0.001)47,54,55.
The conservation of oriT structural properties inside MOB groups might be a consequence of the evolutionary development of the specific relaxation systems. According to our results and the current understanding, one possible way that oriT regions have evolved, is that relaxases in the ancestral state were of lower specificity and targeted multiple existing oriTs48,56. These oriTs evolved and adapted to their particular relaxase, after which the relaxase evolved to optimize interaction and enzymatic function with the best oriT. In some MOB systems, this includes the acquisition of other (auxiliary) proteins to aid the process. A particular relaxase therefore defines a particular oriT as this enables a stable structure of genes, a low number of deletions during conjugation, stable size of plasmids as well as the optimization of levels and functioning of plasmid-coded proteins and timing of their expression8,15,57. However, according to the above process it is also possible that (i) certain mobile elements can carry multiple oriTs58, and (ii) oriT regions might be present on elements lacking relaxases to confer mobility59,60.
According to such oriT evolutionary processes as described above, we hypothesize that relaxation systems with a larger amount of auxiliary proteins, such as MOB F and P, are more mature and optimized than ones with less auxilliary proteins (e.g. MOB Q and V, see Fig. 1). They could have had a more directed or longer evolution, meaning they are evolutionarily older systems. The observations are also supported by the reported characteristics of relaxation systems and conjugative properties of the conjugative elements that carry them. In contrast to the more advanced MOB F and P systems frequently carried by conjugative and larger (>30 kb) plasmids12, simpler MOB Q and V systems are usually carried by mobilizable and not conjugative elements. Therefore they rely on conjugation components (see Introduction) of the host or other plasmids for transfer12. The elements might lack such components due to being smaller (<30 kb) and potentially less evolved, which drives them to be more promiscuous so that they can exploit horizontal gene transfer to endure negative selection pressure. This higher promiscuity relates to simplicity of the oriT system of MOB V, which directly possesses the structural properties required for strand separation and relaxation (Fig. 5: low stability and high amount of destabilizations near nic), whereas the other MOB groups require auxiliary proteins to help them achieve this11. Nevertheless, in plasmids from the group MOB Q both auxiliary proteins and relaxases are known to have a very low DNA-binding specificity (e.g. RSF1010)48 and therefore we also expect that they are more promiscuous.
The results based on conventional nucleotide sequence analysis using evolutionary distance models (p-distance and Kimura) and the low DNA sequence conservation in oriT regions (Fig. 3, Supp. Fig. S3) support our findings on the conservation and evolution of oriT structure within conjugation systems. An important restriction with the sequence based analysis was that oriT sequences were misaligned, resulting in large distances between sequences and the inability to determine the Kimura distance (tendency of pyrimidine or purine substitutions)24 for all sequences, which led to inaccurate clustering (Supp. Fig. S2). Accordingly, with regions that display a high degree of conservation of structures, such as oriT, a more suitable approach would be to align them based on patterns of conservation of structural properties instead of merely nucleotide sequence patterns.
The cause for low classification frequencies of certain conjugative elements (Supp. Table S6), was that most of them were independent representatives of MOB subgroups or belonged to unknown subgroups9,10. Comparison with classification of plasmids according to relaxase amino acid sequence conservation in Barcia et al.10 shows that in our study, the misclassified plasmids differed from other elements also according to the conservation of their cognate relaxases. In the case of plasmid pWWO from MOB subgroup F11, in Barcia et al.10 the three other plasmids in subgroup F11 were clustered together in the same branch based on relaxase classification (bootstrap confidence of 99%), while pWWO was in a separate branch (bootstrap confidence of 99%). Similarly, the plasmid pAB6 from MOB Q1 was clustered separately from the other elements (bootstrap confidence of 100%). In the case of plasmids pTA1060 (MOB subgroup V1) and pIP421 (MOB V4), no possible cause for misclassification could currently be determined, since the phylogeny of all elements of MOB V is currently unavailable10. The results indicate that the phylogeny of oriT subtrates reflects that of their cognate relaxases (initial tests of classification using the whole dataset and MOB subgroups resulted in over 88% accuracy of cross-validations).
Since researchers require fast procedures to identify a plasmids MOB group and transfer range, we implemented the oriT structure-based procedure as a webtool (see Fig. 6). Although based on mere MOB classification we cannot predict the exact receiving host of a plasmid, we can restrict the selection to a range of hosts, where such types of plasmid have been found previosly. Given that the potential host range of a plasmid is not defined only by plasmid transfer, but also by the propensity of the plasmid to stabilize in the subsequent generations of the bacterial host8,10,52, two separate ranges can be distinguished (see Fig. 6): (i) the range of potential transfer hosts, based on the hosts of plasmids used for training the models (Supp. Table S7), and (ii) the range of potential incompatibility and replication (Inc/Rep) types that can help determine the replication host range (Supp. Table S8). Since they define entire transfer systems, MOB groups are one of the factors by which to determine the transfer host range, which is generally wider than the replication host range10,12. In Gammaproteobacteria, the plasmid replication (Rep) types were shown to be much more restrictive (in the plasmids they can amplify) than the MOB types8. However, since MOB groups were shown to include highly conserved distributions of Inc/Rep types8,52 and to describe complete plasmid backbones12,15, they can potentially provide important information on plasmid stability and behaviour in the host. Moreover, studies have shown that plasmid transfer host ranges can also be defined by other components of the conjugation system, such as the T4SS (mating complex) proteins12,52, which will undoubtedly serve as the basis for future improvements.
The significance of our results is that the transfer range of an AMR carrying plasmid can be determined merely by analysis of the structure of the oriT sequence instead of whole relaxase genes. Since they can facilitate binding of relaxases even in trans48,59,60, oriT substrates are the most elementary prerequsites for DNA mobility. Considering that there are potentially more oriT regions than relaxase genes58–60, as well as the algorithmic differences between nucleotide and protein sequence analysis, we presume that the identification and characterization of oriT substrates can potentially greatly improve the accuracy of predictions of plasmid mobility and hosts, over protein-based analyses. Consequently, the present method facilitates development of novel solutions to decrease AMR incidence with antibiotic treatments, since for a given AMR carrying plasmid the potential routes of transfer within its MOB group can guide the optimization of antibiotic treatments that limit the growth of the most frequent hosts.
Electronic supplementary material
Acknowledgements
We thank Prof. Dr. Aleš Belič and Prof. Dr. Tatjana Zrimec for valuable discussions regarding computational methods used in the study. This work was supported by the EU Seventh Framework Programme under grant agreements n° [282881, 261810], the European Social Fund of the EU under grant agreement n° [P-MR-09/124], the Slovenian Research Agency under grant agreements n° [P1-0237, Z2-7257, J4-7640] and the Government of the Russian Federation under grant n° [14.Z50.31.0004] and Saratov State University.
Author Contributions
J.Z. and A.L. designed the experiments, performed the experiments, analyzed the data and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20157-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jan Zrimec, Email: janzrimec@gmail.com.
Aleš Lapanje, Email: lapanje.ales@gmail.com.
References
- 1.Organization, W. H. & others. Antimicrobial resistance: global report on surveillance. (World Health Organization, 2014).
- 2.Leung E, Weil DE, Raviglione M, Nakatani H. The WHO policy package to combat antimicrobial resistance. Bull. World Health Organ. 2011;89:390–392. doi: 10.2471/BLT.11.088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beceiro A, Tomás M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013;26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob. Agents Chemother. 2011;55:3649–3660. doi: 10.1128/AAC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.zur Wiesch PA, Kouyos R, Engelstädter J, Regoes RR, Bonhoeffer S. Population biological principles of drug-resistance evolution in infectious diseases. Lancet Infect. Dis. 2011;11:236–247. doi: 10.1016/S1473-3099(10)70264-4. [DOI] [PubMed] [Google Scholar]
- 8.Garcillán-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol. Rev. 2011;35:936–956. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 9.Francia M, et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Garcillán‐Barcia MP, Francia MV, De La Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 11.D L Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram‐negative bacteria. FEMS Microbiol. Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 12.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guglielmini, J., de la Cruz, F. & Rocha, E. P. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. mss221 (2012). [DOI] [PMC free article] [PubMed]
- 14.Schröder G, Lanka E. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-López R, et al. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 2006;30:942–966. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- 16.Revilla C, et al. Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob. Agents Chemother. 2008;52:1472–1480. doi: 10.1128/AAC.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammar P, et al. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 18.Kolomeisky AB. Physics of protein–DNA interactions: mechanisms of facilitated target search. Phys. Chem. Chem. Phys. 2011;13:2088–2095. doi: 10.1039/C0CP01966F. [DOI] [PubMed] [Google Scholar]
- 19.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2009;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little EJ, Babic AC, Horton NC. Early interrogation and recognition of DNA sequence by indirect readout. Structure. 2008;16:1828–1837. doi: 10.1016/j.str.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas M, et al. Relaxase DNA binding and cleavage are two distinguishable steps in conjugative DNA processing that involve different sequence elements of the nic site. J. Biol. Chem. 2010;285:8918–8926. doi: 10.1074/jbc.M109.057539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carballeira, J. D., González-Pérez, B., Moncalián, G. & de la Cruz, F. A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation. Nucleic Acids Res. gku741 (2014). [DOI] [PMC free article] [PubMed]
- 23.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 783–791 (1985). [DOI] [PubMed]
- 26.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zrimec, J., Kopinč, R., Rijavec, T., Zrimec, T. & Lapanje, A. Band smearing of PCR amplified bacterial 16S rRNA genes: Dependence on initial PCR target diversity. J. Microbiol. Methods (2013). [DOI] [PubMed]
- 29.Olson WK, Gorin AA, Lu X-J, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl. Acad. Sci. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brukner I, Sanchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995;14:1812. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geggier S, Kotlyar A, Vologodskii A. Temperature dependence of DNA persistence length. Nucleic Acids Res. 2011;39:1419–1426. doi: 10.1093/nar/gkq932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zrimec, J. & Lapanje, A. Fast prediction of DNA melting bubbles using DNA thermodynamic stability. IEEE/ACM Trans. Comput. Biol. Bioinform. 12, 1137–1145 (2015). [DOI] [PubMed]
- 34.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 35.Keppel, G. & Wickens, T. D. Simultaneous comparisons and the control of type I errors. Des. Anal. Res. Handb. 4th Ed Up. Saddle River NJ Pearson Prentice Hall P111–130 (2004).
- 36.Mann, H. B. & Whitney, D. R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 50–60 (1947).
- 37.Hall MA, Holmes G. Benchmarking attribute selection techniques for discrete class data mining. IEEE Trans. Knowl. Data Eng. 2003;15:1437–1447. doi: 10.1109/TKDE.2003.1245283. [DOI] [Google Scholar]
- 38.Hall, M. A. Correlation-based feature selection of discrete and numeric class machine learning. (2000).
- 39.Kononenko, I. Estimating attributes: analysis and extensions of RELIEF. in European conference on machine learning 171–182 (Springer, 1994).
- 40.Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 41.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta BBA-Protein Struct. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 42.Hand DJ, Till RJ. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach. Learn. 2001;45:171–186. doi: 10.1023/A:1010920819831. [DOI] [Google Scholar]
- 43.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 44.Frost LS, Ippen-Ihler K, Skurray RA. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 1994;58:162. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai MM, Fu YH, Deonier RC. Intrinsic bends and integration host factor binding at F plasmid oriT. J. Bacteriol. 1990;172:4603–4609. doi: 10.1128/jb.172.8.4603-4609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziegelin G, Pansegrau W, Lurz R, Lanka E. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J. Biol. Chem. 1992;267:17279–17286. [PubMed] [Google Scholar]
- 47.Caryl JA, Thomas CD. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol. Microbiol. 2006;60:1302–1318. doi: 10.1111/j.1365-2958.2006.05188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker EC, Meyer RJ. Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J. Bacteriol. 2003;185:3538–3546. doi: 10.1128/JB.185.12.3538-3546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurenbach B, et al. The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology. 2006;152:637–645. doi: 10.1099/mic.0.28468-0. [DOI] [PubMed] [Google Scholar]
- 50.Lorenzo-Díaz F, et al. The MobM relaxase domain of plasmid pMV158: thermal stability and activity upon Mn2+and specific DNA binding. Nucleic Acids Res. 2011;39:4315–4329. doi: 10.1093/nar/gkr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vedantam G, Knopf S, Hecht DW. Bacteroides fragilis mobilizable transposon Tn5520 requires a 71 base pair origin of transfer sequence and a single mobilization protein for relaxosome formation during conjugation. Mol. Microbiol. 2006;59:288–300. doi: 10.1111/j.1365-2958.2005.04934.x. [DOI] [PubMed] [Google Scholar]
- 52.Shintani, M., Sanchez, Z. K. & Kimbara, K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6 (2015). [DOI] [PMC free article] [PubMed]
- 53.Wong JJ, Lu J, Glover JN. Relaxosome function and conjugation regulation in F‐like plasmids–a structural biology perspective. Mol. Microbiol. 2012;85:602–617. doi: 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- 54.Pansegrau W, Lanka E. Mechanisms of Initiation and Termination Reactions in Conjugative DNA Processing INDEPENDENCE OF TIGHT SUBSTRATE BINDING AND CATALYTIC ACTIVITY OF RELAXASE (TraI) OF IncPα PLASMID RP4. J. Biol. Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 55.Moncalián G, Valle M, Valpuesta JM. & De La Cruz, F. IHF protein inhibits cleavage but not assembly of plasmid R388 relaxosomes. Mol. Microbiol. 1999;31:1643–1652. doi: 10.1046/j.1365-2958.1999.01288.x. [DOI] [PubMed] [Google Scholar]
- 56.Parker C, Becker E, Zhang X, Jandle S, Meyer R. Elements in the co-evolution of relaxases and their origins of transfer. Plasmid. 2005;53:113–118. doi: 10.1016/j.plasmid.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Norman A, Hansen LH, Sørensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Kranenburg R, de Vos WM. Characterization of multiple regions involved in replication and mobilization of plasmid pNZ4000 coding for exopolysaccharide production in Lactococcus lactis. J. Bacteriol. 1998;180:5285–5290. doi: 10.1128/jb.180.20.5285-5290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Brien FG, et al. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res. 2015;43:7971–7983. doi: 10.1093/nar/gkv755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pollet RM, et al. Processing of nonconjugative resistance plasmids by conjugation nicking enzyme of staphylococci. J. Bacteriol. 2016;198:888–897. doi: 10.1128/JB.00832-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furuya N, Komano T. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J. Bacteriol. 1995;177:46–51. doi: 10.1128/jb.177.1.46-51.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cook DM, Farrand SK. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J. Bacteriol. 1992;174:6238–6246. doi: 10.1128/jb.174.19.6238-6246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szpirer CY, Faelen M, Couturier M. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 2001;183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 65.Sut MV, Mihajlovic S, Lang S, Gruber CJ, Zechner EL. Protein and DNA effectors control the TraI conjugative helicase of plasmid R1. J. Bacteriol. 2009;191:6888–6899. doi: 10.1128/JB.00920-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexandrov BS, et al. DNA dynamics play a role as a basal transcription factor in the positioning and regulation of gene transcription initiation. Nucleic Acids Res. 2010;38:1790–1795. doi: 10.1093/nar/gkp1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mihajlovic S, et al. Plasmid r1 conjugative DNA processing is regulated at the coupling protein interface. J. Bacteriol. 2009;191:6877–6887. doi: 10.1128/JB.00918-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams SL, Schildbach JF. TraY and integration host factor oriT binding sites and F conjugal transfer: sequence variations, but not altered spacing, are tolerated. J. Bacteriol. 2007;189:3813–3823. doi: 10.1128/JB.01783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.