Abstract
Background
Intermittent explosive disorder is defined as a recurrent, problematic, and impulsive aggression that affects 3% to 4% of the US population. While behavioral genetic studies report a substantial degree of genetic influence on aggression and impulsivity, epigenetic mechanisms underlying aggression and intermittent explosive disorder are not well known.
Methods
The sample included 44 subjects (22 with a DSM-5 diagnosis of intermittent explosive disorder and 22 comparable subjects without intermittent explosive disorder). Peripheral blood DNA methylome was profiled using the Illumina Infinium HumanMethylation450 Beadchip. Intermittent explosive disorder-associated genome-wide DNA methylation changes were analyzed using the CpGassoc R package, with covariates age, sex, and race being adjusted. A gene-based functional enrichment analysis was performed to identify pathways that were overrepresented by genes harboring highly differentially methylated CpG sites.
Results
A total of 27 CpG sites were differentially methylated in IED participants (P<5.0×10–5), but none reached genome-wide significant threshold. Functional enrichment analysis revealed that genes mapped by these CpG sites are involved in the inflammatory/immune system, the endocrine system, and neuronal differentiation.
Conclusions
Consistent with our previous studies showing an association of inflammatory response with aggressive behavior in intermittent explosive disorder subjects, our gene-based pathway analysis using differentially methylated CpG sites supports inflammatory response as an important mechanism involved in intermittent explosive disorder and reveals other novel biological processes possibly associated with intermittent explosive disorder.
Keywords: IED, DNA methylome, pathway analysis, inflammation
Significance Statement
This is the first genome-wide DNA methylation study to identify DNA methylation markers associated with IED. Although no statistically significant genome-wide findings were obtained, the gene-based functional enrichment analysis identified several biological processes associated with IED, including those involved in the immune/inflammatory system, the endocrine system, and neuronal differentiation. While preliminary, this study supports the association between IED and inflammatory response and reveals novel pathways that may be associated with IED that can then be used for clues for novel targets of intervention in the treatment of impulsive aggressive behavior. In addition, identifying biologically relevant DNA methylation signatures associated with IED may lead to the discovery of biomarkers for impulsive aggression that run across diagnostic conditions. If so, this could allow for treatment interventions in individuals in whom impulsive aggression might not be recognized as a clinical focus.
Introduction
Intermittent explosive disorder (IED), as defined by DSM-5 (American Psychiatric Association, 2013), is characterized by recurrent, problematic, and impulsive aggression. It occurs in 3% to 4% of individuals in the United States (Coccaro et al., 2017) and has been shown to have numerous neurobiological features, including anomalies of neuroanatomy (Coccaro et al., 2015c, 2016c); corticolimbic response to social threat (Coccaro et al., 2007; Cremers et al., 2015; McCloskey et al., 2016); neurotransmitter function (Coccaro et al., 2015a), including peripheral (Coccaro et al., 2014b, 2016d) and central (Coccaro et al., 2014a, 2015b) inflammatory mediators; and social cognition (Coccaro et al., 2009, 2016a, 2016b). Not surprisingly, numerous studies indicate that both genetic and environmental factors contribute to the development of aggression (Coccaro et al., 1997a; Miles et al., 1997; Yeh et al., 2010). However, while the heritability of aggression in adults ranges between 37% and 57%, depending on the type of aggression (Coccaro et al., 1993; Yeh et al., 2010), little is known about the interaction of genetic and environmental influences and how epigenetic mechanisms, which allow the reprogramming of gene expression, are involved in aggression.
Epigenetic mechanisms include structural modification of chromatin, posttranslational histone modifications (such as acetylation and methylation), chemical modification of DNA through methylation or hydroxymethylation of cytosines, as well as expression of interfering noncoding RNAs, including miRNAs and long-noncoding RNAs. These mechanisms allow reprogramming of the genome upon environmental inputs at specific time-points during development. Recently, the study of DNA methylation has been a focus of psychosocial stress and aggression research.
Several studies have examined epigenetic changes associated with aggression or related phenotypes in humans, most of them using candidate gene approaches. Methylation differences have been observed between individuals with antisocial personality disorder from a prisoner population and controls in the monoamine oxidase A (MAOA) gene in blood (Checknita et al., 2015). Hypermethylation of several CpG sites in the serotonin transporter gene (SLC6A4) promoter in T-cells and monocytes has been also observed in adult men with high levels of physical aggression in childhood (Wang et al., 2012). Methylation changes have been reported in candidate genes encoding cytokines and their transcription factors in a study of 8 adult men with a history of chronic physical aggression during childhood (Provencal et al., 2013a). While these studies suggest that DNA methylation is one mechanism associated with aggressive behavior, no prior study has examined individuals with clinically significant impulsive aggression (e.g., IED) in adulthood.
Methods
Participants
Participants included a total of 44 individuals from which 22 had a DSM-5 diagnosis of IED and 22 were comparison participants without any clinically significant history of impulsive aggression. Participants were recruited through public service announcements seeking individuals who reported psychosocial difficulties related to one or more syndromal and/or personality disorders or individuals who had little evidence of psychopathology. Participants were matched for age, sex, and race, and all provided written informed consent as approved by the University of Chicago institutional review board.
Diagnostic Assessment of DSM-5 Diagnoses
All psychiatric and personality disorder diagnoses were made by the DSM-5 criteria (American Psychiatric Association, 2013). Diagnostic interviews were performed by trained masters-level psychologists with good to excellent inter-rater reliability (mean kappa of .84±.05; range:.79 to.93) across mood, anxiety, substance use, intermittent explosive, and personality disorders. Final diagnoses were assigned by “team best estimate” involving psychiatrists/clinical psychologists as previously noted (Coccaro et al., 2012a) using information from (1) Structured Clinical Interview for DSM-5 Syndromal Disorders (First et al., 2014); (2) Structured Interview for DSM Personality Disorders (Pfohl et al., 1997); (3) clinical interview by a research psychiatrist; and (4) review of all available clinical data.
By definition, none of the control participants met DSM-5 criteria for IED in their lifetime. However, 10 control participants had a current or lifetime DSM-5 disorder, while 12 others did not. The frequency of current DSM-5 disorders in the control group was: major depressive disorder (n=1), any anxiety disorder (n=3), any trauma/stress disorder (n=2), and any eating disorder (n=1). The frequency of lifetime DSM-5 disorders in the control group was: major depressive disorder (n=3), any anxiety disorder (n=3), any drug use disorder (n=1), any trauma/stress disorder (n=3), and any eating disorder (n=3). The frequency of personality disorder in the control group was: Cluster A (n=0), Cluster B (n=3), Cluster C (n=2), and PD-Not Otherwise Specified (n=2). By definition, all IED participants met DSM-5 criteria for IED in their lifetime, with 19 (82%) meeting criteria for current IED. The frequency of current DSM-5 disorders in the IED group was: major depressive disorder (n=7), any anxiety disorder (n=4), any trauma/stress disorder (n=2), any eating disorder (n=2), and somatoform disorder (n=1). The frequency of lifetime DSM-5 disorders in the IED group was: major depressive disorder (n=17), any anxiety disorder (n=8), any substance use disorder (n=6), any eating disorder (n=4), any trauma/stress disorder (n=2), any Non-IED impulse control disorder (n=1), obsessive compulsive disorder (n=1), and somatoform disorder (n=1). The frequency of specific personality disorders in the IED group was: Cluster A (n=4), Cluster B (n=14), and Cluster C (n=10). While most of the IED participants were not in psychiatric treatment at time of study (82%, 18/22), nearly all had a history of psychiatric treatment (95%) and 4 (18%) were taking psychotropic medication at time of study (all 4 with an antidepressant, 2 with a benzodiazepine, 1 with lithium, and 1 with a stimulant). Methylation patterns did not differ among all subjects on medications compared all subjects not on medications.
Measures of Aggression, Impulsivity, and Related Variables
Aggression was assessed with the Aggression Scale from Life History of Aggression [LHA (Coccaro et al., 1997b)] and the physical aggression and verbal aggression subscales of the Buss-Perry Aggression Questionnaire [BPAQ (Buss et al., 1992)]. The LHA assesses the number of times a person has engaged in actual aggressive behavior in their life and ranges from 0 to 25. LHA Aggression has good to excellent psychometric properties (Coccaro et al, 1997). BPAQ Aggression assesses aggressive tendencies as a personality trait by questionnaire and also has good to excellent psychometric properties. Impulsivity was assessed with the Life History of Impulsive Behavior [LHIB (Coccaro et al., 2012b)]. As with LHA Aggression, the LHIB assesses the number of times a person has engaged in actual impulsive behavior in their life and has good to excellent psychometric properties. Other assessments included the global assessment of psychosocial function (American Psychiatric Association, 1994). Racial and socioeconomic data, collected by diagnostic assessors, reflected the self-identified characteristics of the subjects.
DNA Specimens
Genomic DNA was extracted from peripheral blood using standard techniques. To prepare specimens for methylation analysis, 500 ng of genomic DNA was treated with bisulfite reagents included in the EZ-96 DNA methylation kit (Zymo Research) according to the manufacturer’s protocol. Unmethylated cytosines were converted to uracils while methylated cytosines remained unchanged. Bisulfite-converted DNA samples were used in the array-based genome-wide DNA methylation assay.
Infinium HumanMethylation450 BeadChip Assay
The Illumina 450K Methylation BeadChip was used in the current investigation. This BeadChip interrogates >450000 methylation sites per sample at single-nucleotide resolution. It covers all designable (96%) RefSeq genes, with 41% of CpG sites in promoter regions, 31% in the gene body, 3% UTRs, and 25% in intergenic regions. The methylation assays were completed at the Genome Center of Northwestern University, a core resource available to academic institutions in Chicago, including the University of Chicago and the University of Illinois at Chicago. The GenomeStudio software (Illumina) was used to generate β values for each CpG site, with β values ranging from 0.0 to 1.0 quantifying the ratio of methylated/unmethylated alleles from fluorescent signals at each CpG site. Raw scanned data were normalized; average β values were recalculated using background intensity measured by negative background probes present on the array. Standard quality control tests and functional normalization were conducted using the Preprocessfunnorm function in the R minfi package. Since DNA methylation patterns can be cell-type specific, we accounted for cell heterogeneity by estimating the relative proportion of 5 different cell types in the whole blood samples (i.e., CD8T, CD4T, NK, B cells, monocytes, and granulocytes) using the estimateCellCounts function from the R minfi package (Houseman et al., 2012; Jaffe and Irizarry, 2014).
Genome-Wide DNA Methylation Association Analysis
To test for association between methylation at CpG sites across the genome and IED diagnosis, we performed the analysis using the CpGassoc command in R software environment (https://cran.r-project.org/web/packages/CpGassoc/index.html). Potential confounding factors, including age, sex, race, and cell type composition, were included in the model to normalize residuals. Genome-wide significant threshold for analysis was set at P<5.0x10–7. To correct for multiple comparison testing, false discovery rate (FDR) was set at 0.05.
To investigate potential gene expression regulatory function of the differentially methylated CpG sites identified, we used the expression-associated Methylated Site (eMS) database (Liu et al., 2013), which examines the association between CpG methylation and gene expression in human monocytes of 1264 participants. To provide genome-wide coverage of gene expression and DNA methylation, they used the Illumina Human HT-12 v4 Expression BeadChip and the Illumina Human Methylation 450 BeadChip, respectively. The dataset includes 11203 potential cis-acting CpG loci whose degree of methylation is associated with gene expression (eMS) at a FDR threshold of 0.001.
Gene-Based Functional Enrichment Analysis
Top differentially methylated CpG sites with a suggestive significance of P<5.0×10–5 were used for gene-based functional enrichment analysis using the MetaCore (Thompson Reuters) software. To adjust for multiple comparisons, the FDR was set at 0.05.
Results
Characteristics of Participants
IED and Control participants did not differ in age, sex, ethnicity, or socio-economic status. As expected, IED participants had lower global assessment of psychosocial function scores but higher LHA Aggression, BPAQ Aggression, and LHIB Impulsivity scores compared with Control participants (Table 1).
Table 1.
Demographic and Behavioral Data of Participants
| Controls (n = 22) |
Intermittent explosive disorder (n = 22) |
P | |
|---|---|---|---|
| Demographic Variables | |||
| Age (y ± SD) | 38.1 ± 5.6 | 36.3 ± 9.3 | .449 |
| Gender (M/F) | 11 / 11 | 12 / 10 | .999 |
| Ethnicity (white / AA / other) | 14 / 4 / 4 | 16 / 2 / 4 | .670 |
| SES score | 45.6 ± 10.4 | 42.4 ± 12.3 | .370 |
| Behavioral variables | |||
| GAF score | 73.2 ± 13.1 | 51.2 ± 10.8 | <.001 |
| LHA aggression | 7.9 ± 5.3 | 18.4 ± 3.7 | <.001 |
| BPAQ aggression | 31.1 ± 9.0 | 42.6 ± 12.4 | .003 |
| LHIB impulsivity | 11.6 ± 5.0 | 39.1 ± 23.5 | <.001 |
Genome-Wide DNA Methylation Changes Associated with IED
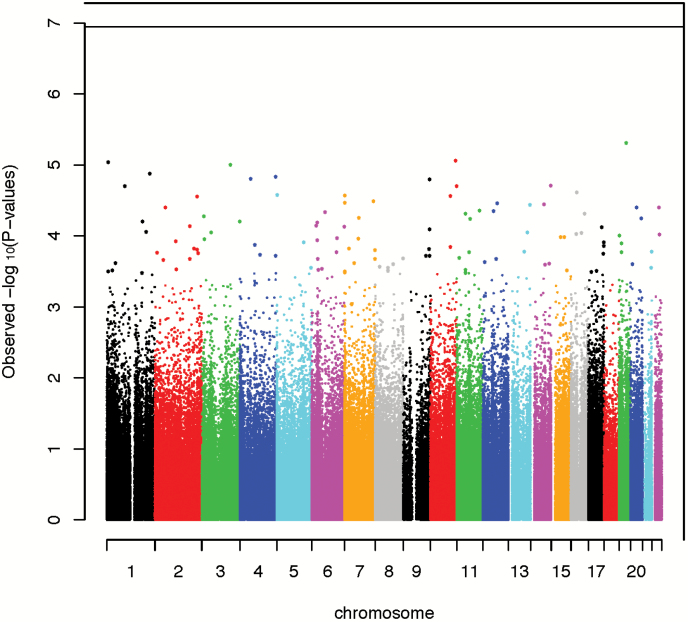
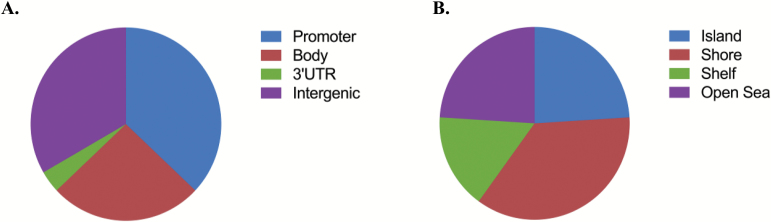
The density distribution of methylation beta-values before and after normalization is shown in Supplementary Figure 1. Figure 1 shows the Manhattan plot depicting all the analyzed CpG with their calculated P values. A total of 27 CpG sites were differentially methylated in IED subjects with a suggestive significance of P<5.0×10–5 (Table 2). None of these sites reached genome-wide significance (GWS; P<5.0×10–7). Quantile-quantile plots show no evidence for inflation or bias in the analysis (supplemental Figure 2; Lambda factor=0.98). The majority of differentially methylated CpG sites showed increased methylation in IED subjects compared with control subjects (supplemental Figure 3) and reside in gene promoter regions (10/27 CpG sites, Figure 2A).
Figure 1.
Manhattan plot for association between DNA methylation and intermittent explosive disorder (IED).
Table 2.
Top Differentially Methylated CpG Sites Associated with IED
| Symbol | Gene name | Chr | Gene location | Illumina ID | F statistic | FDR | P value |
|---|---|---|---|---|---|---|---|
| PTPRN2 | Protein Tyrosine Phosphatase, Receptor Type N2 | 7 | 157996423 | cg03899215 | 30.01 | 0.78 | 5.37E-06 |
| HK1 | Hexokinase 1 | 10 | 71091204 | cg15258080 | 27.88 | 0.78 | 9.56E-06 |
| TYMS | Thymidylate Synthetase PA | 18 | 658602 | cg11726572 | 27.69 | 0.78 | 1.01E-05 |
| CYFIP1 | Cytoplasmic FMR1 Interacting Protein 1 | 15 | 22893147 | cg02594498 | 27.42 | 0.78 | 1.08E-05 |
| PLD2 | Phospholipase D1 | 17 | 4714200 | cg04521626 | 26.37 | 0.78 | 1.45E-05 |
| ALDOA | Aldolase, Fructose- Biphosphate A | 16 | 30075904 | cg01165575 | 26.03 | 0.78 | 1.60E-05 |
| YME1L1 | YME1 Like 1 ATPase PA | 10 | 27443335 | cg23635883 | 25.76 | 0.78 | 1.72E-05 |
| FEZF2 | FEZ Family Zinc Finger 2 | 3 | 62351484 | cg19864138 | 25.71 | 0.78 | 1.75E-05 |
| KDELC2 | KDEL Motif Containing 2 PA | 11 | 108369215 | cg25505476 | 25.03 | 0.78 | 2.13E-05 |
| SAR1A | Secretion Associated Ras Related GTPase 1A PA | 10 | 71929509 | cg05292330 | 24.97 | 0.78 | 2.17E-05 |
| MAD1L1 | MAD1 Mitotic Arrest Deficient Like 1 | 7 | 2078988 | cg06100570 | 24.97 | 0.78 | 2.17E-05 |
| DIABLO | Diablo IAP-Binding Mitochondrial Protein PA | 12 | 122706597 | cg17576580 | 24.27 | 0.78 | 2.66E-05 |
| C6orf136 | Chromosome 6 Open Reading Frame 136 PA | 6 | 30614397 | cg13253439 | 24.00 | 0.78 | 2.88E-05 |
| SORD | Sorbitol Dehydrogenase PA | 15 | 45314933 | cg27073142 | 23.91 | 0.78 | 2.95E-05 |
| EOGT | EGF Domain Specific O-Linked N-Acetylglucosamine Transferase PA | 3 | 69063328 | cg21785536 | 23.87 | 0.78 | 2.99E-05 |
| PRDM2 | PR/SET Domain 2 | 1 | 14145540 | cg16560370 | 23.83 | 0.78 | 3.02E-05 |
| NR1H2 | Nuclear Receptor Subfamily 1 Group H Member 2 PA | 19 | 50879636 | cg13567813 | 23.29 | 0.78 | 3.55E-05 |
| SORD | Sorbitol Dehydrogenase PA | 15 | 45314915 | cg22023531 | 23.14 | 0.78 | 3.71E-05 |
| GBP4 | Guanylate Binding Protein 4 | 1 | 89664407 | cg21365602 | 23.07 | 0.78 | 3.79E-05 |
| TMEM180 | Transmembrane Protein 180 PA | 10 | 104221000 | cg03417317 | 22.99 | 0.78 | 3.88E-05 |
| ADAMTS17 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 17 PA | 15 | 100469152 | cg13132497 | 22.94 | 0.78 | 3.94E-05 |
| TMEM81 | Transmembrane protein 81 | 1 | 205053265 | cg00103209 | 22.67 | 0.78 | 4.27E-05 |
| LOC100505921 | 7 | 8002109 | cg06640047 | 22.65 | 0.78 | 4.30E-05 | |
| CPLX2 | Complexin 2 | 5 | 175224952 | cg04446284 | 22.64 | 0.78 | 4.31E-05 |
| FN3KRP | Fructosamine 3 Kinase Related Protein | 17 | 80673675 | cg23522895 | 22.31 | 0.79 | 4.76E-05 |
| BMPR1A | Bone Morphogenetic Protein Receptor Type 1A | 10 | 88632654 | cg26381210 | 22.25 | 0.79 | 4.85E-05 |
| INHBA | Inhibin Beta A Subunit | 7 | 41742630 | cg11079619 | 22.17 | 0.79 | 4.97E-05 |
Figure 2.
Functional genomic distribution of top differentially methylated CpG sites in intermittent explosive disorder (IED).
We also examined whether top CpG differentially methylated CpG sites have a role in the regulation of gene expression using the eMS dataset. We found that cg04521626 is associated with decreased gene expression of ZMYND15 (Beta = -0.23, P=2.47E-06, FDR=0.0005).
DNA Methylation in Aggression-Related Candidate Genes
We examined IED-associated DNA methylation changes in MAOA and SLC6A4 (2 most well-studied candidate genes for aggression). However, no significant results were obtained.
Effect of Current IED from Past IED on the Differentially Methylated CpG Sites
Mean (±SD) methylation values for the top 27 differentially methylated CpG sites (P<5.0x10–5) (did not differ as a function of current vs past IED, with both IED groups differing from controls (current IED: 0.311±0.002 vs past IED:0.316±0.007 vs controls: 0.299±0.004; F[2,41]=47.61, P=2.04E-11; Tukey HSD: current IED=past IED>controls). Inspection of the data revealed that current IED and past IED participants did not differ at any of the 27 CpG sites examined.
Effect of Psychiatric Comorbidity on the Differentially Methylated CpG Sites
There was no difference in the mean methylation value for the top 27 CpG sites in IED participants as a function of comorbidity with current or lifetime major depressive disorder, lifetime anxiety disorder, substance use disorder, or Cluster B or Cluster C Personality Disorder (Table 3). Inspection of the data revealed that, of the 27 top CpG sites examined, participants with IED differed significantly as a function of comorbidity at only one CpG site (i.e., 17576580) for current major depressive disorder only.
Table 3.
Mean (±SD) Methylation Levels of Top 27 CpG Sites as a Function of Diagnostic Comorbidity
| Type of Comorbidity | ||||||
|---|---|---|---|---|---|---|
| Major depressive disorder current | Major depressive disorder lifetime | Anxiety disorder lifetime | Substance use disorder lifetime | Cluster B personality disorder | Cluster C personality disorder | |
| IED with comorbidity | 0.317 ± 0.007 (n = 7) |
0.316 ± 0.006 (n = 17) |
0.316 ± 0.010 (n = 8) |
0.316 ± 0.007 (n = 6) |
0.314 ± 0.007 (n = 14) |
0.315 ± 0.008 (n = 10) |
| IED without comorbidity | 0.315 ± 0.007 (n = 15) |
0.314 ± 0.010 (n = 5) |
0.315 ± 0.005 (n = 14) |
0.315 ± 0.007 (n = 16) |
0.318 ± 0.006 (n = 8) |
0.316 ± 0.006 (n = 12) |
All comparisons are nonsignificant. Only comorbid disorders affecting at least 5 IED participants were examined.
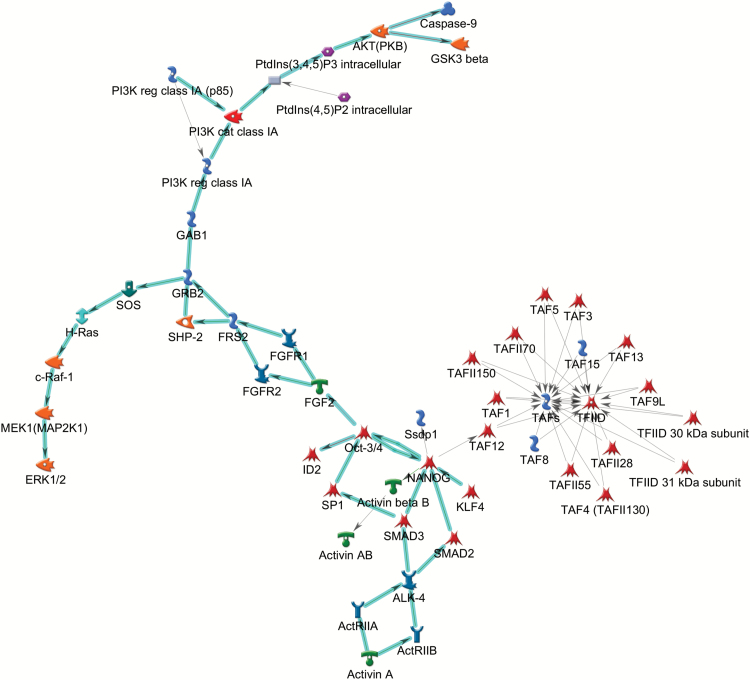
Figure 3.
Top networks associated with top gene ontology (GO) processes genes differentially methylated in intermittent explosive disorder (IED) subjects.
Gene-Based Functional Enrichment Analysis
We included genes mapped by the top differentially methylated CpG sites (P<5.0x10–5; Table 2) in enrichment analyses using MetaCore (Thomson Reuters). Top GO biological processes enriched with such genes included(1)neuronal differentiation,suchasGABAergic neuron differentiation(P=4.71x10–8, FDR=7.05x10–5), striatal neuron differentiation (P=1.59x10–8, FDR=7.05x10–5), and forebrain neuron differentiation (P=1.24x10–5, FDR=5.59x10–4); (2) the immune/inflammation system, such as negative regulation of B cell differentiation (P=3.40x10–7, FDR=7.06x10–5), negative regulation of interferon-gamma biosynthetic process (P=1.28x10–6, FDR=1.42x10–4), negative regulation of cytokine biosynthetic process (P=3.39x10–5, FDR=9.52x10–4), and immune system process (P=4.09x10–5, FDR=1.07x10–3); and (3) the hormonal system, such as progesterone secretion (P=9.97x10–8, FDR=7.06x10–5) and hormone secretion (P=9.36x10–7, FDR=1.11x10–4) (Table 4).
Table 4.
Top 10 Biological GO Processes Associated with IED
| Enrichment by GO Processes | |||
|---|---|---|---|
| Processes | P value | FDR | Ratio |
| GABAergic neuron differentiation | 4.71E-08 | 7.06E-05 | 4/25 |
| Progesterone secretion | 9.97E-08 | 7.06E-05 | 3/7 |
| Striatal medium spiny neuron differentiation | 1.59E-07 | 7.06E-05 | 3/8 |
| Negative regulation of hair follicle development | 2.39E-07 | 7.06E-05 | 3/9 |
| Positive regulation of ovulation | 2.39E-07 | 7.06E-05 | 3/9 |
| Negative regulation of hair cycle | 3.41E-07 | 7.06E-05 | 3/10 |
| Regulation of ovulation | 3.41E-07 | 7.06E-05 | 3/10 |
| Negative regulation of B cell differentiation | 3.41E-07 | 7.06E-05 | 3/10 |
| Negative regulation of follicle-stimulating hormone secretion | 6.24E-07 | 9.39E-05 | 3/12 |
| Steroid hormone secretion | 6.24E-07 | 9.39E-05 | 3/12 |
The top diseases overrepresented by biomarkers associated with genes mapped by the top differentially methylated CpG sites include adenocarcinoma, clear cell (P=2.44x10–7, FDR=1.03x10–4), Klinefelter Syndrome (P=1.97x10–6, FDR=4.16x10–4), and fibroadenoma (P=5.71x10–6, FDR=8.05x10–4) (Table 5).
Table 5.
Top 10 Diseases Associated with IED
| Disease (by biomarkers) | |||
|---|---|---|---|
| Diseases | P value | FDR | Ratio |
| Adenocarcinoma, clear cell | 2.44E-07 | 1.03E-04 | 3/8 |
| Klinefelter Syndrome | 1.97E-06 | 4.16E-04 | 3/15 |
| Fibroadenoma | 5.71E-06 | 8.05E-04 | 3/21 |
| Neoplasms, fibroepithelial | 8.66E-06 | 9.16E-04 | 3/24 |
| Goiter, nodular | 1.39E-05 | 1.10E-03 | 3/28 |
| Sex chromosome disorders of sex development | 1.56E-05 | 1.10E-03 | 3/29 |
| Sex chromosome disorders | 3.56E-05 | 1.59E-03 | 3/38 |
| In-house adverse events | 3.72E-05 | 1.58E-03 | 19/4463 |
| Fibrocystic breast disease | 3.84E-05 | 1.58E-03 | 3/39 |
| Chylomicron retention disease | 4.10E-05 | 1.58E-03 | 2/6 |
Discussion
This is the first genome-wide DNA methylation study to identify DNA methylation markers associated with IED. Although no genome-wide significance findings were obtained, our gene-based functional enrichment analysis identified several biological processes associated with IED. These biological processes are mostly involved in the immune/inflammatory system, the endocrine system, and neuronal differentiation. Regarding the immune/inflammatory system, this is consistent with previous work that reports associations between circulating inflammatory cytokines and aggression in human studies (Kraus et al., 2003; Marsland et al., 2008; McHuthison et al., 1998; Suarez, 2003, 2004). For example, we have reported that the circulating levels of inflammatory cytokines are higher in individuals with IED compared with healthy and psychiatric controls (Coccaro et al., 2014b), and that C-Reactive Protein (CRP), Interleukin-6 (IL-6), and Interleukin-1RAII protein correlate with the Life History of Aggression and/or the Buss-Perry Aggression (Coccaro et al., 2014a, 2014b, 2015b).
These findings were not accounted for by state-trait differences in current vs past IED or by the presence of psychiatric comorbidity in the IED participants. This is important because each IED participant had a history of other non-IED disorders, and the presence of one or more could account for IED-control differences in methylation sites. For example, if the presence of major depressive disorder accounted for IED-control differences in DNA methylation, IED participants with history of major depressive disorder would have had significantly higher methylation levels of CpG sites compared with IED participants without major depressive disorder. However, the mean number of CpG methylation sites did not differ between IED participants with or without major depressive disorder (or other comorbidities). This is consistent with the fact that IED typically begins earlier in life compared with mood, anxiety, or substance use disorders, and it is likely a risk factor for these disorders rather than the converse (Kessler et al., 2006). Subsequent analysis of each of the top 27 CpG sites found no more than one CpG site (over all comorbidities) in which IED differed as a function of comorbidity, a result no greater than chance.
Our findings are consistent with those obtained from previous studies regarding DNA methylation-aggression relationship. Specifically, studies of DNA methylation changes associated with chronic physical aggression in childhood found an elevated level of methylation of genes coding for regulatory elements of transcription factors and cytokines (e.g., interleukin 6) (Provencal et al., 2013a, 2013b). The inflammatory pathways were also reported to be associated with early-lifetime-dependent environmental factors associated with aggressive behavior such as maternal deprivation in rhesus monkeys, child abuse, socioeconomic status, or PTSD (Provencal et al., 2015). Genome-wide studies performed by Provencal and colleagues associated methylation levels of several gene promoters with chronic physical aggression. The most important genes they identified were related to inflammatory and immune response pathways, the serotonergic system (e.g., HTR1D), the dopaminergic system (DAT; SLC63A), the HPA axis (AVPR1A), the glutamatergic system (GRM5) (Provencal et al., 2014), and the stress regulatory genes (NR3C1 and CRHBP) (Guillemin et al., 2014).
Here, we found that biological processes involved in the immune system are also associated with IED. One of our top-ranked CpG sites is located in the guanylate binding protein 4 gene (GBP4), which is involved in interleukin-3 pathway, part of the cytokine signaling in the immune system. We also found an enrichment of the top-ranked CpG sites with the central nervous system, with the strongest association with GABAergic neuronal differentiation. While its role in aggression is not clear in humans, it is thought to be involved in impulsivity and suicidal behavior (Lee et al., 2009; 2011). In mice, activation of GABA-B receptors in the dorsal raphe nucleus increases aggressive behavior (Takahashi et al., 2010). Even though we were not able to assess the gene expression in this dataset, we examined the correlation with gene expression among the top 27 differentially methylated CpG sites identified using the eMS dataset. One CpG site (cg04521626) is associated with decreased gene expression of zinc finger MYND-Type Containing 12 (ZMYND15; Beta=-0.23, P=2.47E-06, FDR=0.0005) gene, involved in spermatogenesis. Further, 10 of the top 27 differentially methylated CpG sites reside in promoter regions, suggesting a functional role in gene regulation.
In a recent genome-wide DNA methylation study of aggressive disposition in a large group of population-based adult twins (van Dongen et al., 2015), 2 CpG sites were identified. One was CpG cg01792876, which is located near the trichorhinophalangeal syndrome I (TRPS1) gene coding for a zinc finger transcription factor that represses GATA-regulated genes. This CpG site is also located in a region that harbors a suggestive SNP association with major depressive disorder (rs2721937, chr8: 116,702,874) (Major Depressive Disorder Working Group of the Psychiatric et al., 2013). Another was CpG cg06092953, which is located between the noncoding RNA PARD6G antisense RNA 1 (PARD6G-AS1) and the activity-dependent neuroprotective protein 2 gene (ADNP2). ADNP2 is highly expressed in the brain (particularly in the cerebral cortex) and involved in cellular survival pathways (Kushnir et al., 2008). The expression of ADNP2 has been linked to schizophrenia (Dresner et al., 2011) and the action of bipolar disorder medications (McEachin et al., 2010). Although none of these sites were identified in our study, one of our top-ranked genes associated with IED, that is, CPLX2, is highly expressed in the brain and associated with cognitive impairment in schizophrenic patients (Begemann et al., 2010; Hass et al., 2015).
This study has a number of strengths and limitations. The major strength of this study is that it includes a participant group well characterized in both clinical diagnosis and behavioral variables of relevance. We have used the new DSM-5 criteria for IED that requires a history of frequent and problematic impulsive aggressive behavior rather than simply using scores from personality assessments that include aggressive related constructs. In addition, we have used specific and well-validated assessments of aggression both as a history of actual aggressive events and as a personality disposition of aggressiveness. This differs from a recent EWAS study (van Dongen et al., 2015) that used a questionnaire assessment containing a heterogeneous group of items reflecting aggressive disposition as well as nonspecific items reflecting negative affect and mood liability. Our study also differs from another study that assessed methylation patterns in young adult males with a childhood, but not necessarily current, history of chronic physical aggression (Provençal et al., 2014). While physical aggression occurs in IED, it is unknown how many individuals with chronic physical aggressive behaviors in childhood continued to display these behaviors in adulthood.
Additional limitations of the study include the small sample size, the inclusion of participants drawn from the community rather than from treatment settings, and the absence of circulating inflammatory markers in these specific participants. First, while the sample size was small, our experimental group was nearly 3 times of that of the study that investigated chronic physical aggression in childhood (Provençal et al., 2014). At this time, a larger sample would have been prohibitive in cost. Given this, we chose to examine the top differentially methylated CpG sites by way of a gene-based pathway analysis. By using this approach, we identified the inflammatory pathway as one of the main networks associated with IED. Accordingly, these data fit nicely with the extant data on the relevance of inflammatory pathways in aggression (Kraus et al., 2003; Suarez, 2003, 2004; Coccaro, 2006, 2014a, 2014b, 2015b, 2016d; Zalcman et al., 2006; Marsland et al., 2008). Second, while none of the IED participants were recruited from clinical treatment sites, nearly all (95%) had a past history of psychiatric treatment (95%) or a history of behavioral disturbance that warranted evaluation/treatment (5%). Thus, these participants are likely similar to those who could have been referred from treatment facilities. Finally, we did not have inflammatory markers in any of the participants in this study, and this limits our ability to assess the relationship between peripheral inflammatory markers and the level of methylation of genes in this study. Future studies are needed to include such data and replicate the current findings in a larger sample.
In conclusion, while preliminary, our study supports the association between IED and inflammatory response and adds to the growing literature of the potential modulatory role of inflammatory pathways in aggressive behavior in humans. It also reveals novel pathways that may be associated with IED, including the neural and endocrine systems. By identifying biologically relevant DNA methylation signatures associated IED, we can discover potential biomarkers that can be used to facilitate the diagnosis of IED in humans.
Supplementary Material
Supplementary data are available at International Journal of Neuropsychopharmacology online.
Statement of Interest
Dr. Coccaro reports being a consultant to and being on the Scientific Advisory Boards of Azevan Pharmaceuticals, Inc. and of Avanir Pharmaceuticals, Inc., and being a current recipient of a grant award from the NIMH. Drs. Montalvo-Ortiz, Zhang, Chen, and Liu report no conflicts of interest regarding this work.
Supplementary Material
Acknowledgments
This work is supported by NIMH grant awards RO1MH60836, RO1MH80108, and RO1MH104673 (E.F.C.) and the Biological Sciences Training Program through 5T32 MH14276 (J.L.M.-O.). Finally, the research was made possible in part by the services of the NUSeq Core Facility, which is supported by the Northwestern University Center for Genetic Medicine, Feinberg School of Medicine, and Shared and Core Facilities of the University’s Office for Research.
References
- American Psychiatric Association (1994)Diagnostic and statistical manual of mental disorders, 4th ed American Psychiatric Association Press: Washington, DC. [Google Scholar]
- American Psychiatric Association (2013)Diagnostic and statistical manual of mental disorders: DSM-5, 5th ed American Psychiatric Press: Washington, DC. [Google Scholar]
- Buss AH, Perry M(1992)The aggression questionnaire. J Pers Soc Psychol 63:452–459. [DOI] [PubMed] [Google Scholar]
- Checknita D, Maussion G, Labonte B, Comai S, Tremblay RE, Vitaro F, Tirecki N, Bertazzo A, Gobbi G, Cote G, Turecki G (2015)Monoamine oxidase A gene promoter methylation and transcriptional downregulation in an offender population with antisocial personality disorder. Brit J Psychiatry 206:216–222. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Bergeman CS, McClearn GE(1993)Heritability of irritable impulsiveness: a study of twins reared together and apart. Psychiatry Res 48:229–242. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Bergeman CS, Kavoussi RJ, Seroczynski AD (1997a) Heritability of aggression and irritability: a twin study of the Buss-Durkee aggression scales in adult male subjects. Biol Psychiatry 41:273–284. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Berman ME, Kavoussi RJ (1997b) Assessment of life history of aggression: development and psychometric characteristics. Psychiatry Res 73:147–157. [DOI] [PubMed] [Google Scholar]
- Coccaro EF.(2006)Association of C-reactive protein elevation with trait aggression and hostility in personality disordered subjects: a pilot study. J Psychiatr Res 40:460–465. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, McCloskey MS, Fitzgerald DA, Phan KL(2007)Amygdala and orbitofrontal reactivity to social threat in individuals with impulsive aggression. Biol Psychiatry 62:168–178. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Noblett KL, McCloskey MS(2009)Attributional and emotional responses to socially ambiguous cues: validation of a new assessment of social/emotional information processing in healthy adults and impulsive aggressive patients. J Psychiatric Research 43:915–925. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Nayyer H, McCloskey MS (2012a) Personality disorder: not otherwise specified evidence of validity and consideration for DSM-5. Comprehensive Psychiatry 53:907–914. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Schmidt-Kaplan CA (2012b) Life history of impulsive behavior: development and validation of a new questionnaire. J Psychiatric Research 46:346–352. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Lee R, Coussons-Read M (2014a) Cerebrospinal fluid and plasma C-reactive protein and aggression in personality-disordered subjects: a pilot study. J Neural Transmission 122:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Lee R, Coussons-Read M (2014b) Elevated plasma inflammatory markers in individuals with intermittent explosive disorder and correlation with aggression in humans. JAMA Psychiatry 71:158–165. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Fanning JR, Phan KL, Lee R (2015a) Serotonin and impulsive aggression. CNS Spectrums 20:295–302. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Lee R, Coussons-Read M (2015b) Cerebrospinal fluid inflammatory cytokines and aggression in personality disordered subjects. Int J Neuropsychopharmacology 3:pyv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Lee R, McCloskey M, Csernansky JG, Wang L (2015c) Morphometric analysis of amygdla and hippocampus shape in impulsively aggressive and healthy control subjects. J Psychiatric Res 69:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Fanning J, Lee R (2016a) Development of a social emotional information processing assessment for adults (SEIP-Q). Aggress Behav 43:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Fanning JR, Keedy SK, Lee RJ (2016b) Social cognition in intermittent explosive disorder and aggression. J Psychiatric Res 83:140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Fitzgerald DA, Lee R, McCloskey MS, Phan KL (2016c) Fronto-limbic morphometric abnormalities in intermittent explosive disorder and aggression. Biol Psychiatry Clin Neurosci Neuroimaging 1:32–38. [DOI] [PubMed] [Google Scholar]
- Coccaro EF, Lee R, Fanning JR, Fuchs D, Goiny M, Erhardt S et al. (2016d) Tryptophan, kynurenine, and kynurenine metabolites: relationship to lifetime aggression and inflammatory markers in human subjects. Psychoneuroendocrinology 71:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Fanning J, Lee R(2017)Intermittent explosive disorder and substance use disorder: analysis of the national comorbidity study - replication sample. J Clinical Psychiatry 78:697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers H, Lee R, Keedy S, Phan KL, Coccaro EF(2015)Effects of escitalopram administration on face processing in intermittent explosive disorder: an fMRI study. Neuropsychopharmacology 41:590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresner E, Agam G, Gozes I(2011)Activity-dependent neuroprotective protein (ADNP) expression level is correlated with the expression of the sister protein ADNP2: deregulation in schizophrenia. Eur Neuropsychopharmacology 21:355–361. [DOI] [PubMed] [Google Scholar]
- First MB, Williams JBW, Gibbon M(2014)Structured clinical interview for DSM-5 patient edition (SCID-P) edn. American Psychiatric Press: Washington, DC. [Google Scholar]
- Guillemin C, Provencal N, Suderman M, Cote SM, Vitaro F, Hallett M et al. (2014)DNA methylation signature of childhood chronic physical aggression in T cells of both men and women. PloS One 9:e86822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus MR, Schafer A, Faller H, Csef H, Scheurlen M(2003)Psychiatric symptoms in patients with chronic hepatitis C receiving interferon alfa-2b therapy. J Clin Psychiatry 64:708–714. [DOI] [PubMed] [Google Scholar]
- Kushnir M, Dresner E, Mandel S, Gozes I(2008)Silencing of the ADNP-family member, ADNP2, results in changes in cellular viability under oxidative stress. J Neurochemistry 105:537–545. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ding J, Reynolds LM, Lohman K, Register TC, De La Fuente A et al. (2013)Methylomics of gene expression in human monocytes. Hum Mol Genet 22:5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GC , Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM et al. (2013)A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 18:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland AL, Prather AA, Petersen KL, Cohen S, Manuck SB(2008)Antagonistic characteristics are positively associated with inflammatory markers independently of trait negative emotionality. Brain Behav Immunity 22:753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey MS, Phan KL, Angstadt A, Fettich KC, Keedy S, Coccaro EF(2016)Amygdala hyperactivation to angry faces in intermittent explosive disorder. J Psychiatric Res 79:34–41. [DOI] [PubMed] [Google Scholar]
- McEachin RC, Chen H, Sartor MA, Saccone SF, Keller BJ, Prossin AR et al. (2010)A genetic network model of cellular responses to lithium treatment and cocaine abuse in bipolar disorder. BMC Syst Biol 4:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHuthison J, Gordon S, Schiff E(1998)Interferon alpha-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med 339:1485–1492. [DOI] [PubMed] [Google Scholar]
- Miles DR, Carey G(1997)Genetic and environmental architecture of human aggression. J Pers Soc Psychol 72:207–217. [DOI] [PubMed] [Google Scholar]
- Pfohl B, Blum N, Zimmerman M,and the University of Iowa Department of Psychiatry (1997)Structured interview for DSM Personality Disorder: SIDP. American Psychiatric Press: Washington DC. [Google Scholar]
- Provencal N, Suderman MJ, Caramaschi D, Wang D, Hallett M, Vitaro F et al. (2013a) Differential DNA methylation regions in cytokine and transcription factor genomic loci associate with childhood physical aggression. PloS One 8:e71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Vitaro F, Szyf M, Tremblay RE (2013b) Childhood chronic physical aggression associates with adult cytokine levels in plasma. PloS One 8:e69481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Guillemin C, Vitaro F, Cote SM, Hallett M et al. (2014)Association of childhood chronic physical aggression with a DNA methylation signature in adult human T cells. PloS One 9:e89839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Booij L, Tremblay RE(2015)The developmental origins of chronic physical aggression: biological pathways triggered by early life adversity. J Exp Biol 218:123–133. [DOI] [PubMed] [Google Scholar]
- Suarez EC.(2003)Joint effect of hostility and severity of depressive symptoms on plasma interleukin-6 concentration. Psychosom Med 65:523–527. [DOI] [PubMed] [Google Scholar]
- Suarez EC.(2004)C-reactive protein is associated with psychological risk factors of cardiovascular disease in apparently healthy adults. Psychosom Med 66:684–691. [DOI] [PubMed] [Google Scholar]
- van Dongen J, Nivard MG, Baselmans BM, Zilhão NR, Ligthart L, BIOS Consortium et al. (2015)Epigenome-wide association study of aggressive behavior. Twin Res Hum Genetics 186:686–698 [DOI] [PubMed] [Google Scholar]
- Wang D, Szyf M, Benkelfat C, Provencal N, Turecki G, Caramaschi D et al. (2012)Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PloS One 7:e39501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh MT, Coccaro EF, Jacobson KC(2010)Multivariate behavior genetic analyses of aggressive behavior subtypes. Behav Genet 40:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalcman SS, Siegel A(2006)The neurobiology of aggression and rage: role of cytokines. Brain Behav Immun 20:507–514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.