Abstract
Each new virus introduced into the human population could potentially spread and cause a worldwide epidemic. Thus, early quantification of epidemic spread is crucial. Real-time sequencing followed by Bayesian phylodynamic analysis has proven to be extremely informative in this respect. Bayesian phylodynamic analyses require a model to be chosen and prior distributions on model parameters to be specified. We study here how choices regarding the tree prior influence quantification of epidemic spread in an emerging epidemic by focusing on estimates of the parameters clock rate, tree height, and reproductive number in the currently ongoing Zika virus epidemic in the Americas. While parameter estimates are quite robust to reasonable variations in the model settings when studying the complete data set, it is impossible to obtain unequivocal estimates when reducing the data to local Zika epidemics in Brazil and Florida, USA. Beyond the empirical insights, this study highlights the conceptual differences between the so-called birth–death and coalescent tree priors: while sequence sampling times alone can strongly inform the tree height and reproductive number under a birth–death model, the coalescent tree height prior is typically only slightly influenced by this information. Such conceptual differences together with non-trivial interactions of different priors complicate proper interpretation of empirical results. Overall, our findings indicate that phylodynamic analyses of early viral spread data must be carried out with care as data sets may not necessarily be informative enough yet to provide estimates robust to prior settings. It is necessary to do a robustness check of these data sets by scanning several models and prior distributions. Only if the posterior distributions are robust to reasonable changes of the prior distribution, the parameter estimates can be trusted. Such robustness tests will help making real-time phylodynamic analyses of spreading epidemic more reliable in the future.
Keywords: tree height, substitution rate, tree prior, molecular epidemiology, start of epidemic
1. Introduction
In February 2016, the WHO declared the current Zika virus (ZIKV) epidemic ongoing in the Americas to be a public health emergency of international concern (World Health Organization 2016). This emergency level was reached because of a possible link between ZIKV infection and an increased number of microcephaly in newborns as well as between ZIKV infection and other neurological disorders such as the Guillian-Barré syndrome (Oehler et al. 2014; Pan American Health Organization and World Health Organization Regional Office for the Americas 2015; Ventura et al. 2016; Zika Situation Report 2016). Although the ZIKV public health emergency ended in November 2016, the ZIKV epidemic remains ‘a highly significant and long-term problem’ according to the WHO (World Health Organization 2016).
Bayesian phylodynamic approaches have proven very helpful in quantifying the spread of the ZIKV outbreak (Faria et al. 2016, 2017; Grubaugh et al. 2017; Metsky et al. 2017). Such analyses revealed that the current epidemic in the Americas most likely goes back to one Asian lineage and was introduced between May and December 2013 (Faria et al. 2016) first in Brazil, from where it spread to other countries in the Americas (Metsky et al. 2017). The latter hypothesis is confirmed by the observation that the epidemic in Florida, USA, seems to have started from several introductions, most of these introduced lineages clustering with Carribean ZIKV sequences, interspersed within the bigger cluster of Brazilian sequences (Grubaugh et al. 2017).
In an emerging epidemic, sequence data might be limited and may not have yet accumulated enough information in order to unequivocally quantify the phylodynamics, i.e. identify the underlying phylogenetic tree and the evolutionary and epidemiological parameters. In fact, the following two aspects play a role in the outcome of such analysis. First, the tree prior may be very important when inferring the phylogeny using a Bayesian framework. In particular, the impact of treating sampling times as given a priori vs. as part of the data has not been investigated in a thorough analysis. Second, the interplay of the tree height and the clock rate prior in the case of limited empirical data may impact the tree inference in an unpredictable fashion. This effect too, has not been explored in detail so far (but see Möller (2017) for a thorough simulation study).
In the present work, we explore the relevance of these two points for the estimation of the tree height based on data from the ZIKV outbreak in the Americas, and compare the results obtained with the Bayesian phylogenetics tool BEAST 2 to simpler regression-based and least squares-based techniques for tree height inference. The tree height can be used to estimate the start of the epidemic. A robust estimate of when the epidemic started is in particular necessary to investigate whether the increase of microcephaly cases in Brazil coincides with the ongoing ZIKV epidemic. Furthermore, we discuss the appropriateness of interpreting the reproductive number—a key epidemiological quantity describing the intensity of epidemic spread—which is co-estimated with the tree height using the so-called birth–death model.
For the remainder of this section, we provide some background regarding phylodynamics, introduce common tree priors, and discuss their conceptual differences. These insights will be important for interpreting the results presented in this paper.
1.1 Phylodynamics and tree priors
The main idea of phylodynamic approaches is to quantify population dynamic parameters based on the phylogenetic tree obtained from sequencing data (Grenfell et al. 2004). In practice, when using a Bayesian framework, the phylogenetic tree is not fixed either, but co-inferred with the population dynamic as well as the evolutionary parameters based on the sequencing data (Drummond and Rambaut 2007; du Plessis and Stadler 2015). More formally, the Bayesian phylodynamic approach aims at inferring the posterior distribution where par is the vector of all parameters of the population dynamic and evolutionary models, seq is the sequencing data, and data denotes other potential sources of data. We can re-write,
The normalizing constant cannot be calculated directly, and thus Markov chain Monte Carlo methods are employed to sample from the posterior distribution . Typically, we assume that seq is independent of data such that . The distribution is referred to as the phylogenetic likelihood and is specified by an evolutionary model, and the distribution P[tree, data|par] is referred to as the phylodynamic likelihood and is specified by a population dynamic model. In the application of phylodynamics to epidemiology, the sequencing data comes from pathogens obtained from different hosts, and the population dynamic model is an epidemiological model. The term is the prior distribution on the parameters of the population dynamic and evolutionary models.
If the sequence data contains a strong phylogenetic signal, the inferred tree topology will be independent of the prior specifications, i.e. the tree topology will be purely determined by differences in the sequences. In addition to the tree topology, we are typically interested in the tree height, i.e. the age of the most recent common ancestor (MRCA) of all samples, and in all other branching times of the tree. The clock rate is the parameter of the evolutionary model that specifies how many substitutions a sequence accumulates per calendar time unit. It thereby translates the branches in the tree from units of substitutions to units of calendar time. If all sequences are sampled at the same time point, we can only estimate the product of the tree height and the clock rate. If the data is sampled through time, i.e. the tips of the tree are at different time points, we can in principle tease the two parameters apart (Korber et al. 2000). However, if there is not enough temporal signal in the data, it might not be possible to separate the clock rate from the tree height robustly. Little temporal signal will result in high sensitivity of the tree height (and all tree branch lengths) to prior assumptions on the clock rate and the tree height.
While we can specify a clock rate prior directly through any probability distribution, we can only indirectly specify the tree height prior through the population dynamic model. In phylodynamics, each population dynamic model is described by a so-called tree prior. There are two classes of tree priors that have proven especially useful: the coalescent (Kingman 1982; Drummond et al. 2005), and the birth–death process (Nee 2006; Stadler 2010). In the following paragraphs, we describe their main assumptions and point out why the tree height cannot be directly controlled through setting of the model priors.
The coalescent describes the dynamics of the population giving rise to the tree backward in time, i.e. going from the tips to the root. It is parameterized by the effective population size. The sampling times of sequences are assumed to be given a priori. Given such a priori sampling times samp and an effective population size Ne as a parameter par, the coalescent induces a distribution of phylogenetic trees and a distribution of tree heights,
Given the sampling times, we therefore indirectly control the tree height via the effective population size.
Birth–death models describe the dynamics of a population forward in time. They are parameterized by the birth (transmission) rate λ, the death (become uninfectious) rate δ, the sampling probability p, and the time of the start of the population (outbreak; also called origin time) T. Note that the reproductive number, a key epidemiological quantity (Anderson and May 1991), can be directly extracted from these parameters by simply dividing the birth rate by the death rate. The birth–death models give rise to a distribution of phylogenetic trees and in particular a distribution of sampling times and tree heights,
Thus, the upper limit of the tree height can be controlled, as it is strictly smaller than the origin time T, while the precise tree height is influenced by the birth, death and sampling parameters.
Notice that while the coalescent conditions on the sampling times when the tree is induced by the population size parameter, in the birth–death model the sampling times along with the tree are induced by the birth–death parameters. This different conceptual usage of the sampling times samp under the two approaches has important implications towards assessing the amount of information in the data regarding the tree height with popular Bayesian phylodynamics softwares. A Bayesian phylogenetic analysis sampling from the posterior distribution of trees tree and parameters par using the coalescent conditions on the sampling times samp:
where is the probability density of a coalescent tree tree given the sampling times samp. On the other hand, the birth–death model uses the number of samples together with the associated sampling times samp and the sequence alignment seq as data:
where is the probability density of the sampling times samp together with the birth–death tree tree.
Common practice for investigating the signal in the data is to run the Bayesian method ‘under the prior’ and then compare the results to those obtained from the full Bayesian analysis. In the strict sense, running an analysis ‘under the prior’ would mean running the analysis when all data is ignored. With the coalescent approach, this means that sequencing data is ignored, while the number and the time of sequence samples is used, as it is not considered to be a part of the data but rather a priori information. In contrast, with the birth–death model, the sequences, the number of sequences as well as the sequence sampling times are ignored when running the Bayesian method ‘under the prior’. This stems from the fact that the birth–death model parameters induce a distribution of the number of sequences as well as the sequence sampling times, meaning any information we have in that respect during the analysis is data.
Many users perform phylodynamic analyses with the software package BEAST 2 (Bouckaert et al. 2014). The graphical user interface BEAUti allows the specification of model priors and input data. It also allows the user to specify if ‘sampling from prior’ should be performed. If this option is chosen, the sequencing data, but not the sampling times, are ignored. For the coalescent models this means that one obtains the prior distributions. However, for the birth–death models this means that one obtains a posterior distribution of trees and parameters using sampling times data, while only the sequencing data is ignored. The analysis with the birth–death model when opting for ‘sampling from prior’ in BEAST 2 is therefore not equivalent to the analysis ‘under the prior’ in the usual Bayesian sense.
2. Methods
2.1 Data sets
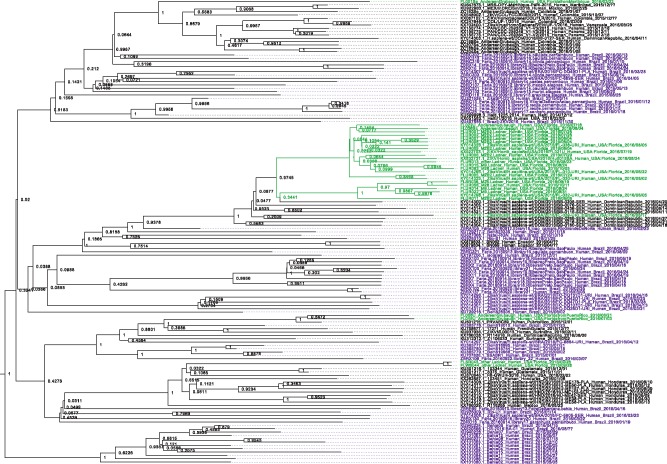
On 3 November 2016 we gathered 252 full genome ZIKV consensus sequences. These consensus sequences stem from various sources: we obtained 185 sequences from GenBank, 33 sequences from the Zibra project (https://github.com/zibraproject/zibraproject.github.io/tree/master/data/consensus), 17 sequences from the Andersen et al. github page (https://github.com/andersen-lab/zika-florida/tree/master/consensus_sequences), and 17 sequences from Ladner et al. github page (https://github.com/jtladner/ZIKA_Florida/tree/master/sequences). From this set, we removed sequences isolated before 2014, sequences isolated from other organisms than humans, sequences with missing date of isolation, i.e. year and/or month information missing, sequences resulting from vertical (mother to child) transmissions and sequences which appeared to be duplicates, i.e. sequences that seemed to have been isolated from the same individual on the same date but had different passaging histories or were sequenced by different labs. This resulted in a set of 139 sequences. We refer to this data set as the ‘ALL’ data set, because it includes sequences from different parts of the Americas. We also separately analysed a subset (sixty-seven sequences) of this full data set which only contained the sequences coming from Brazil (abbreviated as BRAZIL). Additionally, we separately analysed a set of sequences from Florida, USA (twenty-three sequences, abbreviated USA). These sequences formed a monophyletic cluster within the maximum clade credibility (MCC) trees of the ‘ALL’ data set upon removal of one sequence from the Dominican Republic (Fig. 1 and Supplementary Figs S1 and S2, see figure legend for a description of the model settings with which we obtained the MCC tree). For the sake of simplicity, we refer to this cluster as monophyletic in the following.
Figure 1.
MCC tree of the 139 ZIKV sequences included in this study. The posterior clade support is displayed at each branching point. The names of all virus sequences that were isolated in Florida, USA, are highlighted in green and all sequences from Brazil are highlighted in magenta. The cluster of sequences highlighted in green represents the twenty-three strains of the USA data set that form one monophyletic cluster upon exclusion of one non-USA sequence. The MCC tree was obtained using the BDSKY model in BEAST 2 with δ = 18.25, three intervals for Re and a relaxed clock (see Supplementary Figs S1 and S2 for other model parametrizations).
2.2 Phylogenetic and phylodynamic analyses
We aligned the sequences using MUSCLE v3.8.31 (Edgar 2004). We constructed trees with a maximum likelihood (ML) method PhyML v3.1 (Guindon et al. 2010) under the HKY substitution model, estimating the equilibrium frequencies, and starting the ML tree search using ten different random seeds. We assessed the clock-like behaviour of the data using TempEst v1.5 (Rambaut et al. 2016), by looking at the correlation of the sampling times with the root-to-tip divergence, optimising the position of the root of the ML tree by maximising the correlation metric. From the ML trees, we also estimated the time of the MRCA in calendar time units using LSD v0.3 (To et al. 2015), using options -c (constrained mode, i.e. imposing that the node is always older than any of its descendants) and -r as (searching for root node on all branches, using constraints). Finally, we analysed the sequences using the Bayesian phylogenetic software tool BEAST v2.4.2 (Bouckaert et al. 2014). For all analyses, the tip dates were fixed to the dates found in the sequence annotation. For those sequences with the day-of-sampling information missing, we simply set the date to be year/month/15 (total of 6/139 sequences). The results are consistent with those obtained from the analyses where the samples with incomplete sampling date information were excluded.
For the Bayesian analyses, we need to specify two modelling components, namely the sequence evolution model and the tree prior model, which we specified as follows.
2.2.1 Sequence evolution model settings
For all analyses we chose the HKY substitution model as the sequence evolution model with substitution rate fixed to 1, prior and 0 as the lower limit for the κ parameter, and estimated base frequencies and Γ-distributed variation between sites, using four categories, the shape parameter prior being set to Exp(1). We allowed for between-branch rate variation using a relaxed clock model (Drummond et al. 2006). We used the log-normal relaxed clock model with ucldMean prior being in real space. Furthermore, the ucldStdev prior was set to with 0 as the lower limit. This resulted in an effective prior on the clock model’s rate.mean to have mean of 0.001 and median of 0.0001. This prior was chosen to reflect the estimates of previous studies, where the substitution rate was estimated to be between subst/site/year (note that this unit corresponds to ‘substitutions per site and year’ or subst × site−1 × year–1) (Faria et al. 2016) but was suspected to vary between 2.6×10–4 and 4.4×10–3 subst/site/year depending on the host organism (virological.org 2016). We also performed analyses using a strict clock with prior for the rate parameter. Note that in the Bayesian framework, the product of the substitution rate from the substitution model and the clock rate from the clock model is the overall substitution rate of the process. Since we fixed the substitution rate in the substitution model to 1, and we only estimate the clock rate, the clock rate we report effectively refers to the overall substitution rate.
2.2.2 Tree prior model settings
For the tree prior, we used the birth–death skyline model (Stadler et al. 2013) and the coalescent Bayesian skyline plot (Drummond et al. 2005). The birth–death skyline model is a birth–death model with constant parameters within intervals through time while parameters may change across intervals. We used one interval for the become uninfectious rate (i.e. the ‘death’ rate) and fixed its value to 14.26, 18.25, or 23.40 (corresponding to the inverse of the lower 95-percentile, mean and upper 95-percentile of the estimates of the mean ZIKV generation time obtained by Ferguson et al. (2016)). The become uninfectious rate is the inverse of the time period of being infectious. All the values of the become uninfectious rate here are in units per year. Furthermore, we used five equidistant intervals between the first and last sample for the sampling probability. For each interval we set the prior distribution to Beta(1, 700). The sampling probability before the first sample was set to 0. For the effective reproductive number, Re (which is the ‘birth’ rate divided by the ‘death’ rate), we used three, four, or six equidistant intervals between the root and the last sample with LN(0, 1) prior distribution and 0 as the lower and 50 as the upper bound. For the origin parameter, the prior was in real space and lower and upper bounds of 0 and 15, respectively. The coalescent Bayesian skyline model is based on a coalescent with constant effective population sizes within intervals through time while the effective population size may change across intervals. In the coalescent Bayesian skyline plot we set the prior for the effective population size to a Markov-chained distribution, i.e. the prior value of the parameter in the interval follows a Gamma distribution with mean being set to the value in the previous interval. We set the prior distribution of the population size in the first interval to . We set the number of intervals for the effective population size and group size to 3, 4, or 6.
In summary, for the three ZIKV data sets, we performed analyses under relaxed and strict clock models, using the coalescent and birth–death models with a range of settings as summarized in Table 1. We ran the chain for 109 steps both with and without sequencing data (sampling from prior). We sampled parameters every 104 steps and trees every 106 steps. The log files were inspected in Tracer (Rambaut et al. 2014). All parameters in all the runs mixed well (ESS >200) with one exception, namely strict clock, all data, birth–death (BD): 4×Re ∼ LN(0, 1), without sequences, in Supplementary Fig. S3B (tree height and origin had ESS of only 159 and 187, respectively).
Table 1.
Overview of models used in this study.
| Phylodynamic model | Parameter | Value | Evolutionary model | Clock model |
|---|---|---|---|---|
| Birth–death skyline model with serial sampling (BD) | Effective reproductive number (Re) | Estimated; 3, 4, 6 intervals (3 × Re, 4 × Re, 6 × Re) with LN prior with various parameterizations | HKY+Γ | Relaxed clock |
| Become uninfectious rate (δ) |
|
Strict clock | ||
| Sampling probability (P) | Estimated; with prior Beta(1, 700) | |||
| Coalescent Bayesian skyline plot (Coal) | Effective population size (Ne) |
|
HKY+Γ | Relaxed clock Strict clock |
The abbreviations used in Figs 3 and 4 and Supplementary Figs S3 and S4 refer to this scheme. For example, BD: 3×Re∼LN(0, 1), refers to the birth–death skyline model with serial sampling which allows for three intervals for Re, each with LN(0, 1) distributed prior, and the become uninfectious rate δ = 18.25.
For the analyses investigating the evolution of Re over time in Florida, USA, we used the sequences of the Florida monophyletic cluster. We fixed the ucldMean parameter to subst/site/year, which was the mean and the median estimate of the clock rate (rate.mean parameter in BEAST 2) we obtained when analysing the ALL data set without Florida, USA sequences (see Supplementary Fig. S4, w/o Florida). Furthermore, in these analyses we varied the number of intervals for Re to be 3, 4, or 6 and the number of intervals for sampling probability p to be 1 or 5.
Post-processing of Bayesian analyses was done by first discarding the initial 10% of the MCMC samples as burn-in. Results were then analysed in R (R Core Team 2013) using custom-made scripts with the R packages boa (Smith 2007) for calculating the highest posterior density (HPD) intervals, and RColorBrewer (Neuwirth 2014) for plotting. To obtain the lineages-through time (LTT) plot, the R package ape (Paradis, Claude, and Strimmer 2004) was used in combination with the function LTT.plot.gen in the R package TreeSim (Stadler 2011). To extract the node (tree) height from the tree necessary for plotting the LTT along with the Re plots, we used R package phytools (Revell 2012). We used the R package beanplot for plotting the probability densities of time of the MRCA and clock rate estimates (Kampstra 2008). Lastly, MCC trees were obtained using TreeAnnotator v2.3.0 (Rambaut and Drummond 2015), removing initial 10% of samples as burn-in, setting posterior probability limit to 0.1 and reconstructing node height based on Common Ancestor heights criterium. The MCC trees were visualised using FigTree v1.4.0 (Rambaut 2012). The BEAST 2 source files and analysis scripts can be found in the Supplementary Materials and Methods zip file.
3. Results
3.1 Robustness analysis of the tree height and the clock rate
First, we set out to estimate the start of the epidemic and the clock rate of the ZIKV epidemic for (1) the complete data set containing 139 ZIKV sequences sampled in the Americas (ALL), (2) 67 sequences sampled only in Brazil (BRAZIL), and (3) 23 sequences belonging to the locally contained outbreak in Florida, USA (USA). All data sets contain sequentially sampled sequences and therefore should in principle allow for calibration of the molecular clock (i.e. estimating the clock rate) and reliable estimation of the tree height. We used three methods for estimation of the clock rate and the tree height: a regression model correlating sampling time to the root-to-tip divergence (PhyML combined with TempEst), a least squares-based approach (PhyML combined with LSD) and Bayesian phylodynamic approaches (BEAST 2). For the latter, we used two different models, the birth–death skyline model (Stadler et al. 2013) and the coalescent Bayesian skyline plot (Drummond et al. 2005).
The start of the epidemic is parameterized in the BD model as T, and the tree height is the age of the MRCA. As in the Bayesian coalescent (Coal) framework and the two non-Bayesian frameworks, only the tree height rather than T is estimated, we compare the estimated tree height, , under all methods, using it as an approximation for the start of the epidemic, T. In what follows, we will refer to the ‘time of the MRCA’ (tMRCA) as the ‘date when the epidemic started’. This quantity is derived by subtracting the tree height from the date of the most recent tip in the data set. The rationale for comparing non-Bayesian to Bayesian estimates of the tree height (or of the date when the epidemic started) and the clock rate is to assess the amount of information available in the data without adding prior information.
3.1.1 Regression analysis of the tree height and the clock rate
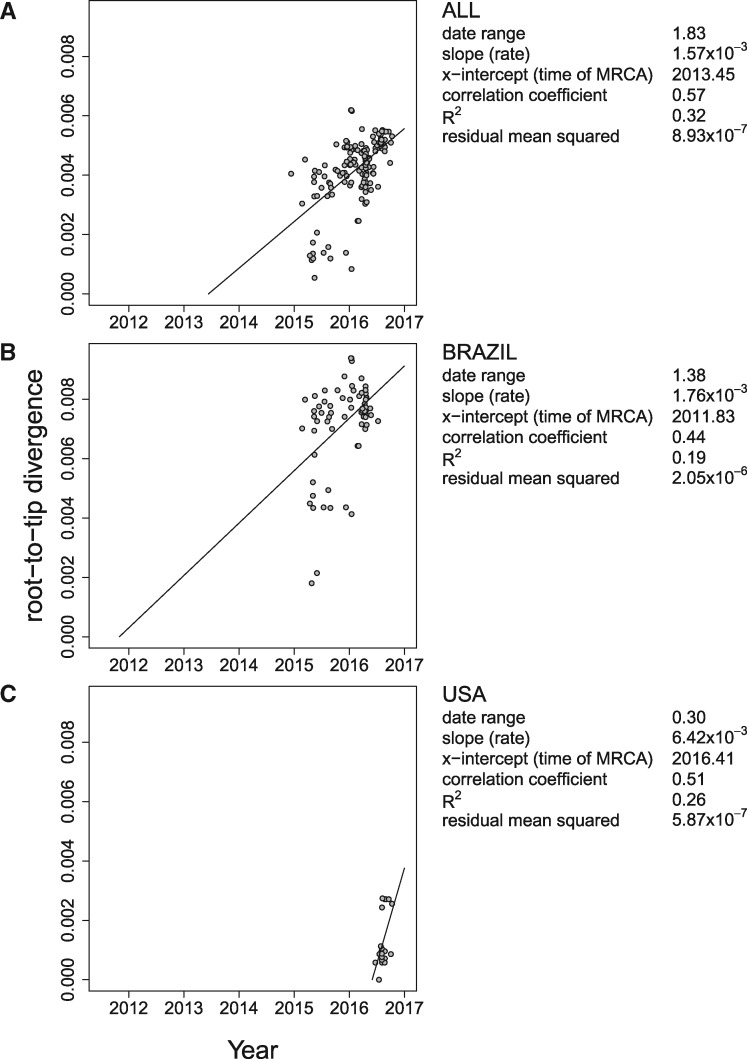
To obtain the estimate of the phylogenetic tree, we applied the ML method PhyML v3.1 (Guindon et al. 2010) to the sequences. We assessed the clock signal in the data using TempEst v1.5 (Rambaut et al. 2016) (Fig. 2). All three data sets show a positive correlation between genetic divergence (root-to-tip divergence) and sampling time, yielding estimates of clock rate of 1.57 × 10–3 subst/site/year for the ALL data set, 1.76 × 10–3 subst/site/year for the BRAZIL data set, and 6.42 × 10–3 subst/site/year for the USA data set. The ALL data set exhibits the strongest association between the genetic divergence and the sampling time (). The BRAZIL and USA data sets show more diffuse patterns with lower R2-values: BRAZIL: , USA: . The samples in the ALL and BRAZIL data sets range over more than 1.37 years whereas the sequences in the USA data set were only collected during less than four months. The x-intercept of the regression line between the calendar time (x-axis) and the root-to-tip divergence (y-axis) can be used as an approximation of the date when the epidemic started. This way, the start of the epidemic in the Americas was estimated to be 12 June 2013. The Brazilian epidemic was estimated to start 1.6 years earlier, i.e. 30 October 2011. The epidemic in Florida was estimated to have started on 31 May 2016, i.e. three years after the tMRCA of the ALL data set (see Fig. 2). These results indicate that the clock signal may not be strong enough in (some of) the data sets, as the ALL data set cannot have a younger tMRCA than any of its sub-data sets. In addition, the estimates for the ALL and USA data sets are very close to the earlier obtained estimates of the introduction of ZIKV into the Americas; however, the estimate for the BRAZIL data set seems to be much older than estimated before (Faria et al. 2016, 2017; Grubaugh et al. 2017).
Figure 2.
TempEst estimates of tree heights and clock rates. The tree height and the clock rate (i.e. slope) estimates obtained using TempEst for the three data sets: (A) the complete data set (ALL), (B) the sequences isolated in Brazil (BRAZIL), and (C) the (monophyletic cluster of) sequences isolated in Florida (USA). The tree for each data set was reconstructed using the ML method.
3.1.2 Least squares analysis of the tree height and the clock rate
The ML tree was also analysed with the least squares dating tool LSD v0.3 (To et al. 2015), in order to estimate the tree height and the clock rate in the ZIKV epidemic. The inferred tree height for the ALL data set resulted in the estimated date when the epidemic started to be 8 January 2012. For the Brazilian data set, the start of the epidemic was estimated to be 28 July 2010. The oldest tree and thus the most ancient start of the epidemic, dating to 18 October 1980 was estimated for the USA data set. The clock rate estimates for the ALL, BRAZIL, and USA data sets were 5.21 × 10–4, 5.62 × 10–4, and 3.46 × 10–5 subst/site/year, respectively. Again, these results indicate that the clock signal is weak in (some of) our data sets.
3.1.3 Bayesian phylodynamic analysis of the tree height and the clock rate
In the next step, we used phylodynamic analysis to overcome the problem of point estimates imposed by PhyML combined with TempEst and LSD. In a nutshell, we used two different models, the birth–death skyline model (Stadler et al. 2013) and the coalescent Bayesian skyline plot (Drummond et al. 2005). The term ‘skyline’ refers to the parameters (birth, death, sampling, or effective population size) changing in a piecewise constant fashion through time. As laid out in the introduction, the two models differ conceptually in how data is used, and as a common practice, the models are run without sequence data as well as with all sequence data to determine how much information the sequence data contains. Therefore, we will first explain the estimation of the tree height and the clock rate without sequences and then include the sequence data.
Since in the BD model the effective reproductive number, the become uninfectious rate, and the sampling probability are simultaneously unidentifiable (Stadler et al. 2013; Boskova, Bonhoeffer, and Stadler 2014), we fixed the become uninfectious rate in all analyses. Ferguson et al. (2016) estimated the inverse of this rate, i.e. the mean ZIKV generation time with its lower and upper 95-percentile. For our main analyses, we used their mean estimate translating to the become uninfectious rate of 18.25, and for the Supplementary analyses we used the upper 95-percentile and the lower 95-percentile of the Ferguson et al. estimates, i.e. we set the become uninfectious rate to and 23.40, respectively.
3.1.3.1 What can we learn about the tree height and the clock rate when ignoring the sequencing data?
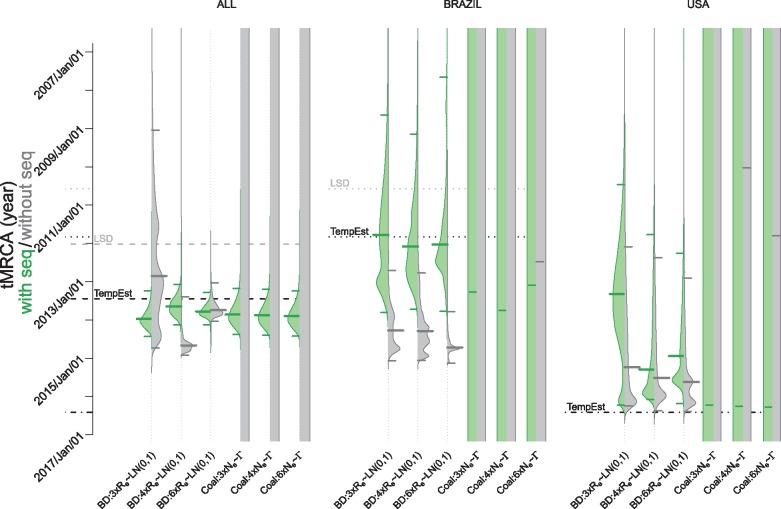
When analysing the data sets using the BD model with information on sampling times and number of samples only, i.e. ignoring the sequence data, we obtain very peaked tree height distributions (gray distributions in Fig. 3). Thus, the number of samples and sampling times contain a lot of information regarding the tree height. The medians of these distributions depend on different model specifications. When analysing the data sets using the Coal model ignoring the sequence data, i.e. running the analysis under the prior, the tree height distributions are very wide as expected since the sampling times are conditioned upon under the coalescent (gray distributions in Fig. 3).
Figure 3.
The effect of addition of sequence data on the tMRCA estimates in the Bayesian analysis. The probability distribution of the estimates of the tMRCA resulting from the analysis with (green) and without (gray) sequences is shown. The figure shows estimates obtained under various model assumptions (labels on the x-axis summarize the models as explained in Table 1) and the three different data sets (header). The median date of MRCA is indicated with a thick solid line and the 95% HPD intervals are marked with thin solid lines. The black dashed, dotted, and dashed-dotted lines represent the tMRCA estimates based on the TempEst analysis (Fig. 2) for the ALL, BRAZIL, and USA data sets, respectively. The gray dashed, dotted, and dashed-dotted lines represent the tMRCA of the LSD estimates for the ALL, BRAZIL, and USA data sets, respectively. The become uninfectious rate in the BD model is set to δ = 18.25, which is the mean estimate in Ferguson et al. (2016). Notice that the median estimates of the tMRCA for the BRAZIL and USA data sets analysed with the Coal model are beyond the limits of the figure, so we state the estimated tree heights below. For the BRAZIL data set, the median tree height estimate of distributions resulting from analyses without sequence data for Coal: 3 × Ne ∼Γ is 1.8 × 106 years, for Coal: 4 × Ne ∼Γ is 1.0 × 106 years and for Coal: 6 × Ne ∼Γ is 3.5 × 105 years. The median tree height estimate of the distribution when sequence data is included for Coal: 3 × Ne ∼Γ is 1026.7 years, for Coal: 4 × Ne ∼Γ is 960.1 years, and for Coal: 6 × Ne ∼Γ is 1039.8 years. For the USA data set, the median tree height estimate of distributions resulting from analyses without sequence data for Coal: 3 × Ne ∼Γ is 2.0 × 106 years, for Coal: 4 × Ne ∼Γ is 1.2 × 106 years, and for Coal: 6 × Ne ∼Γ is 5.3 × 105 years. The median tree height estimate of the distribution when sequence data is included for Coal: 3 × Ne ∼Γ is 460.8 years, for Coal: 4 × Ne ∼Γ is 449.1 years, and for Coal: 6 × Ne ∼Γ is 443.5 years.
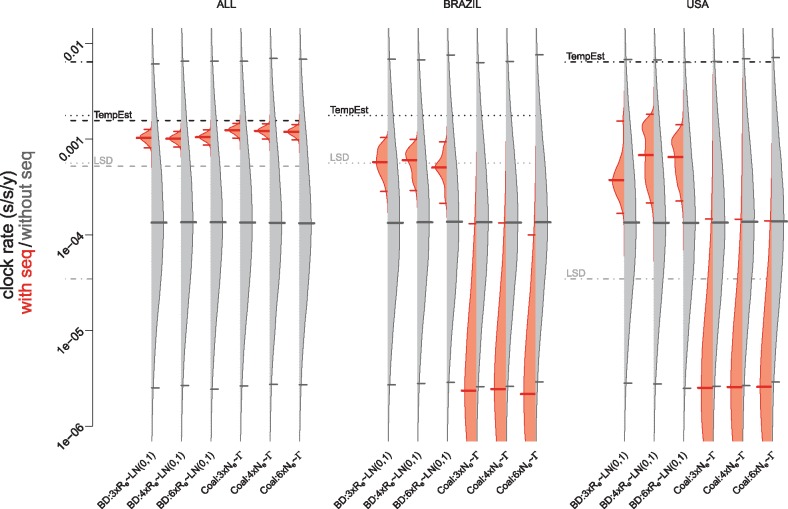
We performed all analyses shown in Fig. 3 using a relaxed molecular clock that allows for variation of the clock rates between the different branches of the phylogenetic tree (Drummond et al. 2006). The clock rate for the relaxed clock model is defined as the mean clock rate averaged over all branches (in BEAST 2, this is the rate.mean parameter). Ignoring the sequences, we obtain the chosen prior distribution for the clock rate under both the BD and the Coal framework as expected (gray distributions in Fig. 4).
Figure 4.
The effect of addition of sequence data on the clock rate estimates in the Bayesian analysis. The probability distribution of the clock rate (rate.mean parameter) estimates resulting from the analysis with (red) and without (gray) sequences is shown. The figure shows estimates obtained under various model assumptions (labels on the x-axis summarize the models as explained in Table 1) and the three different data sets (header). The median clock rate is indicated with a thick solid line and the 95% HPD interval marked with thin solid lines. The black dashed, dotted, and dashed-dotted lines represent the clock rate estimates based on the TempEst analysis (Fig. 2) for the ALL, BRAZIL, and USA data sets, respectively. The gray dashed, dotted, and dashed-dotted lines represent the clock rate of the LSD estimates for the ALL, BRAZIL, and USA data sets, respectively. The become uninfectious rate in the BD model is set to δ = 18.25, which is the mean estimate in Ferguson et al. (2016). The clock rate displayed is in units of s/s/y, i.e. subst/site/year.
3.1.3.2 How much additional information do the sequences contain concerning the tree height and the clock rate?
When performing phylogenetic analyses for the ALL data set including the sequence data, we obtain relatively consistent estimates of the median tree heights and clock rates in all models and model specifications (thick green lines in Fig. 3 and thick red lines in Fig. 4, respectively). The median estimates of the tree heights translate to the estimated start of the epidemic ranging from 25 August 2013 to 22 December 2013. The range of median estimates of the clock rate is subst/site/year.
In the BD analyses with various numbers of intervals for the effective reproductive number Re, all 95% HPD intervals of the tree height include the tree height estimate obtained by TempEst (12 June 2013)—Fig. 3. This finding is robust also when we assume a slower become uninfectious rate, but not for other variations of priors for BD model parameters that we tested, i.e. when assuming a faster become uninfectious rate or when setting a strong prior on very large or an unrealistically low Re in the BD model (Supplementary Fig. S3A, BD: 3×Re∼LN(2, 0.2), δ = 18.25 and BD: 3×Re∼LN(–2, 0.2), δ = 18.25). In the Coal analyses with various numbers of intervals for the effective population size Ne, all 95% HPD intervals of the tree height include the tree height estimate obtained by TempEst (12 June 2013). This finding is robust for different priors on the effective population size (Supplementary Fig. S3A). Only an extremely small coalescent population size priors in the Coal model (Supplementary Fig. S3A, Coal: 3 × Ne ∼LN(–2, 0.2)) produce HPDs not including the TempEst estimates.
Neither the BD nor the Coal 95% HPD intervals of the clock rate estimates contain the TempEst estimate of 1.57 × 10–3 subst/site/year (Fig. 4), although they are in the same order of magnitude (10−3 subst/site/year). This is consistent for all models (Supplementary Fig. S4A). The median clock rates estimated in a Bayesian framework are consistently lower than the TempEst estimate (Fig. 4 and Supplementary Fig. S4A). Only a prior for extremely small coalescent population size pushes the clock rate estimate to a higher rate (Supplementary Fig. S4A, Coal: 3 × Ne ∼LN(–2, 0.2)).
None of the HPD intervals contain the estimates of the tree height and the clock rate provided by LSD for any of the prior settings. The LSD clock rate estimates are consistently lower than the BD and the Coal estimates. The LSD tree height estimates are always above the BD/Coal tree height estimates (see Figs 3 and 4, and Supplementary Figs S3 and S4).
In contrast to the ALL data set, the analysis of the BRAZIL and USA data sets including the sequence data leads to distributions of the tree height and the clock rate that varied strongly amongst the models (Figs 3 and 4 and Supplementary Figs S3A and S4A). In particular, the 95% HPD intervals of the tree height distributions for the BRAZIL and the USA data sets are very broad (Fig. 3). For the Coal model, the posterior distribution remains very flat (i.e. does not change much from the prior). These results indicate that the BRAZIL and USA data sets gathered in November 2016 do not contain a strong enough signal to ultimately estimate the tree height reliably using phylodynamic analysis.
When analysing the BRAZIL and USA data sets separately, we obtain very different estimates for the clock rate from the BD models and the Coal models (Fig. 4). In the BD models, the sequence data is able to bring the clock rate estimates closer to the TempEst estimates although the posterior distributions remain quite diffuse. We do not see a consistent pattern of the clock rate estimates getting closer to the LSD estimates however. Adding sequence data to the Coal models leads to clock rate estimates that are even lower than expected from the prior (Fig. 4) and thus further away from the TempEst and LSD estimates.
In our main analyses we used the relaxed clock model. Further, we repeated the analyses with a strict clock model (Supplementary Figs S3B,C and S4B,C). Employing the strict clock models, we estimated the clock rates and tree heights to be very similar to those inferred by the relaxed clock models.
From our analyses we conclude that the signal contained in the ALL data set is sufficient to relatively consistently estimate the tree height and the clock rate using Bayesian methods, if the priors on the tree height are not too strongly biased towards extremely early or late introductions. Therefore, one can date the introduction of ZIKV into the Americas around late 2013. The posterior distributions of the time of introduction are in agreement with the TempEst analyses. It remains to be investigated though why LSD produce outliers. However, the BRAZIL and the USA data sets do not contain enough information to perform reliable estimation of tree height and clock rate parameters of these sub-epidemics.
Through the inclusion of the sequence data in the analysis, the clock rate and the tree height priors are allowed to interact. If the data contains enough signal, the clock rate and the tree height can be teased apart. However, if the data has a weak signal for calibrating the clock, then both the clock rate and the tree height estimates may be biased, and the nature of this bias would be influenced by the interaction of the two prior distributions.
If there is very little signal in the data for separation of the clock rate and the tree height, only their product can be estimated reliably. The median of the product of the clock rate and the tree height for the BD analyses with three intervals for Re for the ALL data set is 0.00291 (95% HPD=(0.00250, 0.00335)), for BRAZIL 0.00270 (95% HPD=(0.00192, 0.00347)) and for USA 0.00127 (95% HPD=(0.00081, 0.00170)). For the Coal analyses with three intervals for Ne, we obtain following median estimates of this product: ALL 0.00362 (95% HPD=(0.00299, 0.00441)), BRAZIL 0.00239 (95% HPD=(0.00157, 0.00360)), and USA 0.00117 (95% HPD=(0.00079, 0.00163)). For both BD and Coal models, the ALL data set has the highest product, followed by BRAZIL and finally USA. This ordering makes sense: the full data set has an underlying tree as old as, or older than, the tree obtained from any subset of the full data set. Assuming all the data sets share the same clock rate, the product of tree height and clock rate should be largest for the ALL data set. Also, the 95% HPD intervals of this product overlap for the Coal and the BD analyses in all three data sets.
Having in mind that we can estimate this product from our data, we can now understand better the inconsistencies obtained when aiming to separate the clock rate and the tree height using sequencing data. We illustrate the reason for the inconsistencies using the BRAZIL data set. The BD: 3 × Re model produces a peaked estimate of the tree height when the tree prior is combined with the sampling times only (gray distribution in Fig. 3). The median estimate of ≈ 2.2 years (start of epidemic ≈ April 2014) is close to the previously reported estimates of the start of the ZIKV epidemic in the Americas (Faria et al. 2016). So why does adding sequence data shift the estimated date of when the epidemic started further into the past? The median estimate of the product of the clock rate and the tree height is 0.00270 subst/site. If the true clock rate was subst/site/year as estimated by Faria et al. (2016), the tree height would be 2.7 years. Given the last sample in this particular data set was isolated on 10 July 2016, the start of the epidemic would then be ( years) ≈ October 2013. However, our clock rate prior has a median of the rate.mean parameter set to 1.3 × 10–4 subst/site/year, which, when combined with the estimated product of the clock rate and the tree height, would lead to tree height estimates of years, i.e. the start of the epidemic would be ≈ October 1995. Thus, if the sequencing data does not contain enough information to calibrate the clock to the expected 1 × 10–3 subst/site/year, the inferred tree height will increase, i.e. the tMRCA will be pulled up from April 2014 and beyond October 2013, upon inclusion of the sequence data. This insight can explain our large tree height (median estimate of 4.7 years, i.e. the start of the epidemic being ≈ October 2011), and the slow posterior clock rate (median estimate of the rate.mean parameter being 5.8 × 10–4 subst/site/year which is much slower than estimates by other studies (Faria et al. 2016)). The situation for the other prior settings for the BRAZIL and USA data set analysed with the BD model is analogous to what we have just described for the BRAZIL BD: 3 × Re model.
When the BRAZIL data set is analysed under the Coal: 3 × Ne model, the median estimate of the product of the clock rate and the tree height is 0.00239 subst/site, which is very similar to the one obtained with the BD model. Again, if we assume the true clock rate to be subst/site/year, as estimated before (Faria et al. 2016), the tree height would be 2.4 years and the start of the epidemic would be estimated to be in ≈ February 2014. Our clock rate prior has a median of the rate.mean parameter set to 1.4 × 10–4 subst/site/year, leading to estimated median tree height of 17.07 years, i.e. a start of the epidemic being in ≈ June 1999. However, unlike in the BD model, the tree height prior is very diffuse under the coalescent. The median prior tree height is years. So if the data is unable to provide calibration information for the clock rate, the inferred tree height would increase a lot and the tMRCA will be pulled up from February 2014 due to the tree prior. Indeed, when the sequence data is included in the analysis, the median tree height estimate turns out to be 1026 years. The tree height gets pulled down from what would be dictated by the tree prior. This estimate is however still higher than what would be expected under the clock rate prior (or the clock rate estimates in other studies), requiring the clock to be even slower than defined by the prior (median estimate of the rate.mean parameter after inclusion of sequences being subst/site/year). The situation for other prior settings for BRAZIL and also when using the Coal model with the Florida, USA data set is similar.
These findings show that the data sets of BRAZIL and USA sequences do not contain enough information to calibrate the molecular clock and thus fail to infer a proper tree height.
3.2 Estimates of the effective reproductive number for the USA data set
The number of secondary cases induced by one index case, i.e. the number of individuals infected by one infected individual during his/her infectious period, in a totally susceptible population is referred to as the basic reproductive number R0. It is a very important parameter to be estimated at the start of an epidemic. If the epidemic is prone to die out, if R0 > 1 the disease will spread amongst the population (Anderson and May 1991). During an ongoing epidemic, some individuals recover from the disease and gain immunity such that the population is not completely susceptible anymore. Further, the individuals might change their behaviour and thus the number of secondary cases caused by a single infected individual might change. The number of secondary cases caused by a single infected individual at time t is referred to as the effective reproductive number, Re, at time t. Similarly to R0, the effective reproductive number provides important information on whether the epidemic is able to spread further—this is the case if , or whether it is prone to die out—i.e. if .
The effective reproductive number during an ongoing epidemic is directly estimated under the birth–death skyline model as the birth rate divided by the death rate at time t. One of the assumptions of the birth–death skyline model is that the sampling intensity may vary through time but it is distributed uniformly at random across the considered infected population at any point in time. This assumption is violated for the ALL and BRAZIL data sets, since they encompass a big geographic area, with sampling and sequencing efforts being higher in some regions than in others (Faria et al. 2017; Metsky et al. 2017). In contrast, the sequences in the Florida, USA data set were sampled across a smaller area formed by three neighbouring counties (Grubaugh et al. 2017) with the zone of transmission being estimated within these counties for most individuals (only for three of twenty-three sequences included in our data set had an unknown zone of transmission). In addition, the infected individuals in this region seemed to be sampled uniformly at random at any point in time.
As we cannot estimate the clock rate for this data set, we set it to the median (identical to mean) clock rate estimate we obtained from analysing the full data set without the twenty-nine Florida sequences (i.e. the sequences highlighted in green in Fig. 1 and Supplementary Figs S1 and S2). Thus, we set the clock rate to 9 × 10–4 subst/site/year. We show the results in Fig. 5 using six intervals for the birth rate (and thus Re) and assuming a constant sampling probability throughout the period of sample collection. This analysis reveals that for up to the time of the last sample, i.e. until October 2016, the ZIKV spread fastest in Florida in mid-2016 and that the Re dropped below 1 around August 2016. Allowing fewer intervals for the birth rate, meaning only estimating a coarser pattern for Re, yields Re estimates which are averages from the finer six interval analysis, as expected (Supplementary Fig. S5).
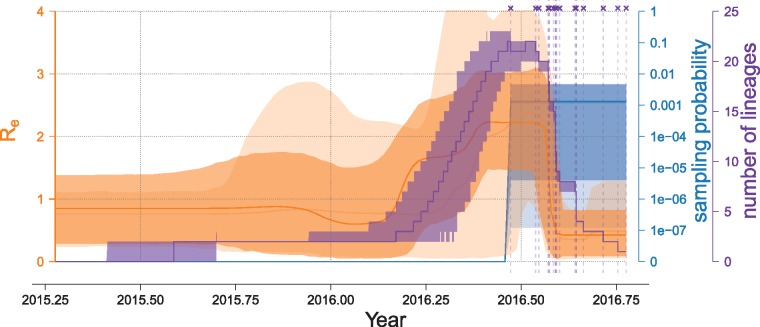
Figure 5.
Birth–death skyline plot based on the USA data set. We used the BDSKY model allowing six intervals for Re and 1 mean value for the sampling probability. Opaque coloring for the effective reproductive number, Re (orange), and the sampling probability (cyan), depict the results of analyses without sequence data, the darker shades display estimates after the addition of sequence data. The vertical dashed magenta lines and crosses indicate the sampling time points of the viral sequences. The magenta curve summarizes the lineages-through-time plot averaged over all MCMC samples without the burn-in (first 10% of samples). For all parameters we show the 95% confidence intervals and the median estimates (solid central line).
We were interested to compare our Re estimates to the prevalence and incidence data reported by Pan American Health Organization (2017) and the Florida Department of Health (2017). However, these statistics on local transmissions only begin on 18 August 2016 and 30 July 2016, respectively. Both sources report a drastic increase in new cases in the first half of October 2016. Since in the Florida cluster we analysed here, the sequences are sampled as early as in June 2016, we observe an increase in Re before the first reports of local transmission from PAHO/Florida Department of Health. Also, our last sample was obtained on 11 October 2016 and we therefore do not capture the potential peak in Re beyond (nor potentially slightly before) this date. However, Grubaugh et al. (2017) provide an estimate of the R0 in the Miami Dade county, one of the three counties the sequence data included in our analyses comes from. Based on the observed local transmission data and the total number of introduction events (travel-associated cases), they estimated the R0 to be 0.5–0.8. In our analyses, once we see the epidemic spread taking off in early March 2016, we estimate median Re of 0.88, quickly rising above 1 as the epidemic progresses. Between end of May and mid-July 2016 we consistently observe : median Re between 2.1 and 2.2 and 95% HPD=(1.1, 3.1). Prior to March 2016 we do not see the epidemic spread and estimate median Re between 0.6 and 0.88 (95% HPD: 0.05–1.8), which would be in line with Grubaugh et al. estimates. The discrepancy between our Re estimates after March 2016 and the R0 estimates of Grubaugh et al. could stem from the estimates of Grubaugh et al. being based on the infected patient count data reported by the public health agencies, which could have potentially missed a lot of mild or asymptomatic ZIKV infections.
4. Discussion
As soon as a new virus starts to spread within the human population, it is important to estimate the potential impact of this epidemic on a global scale. To obtain trustworthy parameter estimates, the sequences must contain enough signal regarding the quantities of interest. Testing the sequence data for a signal on the molecular clock rate is therefore necessary.
4.1 Analyses using point estimate methods
Point estimate methods allow for dating the tree and obtaining an estimate for the molecular clock rate. Here, we explored such estimates using TempEst (Rambaut et al. 2016) and LSD tree dating (Guindon et al. 2010; To et al. 2015). Tools such as TempEst (Rambaut et al. 2016) that visualise the regression of the root-to-tip divergence with the calendar time can be helpful in determining the amount of clock signal in the data. There are only few rules on how exactly these tools should be used. The authors of the TempEst say that ‘[…] the estimation of a negative evolutionary rate indicates that the data set contains little or no temporal signal’ (Rambaut et al. 2016). They also point out that the amount of signal in the data can be assessed visually or through the correlation coefficient, R2, provided by the method. However, guidelines that describe how to exactly perform this visual inspection, or what the reasonable cutoff for R2 is to determine if the information in the data is sufficient to allow for the separation of the clock rate and the tree height signal are lacking. Assessment of the clock signal in the data using this tool is therefore very subjective.
Such regression methods as well as a molecular clock selection approach have been found to lead to wrong estimates of the amount of clock signal in the data. This is especially the case when the sampling periods are too short or when sampling is biased, i.e. when sampling is confounded with the genetic structure in the population (Murray et al. 2016). Repeating the analyses by randomly permuting the sampling dates of the sequences, i.e. doing a Bayesian permutation test (Ramsden et al. 2008), can identify the latter but not the former bias (Duchêne et al. 2015). In contrast, a version of the Bayesian permutation test, the so-called clustered permutation test, which randomises the date assignment between sequences belonging to a monophyletic cluster, rather than doing the assignment completely at random, can deal with both situations when assessing the amount of clock signal in the data (Duchêne et al. 2015).
In our ZIKV data analyses, the tree height estimates from TempEst are largely in agreement with the Bayesian estimates for the ALL data set. Using LSD method, we obtain conflicting tree height estimates compared to the Bayesian and TempEst estimates already for the ALL data set, which may be due to LSD needing stronger clock signal in the data to produce reliable estimates.
Both TempEst and LSD provide only point estimates for the clock rate and the tMRCA parameters. To explore the robustness of these estimates, and to obtain the confidence intervals, indirect approaches such as a bootstrap procedure has to be performed. Furthermore, these estimates are based on a single input tree. In case of limited data, the tree reconstruction method and settings, e.g. evolutionary model, chosen can strongly influence the results of these analyses (compare Fig. 2 where the input three is reconstructed using ML procedure, and Supplementary Fig. S6, where the tree has been reconstructed using another popular method, namely neighbour-joining algorithm (Saitou and Nei 1987).
4.2 Analyses using Bayesian phylodynamic method
Bayesian methods are an alternative to the point estimate methods. In the Bayesian framework, the uncertainty in the tree topology can be naturally integrated out by sampling over many plausible topologies. In addition, the result of a Bayesian inference is a distribution of parameter values, rather than a single point estimate, providing a measure of uncertainty in the estimate. Furthermore, results can easily be tested for robustness across models and across priors for parameters to investigate the amount of signal contained in the data about any particular parameter.
Bayesian phylodynamic analysis requires model choices and decisions on prior settings before analysing the sampled sequence data. Making these choices is not trivial. Despite the fact that prior distribution for in total four parameters, i.e. origin, become uninfectious rate, effective reproductive number and the sampling probability, needs to be specified in the birth–death model, this task may be easier than specifying the prior distribution on the single parameter, i.e. effective population size, of the coalescent model. Epidemiological surveillance data often provide information on the duration of the infection in the patient and also on the sampling probability. Furthermore, assuming Re around 1 is reasonable for most infections. However, there is usually no information on the effective infected population size, which is in fact a different quantity than the infected population size.
A good practice in Bayesian phylodynamics is to first run the analysis without the sequence data and only add the sequence data in a second run. Such analyses can be very revealing about both the amount of information in the sequences and the assumptions of the underlying models. Analysing just the sampling times of ZIKV data sets with the coalescent tree prior led to wide distributions for the tree height parameter. When the sequence data was included, the tree height distributions peaked for the case of the ALL data set but remained very broad for the BRAZIL and the USA sequences only, indicating a lack of signal in the two latter data sets. Even though we assumed a flat prior on the origin, the birth–death model analyses yielded peaked estimates of the tree height already when just the information on the sampling times was included. This is due to the fact that the sampling process is part of the birth–death model outcome. Thus, the sampling times already provide information about the epidemiological dynamics. In this sense, the birth–death models are very similar to classic epidemiology, where the sampling times without sequences are used for estimation of epidemiological parameters.
If the birth–death model is an appropriate model regarding the dynamics of the (sampling and epidemiological) process, sampling times provide useful information regarding the parameters of the process. However, if the birth–death model does not describe the process correctly, a peaked distribution obtained based on sampling times can be misleading, especially if this distribution is shifted away from the true parameter value. When sequence data is combined with such shifted distributions, and the information in the sequences is not strong enough to push the distribution to the correct values, wrong conclusions about important epidemiological parameters may be drawn.
Sampling from an actual prior distribution under the birth–death model involves sampling the number of samples and the sampling times along with all other parameters. One option to obtain such a prior distribution under the birth–death model would be to simulate trees under a given set of epidemiological parameters using, e.g. MASTER (Vaughan and Drummond 2013) or TreeSim package in R (Stadler 2011). If such sampling was carried out, one would obtain prior distributions on all parameters together with prior distribution of the sample number and sampling times. In such a case, if the prior distribution on the origin parameter was specified to be uniform, sampling from the actual prior will produce a uniform prior distribution for the origin parameter. However, the prior distribution for the tree height parameter would be defined by the exact combination of the birth rate, the death rate and the sampling probability parameter.
4.3 Analysis of limited sequence data using Bayesian phylodynamics
Sequence data carry information for two distinct parameters: the tree topology and the clock rate. If the sequences are not divergent enough from one another, the topology will be hard to estimate. If the sequences are sampled close to one another in time, relative to the substitution rate, even if the topology is correctly resolved, it will be hard to tease apart the tree height and the clock (substitution) rate. The former problem is dealt with using the Bayesian analysis, since the tree topology can be treated as a nuisance parameter and is integrated out. When sequences do not contain enough information to calibrate the molecular clock, only the product of the clock rate and the tree height can be estimated reliably. Here, we have shown that once we attempt to decompose this product into the clock rate and tree height estimates using insufficient data, severe problems arise. Sequences with little information on the clock rate may shift the estimates in a wrong direction (compared to the estimates without sequences) as the clock rate and the tree height distributions interact once the sequences are added. This phenomenon has been also recently observed in simulations (Möller 2017).
Only when we are guaranteed that the data contains enough signal for the clock calibration, we can proceed with interpreting phylodynamic parameters. In case of limited data, it is possible to carry out the analyses by fixing one of the parameters: the clock rate or the tree height. Fixing the former is straightforward as the clock rate is a parameter for which a prior is chosen, however, fixing the latter is more complex as the tree height prior distribution is induced by the prior distributions on the model parameters. One may simply fix the MRCA node height, i.e. use a calibration node. However, superimposing of such MRCA node age prior on top of the population dynamic prior can lead to unexpected actual prior distributions (Heled and Drummond 2015). In addition to the technical issue with fixing the clock rate or the tree height parameters in the analyses, availability of reliable estimates also influences which parameter one chooses to fix. Clock rate estimates are often available for many data sets. Fixing this value and assuming the sampling process is unbiased across the population at any point in time, we can infer epidemiological parameters using the birth–death (skyline) model. It has been shown before, that if the sampling process is mis-specified in the birth–death model, e.g. if it is fixed to a high value, while the true value is low, then other epidemiological parameters will be biased too (see effect of misspecification due to parameter correlations in Boskova, Bonhoeffer, and Stadler (2014)). If the sampling process is biased in such a way that certain parts of the transmission tree are sampled more than others, and one fails to capture those variations in the model both the birth–death and the coalescent model may produce biases as both of these models assume that at each point in time, a random sample is taken from the full process.
We performed a phylodynamic analysis on the Florida, USA data set, by fixing the clock rate, and find evidence for the increase in spread of the ZIKV in Florida in mid-2016 followed by the decline in spread after August 2016. We used the birth–death skyline model with up to 6 intervals for Re to analyse the USA data. The reason for not increasing this number further was that we did not want to over-parametrise the model. In particular, the data set only consisted of 23 sequences and including more parameters to estimate in the model would only lead to wider posterior intervals. An ideal alternative to the simple skyline model would be to use a smoothing prior, informing each next interval by the results gathered from the previous one. In the future, we hope such prior will become available for the birth–death skyline model, allowing for more detailed analyses of small data sets such as the ZIKV Florida cluster explored here.
5. Conclusions
Based on our results presented here, we suggest to perform Bayesian phylodynamic analyses during ongoing epidemics with highest care with respect to model robustness. Instead of using just one set of priors to perform an analysis, it is essential to check the robustness of the estimates obtained with the chosen models and different prior settings. If the data has enough phylogenetic and clock signal, the posterior distributions will be consistent over the range of models chosen. Only then, the posterior distributions can reliably reflect important parameters such as the effective reproductive number, the start or the size of the epidemic.
Supplementary Material
Acknowledgements
The authors express their gratitude to Louis du Plessis for helpful discussions and for making his skyline plotting scripts and TreeSlicer class from unreleased BEAST2 package smoothingpriors (https://github.com/laduplessis/smoothingpriors) available. V.B., T.S., and C.M. thank ETH Zürich for funding. T.S. and V.B. were supported by the European Research Council under the Seventh Framework Programme of the European Commission (PhyPD: grant agreement number 335529).
Authors’ contributions
V.B., T.S., and C.M. designed the study and the analyses. V.B. performed the analyses. V.B., T.S., and C.M. wrote the paper.
Supplementary data
Supplementary data are available at Virus Evolution online.
References
- Anderson R. M., May R. M. (1991) Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford University Press. [Google Scholar]
- Boskova V., Bonhoeffer S., Stadler T. (2014) ‘Inference of Epidemiological Dynamics Based on Simulated Phylogenies Using Birth-Death and Coalescent Models’, PLoS Computational Biology, 10: e1003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R. et al. (2014) ‘BEAST 2: A Software Platform for Bayesian Evolutionary Analysis’, PLoS Computational Biology, 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2005) ‘Bayesian Coalescent Inference of past Population Dynamics from Molecular Sequences’, Molecular Biology and Evolution, 22: 1185–92. [DOI] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2006) ‘Relaxed Phylogenetics and Dating with Confidence’, PLoS Biology, 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007) ‘BEAST: Bayesian Evolutionary Analysis by Sampling Trees’, BMC Evolutionary Biology, 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Plessis L., Stadler T. (2015) ‘Getting to the Root of Epidemic Spread with Phylodynamic Analysis of Genomic Data’, Trends in Microbiology, 23: 383–6. [DOI] [PubMed] [Google Scholar]
- Duchêne S. et al. (2015) ‘The Performance of the Date-Randomization Test in Phylogenetic Analyses of Time-Structured Virus Data’, Molecular Biology and Evolution, 32: 1895–906. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N. R. et al. (2016) ‘Zika Virus in the Americas: Early Epidemiological and Genetic Findings’, Science, 352: 345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N. R. et al. (2017) ‘Establishment and Cryptic Transmission of Zika Virus in Brazil and the Americas’, Nature, 546: 406–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N. M. et al. (2016) ‘Countering the Zika Epidemic in Latin America’, Science, 353: 353–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florida Department of Health (2017) <http://www.floridahealth.gov/diseases-and-conditions/zika-virus/index.html> accessed 26 July 2017.
- Grenfell B. T. et al. (2004) ‘Unifying the Epidemiological and Evolutionary Dynamics of Pathogens’, Science, 303: 327–32. [DOI] [PubMed] [Google Scholar]
- Grubaugh N. D. et al. (2017) ‘Genomic Epidemiology Reveals Multiple Introductions of Zika Virus into the United States’, Nature, 546: 401–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of Phyml 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Heled J., Drummond A. J. (2015) ‘Calibrated Birth–Death Phylogenetic Time-Tree Priors for Bayesian Inference’, Systematic Biology, 64: 369–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampstra P. (2008) ‘Beanplot: A Boxplot Alternative for Visual Comparison of Distributions’, Journal of Statistical Software, 28: 1–9.27774042 [Google Scholar]
- Kingman J. F. (1982) ‘On the Genealogy of Large Populations’, Journal of Applied Probability, 19: 27–43. [Google Scholar]
- Korber B. et al. (2000) ‘Timing the Ancestor of the HIV-1 Pandemic Strains’, Science, 288: 1789–96. [DOI] [PubMed] [Google Scholar]
- Metsky H. C. et al. (2017) ‘Zika Virus Evolution and Spread in the Americas’, Nature, 546: 411–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller S. (2016) ‘The Impact of the Tree Prior and Purifying Selection on Estimating Clock Rates during Viral Epidemics', ETH Zurich.
- Murray G. G. et al. (2016) ‘The Effect of Genetic Structure on Molecular Dating and Tests for Temporal Signal’, Methods in Ecology and Evolution, 7: 80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee S. (2006) ‘Birth-Death Models in Macroevolution’, Annual Review of Ecology, Evolution, and Systematics, 37: 1–17. [Google Scholar]
- Neuwirth E. (2014) Rcolorbrewer: Colorbrewer Palettes. R package version 1.1-2.
- Oehler E. et al. (2014) ‘Zika Virus Infection Complicated by Guillain-Barre Syndrome—Case Report, French Polynesia, December 2013’, Eurosurveillance, 19: pii: 20720. [DOI] [PubMed] [Google Scholar]
- Pan American Health Organization (PAHO) (2017) <http://www.paho.org> accessed 9 June 2017.
- Pan American Health Organization, and World Health Organization Regional Office for the Americas (2015) ‘Epidemiological Alert: Neurological Syndrome, Congenital Malformations, and Zika Virus Infection’. Implications for Public Health in the Americas. Technical report. <http://www.paho.org/hq/index.php?option=com_docman&task=doc_download&Itemid=&gid=32405&lang=en>.
- Paradis E., Claude J., Strimmer K. (2004) ‘APE: Analyses of Phylogenetics and Evolution in R Language’, Bioinformatics (Oxford, England), 20: 289–90. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: <http://www.R-project.org/>. [Google Scholar]
- Rambaut A. (2012) Figtree v1.4. Molecular evolution, phylogenetics and epidemiology. Edinburgh, UK: University of Edinburgh, Institute of Evolutionary Biology. <http://tree.bio.ed.ac.uk/software/figtree/>.
- Rambaut A. et al. (2014) Tracer v1.6, Molecular evolution, phylogenetics and epidemiology, Edinburgh. UK: University of Edinbugh, Institute of Evolutionary Biology. <http://beast.bio.ed.ac.uk/tracer>.
- Rambaut A. et al. (2016) ‘Exploring the Temporal Structure of Heterochronous Sequences Using Tempest (Formerly Path-o-Gen)’, Virus Evolution, 2: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A. (2015) Treeannotator v2.3.0. Edinburgh, UK: University of Edinburgh, Institute of Evolutionary Biology and Auckland, NZ: University of Auckland, Department of Computer Science.
- Ramsden C. et al. (2008) ‘High Rates of Molecular Evolution in Hantaviruses’, Molecular Biology and Evolution, 25: 1488–92. [DOI] [PubMed] [Google Scholar]
- Revell L. J. (2012) ‘Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things)’, Methods in Ecology and Evolution, 3: 217–23. [Google Scholar]
- Saitou N., Nei M. (1987) ‘The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees’, Molecular Biology and Evolution, 4: 406–25. [DOI] [PubMed] [Google Scholar]
- Smith B. J. (2007) ‘boa: An R Package for MCMC Output Convergence Assessment and Posterior Inference’, Journal of Statistical Software, 21: 1–37. [Google Scholar]
- Stadler T. (2010) ‘Sampling-through-Time in Birth-Death Trees’, Journal of Theoretical Biology, 267: 396–404. [DOI] [PubMed] [Google Scholar]
- Stadler T. (2011) ‘Simulating Trees with a Fixed Number of Extant Species’, Systematic Biology, 60: 676–84. [DOI] [PubMed] [Google Scholar]
- Stadler T. et al. (2013) ‘Birth–Death Skyline Plot Reveals Temporal Changes of Epidemic Spread in HIV and Hepatitis C Virus (HCV)’, Proceedings of the National Academy of Sciences of the United States of America, 110: 228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T. H., et al. (2015) ‘Fast Dating Using Least-Squares Criteria and Algorithms’, Systematic Biology, 65: 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan T. G., Drummond A. J. (2013) ‘A Stochastic Simulator of Birth–Death Master Equations with Application to Phylodynamics’, Molecular Biology and Evolution, 30: 1480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura C. V. et al. (2016) ‘Zika Virus in Brazil and Macular Atrophy in a Child with Microcephaly’, The Lancet, 387: 228. [DOI] [PubMed] [Google Scholar]
- virological.org (2016). <http://virological.org> accessed 30 Nov 2016.
- World Health Organization (WHO) (2016) Geneva <http://www.who.int/emergencies/zika-virus/quarterly-update-october/en/> accessed 1 Dec 2016.
- World Health Organization (WHO) (2016) ‘Zika Situation Report – Neurological syndrome and congenital anomalies. Technical report’ <http://apps.who.int/iris/bitstream/10665/204348/1/zikasitrep_5Feb2016_eng.pdf?ua=1>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.